Abstract
When using SIR and related models, it is common to assume that the infection rate is proportional to the product of susceptible and infected individuals. While this assumption works at the onset of the outbreak, the infection force saturates as the outbreak progresses, even in the absence of any interventions. We use a simple agent–based model to illustrate this saturation effect. Its continuum limit leads a modified SIR model with exponential saturation. The derivation is based on first principles incorporating the spread radius and population density. We use the data for coronavirus outbreak for the period from March to June, to show that using SIR model with saturation is sufficient to capture the disease dynamics for many jurstictions, including the overall world-wide disease curve progression. Our model suggests the R0 value of above 8 at the onset of infection, but with infection quickly “flattening out”, leading to a long-term sustained sub-exponential spread.
Keywords: COVID-19, SIR models, Saturation
As the coronavirus pandemic has spread and dramatically altered human lives on a global scale, it also generated an intense interest to model the epidemic. These include ODE models (Hu et al., 2013; Bertozzi et al., 2004; Kissler et al., 2020; Humphrey et al., Cojocaru), statistical models (Bertozzi et al., 2004; Xia et al., 2004), agent-based models (McCallum et al., 2001;Ferguson et al., Cuomo-Dannenburget al.), spatial network models (Colizza & Vespignani, 2008; Xia et al., 2004) and PDE models (Gai et al., 2020; Kevrekidis et al., 2005); see also (Chen, 2014, pp. 19–42; McCallum et al., 2001) for reviews. Some of these papers had an outsized influence on public thinking and policy (Kissler et al., 2020; Ferguson et al., Cuomo-Dannenburget al.). At the heart of many of these efforts are the so-called compartmental models, consisting of various classes of individuals and their interactions. The simplest such model is the classic SIR (Susceptiple-Infected-Removed) model. It has the form
| (1) |
where S, I and R is the susceptible, infected and removed (or recovered) population, respectively, and N is the total number of infected individuals. The basic assumption underlying SIR and related models is that the disease spreads when an susceptible individual encounters an infected individual. The probability of this encounter is typically modeled using the law of mass action from chemical literature, where this probability is proportional the product SI of susceptible and infected population, with β being the infection rate. The infected individuals then recover with a recovery rate γ.
Can SIR model by itself capture the coronavirus infection? To answer this question, we took data from the from John Hopkins CSSE database (Dong et al., 2020), which lists the total recorded infections as well as the number of recovered and dead for different countries. We took the period from March 15 to June 15, 2020, corresponding to the initial wave of infection (and mostly after the implementation of the quarantine measures). We used the sum of recovered and dead as a proxy for “removed” population R(t), while I(t) is obtained by substracting by substracting “removed” from the total number of infected. We can then estimate β and γ directly from equation (1) by solving and (where we approximate the derivatives using the finite differences on one-week running averages). This is shown in Fig. 1. We took the data from March 15 to June 15. As can be seen, the recovery rate γ is relatively constant, while the infection rate β has a clear decrease over time. Similar decrease has been noted by others, see e.g. (Ke et al., 2020; Tang et al., 2020).
Fig. 1.
Estimated infection (β) and recovery (γ) rates as a function of time for several jurisdictions during the period of March 15 to June 15, 2020. Note that β is decreasing with time while γ remains relatively constant.
In this note we propose a simple modification of the basic SIR model (1) to incorporate spatial distribution of population, and which can partially explain the decrease of β over time, even without time-dependent changes in parameters. Our implementation also naturally incorporates the population density into the model. We also test the modified model against the COVID data.
We start with the following very simple agent-based model that implicitly incorporates spatial component. Assume that each agent has some “foraging area” characterized by radius r. Agents can only encounter each-other within their respective foraging area. Upon encounter, assume there is a probability p of infection. Assume that a susceptible individual spends on average μ fraction of the time interacting with others (i.e. going shopping, to bars, etc). Finally an, infected individual recovers with a daily rate γ. The matlab implementation of this is shown in Fig. 2.
Fig. 2.
Top: self-contained Matlab code for simulating ABM and its mean-field limit (2). Cut-and-paste to run. Bottom: the output of the simulation.
We now derive the mean-field limit of this agent-based model. Assuming the agents are well mixed, the chances of encountering an infected individual and then getting infected is given by , where A is the total area of the domain. When I infected agents are present, the chances of getting infected is then given by . We also assume that the foraging area is sufficiently small (compared to the total area A) so that 0 < a ≪ 1, in which case we may approximate . As the result, the mean-field limit of this agent-based simulation takes the form
| (2) |
Fig. 2 shows the direct comparison of the agent-based model described above, and the mean-field limit (2); excellent agreement can be seen.
The parameter a can be rewritten in terms of the population density as follows. Write A = N/ρ, where N is the total population and ρ is the population density, so that
| (3) |
This shows that the parameter a is directly proportional to the population density.
Non-pharmaceutical interventions are used to reduce μ, α and increase γ. In the context of coronavirus, the rate of interaction μ is decreased by stay-at-home orders. Social distancing and mask-wearing will have the effect of reducing the exponent α. Finally, isolating infected individuals has the effect of increasing γ.
The model (2) reduces to the classical SIR model (1) for small infection rates I via Taylor expansion , so that β = μα. This shows that the reproduction number R0 = β/γ = ρμpr2π/γ is proportional to the population density ρ. This is consistent with the progress of the epidemic: large cities with high population densities (e.g. New York, Toronto, Montreal…) have been hit much harder than smaller localities with lower population density. As the number of infected I increases, the saturation terms kicks in and eventually stabilize the force of infection, leading to linear rather than exponential growth.
We used nonlinear least-squares method to fit the model (2) to the data for several juristictions. This consists of minimizing the L2 distance to fit both the infected as well as recovered individuals simultaneously:
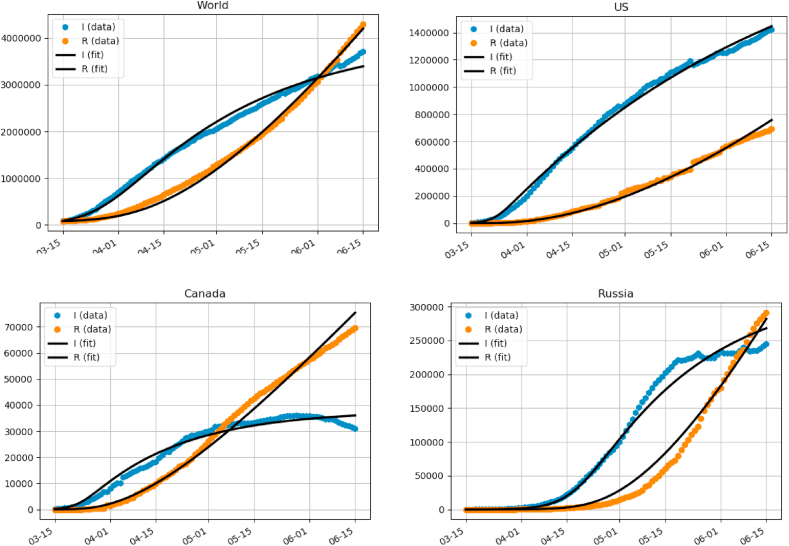
Here, Ij, Rj is the real-world data from John Hopkins database (Dong et al., 2020), and I(t), R(t) is the solution to (2) for given parameters α, β, γ. In this way, we solve all three parameters, without having to assume a-priori values for any of them. We used python’s standard scipy.optimize package to perform the minimization. We also experimented with different distance metrics, such as log distance, which gave very similar results. Fig. 3 shows the fit for several juristictions with a good agreement.
Fig. 3.
Fitting model (2) for several juristictions.
The following table gives the corresponding parameter values for these jurisdictions:
| N | α | μ | γ | |
|---|---|---|---|---|
| World | 7.7 × 109 | 15050 | 1.25 × 10−5 | 0.0232 |
| US | 3.4 × 108 | 5857.8 | 7.84 × 10−5 | 0.0107 |
| Canada | 3.7 × 107 | 13165 | 3.48 × 10−5 | 0.0345 |
| Russia | 1.5 × 108 | 3448.8 | 6.43 × 10−5 | 0.0278 |
These are the countries where the virus is not over the peak yet. The fit is not always great, particularly for the countries that are over the peak and have gotten and virus under control and completely suppressed the new cases, such as Germany, Italy or South Korea.
It is interesting to note that the values of parameters are relatively consistent (same order of magnitude) among the different data sets. Also, the parameter γ is consistent with other estimates in the literature (Bar-On et al., 2020; Bertozzi et al., 2004). For the US dataset, we used US Census to compute the effective population density for urban areas where the majority of population resides (US Census). We obtained an estimate of ρ ≈ 1000 people per km2. This gives us a value of a = pr2π ≈ 5.8 km2. It is hard to disentangle the probability p of the infection from the infection area πr2 per individual. One possibility is to use the average daily driving range estimate, which was reported in (Triplett et al., 2013) to be 75 km involving on average 2.3 trips. This gives r ≈ 32 km and p ≈ 0.0017. However this does not account for other modes of transportation (walking, public transport, etc).
For Canada, we used Canadian Census (Canadian Census Population centres, 2011) which lists 942 Canadian cities and towns having a total area of 23998 km^2 and a total population of about 27 million (out of a total population of 37 million). This gives an effective density estimate for Canadian population of ρ ≈ 1174 people/km^2, yielding a value of a = pr2π ≈ 11.2 km2, about twice that of the US. Assuming similar driving statistics as the US, would yield the same radius r and p of about twice as much as the US.
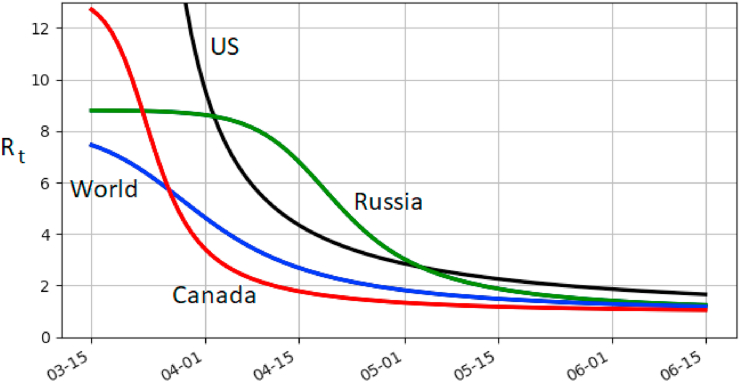
For the model (2), the reproduction number R0 is given by R0 = μα/γ. For world, US, Canada and Russia, R0 is 8.1, 49.13, 13.26 and 8.8, respectively. It is in the higher-end of the current estimates for Covid-19. However, the time-dependent (dynamical) reproduction number, Rt, decreases rapidly due to saturation. It is given by
and is plotted in Fig. 4. Currently US appears to have the highest growth rate with Canada the slowest of the four. Note that Russia had a long period of exponential growth, which is reflected in a relatively long period of near-constant Rt before Rt comes down.
Fig. 4.
Dynamical reproduction number Rt for (2) using the best-fit parameters. It is relatively high at the start of the outbreak but quickly decreases.
We have presented a modified SIR model with saturation. Rather than using a phenomenological saturation (Capasso & Serio, 1978; McCallum et al., 2001), we used agent-based modelling and then derived the mean-field limit continuum limit using basic probability. We were able to fit the model parameters to obtain a good agreement with the disease progression during the period of March 15 to June 15, when quarantine and shutdown policies were mostly in-place.
Fitting the original SIR model (1) without any saturation and without dynamically changing the infection rate β gives rather terrible results as illustrated in Fig. 5. An alternative is to assume that parameters are time-dependent (Bertozzi et al., 2004; Humphrey et al., Cojocaru; Kevrekidis et al., 2005; Dehning et al., 2020). Other models that can reproduce sub-exponential growth include the generalized logistic growth model (Roosa et al., 2020).
Fig. 5.
Fitting basic SIR model (1) versus SIR with saturation (2). Left: world population. Without saturation, the fit is very bad. With saturation, it is relatively good. Right: synthetic data from ABM model. The curve with saturation corresponds to the exact parameters used in ABM simulation without any fitting. The curve without saturation is the “best-fit” parameters and does not accurately capture the peak of infection.
One may ask whether a model with more that just three compartments (such as SEIR, E = exposed, or SAEIR, A = asymptomatic etc) but without saturation can produce a better fit than SIR with saturation? Looking at the data of Fig. 3, the infected cases exhibit what looks like a subex ponential growth. At least in this stage of the epidemic (as of June 2020), only a small fraction of the total population has been infected (worldwide, less than 1%). Without saturation, regardless of the number of compartments used, the model becomes effectively linear when only a small fraction is infected (e.g., in (1)). Linear ODE models generically exhibit exponential growth, so it is impossible to obtain sub-exponential growth without either having saturation, or time-dependent parameters.
We completed the first draft of this paper by the end of June. By July, the US re-entered a period of fast coronavirus growth, mostly due to re-opening of economy. In effect, the parameters “on the ground” have shifted. By the time the paper was accepted in October 2020, a third and a more severe wave is well under way. Fig. 6 shows the estimated β and γ for the US from April to end of October 2020. A second peak in β is visible in mid-July. Then from beginning of September, β starts to grow again with no end in sight as of this writing. This third wave coincided with re-opening of schools, and is the most intense of the three waves so far. One possible explanation is that asymptomatic transmission is increased with kids acting as disease vectors. There is also a possibility of mutation to a more contageous strain. A more complex model with time-dependent infection rate is required to better model the progression of the disease on a longer time-scale. This is left for future work.
Fig. 6.
Estimated infection (β) and recovery (γ) rates for the US, from April 1st to Oct 31, 2020. Second and third “waves” are clearly visible corresponding to the period of growth in β.
Declaration of competing interest
The authors have no conflict of interest.
Handling editor. Dr. J Wu
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. Science forum: Sars-cov-2 (covid-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A. L. Bertozzi, E. Franco, G. Mohler, M. B. Short, D. Sledge, The challenges of modeling and forecasting the spread of covid-19, arXiv preprint arXiv:2004.04741. [DOI] [PMC free article] [PubMed]
- Canadian Census Population centres https://www12.statcan.gc.ca/census-recensement/2011/dp-pd/prof/details/downl oad-telecharger/comprehensive/comp-csv-tab-dwnld-tlchrgr.cfm?Lang=E
- Capasso V., Serio G. A generalization of the kermack-mckendrick deterministic epidemic model. Mathematical Biosciences. 1978;42(1–2):43–61. [Google Scholar]
- Chen D. 2014. Modeling the spread of infectious diseases: A review, analyzing and modeling spatial and temporal dynamics of infectious diseases. [Google Scholar]
- Colizza V., Vespignani A. Epidemic modeling in metapopulation systems with heterogeneous coupling pattern: Theory and simulations. Journal of Theoretical Biology. 2008;251(3):450–467. doi: 10.1016/j.jtbi.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. The Lancet Infectious Diseases. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. https://github.com/CSSEGISandData/COVID-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- N. Ferguson, D. Laydon, G. Nedjati Gilani, N. Imai, K. Ainslie, M. Baguelin, S. Bhatia, A. Boonyasiri, Z. Cucunuba Perez, G. Cuomo-Dannenburg, et al., reportReport 9: Impact of non-pharmaceutical interventions (npis) to reduce covid 19 mortality and healthcare demand.
- Gai C., Iron D., Kolokolnikov T. Localized outbreaks in an sir model with diffusion. Journal of Mathematical Biology. 2020;80(5):1389–1411. doi: 10.1007/s00285-020-01466-1. [DOI] [PubMed] [Google Scholar]
- L. Humphrey, E. W. Thommes, R. Fields, N. Hakim, A. Chit, M. G. Cojocaru, A path out of covid-19 quarantine: An analysis of policy scenarios, medRxiv.
- Hu H., Nigmatulina K., Eckhoff P. The scaling of contact rates with population density for the infectious disease models. Mathematical Biosciences. 2013;244(2):125–134. doi: 10.1016/j.mbs.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Ke R., Sanche S., Romero-Severson E., Hengartner N. medRxiv; 2020. Fast spread of covid-19 in europe and the us suggests the necessity of early, strong and comprehensive interventions. 04.04.20050427v2. [Google Scholar]
- P. Kevrekidis, J. Cuevas-Maraver, Y. Drossinos, Z. Rapti, G. Kevrekidis, Spatial modeling of covid-19: Greece and andalusia as case examples, arXiv preprint arXiv:2005.04527. [DOI] [PubMed]
- Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of sars-cov-2 through the postpandemic period. Science. 2020;368(6493):860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H., Barlow N., Hone J. How should pathogen transmission be modelled? Trends in Ecology & Evolution. 2001;16(6):295–300. doi: 10.1016/s0169-5347(01)02144-9. [DOI] [PubMed] [Google Scholar]
- Dehning J., Zierenberg J., Spitzner F.P., Wibral M., Neto J.P., Wilczek M., Priesemann V. Inferring change points in the spread of covid-19 reveals the effectiveness of interventions. Science. 2020;369(6500) doi: 10.1126/science.abb9789. https://science.sciencemag.org/content/369/6500/eabb9789.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosa K., Lee Y., Luo R., Kirpich A., Rothenberg R., Hyman J., Yan P., Chowell G. Real-time forecasts of the covid-19 epidemic in China from february 5th to february 24th, 2020. Infectious Disease Modelling. 2020;5:256–263. doi: 10.1016/j.idm.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B., Wang X., Li Q., Bragazzi N.L., Tang S., Xiao Y., Wu J. Estimation of the transmission risk of the 2019-ncov and its implication for public health interventions. Journal of Clinical Medicine. 2020;9(2):462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- T. Triplett, R. Santos, S. Rosenbloom, American driving survey: Methodology and year one results, may 2013–may 2014.
- US Census, Urban areas in the United States and Puerto Rico (CPH-T-9) https://www.census.gov/data/tables/time-series/dec/cph-series/cph-t/cph-t-9. html..
- Xia Y., Bjørnstad O.N., Grenfell B.T. Measles metapopulation dynamics: A gravity model for epidemiological coupling and dynamics. The American Naturalist. 2004;164(2):267–281. doi: 10.1086/422341. [DOI] [PubMed] [Google Scholar]