Abstract
Trypanosome U-insertion/deletion RNA editing in mitochondrial mRNAs involves guide RNAs (gRNAs) and the auxiliary RNA editing substrate binding complex (RESC) and RNA editing helicase 2 complex (REH2C). RESC and REH2C stably copurify with editing mRNAs but the functional interplay between these complexes remains unclear. Most steady-state mRNAs are partially edited and include misedited “junction” regions that match neither pre-mRNA nor fully edited transcripts. Editing specificity is central to mitochondrial RNA maturation and function, but its basic control mechanisms remain unclear. Here we applied a novel nucleotide-resolution RNA-seq approach to examine ribosomal protein subunit 12 (RPS12) and ATPase subunit 6 (A6) mRNA transcripts. We directly compared transcripts associated with RESC and REH2C to those found in total mitochondrial RNA. RESC-associated transcripts exhibited site-preferential enrichments in total and accurate edits. REH2C loss-of-function induced similar substrate-specific and site-specific editing effects in total and RESC-associated RNA. It decreased total editing primarily at RPS12 5′ positions but increased total editing at examined A6 3′ positions. REH2C loss-of-function caused site-preferential loss of accurate editing in both transcripts. However, changes in total or accurate edits did not necessarily involve common sites. A few 5′ nucleotides of the initiating gRNA (gRNA-1) directed accurate editing in both transcripts. However, in RPS12, two conserved 3′-terminal adenines in gRNA-1 could direct a noncanonical 2U-insertion that causes major pausing in 3′–5′ progression. In A6, a noncanonical sequence element that depends on REH2C in a region normally targeted by the 3′ half of gRNA-1 may hinder early editing progression. Overall, we defined transcript-specific effects of REH2C loss.
Keywords: kinetoplastid RNA editing, RNA processing, Trypanosoma brucei, editosome, guide RNA, REH2C helicase complex
INTRODUCTION
Species of Trypanosoma and other kinetoplastid protozoa within the protist group Euglenozoa utilize atypical mechanisms of gene expression (Tschudi and Ullu 1994; Simpson and Maslov 1999; Zimmer 2019; Butenko et al. 2020). Several mitochondrially encoded genes give rise to primary mRNA transcripts that are incomplete and packed with stop codons, preventing the formation of a functional open reading frame (ORF). Editosomes create translatable mRNAs encoding a canonical protein sequence through posttranscriptional U-specific insertion/deletion. This maturation process progresses 3′ to 5′ and is directed by guide RNAs (gRNAs). In Trypanosoma brucei, editosomes involve ∼40 proteins and about 1000 gRNAs to create 12 canonical ORFs (Cruz-Reyes et al. 2018a; Aphasizheva et al. 2020). Preedited mRNA and gRNA transcripts are encoded in mitochondria, whereas all known editing proteins are encoded in the nucleus and require import into mitochondria.
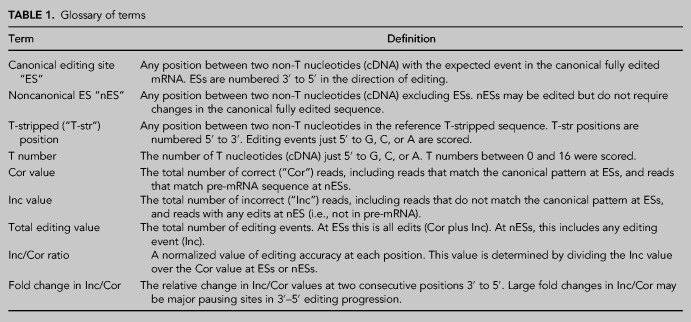
In most cases, multiple gRNAs are required to create a fully edited mRNA transcript (Koslowsky et al. 2013; Cooper et al. 2019). The 3′-most initiating gRNA (gRNA-1) and subsequent gRNAs (gRNA-2, gRNA-n) participate in a relay pathway. A 5′ anchor in the initiating gRNA anneals with pre-mRNA before its guiding sequence directs the editing changes. Completion of a full block of editing by the initiating gRNA enables 5′ anchor annealing by the subsequent gRNA (Koslowsky et al. 1991; Maslov and Simpson 1992). Thousands of canonical editing sites (ESs; see Table 1 for terminology) in mitochondria, are interspersed with noncanonical ESs (nESs). While nESs do not require processing in fully edited transcripts, these positions usually exhibit U-changes in presumed transcript intermediates of editing. This shows that any space between two non-Us has the potential to be edited by U-insertion/deletion, and presumably to be proofread by additional editing attempts. Cognate gRNA annealing with mRNA leads to the creation of canonical “correct sequences” at either ESs (directing precise editing changes) or nESs (preventing editing changes) at these sites. As editing progresses 3′ to 5′, a downstream sequence with correct editing is often followed by partially edited sequence junctions that match neither fully edited or preedited sequence, including ESs and or nESs (Decker and Sollner-Webb 1990; Koslowsky et al. 1991; Simpson et al. 2016). Alternative editing events have been proposed or identified (Ochsenreiter and Hajduk 2006; Koslowsky et al. 2013; Madina et al. 2014; Simpson et al. 2016) and off-targets by gRNAs may be possible (Koslowsky et al. 1991; Sturm et al. 1992; Kirby and Koslowsky 2017; Tylec et al. 2019). However, the possible function of alternative transcripts, particularly those that might create a noncanonical protein sequence, remains to be validated.
TABLE 1.
Glossary of terms
Editosomes include at least three macromolecular assemblies: RNA editing catalytic complex (RECC), RNA editing substrate binding complex (RESC), and RNA editing helicase 2 complex (REH2C). RECC contains 19 stably bound protein subunits, including enzymes for the key editing steps of endonucleolytic RNA cleavage, U addition or removal, and ligation (Rusche et al. 1997; Aphasizhev et al. 2003; Panigrahi et al. 2003a). Functional RECC, purified without RESC and REH2C, catalyzes editing at one or two consecutive substrate sites in vitro (Rusche et al. 1997; Panigrahi et al. 2003b; Carnes and Stuart 2007; Alatortsev et al. 2008). RECC transiently contacts RESC and REH2C via RNA. Purified RECC does not usually contain RNA (Rusche et al. 1997; Golas et al. 2009; Aphasizheva et al. 2014). However, proteins in purified RECC have been cross-linked to RNA in vitro (Sacharidou et al. 2006; Hernandez et al. 2008, 2010). RESC is heterogeneous and its organization is not completely understood, but it contains gRNA and at least 14 protein subunits (Hashimi et al. 2008; Panigrahi et al. 2008; Weng et al. 2008; Ammerman et al. 2012). REH2C includes three protein subunits and ATP-dependent 3′–5′ double-stranded RNA unwinding activity (Madina et al. 2015; Kumar et al. 2016, 2019). The kinetoplastid RNA helicase KREH2 (aka REH2) and zinc-finger protein KH2F1 (aka H2F1) subunits of REH2C promote efficient editing (Hashimi et al. 2009; Kumar et al. 2016). KREH2 is the sole DEAH/RHA type RNA helicase in editosomes (Cruz-Reyes et al. 2018a). Interaction between the trypanosomal editing complexes involves RNA, but contacts with RECC are particularly transient. Purifications of RESC and REH2C include stably associated mRNA (Aphasizheva et al. 2014; Madina et al. 2014; Huang et al. 2015).
The current model of the editosome organization posits that RESC serves as a platform for the assembly of gRNA hybrids with mRNA (or its mRNPs including REH2C) and catalysis by the RECC enzyme complex (Aphasizheva et al. 2014; Madina et al. 2014; Kumar et al. 2016; Dixit et al. 2017; McAdams et al. 2018). The functional interplay between these complexes and how their interactions may affect the editing processivity and accuracy remain unclear: variants of RECC and RESC complexes complicates our understanding of the editing mechanism further (Carnes et al. 2011; Ammerman et al. 2012; Madina et al. 2014, 2015; Huang et al. 2015; McAdams et al. 2018). REH2C interacts weakly with a RESC variant purified via immunoprecipitation of RESC6 (aka MRB3010), a critical protein in 3′ early editing (Ammerman et al. 2011; Madina et al. 2014). Previous studies using quantitative (q)RT-PCR indicated that fully edited mRNAs, including RPS12 and A6, are enriched in RESC6-purified RESC versus REH2C or total mitochondrial RNA (mtRNA) (Madina et al. 2014, 2015). RNAi of the helicase subunit in REH2C decreased that level of fully edited mRNAs in RESC6-purified RESC and appeared to increase editing pausing in the associated transcripts (Madina et al. 2014, 2015). Editing pausing in those studies was examined indirectly using a gel assay of end point RT-PCR products established by the Read laboratory (Ammerman et al. 2010). Whether REH2C affects all or only specific positions in mRNAs was not established in those studies. Amplicon-based RNA-seq studies of mRNA in T. brucei have examined the highly heterogeneous population of partially edited transcripts in the total mtRNA pool at steady state (Simpson et al. 2016, 2017; Carnes et al. 2017; McAdams et al. 2018, 2019; Zimmer et al. 2018; Tylec et al. 2019). Targeted RNA-seq studies by the Read laboratory characterized features of partially edited sequence junctions such as the junction start and end sites and defined intrinsic pause sites (IPS) in 3′–5′ progression. These authors also used RNAi of some RESC proteins to identify the specific effects of these proteins during 3′ early editing and 3′–5′ progression. RNA editing in other kinetoplastids was also examined by RNA-seq (Gerasimov et al. 2018). In this study, we applied a new bioinformatic tool that enables detailed analyses of editing at every position, either ES or nES, and thus can summarize at any position and all positions. Our systematic approach compared transcripts from different samples and examined values of total editing and editing accuracy at each position. We focused on the mRNA that encodes the mitochondrial ribosome protein subunit 12 (RPS12) (Read et al. 1992), and the mRNA that encodes the ATPase subunit 6 (A6) (Bhat et al. 1990). These transcripts are most likely essential in bloodstream and procyclic insect-borne life-cycle stages of T. brucei (Zikova et al. 2009; Dean et al. 2013). Amplicon-based RNA-seq was used to compare edited transcripts associated with RESC or REH2C complexes, and in mtRNA. These studies linked the enrichment of fully edited mRNAs in the RESC6-bound RESC variant to site-preferential changes in total and accurate editing. Our studies also defined REH2C-mediated substrate-specific and site-preferential changes in editing in transcripts associated with RESC and in mtRNA.
RESULTS
Earlier studies in procyclic T. brucei indicated that the proportion of fully edited mRNAs, including RPS12 and A6, is relatively higher in RESC6-purified RESC than in REH2C or mtRNA (Madina et al. 2014, 2015). Those authors also showed that KREH2 affects the level of fully edited mRNAs, as well as the general pattern of editing progression in transcripts associated with RESC6-purified RESC. Because affinity purification of RESC6-tagged RESC included most of the known RESC proteins (Ammerman et al. 2011), hereon, we will refer to RESC6-containing RESC as “RESC,” a presumed fully active variant of this complex (for review, see Cruz-Reyes et al. 2018a). In this study, again focusing on procyclic T. brucei, we asked whether the differential enrichment of edited mRNAs in RESC, and the KREH2-mediated effects on the editing of these transcripts, involve editing changes in all positions or select positions in RPS12 and A6.
To address these questions, we used amplicon-based RNA-seq analyses to identify putative differential editing events at every position in mRNA RPS12 and A6 transcripts at steady state in RESC6-IPs, KREH2-IPs, KH2F1-IPs, and total mtRNA. The immunoprecipitated samples represent the editing complexes RESC and REH2C (including KREH2 and zinc-finger protein KH2F1 subunits) (Kumar et al. 2016). We also applied RNAi of KREH2 or KH2F1 to examine trans effects of the REH2C complex on RESC-associated transcripts. In total, we examined 42 samples and 19 different conditions. The samples used in each figure are indicated in Supplemental Table S1. Pearson's correlation analyses showed no correlation coefficients between the two independent biological replicates of each sample are smaller than 0.9 (Supplemental Table S1), demonstrating strong experimental replicability.
Targeted RNA-seq studies of mRNA RPS12
We examined the RPS12 sequence that starts with edits directed by the initiating gRNA and includes ORF (121 positions) and 5′ UTR (14 positions) regions. We identified all positions in the RPS12 amplicons that may be potentially modified by editing, that is, by numbering each position between two non-T bases (AGC) in the genome sequence from 5′ to 3′. These positions in the reference T-stripped sequence are referred to hereafter as “T-str” positions. We scored all editing events immediately 5′ of each non-T nucleotide after removing the primer sequences comparing RESC6-IPs to other samples (Supplemental Fig. S1; Supplemental Table S2). ESs were numbered from 3′ to 5′. The first two canonical sites, ES1 and ES2, in the 3′ UTR of RPS12 were not examined in this study as they were contained in the reverse PCR primer used to generate the transcript amplicons (Supplemental Fig. S1). Recent studies found alternative 5′ UTR sequences within a strain or between strains, perhaps because of the rapid evolution of this region (Simpson et al. 2016; Kirby and Koslowsky 2017; Cooper et al. 2019). While different 5′ UTR sequences are likely functional, we used a predicted 5′ UTR version, which was present in a relatively high number in all our RPS12 Illumina libraries. The 5′ UTR version reported in most studies (Read et al. 1992; Koslowsky et al. 2013; Simpson et al. 2016) and other possible 5′ UTR variants were present at much lower numbers or missing in our amplicons.
Distribution of editing events and analysis of total editing in RPS12 from mtRNA, REH2C, and RESC
We asked whether the reported ratios of fully edited RPS12 in RESC, REH2C, and mtRNA that were examined using qRT-PCR involves differences in the total number of possible edits among samples. In that case, we also wondered whether those differences would affect all or only some positions in RPS12. To address these questions, we began by scoring the relative distribution of all editing events at ESs and nESs in RPS12 from four different samples: RESC6-IPs, KREH2-IPs, KH2F1-IPs, and mtRNA. We checked the integrity of canonical proteins in RESC, REH2C, and RECC using western blot analyses of these samples (Supplemental Fig. S2A,B; Kumar et al. 2016). Transient RECC enzyme contacts with accessory editing complexes in our pulldowns were detected using radioactive autoadenylylation of an RNA ligase in RECC (Sabatini and Hajduk 1995). Supplemental Table S1 indicates the source of the biological replicates in this analysis, including the IP and mtRNA samples shown in Supplemental Figure S2. Western blots of representative pulldowns using the same affinity-purified antibodies and standard IP conditions in our prior reports (Madina et al. 2014, 2015; Kumar et al. 2016; see Materials and Methods) showed that RESC6-IPs had negligible levels of KH2F1. Similarly, KH2F1-IPs had negligible levels of RESC2 (a protein subunit in RESC6-bound RESC). However, we found a minor amount of KREH2 in the RESC6-IPs. Despite this minor cross-contamination, the current study confirmed the expected enrichment of fully edited mRNA in RESC6-IPs, as shown below. To illustrate the raw data collected in our studies of full or partial editing at ESs, and any editing event at nESs, we included a representative stacked histogram of one of our replicates (Fig. 1). This histogram provided a “snapshot” of RPS12 editing events at steady state with percentages of all editing events at each ES and nES showing a typical overall decrease in a 3′ to 5′ direction (Decker and Sollner-Webb 1990; Koslowsky et al. 1992). All subsequent analyses used the collected information directly comparing independent biological replicates of each sample. Surprisingly, we found that all 5′ positions in the amplified RPS12 sequence contained edits, that is, further upstream of the 5′-most ES reported in most earlier studies of RPS12 editing. Because the boundary in the 5′ UTR in RPS12 remains uncertain, we focused hereon on the ORF region of this transcript. First, we added up all editing events at each position (i.e., excluding preedited sequence reads) and plotted the total editing values (see Table 1 for terminology) along the RPS12 ORF sequence in two independent samples groups: RESC6-IPs, KREH2-IPs, and mtRNA (Fig. 2A) or RESC6-IPs, KH2F1-IPs, and mtRNA (Supplemental Fig. S3A). Each group of samples derived from a distinct cell line (Supplemental Table S1). The first five ESs at the 3′ end of RPS12 in all samples examined exhibited high values of total editing (∼90%–95% of all reads), including the 3′-most position examined (T-str 151, aka ES3), which creates the stop codon. All nESs in RPS12 in our samples carried editing events in at least some of the reads. However, most nESs exhibited relatively fewer editing events than the ESs.
FIGURE 1.
“Snapshot” of a typical data set collected using targeted RNA-seq analysis of RPS12 amplicons in this study. Stacked histogram of editing events at each position in RPS12 in a representative replicate sample of RESC6-IP. The cDNA fragment examined is shown as a reference T-stripped sequence (T-str) spanning the internucleotide positions T-str 17 to 151 (see the canonical fully edited RPS12 ORF and 5′ UTR “B-form” sequence in Supplemental Fig. S1). A canonical editing site (ES) for uridine insertion (Ins) or deletion (Del) is just 5′ to a red or a blue non-T nucleotide, respectively. A noncanonical editing site (nES) is just 5′ to a black non-T nucleotide. The representative histogram shows the percentage of RNA-seq reads at each position: ESs with correct insertion (red) or deletion (blue), and incorrect “partial” editing events (yellow). The remaining reads contain a preedited sequence. Editing events at nESs were scored (black). nESs do not require sequence changes in mature transcripts. However, all nESs at steady state carried editing events in our samples. The start codon (box) includes T-str positions 30–31 and requires editing at T-str 31 (ES72). The stop codon (box) includes T-st 151 (ES3) and requires editing at this position.
FIGURE 2.
Total RNA editing in mtRNA, KREH2-IPs, and RESC6-IPs: (A) Site-by-site, and (B) cumulative analysis in the RPS12 ORF sequence. Total editing at each site is scored as the percentage of RNA-seq reads showing any editing event, either correct or incorrect, at that site. T-str and ES positions, including those forming the start and stop codons are indicated. Each 10 in the T-str sequence is indicated (+). In B, The RPS12 ORF sequence is divided into two halves (T-str positions 31–88 and 89–151, divided by a vertical line). T-str position 51 “ES54” marked by an empty arrowhead (A,B) indicates the first position in a cluster of ESs with increased total editing in the ORF 5′ half in RESC6-IPs. Values and error bars reflect the mean of n = 2 +SD independent biological replicates. Blue bars represent frequent gRNAs for RPS12 in an extensive gRNA transcriptome study (Koslowsky et al. 2013). Differently colored vertical bars represent the boundaries of each editing block. A sliding window analysis was performed. Three windows centered at positions T-str 43, 65, and 94, respectively, are shown. P-values <0.0005 (***), <0.005 (**), <0.05 (*), and >0.05 (NS) comparing RESC6-IP to mtRNA (top tier) or KREH2-IP to mtRNA (bottom tier) were annotated in panel A.
We also noted a higher percentage of editing events in the 5′ half of RPS12 transcripts from RESC6-IPs than from other samples. In contrast, values of total editing in the 3′ half of RPS12 appeared similar between these samples. To assess the statistical significance of the differences between samples, we performed a sliding window analysis (Zivot and Wang 2006) that confirmed our visual inspection. Only the windows in the 5′ half showed significant differences among groups. The transition between significant differences to no differences is found with sliding windows ending around the position 80.
We plotted cumulative values of total editing along RSP12 (Fig. 2B; Supplemental Fig. S3B). To highlight differences along the ORF, we divided the sequence into 5′ and 3′ halves based on our general observations in Figure 2A. Positions T-str 88 (ES38) and T-str 89 (ES39) marked the boundary between the two halves. The cumulative value at T-str 31 (ES72), which creates the start codon, was higher in RESC6-IPs relative to KREH2-IPs and mtRNA, ∼43% and ∼98%, respectively (Fig. 2B). This cumulative value was also higher in RESC6-IPs relative to KH2F1-IPs and mtRNA, ∼44% and ∼51%, respectively (Supplemental Fig. S3B). Differences in the cell lines used (e.g., inoculum quality or growth rate) for each sample group, or our manipulation of the samples, may account for the variations in cumulative values between the two sample groups. Possible differences in REH2C protein subunit stoichiometry, as previously suggested after sedimentation analysis of mitochondrial extracts (Kumar et al. 2019), may also cause cumulative differences between KREH2-IP and mtRNA samples versus KH2F1-IP and mtRNA samples. Notably, total editing in most ESs in the 5′ half (approximately ES45 to ES72) was relatively stable compared to the gradual decline in total editing in the 3′ half. We found significant differences in total edits between samples in a 5′ region that contains a nearly continuous cluster of ESs, beginning at T-str 51 (ES54; arrowhead), including the start codon. At least three gRNAs direct editing along this cluster of ESs in RPS12. Our sliding window analysis of the RPS12 3′ half did not reveal significant differences between samples. In general, the ORF 5′ half provided a useful estimate of the overall differences in cumulative total editing in RPS12 between our samples.
Overall, we found a site-preferential enrichment in total editing in RPS12 transcripts associated with RESC (RESC6-IPs) compared to transcripts associated with REH2C (KREH2-IPs or KH2F1-IPs) or in mtRNA. Significant differences in total editing among samples concentrated in the 5′ half of RPS12, which includes mainly canonical ESs spanning at least four gRNAs. The reported enrichment of fully edited mRNA in RESC (Madina et al. 2014, 2015) may be partly explained by an increased action (i.e., higher frequency of total edits) by the RECC enzyme in the context of RESC. However, this enhanced editing preferentially affects 5′ positions in the RPS12 transcript, which may be better exposed to editing catalysis in the RESC complex. These observations are consistent with the general idea that RESC6-purified RESC contains a complement of proteins and gRNAs, and trans factors that promote editing on associated transcripts.
Analysis of accurate editing using Inc/Cor values at each position of RPS12
We asked if the above preferential changes in total edits at 5′ positions between sample groups may correlate with changes in editing accuracy at those positions, or whether changes in total edits and accurate edits occur at different positions in RPS12. We assessed editing accuracy in RPS12 by using a ratio value at each ES and nES that takes into account the total number of sequences with the correct T number (“Cor” value) expected in the canonical form and the total number of sequences with the incorrect T number (“Inc” value; here defined as different from the expected canonical form) (Supplemental Fig. S4). A normalized ratio value in the form “Inc/Cor” is particularly useful at ESs because close inspection of the individual Cor and Inc values at these positions showed that these two types of values do not always change proportionally between samples (see the closeup panels B and D in Supplemental Fig. S4; Supplemental Table S3). That is, an increase in correct sequences was combined with either a decrease, increase, or minimal change in incorrect sequences. At ESs, the total RNA-seq reads (100%) includes preedited, correct (Cor), and incorrect (Inc) sequences. At nESs, the total RNA-seq reads (100%) includes preedited (Cor) and incorrect (Inc) sequences. The cognate gRNA dictates the canonical Cor value at ESs and nESs. However, the Inc value may reflect misuse or alternative use of a cognate gRNA, or a noncognate gRNA (Decker and Sollner-Webb 1990; Koslowsky et al. 1991; Maslov and Simpson 1992). In general, Inc/Cor values provided a useful indicator of the overall editing accuracy at each position because they take into account possible differences in the accumulation of correct and incorrect sequences, regardless of the total editing or the source of the incorrect edits in different samples.
RESC-associated RPS12 transcripts exhibit particularly accurate editing
Analyses of Inc/Cor values in RPS12 in our samples (Fig. 3A; Supplemental Fig. S5A) revealed a few general features about the accuracy of editing in this transcript. Inc/Cor values varied substantially along RPS12 but did not readily distinguish between insertion and deletion ESs. Inc/Cor values were generally higher in the 3′ half than the 5′ half of RPS12. Also, Inc/Cor values were generally higher at ESs than at nESs and lower at the few C nucleotides (just 3′ of ESs or nESs), potentially forming stabilizing C–G base pairs with gRNA. A few exceptions to these generalities, particularly at a few positions at the 3′ end, are discussed below. Notably, we found a significantly higher editing accuracy in the RPS12 5′ half in RESC6-IPs than in other samples.
FIGURE 3.
RNA editing accuracy in mtRNA, KREH2-IPs, and RESC6-IPs: (A) Site-by-site, and (B) cumulative Inc/Cor analyses in the RPS12 ORF sequence. Inc/Cor values are scored as the percentage of RNA-seq reads with incorrect editing divided by the percentage of RNA-seq reads with correct editing, according to the canonical fully edited RPS12 sequence. For clarity in B, RPS12 is divided into two regions (T-str positions 31–88 and 89–151, respectively), each with a separate plot of cumulative Inc/Cor. Major differences in Inc/Cor between samples include those at T-str positions 48 and 80 (ES57 and ES46, respectively; filled arrowheads). (C) Distribution of high-frequency incorrect edits (Inc) in RESC6-IPs. Color-coded arrowheads indicate high-frequency reads with incorrect editing (>5% to >30%) at those positions. The RPS12 3′ half included most high-frequency reads of incorrect editing (horizontal broken line). Blue bars represent frequent gRNAs for RPS12 as in Figure 2. Values and error bars reflect mean of n = 2 +SD independent biological replicates. Sliding window analyses are included as in Figure 2 comparing RESC6-IP to mtRNA (top tier) or KREH2-IP to mtRNA (bottom tier).
Based on the general observations in Figure 3A above, we divided the ORF region into two halves (i.e., same as in Fig. 2B) and examined our samples in plots of cumulative Inc/Cor values along RPS12. At the start codon, the cumulative Inc/Cor was lower in RESC6-IPs versus KREH2-IPs and mtRNA, by 43% and 60%, respectively (Fig. 3B). Similarly, this cumulative Inc/Cor was lower in RESC6-IPs versus KH2F1-IPs and mtRNA, ∼42% and ∼44%, respectively (Supplemental Fig. S5B). As above, the different cell lines for each group of samples or our manipulations of the samples may account for the variations in cumulative values between the two sample groups. Possible variations in REH2C protein subunit stoichiometry, as suggested above (Kumar et al. 2019), could also cause cumulative differences between KREH2-IP and mtRNA samples versus KH2F1-IP and mtRNA samples. Several prominent spikes in Inc/Cor appeared to contribute to the differences between the cumulative curves of these samples. These spikes primarily included 5′ positions starting at positions T-str 48 (ES57) and T-str 80 (ES46), respectively (filled arrowheads in Fig. 3A,B; Supplemental Figs. S5A,B). Some spikes in Inc/Cor are near, or include, gRNA termini. At least one cluster of spikes, including position T-str 48, is in a region with significant differences between samples (Fig. 3A,B) and spans an overlap of gRNAs (Fig. 3C), suggesting that some spikes in unfaithful editing may be linked to the transition between gRNAs.
In line with the generally lower editing accuracy in the 3′ half than the 5′ half of RPS12, an analysis of incorrectly edited sequences showed that most 3′ positions of RPS12 in RESC6-IPs contained at least one prominent event of incorrect editing at high frequency (i.e., representing between ≥5% and ≥30% of all reads). In contrast, in most 5′ positions in RESC6-Ips, the most prominent events of incorrect editing were relatively less frequent (<5% of all reads) (Fig. 3C).
The above analyses revealed a jagged profile of editing accuracy along RPS12. Prominent spikes of Inc/Cor at common positions in all samples imply intrinsic “hotspots” of inaccurate editing. Moreover, a decrease of these “hotspots” or spikes at 5′ positions in RESC-associated RPS12 would contribute to the known enrichment of fully edited RPS12 in RESC relative to REH2C and total mtRNA (Madina et al. 2014, 2015). Significant differences in editing accuracy were also detected further downstream in some samples. While we found significant changes in both total editing and accurate editing, major shifts in these two processes did not necessarily involve the same positions in RPS12. This discrepancy suggests that changes in editing accuracy are not directly linked to changes in total editing in our samples. However, our analyses of both processes implied an increased RECC function on RESC-associated RPS12.
RNAi of KH2F1 or KREH2 decreased total editing particularly at 5′ positions in RESC-associated RPS12 transcripts
RNAi of KREH2 is known to decrease the level of fully edited RPS12 in RESC (RESC6-IPs) (Madina et al. 2015). We specifically examined RESC-associated RPS12 and asked whether the helicase complex REH2C affects total edits, accurate edits, or both. We also wondered whether the same RPS12 positions exhibiting differential changes in RESC (relative to REH2C or mtRNA) were also affected upon REH2C loss-of-function. To address these questions, we began by examining the possible effects of KH2F1 or KREH2 knockdowns on total editing in RPS12 in RESC6-IPs (Fig. 4). RNAi at days 3 and 4 postinduction caused no major secondary effects on the steady-state level of RESC2 (aka GAP1), a canonical protein in RESC (Supplemental Fig. S2A,B). The impact of the knockdowns on growth was followed over several days (Supplemental Fig. S2C,D). Previous studies using the same RNAi cell lines and conditions found no evident effects on canonical editing proteins at steady state, including RESC2, RESC6, TbRGG2 (aka RESC13), and KREL1 (Madina et al. 2015; Kumar et al. 2016). RNAi of KH2F1 destabilized KREH2 as reported (Kumar et al. 2016). Supplemental Figure S2A,B also shows that the induced RNAi knockdowns effectively reduced KREH2 and KH2F1 in mitochondrial extracts and also the minor cross-contamination of KREH2 in RESC6-IPs.
FIGURE 4.
Total RNA editing in RESC6-IPs upon RNAi of REH2C proteins: (A) KH2F1–RNAi, and (B) KREH2–RNAi. Plots of cumulative total editing in RPS12, as in Figure 2B. The same positions as in Figure 2B showed evident changes in cumulative total editing, including T-str position 51 (ES54; empty arrowhead in A and B). Uninduced (−Tet) and induced (+Tet) at days 3 or 4. Values and error bars reflect mean of n = 2 +SD independent biological replicates. Some error bars are tiny. Sliding window analyses are included comparing −Tet to +Tet day 4 (top tier) or +Tet day 3 (bottom tier).
Cumulative plots showed a significant decrease in total editing on the RPS12 5′ half in RESC6-IPs of both examined knockdowns (Fig. 4). Values at the start codon decreased at days 3 and 4 upon KH2F1–RNAi (∼32% and ∼64%; panel 4A), and KREH2–RNAi (∼13% and ∼39%; panel 4B), respectively. Significant differences between samples were found at the two time points of RNAi induction. The ORF 5′ half in RPS12 provided a useful estimate of the overall impact of these knockdowns on total editing.
Interestingly, major shifts in total editing in the RPS12 5′ half in RESC6-IPs appeared to involve the same 5′ positions upon RNAi (Fig. 4) and in our comparison of RESC6-IPs with other samples (Fig. 2). These 5′ positions included a cluster of ESs (e.g., including position T-str 51 [ES54]) that contributed to the differences in the cumulative plots of total editing between samples.
In summary, REH2C loss-of-function markedly inhibited total editing preferentially at positions in the 5′ half of RPS12 transcripts associated with RESC. Particularly ESs in RPS12 were affected. This phenotype in total editing was confirmed with RNAi against two REH2C protein subunits, KH2F1 and KREH2. The transcript for each protein was tested at two different RNAi time points, and each time point included two independent replicates. Importantly, the same 5′ positions in RESC-associated RSP12 exhibited differential changes in total editing upon REH2C loss-of-function and in our analysis of RESC compared to REH2C and mtRNA. This finding suggested that the observed change in total editing at these positions is physiologically relevant. As aforementioned, several gRNAs are involved in the RPS12 5′ half, so it was unclear whether their location is linked to the observed RNAi effects on total editing.
RNAi of KH2F1 or KREH2 decreased accurate editing particularly at 5′ positions in RESC-associated RPS12 transcripts
We asked whether changes in editing accuracy may contribute to the known decrease of fully edited RPS12 in RESC upon KREH2 depletion (Madina et al. 2015). In that case, we also wondered whether the same RPS12 positions that exhibited differential changes in RESC versus other samples would be affected upon REH2C loss-of-function. We addressed these possibilities by examining Inc/Cor values of RPS12 in RESC6-IPs upon KH2F1 or KREH2–RNAi (Fig. 5).
FIGURE 5.
RNA editing accuracy in RESC6-IPs upon RNAi of REH2C proteins: (A) Site-by-site, and (B,C) cumulative Inc/Cor in RPS12 as in Figure 3. KH2F1–RNAi (A,B) and KREH2–RNAi (C). Values and error bars reflect mean of n = 2 +SD independent biological replicates. Some error bars are tiny. Sliding window analyses are included comparing −Tet to +Tet day 4 (top tier) or +Tet day 3 (bottom tier).
We found a significant decrease in editing accuracy on the RSP12 5′ half of in both knockdowns. Values at the start codon decreased at days 3 and 4 of KH2F1–RNAi (∼62% and ∼139%; panel 5B) and KREH2–RNAi (∼17% and ∼58%; panel 5C), respectively. Notably, major shifts in Inc/Cor involved the same RPS12 positions in RESC6-IPs upon RNAi (Fig. 5) and in our above study comparing RESC6-IPs versus other samples (Fig. 3A,B). As mentioned above, correct and incorrect sequences at ESs do not always change proportionally, in this case, upon RNAi (Supplemental Fig. S6). A close examination of the positions around T-str 48, 80, and 88 (ES57, ES46, and ES39, respectively) revealed that the percentage of correct sequences decreased at these positions upon KH2F1–RNAi at days 3 and 4 postinduction (Supplemental Fig. S7A). In contrast, high-frequency incorrect sequences at these positions appeared relatively unaffected by the knockdown (Supplemental Fig. S7B). This observation suggested that canonical editing at those ESs was affected by the knockdown. Inc/Cor values provided a useful indicator of editing accuracy at each position in our RNAi analyses.
Overall, RNAi of KH2F1 and KREH2 reduced the level of accurate editing in RESC-associated RPS12. However, both knockdowns only exhibited significant effects on the RPS12 5′ half. Notably, the same RPS12 positions exhibited major shifts in accurate editing in two different experimental settings: that is, in RESC upon REH2C loss-of-function, and comparisons of RESC, REH2C, and mtRNA. This finding suggested that changes in accurate editing at those positions are physiologically relevant. As mentioned above, a short cluster of spikes, including position T-str 48, spans an overlap of gRNAs. So, some changes in accuracy may involve the transition between gRNAs. It is also feasible that changes in secondary structure are involved in this region. The analysis of these possibilities will require additional studies. The effects of KH2F1 and KREH2 knockdowns indicated that the REH2C complex promotes accurate editing primarily at 5′ positions in RESC-associated RPS12.
Early editing in RESC-associated RPS12 transcripts
We identified several high-frequency events of incorrect editing in the region spanning gRNA-2 to gRNA-5 (Fig. 3C). In contrast to the region across gRNA-2 to gRNA-5, which is prone to unfaithful editing, most positions covered by gRNA-1, including the 3′-most positions examined, exhibited relatively faithful editing. We reasoned that the dramatic shift in editing accuracy in the gRNA-1/gRNA-2 transition could have a major impact on editing progression. To examine this possibility, we further analyzed early editing events in RPS12 from RESC6-IPs, including adjoining positions showing abrupt Inc/Cor changes from low to high (Fig. 6). The single gRNA-1 isoform (3′ portion), and the most abundant gRNA-2 isoform in an extensive gRNA-seq study in EATRO 164 cells by the Koslowsky laboratory are depicted in Figure 6A. The initiating gRNA-1 anneals to positions T-str 142–151 (including ES3 to ES7 in this depiction). The gRNA-2 anneals to positions T-str 123–148 (including ES4 to ES17) (Koslowsky et al. 2013). A gRNA-1 bearing an identical guide sequence was found in a limited gRNA-seq study in Lister 427 cells used in the current study (Madina et al. 2014) and in annotated DNA minicircles in EATRO 1125 AnTat1.1 cells (Cooper et al. 2019).
FIGURE 6.
RNA editing at the 3′ end of the RPS12 ORF sequence. (A) Alignment of canonical fully edited RPS12 3′ end transcript sequence to cDNA sequence corresponding to gRNA-1 (3′ portion) and gRNA-2. Color-coded arrowheads indicate high-frequency reads with incorrect editing as in Figure 3C. Trapezoids indicate positions with relatively low Inc/Cor <0.001 or ∼0.001–0.005 that anneal to gRNA-1. Positions T-str 136, 142, and 146 have relatively high Inc/Cor ∼0.3–0.6. The gRNA-1 (single isoform) and gRNA-2 (most frequent isoform) identified in T. brucei EATRO 164 were used (Koslowsky et al. 2013). (B) Alignment of gRNA-1 that may account for a high-frequency event of incorrect editing (∼35% of all reads at position T-str 142, marked by a box). Two adenines in the gRNA 3′ end may direct this event. (C) Frequency of incorrectly edited sequences (Inc) at position T-str 142 in RESC6-IPs upon the indicated RNAi at days 0, 3, and 4 postinduction. The total Inc (all incorrect sequences) and the most frequent incorrectly edited sequence (+2U) at position T-str 142 are plotted. (D) Site-by-site Inc/Cor upon KREH2–RNAi across positions T-str 122–151. Positions T-str 136, 142, and 146 are marked. Blue bars depict gRNA-1 (g1) and gRNA-2 (g2). Values and error bars reflect mean of n = 2 +SD independent biological replicates.
The gRNA-1 and gRNA-2 transcripts cover positions with very low Inc/Cor but also positions with relatively high Inc/Cor. The entire RPS12 ORF region examined in RESC6-IPs has seven Inc/Cor values <0.001 (Supplemental Table S4). These include the first four positions examined at the 3′ end (T-str 148–151, including insertion ES3 and ES4, and two nESs). So, over half of the Inc/Cor values <0.001 in the RPS12 ORF region depend on gRNA-1.
Four positions directed by gRNA-1, T-str 143, 144 (deletion ES7), 145, and 147 (insertion ES5), exhibited Inc/Cor values between ∼0.001 and ∼0.005. These four positions represent one-third of all values within this range in the examined ORF region (Supplemental Table S4). In contrast to the low values above, three positions across gRNA-1 and gRNA-2, T-str 136, 142, and 146 (deletion ES6) exhibited relatively high Inc/Cor values. The nES at position T-str 142 had one of the two largest Inc/Cor values in the entire ORF region despite this position involving a stabilizing C–G base pair (i.e., T-str 142 is just 5′ to C). While gRNA-1 directs canonical editing at position T-str 146, gRNA-2 directs canonical editing at positions T-str 136 and 142 (Fig. 6A). However, gRNA-1 may also direct noncanonical editing at T-str 142, as described below (Fig. 6B). An analysis of incorrectly edited reads (Inc values) showed that at positions T-str 136, 142, and 146 the values largely depended on a high-frequency editing event. At position T-str 142 (nES), a 2U-insertion event accounted for ∼35% of all reads (and >90% of the Inc value) at that position in RESC6-IPs (Fig. 6B,C; Supplemental Table S5, panel A). This 2U-insertion was consistently the most frequent event in all our samples. We noted that two adenines in the 3′ terminus of gRNA-1 might direct the noncanonical 2U-insertion at T-str 142 (Fig. 6B). Importantly, these two 3′-terminal adenines are conserved in the sequenced gRNA-1 in Lister 427 cells (used in this study) and EATRO 164 cells, and in two initiating gRNAs in annotated DNA minicircles in EATRO 1125 AnTat1.1 cells (Koslowsky et al. 2013; Madina et al. 2014; Cooper et al. 2019). The conservation of these adenines suggests that they are functionally important.
We specifically examined the fold change in Inc/Cor at the adjacent positions T-str 142 and 143 (i.e., Inc/Cor ∼0.6 and ∼0.001, respectively) in RESC-associated RPS12. The T-str 142/143 transition predicted a >500-fold loss in editing accuracy during 3′–5′ progression. This Inc/Cor fold change was the largest identified between adjoining sites in the ORF region examined (Supplemental Table S4). Consequently, the T-str 142/143 transition may cause a major pause in editing progression. The second and third largest Inc/Cor fold change values were at the T-str 138/139 and T-str 146/147 transitions (>85-fold and >65-fold, respectively). The KH2F1 or KREH2 knockdowns slightly increased both the percentage of the 2U-insertion event and the Inc value at position T-str 142 (Fig. 6C). Accordingly, these two independent RNAi knockdowns increased Inc/Cor at position T-str 142, and the Inc/Cor fold change at the T-str 142/143 transition (Fig. 6D; Supplemental Table S5). The second most frequent incorrect event at T-str 142 (>2%) did not seem affected by these knockdowns (Supplemental Table S5). However, because very few positions differed among samples in our analysis of 3′ editing, we were unable to establish significant differences using a moving window analysis.
Overall, the initiating gRNA-1 exhibits at least two interesting features during RPS12 editing. First, the 5′ end of gRNA-1 directs particularly faithful editing at the first few positions examined in RPS12. This accurate editing directed by gRNA-1 precedes several high-frequency incorrect events across the subsequent four gRNAs. Second, two conserved adenines in the 3′ end of gRNA-1 may direct the most frequent event of noncanonical editing (at position T-str 142) in the examined ORF region. The T-str 142/143 transition may cause a major pause during 3′–5′ editing progression in RPS12. The proposed pausing at the T-str 142/143 transition in RESC-associated RPS12 was exacerbated by independent knockdowns of two REHC proteins in our visual characterization. However, additional studies will be needed to establish the significance of REH2C effects on editing progression at the T-str 142/143 transition. Regardless, major pausing at the T-str 142/143 transition directed by two conserved adenines in the 3′ end of gRNA-1 may be used to restrict the RECC passage into upstream gRNAs.
Targeted RNA-seq studies of mRNA A6
We wondered whether REH2C exhibits site-specific effects on A6 in RESC6-IPs, as we found in RPS12 in our samples. The ORF sequence in A6 is considerably larger than in RPS12 (Bhat et al. 1990). However, prior studies using qRT-PCR and Sanger sequencing of a cDNA fragment across the first editing block suggested that KREH2 affects 3′ early editing in the A6 substrate (Madina et al. 2015). We examined an A6 3′ fragment that includes ORF (78 positions) and UTR (22 positions) sequences (Supplemental Fig. S8). Editing of this 3′ fragment involves the initiating gRNA. As with RPS12, we scored all editing events at each position after removing the primer sequences. While alternative 3′ UTR edited sequences in A6 are likely functional, we adopted a 3′ UTR edited version that was present in a relatively high number in all our A6 Illumina libraries, and is complementary to sequenced or predicted gRNAs. The 3′ UTR edited version reported in most studies (Bhat et al. 1990; Koslowsky et al. 2013) and other possible 3′ UTR variants examined were present at a lower frequency or missing in our amplicons.
KH2F1–RNAi affects total and accurate edits at preferential 3′ positions in RESC-associated A6 transcripts
We only examined the KH2F1 knockdown because this protein stabilizes KREH2, and the individual KH2F1 and KREH2 knockdowns generated similar outcomes on RPS12 editing. As in RPS12, total editing of A6 in 3010-IPs was generally higher in canonical ESs than in nESs. Because most edits occurred in a sequence spanning the first five gRNA transcripts (i.e., covering ∼50% of the amplified region), we focused on that sequence that undergoes early editing in A6 (Fig. 7). The relatively low level of editing events in further upstream positions in the amplicons may be partly due to the fact that our 5′ primer for amplification targets a preedited sequence.
FIGURE 7.

Total RNA edits in A6 associated with RESC6-IPs upon KH2F1–RNAi: (A) Site-by-site, and (B) cumulative analysis A6 mRNA. T-str (71–115) and ES positions, including ES1 and the stop codon (square) are indicated. Positions upstream of T-str 71 are omitted because the small number of edits at those sites have a negligible effect on the cumulative plot. The gRNAs (blue bars) mostly derive from an extensive gRNA transcriptome study (Koslowsky et al. 2013). The edited 3′ UTR region anneals with gRNA-1 and gRNA-2. We used an alternative gRNA-1 transcript. Variants of gRNA-1 with identical guiding capacity were sequenced in strains EATRO 164 and Lister (Koslowsky et al. 2013; Madina et al. 2014) or predicted in sequenced minicircles in strain EATRO 1125 (Cooper et al. 2019). gRNA-2 was predicted in strain EATRO1125. Values and error bars reflect mean of n = 2 +SD independent biological replicates. Sliding window analyses are included comparing −Tet to +Tet day 4 (top tier) or +Tet day 3 (bottom tier).
Surprisingly, the KH2F1 knockdown increased the level of total edits at several 3′ positions of A6 in RESC6-IPs (Fig. 7). The cumulative value at T-str 71 increased at days 3 and 4 of KH2F1–RNAi, ∼75% and ∼84%, respectively. We found significant differences upon RNAi in a short region across positions T-str 102–109, involving gRNA-1 and the gRNA-1/gRNA-2 overlap. The region across positions T-str 86-98, largely including gRNA-3, and the gRNA-2/gRNA-3 overlap, also showed a significant effect upon RNAi.
Despite the observed increase in total edits, the KH2F1 knockdown significantly increased Inc/Cor in 3′ early editing of A6 in 3010-IPs (Fig. 8A,B). This loss in editing accuracy involved the same short region across gRNA-1 and the gRNA-1/gRNA-2 overlap in our analysis of total edits. Six positions, T-str 102, 103, 106, 107, 108, and 109, exhibited major changes in both total editing and editing accuracy upon KH2F1–RNAi, suggesting that the two processes are related in these positions in A6. The cumulative Inc/Cor value at T-str 59 increased at days 3 and 4 of KH2F1–RNAi, ∼64% and ∼120%, respectively.
FIGURE 8.
RNA editing accuracy in A6 from RESC6-IPs upon KH2F1–RNAi: (A) Site-by-site, and (B) cumulative analysis spanning T-str positions 59–115 (i.e., including ES1–34). Positions upstream of T-str 59 are omitted because the sequence reads containing edits became too few (or zero) in one or both replicates to calculate Inc/Cor ratio. Positions across 102–109 are marked. (C) Site-by-site percentage of incorrect (Inc; top) and correct (Cor; bottom) editing reads at the positions marked in A. Values and error bars reflect mean of n = 2 +SD independent biological replicates. Sliding window analyses are annotated comparing −Tet to +Tet day 4 (top tier) or +Tet day 3 (bottom tier) in B and C. (D) Alignment of fully edited A6 3′ end sequence to gRNA-1 (3′ portion) and gRNA-2. Color-coded arrowheads indicate high-frequency reads with incorrect editing, as in Figure 3B. The alternative gRNA-1 sequence (gA6-1.alt) in strain Lister (Madina et al. 2014) and a predicted gRNA-2 (m0_306(II)_gA6_v2 [724–766]) in strain EATRO 1125 (Cooper et al. 2019) were used. (E) Predicted sequence including the most frequent sequences with incorrect editing (highlighted in gray) at six positions across T-str 102–109 marked in A and D.
We wondered whether the Inc/Cor changes at the canonical ESs at positions T-str 102, 103, 106, and 107 were due to changes in incorrect edits, correct edits, or both. Analysis of the Inc value at these positions showed Inc value increases from 5%–15% to 30%–60% upon RNAi (Fig. 8C, upper). We found a similar increase in Inc value at both nESs at T-str positions 108 and 109, suggesting that a common mechanism underlies the accumulation of incorrectly edited sequences at these ESs and nESs across gRNA-1. The region spanning positions T-str 102–109 exhibited some of the highest Inc values in all our A6 samples (Supplemental Table S3). Analyses of the Cor value showed relatively stable Cor values at the ESs at T-str positions 102, 103, 106, 107. This suggested that canonical editing at those ESs was not affected by KH2F1–RNAi. The nESs at T-str positions 108 and 109 showed the expected converse changes relative to their Inc values (Fig. 8C, lower).
As in RPS12, high Inc values in A6 are largely driven by specific high-frequency events of incorrect editing (Fig. 8D). A predicted alternative short sequence, including all high-frequency incorrect edits across T-str positions 102–109 (Fig. 8E) was indeed present in all A6 samples, and its frequency increased upon RNAi. Nevertheless, in all A6 samples in this study, we found only one transcript that contained this alternative short sequence followed by 3′ fully edited sequence. The 5′ sequence in this transcript was preedited. This suggested that this alternative sequence element is usually flanked by preedited or partial edited flanking sequences in our A6 samples.
As in RPS12, we found that A6 in 3010-IPs showed particularly accurate editing in the first few 3′ positions (i.e., T-str 110–115, including ES1–ES5, with Inc/Cor ∼0.002–0.06). Also, as in RPS12, editing accuracy at these 3′ end positions in A6 was similar between samples. However, the accuracy of early 3′ editing in A6 was not as high as in RPS12 in our analyses (Supplemental Table S4). We found moderate fold changes in Inc/Cor in 3′ editing at the transitions T-str 109/110 and 113/114 (both ∼25-fold). Relatively high fold changes in Inc/Cor occurred in the 5′ half of the examined A6 sequence, including at the T-str 64/65 transition (∼88-fold), or even higher further upstream (Supplemental Table S5). However, whether these upstream values are affected by the 5′ primer used to amplify the 3′ fragment of A6 transcripts will need to be clarified by additional studies.
In brief, RESC-associated A6 mRNA showed major site-specific differences in editing accuracy, as we found in RPS12. The first few A6 positions directed by the 5′ half of gRNA-1 exhibited relatively accurate editing and showed no clear differences upon REH2C loss-of-function. However, a cluster of A6 positions normally targeted by the 3′ half of gRNA-1, showed a significant increase in both total editing and high-frequency incorrect editing upon RNAi. The function, if any, of the short alternative A6 element created by these incorrect edits will need to be clarified by additional studies. However, it is feasible that in normal cells, one role of REH2C is to reduce alternative editing events that may hinder A6 canonical editing progression beyond the initiating gRNA-1. We note that REH2C affected ESs with major changes in editing accuracy differently in A6 and RPS12. That is, REH2C loss-of-function largely affected the Inc value in A6 and the Cor value in RPS12 at those ESs.
REH2C loss-of-function effects on RPS12 and A6 transcripts in mitochondrial extracts
The current study described distinct site-preferential REH2C-mediated effects in RPS12 and A6 transcripts associated with RESC. We finally asked whether the total mitochondrial RNA pool exhibits the observed effects in RESC-associated transcripts.
Although RPS12 exhibits a lower level of 5′ edits in mtRNA versus 3010-IPs, both KH2F1 and KREH2 knockdowns caused a decrease in total edits at generally the same positions in mtRNA and RESC6-IPs (Fig. 4; Supplemental Figs. S9 and S10). The same 5′ cluster of positions in RPS12 in all samples exhibited a significant decrease in total editing upon RNAi. The cumulative value at the start codon decreased at days 3 and 4 of KH2F1–RNAi (∼44% and ∼56%; Supplemental Fig. S9) and KREH2–RNAi (∼25% and ∼40%; Supplemental Fig. S10), respectively.
The KH2F1 knockdown showed additional significant differences in total edits at further downstream positions of RPS12 in mtRNA. These other differences may reflect a distinct efficiency between the KH2F1 and KREH2 knockdowns in mtRNA, or the heterogeneous mixture of transcripts either free or associated with proposed variants of RESC and other editing complexes in mitochondria (for review, see Cruz-Reyes et al. 2018a).
The overall location of Inc/Cor spikes in the RPS12 ORF region appears to be the same in mtRNA and RESC6-IPs upon KH2F1–RNAi (Fig. 5; Supplemental Fig. S11). However, the relative extent of these Inc/Cor spikes in the 5′ and 3′ halves of RPS12 differed between mtRNA and RESC6-IPs. This difference may reflect that the immunopurified samples contain a subset of the transcripts found in the total mtRNA pool. The same cluster of 5′ positions, including T-str 48 (ES57), showed significant differences in both KH2F1 or KREH2 knockdowns in mtRNA and RESC6-IPs. In mtRNA, the cumulative value at the start codon decreased at days 3 and 4 of KH2F1–RNAi (∼14% and ∼26%; panel B) and KREH2–RNAi (∼15% and ∼38%; panel C), respectively.
Finally, we performed an Inc/Cor analysis of A6 in mtRNA upon KH2F1–RNAi (Supplemental Fig. S12). We found a conserved location of Inc/Cor spikes in 3′ positions of A6 in mtRNA and RESC6-IPs (Fig. 8; Supplemental Fig. S12). Inc/Cor values in 5′ positions may be affected by the low number of 5′ reads in mtRNA in our samples. The same 3′ region, spanning A6 positions T-str 102-109, showed significant changes in editing accuracy in mtRNA and RESC6-IPs upon KH2F1–RNAi. The cumulative value at T-str 59 decreased at days 3 and 4 of KH2F1–RNAi, ∼56% and ∼67%, respectively.
Despite the enrichment of accurately edited RPS12 and A6 in native RESC pulldowns, we found that significant REH2C-mediated changes in total editing and editing accuracy in these transcripts can be similarly examined in RESC and the total mitochondrial RNA pool.
DISCUSSION
This amplicon-based RNA-seq study examined RPS12 and A6 transcripts in antibody pulldowns of RESC, REH2C, and total mtRNA, using normalized values of total editing and editing accuracy (Inc/Cor). The new bioinformatics tool introduced here enabled nucleotide-resolution analysis of canonical and noncanonical edits at all positions. Table 2 summarizes the general findings in our study, with a focus on the ORF region in RPS12, and a 3′ fragment of A6. We included analyses of 3′ early editing by the initiating gRNA-1 in both transcripts. All previous amplicon-based RNA-seq studies of RNA editing in T. brucei examined total mitochondrial extracts (Simpson et al. 2016, 2017; Carnes et al. 2017; McAdams et al. 2018, 2019; Tylec et al. 2019). However, the presence of multiple RESC variants in mitochondria complicates their functional analysis in vivo (for review, see Cruz-Reyes et al. 2018a). We examined editing patterns in transcripts associated with a variant of RESC purified via RESC6, a critical protein in early editing (Ammerman et al. 2011), and the role of REH2C on these editing patterns. The function of the KREH2 subunit in the REH2C complex is of particular interest. This ATP-dependent RNA helicase of the DEAH/RHA family exhibits a conserved multidomain organization in related helicases from yeast to humans (Cruz-Reyes et al. 2016, 2018b; Kumar et al. 2019). KREH2 is the sole DEAH box helicase in trypanosomal editosomes. However, spliceosomes and ribosome biogenesis require multiple DExH/RHA helicases with a variety of functions (Jarmoskaite and Russell 2014).
TABLE 2.
Summary of findings on total and accurate editing in RPS12 and A6 mRNA transcripts

Analysis of RPS12
Editing pattern and enrichment of fully edited RPS12 in RESC
In RPS12, total editing was higher in ESs than in nES, and decreasing from 3′ end to the mid-region, then remaining relatively stable for most of the 5′ half. This pattern suggested that editing may progress more efficiently along the RPS12 5′ half. Perhaps the stretch of purines in this region facilitates progression.
The editing accuracy varied greatly site-to-site but did not distinguish ESs from nESs. Relatively high accuracy typically involved C-rich regions except in the first few 3′ positions of RPS12. C–G base pairs may help direct and stabilize accurate editing. Some spikes in Inc/Cor revealed discrete areas and specific positions, mostly ESs, along RPS12 that represent “hotspots” of unfaithful editing. Positions with high Inc/Cor >0.3 were 5′ of A's or G's, except for position T-str 142 (discussed below), which is 5′ to a C nucleotide. In general, the 5′ half spanning gRNA-2 to gRNA-5 is more error-prone than the 3′ half of the ORF region.
RPS12 transcripts exhibited higher levels of total editing and accurate editing in RESC than REH2C and mtRNA. Accordingly, our Illumina libraries from RESC6-IPs consistently contained most transcripts with a fully edited ORF region (∼10% of all transcripts) among the samples examined. These findings are in line with our reported qRT-PCR studies showing a relative enrichment of fully edited RPS12 in RESC6-IPs (Madina et al. 2014, 2015). Minor cross-contaminations between native complexes, which are difficult if not impossible to avoid completely in a case where these complexes interact in a dynamic fashion, do not compromise the major conclusions in our studies. However, they might have obscured other, more subtle differences. Observed differences between our samples were significant, in some cases with P-values <0.0005. The differences in fully edited RPS12 between RESC and REH2C may reflect the dynamic nature of editosomes, including possible cycles of REH2C association/dissociation with RESC. The REH2C complex may bind transcripts that require new or proofreading editing, whereas RESC may accumulate transcripts that were acted upon by REH2C. We linked the enrichment of fully edited RPS12 in RESC to site-preferential changes in total editing and accurate editing in the 5′ half of RPS12. The RECC enzyme may act more efficiently on specific 5′ positions of RPS12 in the context of RESC. Studies of the total mtRNA pool by the Read laboratory indicated that 5′ regions in RPS12 are consistently misedited and some misedits may give rise to alternative protein sequences (Simpson et al. 2016). The increased editing accuracy at 5′ positions in RESC-associated RPS12 suggests that putative alternative protein-coding sequences are less likely in RESC. The functional relevance of putative alternative protein-coding sequences derived from the RPS12 locus remains to be validated.
REH2C promotes total and accurate editing in RESC-associated RPS12
KH2F1 and KREH2 knockdowns indicated that the helicase complex REH2C enhances both total editing and accurate editing primarily at 5′ positions in RESC-associated RPS12. Accordingly, these knockdowns reduced the level of fully edited RPS12 associated with RESC in our Illumina libraries. We also note that both knockdowns further reduced the observed minor KREH2 cross-contamination in our RESC6-IPs, mentioned above. As mentioned above, weak RNA-mediated contacts between native forms of RESC6-bound RESC, REH2C, and other editing components reflect a dynamic nature of the RNA-editing holoenzyme (Cruz-Reyes et al. 2018a). Our combined RNAi/RNA-seq approach revealed nucleotide-resolution details on the functional trans effects of REH2C in the editing apparatus.
As aforementioned, major shifts in total editing involved the same positions in RPS12, for example, a cluster of ESs including ES54 in studies with RNAi or comparing RESC, REH2C, and mtRNA. Similarly, major shifts in accurate editing involved the same positions in RPS12 (e.g., including ES46 and ES56). These positions and other discrete areas in RPS12 may be primary control checkpoints affected by REH2C. The helicase REH2C carries 3′–5′ unwinding activity and forms an RNP with mRNA that may hybridize with gRNA-loaded RESC (Kumar et al. 2016). So, the REH2C RNP may remodel the secondary structure of the mRNA–gRNA duplex and/or interaction with the RECC enzyme. Whether the differential editing events on RPS12 transcripts described here occurred while these transcripts were bound to RESC or before RESC binding was not directly established in these studies.
Early editing in RESC-associated RPS12 transcripts
The initiating gRNA-1 in RPS12 (Koslowsky et al. 2013) directs some of the most accurate editing events in RPS12, including at the first four positions examined (i.e., insertion ES3 and ES4 and two nESs). Notably, only one of these positions (nES at T-str 150) involves a stabilizing C–G base pair with gRNA-1, that is, T-str 150 is just 5′ of a C nucleotide. In contrast to the accurate editing in the first few positions, we noted that two adenines at the 3′ end of gRNA-1 would account for the most frequent event (+2U) of incorrect editing in the ORF region. These two adenines occur in identical guide sequences of gRNA-1 transcripts from different T. brucei strains, including the strain in this study (Koslowsky et al. 2013; Madina et al. 2014; Cooper et al. 2019), and may play an important role. The putative +2U event by these conserved adenines also causes the largest drop in accurate editing (i.e., >500-fold change at the T-str 142/143 transition) in the ORF region. The T-str 142/143 transition may serve as a “tap” to modulate editing progression between gRNA-1 and gRNA-2. Importantly, REH2C loss-of-function using RNAi nearly doubled the Inc/Cor fold change at the T-str 142/143 transition despite position T-str 142 involving a stabilizing C–G base pair. The dramatic change in editing accuracy at the gRNA-1/gRNA-2 overlap might also reflect a “seed” mechanism, as in other RNA processes (Kunne et al. 2014), which stabilizes the gRNA-1 hybrid to promote accurate editing initiation. Moreover, a large shift in editing accuracy at the gRNA-1/gRNA-2 transition may also reflect that gRNA-1 is used more often than other gRNAs allowing more proofreading events. We found that most adjoining positions that exhibited a high Inc/Cor fold change in our study matched intrinsic pause sites (IPSs) identified by the Read laboratory in RPS12 transcripts (Supplemental Table S4; Simpson et al. 2016). So, the REH2C editing complex may modulate 3′–5′ editing progression in RESC-associated RPS12 transcripts in at least some of these positions.
Analysis of A6
As in RPS12, our analysis of an A6 3′ fragment showed site-to-site differences in total editing and accurate editing upon REH2C loss-of-function. Total editing was relatively high across the first four gRNAs (∼50% of the examined 3′ fragment) but became quite low in the remaining upstream sequence. The reason for this shift in total edits is currently unclear, but it may reflect that our amplicons included a 5′ primer matching preedited sequence. The initiating gRNA-1 in A6 (Koslowsky et al. 2013) directed relatively accurate editing at the first six positions (i.e., deletion ES1 and ES4, insertion ES2, ES3 and ES5, and one nES), of which two positions involve stabilizing C–G base-pairing with gRNA-1. Editing at these early 3′ positions appeared unaffected by REH2C. However, we found significant REH2C effects in a region that forms a short alternative sequence across the 3′ half of gRNA-1. This 3′ sequence element is created by the most frequent incorrectly edited sequences (spanning positions T-str 102-109) in all A6 samples examined. The creation of this 3′ element may hinder editing progression between the initiating gRNA-1 and gRNA-2, or could generate dead-end editing products.
Interestingly, a folding prediction showed that the addition of this 3′ element to A6 pre-mRNA creates a long intrastrand duplex in a region targeted by the first three to four gRNAs. Such a stable duplex could hinder annealing by early gRNAs. Notably, REH2C loss-of-function increased the frequency of this 3′ element in our samples, suggesting that RNA or RNP remodeling is involved. Analyses of annotated gRNAs in sequenced minicircles (Cooper et al. 2019) did not yield high-quality predicted gRNAs that could account for the A6 3′ element. However, multiple small gRNA pieces could be involved. The source of the 3′ element remains to be defined. Additional studies underway will address these possibilities, including possible changes in secondary structure.
REH2C distinct effects on the interplay between correct and incorrect edits in RPS12 and A6, and possible editing-initiation control mechanisms
Changes in total and accurate editing in RPS12 and A6 sequences examined did not necessarily involve the same positions, although most Inc/Cor spikes in A6 early 3′ editing were associated with large changes in total editing. The discrepancy in the location of total and accurate editing spikes, particularly in RPS12, suggests that the molecular mechanisms controlling these two processes are not necessarily concerted. REH2C showed substrate-specific effects on RPS12 and A6 transcripts. In RPS12, REH2C loss-of-function decreased both total and accurate editing at preferential 5′ positions. In A6, REH2C loss-of-function increased total editing but decreased accurate editing at preferential 3′ positions. Importantly, Inc/Cor spikes at ESs in RPS12 were primarily due to a loss of correctly edited sequences. At these positions, dominant high-frequency incorrectly edited sequences were largely unaffected by RNAi (Supplemental Fig. S7).
In contrast, Inc/Cor spikes in early 3′ positions of A6 were primarily due to an increase of incorrectly edited sequences. Correctly edited sequences at these A6 positions were largely unaffected by RNAi (Fig. 8). This shows a different effect of REH2C on the interplay between correct and incorrect edits in RPS12 and A6.
The control of site-preferential changes in editing may involve the recognition and remodeling of RNA features by the helicase REH2C that carries 3′–5′ unwinding activity (Kumar et al. 2016). Whether the differential editing events on RPS12 and A6 described here occurred while these transcripts were bound to RESC or before RESC binding was not directly established in these studies.
Early 3′ editing in RPS12 and A6 exhibited two contrasting effects. The first few guiding nucleotides in gRNA-1 direct relatively accurate editing that was unaffected by REH2C in both transcripts. However, different mechanisms may slow down early 3′ editing progression. In RPS12, the 3′ end nucleotides of gRNA-1 may direct a highly frequent noncanonical editing event. In A6, high-frequency inaccurate 3′ edits may create an mRNA fold that hinders annealing of early gRNAs as discussed above. Control of early 3′ editing progression by these proposed mechanisms may involve REH2C. The observed changes in A6 may be specific to early 3′ editing, but whether upstream regions in A6 are also affected by REH2C loss-of-function will require analysis of the entire A6 transcript. The current study dissected potential site-preferential checkpoints in REH2C-mediated control in RPS12 and A6 editing.
MATERIALS AND METHODS
Cell culture
Trypanosoma brucei procyclic strain Lister 427 29–13 (Wirtz et al. 1999) was grown axenically in log phase in SDM-79 medium (Brun and Schonenberger 1979) and harvested at a cell density of 1–3 × 107 cells/mL. The transgenic KH2F1–RNAi and KREH2–RNAi cell lines generated in our laboratory were induced with tetracycline (Tet) at 1 µg/mL (Madina et al. 2015; Kumar et al. 2016). Growth curves started at a cell density of 2 × 106 cells/mL, and dilutions were performed every other day. RNAi cell lines were maintained in tetracycline-screened HyClone Fetal Bovine (FBS) (SH30070.03T; GE Healthcare).
Western blots and radioactivity assays
Western blots of KREH2 and KH2F1 (subunits of REH2C), and GRBC2 (alias GAP1; subunit of GRBC), in mitochondrial extracts and immunoprecipitations (IPs) were performed as reported (Hernandez et al. 2010; Kumar et al. 2016). Protein analyses using Amersham ECL western blot detection reagents were performed in an Amersham Imager 600. RNA editing ligase KREL1 (subunit of RECC) was detected by radiolabeled self-adenylation directly on the beads in antibody pulldowns (Sabatini and Hajduk 1995).
Preparation of samples for RNA-seq
The source of the RNA samples used in each figure is in Supplemental Table S1. Independent biological replicates of each sample (n = 2) were prepared from separate cultures, including RNAi-induced and untreated control samples. Mitochondria-enriched extracts were prepared as reported (Kumar et al. 2019). Total RNA from mitochondrial extract (mtRNA) was isolated using TRIzol per manufacturer's instructions. IPs of native RESC6, KREH2, and KH2F1 from the extracts were performed using a standard protocol as reported (Madina et al. 2014; Kumar et al. 2016) with some modifications. Approximately 2 mg of protein was mixed with 1× SUPERase·In RNase inhibitor (Invitrogen), 1× cOmplete Protease inhibitor (Roche), and precleared by passage over Protein A-Sepharose beads (GE Healthcare) that were pretreated with 5% BSA before loading onto antibody-conjugated beads. Affinity-purified peptide antibodies of identical quality and concentration (Bethyl Laboratories) against RESC6, KREH2, and KH2F1 were conjugated to Dynabeads Protein A (10002D; Invitrogen). The conjugated beads were washed five times with 1 mL of wash buffer (25 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 10 mM Mg(OAc)2, 0.1% NP-40). The above affinity-purified antibodies and standard assay conditions typically generate KREH2-IPs and KH2F1-IPs (REH2C complex) with negligible levels of RESC6 (also known as MRB3010) detected by western blot analyses of the pulldowns, and RESC6-IPs samples with low or undetectable levels of KREH2 and KH2F1 (e.g., see Madina et al. 2014, 2015; Kumar et al. 2016). In the current study, RESC6-IPs had no detectable KH2F1 but a small amount of KREH2, while IPs of KH2F1 (in REH2C) had negligible levels of RESC2 (a marker protein subunit in RESC6-bound RESC). Despite minor cross-contaminations in our samples, the current study confirmed our reported relative enrichment of accurate edits in RESC6-bound RESC, and effects of REH2C loss on the level of accurate edits in RESC and total mtRNA (Madina et al. 2014, 2015). To prepare samples for RNA-seq, RNA was extracted by treating the beads with 4U proteinase K (NEB) for 2 h at 55°C, followed by phenol/chloroform extraction and ethanol precipitation. Isolated RNA from the pulldowns or directly from mitochondrial extracts was treated with DNase I, RNase-free (EN0523; Thermo Scientific) before cDNA synthesis with the iScript Reverse Transcription Supermix (Bio-Rad) as described elsewhere (Madina et al. 2014). The cDNA was analyzed by Qubit. RPS12 cDNA was amplified using a forward primer (5′-ctaatacacttttgataacaaac-3′; #952) including genomically encoded 5′ UTR sequence, and a reverse primer (5′-aaaaacatatcttatatctaaatct-3′; #1700) including genomically encoded 3′ UTR sequence and the first two canonical sites, ES1 and ES2, in fully edited RPS12 (Supplemental Fig. S1; Supplemental Table S2). A6 cDNA was amplified using a forward primer 5′-acggcggttttgaaaacac-3′; #1417) including genomically encoded preedited sequence, and a reverse primer (5′-cttatttgatcttattctataactcc-3′; #951) targeting genomically encoded 3′ UTR sequence. The linear range of the PCR reactions was determined by qRT-PCR. To generate samples for MiSeq sequencing, cDNA was amplified by PCR using the same primer concentrations and a number of cycles determined to be in the linear range (i.e., 18 cycles) for each sample. The PCR amplicons were gel-isolated and eluted into 10 µL buffer of 10 mM Tris-HCl, pH 8.0.
Library construction and sequencing
The amplified cDNA described above was analyzed on Tape Station and cleaned using Agencourt AMPure XP magnetic beads (Beckman Coulter). Libraries were made using a modified version of the Illumina Metagenomic Sequencing Library Preparation protocol (15044223 B), replacing the 16S specific primer sequence with RPS12 gene-specific primers. In brief, a 30 µL index PCR reaction was performed to attach Nextera XT indices and Illumina sequence adapters. A 2× KAPA HiFi HotStart Ready mix (12.5 µL) was combined with 0.16 µM of each Illumina primer, and 5 ng of cDNA per PCR reaction (18 cycles). The final libraries were purified in Agencourt AMPure XP magnetic beads (Beckman Coulter). Library concentrations were quantified using the Qubit Fluorometer high-sensitivity double-stranded DNA assay (Thermo Fisher). The size of the amplicons was confirmed on an Agilent TapeStation D1000 tape (Agilent). The libraries were normalized and pooled to a concentration of 4 nM and were sequenced on an Illumina MiSeq V2 250 × 250 sequencing kit to generate approximately 15 million read pairs (Texas A&M Institute of Genome Sciences and Society). We note that the sequence of the RPS12 and A6 genes (i.e., the sequence in the preedited transcripts) were confirmed by amplification of a genomic fragment containing the RPS12 or A6 genes, and direct Sanger sequencing of two independent amplicons for each transcript using forward and reverse primers.
Processing of RNA-seq paired-end reads
Raw sequence data for each sample were assessed for quality using FASTQC software (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and outputs manually inspected. Read pairs were merged where possible using FLASH v2.2.00 (https://ccb.jhu.edu/software/FLASH/; parameters: –max-overlap = 250 –max-mismatch-density = 0.05 -t 12 -z -O -m 35). Merged read pairs were used for subsequent analyses. Gene-specific primers were identified within the merged sequence, and trimmed to remove “external” bases, thereby yielding an amplicon for each merged read pair. Amplicons from each sample were assessed for replicates and collapsed as appropriate to yield a nonredundant set of amplicon sequences for each sample. Sequences were stored in FASTA format, with the sample, sequence, and replicate depth included in the sequence name. “T-stripped” versions of these sequences, in which all T residues were removed, were generated and aligned to T-stripped versions of the target gene sequences (RPS12 and A6) using bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Alignment was performed with parameters—very sensitive -p 20 -f -U; the SAM output was parsed on the fly to exclude all alignments that were not a perfect match with no gaps prior to storage in sorted, indexed BAM format using samtoools (http://www.htslib.org/). The BAM output provides information as to the aligning sequence, alignment start position, and the initial collapsed readset sequence prior to T-stripping. Each non-A|G|C in the pre-T-stripped collapsed readset represented a possible editing event for the collapsed readset: all were assessed irrespective of known model status. Each aligning collapsed readset was processed for each alignment position to determine whether additional bases were present or not in the pre-T-stripped collapsed readset sequence. Content information at each position in the target gene sequence was generated in tab-delimited text format, one line per T-stripped collapsed readset. Subsequently, sample alignment output data were further processed in the R environment (http://www.r-project.org) for summarizing and figure generation purposes.
Calculation of incorrect/correct (Inc/Cor) ratios
Inc/Cor values at the examined positions in RPS12 and A6 were calculated by dividing the total percentage of “incorrectly” edited reads (here defined as different from the expected canonical pattern) by the percentage of “correct” reads (i.e., corresponding to the canonical sequence) at that position (Supplemental Table S3). Values are determined for all ES and nES positions. At nES, correct reads represent preedited sequence that does not require editing at that position. Any editing event at nESs would generate incorrectly edited reads. Correct reads at both ESs and nESs are dictated by cognate gRNA annealing.
Calculation of Inc/Cor fold change values
The fold change of two Inc/Cor values during 3′–5′ progression was calculated by dividing the Inc/Cor value in a 5′ position by the Inc/Cor value in the adjacent 3′ position. Fold change values were calculated for all positions within the RPS12 ORF sequence and an A6 3′ fragment sequence in RESC6-IPs (Supplemental Table S4). Fold change values were also calculated at 3′ positions of RPS12 in RESC6-IPs upon RNAi (Supplemental Table S5). The mean + SD of n = 2 biological replicates was reported.
Statistical analysis
We used sliding window analyses to reveal the differences in the total edits among samples as a function of the position in mRNAs (Zivot and Wang 2006). Along the sequence, we typically used sliding windows with a window size = 20, which means 10 upstream and 10 downstream positions are included in a window for each focal position located at the center of the window. A slightly larger window centered 65 [54–81] (e.g., Fig. 2) was needed to detect significant differences in an area that contains a few scattered spikes primarily in RESC6-IPs. For each window, we pooled all data points located within the same window and used the one-way ANOVA to test the null hypothesis that there is no significant difference between groups. If this null hypothesis is rejected by the P < 0.05, then Tukey's honest significant difference (HSD) procedure is used to identify which two groups are significantly different from each other. P-values <0.0005 (***), <0.005 (**), <0.05 (*), and >0.05 (NS) between samples were annotated. The step size of the sliding windows analysis is 1, which means that we moved the sliding window from upstream to downstream by one position each time until the end of the sequence. Positions at the very beginning and very end have no results because of the boundary effect.
DATA DEPOSITION
Supplemental data is also available on Figshare, DOI 10.6084/m9.figshare.12609686. RNA sequencing data are deposited in NCBI SRA under BioProject ID PRJNA597821.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members at the Institute for Genome Sciences and Society at Texas A&M University for their excellent assistance with deep sequencing. Laurie K. Read kindly provided the anti-serum against GAP1 (GRBC2). We thank Nicholas Savill for checking a data base of annotated gRNAs in the Schnaufer laboratory for possible alignments against an A6 element described in this study, and Casey Hughes Lago for reading the manuscript. We are very grateful to Isaac Chavez, Pedro Monagas, Jana Gomez, and Elizabeth Alvarez Hernandez for their kind assistance in the preparation of excel files and charts. This work was supported by the National Science Foundation (1616845 to J.C.-R.); TAMU T3 award (to J.C.-R.); TAMU X-grant (to X.Z., J.J.C., J.C.-R.); and UK Medical Research Council (MR/L019701/1 to A.S.). Funding for open access charge: National Science Foundation (1616845).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.076513.120.
REFERENCES
- Alatortsev VS, Cruz-Reyes J, Zhelonkina AG, Sollner-Webb B. 2008. Trypanosoma brucei RNA editing: coupled cycles of U deletion reveal processive activity of the editing complex. Mol Cell Biol 28: 2437–2445. 10.1128/MCB.01886-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK. 2010. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA 16: 2239–2251. 10.1261/rna.2285510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Hashimi H, Novotna L, Cicova Z, McEvoy SM, Lukes J, Read LK. 2011. MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA 17: 865–877. 10.1261/rna.2446311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Downey KM, Hashimi H, Fisk JC, Tomasello DL, Faktorova D, Kafkova L, King T, Lukes J, Read LK. 2012. Architecture of the trypanosome RNA editing accessory complex, MRB1. Nucleic Acids Res 40: 5637–5650. 10.1093/nar/gks211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. 2003. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J 22: 913–924. 10.1093/emboj/cdg083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Zhang L, Wang X, Kaake RM, Huang L, Monti S, Aphasizhev R. 2014. RNA binding and core complexes constitute the U-insertion/deletion editosome. Mol Cell Biol 34: 4329–4342. 10.1128/MCB.01075-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Alfonzo J, Carnes J, Cestari I, Cruz-Reyes J, Goringer HU, Hajduk S, Lukes J, Madison-Antenucci S, Maslov DA, et al. 2020. Lexis and grammar of mitochondrial RNA processing in Trypanosomes. Trends Parasitol 36: 337–355. 10.1016/j.pt.2020.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat GJ, Koslowsky DJ, Feagin JE, Smiley BL, Stuart K. 1990. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell 61: 885–894. 10.1016/0092-8674(90)90199-O [DOI] [PubMed] [Google Scholar]
- Brun R, Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop 36: 289–292. [PubMed] [Google Scholar]
- Butenko A, Opperdoes FR, Flegontova O, Horak A, Hampl V, Keeling P, Gawryluk RMR, Tikhonenkov D, Flegontov P, Lukes J. 2020. Evolution of metabolic capabilities and molecular features of diplonemids, kinetoplastids, and euglenids. BMC Biol 18: 23 10.1186/s12915-020-0754-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Stuart KD. 2007. Uridine insertion/deletion editing activities. Methods Enzymol 424: 25–54. 10.1016/S0076-6879(07)24002-9 [DOI] [PubMed] [Google Scholar]
- Carnes J, Soares CZ, Wickham C, Stuart K. 2011. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J Biol Chem 286: 19320–19330. 10.1074/jbc.M111.228965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, McDermott S, Anupama A, Oliver BG, Sather DN, Stuart K. 2017. In vivo cleavage specificity of Trypanosoma brucei editosome endonucleases. Nucleic Acids Res 45: 4667–4686. 10.1093/nar/gkx116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Wadsworth ES, Ochsenreiter T, Ivens A, Savill NJ, Schnaufer A. 2019. Assembly and annotation of the mitochondrial minicircle genome of a differentiation-competent strain of Trypanosoma brucei. Nucleic Acids Res 47: 11304–11325. 10.1093/nar/gkz928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Mooers BH, Abu-Adas Z, Kumar V, Gulati S. 2016. DEAH-RHA helicase•Znf cofactor systems in kinetoplastid RNA editing and evolutionarily distant RNA processes. RNA Dis 3: e1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Mooers BHM, Doharey PK, Meehan J, Gulati S. 2018a. Dynamic RNA holoeditosomes with subcomplex variants: insights into the control of trypanosome editing. WIREs RNA 9: e1502 10.1002/wrna.1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Mooers BHM, Kumar K, Doharey PK, Meehan J, Chaparro L. 2018b. Control mechanisms of the holo-editosome in trypanosomes. In RNA metabolism in mitochondria (ed. Cruz-Reyes J, Gray M), Vol. 34, pp. 125–144. Springer, Berlin. [Google Scholar]
- Dean S, Gould MK, Dewar CE, Schnaufer AC. 2013. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc Natl Acad Sci 110: 14741–14746. 10.1073/pnas.1305404110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Sollner-Webb B. 1990. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell 61: 1001–1011. 10.1016/0092-8674(90)90065-M [DOI] [PubMed] [Google Scholar]
- Dixit S, Muller-McNicoll M, David V, Zarnack K, Ule J, Hashimi H, Lukes J. 2017. Differential binding of mitochondrial transcripts by MRB8170 and MRB4160 regulates distinct editing fates of mitochondrial mRNA in Trypanosomes. MBio 8: e02288-02216 10.1128/mBio.02288-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimov ES, Gasparyan AA, Kaurov I, Tichy B, Logacheva MD, Kolesnikov AA, Lukes J, Yurchenko V, Zimmer SL, Flegontov P. 2018. Trypanosomatid mitochondrial RNA editing: dramatically complex transcript repertoires revealed with a dedicated mapping tool. Nucleic Acids Res 46: 765–781. 10.1093/nar/gkx1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golas MM, Bohm C, Sander B, Effenberger K, Brecht M, Stark H, Goringer HU. 2009. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J 28: 766–778. 10.1038/emboj.2009.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J. 2008. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA 14: 970–980. 10.1261/rna.888808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Cicova Z, Novotna L, Wen YZ, Lukes J. 2009. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA 15: 588–599. 10.1261/rna.1411809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Panigrahi A, Cifuentes-Rojas C, Sacharidou A, Stuart K, Cruz-Reyes J. 2008. Determinants for association and guide RNA-directed endonuclease cleavage by purified RNA editing complexes from Trypanosoma brucei. J Mol Biol 381: 35–48. 10.1016/j.jmb.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Madina BR, Ro K, Wohlschlegel JA, Willard B, Kinter MT, Cruz-Reyes J. 2010. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem 285: 1220–1228. 10.1074/jbc.M109.051862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Faktorova D, Krizova A, Kafkova L, Read LK, Lukes J, Hashimi H. 2015. Integrity of the core mitochondrial RNA-binding complex 1 is vital for trypanosome RNA editing. RNA 21: 2088–2102. 10.1261/rna.052340.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. 2014. RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem 83: 697–725. 10.1146/annurev-biochem-060713-035546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby LE, Koslowsky DJ. 2017. Mitochondrial dual-coding genes in Trypanosoma brucei. PLoS Negl Trop Dis 11: e0005989 10.1371/journal.pntd.0005989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowsky DJ, Bhat GJ, Read LK, Stuart K. 1991. Cycles of progressive realignment of gRNA with mRNA in RNA editing. Cell 67: 537–546. 10.1016/0092-8674(91)90528-7 [DOI] [PubMed] [Google Scholar]
- Koslowsky DJ, Goringer HU, Morales TH, Stuart K. 1992. In vitro guide RNA/mRNA chimaera formation in Trypanosoma brucei RNA editing. Nature 356: 807–809. 10.1038/356807a0 [DOI] [PubMed] [Google Scholar]
- Koslowsky DJ, Sun Y, Hindenach J, Theisen T, Lucas J. 2013. The insect-phase gRNA transcriptome in Trypanosoma brucei. Nucleic Acids Res 42: 1873–1886. 10.1093/nar/gkt973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Madina BR, Gulati S, Vashisht AA, Kanyumbu C, Pieters B, Shakir A, Wohlschlegel JA, Read LK, Mooers BH, et al. 2016. REH2C helicase and GRBC subcomplexes may base pair through mRNA and small guide RNA in kinetoplastid editosomes. J Biol Chem 291: 5753–5764. 10.1074/jbc.M115.708164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Doharey PK, Gulati S, Meehan J, Martinez MG, Hughes K, Mooers BHM, Cruz-Reyes J. 2019. Protein features for assembly of the RNA editing helicase 2 subcomplex (REH2C) in Trypanosome holo-editosomes. PLoS One 14: e0211525 10.1371/journal.pone.0211525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunne T, Swarts DC, Brouns SJ. 2014. Planting the seed: target recognition of short guide RNAs. Trends Microbiol 22: 74–83. 10.1016/j.tim.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Madina BR, Kumar V, Metz R, Mooers BH, Bundschuh R, Cruz-Reyes J. 2014. Native mitochondrial RNA-binding complexes in kinetoplastid RNA editing differ in guide RNA composition. RNA 20: 1142–1152. 10.1261/rna.044495.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madina BR, Kumar V, Mooers BH, Cruz-Reyes J. 2015. Native variants of the MRB1 complex exhibit specialized functions in kinetoplastid RNA editing. PLoS One 10: e0123441 10.1371/journal.pone.0123441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov DA, Simpson L. 1992. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell 70: 459–467. 10.1016/0092-8674(92)90170-H [DOI] [PubMed] [Google Scholar]
- McAdams NM, Simpson RM, Chen R, Sun Y, Read LK. 2018. MRB7260 is essential for productive protein-RNA interactions within the RNA editing substrate binding complex during trypanosome RNA editing. RNA 24: 540–556. 10.1261/rna.065169.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams NM, Harrison GL, Tylec BL, Ammerman ML, Chen R, Sun Y, Read LK. 2019. MRB10130 is a RESC assembly factor that promotes kinetoplastid RNA editing initiation and progression. RNA 25: 1177–1191. 10.1261/rna.071902.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreiter T, Hajduk SL. 2006. Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep 7: 1128–1133. 10.1038/sj.embor.7400817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP. 2003a. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J Am Soc Mass Spectrom 14: 728–735. 10.1016/S1044-0305(03)00126-0 [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. 2003b. Identification of novel components of Trypanosoma brucei editosomes. RNA 9: 484–492. 10.1261/rna.2194603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. 2008. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol Cell Proteomics 7: 534–545. 10.1074/mcp.M700430-MCP200 [DOI] [PubMed] [Google Scholar]
- Read LK, Myler PJ, Stuart K. 1992. Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei. J Biol Chem 267: 1123–1128. [PubMed] [Google Scholar]
- Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J 16: 4069–4081. 10.1093/emboj/16.13.4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini R, Hajduk SL. 1995. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J Biol Chem 270: 7233–7240. 10.1074/jbc.270.13.7233 [DOI] [PubMed] [Google Scholar]
- Sacharidou A, Cifuentes-Rojas C, Halbig K, Hernandez A, Dangott LJ, De Nova-Ocampo M, Cruz-Reyes J. 2006. RNA editing complex interactions with a site for full-round U deletion in Trypanosoma brucei. RNA 12: 1219–1228. 10.1261/rna.2295706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Maslov DA. 1999. Evolution of the U-insertion/deletion RNA editing in mitochondria of kinetoplastid protozoa. Ann NY Acad Sci 870: 190–205. 10.1111/j.1749-6632.1999.tb08879.x [DOI] [PubMed] [Google Scholar]
- Simpson RM, Bruno AE, Bard JE, Buck MJ, Read LK. 2016. High-throughput sequencing of partially edited trypanosome mRNAs reveals barriers to editing progression and evidence for alternative editing. RNA 22: 677–695. 10.1261/rna.055160.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RM, Bruno AE, Chen R, Lott K, Tylec BL, Bard JE, Sun Y, Buck MJ, Read LK. 2017. Trypanosome RNA editing mediator complex proteins have distinct functions in gRNA utilization. Nucleic Acids Res 45: 7965–7983. 10.1093/nar/gkx458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm NR, Maslov DA, Blum B, Simpson L. 1992. Generation of unexpected editing patterns in Leishmania tarentolae mitochondrial mRNAs: misediting produced by misguiding. Cell 70: 469–476. 10.1016/0092-8674(92)90171-8 [DOI] [PubMed] [Google Scholar]
- Tschudi C, Ullu E. 1994. Trypanosomatid protozoa provide paradigms of eukaryotic biology. Infect Agents Dis 3: 181–186. [PubMed] [Google Scholar]
- Tylec BL, Simpson RM, Kirby LE, Chen R, Sun Y, Koslowsky DJ, Read LK. 2019. Intrinsic and regulated properties of minimally edited trypanosome mRNAs. Nucleic Acids Res 47: 3640–3657. 10.1093/nar/gkz012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. 2008. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell 32: 198–209. 10.1016/j.molcel.2008.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101. 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- Zikova A, Schnaufer A, Dalley RA, Panigrahi AK, Stuart KD. 2009. The F0F1-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog 5: e1000436 10.1371/journal.ppat.1000436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer SL. 2019. Revisiting trypanosome mitochondrial genome mysteries: broader and deeper. Trends Parasitol 35: 102–104. 10.1016/j.pt.2018.09.006 [DOI] [PubMed] [Google Scholar]
- Zimmer SL, Simpson RM, Read LK. 2018. High throughput sequencing revolution reveals conserved fundamentals of U-indel editing. Wiley Interdiscip Rev RNA 2018: e1487 10.1002/wrna.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivot E, Wang J. 2006. Rolling analysis of time series. In Modeling financial time series with S-PLUS, pp. 313–360. Springer, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.