Abstract
Coronavirus EndoU inhibits dsRNA-activated antiviral responses; however, the physiologic RNA substrates of EndoU are unknown. In this study, we used mouse hepatitis virus (MHV)-infected bone marrow–derived macrophage (BMM) and cyclic phosphate cDNA sequencing to identify the RNA targets of EndoU. EndoU targeted viral RNA, cleaving the 3′ side of pyrimidines with a strong preference for U↓A and C↓A sequences (endoY↓A). EndoU-dependent cleavage was detected in every region of MHV RNA, from the 5′ NTR to the 3′ NTR, including transcriptional regulatory sequences (TRS). Cleavage at two CA dinucleotides immediately adjacent to the MHV poly(A) tail suggests a mechanism to suppress negative-strand RNA synthesis and the accumulation of viral dsRNA. MHV with EndoU (EndoUmut) or 2′–5′ phosphodiesterase (PDEmut) mutations provoked the activation of RNase L in BMM, with corresponding cleavage of RNAs by RNase L. The physiologic targets of EndoU are viral RNA templates required for negative-strand RNA synthesis and dsRNA accumulation. Coronavirus EndoU cleaves U↓A and C↓A sequences (endoY↓A) within viral (+) strand RNA to evade dsRNA-activated host responses.
Keywords: coronavirus, endoribonuclease, innate immunity, dsRNA, mouse hepatitis virus
INTRODUCTION
Viruses in the order Nidovirales express a virus-encoded endoribonuclease, NendoU (Ivanov et al. 2004). NendoU is unique to nidoviruses (Gorbalenya et al. 2006), including viruses of the Coronaviridae and Arteriviridae families. Nidoviruses that express NendoU have vertebrate hosts, whereas nidoviruses of crustaceans (Roniviridae), and RNA viruses outside the Nidovirales order, do not encode this protein. The precise role(s) of NendoU in virus replication remain enigmatic; however, significant progress has been made in recent years to elucidate the contributions of NendoU to virus replication and pathogenesis. The SARS-CoV-2 pandemic underscores the importance of understanding host–pathogen interactions, including the immunomodulatory functions of EndoU (Wu et al. 2020).
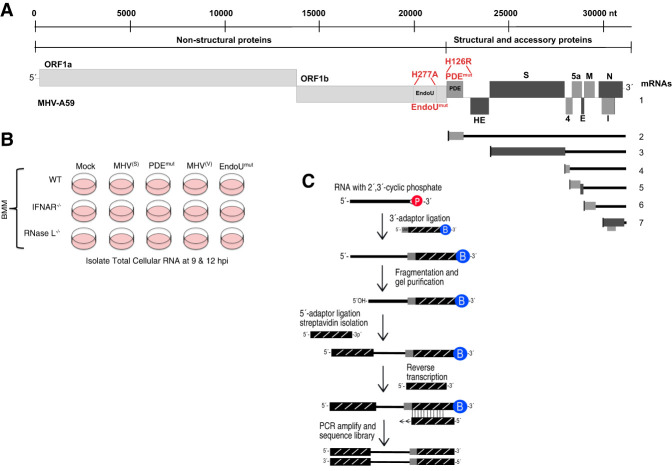
Arterivirus (nsp11) and coronavirus (nsp15) EndoU proteins have been characterized by genomic (Gorbalenya et al. 2006), structural (Ricagno et al. 2006; Zhang et al. 2017, 2018) and biochemical studies (Ricagno et al. 2006; Bhardwaj et al. 2008; Nedialkova et al. 2009). EndoU is encoded near the 3′ end of ORF1b (Fig. 1A, schematic of the MHV genome; Gorbalenya et al. 2006). Mouse hepatitis virus (MHV), a well-studied coronavirus, has a single-stranded positive-sense RNA genome 31.1 kb in length. MHV RNA, like other coronaviruses, is 5′ capped and 3′ polyadenylated. Upon infection, the ORF1a and ORF1b regions of MHV RNA are translated into two polyproteins (ORF1a and ORF1ab) through a frameshifting mechanism (Bredenbeek et al. 1990). MHV proteins nsp1–nsp16 are produced via proteolytic processing of the ORF1a and ORF1ab polyproteins. EndoU is the nsp15 protein of MHV (Fig. 1A, schematic of MHV RNA genome). Other proteins from the ORF1a/1b region of the RNA genome include viral proteases and components of the viral replicase (nsp12 is the RdRP, nsp13 is a helicase, nsp14 is a 3′ → 5′ exonuclease, and an N7-methyltransferase, and nsp16 is a 2′-O-methyltransferase). An H277A mutation in nsp15 greatly reduces the catalytic activity of EndoU (Fig. 1A, EndoUmut; Kang et al. 2007).
FIGURE 1.
Coronavirus RNA genome and experimental approach. (A) MHV RNA genome highlighting two mutations: His to Arg mutation in the MHV phosphodiesterase domain active site (PDEH126R), and a His to Ala mutation in the MHV EndoU domain active site (EndoUH277A) (Roth-Cross et al. 2009; Kindler et al. 2017). MHV proteins are categorized as nonstructural (light gray), accessory (dark gray), and structural (black). Subgenomic mRNAs 2–7, produced during infection, are illustrated. (B) Bone marrow–derived macrophage (BMM) from wt, IFNAR−/−, and RNase L−/− mice were mock-infected or infected with wt MHV (MHV(S) and MHV(V)), the PDEmut, or EndoUmut for 9 and 12 h (Zhao et al. 2012; Kindler et al. 2017), after which total cellular RNA was isolated for cyclic phosphate sequencing. (C) Schematic of cyclic phosphate sequencing; protocol adapted from Schutz et al. (2010).
Coronavirus RNA replication and RNA transcription are mediated by the replicase expressed from the ORF1a/1b region of the genome (Fig. 1A) with assistance of the nucleocapsid protein (Sawicki et al. 2007). Both RNA replication and RNA transcription occur within membrane-anchored replication organelles in the cytoplasm of infected cells (Knoops et al. 2008; van Hemert et al. 2008; van der Hoeven et al. 2016). MHV RNA replication involves negative-strand RNA synthesis, wherein the positive-strand viral RNA genome is copied into a genome-length negative-strand RNA intermediate, which is subsequently used as a template to make new positive-strand RNA genomes. MHV RNA transcription involves the synthesis of subgenomic (sg) negative-strand RNAs from the viral RNA genome via discontinuous transcription mechanisms and subsequent synthesis of sg mRNAs (Sethna et al. 1989; Sawicki and Sawicki 1990). Intergenic transcriptional regulatory sequences (TRS) within MHV RNA guide discontinuous transcription mechanisms (Sola et al. 2015), leading to the production of sg negative-strand RNAs, which function as templates for the synthesis of sg mRNAs. A nested set of 3′ coterminal sg mRNAs (sg mRNA2 to sg mRNA7) is used to express each of the remaining viral proteins (phosphodiesterase [PDE] from sg mRNA2a, spike [S] from sg mRNA3, and so forth) (Fig. 1A). Hemagglutinin-esterase (HE) is an unexpressed pseudogene in MHV A59 because of a TRS mutation that prevents the expression of mRNA2b, as well as a nonsense mutation at codon 15 (Luytjes et al. 1988; Shieh et al. 1989; Kazi et al. 2005; Lissenberg et al. 2005). EndoU colocalizes with viral RNA replication and RNA transcription machinery at membrane-anchored replication organelles (Deng et al. 2017; V'Kovski et al. 2019). Colocalization of EndoU with viral RNA synthesis machinery may influence the RNAs targeted by EndoU. Furthermore, coronavirus nsp16, a 2′-O-ribose-methyltransferase (2′-O-MT), could potentially modify RNA substrates to make them resistant to cleavage by EndoU (Ivanov et al. 2004). These studies suggest that viral RNA stability may be regulated by nsp15 (EndoU) and nsp16 (2′-O-MT).
Intriguingly, neither EndoU (nsp15) nor 2′-O-MT (nsp16) enzyme activities are required for virus replication in transformed cells in culture (Kang et al. 2007; Ulferts and Ziebuhr 2011; Zust et al. 2011); rather, these enzymes counteract dsRNA-activated antiviral responses (Zust et al. 2011; Deng et al. 2017; Kindler et al. 2017). EndoU catalytic activity inhibits the activation of dsRNA-dependent antiviral innate immune pathways (Deng et al. 2017; Kindler et al. 2017), including type I and type III IFN responses, PKR, and OAS–RNase L (Deng et al. 2019). EndoU-deficient viruses can replicate in IFNAR−/− cells or cells lacking PKR and RNase L (PKR−/− and RNase L−/−) (Deng et al. 2017, 2019; Kindler et al. 2017). In addition to EndoU, coronavirus NS2, a 2′–5′ PDE, inhibits the activation of RNase L (Zhao et al. 2011, 2012, 2013). Thus, there are two pathways by which MHV inhibits the activation of OAS–RNase L, suggesting this pathway is crucial for antiviral defense. Whereas coronavirus EndoU inhibits dsRNA-activated antiviral responses within virus-infected cells, it is unclear how it achieves this because the physiologically relevant targets of EndoU have not been defined.
In this study, we used MHV-infected bone marrow–derived macrophage (BMM) and cyclic phosphate cDNA sequencing to identify the host and viral RNA targets of EndoU. Cyclic phosphate cDNA sequencing reveals the location and frequency of endoribonuclease cleavage sites within host and viral RNAs (Schutz et al. 2010; Cooper et al. 2014, 2015; Donovan et al. 2017). We exploited wild-type and mutant forms of MHV (wt MHV, PDEmut, and EndoUmut) along with wild-type and mutant forms of BMM (wt BMM, IFNAR−/−, and RNase L−/−) to distinguish between EndoU-dependent cleavage sites and RNase L–dependent cleavage sites within host and viral RNAs.
RESULTS
Products of cleavage by coronavirus EndoU have 2′,3′-cyclic phosphate termini (Ivanov et al. 2004), effectively marking the location of cleavage within host and viral RNAs. Thus, in this study, we used cyclic phosphate cDNA sequencing to monitor the frequency and location of endoribonuclease cleavage sites in RNA from MHV-infected bone marrow–derived macrophage (BMM) (Fig. 1). Wild-type and mutant MHVs (wt MHV(V), wt MHV(S), PDEmut, and EndoUmut) along with BMM derived from wild-type and particular knockout C57BL/6 mice (wt, IFNAR−/−, and RNase L−/− BMM) were used to distinguish between EndoU-dependent cleavage sites and RNase L–dependent cleavage sites (Fig. 1B). A pair of wt and mutant viruses derived from each isolate were used (Fig. 1A,B): wt MHV from Susan Weiss’ laboratory designated MHV(S) and a phosphodiesterase mutant designated PDEmut (Roth-Cross et al. 2009; Zhao et al. 2011, 2012, 2013), wt MHV from Volker Thiel's laboratory designated MHV(V), and an EndoU mutant designated EndoUmut (Kindler et al. 2017). We chose to use RNA samples from 9 and 12 hpi based on a previous study of wt MHV and EndoUmut-infected BMM cells (Kindler et al. 2017). These are sequential time points when coronavirus NS2 PDE and nsp15 EndoU activities prevent dsRNA-dependent antiviral responses (Kindler et al. 2017), including the OAS/RNase L pathway (Zhao et al. 2011, 2012, 2013). Furthermore, virus replication is nearing completion by the later time point, ensuring abundant amounts of viral RNA. Abundant amounts of viral RNA make it more likely that endoribonuclease cleavage sites will be readily detected and easily comparable from one condition to another. Under these experimental conditions (Fig. 1B), we expect that RNase L activity will be increased within PDEmut-infected and EndoUmut-infected wt BMM, as compared to MHV(S)-infected and MHV(V)-infected wt BMM. Furthermore, we expect that EndoU activity will be evident within MHV(S)-infected and MHV(V)-infected BMM, as compared to EndoUmut-infected BMM.
Cyclic phosphate cDNA libraries were prepared using total cellular RNA (Fig. 1C). The RNA ligase RtcB was used to ligate a 3′ adaptor to RNA fragments containing a cyclic phosphate. The 3′ adaptor has a biotin moiety and a unique molecular identifier to enumerate cleavage sites (Kivioja et al. 2011). A 5′ adaptor was ligated to the RNA samples, followed by reverse transcription, PCR amplification, and Illumina sequencing. Analysis of DNA sequences revealed the frequency and location of endoribonuclease cleavage sites in host and viral RNAs. Figures 2–7 correspond to data from the experiment outlined here (Fig. 1). Replicate data from infections by wt and mutant MHV (wt MHV(S), PDEmut, and EndoUmut) in wt and RNase L−/− BMM yield similar outcomes (Supplemental Figs. S9 and S10).
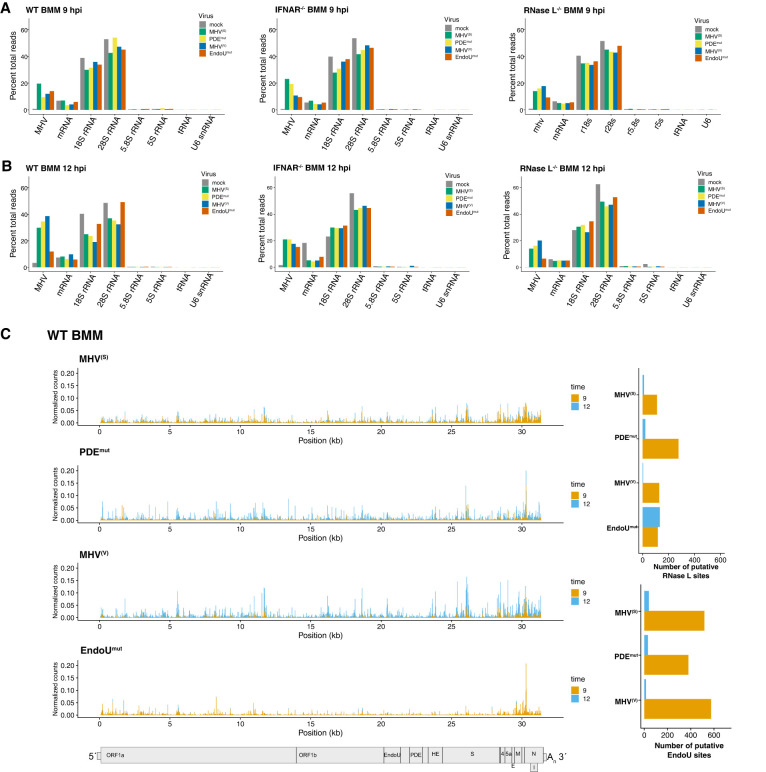
FIGURE 2.
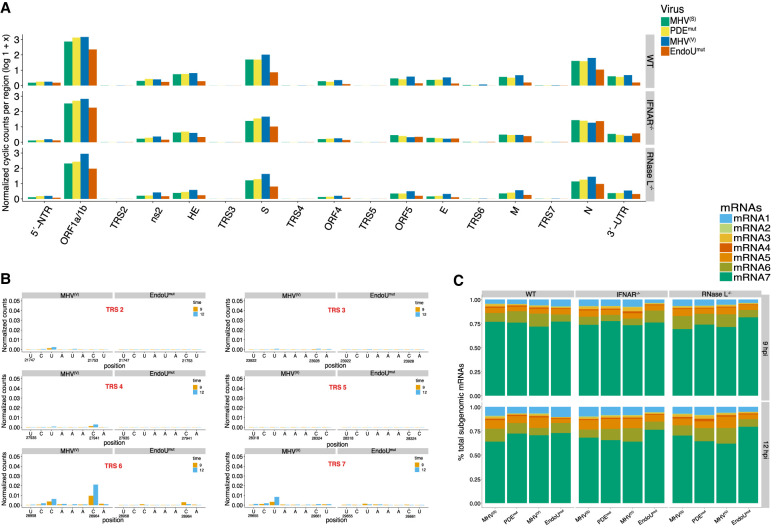
Endoribonuclease cleavage in host and viral RNAs. (A,B) Relative amounts of endonucleolytic cleavage in host and viral RNAs. Normalized cyclic phosphate cDNA reads (reads at each position/total reads in library) mapped to host and viral RNAs at 9 and 12 hpi in wt, IFNAR−/−, and RNase L−/− bone marrow macrophages (BMMs). (C) Frequency and location of cleavage sites in MHV RNA. Normalized cyclic phosphate cDNA reads captured at each position along the MHV genomic RNA at 9 and 12 hpi from MHV(S)-, MHV(V)-, PDEmut-, and EndoUmut-infected wt BMM. Putative cleavage sites attributed to EndoU or RNase L were calculated from RNase L– or EndoU-dependent signal generated by subtracting signal from each captured position that occurs in the absence of either enzyme (RNase L−/− BMM or during EndoUmut infection). These data were then filtered for sites with reads representing at least 0.01% of total reads in the library. At each of these positions, the log2-fold change in signal when either RNase L or EndoU were absent was calculated and sites with ≥2.5-fold change were designated putative RNase L or EndoU sites.
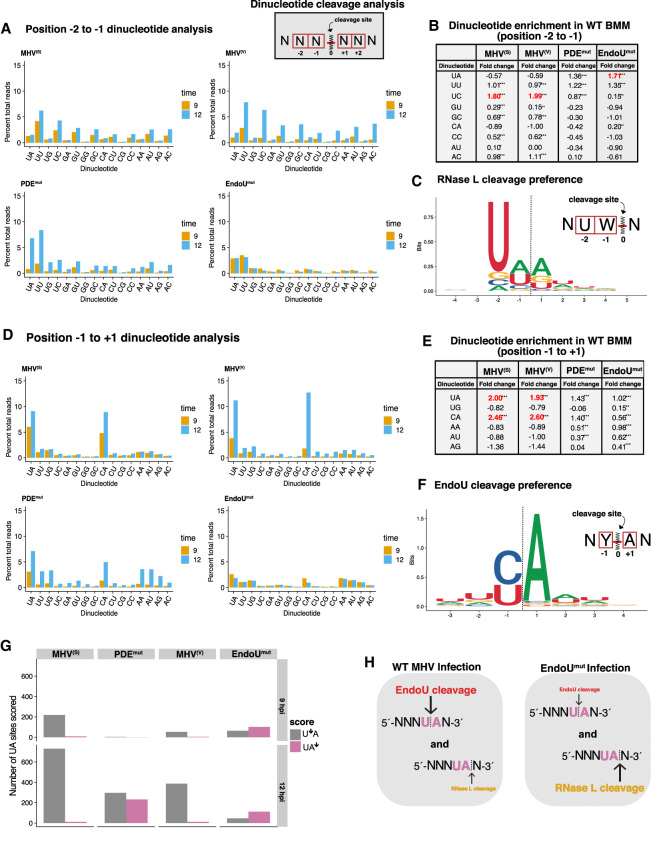
FIGURE 3.
Sequence specificity of cleavage sites in MHV RNA. (A,D) Dinucleotide specificity analysis for cleavage in MHV RNA by percent total cDNA reads captured at each 3′-dinucleotide in wt BMM at 9 and 12 hpi for (A) Dinucleotide analysis for positions −2:−1 and (D) dinucleotide analysis for positions −1:+1 from captured cleavage position (0 position). (B,E) Dinucleotide enrichment for dinucleotide positions from −2:−1 (B) or −1:+1 (E) for each condition of viral infection at 12 hpi in wt BMM by comparing the frequency of dinucleotide capture in experimental conditions to the frequency of occurrence for each dinucleotide in the MHV RNA sequence (control). Significant enrichment was determined by adjusted P-value (q) for fold change (log2[experiment/control]). <0.02*, <0.0001**, <1 × 108***. Only dinucleotides with positive enrichment are shown. (C,F) Sequence logos for the six bases surrounding the cleavage site for position −2:−1 (C) or −1:+1 (F). Logos generated from the top 1% of either RNase L (215 sites) or EndoU-dependent cleavages (306 sites). (G) UA cleavage scoring analysis. All UA sequences in the MHV RNA with ≥30 cyclic phosphate counts in either the UA↓ or U↓A cleavage position were compared by calculating the ratio of normalized counts (UA↓ counts/U↓A counts). Ratios >1 were scored as UA↓ (RNase L) sites and ratios <1 were scored as U↓A sites (EndoU) and total number of scored sites for either position are shown for each condition of viral infection in wt BMM at 9 and 12 hpi. (H) Model of EndoU and RNase L interaction at UA sites in MHV RNA.
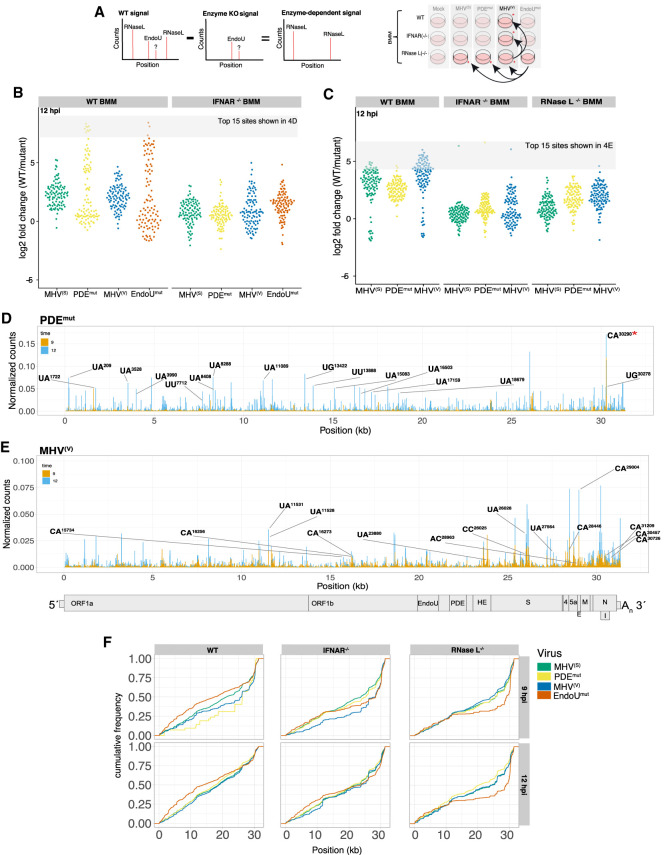
FIGURE 4.
RNase L–dependent and EndoU-dependent cleavage sites in MHV RNA. (A) Schematic outline of analysis to identify EndoU/RNase L–dependent cyclic phosphate reads. (B,C) Fold change values for the top 100 RNase L–dependent or EndoU-dependent cleavage sites. Fold change in cyclic phosphate signal when comparing wt or IFNAR(−/−) BMM infected with MHV(S), MHV(V), PDEmut, and EndoUmut virus to RNase L−/− BMM (B) or MHV(S), MHV(V), PDEmut virus to infection with EndoUmut virus across all cell types (C) displayed as a violin scatterplot. Log2-fold change in the absence of RNase L activity (B) or in the absence of EndoU activity (C) was calculated for each position in the MHV RNA. Fold change values for the top 100 RNase L–dependent or EndoU-dependent sites were compared in wt and IFNAR−/− BMM under conditions of infection with MHV(S), MHV(V), PDEmut, and EndoUmut virus at 12 hpi (B) or in all cell types across conditions of infection with MHV(S), MHV(V), PDEmut virus at 12 hpi. (D) Frequency and location of RNase L–dependent cleavage sites in MHV RNA. Cyclic phosphate counts at each position in the viral genome were normalized by removing signal that occurred in the absence of RNase L, which emphasizes sites that are RNase L–dependent in wt BMM infected with MHV(S), and PDEmut at 9 and 12 hpi. Labeled positions and dinucleotides (−2 base:−1 base) on the graph of PDEmut represent the top 15 RNase L–dependent cleavage sites (from B) with the greatest fold change in RNase L activity (*site with robust cleavage without canonical RNase L dinucleotide preference and independent of EndoU activity; not identified as top site by RNase L–fold-change analysis). (E) Frequency and location of EndoU-dependent cleavage sites in MHV RNA. Cyclic phosphate counts at each position in the viral genome were normalized by removing signal that occurred in the absence of EndoU, which emphasizes sites that are EndoU-dependent and RNase L–independent in RNase L−/− BMM infected with wt MHV(V) at 9 and 12 hpi. Labeled positions and dinucleotides (−1 base:+1 base) represent the top 15 EndoU-dependent cleavage sites with the greatest fold change in EndoU activity (from C). (F) Cumulative distribution of normalized counts by position of MHV genome for every position with ≥10 cyclic phosphate counts across all cell types and infection conditions.
FIGURE 5.
Abundance of cyclic phosphate ends by MHV genomic region and MHV mRNA abundance. (A) Normalized cyclic phosphate counts per MHV genomic region in wt, IFNAR−/−, and RNase L−/− BMM across all conditions of viral infection at 12 hpi. Transcriptional regulatory sequences (TRSs) are numbered by their associated mRNA (2–7). Other MHV genomic regions are labeled as shown in Figure 1A. (B) Frequency and location of cleavage in the MHV TRS elements in wt BMM during infection with MHV(V) and EndoUmut at 12 hpi. The x-axis includes the sequence and position of the six-base MHV TRS elements. (C) Normalized RNA-seq counts (sum of MHV sg mRNA/sum of all MHV mRNAs) of MHV sg mRNAs detected in wt, IFNAR−/−, and RNase L−/− BMM across all conditions of viral infection at 9 and 12 hpi.
FIGURE 6.
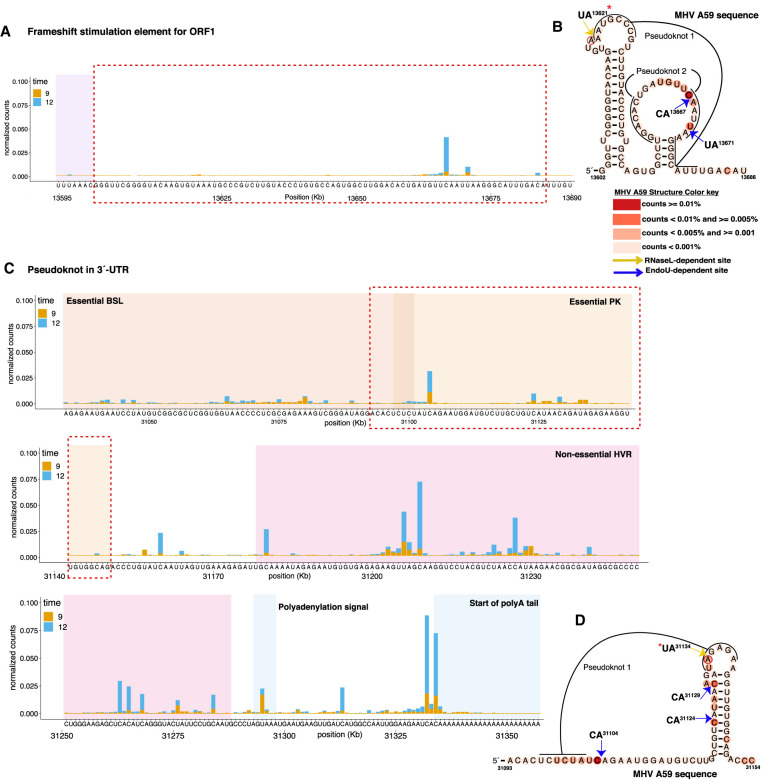
MHV secondary structures associated with RNase L–dependent and EndoU-dependent cleavage sites. (A,C) Nucleotide resolution graphs displaying normalized counts by position for the regions encompassing secondary structure predictions. (B,D) Secondary structures of frameshift stimulation element (B) and MHV 3′-UTR pseudoknot (D), generated using available consensus alignment and the R-scape program (Rivas et al. 2017). MHV A59 sequence mapped to consensus secondary structures using available covariation model and the Infernal program (Nawrocki and Eddy 2013). Base coloring of MHV A59 sequence based on normalized cDNA reads as indicated in key for 12 hpi in wt BMM infected with MHV(V). *Base RNase L–dependent cleavage activity is increased in PDEmut or EndoUmut infection as compared to MHV(V) infection.
FIGURE 7.
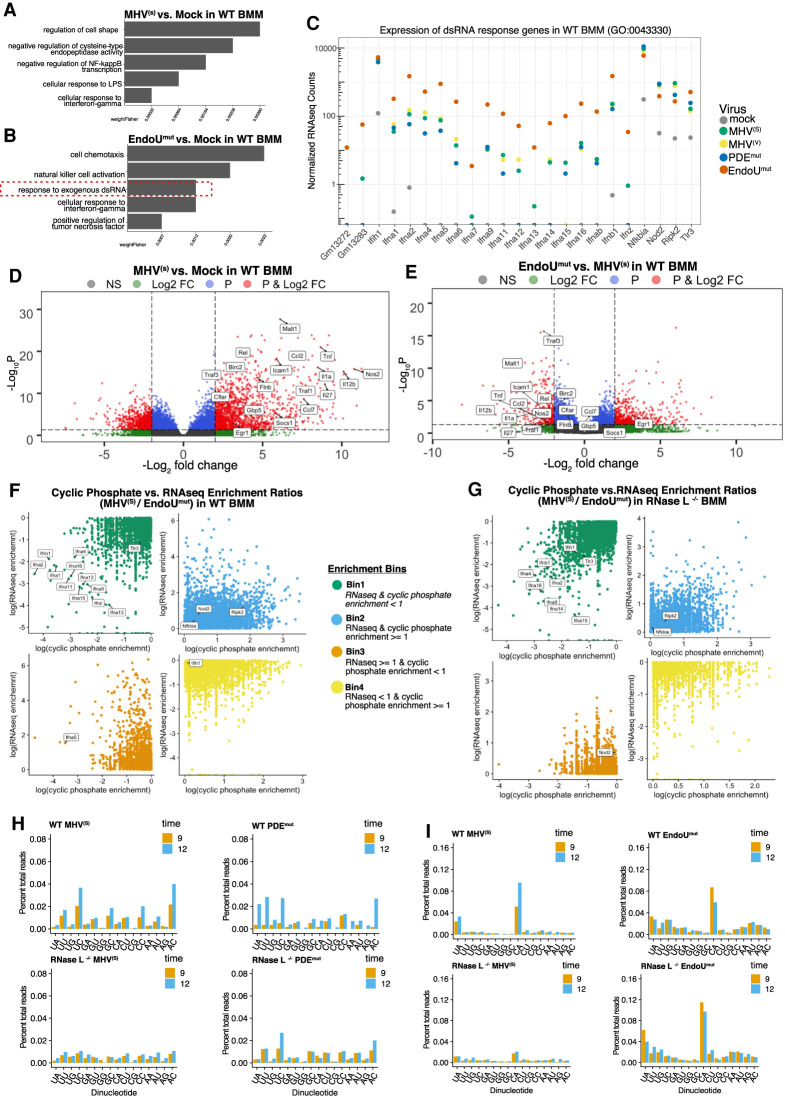
Endoribonuclease cleavage of cellular RNAs and changes in host gene expression. (A,B) Gene ontology (GO) analysis of host gene expression during MHV infection. Categories of biological processes enriched with significantly up-regulated genes (P < 0.01, log2FC > 2) from (A) MHV(s)-infected or (B) EndoUmut-infected wt BMM. The top five significantly enriched categories (weightFisher < 0.01) are shown. (C) Expression of host genes in GO category “response to exogenous dsRNA.” Expression (log10-normalized counts) of genes in the GO category “response to exogenous dsRNA” for wt BMM at 12 hpi. (D,E) Volcano plots of changes in host gene expression during MHV infection. (C) Plot comparing MHV(s)-infected and mock-infected wt BMM and (D) comparing EndoUmut-infected and MHV(s)-infected wt BMM. Host genes were considered significantly differentially expressed at FDR < 0.05 and logFC > 2 (up-regulated) or logFC < −2 (down-regulated). (F,G) Relationship between cyclic phosphate and RNA-seq enrichment scores. An enrichment ratio was calculated for all mRNAs using the total sum of cyclic phosphate or RNA-seq normalized counts in MHV(S) infected samples/total sum of cyclic phosphate or RNA-seq normalized counts in EndoUmut-infected samples at 9 and 12 hpi in wt and RNase L−/− BMM (enrichment score = [MHV(S)/[EndoUmut]). Genes were assigned to bins as follows: bin 1 = cyclic phosphate and RNA-seq enrichment ratio <1, bin 2 = cyclic phosphate and RNA-seq enrichment ratio ≥1, bin 3 = cyclic phosphate ratio <1 and RNA-seq ratio ≥1, bin 4 = cyclic phosphate ratio ≥1 and RNA-seq ratio <1. Each bin includes genes assigned from 9 and 12 hpi and highlighted genes represent those identified in the dsRNA response GO category (Fig. 7C). (H,I) Dinucleotide specificity analysis for cleavage of transcripts involved in the dsRNA response (Fig. 7C) during infection with MHV(s) and PDEmut for positions −2:−1 (H) or MHV(s) and EndoUmut for positions −1:+1 (I). Percent of cleavage at each 3′-dinucleotide calculated relative to the total cDNA reads aligned to the mm10 transcriptome per library in wt and RNase L−/− BMM at 9 and 12 hpi.
Endoribonuclease cleavage sites in host and viral RNAs
Endoribonuclease cleavage sites were detected in host and viral RNAs at 9 (Fig. 2A) and 12 hpi (Fig. 2B). The frequency of cleavage sites in individual RNAs was normalized to percent total cDNA reads in each library, allowing for quantitative comparisons between individual RNAs in each sample and between RNAs across distinct samples. The vast majority of cleavage sites were detected in MHV RNA, cellular mRNA, and ribosomal RNAs (18S, 28S, 5.8S, and 5S rRNAs), with a smaller portion of cleavage sites in tRNAs and U6 snRNA (Fig. 2A,B).
Cleavage sites in MHV RNA were found predominantly in the positive strand of viral RNA, ranging from 10% to 40% of all cleavage sites in each library (Fig. 2A,B), with cleavage in all regions of the MHV genome (Fig. 2C). Very few reads were detected in the MHV negative-strand RNA (Supplemental Figs. S1B and S12C,D). Cleavage of MHV RNA in the ORF1a/1b region can be definitively attributed to cleavage of the MHV RNA genome (aka mRNA1). However, because MHV sg mRNAs 2–7 are 3′ coterminal with the MHV genome (Fig. 1A), cleavage detected in MHV RNA downstream from the ORF1a/1b region could be in the MHV RNA genome (mRNA1) or in any of the MHV sg mRNAs (mRNAs 2–7).
The relative amount of host RNAs captured at 9 and 12 hpi were similar across all cell types and in agreement with capture frequencies from uninfected and virus-infected cells previously reported (Schutz et al. 2010; Cooper et al. 2014, 2015). Data from an independent experiment revealed similar outcomes, with 10% to 30% of all cleavage sites in MHV RNA, 5% to 10% of cleavage sites in cellular mRNA, and more than 60% of cleavage sites in ribosomal RNAs (Supplemental Fig. S8A).
Cleavage sites in cellular RNAs can be attributed to specific endoribonucleases in some cases, but not others. For instance, U6 snRNA had 3′-terminal cyclic phosphates (Supplemental Fig. S1A) attributed to the nucleolytic activity of C16orf57/USB1 (Mroczek et al. 2012; Shchepachev et al. 2012; Cooper et al. 2014). Ribosomal RNAs accounted for ∼50%–80% of the cleavage sites detected in each library (Fig. 2A,B). The majority of cleavage sites within rRNAs are the result of unspecified endoribonucleases, along with some RNase L–dependent cleavage sites (Cooper et al. 2014, 2015). Cellular mRNAs accounted for ∼5% of endoribonuclease cleavage sites in each cDNA library (Fig. 2A,B). As described below, we attribute cleavage sites in host and viral RNAs to specific endoribonucleases.
Frequency, location, and sequence specificity of cleavage sites in MHV RNA
Metal-ion-independent endoribonucleases have characteristic specificities (e.g., RNase A family members [RNase 1–8] cleave RNA 3′ of pyrimidines while RNase L cleaves RNA 3′ of UpN↓ dinucleotides [UA↓, UU↓ > UG↓]) (Floyd-Smith et al. 1981; Wreschner et al. 1981; Yang 2011; Cooper et al. 2014). EndoU is reported to cleave RNA 3′ of pyrimidines in vitro (Ivanov et al. 2004; Bhardwaj et al. 2006; Nedialkova et al. 2009); however, physiologically relevant targets of EndoU have not been defined.
We detected endoribonuclease cleavage sites throughout MHV RNA, under all experimental conditions (Fig. 2C; Supplemental Fig. S2). The frequency of cleavage at each base of MHV RNA ranged from ∼0.00% to 0.2% of all cDNA reads in each library (Fig. 2C; Supplemental Fig. S2, y-axis). Peaks of cleavage approaching 0.2% of all cDNA reads in each library (corresponding to 1 in 500 cleavage sites across all RNAs in each cDNA library) are present at particular sites in the N gene open reading frame, near the 3′ terminus of MHV RNA (Fig. 2C, wt BMM, PDEmut, and EndoUmut). Typically, when measurable cleavage was detected at a particular base in MHV RNA at 9 hpi, measurable cleavage was also detected at that same site at 12 hpi, often with increased abundance (Fig. 2C; Supplemental Fig. S2, overlapping orange and blue lines at each base for 9 and 12 hpi).
The sequence specificity of cleavage sites in MHV RNA revealed profound differences in the endoribonuclease activities present within wt BMM cells infected with wt and mutant viruses (Fig. 3). Distinct RNase L–dependent and EndoU-dependent cleavage specificities were evident (Fig. 3). The sequence specificity of endoribonuclease cleavage sites was assessed in two registers: positions −2 to −1 of cleavage (Fig. 3A–C) and positions −1 to +1 of cleavage (Fig. 3D–F). wt MHV RNA was cleaved 3′ of pyrimidines in wt BMM (Fig. 3A, MHV(S) and MHV(V)), with a notable preference for cleavage between U↓A and C↓A sequences (Fig. 3D–F). This pattern of pyrimidine-specific cleavage between U↓A and C↓A sequences was lost in EndoUmut-infected wt BMM (Fig. 3A,D). Similar patterns of cleavage were evident in an independent experiment (Supplemental Fig. S9C,D).
Dinucleotide enrichment, a measurement comparing the frequency of cleavage at each dinucleotide to the frequency of each dinucleotide in the MHV genomic RNA, showed that U↓A and C↓A sequences were the only sequences with positively enriched cleavage in wt MHV-infected wt BMM (Fig. 3E, adjusted P-value (q) for fold change [log2(experiment/control)] of <1 × 108***). Dinucleotide enrichment and de-enrichment data for all dinucleotides at 9 and 12 hpi are available as supplemental data (Supplemental Tables S1 and S2). These data indicate that EndoU cleaved MHV RNA at U↓A and C↓A sequences.
RNase L activity was also evident within MHV-infected wt BMM (Fig. 3A–C). RNase L activity, with characteristic cleavage predominantly after UA↓ and UU↓ dinucleotides, was significantly increased in both PDEmut-infected and EndoUmut-infected wt BMM (Fig. 3A). Dinucleotide enrichment showed that UA↓, UU↓, and UC↓ sequences were positively enriched cleavage sites in PDEmut-infected and EndoUmut-infected wt BMM (Fig. 3B, adjusted P-value [q] for fold change [log2(experiment/control)] of <1 × 108***). In IFNAR−/− and RNase L−/− BMM, the robust cleavage at UA↓, UU↓, and UC↓ sequences decreased and pyrimidine-specific cleavage dominated, especially in PDEmut-infected cells (Supplemental Fig. S4A,B). These data indicate that RNase L cleaved MHV RNA after UA↓, UU↓, and UC↓ sequences, consistent with other studies (Floyd-Smith et al. 1981; Wreschner et al. 1981; Cooper et al. 2014, 2015).
The distinct specificity of cleavage for RNase L (UA↓, UU↓, and UC↓ sequences) and EndoU (U↓A and C↓A sequences) allowed us to compare the relative amounts of each enzyme activity in the various experimental conditions. MHV RNAs were cleaved predominantly by EndoU activity within MHV(S)-infected and MHV(V)-infected BMM (Fig. 3A,D). MHV RNA was cleaved by both RNase L and EndoU activities within PDEmut-infected wt BMM while MHV RNA was cleaved predominantly by RNase L activity within EndoUmut-infected wt BMM (Fig. 3A,D). The activation of RNase L within PDEmut-infected and EndoUmut-infected wt BMM was expected, as these viral proteins coordinately block the OAS–RNase L pathway (Zhao et al. 2012, 2013; Deng et al. 2017; Kindler et al. 2017). Dinucleotide analysis of positions downstream from cleavage sites confirmed a strong preference for adenine 3′ of the cleavage positions in MHV RNA in wt BMM (Supplemental Fig. S3A,B). When EndoU was inactivated within EndoUmut-infected cells, the strong preference for adenine 3′ of cleavage positions in MHV RNA was dramatically reduced, but not entirely eliminated in wt BMM (Supplemental Fig. S3A), IFNAR−/− BMM (Supplemental Fig. S4A) and RNase L−/− BMM (Supplemental Fig. S4B). The residual cleavage of MHV RNA within EndoUmut-infected RNase L−/− BMM is likely due to angiogenin or another RNase A family member, as these enzymes are present within macrophage and they share a predilection for cleavage at U↓A and C↓A sequences (Harper et al. 1990; Shapiro and Vallee 1991; Hofsteenge et al. 1998; Schwartz et al. 2018).
We identified cyclic phosphate cDNAs dependent on the presence of either RNase L or EndoU and then used fold change to identify and assign specific sites as RNase L or EndoU targets (Fig. 2C; Supplemental Fig. S5). We determined how many of these sites could be assigned to either endoribonuclease for each experimental condition (Fig 2C; Supplemental Fig. S2). EndoU cleaved MHV RNA at both 9 and 12 hpi in all three cell types, with increased amounts of cleavage at 12 hpi as compared to 9 hpi (Fig. 2C; Supplemental Fig. S2A,B). MHV RNA was cleaved by RNase L activity at both 9 and 12 hpi in wt BMM, with exacerbated amounts of RNase L activity in PDEmut-infected and EndoUmut-infected wt BMM, as expected. In EndoUmut-infected wt BMM, there were nearly equal numbers of cleavage sites assigned to RNase L at 9 and 12 hpi, which was not observed in any other condition (Fig. 2C). By comparison with wt BMM, less RNase L–dependent cleavage was detected in IFNAR−/− BMM (Supplemental Fig. S2A), consistent with reduced OAS expression and reduced RNase L activity in IFNAR−/− BMM (Birdwell et al. 2016). Additionally, the number of sites assigned to EndoU in IFNAR−/− and RNase L−/− BMM was less than that observed in wt BMM, suggesting that EndoU activity was altered in the absence of IFN signaling and innate immune effectors (Supplemental Fig. S2A,B). We attributed the majority of endoribonuclease cleavage sites within MHV RNA to either EndoU (U↓A and C↓A sequences) or RNase L (UA↓, UU↓, and UC↓ sequences) activities (Figs. 2 and 3); however, undefined enzymes cleaved MHV RNA within EndoUmut-infected RNase L−/− BMM (Supplemental Fig. S2B). As mentioned above, the residual cleavage of MHV RNA within EndoUmut-infected RNase L−/− BMM was likely due to angiogenin or another RNase A family member, as these enzymes are present within macrophage and they share a predilection for cleavage at U↓A and C↓A sequences (Harper et al. 1990; Shapiro and Vallee 1991; Hofsteenge et al. 1998; Schwartz et al. 2018). The patterns and amounts of EndoU-dependent and RNase L–dependent cleavage in MHV RNA were consistent from one experiment (Figs. 2 and 3) to another (Supplemental Fig. S9B–F).
It is intriguing to note that EndoU and RNase L share a common substrate dinucleotide, UA. Furthermore, we can distinguish between cleavage of UA by EndoU and RNase L as these enzymes cleave the UA sequence at distinct sites: EndoU cleaves between U↓A sequences, whereas RNase L cleaves after UA↓ dinucleotides (Fig. 3H). We found hundreds of UA sequences in MHV RNA cleaved by both EndoU and RNase L (Fig. 3G). EndoU activity predominated in MHV(S)-infected and MHV(V)-infected wt BMM at 9 and 12 hpi (Fig. 3G, MHV(S) and MHV(V)). Yet in PDEmut-infected wt BMM, either EndoU or RNase L cleaved about half of the UA sequences that were targeted by both enzymes (Fig. 3G; Supplemental Fig. S9F, PDEmut). EndoU cleaved to a greater extent about half of the shared sites, whereas RNase L cleaved another half to a greater extent (Fig. 3G; Supplemental Fig. S9F, PDEmut). Thus, while EndoU and RNase L have overlapping sequence specificity and share common UA targets within MHV RNAs, these enzymes do not tend to cleave the same molecule at the same site at any one moment in time. Our data show that the majority of cleavage of MHV RNA was from EndoU rather than RNase L during wt MHV infections (Fig. 3G; Supplemental Fig. S9F, wt); however, when the MHV PDE was mutated, a much larger proportion of cleavage events in viral RNA were from RNase L (Fig. 3G; Supplemental Fig. S9F, PDEmut).
Taken together, these data indicate that EndoU and RNase L cleaved MHV RNA within infected BMMs. The majority of endoribonuclease cleavage sites within MHV RNA were attributed to either EndoU (U↓A and C↓A sequences) or RNase L (UA↓, UU↓, and UC↓ sequences) activities (Figs. 3 and 4). However, data from EndoUmut-infected RNase L−/− BMM (Supplemental Fig. S2B) indicate that viral RNA was cleaved by other undefined endoribonucleases as well. Furthermore, when MHV NS2 PDE or nsp15 EndoU were inactivated by mutations, RNase L activity was much greater, with increased cleavage of MHV RNA by RNase L. Thus, both MHV NS2 PDE and nsp15 EndoU activities prevent MHV RNA cleavage by the dsRNA-activated OAS/RNase L pathway, confirming our previous reports (Zhao et al. 2012, 2013; Kindler et al. 2017).
RNase L–dependent and EndoU-dependent cleavage sites in MHV RNA
A fold-change analysis was used to compare the magnitudes of RNase L–dependent and EndoU-dependent cleavage at each base of MHV RNA across experimental conditions (Fig. 4A). By subtracting endoribonuclease cleavage events detected for each virus in RNase L−/− BMM, we identified the top 100 RNase L–dependent cleavage sites in MHV RNA (Fig. 4B). By subtracting the endoribonuclease cleavage events detected for the EndoUmut, we identified the top 100 EndoU-dependent cleavage sites in MHV RNA (Fig. 4C).
RNase L–dependent sites in MHV RNA were cleaved at the greatest magnitudes in PDEmut-infected and EndoUmut-infected wt BMM (Fig. 4B). RNase L–dependent cleavage of MHV RNA was substantially lower in IFNAR−/− cells, as expected (Birdwell et al. 2016), especially that associated with infections by the PDEmut and EndoUmut (Fig. 4B). The top 15 RNase L–dependent cleavage sites in MHV RNA were at UA↓, UU↓, and UG↓ dinucleotides distributed across the viral genome, with a clustering of sites within the first 2/3 of the genome (Fig. 4D). Cleavage downstream from the ORF1a/1b region could be in the MHV RNA genome (mRNA1) or any of the sg mRNAs (mRNAs 2–7). Magnitudes of cleavage at each of these sites ranged from 0.05% to 0.08% of all cleavage sites in each cDNA library (∼1/2000 cleavage sites in the cDNA library). Together, these top 15 cleavage sites in MHV RNA accounted for ∼1% of all cleavage sites in this cDNA library, across all host and viral RNAs. These data indicate that RNase L cleaved coronavirus RNA most efficiently at a relatively small number of sites.
EndoU-dependent cleavage sites in MHV RNA were evident in wt, IFNAR−/−, and RNase L−/− BMMs; however, EndoU cleaved MHV RNA to a greater extent in wt BMM (Fig. 4C). Subdued magnitudes of EndoU-dependent cleavage of MHV RNA were observed at 12 hpi in IFNAR−/− and RNase L−/− cells, as compared to wt BMM, suggesting a potential functional interaction between EndoU and dsRNA-activated host responses, or RNase L in particular. Additionally, most of the sites with EndoU-dependent cleavage activity had similar magnitudes of change, leading to a uniform distribution of sites across all conditions, excluding a few outliers. The top 15 EndoU-dependent cleavage sites in MHV RNA were at C↓A and U↓A sequences distributed to a greater extent in the last 2/3 of the viral genome (Fig. 4E).
We examined the cumulative distribution of cleavage in MHV RNA, across all conditions (Fig. 4F; Supplemental Fig. S9E). In this analysis, we plotted the overall accumulation of cyclic phosphate reads as a function of position along the MHV genomic RNA (Fig. 4F; Supplemental Fig. S9E). Cleavage downstream from the ORF1a/1b region could be in the MHV RNA genome (mRNA1) or sg mRNAs (mRNAs 2–7). Because RNase L–dependent cleavage sites (Fig. 4D) and EndoU-dependent cleavage sites (Fig. 4E) were distributed across the MHV RNA in wt BMM, cumulative cleavage increased from 0% at the 5′ end of the genome to 100% at the 3′ end, with a slope of ∼45° for MHV(S) and MHV(V) in wt BMM (Fig. 4F, wt BMM, green and blue lines for MHV(S) and MHV(V)). In EndoUmut-infected wt BMM, cleavage of MHV RNA increased in the ORF1a and ORF1b regions of the genome as compared to MHV(S) and MHV(V), shifting the slope of cumulative cleavage to the left (Fig. 4F, wt BMM, red line for EndoUmut). In contrast, when both EndoU and RNase L activities were absent, as in EndoUmut-infected RNase L−/− BMM, cleavage of MHV RNA was substantially reduced across most of the genome, with a spike of EndoU- and RNase L–independent cleavage near the 3′ UTR (Fig. 4F, RNase L−/− BMM, red line for EndoUmut). Note how the slope of the line for EndoUmut goes from ∼50% to 100% of cumulative cleavage between nucleotides 30,000 and 31,344. This indicates that endoribonucleolytic cleavage was much more pronounced near the 3′ terminus of MHV RNA in EndoUmut-infected IFNAR−/− and RNase L−/− BMM, as compared to wt BMM. These data indicate that EndoUmut MHV RNA was cleaved at very different magnitudes from one end to the other in wt BMM versus that in RNase L−/− BMM, with increased relative amounts of cleavage between nts 1 and 20,000 in wt BMM, less cleavage between nts 1 and 30,000 in RNase L−/− BMM, and a spike in cumulative cleavage near the 3′ terminus in RNase L−/− BMM.
These data also indicate that EndoU and RNase L account for a substantial amount of the cumulative cleavage in the orf1a and orf1b regions of the MHV RNA genome. MHV RNA was cleaved to a greater extent within orf1a and orf1b in wt BMM, especially when EndoU was disabled (Fig. 4F, red line for EndoUmut shifts to the left in wt BMM). Conversely, MHV RNA was cleaved to a lower extent within orf1a and orf1b in RNase L−/− BMM, especially when EndoU was disabled (Fig. 4F, red line for EndoUmut shifts to the right in RNase L−/− BMM). When EndoU and RNase L activities were absent, as in EndoUmut-infected RNase L−/− BMM, the residual cleavage of MHV RNA by unspecified endoribonucleases occurred predominantly near the 3′ terminus of the viral RNA (Fig. 4F, RNase L−/− cells, EndoUmut curve in red). This cleavage of MHV RNA near the 3′ terminus could be in the MHV RNA genome (mRNA1), in any of the MHV sg mRNAs (mRNAs 2–7), or both.
Endoribonuclease cleavage sites in distinct MHV RNA sequences and structures
We next examined the frequency of endoribonuclease cleavage in distinct regions of MHV RNA (Fig. 5; Supplemental Fig. S8). The cumulative amounts of cleavage in each region of MHV RNA were plotted unadjusted (Fig. 5A), or adjusted for size (Supplemental Fig. S8A), RNA abundance (Supplemental Fig. S8B), or both size and RNA abundance (Supplemental Fig. S8C). Cleavage was detected in every region of MHV RNA, from the 5′ NTR to the 3′ NTR, including relatively small TRS sequences (Fig. 5A,B). The vast majority of cleavage events occurred in 1a/1b, S and N open reading frames (Fig. 5A). When adjusted for MHV RNA abundance, cleavage was most frequent in the ORF 1a/1b region and the ns2, HE, and S ORFs (Supplemental Fig. S8B). Furthermore, with adjustments for size and abundance (Supplemental Fig. S8B,C), one can see that some of the TRS elements were targeted for cleavage at frequencies similar to that observed in Orf1a/1b. Thus, although TRS sequences are quite small, they can be cleaved just as frequently as RNA sequences in other regions of MHV RNA. Intriguingly, TRS6 was targeted more frequently (by EndoU) than other regions of MHV RNA, including other TRS elements (Fig. 5B). TRS6, with a UCCAAAC sequence, is distinct from other TRS elements, which possess UCUAAAC sequences. We detected the most robust EndoU-dependent cleavages at C↓A and U↓A dinucleotides of TRS elements 4, 6, and 7 (Fig. 5B). In TRS elements 4 and 6, cleavage at the very 3′ end of the TRS sequence was dependent on the presence of a downstream adenine outside of the TRS sequence (Fig. 5B). Interestingly, the upstream C↓A cleavage site in TRS 6 (Fig. 5B) relies on one of the single-nucleotide polymorphisms (28,960 T > C) that we detected in the viral genomes (Supplemental Table S3). In vitro studies using purified EndoU show cleavage of a U↓A dinucleotide within a TRS substrate (Kang et al. 2007). Our data indicate that C↓A and U↓A dinucleotides of TRS elements are physiologic targets of EndoU.
RNA-seq was used to measure the abundance of MHV RNA in all experimental conditions (Supplemental Fig. S6). MHV RNA was abundant in all samples from virus-infected cells, with similar amounts of MHV RNA across conditions, but for EndoUmut-infected wt BMM at 9 and 12 hpi (Supplemental Fig. S6A). Decreased amounts of EndoUmut RNA in wt BMM (Supplemental Fig. S6A) correlated with decreased virus replication in EndoUmut-infected wt BMM at 9 and 12 hpi (Kindler et al. 2017). RNA-seq reads were detected across the MHV RNA genome, with the most abundant reads corresponding to leader sequences at the 5′ end of the genome and sg mRNA sequences at the 3′ end of the genome (Supplemental Fig. S6B). Our initial RNA-seq analysis (Supplemental Fig. S6) did not estimate the abundance of any specific subgenomic mRNA; it estimates the abundance of all captured viral positive sense RNAs. We determined the abundance of individual sg mRNAs by identifying “chimeric” RNA-seq reads spanning a leader/body junction, similar to a method described in a previous study (Irigoyen et al. 2016). Consistent with this and other studies, MHV mRNA7 was most abundant, accounting for 70% to 80% of MHV mRNAs (Fig. 5C; Supplemental Fig. S10C). MHV mRNAs 1–7 were present in all conditions, with some changes in relative amounts from one condition to another (Fig. 5C; Supplemental Fig. S10C). MHV mRNA1 (genomic RNA) was increased proportionally to other MHV mRNAs in EndoUmut-infected wt BMM at 12 hpi. MHV mRNA 7 was increased slightly relative to other MHV mRNAs at 12 hpi in EndoUmut-infected IFNAR−/− BMM and RNase L−/− BMM. Overall, the relative ratios of genomic to sg mRNAs were approximately the same across cell types when comparing wt MHV and EndoUmut infections, consistent with a previous study (Kang et al. 2007). Remarkably, MHV RNA abundance did not correlate with the frequency of cyclic phosphate reads in viral RNA (Supplemental Fig. S12A,B). Altogether, these data indicate that MHV RNA replication was able to produce each of the MHV mRNAs in proportional amounts, despite considerable changes in endoribonuclease activity from one condition to another.
Endoribonuclease cleavage sites were detected in functional RNA sequences and structures, including the Orf1a/1b frameshift element and the MHV 3′ NTR (Fig. 6). The Orf1a/1b frameshift element contains both RNase L–dependent and EndoU-dependent cleavage sites (Fig. 6A,B). Likewise, the MHV 3′ NTR contains both RNase L–dependent and EndoU-dependent cleavage sites (Fig. 6C,D). The MHV 3′ NTR spans nucleotide 31,034, adjacent to the N stop codon, to nucleotide 31,334, adjacent to the poly(A) tail (Fig. 6C). Functional RNA sequences and structures within the 3′ NTR include an essential bulged stem–loop (nts 31,034–31,100), an essential pseudoknot (nts 31,101–31,150), a nonessential hypervariable region (HVR) (nts 31,179–31,288), a polyadenylation signal (nts 31,293–31,298), and a poly(A) tail (Goebel et al. 2007; Zust et al. 2008; Peng et al. 2016). A number of EndoU-dependent cleavage sites were detected within the 3′ NTR, including prominent cleavage sites immediately adjacent to the poly(A) tail (Fig. 6C, 31332C↓AC↓A31335). Together, these two cleavage sites account for ∼0.15% of all cleavage sites in the cDNA library for the wt MHV in wt BMM at 12 hpi, corresponding to ∼1/677 cleavage sites in the entire cDNA library.
When EndoU was inactivated by an H277A mutation, the cleavage of MHV RNA at the 31332C↓AC↓A31335 sequences adjacent to the poly(A) tail was dramatically reduced, but not entirely eliminated, in wt BMM (Supplemental Fig. S7A). Furthermore, there was EndoU-independent cleavage of MHV RNA at the 31332C↓AC↓A31335 sequence in IFNAR−/− BMM (Supplemental Fig. S7B) and RNase L−/− BMM (Supplemental Fig. S7C). Cleavage of MHV RNA at the 31332C↓AC↓A31335 sequences adjacent to the poly(A) tail was notable whether unadjusted (Supplemental Fig. S7A–C) or adjusted for RNA abundance (Supplemental Fig. S7D–F). These data indicate that the 31332C↓AC↓A31335 sequence in MHV RNA was susceptible to both EndoU-dependent and EndoU-independent cleavage. The EndoU-dependent cleavage of the 31332C↓AC↓A31335 sequence in MHV RNA was substantially greater than the EndoU-independent cleavage in wt BMM (Supplemental Fig. S7A); however, substantial amounts of EndoU-independent cleavage were detected at the 31332C↓AC↓A31335 sequence in IFNAR−/− BMM (Supplemental Fig. S7B) and RNase L−/− BMM (Supplemental Fig. S7C).
Changes in host gene expression and endonucleolytic cleavage of cellular RNAs
Because MHV EndoU activity is associated with evasion of dsRNA-dependent host responses (Deng et al. 2017; Kindler et al. 2017; Volk et al. 2020) and cellular RNAs were cleaved under the conditions of our experiments (Fig. 2A,B), we examined host gene expression (Fig. 7A–C) and characterized endonucleolytic cleavage of cellular RNAs (Fig. 7D–I).
Double-stranded RNA (dsRNA)-dependent host gene expression was increased within MHV-infected wt BMM (Fig. 7A–C). We performed gene ontology (GO) analysis to identify enriched gene expression pathways within MHV-infected wt BMM (Fig. 7A; Alexa and Rahnenfuhrer 2019). Up-regulated and differentially expressed genes in MHV(S)-infected wt BMM corresponded to a number of biological processes involved in innate, inflammatory, and adaptive immune response pathways (Fig. 7A; Supplemental Fig. S14B). Increased host gene expression within EndoUmut-infected wt BMM corresponded to similar groups of host genes, with a notable addition, response to exogenous dsRNA (Fig. 7B; Supplemental Fig. S14C). Thus, GO analysis indicated that host gene expression associated with the response to exogenous dsRNA was specifically activated in EndoUmut-infected wt BMM, as compared to MHV(S)-infected wt BMM (Fig. 7A,B; Supplemental Fig. S14B,C).
Because GO analysis implicated a “response to exogenous dsRNA,” we examined the magnitudes of expression for each host gene in this gene ontology group: GM13272, GM13283, IFN-α genes, IFN-β, IFN-Z, Nfkbia, Nod2, Ripk2, and Tlr3 (Fig. 7C). We compared magnitudes of expression in mock-infected, MHV(S)-infected, MHV(V)-infected, PDEmut-infected, and EndoUmut-infected wt BMM at 12 hpi (Fig. 7C). Host gene expression associated with response to dsRNA increased by 100- to 1000-fold in MHV-infected wt BMM as compared to mock-infected cells, with even larger 1000- to 10,000-fold increases in EndoUmut-infected wt BMM (Fig. 7C). Thus, host genes associated with response to dsRNA were notably increased in MHV-infected BMM, with the greatest increases occurring within EndoUmut-infected wt BMM (Fig. 7C).
We further examined the other enriched pathways, many of which belong to inflammatory and cytokine signaling. We used Volcano plots to compare all differentially expressed genes during infection with MHV(S)-infected cells relative to mock infection (Fig. 7D) or EndoUmut-infected cells relative to MHV(S)-infection in wt BMM (Fig. 7E; Blighe et al. 2019). The Volcano plots show the expression of many genes increasing by 22- to 210-fold/fourfold to 1024-fold, including the highlighted transcripts, which represent the top 20 most differential expressed (P < 0.05) transcripts from the enriched GO terms: cellular response to tumor necrosis factor, cytokine biosynthesis, cellular response to interferon-γ, and positive regulation of NF-κB (Fig. 7D). We observed that many of these transcripts are down-regulated in EndoUmut-infected cells relative to the MHV(S) infection (Fig. 7E). This pattern was also observed when comparing expression of all the transcripts involved in cytokine biosynthesis (Supplemental Fig. S14D) and NF-κB-positive regulation (Supplemental Fig. S14E) during wt and mutant virus infection in wt and RNase L−/− BMM. In wt BMM, many of the genes involved in inflammation and cytokine signaling were down-regulated during infection with PDEmut and, to a greater extent, EndoUmut relative to wt MHV (Supplemental Fig. S14D,E).
Altogether, these data indicate that dsRNA-dependent host responses were exacerbated within MHV-infected cells, especially in EndoUmut-infected wt BMM. These data are consistent with recent studies from the Baker laboratory (Volk et al. 2020).
Do changes in host gene expression arise due to changes in endonucleolytic cleavage of cellular mRNAs? To address the relationship between host gene expression and cleavage of cellular mRNAs, we sorted cellular mRNAs into four categories based on their abundance (RNA-seq) and their frequency of endonucleolytic cleavage (normalized cyclic phosphate counts), comparing one experimental condition to another (Fig. 7F,G; Supplemental Fig. S15). These analyses identify cellular mRNAs with changes (increases or decreases) in both abundance and cleavage frequency between experimental conditions (e.g., conditions with and without EndoU or RNase L activities).
When comparing MHV(S)- and EndoUmut-infected wt BMM, conditions with and without MHV EndoU activity, we found the majority of the cellular mRNAs from the dsRNA response pathway in bin 1 (Fig. 7F, locations of dsRNA response mRNAs annotated in bins 1–4). The cellular mRNAs in bin 1, including the majority of dsRNA response transcripts, increased in abundance and increased in cyclic phosphate reads in the absence of EndoU activity, as compared to the presence of EndoU activity. An increase in cyclic phosphate reads in the mRNAs in bin 1, in the absence of EndoU, is not consistent with EndoU targeting these mRNAs. In RNase L−/− BMM, mRNAs from the dsRNA response pathway either move out of bin 1 completely or become de-enriched within bin 1 (move up the x-axis [e.g., Nod1, Ripk2]), suggesting that RNase L activity is contributing to the cyclic phosphate reads in these mRNAs (Fig. 7F,G; Supplemental Fig. S15A,B). We might expect potential EndoU targets in bin 4, where cellular mRNAs have decreased mRNA abundance and increased cyclic phosphate counts in the presence of EndoU activity; however, we did not detect dsRNA response mRNAs in bin 4. Interestingly, many of the genes involved in inflammatory and cytokine signaling (Fig. 7D,E) occupy bin 2, whereas dsRNA response transcripts occupy bin 1 (Supplemental Fig. S15). Thus, EndoU activity does not appear to directly regulate the abundance of cellular mRNAs associated with either of these important host response pathways.
We also examined the specificity of endonucleolytic cleavage events in cellular mRNAs to attribute cleavage to one or another endoribonuclease. dsRNA host response mRNAs (Fig. 7H,I), like host mRNAs in the aggregate (Supplemental Fig. S16), were cleaved at dinucleotides we attribute to cellular endoribonuclease activities: RNase A (U↓A and C↓A) and RNase L (UA↓ and UU↓) (Fig. 7H,I). Furthermore, cleavage of dsRNA host response mRNAs at U↓A and C↓A sequences, like host mRNAs in the aggregate (Supplemental Fig. S16), were unaffected by an EndoU mutation (Fig. 7I). The cleavage of cellular mRNAs by cellular endoribonucleases at U↓A and C↓A sequences is consistent with other reports (Peach et al. 2015; Shigematsu et al. 2019).
Altogether, these data indicate that MHV EndoU does not cleave dsRNA host response mRNAs in a manner that would affect their overall abundance. Rather, transcriptional regulation is likely responsible for increased expression of dsRNA host response mRNAs.
rRNA
Because RNase L cleaves 18S rRNA at specific sites in human cells (Cooper et al. 2014, 2015), we examined RNase L–dependent cleavage of 18S rRNA within MHV-infected murine BMMs (Supplemental Fig. S11). Four RNase L–dependent cleavage sites were clearly evident in 18S rRNA: UU542, UU543, UU771, and UA772. These sites, on the surface of 18S ribosomal subunits, are analogous to RNase L–dependent cleavage sites in human 18S subunits (Cooper et al. 2014, 2015). 18S rRNA was cleaved at these sites to a significant magnitude in PDEmut-infected and EndoUmut-infected wt BMM (Supplemental Fig. S11). Thus, as in human cells (Cooper et al. 2014, 2015), RNase L targets 18S rRNA for cleavage at precise sites in murine cells. Furthermore, RNase L activity was specifically increased within PDEmut-infected and EndoUmut-infected wt BMM, as compared to MHV(S)-infected and MHV(V)-infected wt BMM. Although RNase L–dependent cleavage sites in rRNA were easily detected (Supplemental Fig. S11), EndoU-dependent cleavage sites in rRNA were not detected (Supplemental Fig. S11D). These data show the dsRNA-dependent OAS/RNase L pathway was significantly activated in PDEmut- and EndoUmut-infected wt BMM, and excluded rRNAs as targets of EndoU.
Cellular endoribonucleases
The cleavage of MHV RNA within EndoUmut-infected RNase L−/− BMM (Supplemental Figs. S2B, S4B, and S7C,F) and the cleavage of cellular mRNAs within mock-infected cells (Supplemental Fig. S16) provoked our consideration of other cellular endoribonucleases. We hypothesized that pyrimidine-specific cleavage of MHV RNA within EndoUmut-infected RNase L−/− BMM might be due to one or another RNase A family enzyme (Schwartz et al. 2018). We also considered T2 endoribonucleases based on their reported contributions to TLR8 activation (Greulich et al. 2019). Consequently, we examined the expression of RNases 4 and 5 (angiogenin) and RNases T2A and T2B (Supplemental Fig. S13). Changes in magnitudes of RNase 4 and 5 expression were observed, with ∼10-fold decreased expression in MHV(S)-infected and MHV(V)-infected wt BMM as compared to mock-infected wt BMM (Supplemental Fig. S12A,B). Decreased expression of RNases 4 and 5 was not as strong in PDEmut-infected wt BMM, and very little decrease in expression was observed in EndoUmut-infected wt BMM. Similar changes in expression of RNases 4 and 5 were observed in IFNAR−/− BMM and RNase L−/− BMM, with significantly decreased expression in MHV(S)-infected and MHV(V)-infected cells and a more limited decrease in EndoUmut-infected cells (Supplemental Fig. S12C). Because RNases 4 and 5 share a complex dual promoter (Dyer and Rosenberg 2005), with alternative splicing leading to the expression of either RNase 4 or RNase 5, coordinate increases and decreases in their expression was not unexpected. These data reinforce our suspicion regarding the residual pyrimidine-specific cleavage of MHV RNAs within EndoUmut-infected RNase L−/− BMM.
In contrast to expression of RNase 4 and 5, changes in magnitudes of expression of RNases T2A and T2B were relatively small within MHV-infected cells, with a tendency for slightly increased expression (Supplemental Fig. S12C,D). RNase T2 cleaves RNA within endosomes and lysosomes, targeting purine:uridine dinucleotides, R↓U (Greulich et al. 2019). The residual purine-specific cleavage of MHV RNA within EndoUmut-infected RNase L−/− BMM might be associated with RNase T2 activity; however, our experiments do not definitively address this possibility.
DISCUSSION
We address a key question in the coronavirus field (Deng and Baker 2018): What is the natural target of EndoU? Coronavirus EndoU inhibits dsRNA-activated antiviral responses in infected cells (Kindler et al. 2017); however, it is not clear how EndoU does this because its physiologic RNA substrates are unknown. In this study, we used MHV-infected, bone marrow–derived macrophage (BMM) and cyclic phosphate cDNA sequencing to identify the RNA targets of EndoU.
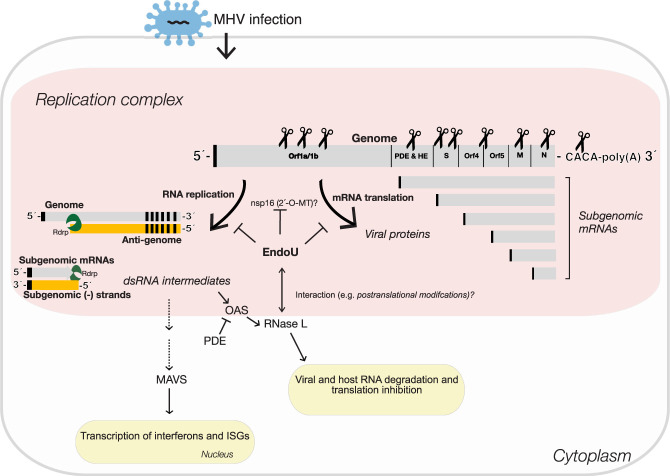
We found that EndoU targeted MHV RNA within infected cells, cleaving viral RNA on the 3′ side of pyrimidines with a strong preference for cleavage between U↓A and C↓A sequences (endoY↓A) (Fig. 3). This cleavage specificity from MHV-infected cells is consistent with that of purified EndoU (Bhardwaj et al. 2006; Kang et al. 2007) and RNase A (Harper et al. 1990; Hofsteenge et al. 1998), enzymes that are functionally and structurally related to one another (Bhardwaj et al. 2008). EndoU cleavage was detected in every region of MHV RNA, from the 5′ NTR to the 3′ NTR, including relatively small TRS sequences (Figs. 4E, 5, and 6). Because MHV RNA is a template for both viral mRNA translation and viral RNA replication, cleavage by EndoU could inhibit both of these biosynthetic processes (Fig. 8). Intriguingly, MHV TRS sequences contain EndoU target sequences (C↓A and U↓A sequences) (Fig. 5B). TRS6, which was targeted more frequently by EndoU than other TRS elements, contains a C↓A target sequence rather than a U↓A sequence. We postulate that EndoU cleaves MHV RNA in a regulated manner, to inhibit negative-strand RNA synthesis, thereby preventing the accumulation of viral dsRNA (Fig. 8). Nsp16 (2′-O-MT) could regulate EndoU-mediated cleavage of MHV RNA by methylating C↓A and U↓A sequences (Ivanov et al. 2004).
FIGURE 8.
EndoU targets in MHV RNA. MHV RNA was targeted for cleavage by EndoU within infected BMM. MHV RNA was cleaved by EndoU in all regions of the genome, at C↓A and U↓A sequences. Because MHV RNA is a template for both viral mRNA translation and viral RNA replication, cleavage by EndoU could inhibit both of these biosynthetic processes. Intriguingly, MHV TRS sequences contain EndoU target sequences (C↓A and U↓A sequences). TRS6, which was targeted more frequently by EndoU than other TRS elements, contains a C↓A target sequence rather than a U↓A sequence. We postulate that EndoU cleaves MHV RNA in a regulated manner, to inhibit negative-strand RNA synthesis, thereby inhibiting the accumulation of viral dsRNA. Nsp16 (2′-O-MT) could regulate EndoU-mediated cleavage of MHV RNA by methylating C↓A and U↓A sequences. EndoU and RNase L cleave an overlapping set of UA sequences within MHV, suggesting a functional interplay between host and viral endoribonucleases.
How does EndoU inhibit double-stranded RNA-activated antiviral responses?
Coronavirus EndoU inhibits the activation of multiple host dsRNA sensors, including MDA5, OAS, and PKR (Deng et al. 2017, 2019; Kindler et al. 2017). dsRNA-activated OAS/RNase L and PKR pathways restrict the replication of EndoU-deficient coronaviruses (Kindler et al. 2017). Because EndoUmut-infected cells had increased accumulation of dsRNA, Kindler et al. (2017) concluded that EndoU functions as a viral RNA decay pathway to evade dsRNA-activated antiviral host cell responses. Consistent with this idea, Hackbart et al. (2020) report that EndoU targets poly(U) sequences at the 5′ end of viral negative-strand RNA. Another report suggests that EndoU might control the localization of viral dsRNA within cells, perhaps maintaining dsRNA within membranous RNA replication complexes (Deng et al. 2017). Our data suggest a third possibility, that EndoU targets MHV RNA to limit the synthesis of dsRNA (Fig. 8): EndoU-dependent cleavages were detected throughout MHV RNA (Figs. 4E, 5, and 6), including the ORF1a/1b regions, indicating that EndoU destroys the template for negative-strand RNA synthesis, precluding the formation of dsRNA, rather than acting on dsRNA. Cyclic phosphate cDNA sequencing detected large amounts of cleavage in MHV (+) strand (Fig. 2; Supplemental Figs. S2 and S9A) and vanishingly little cleavage in MHV (−) strand (Supplemental Figs. S1B and S12C,D). Cyclic phosphate cDNA sequencing can readily detect cleavage sites in both (+) and (−) strands of viral RNA (Cooper et al. 2014, 2015); however, cleavage of poly(U) sequences at the 5′ end of MHV negative-strand RNA cannot be detected because the resulting cyclic phosphate RNA fragments are too small (<20 bases long) and they are homopolymeric, preventing detection by our sequencing and bioinformatics pipelines. While it is possible that EndoU targets poly(U) sequences, the specificity of EndoU for C↓A and U↓A sequences in vivo (Fig. 3) is inconsistent with poly(U) substrates being physiologically relevant. Furthermore, purified EndoU (Bhardwaj et al. 2006) and RNase A (Harper et al. 1990; Hofsteenge et al. 1998) readily target UA sequences within heteropolymeric substrates. Thus, we conclude that EndoU targets MHV (+) strand RNA to prevent the synthesis of dsRNA (Fig. 8). Nonetheless, potential RNA substrates in (+) and (−) strands are not mutually exclusive. EndoU-dependent cleavage of the CACA sequences at the 3′ end of the (+) strand and the poly(U) at the 5′ end of the (−) strand could occur coordinately, as both are colocalized adjacent to one another at the same end of dsRNA products. When EndoU was mutated, we detected the activation of the dsRNA-dependent OAS/RNase L pathway (Figs. 3 and 4; Supplemental Fig. S11) and increased host gene expression associated with response to dsRNA (Fig. 7). These data, like other reports (Deng et al. 2017, 2019; Kindler et al. 2017; Volk et al. 2020), indicate EndoU inhibits the activation of dsRNA sensors.
EndoU cleaved MHV RNA in every region of the genome (Figs. 4E, 5, and 6). Because MHV RNA is a template for both viral mRNA translation and viral RNA replication, cleavage by EndoU could inhibit both of these biosynthetic processes (Fig. 8). Cleavage of the viral genome (mRNA 1) would reduce the expression of the viral replicase. Coronavirus RNA synthesis requires ongoing expression of the viral replicase, with negative-strand RNA synthesis being most dependent on new replicase expression (Sawicki and Sawicki 1986). Substantial amounts of EndoU-dependent cleavage were detected in orfs 1a and 1b, especially within wt BMM (Fig. 4), potentially limiting the expression of replicase. Cleavage of MHV genomic RNA (aka mRNA 1), the template for both genomic and subgenomic negative-strand RNA synthesis (Sawicki et al. 2007), would also prevent the synthesis of dsRNA products (Fig. 8). EndoU-mediated cleavage of the tandem CA sequences adjacent to the MHV RNA poly(A) tail is most intriguing in this regard (Fig. 6C; Supplemental Fig. S7). A single or tandem CA sequence is conserved at the 3′ NTR/poly(A) junction of representative α-, β-, γ- and δ-coronaviruses (Supplemental Table S4), present in both genomic and sg mRNAs, and maintained within conserved secondary RNA structures (Zust et al. 2008). Endonucleolytic cleavage of this site would have important phenotypic effects, as highlighted in our model (Fig. 8). Importantly, we detect cleavage of the C↓A sequence at the 3′ NTR/poly(A) junction by EndoU as well as cellular endoribonucleases, presumably RNase A family members. Intriguingly, coronavirus nsp8 is reported to have 3′-terminal adenylyltransferase activity (Tvarogova et al. 2019), which, in theory, could restore a poly(A) tail on viral RNA following endonucleolytic cleavage of the C↓A sequence at the 3′ NTR/poly(A) junction. Furthermore, EndoU colocalizes with the polymerase and its cofactors (Athmer et al. 2017). The coronavirus polymerase, nsp12, with nsp7 and 8 cofactors (Kirchdoerfer and Ward 2019), initiates negative-strand RNA synthesis on the poly(A) tail of genomic RNA, leading to the synthesis of poly(U) at the 5′ end of negative-strand RNA (Sola et al. 2015). Colocalization of EndoU with nsp8 and the coronavirus polymerase enable countervailing enzymatic activities that could inhibit or promote negative-strand RNA synthesis at the point of initiation. This provides a theoretically appealing mechanism for EndoU and other endoribonucleases to regulate the synthesis of dsRNA (Fig. 8).
MHV RNA was cleaved by one or more unspecified endoribonucleases in EndoUmut-infected RNase L−/− BMM. Thus, in addition to EndoU- and RNase L–dependent cleavage of MHV RNA, we observed EndoU- and RNase L–independent cleavage of MHV RNA (Fig. 2; Supplemental Fig. S9). By using fold-change analyses between wt and mutant conditions, we attributed the majority of endoribonucleolytic cleavage sites in MHV RNA to EndoU activity and RNase L activity (Fig. 4); however, a substantial amount of cleavage in MHV RNA persisted in EndoUmut-infected RNase L−/− BMM (Fig. 2A,B, EndoUmut, red bars). More than 5% of the cyclic phosphates in EndoUmut-infected RNase L−/− BMM RNA samples were in MHV RNA (Fig. 2A,B, EndoUmut, red bars). This EndoU- and RNase L–independent cleavage of MHV RNA occurred predominantly at UA and CA dinucleotides (Fig. 3D); especially within IFNAR−/− (Supplemental Fig. S4A, position −1 to +1) and RNase L−/− (Supplemental Fig. S4B, position −1 to +1) BMM. Thus, the EndoU- and RNase L–independent cleavage of MHV RNA exhibited a nucleotide specificity similar to that of EndoU-dependent cleavage. It is possible that the H277A mutation in EndoU fails to completely inhibit endoribonuclease activity; however, we suspect that RNase A family members are responsible for this residual EndoU-independent cleavage of MHV RNA at U↓A and C↓A sequences. RNase A family enzymes are expressed in macrophage (Schwartz et al. 2018) and they cleave RNA at U↓A and C↓A sequences (Shapiro and Vallee 1991; Hofsteenge et al. 1998). EndoU-independent cleavage of MHV RNA at the 31332C↓AC↓A31335 sequence was evident in wt BMM (Supplemental Fig. S7A), IFNAR−/− BMM (Supplemental Fig. S7B), and RNase L−/− BMM (Supplemental Fig. S7C). The expression of RNases 4 and 5 (Supplemental Fig. S13A,B) is consistent with residual cleavage at the 31332C↓AC↓A31335 sequences in EndoUmut-infected BMM (Supplemental Fig. S7). These data indicate that the 31332C↓AC↓A31335 sequence in MHV RNA was susceptible to both EndoU-dependent and EndoU-independent cleavage. The atomic structure of EndoU revealed an RNase A-like catalytic domain (Ricagno et al. 2006); however, we did not anticipate the degree of overlap in substrate specificity observed for EndoU-dependent and EndoU-independent (presumably RNase A family) enzymes within BMM. Additional experiments will be required to address the identity and functional significance of the EndoU-independent (presumably RNase A family) enzymes within BMM.
EndoU activity and changes in host gene expression
MHV EndoU activity is associated with evasion of dsRNA-dependent host responses (Deng et al. 2017; Kindler et al. 2017; Volk et al. 2020). Host gene expression associated with response to dsRNA increased by 100- to 1000-fold in MHV-infected wt BMM as compared to mock-infected cells, with even larger 1000- to 10,000-fold increases in EndoUmut-infected wt BMM (Fig. 7C). How does EndoU activity reduce the expression of cellular mRNAs associated with the dsRNA host response? Does EndoU cleave these mRNAs or does it inhibit the accumulation of viral dsRNA? If EndoU is localized within membranous RNA replication complexes, as reported (Athmer et al. 2017; V'Kovski et al. 2019), it likely has the potential to cleave viral RNA to regulate negative-strand RNA synthesis while sparing cellular mRNAs from cleavage.
When comparing MHV(S)- and EndoUmut-infected wt BMM (Fig. 7F), conditions with and without MHV EndoU activity, we found the majority of the cellular mRNAs from the dsRNA response were not directly cleaved by EndoU. dsRNA host response mRNAs (Fig. 7H,I), like host mRNAs in the aggregate (Supplemental Fig. S16), were cleaved at dinucleotides we attribute to cellular endoribonuclease actvities: RNase A (U↓A and C↓A) and RNase L (UA↓ and UU↓) (Fig. 7H,I). Furthermore, cleavage of dsRNA host response mRNAs at U↓A and C↓A sequences, like host mRNAs in the aggregate (Supplemental Fig. S16), were unaffected by an EndoU mutation (Fig. 7I). Altogether, these data indicate that MHV EndoU does not cleave dsRNA host response mRNAs in a manner that would affect their overall abundance. Rather, our data suggest that MHV EndoU cleaves viral RNA to inhibit the accumulation of viral dsRNA, thereby inhibiting dsRNA-dependent host transcription (Fig. 8).
Does nsp16 (2′-O-MT) regulate EndoU?
How is EndoU activity regulated to avoid unwanted cleavage events? This is an important question because MHV RNA integrity is critical for viral mRNA translation and viral RNA replication (Fig. 8). When EndoU cleaves MHV RNA, it must do so in a regulated manner to avoid self-destruction. Some amount of intact and functional MHV genomic RNA must be maintained within infected cells to sustain an infection. One factor thought to regulate EndoU is nsp16, a 2′-O-methyltransferase (Ivanov et al. 2004).
When EndoU was first characterized, Ivanov and colleagues demonstrated that EndoU-mediated cleavage of RNA substrates was prevented by 2′-O-methylation (Ivanov et al. 2004). They also highlighted the modular nature of viral evolution, drawing attention to the side-by-side nature of nsp15 (EndoU) and nsp16 (2′-O-MT) within nidovirus genomes, suggesting a functional interplay between the two enzymes (Ivanov et al. 2004). 2′-O-methyltransferases have been functionally characterized in two families of positive-strand RNA viruses, coronaviruses (Zust et al. 2011; Menachery et al. 2014) and flaviviruses (Ray et al. 2006; Daffis et al. 2010; Dong et al. 2012; Szretter et al. 2012). One function of these enzymes is to methylate the adenosine of 5′ cap structures in viral mRNAs (Ray et al. 2006), to evade the antiviral activity of IFIT1 (Daffis et al. 2010; Zust et al. 2011; Dong et al. 2012). Whether these enzymes can methylate other residues throughout viral RNA is less certain; however, 2′-O-methyltransferases are reported to inhibit the recognition of viral dsRNA by MDA5 (Zust et al. 2011). It is intriguing to note that EndoU cleavage sites (C↓A and U↓A sequences) contain adenosine. 2′-O-methylation of the pyrimidine at cleavage sites would prevent cleavage of viral RNA because the 2′-hydroxyl of ribose is the nucleophile responsible for attacking the phosphodiester backbone (Yang 2011). Whether 2′-O-methylation of adenosine can prevent EndoU-mediated cleavage of C↓A and U↓A sequences remains to be determined; however, some amount of intact MHV genomic RNA must be maintained within infected cells to sustain an infection.
RNA-seq showed that MHV RNAs were abundant (Supplemental Fig. S6) and there were proportional amounts of each MHV mRNA within infected cells (Fig. 5C) despite profound changes in endoribonuclease activity from one condition to another. Thus, neither EndoU nor RNase L activities were associated with extreme changes in the proportions of one MHV mRNA to another. Rather, relatively subtle changes in MHV mRNA1–7 proportions were observed. These data suggest that EndoU and RNase L activities modulate MHV RNA abundance during infections, but do not contribute to extreme changes in the relative amounts of one MHV mRNA to another. In contrast, the absence of EndoU activity during MHV infection leads to profound increases in host gene expression associated with response to dsRNA (Fig. 7A–E), despite the activation of RNase L activity. During a dsRNA-activated stress response (Burke et al. 2019; Rath et al. 2019), expression and translation of cellular mRNAs occurs in the context of activated RNase L despite the ongoing degradation of cellular mRNAs by RNase L. Ongoing expression and translation of MHV mRNAs likely occurs in the context of EndoU or RNase L activities in the same manner. When preexisting host or viral mRNAs are cleaved by EndoU or RNase L activities, new MHV mRNAs are synthesized, refreshing the pool of viral mRNAs. Thus, although RNase L activity restricts virus replication (Zhao et al. 2012), MHV replication can clearly tolerate and benefit from EndoU activity.
Do EndoU and RNase L coregulate MHV RNA gene expression and replication?
Importantly, EndoU and RNase L share a common cleavage site, UA (Fig. 3). Furthermore, we can distinguish between EndoU-dependent and RNase L–dependent cleavage of UA sequences because EndoU cleaves between U↓A dinucleotides, whereas RNase L cleaves after UA↓ dinucleotides. Under some conditions, such as MHV(S)-infected and MHV(V)-infected wt BMM, UA sequences in viral RNA were cleaved predominantly by EndoU (Fig. 3G). Under other circumstances, UA sequences in MHV RNA were cleaved predominantly by RNase L, as in PDEmut-infected and EndoUmut-infected wt BMM (Fig. 3G). In both cases, regardless of whether the host or viral endoribonuclease cleaves MHV RNA, the consequence will be an inhibition in viral mRNA translation and an inhibition in viral RNA replication (Fig. 8). It is interesting to see that both a host and a viral endoribonuclease have the capacity to inhibit magnitudes of MHV gene expression and replication by targeting a common set of UA sequences within the viral genome. It is also interesting that EndoU activity was subdued within IFNAR−/− and RNase L−/− cells, as if EndoU activity was modulated by RNase L activity (Fig. 4C). Together, these results suggest an interesting interplay between EndoU- and dsRNA-activated host responses (Fig. 8).
Summary
We addressed a key question in the field (Deng and Baker 2018): What is the natural target of coronavirus EndoU? We find that EndoU targets MHV RNA within infected cells, cleaving viral RNA on the 3′ side of pyrimidines with a strong preference for cleavage between U↓A and C↓A sequences (endoY↓A). We postulate that EndoU cleaves MHV RNA in a regulated manner, to inhibit negative-strand RNA synthesis, reducing the accumulation of viral dsRNA, while ensuring continuing virus replication (Fig. 8). By regulating the synthesis and accumulation of viral dsRNA, coronaviruses can evade double-stranded RNA-activated antiviral responses within infected cells (Deng et al. 2017, 2019; Kindler et al. 2017; Deng and Baker 2018).
MATERIALS AND METHODS
Viruses
Wild-type mouse hepatitis virus A59 from Volker Thiel (MHV(V)) (Thiel et al. 2001; Coley et al. 2005; Eriksson et al. 2008) and Susan Weiss (MHV(S)) (Roth-Cross et al. 2009) were used, along with a mutant derivative of each. An H277A mutation in nsp15 rendered an EndoU-deficient mutant (EndoUmut) from MHV(V) (Kindler et al. 2017). An H126R mutation in NS2 rendered a phosphodiesterase mutant (PDEmut) from MHV(S) (Roth-Cross et al. 2009).
Murine bone marrow–derived macrophages
Bone marrow–derived macrophage (BMM) from wt, IFNAR−/−, and RNase L−/− C57BL/6 mice were obtained as previously described (Kindler et al. 2017). Progenitor cells were isolated from the hind limbs of 8- to 12-wk-old mice, passed through a cell strainer and RBCs were lysed using 1 mL of lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM EDTA). Cells were washed 3× with PBS and cultured in macrophage medium (Iscove's Modified Dulbecco's Medium, 5%–10% M-CSF [L929-supernatant], 0.1% 50 mM 2-mercaptoethanol). Adherent BMM were harvested at 7 dpi.
Virus infection
BMM were infected with MHV(S), MHV(V), EndoUmut, and PDEmut at an MOI of 1 PFU per cell at 37°C as previously described (Kindler et al. 2017). At 9- and 12-h postinfection (hpi), supernatant was harvested for virus titration and cells were lysed in TRIzol (Invitrogen). MHV in the supernatant was quantified by standard plaque assay on L2 cells.
Cyclic phosphate cDNA sequencing
Total RNA was extracted from cell lysates and split equally for cyclic phosphate and RNA-seq library preparations. Cyclic phosphate cDNA libraries were prepared by DNase treating the total RNA for 30 min followed by ethanol precipitation with 20 µg of glycogen and ligation with 50 µM 3′ linker (Table 1) in 30 µL final volume. The ligation reactions were conducted using 15 pmol of RtcB ligase (NEB), 1× RtcB buffer (NEB), 100 µM GTP, 1 mM MnCl2, and 20 units of RNase inhibitor (Enzymatics) at 37°C for 2 h. Samples were ethanol precipitated with 20 µg of glycogen and resuspended in 10 µL of RNase-free H2O for chemical fragmentation (Ambion Fragmentation Reagent) at 65°C for 4 min. Samples were then denatured in 1 volume of stop dye (95% formamide, 0.01% xylene cyanol/bromophenol blue), heated to 65°C for 5 min and separated on a 6% polyacrylamide TBE–urea gel. Gels were stained with SYBR Gold (Invitrogen) and visualized to excise RNA larger than adaptor (∼100–1000 bp). RNA was eluted from the gel slices with 2 h incubation at 40°C in 0.3 M sodium acetate, pH 5.2, 1 mM EDTA, pH 8.0 followed by gentle mixing overnight at 4°C. Eluted RNA was recovered by ethanol precipitation with 20 µg of glycogen and resuspended in 12 µL of RNase-free H2O. RNA was ligated to 50 µM 5′ linker (Table 1) in 20 µL final volume. The ligation reactions were conducted using 15 pmol of RtcB (NEB), 1× RtcB buffer (NEB), 100 µM GTP, and 1 mM MnCl2, 20 units of RNase inhibitor (Enzymatics) at 37°C for 2 h followed by ethanol precipitation with 20 µg of glycogen and resuspended in 100 µL of RNase-free H2O. Ligated RNAs were purified using 25 µL of magnetic Streptavidin beads (Invitrogen) washed three times with 100 µL of B&W buffer (5 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 1 M NaCl) supplemented with 0.1% Tween 20, twice with 100 µL of solution A (0.1 M RNase-free NaOH, 0.05 M RNase-free NaCl), and twice with 100 µL of solution B (0.1 M RNase-free NaCl). Washed beads were resuspended in 2× B&W buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 2 M NaCl) with 20 units of RNase inhibitor (Enzymatics), and the RNA solution was added to the beads and incubated with rotation for 15 min at room temperature. After incubation, the beads were washed three times with 100 µL of 1× B&W buffer before resuspending the beads in 20 µL of 25 mM biotin in elution buffer (Omega Bio-Tek). The beads were incubated at room temperature for 15 min with occasional mixing. After binding the beads to the magnet, the supernatant was collected. The elution process was repeated once for a final volume for 40 µL of eluted RNA. cDNA was prepared using 5 µM of an Illumina-compatible primer complementary to the 3′ linker (Table 1), 20 µL of eluted RNA, and ProtoScript II RT (NEB). An amount of 10 µL of cDNA was PCR amplified for 18 cycles with Illumina TruSeq primers (Table 1) and Phusion DNA polymerase. PCR reactions were purified with AMPure XP beads (Beckman Coulter). Indexed libraries were quantified by Qubit (Invitrogen). Library quality was assessed on a 4200 TapeStation System instrument (Agilent Technologies) using a D100 ScreenTape assay, mixed to a final concentration of 1–10 nM, and sequenced on an Illumina HiSeq in a 50-cycle run.
TABLE 1.
Linkers and primers for cyclic phosphate cDNA library
Stranded RNA-seq
Total RNA was enriched for polyadenylated mRNA using oligo(dT) magnetic beads (Ambion). cDNA was generated from the enriched polyA+ mRNAs after fragmentation in 2.2× SuperScript IV reverse transcriptase buffer at 94°C for 3 min. After immediately cooling on ice, RT reaction with SuperScript IV RT (Thermo Fisher Scientific) was performed per manufacture's recommendations with 150 ng of random primers (Thermo Fisher Scientific) in 20 µL final volume. cDNA:RNA hybrids were purified using MyOne Silane beads (Thermo Fischer Scientific) per manufacture's recommendations and eluted in 18 µL of RNase-free H2O. Second-strand cDNA was then generated using RNase H and E. coli DNA Polymerase (Enzymatics) with dUTP incorporation (1× NEB buffer 2, 100 µM dATP, dCTP, dGTP, 200 µM dUTP, 2.5 units of RNase H, 30 units of DNA polymerase) in 100 µL final volume at 15°C for 2.5 h. cDNA was purified with Silane beads and eluted in 52 µL of RNase-free H2O as input for end repair reaction using an End Repair Module (NEB) following manufacturer's recommendations. A tailing reaction (50 µL final volume) performed with Klenow fragment (minus 3′–5′ exonuclease activity, Enzymatics) and end repaired Silane-purified cDNA eluted in 32 µL of RNase-free H2O (1× NEB buffer 2, 200 µM dATP, 15 units Klenow fragment) at 37°C for 30 min. Reaction products were purified with 1.8× AMPure XP beads (Beckman Coulter) and eluted in 10 µL of RNase-free H2O. Purified cDNA was ligated to 40 nM of annealed Illumina TruSeq Universal adaptors (Table 2) in 50 µL final volume reaction for 30 min at 25°C (40 nM adaptors, 1× Rapid Ligation Buffer [Enzymatics], 3000 units of T4 DNA ligase [Enzymatics]). Reaction products were purified with AMPure XP beads and eluted in 12 µL of RNase-free H2O. USER enzyme (NEB) was used to degrade the dUTP-containing strand by adding 1 unit of USER to purified cDNA and incubating for 30 min at 37°C. Reactions were used directly in PCR amplification with Illumina TruSeq primers and Phusion DNA polymerase with 10 µL of input for 18 cycles. Libraries were size selected from 200 to 700 bp using AMPure XP beads, quantified by Qubit (Invitrogen), and mixed to a final concentration of 4 nM. Library quality was assessed on a 4200 TapeStation System Instrument (Agilent Technologies) using a D100 ScreenTape assay and sequenced on an Illumina NovaSEQ 6000 in a paired end 150 cycle run.
TABLE 2.
Illumina TruSeq Universal Adaptors
Computational analyses of next-generation sequencing data
Processing and analysis of cyclic phosphate cDNA libraries
Unique molecular identifier (UMI) sequences were extracted and added to FASTQ reads using UMI-tools (v0.5.4) (Smith et al. 2017). Only read 1 was used from the second experiment, to adhere with the analysis process applied for experiment 1. FASTQ reads were then aligned to the MHV genome alone (GenBank accession: NC_001846.1) and a combined reference including the MHV genome, Mus musculus rRNA, and U6 snRNA references (GenBank accession numbers: NR_003278.3, NR_003279.1, NR_003280.2, NC_000074.6, NR_003027.2), and annotated ORFs from the Mouse ORFeome collection (MGC full-cds collection for Mus musculus) using Bowtie version 2 (v2.3.2) (Langmead and Salzberg 2012). Aligned reads were de-duplicated using UMI-tools to remove PCR duplicated reads. De-duplicated reads were converted to genomecov format using BEDTools (v2.26.0) to report the number of reads at each single-base cleavage position, including for sense and antisense sequences for the MHV aligned reads (Quinlan and Hall 2010). Reads at each cleavage position were normalized by library size. Cyclic phosphate libraries we also aligned separately to the Mus musculus complete transcriptome reference (Gencode GRCm38.p6) using Bowtie2, de-duplicated, and converted to genomecov format, to capture potential cleavage of transcript isoforms and other noncoding RNAs.
To identify a signal dependent on the presence of a specific endoribonuclease, normalized counts at each cleavage position in RNase L−/− or EndoUmut libraries were subtracted from the signal in libraries with wild-type RNase L or EndoU activity, to remove signal that occurred in the absence of either endoribonuclease. The difference in cleavage activity at each position in the the absence of RNase L or EndoU was determined by calculating the log2-fold change. The frequency of cleavage at particular dinucleotides was determined by quantifying the sum of reads assigned to each of the 16 possible dinucleotides divided by total number of aligned reads in the library. Dinucleotide enrichment was determined by calculating the frequency of cleavage at each dinucleotide in the MHV genomic sequence and determining the log2-fold enrichment of the observed (experimental) frequencies compared to the expected (background) frequencies. Significance of enrichment was calculated using the Fisher's exact test to compare the odds ratio of obtaining a specific dinucleotide in the expected data to the observed data.
RNA-seq alignment, annotation, and differential expression analysis
Illumina adaptor sequences were trimmed from FASTQ reads using Cutadapt (v1.16) and sequences shorter than 20 nucleotides were discarded (Martin 2011). Trimmed reads were aligned to a combined MHV (GenBank accession: NC_001846.1) and Mus musculus genome reference (Ensembl GRCm38.p6) and bedGraph coverage files were generated from each alignment using STAR (v2.7.1a) (Dobin and Gingeras 2002). Read fragments were assigned and counted using featureCounts (from subread v1.6.2) and a combined MHV and Mus musculus GTF (Ensembl GRCm38.p6) file for gene annotation (Liao et al. 2014). The MHV GTF file included the genomic positions of the combined ORF1a/b nonstructural proteins and each of the structural and accessory proteins. Gene counts were normalized using DESeq2 media of ratios method to account for sequencing depth and RNA composition (Love et al. 2014). For downstream differential expression analysis, trimmed reads were also aligned to the Mus musculus complete transcriptome reference (Gencode GRCm38.p6) using Salmon (v0.14.1) (Patro et al. 2017). Transcript abundance files were used for differential expression analysis with DESeq2 after importing with tximport and counts normalized by the media of ratios method were used for data visualization (Soneson et al. 2015). Genes with an FDR < 0.05 were called significant and used to generate Volcano plots with the EnhancedVolcano package and z-transformed counts were used to generate heatmaps with the ComplexHeatmap package (Gu et al. 2016; Blighe et al. 2019). For gene functional category enrichment analyses, topGO was used to determine significant enrichment (weightFish/P > 0.01) by using nondifferentially expressed genes (<2 or <−2 log2-fold change and FDR < 0.01) as the background to determine the categories enriched in differentially expressed genes. topGO uses conditional enrichment analysis, which takes the nested structure of GO terms into account to reduce redundancy in enrichment results (Alexa and Rahnenfuhrer 2019). Heatmaps were generated using pheatmap and variance stabilized count data from DESeq2 analysis, with data centered to 0. Genes were selected from enriched GO categories (Kolde 2015).
Motif analysis
To visualize the cleavage sequence preferences for RNase L and EndoU, the top 1% of either RNase L– or EndoU-dependent sites from subtractive analysis, as described above, were selected. Using BEDTools, 3 bp were added upstream and downstream from the selected positions and a FASTA file was generated from the 6-bp sequences. Meme was used to determine the sequence preference enrichment and graphed using ggseqlogo (Bailey et al. 2009; Wagih 2017).
UA scoring
UA sequences in the MHV genome were designated as predominantly cleaved 3′ of U (consistent with EndoU targeting) or A (consistent with RNase L targeting). All UA dinucleotides in the MHV genome with >30 cyclic phosphate counts at either position of cleavage in the dinucleotide were selected and the ratio of normalized counts in each position was calculated (RNase L/EndoU). If the ratio was >1, the position was scored as a UA↓ site and if the ratio was <1, the position was scored as a U↓A.
Regional MHV cleavage analysis and abundance normalization
The normalized counts in each MHV genomic region (all the genes and ORFs shown in Figure 1A, in addition to the 5′ and 3′ UTR and body TRS regions) were summed to calculate the total cyclic phosphate reads per region. Size correction was performed by dividing the sum of cyclic phosphate counts in each region by the length of the region in bases. To normalize the cyclic phosphate data by RNA abundance, stranded bedGraph files were generated from the bam files produced by STAR alignment of the RNA-seq libraries. At each position with both cyclic phosphate and RNA-seq data in the MHV genome, the cyclic phosphate counts were divided by the normalized (reads per million mapped reads) RNA-seq counts to generate a normalized value for cyclic phosphate RNA abundance.
Cyclic phosphate and RNA-seq enrichment analysis
Normalized cyclic phosphate and RNA-seq counts (normalized counts from DESeq2) were summed per protein-coding mRNA that aligned to the mm10 transcriptome. For each mRNA, a ratio was calculated to determine enrichment in cyclic phosphate and RNA-seq counts relative to EndoUmut infection (total cyclic phosphate or RNA-seq reads in wt/total cyclic phosphate or RNA-seq reads in EndoUmut). Each mRNA was assigned to a bin based on the enrichment ratio for cyclic phosphate and RNA-seq abundance: bin 1 = cyclic phosphate and RNA-seq ratios <1, bin 2 cyclic phosphate, and RNA-seq ratios ≥1, bin 3 = cyclic phosphate ratio <1 and RNA-seq ratio ≥1, bin 4 = cyclic phosphate ratio ≥1, and RNA-seq ratio <1.
TRS analysis
In the above analyses, RNA-seq reads mapping to the viral genome were not distinguished by alignment to genomic RNA or subgenomic mRNAs. To assign RNA-seq reads to subgenomic mRNAs, we used an analysis similar to that described in Irigoyen et al. (2016) to identify leader/body chimeric reads (subgenomic mRNAs). We parsed the bam files generated from STAR alignment of the RNA-seq libraries to the combined mouse and MHV genome for reads containing the 11 nt of the leader sequence, UUUAAAUCUAA (GenBank accession: NC_001846.1, nt 54–65) before the leader TRS sequence. The positions in the reference where the read alignment starts and ends after the leader sequence were extracted to obtain an interval of alignment for the sequence downstream from the leader/body transition. The intervals for each chimeric read were intersected with the intervals of each canonical subgenomic mRNA using valr (Riemondy et al. 2017), with the requirement of at least 30 nt of overlap, to assign each chimeric read to an mRNA: 65 to 21,746 (mRNA 1), 21,747 to 23,921 (mRNA 2), 23,922 to 27,934 (mRNA 3), 27,935 to 28,317 (mRNA 4), 28,318 to 28,959 (mRNA 5), 28,958 to 29,654 (mRNA 6), 29,655 to 31,334 (mRNA 7) (Riemondy et al. 2017). The number of reads assigned to each mRNA were counted and normalized to either the total sum of mRNAs per library or reads per million.
SNP analysis
Variant calling analysis was performed using bcftools (v1.9) to generate genotype likelihoods from the RNA-seq bam files for MHV aligned reads, followed by SNP calling/indel calling to generate VCF files (Li 2011). The generated VCF files were filtered using bcftools with parameters -s LOWQUAL -e %QUAL<30 || DP>20′ to identify low quality sites with less than 20 quality score or 30 bp of read depth.
Bioinformatics pipeline
Code for all described analyses are available at https://github.com/hesselberthlab/endoU in the form of scripts, a data processing pipeline, and analysis package.
DATA DEPOSITION
Raw and processed sequencing data are available at NCBI GEO: GSE147852.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Barton and Weiss labs for critical evaluation of the manuscript. We thank Evan Lester for assistance with bioinformatics analyses. This work was supported by Public Health Service grants from the National Institutes of Health (F30 AI140615 to R.A., R01 AI104887 and R01 AI140442 to S.R.W., R35 GM119550 to J.R.H., and R01 AI042189 to D.J.B.) and the Swiss National Science Foundation (SNF project 149784).
Author contributions: R.A. contributed to experimental design, experimental procedures, bioinformatics, data analysis, data curation, interpretation of data, and manuscript preparation. Y.L. contributed to experimental design, experimental procedures, and interpretation of data. E.K. contributed to experimental design and experimental procedures. D.A.C. contributed to the methodology and the pilot study. M.R. contributed to experimental procedures. V.T. contributed to experimental design, project administration, funding acquisition, and data interpretation. S.R.W. contributed to experimental design, project administration, funding acquisition, and data interpretation. J.R.H. contributed to experimental design, project administration, funding acquisition, data interpretation, and manuscript preparation. D.J.B. contributed to experimental design, project administration, funding acquisition, data interpretation, and manuscript preparation.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.076604.120.
REFERENCES
- Alexa A, Rahnenfuhrer J. 2019. topGO: Enrichment analysis for gene ontology (R package version 2.37.0). Bioconductor, https://www.bioconductor.org
- Athmer J, Fehr AR, Grunewald M, Smith EC, Denison MR, Perlman S. 2017. In situ tagged nsp15 reveals interactions with coronavirus replication/transcription complex-associated proteins. MBio 8: e02320 10.1128/mBio.02320-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K, Sun J, Holzenburg A, Guarino LA, Kao CC. 2006. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol 361: 243–256. 10.1016/j.jmb.2006.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K, Palaninathan S, Alcantara JM, Yi LL, Guarino L, Sacchettini JC, Kao CC. 2008. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem 283: 3655–3664. 10.1074/jbc.M708375200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdwell LD, Zalinger ZB, Li Y, Wright PW, Elliott R, Rose KM, Silverman RH, Weiss SR. 2016. Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal Oas gene expression and independent of virus-induced interferon. J Virol 90: 3160–3172. 10.1128/JVI.03036-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blighe K, Rana S, Lewis M. 2019. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling (R package version). https://github.com/kevinblighe/EnhancedVolcano
- Bredenbeek PJ, Pachuk CJ, Noten AF, Charite J, Luytjes W, Weiss SR, Spaan WJ. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res 18: 1825–1832. 10.1093/nar/18.7.1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM, Moon SL, Matheny T, Parker R. 2019. RNase L reprograms translation by widespread mRNA turnover escaped by antiviral mRNAs. Mol Cell 75: 1203–1217.e5. 10.1016/j.molcel.2019.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley SE, Lavi E, Sawicki SG, Fu L, Schelle B, Karl N, Siddell SG, Thiel V. 2005. Recombinant mouse hepatitis virus strain A59 from cloned, full-length cDNA replicates to high titers in vitro and is fully pathogenic in vivo. J Virol 79: 3097–3106. 10.1128/JVI.79.5.3097-3106.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DA, Jha BK, Silverman RH, Hesselberth JR, Barton DJ. 2014. Ribonuclease L and metal-ion-independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic Acids Res 42: 5202–5216. 10.1093/nar/gku118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DA, Banerjee S, Chakrabarti A, Garcia-Sastre A, Hesselberth JR, Silverman RH, Barton DJ. 2015. RNase L targets distinct sites in influenza A virus RNAs. J Virol 89: 2764–2776. 10.1128/JVI.02953-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468: 452–456. 10.1038/nature09489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Baker SC. 2018. An “old” protein with a new story: coronavirus endoribonuclease is important for evading host antiviral defenses. Virology 517: 157–163. 10.1016/j.virol.2017.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci 114: E4251–E4E60. 10.1073/pnas.1618310114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, van Geelen A, Buckley AC, O'Brien A, Pillatzki A, Lager KM, Faaberg KS, Baker SC. 2019. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J Virol 93: e02000 10.1128/JVI.02000-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR. 2002. Mapping RNA-seq reads with STAR.29, 11.14.1-11.14.19. Wiley, Hoboken, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, et al. 2012. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog 8: e1002642 10.1371/journal.ppat.1002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Rath S, Kolet-Mandrikov D, Korennykh A. 2017. Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23: 1660–1671. 10.1261/rna.062000.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer KD, Rosenberg HF. 2005. The mouse RNase 4 and RNase 5/ang 1 locus utilizes dual promoters for tissue-specific expression. Nucleic Acids Res 33: 1077–1086. 10.1093/nar/gki250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson KK, Makia D, Thiel V. 2008. Generation of recombinant coronaviruses using vaccinia virus as the cloning vector and stable cell lines containing coronaviral replicon RNAs. Methods Mol Biol 454: 237–254. 10.1007/978-1-59745-181-9_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd-Smith G, Slattery E, Lengyel P. 1981. Interferon action: RNA cleavage pattern of a (2′–5′)oligoadenylate-dependent endonuclease. Science 212: 1030–1032. 10.1126/science.6165080 [DOI] [PubMed] [Google Scholar]
- Goebel SJ, Miller TB, Bennett CJ, Bernard KA, Masters PS. 2007. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J Virol 81: 1274–1287. 10.1128/JVI.00803-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117: 17–37. 10.1016/j.virusres.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich W, Wagner M, Gaidt MM, Stafford C, Cheng Y, Linder A, Carell T, Hornung V. 2019. TLR8 is a sensor of RNase T2 degradation products. Cell 179: 1264–1275.e13. 10.1016/j.cell.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Hackbart M, Deng X, Baker SC. 2020. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci 117: 8094–8103. 10.1073/pnas.1921485117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Fox EA, Shapiro R, Vallee BL. 1990. Mutagenesis of residues flanking Lys-40 enhances the enzymatic activity and reduces the angiogenic potency of angiogenin. Biochemistry 29: 7297–7302. 10.1021/bi00483a020 [DOI] [PubMed] [Google Scholar]
- Hofsteenge J, Moldow C, Vicentini AM, Zelenko O, Jarai-Kote Z, Neumann U. 1998. A single amino acid substitution changes ribonuclease 4 from a uridine-specific to a cytidine-specific enzyme. Biochemistry 37: 9250–9257. 10.1021/bi9803832 [DOI] [PubMed] [Google Scholar]
- Irigoyen N, Firth AE, Jones JD, Chung BY, Siddell SG, Brierley I. 2016. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog 12: e1005473 10.1371/journal.ppat.1005473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, Ziebuhr J. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci 101: 12694–12699. 10.1073/pnas.0403127101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Bhardwaj K, Li Y, Palaninathan S, Sacchettini J, Guarino L, Leibowitz JL, Kao CC. 2007. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J Virol 81: 13587–13597. 10.1128/JVI.00547-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi L, Lissenberg A, Watson R, de Groot RJ, Weiss SR. 2005. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J Virol 79: 15064–15073. 10.1128/JVI.79.24.15064-15073.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E, Gil-Cruz C, Spanier J, Li Y, Wilhelm J, Rabouw HH, Zust R, Hwang M, V'Kovski P, Stalder H, et al. 2017. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog 13: e1006195 10.1371/journal.ppat.1006195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer RN, Ward AB. 2019. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10: 2342 10.1038/s41467-019-10280-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Vaharautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. 2011. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 9: 72–74. 10.1038/nmeth.1778 [DOI] [PubMed] [Google Scholar]
- Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6: e226 10.1371/journal.pbio.0060226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. 2015. pheatmap: pretty heatmaps. University of Tartu, Tartu, Estonia. https://www.rdocumentation.org/packages/pheatmap/versions/1.0.12 [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lissenberg A, Vrolijk MM, van Vliet AL, Langereis MA, de Groot-Mijnes JD, Rottier PJ, de Groot RJ. 2005. Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J Virol 79: 15054–15063. 10.1128/JVI.79.24.15054-15063.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W, Bredenbeek PJ, Noten AF, Horzinek MC, Spaan WJ. 1988. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology 166: 415–422. 10.1016/0042-6822(88)90512-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 1–3. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Menachery VD, Yount BL Jr, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J Virol 88: 4251–4264. 10.1128/JVI.03571-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek S, Krwawicz J, Kutner J, Lazniewski M, Kucinski I, Ginalski K, Dziembowski A. 2012. C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3′ end modification. Genes Dev 26: 1911–1925. 10.1101/gad.193169.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova DD, Ulferts R, van den Born E, Lauber C, Gorbalenya AE, Ziebuhr J, Snijder EJ. 2009. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J Virol 83: 5671–5682. 10.1128/JVI.00261-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach SE, York K, Hesselberth JR. 2015. Global analysis of RNA cleavage by 5′-hydroxyl RNA sequencing. Nucleic Acids Res 43: e108 10.1093/nar/gkv536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YH, Lin CH, Lin CN, Lo CY, Tsai TL, Wu HY. 2016. Characterization of the role of hexamer AGUAAA and poly(A) tail in coronavirus polyadenylation. PLoS ONE 11: e0165077 10.1371/journal.pone.0165077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S, Prangley E, Donovan J, Demarest K, Wingreen NS, Meir Y, Korennykh A. 2019. Concerted 2-5A-mediated mRNA decay and transcription reprogram protein synthesis in the dsRNA response. Mol Cell 75: 1218–1228.e6. 10.1016/j.molcel.2019.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 80: 8362–8370. 10.1128/JVI.00814-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S, Egloff MP, Ulferts R, Coutard B, Nurizzo D, Campanacci V, Cambillau C, Ziebuhr J, Canard B. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci 103: 11892–11897. 10.1073/pnas.0601708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemondy KA, Sheridan RM, Gillen A, Yu Y, Bennett CG, Hesselberth JR. 2017. valr: reproducible genome interval analysis in R. F1000Res 6: 1025 10.12688/f1000research.11997.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Clements J, Eddy SR. 2017. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods 14: 45–48. 10.1038/nmeth.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross JK, Stokes H, Chang G, Chua MM, Thiel V, Weiss SR, Gorbalenya AE, Siddell SG. 2009. Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J Virol 83: 3743–3753. 10.1128/JVI.02203-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol 57: 328–334. 10.1128/JVI.57.1.328-334.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol 64: 1050–1056. 10.1128/JVI.64.3.1050-1056.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J Virol 81: 20–29. 10.1128/JVI.01358-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz K, Hesselberth JR, Fields S. 2010. Capture and sequence analysis of RNAs with terminal 2′,3′-cyclic phosphates. RNA 16: 621–631. 10.1261/rna.1934910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L, Cohen A, Thomas J, Spencer JD. 2018. The immunomodulatory and antimicrobial properties of the vertebrate ribonuclease a superfamily. Vaccines (Basel) 6: 76 10.3390/vaccines6040076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna PB, Hung SL, Brian DA. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci 86: 5626–5630. 10.1073/pnas.86.14.5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Vallee BL. 1991. Interaction of human placental ribonuclease with placental ribonuclease inhibitor. Biochemistry 30: 2246–2255. 10.1021/bi00222a030 [DOI] [PubMed] [Google Scholar]
- Shchepachev V, Wischnewski H, Missiaglia E, Soneson C, Azzalin CM. 2012. Mpn1, mutated in poikiloderma with neutropenia protein 1, is a conserved 3′-to-5′ RNA exonuclease processing U6 small nuclear RNA. Cell Rep 2: 855–865. 10.1016/j.celrep.2012.08.031 [DOI] [PubMed] [Google Scholar]
- Shieh CK, Lee HJ, Yokomori K, La Monica N, Makino S, Lai MM. 1989. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J Virol 63: 3729–3736. 10.1128/JVI.63.9.3729-3736.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu M, Morichika K, Kawamura T, Honda S, Kirino Y. 2019. Genome-wide identification of short 2′,3′-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet 15: e1008469 10.1371/journal.pgen.1008469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Heger A, Sudbery I. 2017. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res 27: 491–499. 10.1101/gr.209601.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Almazan F, Zuniga S, Enjuanes L. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2: 265–288. 10.1146/annurev-virology-100114-055218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4: 1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter KJ, Daniels BP, Cho H, Gainey MD, Yokoyama WM, Gale M Jr, Virgin HW, Klein RS, Sen GC, Diamond MS. 2012. 2′-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog 8: e1002698 10.1371/journal.ppat.1002698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V, Herold J, Schelle B, Siddell SG. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol 82: 1273–1281. 10.1099/0022-1317-82-6-1273 [DOI] [PubMed] [Google Scholar]
- Tvarogova J, Madhugiri R, Bylapudi G, Ferguson LJ, Karl N, Ziebuhr J. 2019. Identification and characterization of a human coronavirus 229E nonstructural protein 8-associated RNA 3′-terminal adenylyltransferase activity. J Virol 93: e00291 10.1128/JVI.00291-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulferts R, Ziebuhr J. 2011. Nidovirus ribonucleases: structures and functions in viral replication. RNA Biol 8: 295–304. 10.4161/rna.8.2.15196 [DOI] [PubMed] [Google Scholar]
- van der Hoeven B, Oudshoorn D, Koster AJ, Snijder EJ, Kikkert M, Barcena M. 2016. Biogenesis and architecture of arterivirus replication organelles. Virus Res 220: 70–90. 10.1016/j.virusres.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog 4: e1000054 10.1371/journal.ppat.1000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P, Gerber M, Kelly J, Pfaender S, Ebert N, Braga Lagache S, Simillion C, Portmann J, Stalder H, Gaschen V, et al. 2019. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. eLife 8: e42037 10.7554/eLife.42037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk A, Hackbart M, Deng X, Cruz-Pulido Y, O'Brien A, Baker SC. 2020. Coronavirus endoribonuclease and deubiquitinating interferon antagonists differentially modulate the host response during replication in macrophages. J Virol 94: e00178 10.1128/JVI.00178-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O. 2017. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33: 3645–3647. 10.1093/bioinformatics/btx469 [DOI] [PubMed] [Google Scholar]
- Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. 1981. Interferon action–sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature 289: 414–417. 10.1038/289414a0 [DOI] [PubMed] [Google Scholar]
- Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, et al. 2020. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27: 325–328. 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. 2011. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys 44: 1–93. 10.1017/S0033583510000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Li X, Deng Z, Chen Z, Liu Y, Gao Y, Wu W, Chen Z. 2017. Structural biology of the arterivirus nsp11 endoribonucleases. J Virol 91: e01309 10.1128/JVI.01309-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li L, Yan L, Ming Z, Jia Z, Lou Z, Rao Z. 2018. Structural and biochemical characterization of endoribonuclease Nsp15 encoded by middle east respiratory syndrome coronavirus. J Virol 92: e00893 10.1128/JVI.00893-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Rose KM, Elliott R, Van Rooijen N, Weiss SR. 2011. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J Virol 85: 10058–10068. 10.1128/JVI.05075-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11: 607–616. 10.1016/j.chom.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Birdwell LD, Wu A, Elliott R, Rose KM, Phillips JM, Li Y, Grinspan J, Silverman RH, Weiss SR. 2013. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J Virol 87: 8408–8418. 10.1128/JVI.00769-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Miller TB, Goebel SJ, Thiel V, Masters PS. 2008. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3′ end of the mouse coronavirus genome. J Virol 82: 1214–1228. 10.1128/JVI.01690-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12: 137–143. 10.1038/ni.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.