Abstract
In addition to adenosine-to-inosine RNA editing activities, ADAR1 has been shown to have various RNA editing-independent activities including modulation of RNAi efficacy. We previously reported that ADAR1 forms a heterodimer complex with DICER and facilitates processing of pre-miRNAs to mature miRNAs. In addition to miRNA synthesis, DICER is involved in processing of long dsRNAs into small RNAs (endo-siRNAs). Generation of retrotransposon-derived endo-siRNAs by DICER and their functions in regulation of transcripts in mouse oocytes has been previously reported. However, the synthesis and functions of endo-siRNAs in somatic cells remain largely unknown. Here, we report that ADAR1 together with DICER generates endogenous small RNAs, Alu endo-siRNAs by cleaving long double-stranded regions of inverted Alu repeats. We identified AGO2-loaded Alu endo-siRNAs, which are highly expressed in commonly used cell lines. These Alu endo-siRNAs carrying both sense and antisense Alu sequences seem to target a set of genes containing a single Alu sequence, either antisense or sense, respectively, within their 3′UTR. In silico screening identified potential RNA silencing target genes for these Alu endo-siRNAs. We present results of a proof-of-concept experiment, in which sense Alu endo-siRNAs derived from AluSz and AluJr family elements target CUB Domain Containing Protein 1 mRNAs containing an antisense copy of AluJb in their 3′UTRs and consequently induce apoptosis in HeLa cells. Our results clearly indicate that Alu endo-siRNAs are functional also in somatic cells.
Keywords: RNA editing, ADAR1, DICER, RNAi, Alu, siRNA
INTRODUCTION
RNA editing that converts adenosine to inosine (A-to-I RNA editing) specifically in double-stranded RNAs (dsRNAs) is catalyzed by adenosine deaminases acting on RNA (ADARs) (Mannion et al. 2015; Nishikura 2016; Walkley and Li 2017; Eisenberg and Levanon 2018). Three ADAR gene family members (ADAR1–3) have been identified in vertebrates. Members of the ADAR gene family share common structural features, such as the presence of multiple dsRNA binding domains (dsRBDs) and a separate deaminase domain (Lai et al. 1995; Macbeth et al. 2005). Although both ADAR1 (ADAR) and ADAR2 (ADARB1) are catalytically active enzymes, catalytic activities of ADAR3 (ADARB2) have not been shown yet. As the translation machinery reads inosine as guanosine, A-to-I RNA editing of neurotransmitter and ion channel gene transcripts result in recoding and diversification of their functions (Hood and Emeson 2012). For instance, the physiologically important recoding-type editing target site of ADAR2, the Q/R site of α-amino-3-hydroxy-5-methyl-isoxazole-4-propionate (AMPA) glutamate receptor GluR-2 mRNAs, has been reported (Higuchi et al. 2000; Hood and Emeson 2012). In contrast, a wide range of ADAR1 functions have been reported (Mannion et al. 2015; Nishikura 2016). There are two ADAR1 isoforms, a full-length interferon-inducible ADAR1p150 and a shorter and constitutive ADAR1p110 truncated at the amino terminus. ADAR1p110 mainly localizes in the nucleus, whereas ADAR1p150 is mostly detected in the cytoplasm (Patterson and Samuel 1995). The inactivation of ADAR1 in mice leads to an embryonic lethal phenotype because of widespread apoptosis, revealing that requirement of ADAR1 functions for development (Hartner et al. 2004; Wang et al. 2004). Although ADAR1 edits select protein coding sequences (Song et al. 2016) as well as certain microRNA precursors (Yang et al. 2006; Kawahara et al. 2007; Iizasa et al. 2010), noncoding sequences consisting of inverted repeats of retrotransposon elements such as Alu and SINE are the most frequent targets of ADAR1 (Bazak et al. 2014; Porath et al. 2014; Sakurai et al. 2014; Tan et al. 2017). Studies by several independent groups revealed that aberrant activation of the dsRNA-sensing mechanism mediated by the MDA5–MAVS–IFN pathway and consequent interferon (IFN) and inflammatory responses underlie the embryonic lethal phenotype of Adar1 null mice (Mannion et al. 2014; Liddicoat et al. 2015; Pestal et al. 2015). The cytoplasmic ADAR1p150 isoform specifically suppresses this dsRNA-sensing pathway by hyper-editing SINE and Alu dsRNAs present in 3′UTRs of certain mRNAs (Mannion et al. 2014; Liddicoat et al. 2015; Pestal et al. 2015; Ahmad et al. 2018). Interestingly, a more recent study indicates the presence of currently unknown non-Alu long dsRNAs as alternate targets of ADAR1p150, rather than 3′UTR Alu dsRNAs (Barak et al. 2020). Regardless of the exact triggers of the dsRNA-sensing mechanism, loss of this particular function of ADAR1 has been linked to the severe autoimmune disease Aicardi–Goutières syndrome (AGS) (Rice et al. 2012; Pestal et al. 2015) and underlies the resistance developed in certain tumors to PD-1 immune checkpoint blockade-based immunotherapy (Ishizuka et al. 2019). These ADAR1 functions described depend on its A-to-I RNA editing activities, which require homodimerization through its dsRBD3 (Cho et al. 2003; Valente and Nishikura 2005; Ota et al. 2013).
Furthermore, several RNA editing-independent functions of ADAR1 have also been reported. The normally nuclear-localized ADAR1p110 moves to the cytoplasm upon phosphorylation by MAP kinases and promotes survival of stressed cells by protecting antiapoptotic gene transcripts from Staufen1-mediated mRNA decay (Sakurai et al. 2017). This stress response function of ADAR1p110 has been shown to be RNA editing-independent (Sakurai et al. 2017). We found that ADAR1 forms a complex with DICER, a member of the RNase III gene family and an essential enzyme involved in miRNA processing and the RNA interference (RNAi) mechanism (Ota et al. 2013). ADAR1 in the complex increases Vmax of the DICER pre-miRNA cleavage reaction and promotes loading of miRNA onto RISC, identifying a new role of ADAR1 in miRNA processing and RNAi mechanisms and revealing a stimulative interaction between RNA editing and RNAi (Nishikura et al. 2013; Ota et al. 2013). The amino-terminus half of the DICER DExD box RNA helicase domain and the ADAR1 dsRBD2 are required for formation of the DICER/ADAR1 heterodimer complex (Ota et al. 2013). This RNAi function of ADAR1 is also RNA editing-independent (Ota et al. 2013). Initial detection of the DICER/ADAR1 complex formation in developing mouse embryos and also in HeLa cells has been recently extended to oral squamous carcinoma cells (Liu et al. 2019a) and mouse cardiomyocytes undergoing viral myocarditis (Zhang et al. 2019). Interestingly, involvement of DICER in synthesis of endo-siRNAs from retrotransposon-related repetitive elements such as Alu and 7SL has been reported (Watanabe et al. 2008; Ren et al. 2012). In this study, we investigated in vitro processing of Alu dsRNAs to Alu endo-siRNAs by DICER/ADAR1 complexes. We found that ADAR1 significantly promotes processing of Alu dsRNA to Alu siRNAs. Furthermore, we identified various AGO2-bound endogenous Alu siRNAs likely to be processed by DICER and ready to act on their target genes in vivo in HEK293T cells. We here report targeting by such Alu endo-siRNAs of CUB Domain Containing Protein 1 (CDCP1) mRNAs containing an antisense copy of AluJb in their 3′UTRs (Gong and Maquat 2011), and consequent induction of apoptosis, thus demonstrating Alu endo-siRNA-mediated gene silencing in somatic cells.
RESULTS
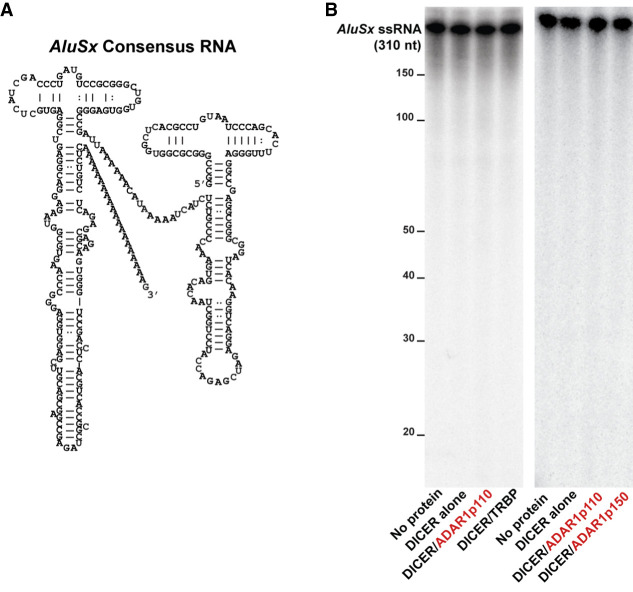
No cleavage of Alu RNAs by DICER alone or DICER/ADAR1 complexes
It has been reported that DICER cleaves cytotoxic Alu RNAs, ∼300 nt in length, transcribed by RNA polymerase III (Fig. 1A) and prevents their accumulation and consequent induction of apoptosis in retinal pigmented epithelium (RPE) cells. Deficiency in this particular DICER function has been suggested to underlie age-related macular degeneration (AMD), a leading cause of blindness (Kaneko et al. 2011; Tarallo et al. 2012). However, several studies arguing against this AMD model have also been reported (Sundermeier et al. 2017; Kosmidou et al. 2018). Furthermore, processing of 7SL RNA, also transcribed by RNA polymerase III and considered as an ancestral RNA of Alu RNAs, into small RNAs by DICER has been suggested (Ren et al. 2012). To reexamine whether DICER indeed cleaves Alu RNAs and also to see whether ADAR1 in any way facilitates such DICER activity, we conducted in vitro assays using DICER/ADAR1 and DICER/TRBP (TARBP2) complexes as well as preparation of highly purified DICER alone and AluSx RNAs (302 nt in length). Differentially epitope tagged FLAG–DICER/HAT–ADAR1 and FLAG–DICER/HAT–TRBP complexes were purified by the baculovirus coexpression system and sequential epitope-based affinity chromatography purification as described previously (Ota et al. 2013), whereas a uniformly 32P-labeled Alu RNA containing a single copy of the sense AluSx sequence was prepared by in vitro transcription. We found that a single copy sense Alu RNA was resistant to cleavage by DICER alone in contrast to the previous report (Kaneko et al. 2011). Furthermore, DICER/ADAR1p110 and DICER/ADAR1p150 complexes or DICER/TRBP complexes all could not cleave Alu RNAs (Fig. 1B). In fact, we detected some degradation of Alu RNAs by partially purified DICER protein preparations (data not shown) prior to establishment of our DICER protein purification protocol. Thus, the previously reported in vitro processing of Alu and 7SL RNAs into small fragmented RNAs by DICER could be due to contaminating RNases in the DICER preparation. Alternatively, the Alu or 7SL RNAs used for previous in vitro dicing assays might be contaminated by their antisense strand RNAs, which are often generated during in vitro transcription of sense strand RNAs. Synthesis of such antisense strand RNAs would result in the formation of completely complementary double-stranded Alu and 7SL RNAs: perfect substrates for DICER. We conclude that Alu RNAs transcribed by RNA polymerase III are not natural substrates of DICER, perhaps because of their rather short stem structures with many mismatched base pairs (Fig. 1A).
FIGURE 1.
DICER does not cleave single Alu RNAs. (A) Secondary structure of single AluSx RNA. (B) The DICER cleavage reaction was done at 37°C for 60 min with 0.15 nM of AluSx RNA and 1.5 nM of DICER alone or various DICER complexes.
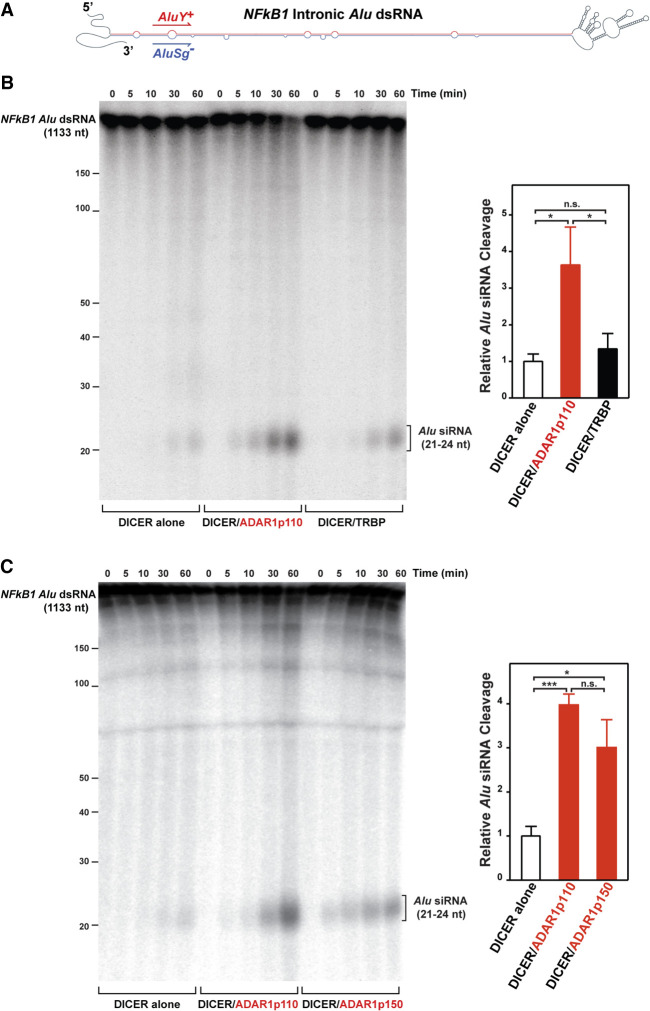
ADAR1 promotes processing of siRNAs from Alu dsRNAs by DICER
In addition to cleavage of pre-miRNAs to mature miRNAs, DICER also cleaves long dsRNAs into siRNAs (Zhang et al. 2002; Lee et al. 2004). We next investigated whether DICER processes Alu dsRNA stems of long hairpin-like structures consisting of inverted repeats of sense and antisense Alu sequences and various sizes of connecting loop (Fig. 2A). We first tested a uniformly 32P-labeled Alu dsRNA hairpin structure consisting of a sense AluY+, an antisense AluSg−, and a 533 nt long loop (Fig. 2A), which is embedded within the intron 16 of the NFκB1 gene (Kawahara and Nishikura 2006). We found that DICER alone cleaved Alu dsRNA into 21–24 nt long small RNAs (Alu siRNAs). Most interestingly, both ADAR1p110 and ADAR1p150 as DICER/ADAR1 complexes promoted DICER cleavage of Alu dsRNA to Alu siRNAs by approximately three- to fourfold (Fig. 2B,C). In contrast, TRBP had very little effect on the DICER activity of Alu dsRNA cleavage (Fig. 2B). We also tested an additional Alu dsRNA hairpin structure consisting of a sense AluSp+, an antisense AluSp−, and a 70 nt long loop (Supplemental Fig. S1A), which is in the 3′UTR of NICN1 gene (Chen et al. 2008). Complementarity of the NICN1 Alu dsRNA stem formed by sense and antisense of the same Alu element subfamily (AluSp) is much higher than that of the NFκB1 Alu dsRNA, and the loop size of the NICN1 Alu hairpin structure is much smaller than that of the NFκB1 Alu hairpin. In spite of these differences, we obtained similar results: The DICER/ADAR1 complex cleaved Alu dsRNA more efficiently and generated more Alu siRNAs than DICER alone (Supplemental Fig. S1B). ADAR1 promotion of DICER activities to cleave pre-miRNAs to mature miRNAs has been reported to be independent of ADAR1 A-to-I editing activities (Ota et al. 2013). Generation of Alu siRNAs by the DICER complex with an ADAR1p110–E912A editing defective mutant was as effective as that of the DICER/ADAR1p110–WT complex (Supplemental Fig. S1C), indicating that ADAR1 promotion of DICER cleavage of Alu dsRNA is also independent of the A-to-I editing activities of ADAR1.
FIGURE 2.
ADAR1 augments the DICER cleavage reaction rate for NFκB1 Alu dsRNAs and increases Alu siRNA production. (A) Secondary structure of NFκB1 intronic Alu dsRNA. (B,C) The time course analysis of DICER cleavage. The DICER reaction was done at 37°C with 0.15 nM of NFκB1 Alu dsRNA and 1.5 nM of DICER alone or DICER complexes for various times. The reaction products were fractionated by 10% Urea-PAGE. The Alu siRNA cleavage efficiencies were determined at the 30 min time point. Data are shown as mean ± SD. (n = 3 technical replicates). (*) P < 0.05; (***) P < 0.001; n.s. not significant by two-tailed Student's t-test.
Alu siRNAs (∼21–24 nt) generated by cleavage of the NFκB1 Alu dsRNA by the DICER/ADAR1p110 complex (Fig. 2C, 60 min time point) were gel-purified and subjected to high-throughput sequencing (small RNA-seq) (Supplemental Fig. S2). Although we found that small RNAs were generated from both sense and antisense strands of the Alu hairpin stem region, their read numbers and patterns were not those expected from symmetrical processive cleavage of the completely matched dsRNA (Supplemental Fig. S2). We noticed no small RNA reads corresponding to certain regions containing internal loops and bulges. These results may indicate that DICER/ADAR1 complexes bind Alu dsRNA stems all along their length, not merely at the ends, and cleave them wherever bound, instead of acting processively in one direction.
A-to-I edited Alu dsRNAs are also cleaved by the DICER/ADAR1 complex
The most frequent ADAR1 target for A-to-I editing is Alu dsRNA (Porath et al. 2014; Sakurai et al. 2014; Tan et al. 2017), and editing may affect their cleavage by DICER. Typical Alu hairpin stems contain both matched A:U and mismatched A–C base pairs (Fig. 2A; Supplemental Fig. S1A). In addition to adenosines of matched A:U base pairs, adenosines of mismatched A–C base pairs are known to be edited by ADAR1. ADAR1-mediated editing of adenosine residues of A:U matched base pairs results in I:U wobble base pairs, which decreases double-strandedness of Alu hairpin stems. On the other hand, editing of A–C mismatched base pairs results in I:C matched base pairs, which increases double-strandedness. Therefore, we next examined the effects of A-to-I editing on DICER cleavage of Alu dsRNAs. We first edited in vitro NFκB1 and NICN1 Alu dsRNA hairpins as much as possible using purified FLAG-ADAR1p110 proteins (Supplemental Fig. S3A) prior to in vitro dicing assay. Interestingly, we found very little effects of A-to-I editing: Both unedited and edited Alu dsRNAs were cleaved by the DICER/ADAR1p110 complex to siRNAs equally well (Supplemental Fig. S3B). Possibly due to the offsetting effects of A:U editing and A–C editing, A-to-I editing does not change significantly the overall double-strandedness of most Alu hairpins and consequently has very little effects on their DICER cleavage, in contrast to the reported antagonistic interaction of A-to-I editing and RNAi on long dsRNAs in C. elegans (Wu et al. 2011).
In vivo identification of Alu siRNAs and their targets in commonly used cell lines
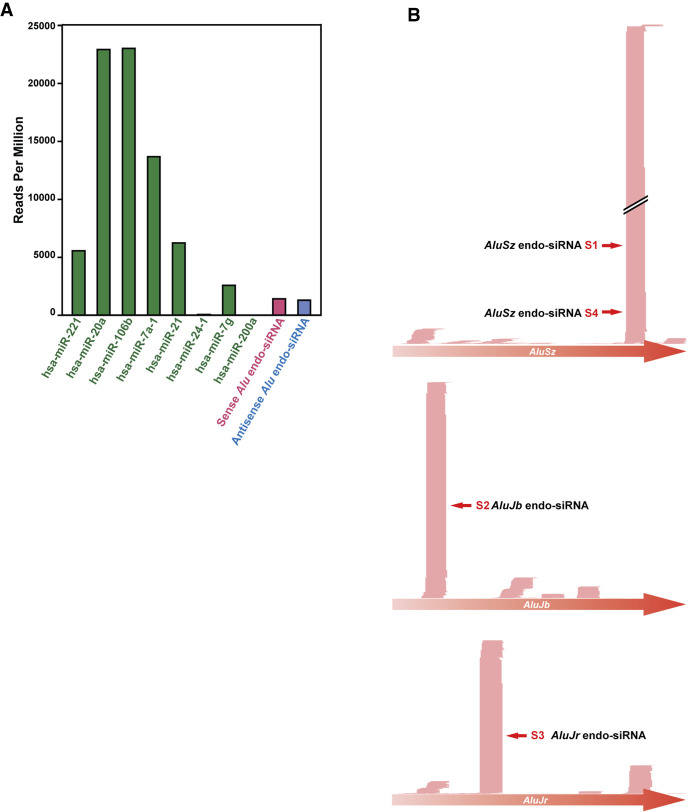
We previously investigated expression of miRNAs in HeLa cells by small RNA-seq analysis (GSM1057798) (Ota et al. 2013). We reexamined our small RNA-seq data and found that both sense and antisense strands of Alu-endo siRNAs ranging from 19 to 33 nt with a peak ranging from 21 to 24 nt in length can be detected in HeLa cells (Fig. 3A), although their expression levels are much lower than those of miRNAs: Alu siRNAs accounted for only 0.3% of total miRNAs. In vivo detection of Alu siRNAs at a slightly higher level (1.2% of total miRNAs) has been also reported in HepG2.2.15 cells (Ren et al. 2012). We annotated HeLa Alu siRNAs to various Alu subfamily consensus sequences, which resulted in sense and antisense strand read mapping patterns expected for DICER cleavage products, as shown for three examples: AluY, AluSz, and AluJb subfamilies (Fig. 3B). In addition, profiling of AGO2-bound small RNAs has been reported in HEK293T cells (GSM1334330) (Rybak-Wolf et al. 2014). These AGO2-bound small RNAs are considered as productive DICER cleavage products ready to be engaged in RNAi (Rybak-Wolf et al. 2014). We examined those published small RNA-seq data and confirmed that AGO2-bound sense and antisense Alu endo-siRNAs can be detected in HEK293T cells (Supplemental Table S3), although their expression levels were much lower than those of AGO2-bound miRNAs (only 0.09% of AGO2-bound miRNAs) (Fig. 4A). These results indicate that Alu endo-siRNAs are produced and also that some of them may be functional, targeting cellular genes via RNA interference in commonly used somatic cell lines such as HeLa and HEK293T.
FIGURE 3.
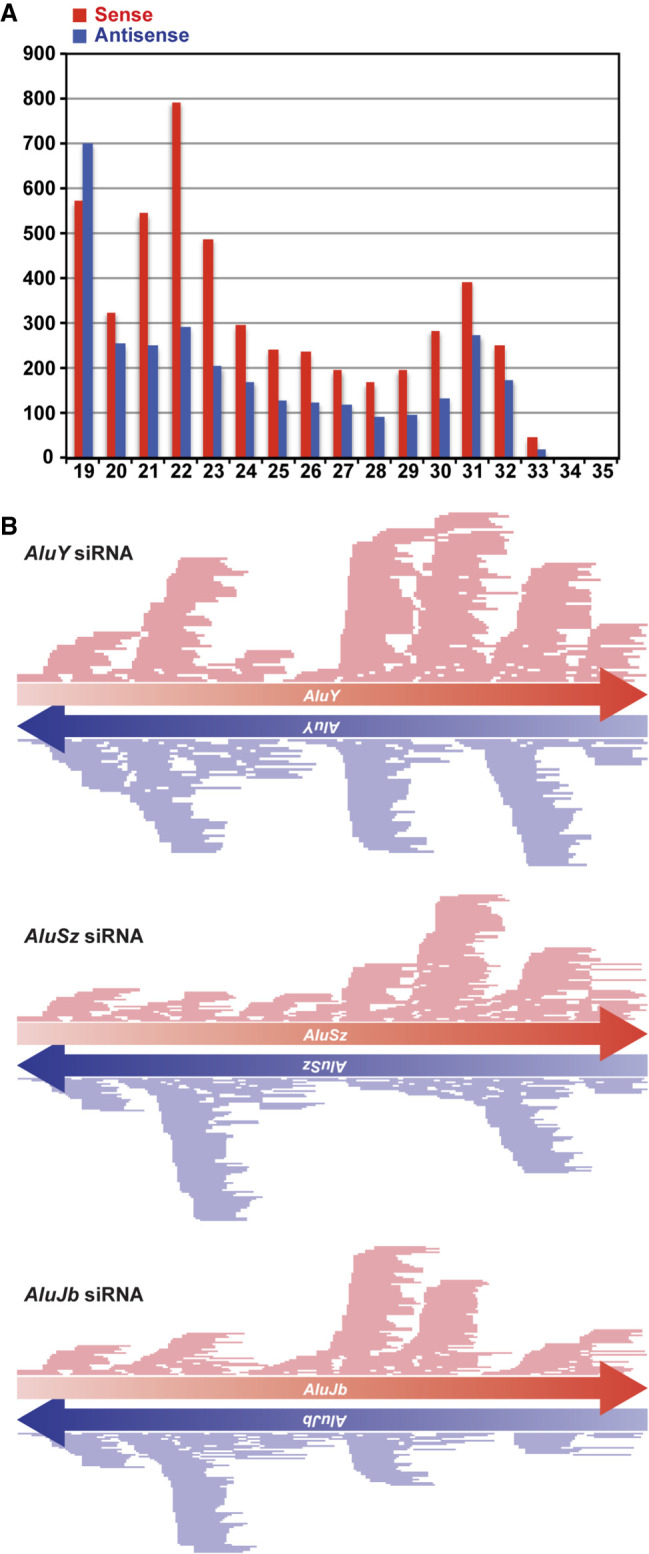
Alu siRNA expression in HeLa cells. (A) The length distribution of Alu-derived small RNAs in HeLa cells. y-axis: counts per million reads; x-axis: base length. (B) Distribution of Alu siRNAs corresponding to sense and antisense strands of consensus AluY, AluSz, and AluJb sequences (UCSC genome database). RNAs of 19 to 24 nt in length were used as siRNAs.
FIGURE 4.
Alu siRNAs are loaded onto AGO2. (A) The read counts per million reads of various mature miRNAs and Alu siRNAs in small RNAs immunoprecipitated with AGO2 antibodies (GSM1334330). (B) Distribution of AGO2-bound Alu siRNAs corresponding to sense strands of AluSz (chr2:5532106–5532415), AluJb (chr1:236404589–236404810), and AluJr (chr1:227459304–227459613).
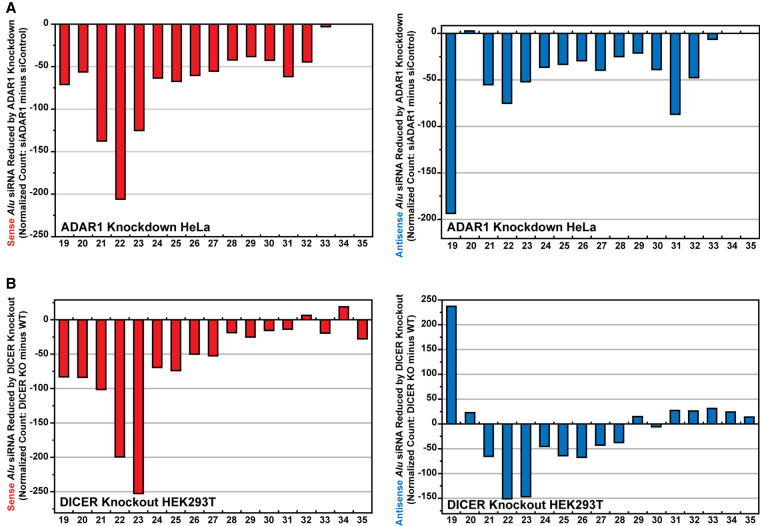
To evaluate the in vivo effects of ADAR1 and DICER on synthesis of these Alu siRNAs, we examined Alu siRNA reads in ADAR1 knockdown HeLa cells (GSM105779) (Ota et al. 2013) in comparison to control cells. We found that both sense and antisense Alu siRNA read counts, peaking at 21–23 (major peak) and 30–32 (minor peak) nucleotides in length, are reduced significantly by ADAR1 depletion (Fig. 5A). As expected, DICER depletion resulted also in significant reduction of both sense and antisense Alu siRNA reads, peaking at 21–23 nt in length, in DICER knockout HEK293T cells (Fig. 5B; Bogerd et al. 2014). These results confirmed the in vivo contributions of ADAR1 and DICER to genesis of Alu endo-siRNAs in somatic cell lines.
FIGURE 5.
Reduced expression of Alu siRNAs in ADAR1 or DICER depleted cells. (A) The length distribution of read numbers of sense and antisense Alu siRNAs reduced by ADAR1 knockdown in HeLa cells. (B) The length distribution of read numbers of sense and antisense Alu siRNAs reduced by DICER knockout in HEK293T cells. Read numbers were first normalized: The read number of Alu siRNAs was divided by the read number excluding miRNA and Alu siRNAs from the total small RNA read number and multiplied by one million. Then, the normalized Alu siRNA read number of ADAR1 knockdown (siADAR1 HeLa) or DICER knockout (DICER null HEK293T) cells was subtracted from that of control cells (siControl HeLa or WT HEK293T), and the difference was plotted on the y-axis. A negative number on the y-axis indicates a decrease in the normalized number of reads after ADAR1 knockdown or DICER knockout. The x-axis is the number of bases in the siRNA sequence.
Annotation of AGO2-bound sense and antisense Alu siRNAs identified in HEK293T cells (Fig. 4A) to known Alu subfamily sequences revealed the presence of several high read Alu siRNAs originating from certain Alu subfamily members such as AluSz, AluJb, and AluJr for sense Alu siRNAs (Fig. 4B) and AluSp, AluYj4, AluYf1, and AluSx for antisense Alu siRNAs (Supplemental Fig. S4). Whereas sense Alu siRNAs seemed to be generated from several separate regions of each Alu subfamily sequence (Fig. 4B), antisense Alu siRNAs appeared to originate from a select region of the antisense strand Alu sequences (Supplemental Fig. S4), possibly indicating a preference of AGO2 binding to Alu siRNAs carrying a specific sequence.
Sense Alu siRNAs target CDCP1 and induce apoptosis in HeLa cells
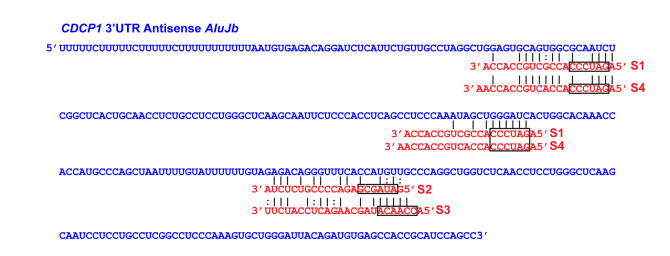
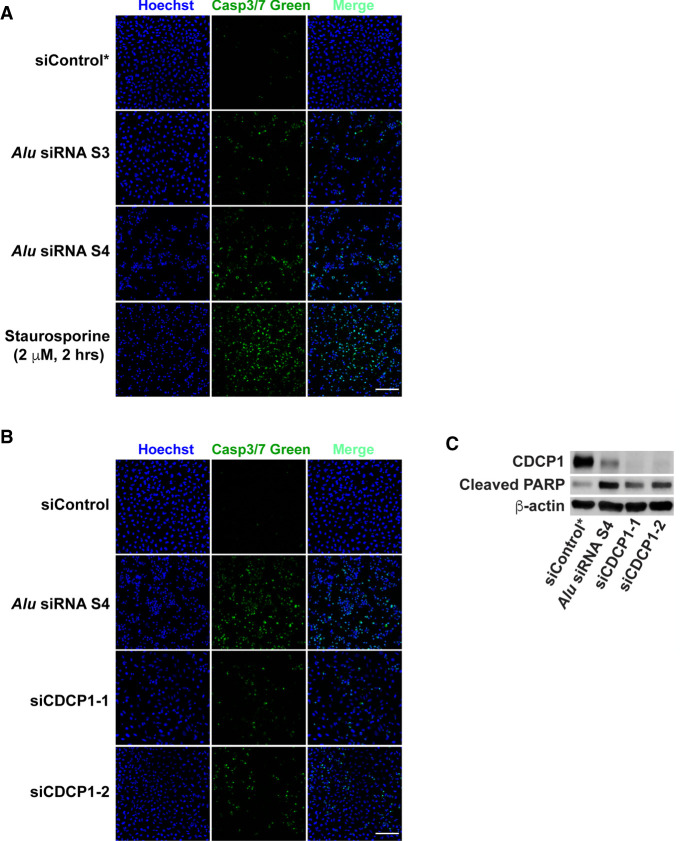
Although binding to AGO2 (Rybak-Wolf et al. 2014) and high expression levels of certain Alu endo-siRNAs (Supplemental Table S3) indicate that they could certainly be engaged in RNA interference-mediated silencing of target genes, it is not known whether they are indeed functional in somatic cell lines. To obtain evidence for functionality of Alu endo-siRNAs in vivo, we chose four sense Alu siRNAs, S1 (AluSz), S2 (AluJb), S3 (AluJr), and S4 (AluSz) (Fig. 4B) and looked for their potential target genes containing a single copy of the antisense Alu in their 3′UTR regions. Using the UCSC Human Genome Database and HeLa App (Liu et al. 2019b), we first identified 155 genes, which are expressed at relatively high levels and contain a single copy of 3′UTR antisense Alu sequences in HeLa cells (Supplemental Table S4A). Similarly, we also identified 165 genes containing a single copy of 3′UTR sense Alu sequences (Supplemental Table S4B), which could be potential targets of antisense Alu siRNA, AS1–AS4 (Supplemental Fig. S4). Among the candidate genes containing a single copy of 3′UTR antisense Alu sequences, we further selected the genes known to regulate apoptosis, which can be easily determined as a phenotype of gene silencing (Supplemental Table S4A). To this end, we picked CUB Domain Containing Protein 1 (CDCP1) containing a single antisense copy of AluJb subfamily sequence within its 3′UTR region (Scherl-Mostageer et al. 2001) as a test target gene for sense Alu siRNAs, S1–S4 (Supplemental Table S4A, highlighted in yellow). CDCP1, a transmembrane protein overexpressed in many types of cancers, suppresses apoptosis and thereby promotes their metastasis (Uekita et al. 2007; Deryugina et al. 2009). We transfected separately four different sense Alu siRNAs (S1–S4) into HeLa cells. Their silencing effects were determined by western blotting analysis, which revealed that all four sense Alu siRNAs indeed suppressed expression of CDCP1. However, their silencing effects varied: S1, S3, and S4 were very effective, whereas S2 was least effective (Fig. 6A). The difference is likely a result of differences in their seed sequence complementarity to target site sequences within the 3′UTR antisense AluJb of CDCP1 mRNAs (Fig. 7). Furthermore, we found that S1 and S4 Alu siRNAs are more effective because they have two target sites, especially the second site with perfect complementarity between their seed sequences and target sequences (Fig. 7). Their silencing effects were completely dependent on AGO2, since depletion of AGO2 abolished their silencing effects on the expression of CDCP1 (Fig. 6B). As expected by its already known antiapoptotic function, suppression of CDCP1 by Alu siRNA S3 and S4 (Fig. 8A) as well as two separate siCDCP1 RNAs targeting the CDCP1 coding region (Fig. 8B) resulted in induction of significant apoptosis, determined by fluorescence microscopy detection of activated Caspase-3/7 (Fig. 8A,B) and also by western blotting analysis for cleaved PARP fragments (Fig. 8C). These results certainly proved that Alu siRNAs are functional and capable of silencing their target gene via AGO2-dependent RNAi mechanism.
FIGURE 6.
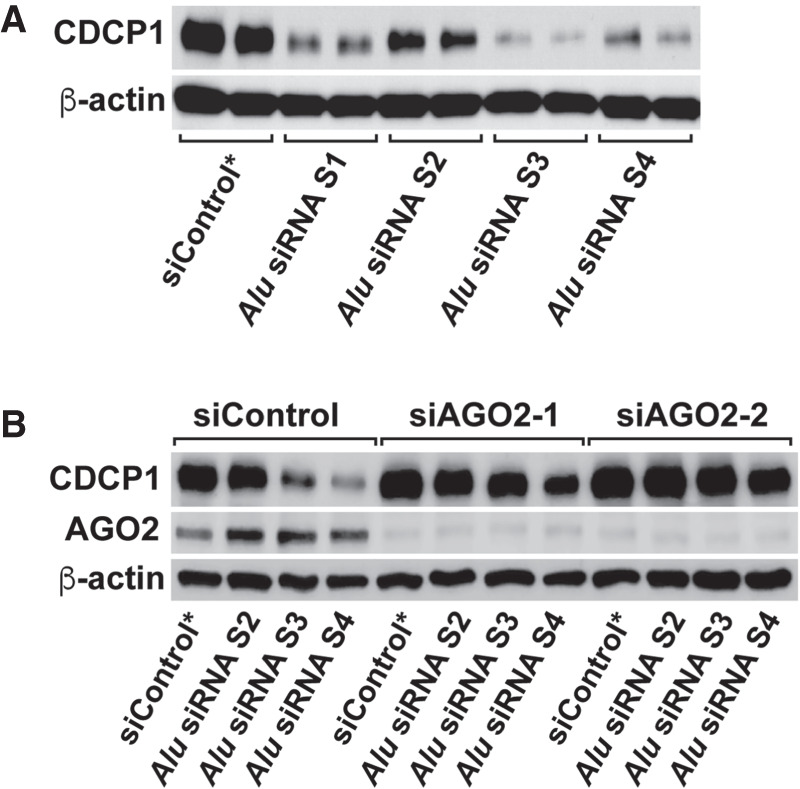
Alu siRNAs target CDCP1 in an AGO2-dependent manner. (A) Western blotting analysis of CDCP1 (full-length 135 kDa bands) in Alu siRNA-transfected HeLa cells at 72 h post-transfection. β-actin was used as a loading control. (B) The CDCP1 levels in Alu siRNA-transfected AGO2 knockdown cells were evaluated by western blotting. HeLa cells were treated with AGO2 siRNA twice. Alu siRNA and the second AGO2 siRNA were transfected into HeLa cells 48 h after the first AGO2 siRNA treatment.
FIGURE 7.
Predicted binding sites of Alu siRNAs in the 3′UTR of CDCP1 mRNAs. One putative target site of Alu siRNA-S2 or -S3 is located in the antisense AluJb located in the CDCP1 3′UTR. Two potential binding sites of Alu siRNA-S1 or -S4 are located in the CDCP1 3′UTR. Black box: seed sequence. The AluJb in the CDCP1 3′UTR located at chr3:45083628–45083942. S2 and S3 Alu siRNAs most likely originated from their target site regions of the corresponding sense Alu strand. Similarly, S1 and S4 Alu siRNAs likely originated from their first (upstream) target site regions of the corresponding sense Alu strand.
FIGURE 8.
Alu siRNAs induce apoptosis. (A) Fluorescence microscopic detection of apoptosis induced in Alu siRNA-transfected HeLa cells in comparison to the cells treated with a known inducer of apoptosis, Staurosporine (2 µM, 2 h). Apoptosis was evaluated with the CellEvent Caspase-3/7 Green apoptosis-detection system (Life Technologies). (B) Apoptosis induced in CDCP1 knockdown HeLa cells was also evaluated using the CellEvent Caspase-3/7 Green apoptosis-detection system. Scale bars, 200 µm. (C) The extent of apoptosis induced by Alu siRNA transfection or CDCP1 knockdown was analyzed by western blotting of cleaved poly(ADP-ribose) polymerase (PARP). CDCP1 bands represent the full-length protein (135 kDa), whereas PARP bands represent the cleaved protein (89 kDa).
DISCUSSION
It has been reported that there are 333 human genes that contain inverted Alu repeats within their 3′UTRs (Chen et al. 2008). These 3′UTR Alu dsRNAs, if not edited by the cytoplasmic ADAR1p150, activate the MDA5–MAVS–IFN signaling pathway (Mannion et al. 2014; Liddicoat et al. 2015; Pestal et al. 2015). Furthermore, loss of this particular ADAR1 function in suppression of MDA5–MAVS–IFN signaling underlies AGS pathogenesis caused by ADAR1 mutations (Rice et al. 2012; Mannion et al. 2014) and also the resistance of tumors to immune checkpoint blockade (Ishizuka et al. 2019). In this study, we showed that the long stem regions of Alu dsRNAs, despite their imperfect complementarity, can be cleaved by DICER, generating 21–24 nt length Alu siRNAs. Furthermore, we found that ADAR1 promotes this DICER activity for processing of Alu siRNAs by forming the DICER/ADAR1 complex, as we previously reported for processing of pre-miRNAs to mature miRNAs (Ota et al. 2013). Interestingly, TRBP had almost no DICER-promoting effects for processing of Alu dsRNAs to Alu siRNAs. The exact mechanism for how ADAR1 promotes DICER activities to process Alu dsRNAs to Alu siRNAs is currently not known. However, we previously reported that ADAR1 forms a heterodimer complex with DICER via its second dsRNA binding domain and the amino-terminal DExD helicase domain of DICER (Ota et al. 2013). Autoinhibitory function of this DExD helicase domain in the DICER activity for processing of long dsRNAs but not miRNA precursors has been reported: Deletion of DExD helicase domain increased dicing rate by eightfold (Ma et al. 2008). It is believed that the helicase domain affects the overall folding and prevents formation of the most effective structure of DICER (Ma et al. 2008). In fact, a Dicer isoform (Dicero) lacking the amino-terminal DExD helicase domain, expressed only in oocytes, is highly active in processing endo-siRNAs from long dsRNAs (Flemr et al. 2013). Taken together, the interaction of ADAR1 with the DICER DExD helicase domain likely masks its autoinhibitory function, thereby allowing DICER to process Alu dsRNAs more efficiently into Alu siRNAs.
Expression of endo-siRNAs processed by DICER from long dsRNAs consisting of repetitive sequences of retrotransposons and pseudogene transcripts has been reported in mouse oocytes (Tam et al. 2008; Watanabe et al. 2008; Flemr et al. 2013). Furthermore, function of endo-siRNAs in RNAi-mediated regulation of oocyte transcripts and its essential role in meiosis have been indicated (Murchison et al. 2007; Liu et al. 2010; Flemr et al. 2013). For instance, oocyte gene transcripts containing 3′UTR SINEs seem to be silenced by SINE siRNAs processed by DICER as a part of the mechanism to regulate retrotransposon activities (Murchison et al. 2007). Interestingly, detection of ADAR1p110 at high levels in oocytes, zygotes, and very early embryos (2–4 cell stages) but rapid elimination at preimplantation stages have been reported with mouse (Garcia-Lopez et al. 2013) and also with human (Qiu et al. 2016). Intriguingly, the DICER expression pattern in maturing oocytes is very similar to that of ADAR1: intense expression in germinal vesicles and rapid decrease at 2–4 cell stage embryos (Murchison et al. 2007). A significant increase in the levels of transcripts derived from retrotransposons is detected in Dicer null oocytes due to deficiency in generation of endo-siRNAs (Murchison et al. 2007; Flemr et al. 2013). One possible function of ADAR1 in oocytes may be to promote the DICER activity for processing of endo-siRNAs from repetitive elements such as Alu and thereby regulate retrotransposon activities in female germ cells. Deficiency in this regulation appears to underlie failure of meiosis resulting from defective spindle formation and chromosome congression detected in Dicer null mouse oocytes (Murchison et al. 2007).
By analyzing small RNA databases, we found that Alu siRNAs, although at low levels, are expressed in commonly used somatic cell lines such as HeLa and HEK293T. Furthermore, we identified several highly expressed AGO2-bound sense and antisense Alu siRNA, which are likely to be processed by DICER from Alu dsRNAs consisting of specific Alu subfamily member sequences. We demonstrated, as our proof-of-concept experiment, that these Alu siRNAs can be actively engaged in RNAi-mediated silencing of a select target gene CDCP1: Suppression of CDCP1 resulted in induction of apoptosis in HeLa cells. CDCP1 has been shown to promote survival of several different types of cancer cells by activating MYC, AKT, and Wnt gene pathways (Majem et al. 2019). Interestingly, infraexpression of CDCP1 targeting miRNAs miR-654 and miR-218 in ovarian and lung cancers, respectively, has been reported (Chiu et al. 2015; Majem et al. 2019). Furthermore, using miR-654 as a therapeutic to target CDCP1 and treat ovarian cancers has been proposed (Majem et al. 2019). Our results indicate that Alu siRNAs, very effective CDCP1 targeting small non-coding RNAs, can also be used for treatment of certain cancers.
Although differential expression of ADAR1p150 in the cytoplasm and ADAR1p110 in the nucleus has been previously reported (Patterson and Samuel 1995), they can be detected also in the reciprocal locations in certain somatic cell lines such as HeLa and HEK293T. Furthermore, we previously reported the nuclear export of ADAR1p110 to the cytoplasm regulated by MAP kinases during stress response (Sakurai et al. 2017). Although DICER is thought to be engaged in processing of pre-miRNAs in the cytoplasm, import of DICER phosphorylated in response to DNA damage to the nucleus has been also reported (Burger et al. 2017). Accordingly, it is currently not clear where processing of Alu siRNAs takes place within the cell, nor the relative contribution of ADAR1p110 versus ADAR1p150 as the DICER partner in vivo. These issues also muddle the source of Alu dsRNAs to be processed to Alu siRNAs: nuclear localized intronic Alu dsRNAs versus cytoplasmic Alu dsRNAs of mRNA 3′UTRs. These issues remain to be addressed in future studies.
In differentiated somatic cells, long dsRNAs induce the interferon (IFN) response via the MDA5-mediated dsRNA-sensing mechanism in contrast to in oocytes and embryonic stem cells where they enter into the RNA interference pathway and are processed to siRNAs by DICER. ADAR1 may contribute to control the potential of long dsRNAs for induction of the MDA5–MAVS–IFN pathway by two ways: introducing extensive A-to-I editing into dsRNAs and thereby suppress MDA5 binding in differentiated somatic cells or promoting the DICER activity and processing dsRNAs to siRNAs in oocytes and ES cells where the IFN pathway is absent. Interestingly, elimination of the IFN pathway components such as MAVS restored the dsRNA-mediated RNA interference pathway in mouse somatic cells, indicating that the dsRNA RNA interference pathway is present also in somatic cells but dominated by the IFN response pathway (Maillard et al. 2016). In this study, we demonstrated that Alu siRNAs most likely processed by DICER are generated even in somatic cells such as HeLa and HEK293T, although at very low levels. It has been recently shown that many tumors elevate ADAR1 expression levels and consequently suppress activation of the dsRNA-sensing mechanism mediated by the MDA5–MAVS–IFN pathway, which in turn makes them resistant to immune checkpoint blockade (Ishizuka et al. 2019). It is interesting to know whether ADAR1 also contributes to suppression of IFN responses and development of resistance to immune surveillance in certain tumors by facilitating degradation, rather than by A-to-I editing, of long dsRNAs.
MATERIALS AND METHODS
Plasmid construction
The pFastBac-DICER-FLAG plasmid used for recombinant protein purification was prepared by PCR cloning using a FLAG–DICER plasmid (Chendrimada et al. 2005). PCR primers used are listed in Supplemental Table S1. Preparation of baculovirus constructs for HAT–ADAR1p110–WT, HA–ADAR1p110–E912A, and His–ADAR1p150–WT was described previously (Lai et al. 1995; Cho et al. 2003). The AluSx sequence was synthesized by Gibson assembly and cloned into pBlueScript II KS (+). Oligonucleotides used for construction of various plasmids are listed in Supplemental Table S1. pBS-NFkB-dsAlu plasmid contains a 1457 bp fragment of an inverted Alu repeat of NFκB intron. The inverted Alu repeat located at chr1:11102837–11267747. The DNA fragment was PCR-amplified using human genomic DNA and PCR primers Bam-NFKBFW2 and Xho-NFKBDW2. Bam-NFKBFW2 contained a BamHI recognition site, and Xho-NFKBDW2 contained a XhoI recognition site. The PCR products were digested with BamHI and XhoI, then inserted into pBluescript II KS (−) vector (Kawahara and Nishikura 2006). pcDNA3.2-NICN1-dsAlu plasmid contained an 895 bp fragment of an inverted Alu repeat of NICN1 3′UTR. The inverted Alu repeat in NICN1 3′-UTR located at chr3:49422946–49429326. The DNA fragment was PCR-amplified using human genomic DNA and PCR primers NICN1 1stPCR and NICN1 2ndPCR. S1 tag DNA fragment was synthesized by PCR amplification. The PCR products were digested with SalI and inserted into pENTR/D-TOPO, and then transferred to pcDNA3.2/V5/GW/CAT vector by Gateway recombination.
Protein purification
The recombinant proteins were purified with a TALON metal resin (Clontech) and/or an anti-FLAG M2 affinity or an anti-HA antibody bead (SIGMA) as described previously (Ota et al. 2013).
In vitro transcription
Uniformly 32P-labeled Alu hairpin dsRNA substrates consisting of a partially double-stranded stem made from inverted sense and antisense Alu sequences and a loop and a single-stranded Alu RNA substrate were prepared by in vitro transcription as described previously (Kawahara and Nishikura 2006). pBS-ssAlu, pBS-NFkB-dsAlu or pcDNA3.2-NICN1-dsAlu were linearized with SwaI, XhoI, or XbaI, respectively, and then transcribed by T7 polymerase (Promega) in the presence of 32P-UTP or -ATP at 37°C for 120 min (Dabiri et al. 1996). After DNase I (Promega) treatment, RNAs were purified by denaturing gel electrophoresis and dissolved in annealing buffer containing 10 mM Tris HCl (pH 7.6), 50 mM NaCl. RNAs were incubated at 80°C for 5 min and slowly cooled to room temperature.
DICER cleavage assay
In vitro DICER cleavage assays were done in a 45 µL reaction mixture containing 20 mM PIPES (pH 6.2), 1.5 mM MgCl2, 80 mM NaCl, 1 mM dithiothreitol (DTT), 0.05% NP40, 10% glycerol, 3U/µL RNasin plus inhibitor (Promega), 10 mM EGTA, 1.5 nM DICER, and 0.15 nM substrate RNA. Reaction mixtures were incubated at 37°C, and 7.5 µL aliquots were taken after 0, 5, 10, 30, and 60 min. The reactions were stopped by adding an equal volume of gel loading buffer (80% formamide, 20% glycerol, 0.025% BPB). After heating at 80°C for 10 min, samples were analyzed by 10% Urea-PAGE and quantified using Molecular Dynamics and ImageJ (Ota et al. 2013).
Analysis of dsRNA structure
Secondary dsRNA structure was calculated using Mfold (Zuker 2003).
Bioinformatics analysis of small RNA-seq data
We analyzed GSM1057798 (total small RNAs from siControl HeLa cell), GSM1057799 (total small RNAs from siADAR1 HeLa cell), GSM1370372 (total small RNAs from HEK293T cell), GSM1370373 (total small RNAs from DICER knockout HEK293T cell), and GSM1334330 (AGO2-loaded small RNAs from HEK293T cell). We followed a multistep approach to align and annotate small RNA reads as follows: (i) removal of adaptor nucleotides; (ii) removal of siRNA sequences; (iii) alignment to ribosomal RNA sequences; (iv) alignment to transfer RNA sequences; (v) alignment to miRNA sequences; (vi) alignment to repetitive sequences; (vii) alignment to snoRNA sequences; (viii) alignment to RefSeq sequences; and (ix) alignment to human genome sequence. Alignment was performed using the Bowtie program allowing a maximum of two mismatches. One read reported one valid alignment. Unaligned reads were mapped to the next reference sequences. Bioinformatics analysis was performed on the Galaxy platform (Afgan et al. 2018).
More specifically: (i) Removal of adaptor: The 3′ adaptor sequences (GSM1057798, GSM1057799, GSM1370372, and GSM1370373: 5′-TGGAATTCTCGGGTGCCAAGGAACTCCAGTCAC-3′, GSM1334330: 5′-TCTCGTATCGTATGCCGTCTTCTGCTTG-3′) were first removed from the sequences. Trimmed reads of length 19 to 35 nt were subjected to further analysis. (ii) Removal of exogenous siRNA sequences: The reads containing the control siRNA and ADAR1 siRNA sequences used were removed. Reads with 5′-CGTACGCGGAATACTTCGAAG-3′, 5′-TCGAAGTATTCCGCGTACGAT-3′, 5′-CCGCCATCATTATGAAAAAAG-3′ and 5′-TTTTTCATAATGATGGCGGAT-3′ sequences were removed. (iii) Alignment to ribosomal RNAs: The human ribosomal RNA sequences were downloaded from Silva (https://www.arb-silva.de) (Pruesse et al. 2007). (iv) Alignment to transfer RNAs: The human transfer RNA sequences were downloaded from GtRNAdb (http://gtrnadb.ucsc.edu/index.html) (Chan and Lowe 2009). (v) Alignment to miRNA: The human miRNA sequences were downloaded from miRBase (http://www.mirbase.org/ftp.shtml) (Griffiths-Jones et al. 2006). (vi) Alignment to repetitive sequences: The human repetitive sequences (rmsk file) were downloaded from the UCSC genome browser (https://hgdownload.soe.ucsc.edu/downloads.html). The Rmsk file contains Alu subfamily sequences. (vii) Alignment to snoRNAs: The human snoRNA sequences were downloaded from snoRNABase (https://www-snorna.biotoul.fr/index.php) (Lestrade and Weber 2006). (viii) Alignment to RefSeq sequences: The human RefSeq sequences were downloaded from the UCSC genome browser (https://hgdownload.soe.ucsc.edu/downloads.html). (ix) Alignment to human genome: The unaligned reads from the previous step were finally aligned to the hg38 human genome.
The alignment data were visualized by the Integrative Genomics Viewer.
Selection of genes containing a single Alu sequence within their 3′UTR
Coding genes with known 3′UTR regions were tested for Alu sequences and genes with exactly one Alu repeat in the forward orientation (sense) to the gene or exactly one Alu in reverse orientation (antisense) to the gene were reported. Highly expressed genes in HeLa cells (CCL2) were picked using HeLa App (proteomics: >log10 4.5). The presence of articles related to apoptosis was examined using a keyword search of the abstract in PubMed.
Cell culture, small RNA transfection
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (GEMINI) and penicillin/streptomycin at 37°C in 5% CO2. HeLa cells were free from mycoplasma contamination. Small-RNA transfection experiments were performed with Lipofectamine RNAiMax (Life Technologies) with a 2 nM final RNA concentration. All small-RNAs used in this study are listed in Supplemental Table S1. Silencer Select siRNAs for human CDCP1(s35060 and s35061) and Negative Control siRNA (s4390844) were purchased from Life Technologies (Supplemental Table S1).
Western blotting analysis and antibodies used
Cell lysates were prepared in Laemmli buffer (Boston BioProducts) containing benzonase nuclease (Sigma), Complete EDTA-free protease-inhibitor cocktail (Roche), and PhosStop phosphatase-inhibitor cocktail (Roche), and fractionated by 4%–15% SDS–PAGE. Proteins were transferred to Immobilon-P nylon membranes (Millipore). Membranes were blocked with 1% Blocker BSA (Life Technologies) and incubated with primary antibodies (Supplemental Table S2) overnight at 4°C. After incubation with secondary antibodies, membranes were developed with ECL (GE Healthcare).
In vitro RNA editing
The A-to-I RNA editing reaction of ADAR1p110 was described previously (Wagner et al. 1989).
Small RNA-seq analysis of in vitro dicing products
In vitro DICER cleavage assays for sequencing were done in a 100 µL reaction mixture containing 20 mM PIPES (pH 6.2), 1.5 mM MgCl2, 80 mM NaCl, 1 mM dithiothreitol (DTT), 0.05% NP40, 10% glycerol, 3 U/mL RNasin plus (Promega), 10 mM EGTA, 1.5 nM DICER/ADAR1p110-WT complexes, and 3 nM NFκB1 dsAlu RNA. DICER cleavage products were purified from 10% acrylamide denaturing gel and ligated with 5′ and 3′ adaptors. 3′ cloning adaptors were ligated to the cleavage products using T4 RNA ligase 2-truncated K227Q in a reaction buffer containing NEB ligation buffer, 15% PEG8000, and 1U/µL RNasin plus inhibitor. After denaturing gel purification, 5′ cloning adaptors were ligated to the purified RNAs using T4 RNA ligase1 in a reaction buffer containing NEB ligation buffer, 20% PEG8000, 1U/µL RNasin plus inhibitor, 20 µM ATP, and 10% DMSO. RNA ligation reactions were incubated at 22°C for 120 min. After denaturing gel purification, the ligated RNAs were reverse-transcribed using the RT-primer and SuperScript III (Life Technologies). The reverse-transcribed products were amplified using first and second PCR primers and AccuPrime high fidelity Taq DNA polymerase (Life Technologies). After acrylamide gel purification, PCR products were sequenced by Ion PGM (316 chip, 400 bp) (Life Technologies). Bioinformatics analysis: The 5′ and 3′ adaptor sequences (5′-TGGAATTCTCGGGCACCAAGGT-3′, 5′-ACGCTGGAATTCGCGGTTAAA-3′) were removed from the sequences. Trimmed reads were then mapped to NFκB1 intronic inverted Alu sequence. Alignment was performed using the Bowtie program without any mismatch. Bioinformatics analysis was performed on the Galaxy platform (Afgan et al. 2018).
Apoptosis analysis: Caspase-3/7 activity and cleaved PARP detection
Apoptotic cells were analyzed by fluorescence microscopy using CellEvent Caspase-3/7 Green apoptosis-detection system (Life Technologies), which utilizes a fluorogenic substrate for activated Caspase-3 and -7. Synthetic siRNAs corresponding to the human CDCP1 mRNA coding region or Alu sense siRNAs (S3–S4) (Supplemental Table S1) were used for knockdown of the CDCP1 gene. Staurosporine (Cell Signaling Technology) treatment at 2 µM for 2 h was used as positive control. DNA was counterstained with Hoechst 33342 (Life Technologies). Apoptosis levels were also determined by western blotting analysis using cleaved PARP (Asp214) antibody (Supplemental Table S2).
Statistical analysis
Image quantification was performed with ImageJ software. Data are presented as means ± SD. Two-tailed t-tests were conducted, and the minimum level for significance was P < 0.05.
DATA DEPOSITION
Dicing product sequencing data has been deposited in the Sequence Read Archive under series accession number SRR10824153.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank John M. Murray for critical reading of the manuscript, Yukio Kawahara for plasmid construction, and Hiromitsu Ota for assistance with protein purification. We also thank the Protein Expression, Genomics, and Bioinformatics Shared Facilities of The Wistar Cancer Center, which are supported by the National Institutes of Health, National Cancer Institute (NIH/NCI; P30 CA010815), for the services provided. The work was supported by grants from the National Institutes of Health (GM040536, CA175058, and GM130716), the Macula Vision Research Foundation, Emerson Collective, and the Japan Society for the Promotion of Science (JSPS S-13204) to K.N. Y.S. was supported in part by a fellowship from the Japan Society for the Promotion of Science (JSPS 2017-19).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.076745.120.
Freely available online through the RNA Open Access option.
REFERENCES
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, et al. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46: W537–W544. 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang CZ, Hur S. 2018. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172: 797–810.e713. 10.1016/j.cell.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak M, Porath HT, Finkelstein G, Knisbacher BA, Buchumenski I, Roth SH, Levanon EY, Eisenberg E. 2020. Purifying selection of long dsRNA is the first line of defense against false activation of innate immunity. Genome Biol 21: 26 10.1186/s13059-020-1937-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24: 365–376. 10.1101/gr.164749.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. 2014. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA 20: 923–937. 10.1261/rna.044545.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K, Schlackow M, Potts M, Hester S, Mohammed S, Gullerova M. 2017. Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage. J Cell Biol 216: 2373–2389. 10.1083/jcb.201612131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37: D93–D97. 10.1093/nar/gkn787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG. 2008. Alu element-mediated gene silencing. EMBO J 27: 1694–1705. 10.1038/emboj.2008.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744. 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu KL, Kuo TT, Kuok QY, Lin YS, Hua CH, Lin CY, Su PY, Lai LC, Sher YP. 2015. ADAM9 enhances CDCP1 protein expression by suppressing miR-218 for lung tumor metastasis. Sci Rep 5: 16426 10.1038/srep16426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DS, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. 2003. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem 278: 17093–17102. 10.1074/jbc.M213127200 [DOI] [PubMed] [Google Scholar]
- Dabiri GA, Lai F, Drakas RA, Nishikura K. 1996. Editing of the GLuR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J 15: 34–45. 10.1002/j.1460-2075.1996.tb00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryugina EI, Conn EM, Wortmann A, Partridge JJ, Kupriyanova TA, Ardi VC, Hooper JD, Quigley JP. 2009. Functional role of cell surface CUB domain-containing protein 1 in tumor cell dissemination. Mol Cancer Res 7: 1197–1211. 10.1158/1541-7786.MCR-09-0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. 2018. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat Rev 19: 473–490. 10.1038/s41576-018-0006-1 [DOI] [PubMed] [Google Scholar]
- Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. 2013. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155: 807–816. 10.1016/j.cell.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez J, Hourcade Jde D, Del Mazo J. 2013. Reprogramming of microRNAs by adenosine-to-inosine editing and the selective elimination of edited microRNA precursors in mouse oocytes and preimplantation embryos. Nucleic Acids Res 41: 5483–5493. 10.1093/nar/gkt247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144. 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. 2004. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279: 4894–4902. 10.1074/jbc.M311347200 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81. 10.1038/35017558 [DOI] [PubMed] [Google Scholar]
- Hood JL, Emeson RB. 2012. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr Top Microbiol Immunol 353: 61–90. 10.1007/82_2011_157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L, et al. 2010. Editing of Epstein–Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem 285: 33358–33370. 10.1074/jbc.M110.138362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, Miller BC, Du PP, Yates KB, Dubrot J, et al. 2019. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565: 43–48. 10.1038/s41586-018-0768-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471: 325–330. 10.1038/nature09830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Nishikura K. 2006. Extensive adenosine-to-inosine editing detected in Alu repeats of antisense RNAs reveals scarcity of sense-antisense duplex formation. FEBS Lett 580: 2301–2305. 10.1016/j.febslet.2006.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. 2007. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140. 10.1126/science.1138050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidou C, Efstathiou NE, Hoang MV, Notomi S, Konstantinou EK, Hirano M, Takahashi K, Maidana DE, Tsoka P, Young L, et al. 2018. Issues with the specificity of immunological reagents for NLRP3: implications for age-related macular degeneration. Sci Rep 8: 461 10.1038/s41598-017-17634-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Drakas R, Nishikura K. 1995. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J Biol Chem 270: 17098–17105. 10.1074/jbc.270.29.17098 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. 10.1016/S0092-8674(04)00261-2 [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162. 10.1093/nar/gkj002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349: 1115–1120. 10.1126/science.aac7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Tang Y, He Z, Rosenwaks Z. 2010. Dicer is a key player in oocyte maturation. J Assist Reprod Genet 27: 571–580. 10.1007/s10815-010-9456-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fu Y, Huang J, Wu M, Zhang Z, Xu R, Zhang P, Zhao S, Liu L, Jiang H. 2019a. ADAR1 promotes the epithelial-to-mesenchymal transition and stem-like cell phenotype of oral cancer by facilitating oncogenic microRNA maturation. J Exp Clin Cancer Res 38: 315 10.1186/s13046-019-1300-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mi Y, Mueller T, Kreibich S, Williams EG, Van Drogen A, Borel C, Frank M, Germain PL, Bludau I, et al. 2019b. Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat Biotechnol 37: 314–322. 10.1038/s41587-019-0037-y [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. 2008. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol 380: 237–243. 10.1016/j.jmb.2008.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539. 10.1126/science.1113150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard PV, Van der Veen AG, Deddouche-Grass S, Rogers NC, Merits A, Reis e Sousa C. 2016. Inactivation of the type I interferon pathway reveals long double-stranded RNA-mediated RNA interference in mammalian cells. EMBO J 35: 2505–2518. 10.15252/embj.201695086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majem B, Parrilla A, Jimenez C, Suarez-Cabrera L, Barber M, Marin A, Castellvi J, Tamayo G, Moreno-Bueno G, Ponce J, et al. 2019. MicroRNA-654-5p suppresses ovarian cancer development impacting on MYC, WNT and AKT pathways. Oncogene 38: 6035–6050. 10.1038/s41388-019-0860-0 [DOI] [PubMed] [Google Scholar]
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, McLaughlin PJ, et al. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 9: 1482–1494. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion N, Arieti F, Gallo A, Keegan LP, O'Connell MA. 2015. New insights into the biological role of mammalian ADARs; the RNA editing proteins. Biomolecules 5: 2338–2362. 10.3390/biom5042338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. 2007. Critical roles for Dicer in the female germline. Genes Dev 21: 682–693. 10.1101/gad.1521307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. 2016. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 17: 83–96. 10.1038/nrm.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K, Sakurai M, Ariyoshi K, Ota H. 2013. Antagonistic and stimulative roles of ADAR1 in RNA silencing. RNA Biol 10: 1240–1247. 10.4161/rna.25947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K. 2013. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153: 575–589. 10.1016/j.cell.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JB, Samuel CE. 1995. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol 15: 5376–5388. 10.1128/MCB.15.10.5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. 2015. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43: 933–944. 10.1016/j.immuni.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porath HT, Carmi S, Levanon EY. 2014. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun 5: 4726 10.1038/ncomms5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Li W, Xiong H, Liu D, Bai Y, Wu K, Zhang X, Yang H, Ma K, Hou Y, et al. 2016. Single-cell RNA sequencing reveals dynamic changes in A-to-I RNA editome during early human embryogenesis. BMC Genomics 17: 766 10.1186/s12864-016-3115-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren YF, Li G, Wu J, Xue YF, Song YJ, Lv L, Zhang XJ, Tang KF. 2012. Dicer-dependent biogenesis of small RNAs derived from 7SL RNA. PLoS One 7: e40705 10.1371/journal.pone.0040705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. 2012. Mutations in ADAR1 cause Aicardi–Goutieres syndrome associated with a type I interferon signature. Nat Genet 44: 1243–1248. 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, Rajewsky N. 2014. A variety of dicer substrates in human and C. elegans. Cell 159: 1153–1167. 10.1016/j.cell.2014.10.040 [DOI] [PubMed] [Google Scholar]
- Sakurai M, Ueda H, Yano T, Okada S, Terajima H, Mitsuyama T, Toyoda A, Fujiyama A, Kawabata H, Suzuki T. 2014. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res 24: 522–534. 10.1101/gr.162537.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Shiromoto Y, Ota H, Song C, Kossenkov AV, Wickramasinghe J, Showe LC, Skordalakes E, Tang HY, Speicher DW, et al. 2017. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat Struct Mol Biol 24: 534–543. 10.1038/nsmb.3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl-Mostageer M, Sommergruber W, Abseher R, Hauptmann R, Ambros P, Schweifer N. 2001. Identification of a novel gene, CDCP1, overexpressed in human colorectal cancer. Oncogene 20: 4402–4408. 10.1038/sj.onc.1204566 [DOI] [PubMed] [Google Scholar]
- Song C, Sakurai M, Shiromoto Y, Nishikura K. 2016. Functions of the RNA editing enzyme ADAR1 and their relevance to human diseases. Genes (Basel) 7: 129 10.3390/genes7120129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier TR, Sakami S, Sahu B, Howell SJ, Gao S, Dong Z, Golczak M, Maeda A, Palczewski K. 2017. MicroRNA-processing enzymes are essential for survival and function of mature retinal pigmented epithelial cells in mice. J Biol Chem 292: 3366–3378. 10.1074/jbc.M116.770024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538. 10.1038/nature06904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, et al. 2017. Dynamic landscape and regulation of RNA editing in mammals. Nature 550: 249–254. 10.1038/nature24041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al. 2012. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149: 847–859. 10.1016/j.cell.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uekita T, Jia L, Narisawa-Saito M, Yokota J, Kiyono T, Sakai R. 2007. CUB domain-containing protein 1 is a novel regulator of anoikis resistance in lung adenocarcinoma. Mol Cell Biol 27: 7649–7660. 10.1128/MCB.01246-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L, Nishikura K. 2005. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol 79: 299–338. 10.1016/S0079-6603(04)79006-6 [DOI] [PubMed] [Google Scholar]
- Wagner RW, Smith JE, Cooperman BS, Nishikura K. 1989. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci 86: 2647–2651. 10.1073/pnas.86.8.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Li JB. 2017. Rewriting the transcriptome: adenosine-to-inosine RNA editing by ADARs. Genome Biol 18: 205 10.1186/s13059-017-1347-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279: 4952–4961. 10.1074/jbc.M310162200 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543. 10.1038/nature06908 [DOI] [PubMed] [Google Scholar]
- Wu D, Lamm AT, Fire AZ. 2011. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat Struct Mol Biol 18: 1094–1101. 10.1038/nsmb.2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. 2006. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21. 10.1038/nsmb1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885. 10.1093/emboj/cdf582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gao X, Hu J, Xie Y, Zuo Y, Xu H, Zhu S. 2019. ADAR1p150 forms a complex with dicer to promote miRNA-222 activity and regulate PTEN expression in CVB3-induced viral myocarditis. Int J Mol Sci 20: 407 10.3390/ijms20020407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.