Abstract
Stress Granules (SGs) are membraneless cytoplasmic RNA granules, which contain translationally stalled mRNAs, associated translation initiation factors and multiple RNA-binding proteins (RBPs). They are formed in response to various stresses and contribute to reprogramming of cellular metabolism to aid cell survival. Because of their cytoprotective nature, association with translation regulation and cell signaling, SGs are an essential component of the integrated stress response pathway, a complex adaptive program central to stress management. Recent advances in SG biology unambiguously demonstrate that SGs are heterogeneous in their RNA and protein content leading to the idea that various SG subtypes exist. These SG variants are formed in cell type- and stress-specific manners and differ in their composition, dynamics of assembly and disassembly, and contribution to cell viability. As aberrant SG dynamics contribute to the formation of pathological persistent SGs that are implicated in neurodegenerative diseases, the biology of different SG subtypes may be directly implicated in neurodegeneration. Here, we will discuss mechanisms of SG formation, their subtypes, and potential contribution to health and disease.
Keywords: Stress granules, Neurodegeneration, RBPs, ALS, FTD, TDP-43, C9orf72 , Tau
Introduction
Ribonucleoprotein (RNP) granules are membraneless organelles that are indispensable for cellular metabolism. They are distributed diffusely throughout the cell and regulate gene expression under normal and stress conditions. Recent finding also implicates RNP granules in cell-type specific transcriptional programing that drives RNA synthesis dictated by dynamic physiological demands [1]. Components of RNA granules characteristically promote multivalent interactions to achieve higher order aggregation into sub-cellular non-membrane bound compartments important for assembly and function of RNPs [2]. Their content and sub-cellular location define their function. Stress granules (SGs) assemble during conditions of cellular stress in the cytoplasm to mediate pro-survival adaptive response. Many SG-associated RBPs are highly conserved from yeast to human [2, 3]. SG assembly minimizes cellular energy demands and directs the resources to maintain ribostasis and proteostasis. Also, their disassembly upon removal of stress is essential to restore normal cellular metabolism. In addition to the stress response, SGs are also involved in signaling pathways and modulation of viral infections [4, 5].
The non-dividing property and characteristic morphology and physiology of neurons make them highly susceptible to aggregation of selected SG components that may mature to pathological aggregates. These pathological aggregates drive disease progression in amyotrophic lateral sclerosis (ALS), frontal temporal dementia (FTD), Alzheimer’s disease (AD) and other neurodegenerative diseases [6, 7]. Disease-associated RBPs like TDP-43, FUS, Tau, etc. are SG components, in which mutations alter their functional and structural properties [8]. It has been proposed that chronic accumulation and maturation of mutant proteins in cytoplasmic foci in neurons cause insoluble, amorphous, and cytotoxic SGs that promote neuronal degeneration and apoptosis [9, 10]. This review examines the mechanism of SG dynamics, subtypes of SGs, and their implication in neurodegenerative diseases.
RNA granules in different cellular compartments
RNA granules were first identified as dark staining polar ‘Germ granules’ involved in differentiation and development in small insects [11, 12]. Since then, these RNA-containing non-membranous organelles have been discovered and described in somatic cells as RNP granules associated with the regulation of post-transcriptional gene expression. These ubiquitous RNPs are indispensable for RNA metabolism and are enriched in proteins involved in RNA processing, transport, translation, storage, and decay [2, 12]. The first discovered germ granules have been extensively studied in Drosophila, Caenorhabditis elegans and zebra fish. These germline foci contain non-coding RNAs (ncRNAs), mRNAs and proteins associated with germline development and stem cell maintenance [13]. Cajal bodies were the next RNA-rich foci identified after germ granules named after their discoverer Ramòn Cajal [14]. These are nuclear RNA granules enriched in small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) and small Cajal-specific RNAs (scaRNAs) [15]. Cajal bodies are associated with RNA transcription, processing and the assembly of other snRNP complexes [14].
Stress granules (SGs) and processing bodies (PBs) are two common cytoplasmic membraneless foci associated with mRNA metabolism. SGs are enriched with polyadenylated mRNAs, while mRNAs with short poly(A) tails are associated with PBs. These two types are proposed to serve diverse functions in cell signaling and the stress response [16]. Their role in reprogramming gene expression and disease is discussed in the later part of this review. Neuronal granules (NGs) are a specific class of cytoplasmic RNA granules associated with spaciotemporal gene expression in both developing and mature neurons. The sub-cellular localized translation associated with NGs is the determinant of development, synapse formation, neuronal plasticity and memory formation [17, 18]. NG associated RBPs also significantly overlap with SG components [18]. Both nuclear and cytoplasmic RNPs modulate RNA turnover and protein synthesis in response to developmental and environmental cues [19]. A common feature of these membraneless granules is that they are dynamic and constantly exchange molecules with the surrounding cytoplasm or nucleoplasm.
SGs and PBs in translational control
SGs and PBs are two well studied RNPs in context stress-induced reprograming of protein synthesis. SGs are phase-dense RNPs generated consequent to the global translational inhibition during stress response. These spatially condensed cytoplasmic foci are enriched in polyadenylated mRNAs, 40S subunits, translation initiation factors (eIF2, eIF3, eIF4A, eIF4G and eIF4E) and RBPs like PABP1, G3BP1/2, TIA-1/R, FMRP, FXR1 and CPEB [3, 20]. The presence of polyadenylated mRNAs, 40S subunits and initiation factors hints at selective sequestration of majority of cellular mRNAs into SGs, and exclusion from SGs of other mRNAs needed for the stress response. SGs reversibly sequester translationally stalled housekeeping transcripts to maintain transcriptome homeostasis, minimize energy expenditure and divert metabolic resources to repair stress-induced damage [5, 21]. PBs are another stress stimulated cytoplasmic RNPs, which unlike SGs are enriched with deadenylated transcripts and components of mRNA decay machinery [22, 23]. Previously described as ‘XRN1 foci’, PBs contain components of the decapping complex (Dcp1/2, Rap55/Lsm14, DDX6/RCK1, CCR4-Not1 complex) and also contain components of nonsense mediated mRNA decay (NMD) like Upf1/2, in addition to the 3′ exonuclease Xrn1 [22]. Certain RBPs that are found in both SGs and PBs are implicated as sites of mRNA triage employing distinct active components and mechanisms [3]. Unlike SGs, PBs also exist under normal conditions but their number and size increases with stress. Kroschwald et al. used chemical 1,6-hexanediol to differentiate between liquid-like and solid-like cell aggregates. The work in yeast cells revealed that PBs behave more like liquid droplets while SGs are more solid-like amorphous protein aggregates [24]. However, in mammalian cells SGs have liquid-like characteristics different from yeast [24]. This could be due a to a different transcriptomic and proteomic landscape. Both SGs and PBs exist in dynamic equilibrium with active translation and are dependent on relative concentration of non-ribosome associated transcripts in the cytoplasm [23, 25]. Inhibition of translation initiation under conditions of cellular stress results in the accumulation of pre-initiation complexes (PICs) and non-polysome associated transcripts in the cytoplasm which favors SG and PB assembly. Drugs like cycloheximide and emetine that ‘lock’ the ribosomes on mRNAs promote SG and PB disassembly, while premature chain terminating drugs like puromycin that release ribosomes from mRNAs promote assembly of these granules [26, 27]. Interestingly, formation of these two mRNP granules is not a requirement for global translation inhibition under stress [23]. For the purpose of this review, we will focus only on SGs, as they are more relevant type of RNA granules linking adaptive stress response to neurodegenerative disorders.
Liquid–liquid phase separations in RNP granule dynamics
RNA granules assemble as distinct dense liquid phase sub-compartments formed by liquid–liquid phase separation (LLPS) [28, 29]. The liquid–liquid phase separations are the result of both specific and promiscuous RNA:RNA, RNA:protein and protein–protein interactions that are multivalent, weak and transient (Fig. 1A) [28, 30]. The use of biotinylated isoxazole (b-isox) to selectively precipitate the proteins associated with the RNA granules has helped identify the RNA granule components. RNA plays a central role in core recruitment of proteins to RNPs in general. While over 1500 RBPs have been identified, RNA promotes critical interactions in both cis and trans to assemble RNPs [3, 31]. Moreover, under physiological buffer conditions RNAs are capable of self-assembly in vitro [32]. This intrinsic property of RNA is essential for its assembly into phase-separated foci, which is assisted by RBPs. Neuropathology-associated RNA repeat expansions have been shown to promote assembly of pathogenic granules independent of stress. The direct correlation between length of RNA repeat expansions and disease severity points to a vital role of RNA in pathogenic RNP assembly and disease progression [10, 33].
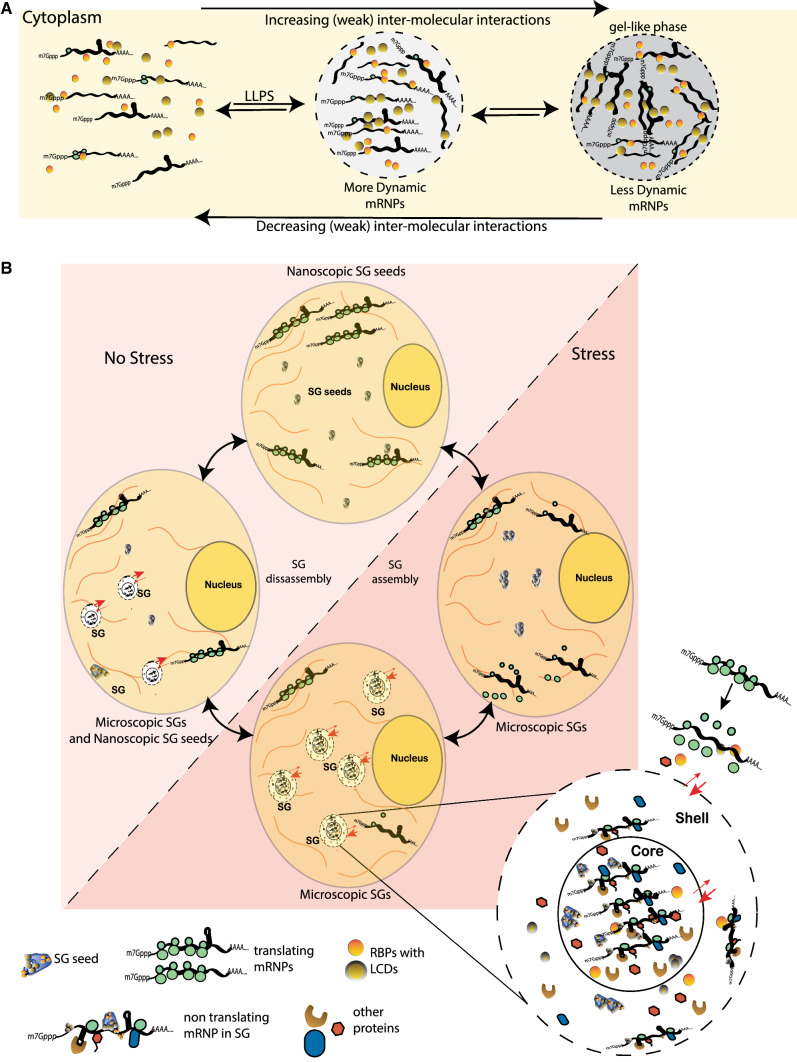
Fig. 1.
A Individual RNPs reversibly assemble into cytoplasmic foci triggered by physiological and environmental stimuli. The ‘liquid–liquid’ phase separations (LLPs) into phase-dense foci are driven by weak and transient intermolecular interactions that are enriched in RBPs containing LCDs and IDPRs. Persistent RNP-inducing stimuli promote interactions between assembled RNPs to form denser, less liquid–liquid, ‘gel-like’ phase. B Cellular stress induces cytoplasmic nanoscopic SG seeds, enriched in RBPs, via electrostatic interactions. The consequent inhibition of protein synthesis reversibly recruits non-translating mRNPs and additional RNA binding proteins to SG foci. The growth of phase separation results in the formation of mature and microscopic biphasic SGs consisting of more stable, less dynamic ‘core’ surrounded by less stable, more dynamic ‘shell’. The assembled SGs are in dynamic equilibrium with the surrounding polysomes. Removal of stress leads to resumption of protein synthesis and disassembly of SGs. The mRNPs are targeted for translation, thus causing the SGs to reduce in size and number to form nanoscopic SG seeds
Proteins enriched in these RNP foci are commonly characterized by the presence of a defined RNA-binding domain (e.g., RNA recognition motif (RRM)) and unstructured intrinsically disordered protein regions (IDPRs) and closely related low-complexity domains (LCDs) [34, 35]. The low-complexity domains are enriched in amino acids like alanine, glycine, lysine, arginine, glutamine, proline and serine which promote disorder [28]. The flexible LCDs and IDPRs are capable of aggregation and templating upon interacting with self and other molecules, especially RNA. Several SG components like G3BP1/2 and TIA-1/R have LCDs that contribute to phase transition to form dense cytoplasmic RNP foci [36, 37]. Interestingly, many small ribosomal subunit proteins are predicted to contain LCDs. They may facilitate LLPS and/or recruit SG nucleators like G3BP, which can directly interact with the 40S ribosomal subunit through its RGG motif and trigger assembly into larger and more stable mRNPs [16, 38, 39]. Several neurodegeneration-associated proteins such as TDP-43, FUS, TAF15, EWSR1 and others also contain prion-like domains (PrLDs) which are LCDs with amino acid motif similar to prion domain in yeast [40, 41]. These LCD-containing proteins also promote phase separation and pathogenic accumulation of protein:RNA aggregates in the nucleus and cytoplasm [41].
SG assembly and dynamics
SG assembly is triggered by bulk inhibition of translation initiation. This stress-mediated translational repression can be either phospho-eIF2α (p-eIF2α) dependent or independent. P-eIF2α mediated suppression of protein synthesis occurs as a result of phosphorylation of its amino acid serine at position 51 (Ser51) by one of the four stress-specific kinases (HRI, PERK, GCN2 and PKR). Phosphorylation at this position inhibits GTP exchange of the eIF2α/GTP/tRNAiMet ternary complex and prevents the tRNAiMet delivery to the 40S ribosomal subunit [42, 43]. One well-studied mechanism of p-eIF2α independent inhibition of translation under stress in through the mTOR (mammalian target of rapamycin) pathway. Under normal growth conditions mTOR constitutively phosphorylates eIF4E-binding proteins (4E-BPs) preventing their association with cap-associated eIF4E. Nutrient starvation and other metabolic stresses inactivate mTOR resulting in non-phosphorylated 4E-BPs binding to eIF4E and displacing eIF4G/eIF4A from the mRNA cap binding eIF4F complex, thereby inhibiting cap-dependent translation [44, 45]. Consequently, stress-mediated translational repression, whether p-eIF2α dependent or independent, triggers polysome disassembly and accumulation of non-translating mRNPs in the cytoplasm [46].
It is hypothesized that, under conditions of normal growth ‘nanoscopic’ cytoplasmic SG seeds consisting of RPBs with LCDs/IDPRs exist in equilibrium with surrounding RNPs. Stress-induced repression of translation initiation stimulates assembly into phase-dense ‘microscopic’ foci [38].The mRNAs released from polysomes and translation associated factors are actively targeted to these cytoplasmic foci. Such compartmentalization is aided by pre-assembled SG seeds (Fig. 1B). Continued nucleation of the mRNP foci results in the growth of phase-separated foci-enriched in RBPs which is promoted by weak and transient intermolecular interactions. These intermolecular interactions are dynamic in nature, which is an intrinsic property of LCD/IDPR-containing proteins [5, 38]. Simultaneous assembly of less dynamic mRNP phase called ‘core’ surrounded by more dynamic ‘shell’ results into the formation of large mature biphasic SGs. These SG ‘cores’ are sites of higher concentration of RNA and proteins surrounded by less concentrated components in the ‘shell’. Intermolecular interactions between components of the ‘shell’ are weaker contributing to their less stable and more dynamic nature [47]. According to the ‘core first’ hypothesis, formation of large mature SGs results from fusion of mature cores, i.e. core assembly precedes SG maturation [48]. The ‘core first’ model of mRNP assembly was confirmed by proximity labelling and sequencing approaches under heat shock induced stress in HEK293T cells where increased interactions between components was observed during early stages of granule formation. Evidently, interactions between ‘core’ proteins occur independent of stress [49]. Super-resolution microscopy approaches followed by validation with fluorescence recovery after photobleaching (FRAP) suggest that cores are dense, less liquid phase separated sub-compartments surrounded by a less stable ‘shell’ [47]. However, both ‘core’ and ‘shell’ are in dynamic equilibrium with each other and with the surrounding cytoplasmic mRNPs [47, 50].
SGs are dynamic by virtue of the protein component capacity to rapidly transition between phases. This characteristic enables a rapid, transient and reversible response to stress. FRAP analysis unambiguously shows that SG proteins like TIA-1, G3BP1, TTP and CPEB have a residence time in SGs in the range of 10–30 s, while less dynamic proteins like PABP, FXR1 and FMRP are recovered 30 s to few minutes post photobleaching [16, 51]. SG dynamics, as a function of protein exchange rates, is dependent on the cellular energy status, rates of translation reinitiation, protein and RNA modification profiles, chaperon activity and clearance by autophagy. Removal of stress favors resumption of protein synthesis, decreasing the pool of untranslating mRNPs and promoting translation of mRNPs released from SGs, thereby triggering their disassembly (Fig. 1B) [5, 38]. Recent work suggests that SG protein recruitment and rate of exchange is energy reliant. Activity of ATP-dependent MCM and RVB helicase complexes, conserved in yeast and mammals, promotes SG assembly [50]. Depleting cellular ATP by using inhibitors of glycolysis or oxidative phosphorylation decreases G3BP exchange thereby affecting its dynamics [50]. Interestingly, hypomorphic mutations in mammalian SG-associated ATPases cause faster disassembly of SGs than wild-type upon removal of stress, probably by inhibiting ATP-dependent stable interactions between mRNPs in the phase-dense foci [48, 50]. However, the exact mechanisms of ATP-mediated modulation of SG dynamics is still unknown.
Remodeling of mRNPs by protein chaperons also affects SG dynamics. Mammalian SG components Hsp70 and Hsp40 are protein chaperons that modulate formation and resolution of granules. Functional defects in these chaperons delay assembly and reinitiation of translation [50, 52]. In yeast, decreased activity of the CCT chaperonin complex results in increased SG formation under conditions of heat shock [50, 53, 54]. ATP-dependent disaggregase complexes are also essential for maintaining the mRNP integrity and identity. Yeast disaggreases Hsp110 and most importantly Hsp104 play an essential role in the disassembly of SGs upon removal of acute stress. Mammalian cells do not express the homolog of yeast Hsp104, which could also explain the differences in yeast and mammalian SG physical states [24].
Protein and RNA modifications that impact intermolecular mRNP interactions affect residence of SG protein components. Post-translational modification of several RBPs near or within the LCDs/IDPRs affect their association with granules (reviewed in [38]). For example, G3BP methylation represses while demethylation promotes SG assembly in vitro and in cells [55]. O-linked N-acetylglucoseamine (O-GlcNac) modification of several translation-associated proteins promotes their recruitment to mRNPs [38, 56]. The rate of clearance by autophagy also governs SG dynamics. Mutations that reduce ubiquitination and clearance of SG-associated proteins result in less dynamic and more persistent granules that underlie neuropathology [57–59]. Post-transcriptional mRNA modification also aids SG assembly and contributes to adaptive translation. For example, oxidative stress induces reversible methylation at position N6 of adenosines (m6A) in the 5′UTR of mRNAs. This dynamic m6A modification facilitates recruitment to SGs through the mammalian reader protein YTHDF3 which selectively recognizes these 5′UTR modifications [60].
Proteomic and transcriptomic heterogeneity among SG subtypes
Proteomic heterogeneity
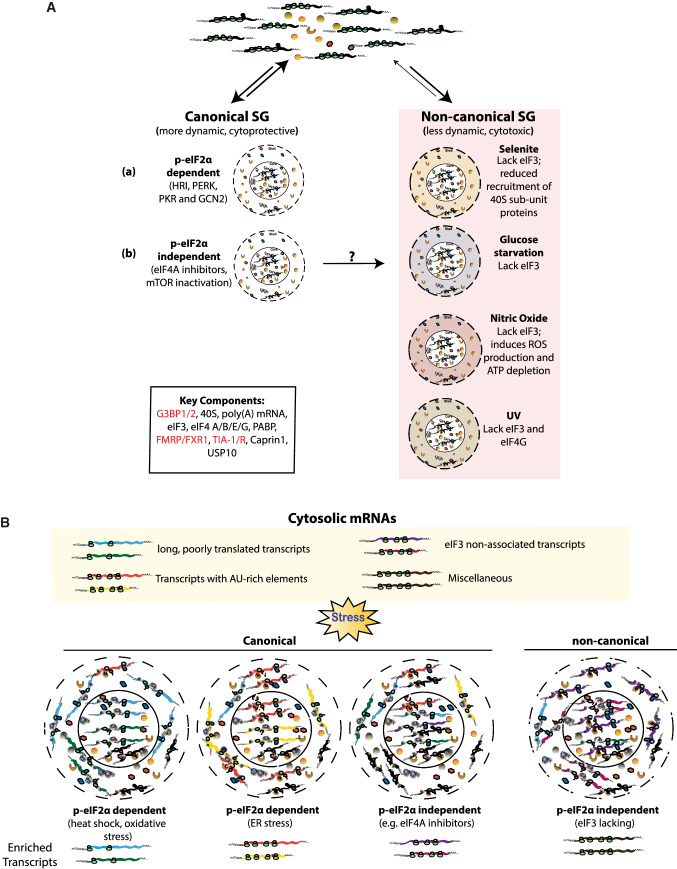
SGs are compositionally heterogenous and serve diverse function under different stresses. Both p-eIF2α-dependent (formed by sodium arsenite or heat shock treatment) and independent (formed by eIF4A inhibitors or mTOR inactivation) canonical SGs are enriched in PICs, 40S, initiation factors and RBPs, but lack eIF2 and eIF5 [5, 16]. These canonical SGs are dynamic and rapidly resolve upon removal of stress to reinitiate translation, therefore, playing a pro-survival cytoprotective role. Inhibitors of eIF4A helicase (like rocaglamide A) induce p-eIF2α independent non-canonical granules that recruit less poly adenylated mRNA, but include eIF2 and eIF5 [61]. Chemotherapy drugs like selenite target translational initiation by inactivating mTOR that promotes formation of eIF4E:4EBP1 complexes. The resulting non-canonical SGs lack eIF3 [38, 62]. eIF3 negative non-canonical granules, independent of eIF2α phosphorylation, are also induced under energy starvation and UV exposure. These non-canonical SGs are less dynamic, pro-apoptotic and cytotoxic to cells (Fig. 2A) [61, 63]. Thus, the stress-specific heterogeneity of SGs may serve as a template for stress-specific response. However, the differential recruitment of specific mRNAs and protein co-factors to each sub-type is a largely unexplored aspect of stress-specific SGs.
Fig. 2.
Stress-specific proteomic (A) and transcriptomic (B) heterogeneity defines SG sub-types and dynamics. Stress induced canonical stress granules can be either (a) p-eIF2α dependent triggered through stress-activated eIF2α kinases (GCN2, HRI, PERK or PKR) or (b) p-eIF2α independent (by mTOR activation or eIF4A inhibitors). Global translational repression prompts reversible recruitment of non-translating mRNPs and other proteins to SGs. Key SG components include poly(A) mRNAs, translation machinery components and RBPs (shown in red). The canonical stress granules are more dynamic and cytoprotective in nature. p-eIF2α independent SGs induced by glucose starvation, chemotherapeutic drugs, UV and other xenobiotic agents usually lack one or more key components of the canonical SGs. A common feature of non-canonical p-eIF2α independent SGs, which are less dynamic and cytotoxic, is the absence of translation initiation factor eIF3 (right panel, shadowed). B The stress-specific transcriptomic heterogeneity among SG is a function of differential association of mRNAs with RBPs. p-eIF2α dependent heat shock and oxidative stresses promote increased targeting of long poorly translated mRNAs (blue and green) to SGs, while ER stress causes preferential association of mRNAs with AU-rich elements (red and yellow) to SGs. At the same time, p-eIF2α independent non-canonical granules are depleted of mRNAs associated with eIF3 (purple)
The diversity of SG components can be attributed to their biphasic nature [50].The relative concentration of some components like G3BP1/2 and poly(A) mRNA is higher in the core than in the shell. The shell components are more dynamic and shuttle rapidly between SGs and the cytoplasm [64]. According to proteomic studies, the LCD domain containing protein G3BP1/2 is 30-fold more concentrated in the core and is capable of recruiting several proteins and poly(A) mRNAs [50]. Recruitment of poly(A) mRNA to the core as a stable sub-compartment may be important for assembling larger and more stable mRNPs for an efficient response to environmental cues. Mass spectrometry analysis of isolated G3BP-associated cores from arsenite or heat shock treated U2OS cells by Jain et al. catalogued 317 proteins associated with the resulting mRNPs. These included novel RBPs, components of translational machinery and unexpected proteins involved in metabolism and cell signaling [50].
Proximity labeling-based proteomic approaches like APEX and BioID utilize transiently expressed SG component fused with ascorbate peroxidase (APEX) or biotin ligase (BioID) as bait to covalently tag proteins in the close proximity with biotin [65, 66]. The biotinylated proteins can be affinity purified and identified by mass spectrometry analysis. The APEX approach is rapid, labeling proteins in minutes rather than hours like BioID [67]. These proximity labeling approaches provide a high-resolution method to profile previously unknown transient interacting partners in dynamic and compartmentalized mRNP granules. Application of these methods, with some limitations, have identified > 150 novel SG interactors [68, 69]. The validated hits are enriched for RBPs consisting of higher percentage of LCDs as compared to the background proteome. The studies greatly highlight the stress- and cell type-specific diversity in the SG granules that is driven by heterotypic multivalent interactions. The quantitative proteomic analysis of the SG-APEX data revealed that a network of SG protein interactions already exists in unstressed cells that enables assembly of large SGs under condition of stress [38, 68]. Markmiller et al. also applied APEX proximity labelling method to fly ALS/FTD in vivo models [68]. This study showed that a major sub-set of cytotoxic and pro-apoptotic SGs are enriched in mutant RBPs FUS and TDP-43, both implicated in neuropathogenesis.
Transcriptomic heterogeneity
High-resolution transcriptomic approaches combined with proteomics indicate stress- and cell-specific variation in targeting of RNA to SGs (Fig. 2B). Khong et al. performed RNA-seq analysis on SG cores purified from yeast and mammalian cells exposed to oxidative stress. The data suggest that > 95% of yeast and > 78% of mammalian RNA content of SGs is mRNAs, with ~ 10–12% of bulk cytosolic RNA (both mRNA and ncRNA) localized to SGs [70]. Interestingly, for 185 genes more than 50% of their cytosolic mRNAs were quantitatively enriched in SGs. Padrón et al. employed APEX-seq analysis in HEK293T cells using APEX2-fused eIF4A1, a DEAD-box RNA helicase and a conserved SG component, and showed that targeting of mRNAs to SGs is stress-dependent. For example, poorly translating, longer transcripts were enriched in SGs in cells heat-shocked over longer duration. This set of mRNAs enriched in SGs significantly overlaps with the ones identified by Khong et al. in SGs of U2OS cells exposed to another p-eIF2α dependent arsenite stress [49]. Thus, length and association with ribosomes determine preferential localization of mRNAs to p-eIF2α-induced canonical SGs. However, these two factors did not corelate with mRNAs targeted to SGs induced by p-eIF2α independent hippuristanol treatment [49]. Another study suggests that transcripts containing AU-rich elements (AREs) are more likely to be target to SGs under endoplasmic reticulum (ER) stress [71]. These studies underscore stress-specific transcriptomic heterogeneity of mRNPs associated with SG formation.
It is important to highlight that the RBPs in consideration, in general, do not have a well-defined RNA binding motif and bind RNA promiscuously. However, certain RBPs enriched in SGs exhibit preferential RNA binding that may further contribute to their transcriptomic heterogeneity. Examples of such RBPs are structurally related TIA-1 and TIAR proteins. These are general translational repressors and SG proteins [72, 73]. These proteins shuttle between the nucleus and cytoplasm and are involved in RNA metabolism on the level of transcription, alternative splicing, translational regulation and mRNA silencing [74]. Both of these proteins have three RNA recognition motifs (RRM 1–3) and a C-terminal prion-related domain (PRD) [75, 76]. The RNA recognition motif 2 (RRM2) is implicated in binding U-rich transcripts and contribute to translational repression by binding to mRNAs with 3′UTRs enriched in AU-rich elements (AREs) [77, 78]. These ARE containing transcripts are implicated in the immune response and proliferation [77]. Moreover, Damgaard et al. showed that TIA-1/R also associate with mRNAs with a 5′ terminal oligopyrimidine (5′TOP) motif through RNA immunoprecipitation experiments. 5′TOP transcripts predominantly include mRNAs encoding ribosomal proteins and translation factors [79]. These mRNAs are characterized by relatively short unstructured 5′UTRs with 4–15 nucleotide CU-rich element [72]. P-eIF2 mediated translational repression under conditions of stress causes translocation of TIA-1 to the cytoplasm, where it escorts associated mRNAs and non-canonical pre-initiation complexes to SGs [26, 80]. Interestingly, translocation of TIA-1 to the cytoplasm is not required for SG assembly, but its availability greatly impacts kinetics and recruitment of mRNA to SGs [81]. TIA-1/R bind to translationally stalled 5′TOP mRNPs and target them to SGs under conditions of amino acid starvation [79, 82]. In another study, Lee et al. used transcriptome wide photoactivable ribonucleoside enhanced crosslinking and immunoprecipitation (PAR-CLIP) analysis to identify mRNAs bound to eIF3, a translation initiation factor that is commonly present in canonical but absent in non-canonical cytotoxic SGs. The PAR-CLIP technique uses UV to covalently crosslink RBPs with their target RNAs that have been labelled with photoactivable ribonucleosides. The cross-linked RNA–protein complexes are immunoprecipitated using an anti-body against the protein of interest followed by transcriptomic approaches to investigate target mRNAs [83]. The data suggest that eIF3 preferentially binds a sub-set of protooncogenic transcripts at their 5′UTRs, with the eIF3d subunit having m7GTP cap binding activity [84]. The differential binding of eIF3 may further contribute to compositional and functional differences among SGs. Intriguingly, eIF3 targets JUN, a proto-oncogene and B-cell translocation gene 1 (BTG1), a tumor suppressor gene, both of which have contrasting effects on protein synthesis [84]. However, the pro-apoptotic nature of eIF3-lacking non-canonical SGs is still unclear. These characteristics may underlie the heterogenous nature of SGs. To add to this, the suggested compartmental nature of mRNPs could function as a template for differential stress-specific modulation of the transcriptome.
It is important to point out that the existing empirical methods to elucidate the components and dynamics of mRNPs have been applied to conventional cell lines used to study the stress response. The stress- and cell-specific heterogeneity of mRNPs may extend beyond what is known with implications for metabolism, physiology and pathogenesis. The remainder of the review discusses how genetic mutations and, altered SG dynamics and subtypes are implicated in neuropathology.
Characteristics of pathogenic RNA granules in neuropathies
Neurons are non-dividing highly polarized cells that dynamically adapt to external physiological stimuli. Moreover, they are long-lived cells and can vary in length from few millimeters to over a meter. This varied and unique morphology requires spatial regulation of translation to meet local physiological demands. Compartmentalized protein synthesis driven by localized mRNPs is essential for maintaining neuronal physiology, synaptic malleability and memory formation [85]. These specialized mRNPs are important for local protein supply and plasticity [85, 86]. Besides regulating protein synthesis, neuronal cytoplasmic mRNP granules are also used for transporting mRNAs along microtubules in both axons and dendrites. Interestingly, RNP foci containing SG related proteins have been implicated in local translational control [87, 88]. Thus, characteristic cytoarchitecture, lifespan and specialized metabolic demands make neurons more vulnerable to chronic stress-mediated dysregulation of mRNP dynamics and function.
Classically, so called ‘protein aggregates’ associated with neurogenerative disorders have been attributed to defective protein metabolism and misfolding. Recent work links several RNA-binding proteins enriched in these pathogenic mRNPs to disease progression. The neurodegenerative related ‘pathogenic’ proteins have mutations in their LCDs/IDPRs/RNA-binding motifs resulting in aggregation prone RBPs. This leads to aberrant phase transitions to form non-canonical mRNPs with persistent accumulation leading to chronic neurological insult.
There is increasing genetic and biochemical evidence that implicates defective mRNP accumulation in several neurodegenerative disorders. Despite a wide range of clinical and pathological presentations, a common hallmark of age-independent neurological disorders is ‘pathogenic’ accumulation of RNA–protein aggregates in neurons of the central nervous system (CNS) [89, 90]. In some cases, genetic studies attribute the inappropriate accumulation of proteins in neurons to mutations in RBPs, most of which overlap with SG proteins. Often, the mutations are in the LCDs of these proteins [91]. LCDs, as discussed above, confer conformational plasticity which is essential for promoting transient interactions between neighboring molecules that drive the assembly and disassembly dynamics of mRNP granules. Some mutant SG-associated proteins have an increased tendency for phase separation and are capable of self-propagating in a prion-like mechanism both in vitro and in vivo independent of stress. Ataxin1 was the first RBP linked to neurodegeneration [92]. The CAG repeat expansion mutation in the coding region results in polyglutamine tract in the LCD of Ataxin1 that promotes its accumulation in cytoplasmic foci and development of spinocerebellar ataxia type 1 (SCA1) [93]. Mackenzie et al. identified five ALS/FTD linked mutations in the LCD of the SG core protein TIA-1. These TIA-1 mutant proteins showed an increased tendency for phase separation and were enriched in cytotoxic, large, hyaline and TDP-43-positive inclusions [94]. Similarly, mutations within the PrLDs of other RBPs like hnRNPA2/B1, TAF15 and EWS also promote their localization to pathological inclusions [95, 96]. Such mutations cause assembly of less dynamic, chronic, more fibrillar and persistent SG subtypes that compromise RNA metabolism, protein quality control and promote apoptosis.
A general consequence of the chronic and persistent mRNP granules, linked to neuropathies, is sequestration of specific RNA-binding proteins in nuclear and cytoplasmic foci. The sequestration and obvious mis-localization of RBPs in pathogenic granules may compromise their cellular function with implications on RNA processing, transport and translation. For example, sequestration of MBNL1 and CUGB1 proteins in nuclear foci linked to myotonic dystrophy types 1 and 2 (DM1 and DM2) alters the RNA splicing and transcriptional profiles that correlates with cognitive decline and clinical phenotypes in mouse models [97, 98]. Overexpression of these proteins partially rescues the splicing and transcriptional defects and mitigates the pathology [99, 100]. Similarly, the CGG repeat expansion in the 5′UTR of FMR-1 mRNA is the main driver of Fragile X tremor syndrome (FXTAS) associated with the appearance of specific RNA foci. The FMR-1 mRNA-induced RNA foci sequester CUGBP1, Purα and hnRNPA2/B1 proteins compromising their cellular function [101, 102].
Another consequence of altered dynamics of persistent neuronal SGs is the impairment of autophagy. Autophagy pathways are shown to assist in clearance of SGs, which contributes to their pro-survival nature [41, 103]. Recent evidence suggests that ubiquitinated proteins in SGs are targeted for autophagy in an ATP-dependent manner, which contributes to their removal and reinitiation of cellular translation [104, 105]. Several mutations in autophagy adaptor proteins (TBK1, OPTN1 and SQSTM1), that deliver ubiquitinated proteins to autophagosomes for clearance, are linked to neuropathologies including ALS and Huntington’s disease (HD) [106–108]. These proteins exhibit an increased abundance in pathogenic aggregates in patient neurons, an indicator of impaired autophagy [58, 108]. Age-associated gradual decline in autophagy may result in a decrease in the clearance of SG granules after the removal of stress. However, age-independent disease-prone increased accumulation of large fibrillated cytoplasmic aggregates overwhelms the autophagy machinery and promotes degenerative pathology [109, 110]. This might be due to the less dynamic nature of the pathogenic aggregates that affects their effective targeting. For example, the ALS-linked pathogenic FUS-(R521C) mutant, but not wild-type protein, preferentially localizes to SGs in mouse cortical neurons when exposed to oxidative stress. Interestingly, FUS-positive SGs also accumulate in autophagy-deficient neurons in the absence of any stress [111]. Accumulation of mutant-FUS positive persistent SGs results in reduced autophagic flux and impaired clearance of autophagosomes [111, 112].
Role of RBPs
Biology of RBPs plays an essential role in achieving specific physiological tasks in long neurons through localized mRNPs. Gradual time-dependent dysregulation of cytoplasmic translocation, autophagy and altered local supply of RBPs contributes to aggregation of defective, large and chronic inclusions responsible for age-associated slow cognitive decline. The disease-associated mutations in RBPs significantly accelerates this progression towards more fibrillar amyloid-like protein rich cytoplasmic foci in patients (Table 1).
Table 1.
Mutations and SG phenotypes for RBPs associated with neurological disorders
| Protein | Mutations | Domain | SG phenotype | References |
|---|---|---|---|---|
| TDP-43 | A90V | NLS | Cytoplasmic mis-localization to SGs | [193, 194] |
| D169G | RRM1 | Decrease Ubiquitination in cytoplasmic and nuclear inclusions | [195, 196] | |
| K263E | RRM2 | |||
| A315T, G335D, M337V, Q343R N345K and R361S | Glycine rich LCD | Promote phase separation; more fibrillar granules | [197–199] | |
| FUS | G156E | PrLD | Increased self-templating capacity, defective RNA binding | [200] |
| R244C | Glycine rich LCD | Defective RNA binding | [200] | |
| R495X | RGG | Increased targeting to SGs | [201] | |
| H517Q, R521G/C, R522G and P525L | NLS | Cytoplasmic mis-localization and increased accumulation in perinuclear SGs | [201–203] | |
| HNRNPA1 | D262V/N, D314V and N267S | PrLD | Increased self-templating capacity | [95, 204, 205] |
| F273L, M276L and F281L | NLS (or TPNO-1 binding) domain | Cytoplasmic mis-localization and increased targeting to SGs | [118] | |
| HNRNPA2/B1 | D290V | PrLD | Increased amyloidogenic cytoplasmic inclusions | [95] |
| EWSR1 | P522L and G511A | RGG | Increased cytoplasmic localization, Increased self-aggregation | [206, 207] |
| TAF15 | M368T, G391E, R408C and G473E | RGG | Cytoplasmic mis-colalization, increased targeting to SGs | [207, 208] |
| TIA-1/R | P362L, A381T and E384K | LCD | Increased targeting to SGs | [94] |
| C9orf72 | (G4C2) hexanucleotide expansions in intron 1 | N/A | G2C2-RNA repeats and arginine-rich dipeptide repeats promote phase separation and maturation of SGs to amyloid-like inclusions | [59, 164] |
Several neurodegenerative phenotypes are thought to proceed through the SG pathway. The aggregation of mutated proteins promotes both stress-dependent and -independent SG assembly and triggers their transition to chronic pathological inclusions. In several cases, mutations in a single RBP is sufficient to significantly change in its subcellular localization, promoting assembly of pathogenic SG-like aggregates, which perturb many aspects of RNA and protein metabolism. For instance, wild-type variants of the ALS-linked proteins TDP-43 and FUS have pre-dominantly nuclear function and harbor a nuclear localization signal (NLS) [113, 114]. Several mutations in their NLS result in their mis-localization and increased local concentration in the cytoplasm. These proteins have PrLDs that promote multimerization and buildup of amyloid-like granules [115, 116]. Specifically, a pathogenic mutation in the C-terminal β-domain of TDP-43 (A315T) results in its cytoplasmic mis-localization and increased detection in cytoplasmic pathological inclusions [117]. Similarly, ALS/FTD-linked mutations, in the LCD domain of TIA-1, P362L, A381T and E384K, cause increased mis-localization to the cytoplasm and recruitment to SGs (Table 1) [94]. In multiple sclerosis (MS), single-nucleotide variants (SNVs) within the NLS, also called the transportin-1 (TPNO-1) binding domain, of RBP hnRNPA1 result in its cytoplasmic mis-localization and increased targeting to SGs. This promotes cellular apoptosis that contributes to the pathogenesis of MS [118].
Dysregulation of nucleocytoplasmic transport also contributes to altered mRNP dynamics and promotes cytoplasmic aggregation in several neurodegenerative diseases. Some nucleocytoplasmic factors localize to SGs under conditions of stress or via interactions with mutant proteins. This results in cytoplasmic mis-localization of several nuclear RBPs like TDP-43, FUS, TAF15, EWSR1, hnRNPA1 and hnRNPA2. Thus, increasing the cytoplasmic concentration of proteins that have higher tendency to promote LLPS contribute to transition of SGs to pathogenic aggregates [119, 120]. Defective nucleocytoplasmic transport and SG assembly are hallmark of C9orf72-mediated ALS/FTD pathogenesis. Increased expression of ALS/FTD associated toxic dipeptide repeats (DPRs), like poly-GA and poly-GR, cause cytoplasmic mis-localization and aggregation of several nuclear transport receptors (NTRs) and TDP-43 in drosophila models [121]. Intriguingly, this also results in increased DPR translation [121]. Overexpression of nuclear import factors or knockdown of nuclear export factors have been shown to restore nuclear RBP localization and alleviate neuronal toxicity mediated by disease-linked FUS [120, 122].
Toxic multimerization of disease associated RBPs alter the local transcriptome and proteome pertaining to axonal and synaptic pathology. Increased targeting of FMRP to toxic cytoplasmic mRNP granules results in transport granule dysfunction, compromised local translation and defective dendritic branching [123, 124]. Frontotemporal lobar degeneration (FTLD) associated TDP-43 aggregates in dendrites are enriched in transcripts involved in synaptic function [125, 126]. This targeting of synaptic mRNAs perturbs their activity-dependent localization and expression in FTLD patient neurons [127]. Mis-localization of several axonal transcripts in TDP-43, FUS and C9-ALS/FTD associated cytoplasmic aggregates results in their decreased expression and compromised local function [123]. For example, increased accumulation of MAP1B and Nef-L in C9-ALS/FTD aggregates causes instability of microtubules at axon terminals and synaptic defects [128, 129].
Several mutations in RBPs and/or post-translational modifications also confer gain of function. Mutations in some domains increase the propensity of intermolecular associations to form more dense aggregates. For example, acetylated and hyper-phosphorylated forms of TDP-43 have been detected in insoluble pathological inclusions [130, 131]. This defective acetylation of lysines and hyper-phosphorylation at serine residues within the RRM may increase their nucleic acid binding capacity and aggregate ‘seeds’ to stimulates intermolecular interactions among SG components. Phosphorylated mutant A315T(p), outside of the RRM, has increased aggregation capacity and accelerated fibril formation as compared to wild-type in vitro [132]. The TDP-43 interactome includes several SG-associated eukaryotic initiation factors (eIF4G, eIF3B and eIF4A1) [133]. Interaction with these initiation factors and bona fide SG components may assist in the association of cytosolic mutant protein with translational machinery (e.g., ribosomes) to assemble in cytotoxic aggregates. Similarly, an altered phosphorylation profile of Tau protein is directly implicated in Alzheimer’s disease (AD) and other tauopathies [7, 134]. Tau is a microtubule-associated protein that plays an essential role in microtubule assembly and axoplasmic transport in neurons. Phosphorylation of amino acid residues within the microtubule interaction domain is essential for tau function and microtubule polymerization [135–137]. Location of tau and its phosphorylation levels determines its function. Tau is localized to axons under normal physiological conditions [135, 138]. Glutamatergenic stimulation of neural ionotropic receptors, AMPA and NMDA, stimulates localized translation and hyperphosphorylation of tau in the somatodendritic compartment. This results in tau mis-localization away from axon and its accumulation in neurofibrillary tangles (NFTs), one of the hallmarks of AD pathology [139]. AD therapies that relieve aberrant activation of these glutamate receptors have shown to attenuate tau phosphorylation and improve synaptic function [140]. Persistent SGs also stimulate tau phosphorylation and aggregation representing a potential stress-driven mechanism of maturation to pathogenic SGs [141].
Chronic stress and high glucocorticoid (GC) levels can cause hyper-phosphorylation of tau outside this domain leading to its mis-localization to the soma and dendrites rather than axons [142]. Hyper-phosphorylation promotes stress-independent interactions with cytoplasmic SG proteins TIA-1 and TTP that in turn modulate the formation of SGs and NFTs [7, 142]. Vanderweyde et al. showed that pseudo-phosphorylated tau mutant was targeted more efficiently and resulted in larger SGs than phospho-null tau mutant [141]. Recent work also suggests that mis-localized hyper-phosphorylated tau interacts with cytoplasmic translational apparatus and many SG associated RBPs. Mass spectroscopy approaches applied to human neuroblastoma and AD mouse models revealed that tau preferentially interacts with 60S subunit proteins, several initiation factors, heteronuclear RNPs and several other SG proteins besides TIA-1 and TTP [143, 144]. Tau association with ribosomes and inhibition of global protein synthesis was co-relative to decreased synaptic function and memory loss in AD patients [145]. The tau interactome, therefore, indicates a much wider role of tau in stress-mediated translational control. It is hypothesized that a combination of disease-linked RBP mutations and hyperphosphorylation of tau due to chronic stress causes translational repression and formation of persistent SGs.
Tau pathophysiology is greatly enhanced by its interactions with TIA-1, which promotes SG formation and repression of translation [141]. Decreased expression of TIA-1 prevents tau misfolding and pathogenic tau accumulation in cytoplasmic inclusions in cell culture and mouse models [137, 144]. This highlights an important role of RBPs and a potential role of SGs in tau-mediated neurodegeneration. Reducing levels of endogenous TIA-1 in specific tau mice model reduced pathogenic SG assembly, decreased mis-localization of TIA-1 and improved cognitive function and lifespan. Interestingly, knockdown of TIA-1 did not decrease the number of NFTs [137].
Angiogenin (ANG) is an RBP and its biological roles include angiogenesis, cell proliferation and neuroprotection [146, 147]. It is a stress responsive ribonuclease of the RNase A superfamily that cleaves tRNAs under conditions of stress to generate stress-induced tRNA halves called tRNA-derived stress-induced RNAs (tiRNAs) [146, 148]. The neuroprotective function of ANG is attributed to its RNase activity, while tiRNAs have been implicated in promoting cell survival and inhibiting apoptosis under conditions of stress [149]. Recent work identified two bioactive tiRNAs involved in pro-survival stress response. The 5′ terminal oligo guanidine (5′TOG) motif containing 5′tiRNAAla and 5′tiRNACys are capable of forming G-quadruplexes (G4s) that are non-canonical structures composed of G-quartets [148, 150]. These G4 containing tiRNAs repress translation by displacing the eIF4F complex from 5′ mRNA cap structure and promote SG assembly in a p-eIF2α independent manner [151]. Certain ANG induced tiRNAs bind directly to cytochrome C (Cyt C) under conditions of stress to inhibit apoptosis [152, 153]. Several ALS/PD associated ANG mutations result in RNase loss of functions resulting in decreased production of tiRNAs compromising its cytoprotective role [146, 154].
RNA-dominant proteinopathies
RNA, a dominant component of SGs, is itself involved in myriad neuronal dysfunction. Several ‘RNA binding proteinopathies’ have been identified as repeat expansions that drive phase separations in myotonic dystrophies and other degenerative pathologies. These nucleotide repeats confer cytotoxicity by inappropriate redistribution and sequestration of selected RBPs [155, 156]. Spinocerebellar ataxia type 10 (SCA10) associated pentanucleotide AUUCU repeat expansions in 3′UTR of E46L mRNA drives their toxic cytosolic accumulation [157, 158]. These cytoplasmic inclusions preferentially recruit hnRNP K, in addition to other RBPs. The hnRNP K loss-of-function leads to mitochondrial dysfunction and activation of apoptosis in the SCA10 mouse model. Over-expression of hnRNP K rescues SCA10 neuronal cells from cell death due to SCA10 pentanucleotide expansion mutation [158]. Another example of RNA repeat mutation conferring cellular degeneration is that of CAG expansion in certain of degenerative pathologies [159]. These tri-nucleotide repeats within the coding region results in increased expression of poly-glutamine rich (polyQ) Ataxin 3 [160]. The polyQ rich proteins are aggregation prone, accumulating into cytotoxic nuclear inclusions (NI) linked to SCA3 [160, 161]. The CAG repeat mRNA is capable of forming hairpin structures and recruiting RBPs in a length-dependent manner into cytoplasmic foci, related to HD pathology [159, 162]. However, the CAG tri-nucleotide cytoplasmic interactome and the mechanism of pathogenesis are still unclear.
The most common genetic cause of ALS/FTD is GGGGCC (G4C2) hexanucleotide repeat expansion in the intron 1 of C9orf72 gene [163]. The repeat length threshold of ≥ 24 repeats is considered pathology-prone [164]. These hexanucleotide repeats are bidirectionally transcribed as sense (rG4C2) and anti-sense (rC4G2) transcripts [165]. Both sense and anti-sense transcripts stimulate non-canonical RAN translation of toxic DPRs; poly-GA, poly-GR and poly-GP from sense transcripts and poly-PA and poly-PR from anti-sense transcripts [166–168]. The G4C2 RAN translation involves canonical scanning and initiation at near-cognate CUG codon where the poly-DPRs are synthesized by sequence-dependent frameshifting [169]. The (rG4C2) repeat sequence is capable of forming higher order hairpin and G-quadruplex structures capable of interacting with several RBPs including SG proteins [170]. Both (rG4C2) and DPRs are detected in nuclear and cytoplasmic pathogenic inclusions in patient neurons and glia. Fay et al. showed that (rG4C2) repeats can promote assembly of RNA granules in repeat length- and structure-dependent manner both in vitro and in vivo independent of stress [164]. Moreover, (rG4C2) cytoplasmic foci partially resemble SGs as they recruit small ribosomal subunits, translational initiation factors and several core SG proteins [5, 164]. The arginine rich DPRs, poly-GR and poly-PR confer toxicity independent of (rG4C2) repeats in patient cells [171]. Nuclear and cytosolic accumulation of these toxic DPRs represses global translation, inhibits nucleocytoplasmic transport, disrupts rRNA processing and localizes to TDP-43 enriched stress granules. Mass spectrometric approaches to characterize DPR associated proteins identify significant overlap between poly-GR and poly-PR interactomes that include several SG associated RBPs [59, 172, 173]. However, the transcriptomic profiles of (rG4C2) and DPR rich inclusions are unexplored. Some evidence suggests that the arginine-rich dipeptides localize to the fibrillar cores of mRNP granules and support formation of insoluble aggregates [174]. The above studies support either RNA- or DPR-mediated toxicity in ALS/FTD pathogenesis. However, a common observation of this RNA and/or protein-mediated toxic gain of function is the disruption of mRNP dynamics to promote assembly of less dynamic, persistent and pathogenic protein aggregates.
Conclusions: therapeutic approaches towards SG-mediated neuropathies
Pathomechanistic studies of RBP mutations and modification linked to specific neurodegenerative and neuromuscular disorders provide potential therapeutic opportunities for pharmacological intervention (Fig. 3). The dominant approach is to target the RBP pathways that mediate pathogenic oligomerization, misfolding and aggregation. Overexpression of wild-type non-pathogenic RBP or knockdown of pathogenic RBPs with small molecule inhibitors or anti-sense oligos (ASOs) can modulate their recruitment into specific subcellular compartments, which is known to ameliorate toxicity [141, 175]. These approaches have been shown to partially rescue the disease phenotype by reducing the number and size of cytotoxic foci in several disease models. For example, ASO therapy to reduce Ataxin2 levels in TDP-43/ALS fly and mice models decreased amount and number of TDP-43 pathogenic aggregates. This ASO treatment significantly slowed disease progression and increased lifespan of the TDP-43 ALS mouse model [175]. ASO therapy also seems promising in targeting the (G4C2) hexanucleotide repeats in C9orf72 ALS/FTD pathology. Targeting these RNA repeats is expected to reduce the size and number of pathogenic RNA foci and the levels of toxic DPRs. Another broad therapeutic approach is the use of small-molecule inhibitors to target components of SG assembly and disassembly. Drugs that inhibit the action of eIF2α kinases reduce pathology primarily due to decrease in phosphorylation levels of eIF2α [176, 177]. However, pleiotropic effects outside translation inhibition may not have favorable outcomes in humans. Pharmacological induction of certain chaperons and disaggregases that prevent protein misfolding and aggregation have been shown to reduce degeneration rates in vulnerable neurons [178]. Overexpression of disaggregases like Hsp110, Hsp40 and Hsp70 may help overcome proteostasis and counter its toxic effects [179]
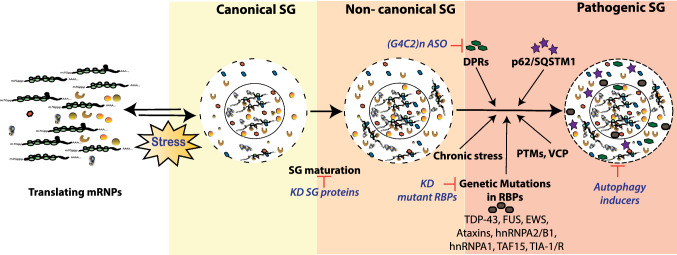
Fig. 3.
Maturation of physiological SGs into pathogenic aggregates is mainly driven by chronic stress and mutant RBPs and/or RNA repeats, with progressive impairment of their physiological clearance. Stress mediated assembly of untranslating mRNPs into microscopic canonical, dynamic and less stable SGs is assisted by RNA and RBPs. Specific stress conditions result in the formation of non-canonical cytotoxic SG subtypes. Genetic mutations in LCDs/IDPRs/ RNA-binding motifs of RBPs (TDP-43, FUS, EWS, Ataxins, hnRNPA1, hnRNPA2/B1TAF15 and TIA-1/R; see Table 1) and/or their post-translational modifications (PTMs) further shift maturation of SGs into non-canonical, less dynamic and cytotoxic SG subtypes. Further, clearance of these SGs is reduced by inhibition of autophagy and/or mutation in the autophagy associated factor VCP (valosin-containing protein). Chronic localization of disease-linked defective RBPs, ALS/FTD-associated dipeptide repeats (DPRs) and components of sequestosome (e.g. sequestosome protein 1 or p62/SQSTM1) promote transition to insoluble, more fibrillar and persistent pathological SGs. Points of therapeutic interventions (shown in red) target SG assembly, maturation and clearance. ASO anti-sense oligo(s), KD knock-down
Autophagy, as discussed earlier, may play a direct role in solubilization of SGs and their subtypes. Its activation has been shown to confer neuroprotection in certain ALS, HD and AD disease models by enhancing clearance of pathogenic cytoplasmic aggregates and attenuation of cellular apoptosis [180–183]. Autophagy inducers can be mTOR-dependent or mTOR-independent. Hyperactivation of mTOR in AD neurons results in decreased autophagy and increased production of amyloid-β-peptide fragments and aggregation into plaques. This also results in hyperphosphorylation of tau and deposition in cytoplasmic NFTs [184, 185]. Pharmacological inhibition of mTOR in two mouse models of AD resulted in activation of autophagy and decreased accumulation of Aβ plaques and phosphorylated tau in neurons [186]. Rapamycin is one of the mTOR dependent, non-ATP competitive inducers that mediates neuroprotection via inhibition of mTORC1 and mTORC2 complexes simultaneously [182, 187]. Although the mechanism of mTOR-independent autophagy activators like Trehalose in neuroprotection is unclear [188], Trehalose dosage in mouse models of ALS, AD and PD decreased autophagic flux and mitigated pervasive symptoms [189–192].
In conclusion, several studies confirm that differential proteomic and transcriptomic content drives the transition of stress granules from physiological subtypes to pathogenic aggregates. While the role of mutant proteins has been greatly explored, investigations of differential RNA content of disease specific cytotoxic aggregates are still pending. Knowledge of distinct transcriptomic and proteomic profiles associated with specific neuropathy will present additional therapeutic opportunities.
Funding
Funding was provided by National Institutes of Health (R01 GM126150).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vivek M. Advani, Email: vadvani@bwh.harvard.edu
Pavel Ivanov, Email: pivanov@rics.bwh.harvard.edu.
References
- 1.Hnisz D, Shrinivas K, Young RA, et al. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Treeck B, Parker R. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell. 2018;174:791–802. doi: 10.1016/j.cell.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fay MM, Anderson PJ. The role of RNA in biological phase separations. J Mol Biol. 2018;430:4685–4701. doi: 10.1016/j.jmb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol. 2018 doi: 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolozin B. Physiological protein aggregation run amuck: stress granules and the genesis of neurodegenerative disease. Discov Med. 2014;17:47–52. [PMC free article] [PubMed] [Google Scholar]
- 7.Wolozin B, Ivanov P. Stress granules and neurodegeneration. Nat Rev Neurosci. 2019;20:649–666. doi: 10.1038/s41583-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeynaems S, Alberti S, Fawzi NL, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan VH, Fawzi NL. Physiological, pathological, and targetable membraneless organelles in neurons. Trends Neurosci. 2019;42:693–708. doi: 10.1016/j.tins.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voronina E, Seydoux G, Sassone-Corsi P. RNA granules in germ cells subject collections. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao M, Arkov AL. Next generation organelles: structure and role of germ granules in the germline. Mol Reprod Dev. 2013;80:610–623. doi: 10.1002/mrd.22115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 15.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA. 2013;4:17–34. doi: 10.1002/wrna.1139. [DOI] [PubMed] [Google Scholar]
- 16.Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moujaber O, Stochaj U. Cytoplasmic RNA granules in somatic maintenance. Gerontology. 2018;64:485–494. doi: 10.1159/000488759. [DOI] [PubMed] [Google Scholar]
- 18.Shigeoka T, Jung H, Jung J, et al. Dynamic axonal translation in developing and mature visual circuits. Cell. 2016;166:181–192. doi: 10.1016/j.cell.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mittag T, Parker R. Multiple modes of protein–protein interactions promote RNP granule assembly. J Mol Biol. 2018;430:4636–4649. doi: 10.1016/j.jmb.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Treeck B, Parker R. Principles of stress granules revealed by imaging approaches. Cold Spring Harb Perspect Biol. 2019;11:a033068. doi: 10.1101/cshperspect.a033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavut A, Raveh D. Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet. 2012;8:e1002527. doi: 10.1371/journal.pgen.1002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker R, Sheth U. P Bodies and the control of mRNA Translation And Degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroschwald S, Maharana S, Mateju D, et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife. 2015;4:e06807. doi: 10.7554/eLife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4:a012286–a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kedersha N, Cho MR, Li W, et al. Dynamic shuttling of Tia-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moutaoufik MT, El Fatimy R, Nassour H, et al. UVC-induced stress granules in mammalian cells. PLoS One. 2014;9:e112742. doi: 10.1371/journal.pone.0112742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyman AA, Weber CA, Jülicher F. Liquid–liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 29.Shin Y, Brangwynne C. Liquid phase condensation in cell physiology and disease. Science. 2017;357(6357):eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 30.Li P, Banjade S, Cheng H-C, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Treeck B, Protter DSW, Matheny T, et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci USA. 2018;115:2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulson H. Repeat expansion diseases. Handb Clin Neurol. 2018;147:105–123. doi: 10.1016/B978-0-444-63233-3.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato M, Han TW, Xie S, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/J.CELL.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han TW, Kato M, Xie S, et al. Cell-free Formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Uversky VN. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 37.Kedersha NL, Gupta M, Li W, et al. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. J Cell Biol. 2016;215:313–323. doi: 10.1083/jcb.201609081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber SC, Brangwynne CP. Getting RNA and Protein in Phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Harrison AF, Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem J. 2017;474:1417–1438. doi: 10.1042/BCJ20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monahan Z, Shewmaker F, Pandey UB. Stress granules at the intersection of autophagy and ALS. Brain Res. 2016;1649:189–200. doi: 10.1016/j.brainres.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wek RC. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol. 2018;10:a032870. doi: 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Advani VM, Ivanov P. Translational control under stress: reshaping the translatome. BioEssays. 2019;41:1900009. doi: 10.1002/bies.201900009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/MCB.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivanov P, Kedersha N, Anderson P. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol. 2019;11:a032813. doi: 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wheeler JR, Matheny T, Jain S, et al. Distinct stages in stress granule assembly and disassembly. Elife. 2016 doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padrón A, Iwasaki S, Ingolia NT. Proximity RNA labeling by APEX-Seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol Cell. 2019;75:875–887.e5. doi: 10.1016/J.MOLCEL.2019.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jain S, Wheeler JR, Walters RW, et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bley N, Lederer M, Pfalz B, et al. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 2015;43:e26–e26. doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazroui R, Di Marco S, Kaufman RJ, Gallouzi I-E. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18:2603–2618. doi: 10.1091/mbc.e06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arimbasseri AG, Blewett NH, Iben JR, et al. RNA Polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genet. 2015;11:e1005671. doi: 10.1371/journal.pgen.1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cherkasov V, Hofmann S, Druffel-Augustin S, et al. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23:2452–2462. doi: 10.1016/J.CUB.2013.09.058. [DOI] [PubMed] [Google Scholar]
- 55.Tsai W-C, Gayatri S, Reineke LC, et al. Arginine demethylation of G3BP1 promotes stress granule assembly. J Biol Chem. 2016;291:22671–22685. doi: 10.1074/jbc.M116.739573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohn T, Kedersha N, Hickman T, et al. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walters RW, Muhlrad D, Garcia J, Parker R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA. 2015;21:1660–1671. doi: 10.1261/rna.053116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cipolat Mis MS, Brajkovic S, Frattini E, et al. Autophagy in motor neuron disease: key pathogenetic mechanisms and therapeutic targets. Mol Cell Neurosci. 2016;72:84–90. doi: 10.1016/J.MCN.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 59.Lee K-H, Zhang P, Kim HJ, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–788.e17. doi: 10.1016/J.CELL.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders M, Chelysheva I, Goebel I, et al. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci Alliance. 2018;1:e201800113. doi: 10.26508/lsa.201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aulas A, Fay MM, Lyons SM, et al. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci. 2017;130:927–937. doi: 10.1242/jcs.199240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujimura K, Sasaki AT, Anderson P. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 2012;40:8099–8110. doi: 10.1093/nar/gks566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reineke LC, Neilson JR. Differences between acute and chronic stress granules, and how these differences may impact function in human disease. Biochem Pharmacol. 2019;162:123–131. doi: 10.1016/j.bcp.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guzikowski AR, Chen YS, Zid BM. Stress-induced mRNP granules: form and function of processing bodies and stress granules. Wiley Interdiscip Rev RNA. 2019;10:e1524. doi: 10.1002/wrna.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhee H-W, Zou P, Udeshi ND, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen C-L, Perrimon N. Proximity-dependent labeling methods for proteomic profiling in living cells. Wiley Interdiscip Rev Dev Biol. 2017 doi: 10.1002/WDEV.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Markmiller S, Soltanieh S, Server KL, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Youn J-Y, Dunham WH, Hong SJ, et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell. 2018;69:517–532.e11. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 70.Khong A, Matheny T, Jain S, et al. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell. 2017;68:808–820.e5. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Namkoong S, Ho A, Woo YM, et al. Systematic characterization of stress-induced RNA granulation. Mol Cell. 2018;70:175–187.e8. doi: 10.1016/j.molcel.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol Cell Biol. 1994;14:3822–3833. doi: 10.1128/MCB.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davuluri RV. CART classification of human 5′ UTR sequences. Genome Res. 2000;10:1807–1816. doi: 10.1101/gr.GR-1460R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rayman JB, Kandel ER. TIA-1 is a functional prion-like protein. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kawakami A, Tian Q, Duan X, et al. Identification and functional characterization of a TIA-1-related nucleolysin. Proc Natl Acad Sci USA. 1992;89:8681–8685. doi: 10.1073/pnas.89.18.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 77.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 78.Gueydan C, Droogmans L, Chalon P, et al. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J Biol Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 79.Damgaard CK, Lykke-Andersen J. Translational coregulation of 5’TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25:2057–2068. doi: 10.1101/gad.17355911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 81.Bounedjah O, Desforges B, Wu T-D, et al. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 2014;42:8678–8691. doi: 10.1093/nar/gku582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ivanov P, Kedersha N, Anderson P. Stress puts TIA on TOP. Genes Dev. 2011;25:2119–2124. doi: 10.1101/gad.17838411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Danan C, Manickavel S, Hafner M. PAR-CLIP: a method for transcriptome-wide identification of RNA binding protein interaction sites. Methods Mol Biol. 2016;1358:153–173. doi: 10.1007/978-1-4939-3067-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee ASY, Kranzusch PJ, Cate JHD. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holt CE, Martin KC, Schuman EM. Local translation in neurons: visualization and function. Nat Struct Mol Biol. 2019;26:557–566. doi: 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]
- 86.Rangaraju V, tom Dieck S, Schuman EM. Local translation in neuronal compartments: how local is local? EMBO Rep. 2017;18:693–711. doi: 10.15252/embr.201744045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shiina N, Shinkura K, Tokunaga M. A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J Neurosci. 2005;25:4420–4434. doi: 10.1523/JNEUROSCI.0382-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barbee SA, Estes PS, Cziko A-M, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/J.NEURON.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elbaum-Garfinkle S. Matter over mind: liquid phase separation and neurodegeneration. J Biol Chem. 2019;294:7160–7168. doi: 10.1074/jbc.REV118.001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aguzzi A, O’Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 91.Bourdenx M, Koulakiotis NS, Sanoudou D, et al. Protein aggregation and neurodegeneration in prototypical neurodegenerative diseases: examples of amyloidopathies, tauopathies and synucleinopathies. Prog Neurobiol. 2017;155:171–193. doi: 10.1016/j.pneurobio.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 92.Orr HT, Chung M, Banfi S, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 93.Banfi S, Servadio A, Chung MY, et al. Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nat Genet. 1994;7:513–520. doi: 10.1038/ng0894-513. [DOI] [PubMed] [Google Scholar]
- 94.Mackenzie IR, Nicholson AM, Sarkar M, et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95:808–816.e9. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim HJ, Kim NC, Wang Y-D, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Svetoni F, Frisone P, Paronetto MP. Role of FET proteins in neurodegenerative disorders. RNA Biol. 2016;13:1089–1102. doi: 10.1080/15476286.2016.1211225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Osborne RJ, Lin X, Welle S, et al. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Timchenko NA, Cai ZJ, Welm AL, et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 99.Ward AJ, Rimer M, Killian JM, et al. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kanadia RN, Shin J, Yuan Y, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sofola OA, Jin P, Qin Y, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jin P, Duan R, Qurashi A, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vidal RL, Matus S, Bargsted L, Hetz C. Targeting autophagy in neurodegenerative diseases. Trends Pharmacol Sci. 2014;35:583–591. doi: 10.1016/j.tips.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 104.Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 106.Wong YC, Holzbaur ELF. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci USA. 2014;111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schwab C, Yu S, McGeer EG, McGeer PL. Optineurin in Huntington’s disease intranuclear inclusions. Neurosci Lett. 2012;506:149–154. doi: 10.1016/J.NEULET.2011.10.070. [DOI] [PubMed] [Google Scholar]
- 108.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/J.CELL.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Coady TH, Manley JL. ALS mutations in TLS/FUS disrupt target gene expression. Genes Dev. 2015;29:1696–1706. doi: 10.1101/gad.267286.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qiu H, Lee S, Shang Y, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Investig. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ryu H-H, Jun M-H, Min K-J, et al. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging. 2014;35:2822–2831. doi: 10.1016/j.neurobiolaging.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 112.Lee J-A. Autophagy manages disease-associated stress granules. Oncotarget. 2015;6:30421. doi: 10.18632/oncotarget.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dormann D, Haass C. TDP-43 and FUS: a nuclear affair. Trends Neurosci. 2011;34:339–348. doi: 10.1016/j.tins.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 116.Acosta JR, Goldsbury C, Winnick C, et al. Mutant human FUS is ubiquitously mislocalized and generates persistent stress granules in primary cultured transgenic zebrafish cells. PLoS One. 2014;9:e90572. doi: 10.1371/journal.pone.0090572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barmada SJ, Skibinski G, Korb E, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee S, Levin M. Novel somatic single nucleotide variants within the RNA binding protein hnRNP A1 in multiple sclerosis patients. F1000Research. 2014;3:132. doi: 10.12688/f1000research.4436.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang K, Daigle JG, Cunningham KM, et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell. 2018;173:958–971.e17. doi: 10.1016/j.cell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo L, Kim HJ, Wang H, et al. Nuclear-import receptors reverse aberrant phase transitions of rna-binding proteins with prion-like domains. Cell. 2018;173:677–692.e20. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Solomon DA, Stepto A, Au WH, et al. A feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-α mediates C9orf72-related neurodegeneration. Brain. 2018;141:2908–2924. doi: 10.1093/brain/awy241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Steyaert J, Scheveneels W, Vanneste J, et al. FUS-induced neurotoxicity in Drosophila is prevented by downregulating nucleocytoplasmic transport proteins. Hum Mol Genet. 2018;27:4103–4116. doi: 10.1093/hmg/ddy303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khalil B, Morderer D, Price PL, et al. mRNP assembly, axonal transport, and local translation in neurodegenerative diseases. Brain Res. 2018;1693:75–91. doi: 10.1016/j.brainres.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Burguete AS, Almeida S, Gao F-B, et al. GGGGCC microsatellite RNA is neuritically localized, induces branching defects, and perturbs transport granule function. Elife. 2015;4:e08881. doi: 10.7554/eLife.08881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Narayanan RK, Mangelsdorf M, Panwar A, et al. Identification of RNA bound to the TDP-43 ribonucleoprotein complex in the adult mouse brain. Amyotroph Lateral Scler Front Degener. 2013;14:252–260. doi: 10.3109/21678421.2012.734520. [DOI] [PubMed] [Google Scholar]
- 126.Sephton CF, Cenik C, Kucukural A, et al. Identification of neuronal RNA Targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Endo R, Takashima N, Nekooki-Machida Y, et al. TAR DNA-binding protein 43 and disrupted in schizophrenia 1 coaggregation disrupts dendritic local translation and mental function in frontotemporal lobar degeneration. Biol Psychiatry. 2018;84:509–521. doi: 10.1016/j.biopsych.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Coyne AN, Siddegowda BB, Estes PS, et al. Futsch/MAP1B mRNA is a translational target of TDP-43 and is neuroprotective in a drosophila model of amyotrophic lateral sclerosis. J Neurosci. 2014;34:15962–15974. doi: 10.1523/JNEUROSCI.2526-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alami NH, Smith RB, Carrasco MA, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cohen TJ, Hwang AW, Restrepo CR, et al. An acetylation switch controls TDP-43 function and aggregation propensity. Nat Commun. 2015;6:5845. doi: 10.1038/NCOMMS6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen Y, Cohen TJ. Aggregation of the nucleic acid-binding protein TDP-43 occurs via distinct routes that are coordinated with stress granule formation. J Biol Chem. 2019;294:3696–3706. doi: 10.1074/jbc.RA118.006351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu M, Zhu L, Liu J, et al. Characterization of β-domains in C-terminal fragments of TDP-43 by scanning tunneling microscopy. J Struct Biol. 2013;181:11–16. doi: 10.1016/j.jsb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vanderweyde T, Yu H, Varnum M, et al. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J Neurosci. 2012;32:8270–8283. doi: 10.1523/JNEUROSCI.1592-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Binder LI, Frankfurter A, Rebhun LI. The distribution of tau in the mammalian central nervous system. J Cell Biol. 1985;101:1371–1378. doi: 10.1083/jcb.101.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hoover BR, Reed MN, Su J, et al. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron. 2010;68:1067–1081. doi: 10.1016/j.neuron.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Apicco DJ, Ash PEA, Maziuk B, et al. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat Neurosci. 2018;21:72–80. doi: 10.1038/s41593-017-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Trojanowski JQ, Schuck T, Schmidt ML, Lee VM. Distribution of tau proteins in the normal human central and peripheral nervous system. J Histochem Cytochem. 1989;37:209–215. doi: 10.1177/37.2.2492045. [DOI] [PubMed] [Google Scholar]
- 139.Kobayashi S, Tanaka T, Soeda Y, et al. Local somatodendritic translation and hyperphosphorylation of tau protein triggered by AMPA and NMDA receptor stimulation. EBioMedicine. 2017;20:120–126. doi: 10.1016/J.EBIOM.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Danysz W, Parsons CG. Alzheimer’s disease, β-amyloid, glutamate, NMDA receptors and memantine-searching for the connections. Br J Pharmacol. 2012;167:324–352. doi: 10.1111/j.1476-5381.2012.02057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vanderweyde T, Apicco DJ, Youmans-Kidder K, et al. Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 2016;15:1455–1466. doi: 10.1016/j.celrep.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Silva JM, Rodrigues S, Sampaio-Marques B, et al. Dysregulation of autophagy and stress granule-related proteins in stress-driven Tau pathology. Cell Death Differ. 2019;26:1411–1427. doi: 10.1038/s41418-018-0217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gunawardana CG, Mehrabian M, Wang X, et al. The human tau interactome: binding to the ribonucleoproteome, and impaired binding of the proline-to-leucine mutant at position 301 (P301L) to chaperones and the proteasome. Mol Cell Proteom. 2015;14:3000–3014. doi: 10.1074/mcp.M115.050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maziuk BF, Apicco DJ, Cruz AL, et al. RNA binding proteins co-localize with small tau inclusions in tauopathy. Acta Neuropathol Commun. 2018;6:71. doi: 10.1186/s40478-018-0574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Meier S, Bell M, Lyons DN, et al. Pathological tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. J Neurosci. 2016;36:1001–1007. doi: 10.1523/JNEUROSCI.3029-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lyons SM, Fay MM, Akiyama Y, et al. RNA biology of angiogenin: current state and perspectives. RNA Biol. 2017;14:171–178. doi: 10.1080/15476286.2016.1272746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ivanov P, Emara MM, Villen J, et al. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ivanov P, O’Day E, Emara MM, et al. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci. 2014;111:18201–18206. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lyons SM, Achorn C, Kedersha NL, et al. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 2016;44:6949–6960. doi: 10.1093/nar/gkw418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Suryanarayana T, Uppala JK, Garapati UK. Interaction of cytochrome c with tRNA and other polynucleotides. Mol Biol Rep. 2012;39:9187–9191. doi: 10.1007/s11033-012-1791-9. [DOI] [PubMed] [Google Scholar]
- 153.Saikia M, Jobava R, Parisien M, et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol Cell Biol. 2014;34:2450–2463. doi: 10.1128/MCB.00136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Steidinger TU, Standaert DG, Yacoubian TA. A neuroprotective role for angiogenin in models of Parkinson’s disease. J Neurochem. 2011;116:334–341. doi: 10.1111/j.1471-4159.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gallo J-M, Jin P, Thornton CA, et al. The role of RNA and RNA processing in neurodegeneration. J Neurosci. 2005;25:10372–10375. doi: 10.1523/JNEUROSCI.3453-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Todd PK, Paulson HL. RNA mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2009 doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Matsuura T, Yamagata T, Burgess DL, et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- 158.White MC, Gao R, Xu W, et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCδ to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McLaughlin BA, Spencer C, Eberwine J. CAG trinucleotide RNA repeats interact with RNA-binding proteins. Am J Hum Genet. 1996;59:561–569. [PMC free article] [PubMed] [Google Scholar]
- 160.Li L-B, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Warrick JM, Paulson HL, Gray-Board GL, et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 162.de Mezer M, Wojciechowska M, Napierala M, et al. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fay MM, Anderson PJ, Ivanov P. ALS/FTD-associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 2017;21:3573–3584. doi: 10.1016/j.celrep.2017.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Van Mossevelde S, van der Zee J, Cruts M, Van Broeckhoven C. Relationship between C9orf72 repeat size and clinical phenotype. Curr Opin Genet Dev. 2017;44:117–124. doi: 10.1016/j.gde.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 166.Zu T, Liu Y, Banez-Coronel M, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mori K, Arzberger T, Grässer FA, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 168.Gendron TF, Bieniek KF, Zhang Y-J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tabet R, Schaeffer L, Freyermuth F, et al. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat Commun. 2018;9:152. doi: 10.1038/s41467-017-02643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Balendra R, Isaacs AM. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat Rev Neurol. 2018;14:544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.DeJesus-Hernandez M, Finch NA, Wang X, et al. In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers. Acta Neuropathol. 2017;134:255–269. doi: 10.1007/s00401-017-1725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hartmann H, Hornburg D, Czuppa M, et al. Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity. Life Sci Alliance. 2018;1:e201800070. doi: 10.26508/lsa.201800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Moens TG, Niccoli T, Wilson KM, et al. C9orf72 arginine-rich dipeptide proteins interact with ribosomal proteins in vivo to induce a toxic translational arrest that is rescued by eIF1A. Acta Neuropathol. 2019;137:487–500. doi: 10.1007/s00401-018-1946-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Babić Leko M, Župunski V, Kirincich J, et al. Molecular mechanisms of neurodegeneration related to C9orf72 hexanucleotide repeat expansion. Behav Neurol. 2019;2019:1–18. doi: 10.1155/2019/2909168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Becker LA, Huang B, Bieri G, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ma T, Trinh MA, Wexler AJ, et al. Suppression of eIF2α kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat Neurosci. 2013;16:1299–1305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Moreno JA, Radford H, Peretti D, et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shen H-Y, He J-C, Wang Y, et al. Geldanamycin induces heat shock protein 70 and protects against MPTP-induced dopaminergic neurotoxicity in mice. J Biol Chem. 2005;280:39962–39969. doi: 10.1074/jbc.M505524200. [DOI] [PubMed] [Google Scholar]
- 179.Shorter J. Designer protein disaggregases to counter neurodegenerative disease. Curr Opin Genet Dev. 2017;44:1–8. doi: 10.1016/j.gde.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Menzies FM, Garcia-Arencibia M, Imarisio S, et al. Calpain inhibition mediates autophagy-dependent protection against polyglutamine toxicity. Cell Death Differ. 2015;22:433–444. doi: 10.1038/cdd.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sarkar S, Krishna G, Imarisio S, et al. A rational mechanism for combination treatment of Huntington’s disease using lithium and rapamycin. Hum Mol Genet. 2008;17:170–178. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]
- 182.Staats KA, Hernandez S, Schönefeldt S, et al. Rapamycin increases survival in ALS mice lacking mature lymphocytes. Mol Neurodegener. 2013;8:31. doi: 10.1186/1750-1326-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Uddin MS, Stachowiak A, Al MA, et al. Autophagy and Alzheimer’s disease: from molecular mechanisms to therapeutic implications. Front Aging Neurosci. 2018;10:04. doi: 10.3389/fnagi.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Caccamo A, Maldonado MA, Majumder S, et al. Naturally secreted amyloid-β increases mammalian target of rapamycin (mTOR) activity via a PRAS40-mediated mechanism. J Biol Chem. 2011;286:8924–8932. doi: 10.1074/jbc.M110.180638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Caccamo A, Majumder S, Richardson A, et al. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: effects on cognitive impairments. J Biol Chem. 2010;285:13107–13120. doi: 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Spilman P, Podlutskaya N, Hart MJ, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-β levels in a mouse model of Alzheimer’s disease. PLoS One. 2010;5:e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Investig. 2011;121:1231–1241. doi: 10.1172/JCI44145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lee H-J, Yoon Y-S, Lee S-J. Mechanism of neuroprotection by trehalose: controversy surrounding autophagy induction. Cell Death Dis. 2018;9:712. doi: 10.1038/s41419-018-0749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Castillo K, Nassif M, Valenzuela V, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–1320. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 190.Rodríguez-Navarro JA, Rodríguez L, Casarejos MJ, et al. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis. 2010;39:423–438. doi: 10.1016/j.nbd.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 191.Du J, Liang Y, Xu F, et al. Trehalose rescues Alzheimer’s disease phenotypes in APP/PS1 transgenic mice. J Pharm Pharmacol. 2013;65:1753–1756. doi: 10.1111/jphp.12108. [DOI] [PubMed] [Google Scholar]
- 192.Sarkar S, Davies JE, Huang Z, et al. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 193.Wobst HJ, Wesolowski SS, Chadchankar J, et al. Cytoplasmic relocalization of TAR DNA-binding protein 43 is not sufficient to reproduce cellular pathologies associated with ALS in vitro. Front Mol Neurosci. 2017;10:46. doi: 10.3389/fnmol.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pesiridis GS, Lee VM-Y, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kim SH, Shi Y, Hanson KA, et al. Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J Biol Chem. 2009;284:8083–8092. doi: 10.1074/jbc.M808064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hans F, Eckert M, von Zweydorf F, et al. Identification and characterization of ubiquitinylation sites in TAR DNA-binding protein of 43 kDa (TDP-43) J Biol Chem. 2018;293:16083–16099. doi: 10.1074/jbc.RA118.003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jiang L-L, Zhao J, Yin X-F, et al. Two mutations G335D and Q343R within the amyloidogenic core region of TDP-43 influence its aggregation and inclusion formation. Sci Rep. 2016;6:23928. doi: 10.1038/srep23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schmidt HB, Rohatgi R. In vivo formation of vacuolated multi-phase compartments lacking membranes. Cell Rep. 2016;16:1228–1236. doi: 10.1016/j.celrep.2016.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.McDonald KK, Aulas A, Destroismaisons L, et al. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- 200.Patel A, Lee HO, Jawerth L, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/J.CELL.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 201.Bosco DA, Lemay N, Ko HK, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shelkovnikova TA, Robinson HK, Connor-Robson N, Buchman VL. Recruitment into stress granules prevents irreversible aggregation of FUS protein mislocalized to the cytoplasm. Cell Cycle. 2013;12:3194–3202. doi: 10.4161/cc.26241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lenzi J, De Santis R, de Turris V, et al. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis Model Mech. 2015;8:755–766. doi: 10.1242/dmm.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gui X, Luo F, Li Y, et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun. 2019;10:2006. doi: 10.1038/s41467-019-09902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shorter J, Taylor JP. Disease mutations in the prion-like domains of hnRNPA1 and hnRNPA2/B1 introduce potent steric zippers that drive excess RNP granule assembly. Rare Dis. 2013;1:e25200. doi: 10.4161/rdis.25200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kapeli K, Martinez FJ, Yeo GW. Genetic mutations in RNA-binding proteins and their roles in ALS. Hum Genet. 2017;136:1193–1214. doi: 10.1007/s00439-017-1830-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Couthouis J, Hart MP, Erion R, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2899–2911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Couthouis J, Raphael AR, Daneshjou R, Gitler AD. Targeted exon capture and sequencing in sporadic amyotrophic lateral sclerosis. PLoS Genet. 2014;10:e1004704. doi: 10.1371/journal.pgen.1004704. [DOI] [PMC free article] [PubMed] [Google Scholar]