Abstract
Repeat-associated non-AUG translation of expanded CGG repeats (CGG RAN) from the FMR1 5’ UTR produces toxic proteins that contribute to neurodegeneration in Fragile X-associated Tremor/Ataxia Syndrome (FXTAS). Here we describe how unexpanded CGG repeats and their translation play conserved roles in regulating FMRP synthesis. In neurons, CGG RAN acts as an inhibitory upstream open reading frame to suppress basal FMRP production. Activation of mGluR5 receptors enhances FMRP synthesis. This enhancement requires both the CGG repeat and CGG RAN initiation sites. Using non-cleaving antisense oligonucleotides (ASOs), we selectively blocked RAN translation. This ASO blockade enhanced endogenous human neuronal FMRP expression. In human and rodent neurons, RAN blocking ASOs suppressed repeat toxicity and prolonged survival. These findings delineate a native function for CGG repeats and RAN translation in regulating basal and activity-dependent FMRP synthesis and demonstrate the therapeutic potential of modulating CGG RAN translation in fragile X-associated disorders.
Introduction:
Nucleotide repeat expansions (NREs) cause more than 40 neurological disorders1,2. Large transcribed repeats outside of AUG-initiated open reading frames (ORFs) can elicit neuronal dysfunction as RNA by binding to and sequestering specific RNA-binding proteins1. NREs outside of ORFs are also sometimes translated via repeat associated non-AUG (RAN) translation to produce toxic proteins that accumulate in patient neurons and contribute to neurodegeneration in model systems1,3,4.
CGG trinucleotide repeat expansions in the 5’ UTR of the Fragile X gene, FMR1 were the first published disease causing NRE5. Repeat expansions from the normal size of ~30 CGGs to greater than 200 repeats (a “full mutation”) transcriptionally silence the FMR1 locus and lead to loss of the fragile X protein, FMRP6. This causes Fragile X Syndrome (FXS), the most common monogenic cause of autism and intellectual disability. In contrast, “Premutation” (55–200) sized CGG repeat expansions cause Fragile X-associated Tremor/Ataxia Syndrome (FXTAS), a clinically distinct age-related neurodegenerative disorder characterized by gait difficulties, action tremor, and dementia7. Premutation-length repeats are transcribed into mRNA at high levels and induce ubiquitinated intranuclear neuronal inclusions and atrophy in FXTAS patient brains8,9. Expanded CGG repeats exhibit somatic mosaicism in both repeat size and methylation, leading sometimes to transcription of full mutation sized repeats9. These unmethylated full mutation (UFM) repeats impair FMRP synthesis and can trigger symptoms typically seen in FXS9–11. UFM cases can also develop neuronal inclusions and FXTAS symptoms in later life11,12. FXS, UFM cases, FXTAS, and Fragile X-associated premature ovarian insufficiency (FXPOI) are collectively referred to as Fragile X-associated disorders.
Originally described at CAG repeats in the SCA8 locus, RAN translation occurs in multiple repeat expansion disorders, including C9orf72-associated amyotrophic lateral sclerosis and Huntington’s disease4. Expanded CGG repeats in FMR1 mRNA are translated in the absence of an AUG start codon3. This non-AUG initiated translation (CGG RAN) occurs in all three potential reading frames in reporter systems, producing homopolymeric poly-glycine (+1 reading frame relative to the AUG of FMRP, +1CGG RAN, FMRpolyG), poly-alanine (+2 reading frame, +2CGG RAN, FMRpolyA), and poly-arginine (+0 reading frame, +0CGG RAN, FMRpolyR) products3,13. Translation of the most abundant RAN product, FMRpolyG, creates an upstream open reading frame (uORF) that terminates after the AUG start codon of FMRP3. FMRpolyG accumulates in intranuclear neuronal inclusions in FXTAS patients3,14,15. CGG RAN translation in general and FMRpolyG production in particular is required for CGG repeat associated toxicity in most model systems. Enhancing FMRpolyG production exacerbates CGG repeat associated toxicity3 and preventing FMRpolyG production markedly suppresses CGG repeat toxicity in Drosophila and transgenic mice3,14.
Despite their roles in disease pathogenesis, we know little about the potential normal functions of transcribed repetitive elements. Simple tandem repeats make up a significant fraction of the human transcriptome, with thousands of microsatellites residing within coding regions, introns or 5’ and 3’ UTRs of nervous system expressed genes2,16. Microsatellite repeats influence both gene expression and function2 and their instability makes them candidates for rapid evolutionary selection16. Whether repetitive elements influence neuronal function and plasticity is largely unexplored.
The FMR1 CGG repeat is conserved among mammals and selectively expanded in primates17. Outside of the repeat itself, the 5’ leader sequence is almost invariant in humans18. Intriguingly, CGG RAN occurs readily from reporters at the normal repeat size in the human population (25–30 repeats)3,13. CGG RAN initiation sites are detectable by ribosome profiling in both humans and mice in the absence of repeat expansion3,19,20. Based on these features, we evaluated whether CGG repeats and their translation might regulate FMRP synthesis. Here we show that CGG RAN translation from normal repeat sizes function as a regulatory uORF to control basal and activity-dependent FMRP synthesis. Using reporters and non-cleaving antisense oligonucleotides (ASOs) to sterically hinder CGG RAN initiation, we demonstrate that blocking CGG RAN or removing the CGG repeat enhances endogenous FMRP synthesis in human cells and rodent neurons. CGG RAN blocking ASOs suppressed RAN translation and neurotoxicity while enhancing FMRP synthesis in patient neurons with a transcribed, full mutation-sized CGG repeat. This enhanced rate of basal FMRP production precludes further upregulation of FMRP synthesis in response to metabotropic glutamate receptor activation, a critical step in regulated local protein synthesis. Overall, these findings define a new physiological role for CGG RAN and CGG repeats, and suggest a potential therapeutic strategy based on selective inhibition of CGG RAN translation in fragile X disorders.
Results:
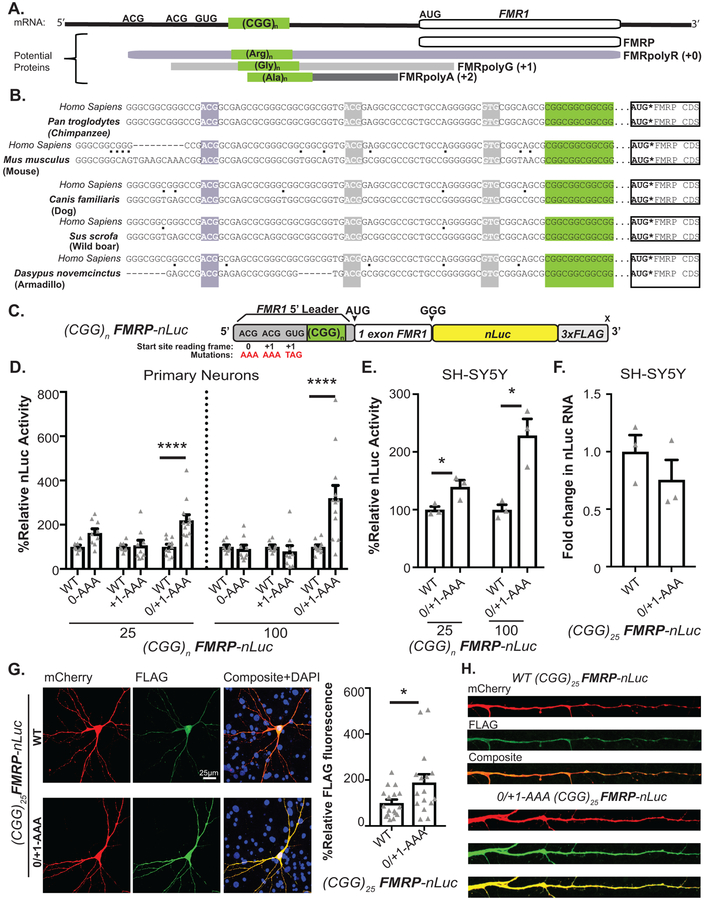
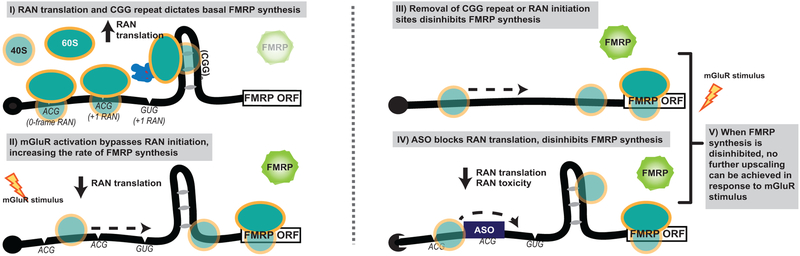
CGG RAN initiation occurs predominantly at one of three specific near-cognate codons just upstream of the repeat13. +0CGG RAN initiates at an ACG codon 60 nucleotides upstream of the repeat to generate an N-terminal polyarginine extension on FMRP (Fig 1A, mauve). This product is not readily detected endogenously or from reporters3,13. +1CGG RAN initiates at one of two near-cognate codons (ACG or GUG) 33 and 9 nucleotides upstream of the repeat and terminates after the AUG start codon of FMRP, creating an overlapping uORF3,14(Fig 1A, gray). All three CGG RAN start sites are conserved in reading frame relative to the CGG repeat and to the AUG of the FMRP ORF in all placental mammals (Fig 1B)14. The mean vertebrate phyloP score of the +0CGG RAN ACG initiation codon is 0.39, and scores for the +1CGG RAN ACG and GUG initiation codons are 1.60 and 1.48, respectively. The average phyloP score for the FMR1 5’ UTR is 1.23 (excluding the CGG repeat), whereas the mean phyloP score for the 5’ UTR of the FMR1 homolog FXR1 is 0.49. This high degree of conservation for each initiation site and their relationships to FMRP suggests potentially native functions for CGG RAN17,18.
Figure 1: CGG RAN translation impedes FMRP translation in neurons.
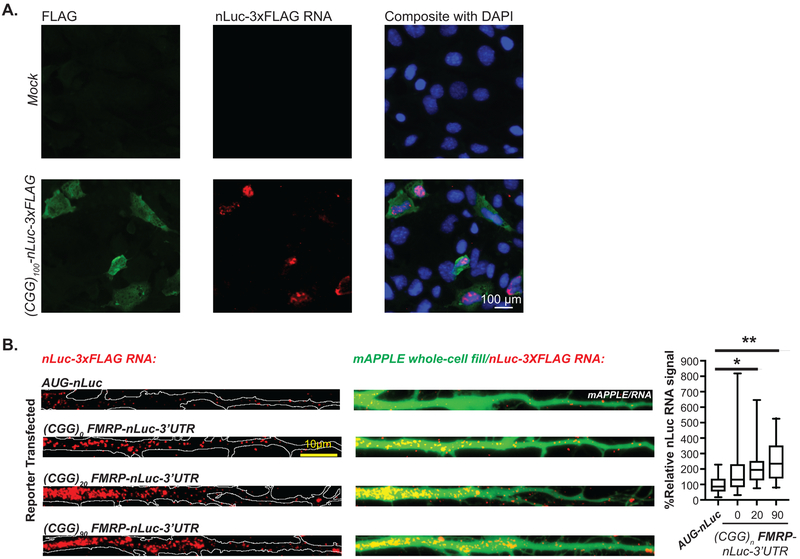
A) FMR1 mRNA support synthesis of 4 different proteins: Initiation at the AUG codon below the 5’ leader is used to produce FMRP (white). RAN translation initiates at three near-AUG codons in the 5’ leader generate FMRpolyR (+0, creating an N-terminal extension on FMRP (slate), and FMRpolyG (+1, creating an overlapping uORF(grey). Initiation within the repeat itself generates FMRpolyA (+2, creating a uORF that terminates prior to AUG of FMRP, charcoal). B) Interspecies conservation of start sites for +0CGG RAN (slate) and +1CGG RAN (grey) and their reading frames relative to CGG repeat (green) and FMRP ORF(white). C) FMRP reporter schematic: Nanoluciferase (nLuc, yellow) start codon is mutated to GGG and fused in-frame with the first coding exon of FMR1 (white). Mutation of one (0-AAA or +1-AAA) or all (0/+1-AAA) initiation codons in the 5’ leader (gray) indicated. D) (CGG)n FMRP-nLuc activity with or without mutated CGG RAN initiation sites in rat hippocampal neurons (CGG25: WT n=8, +0-AAA n=9, WT n=8, +1-AAA n=9, WT n=11, +0/+1-AAA n=12, p=0.001; CGG100: WT n=8, +0-AAA n=9, WT n=8, +1-AAA n=9, WT n=11, +0/+1-AAA n=12, p<0.001). E) FMRP-nLuc reporters in SH-SY5Y cells (n=3/group; CGG25: p=0.04, CGG100: p=0.01). F) FMRP reporter mRNA in SH-SY5Y cells by RT-qPCR (n=3; p=0.15). G) Representative ICC images from 3 independent experiments for (CGG)25 FMRP-nLuc-Flag (green) in neurons co-expressing mCherry (red). Right: Signal from indicated (CGG)25 reporters, normalized to mCherry (WT: n=17, 0/+1AAA: n=16, where “n” is the mean CTCF signal from 5 neurons; p=0.02). H) Representative straightened dendrites expressing indicated reporters from three independent experiments. Panel D: Two-way ANOVA with multiple comparisons within repeat groups. Panels E, F: Two sided unpaired Student’s t-test. Panel H: Two sided Mann Whitney U test. *p<0.05, ****p<0.001. Graphs are mean +/− S.E.M.
CGG RAN impairs neuronal FMRP synthesis.
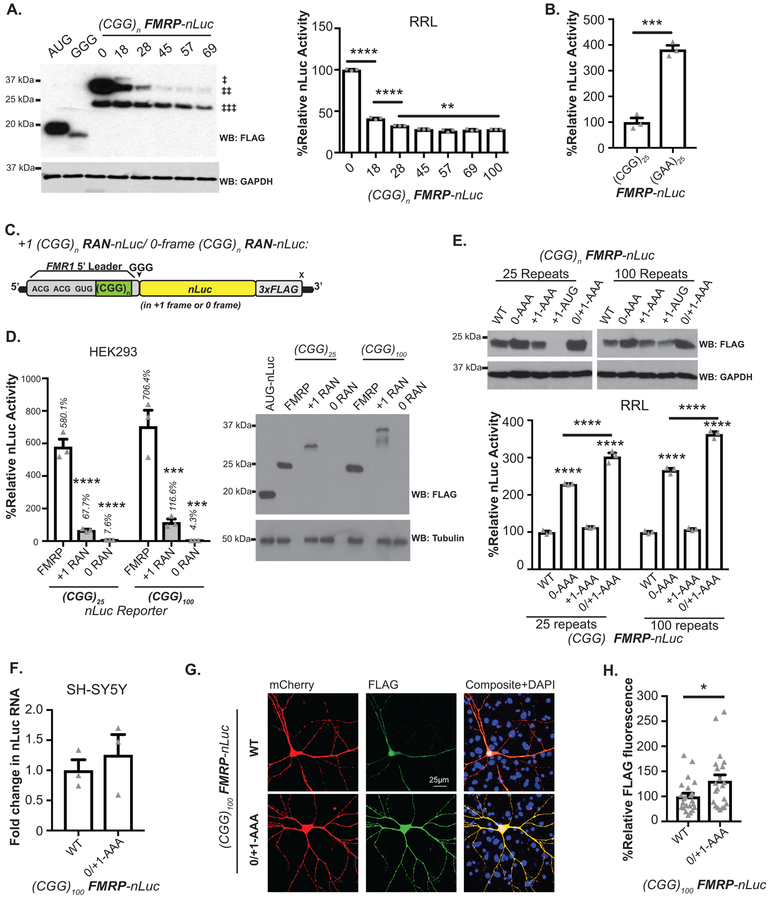
Overlapping uORFs can impair translation of downstream open reading frames21. To test whether CGG RAN and CGG repeats impair FMRP synthesis, we generated a series of FMRP reporters that included the full 5’ UTR and first exon of FMR1 fused to a mutant form of nanoluciferase (nLuc) lacking a start codon. A 3XFLAG sequence allowed for detection by western blot (Fig. 1C). FMRP reporter synthesis decreased with increasing CGG repeat number in rabbit reticulocyte lysates (RRL) (Extended Data 1A)22,23. In contrast, replacing the CGG repeat with an unstructured GAA repeat increased FMRP reporter abundance (Extended Data 1B).
CGG RAN translation requires a 5’ M7G cap on the mRNA, the cap-binding protein eIF4E and the RNA helicase eIF4A, consistent with a cap-dependent and ribosomal scanning dependent initiation mechanism13. CGG RAN translation is also typically more efficient at larger CGG repeat sizes13. We therefore compared FMRP and CGG RAN translation reporter efficiencies at normal and expanded repeats (Extended Data 1C)13. FMRpolyG reporter production was 11.7% as efficient at 25 CGGs and 16.7% as efficient at 90 CGG repeats as FMRP reporter production (Extended Data 1C). In contrast, +0CGG RAN products were detected in vitro in RRL, but +0CGG RAN signal was less than 1% of the FMRP reporter signal and was not detectable by immunoblot in HEK293 cells (Extended Data 1D)13.
To determine the effect of CGG RAN events on FMRP synthesis in neurons, we serially or simultaneously mutated the +0CGG RAN (ACG) and +1CGG RAN (ACG and GUG) initiation sites in FMRP reporters (Fig 1C). Blocking CGG RAN initiation in the +0- or +1 frame (0-AAA or +1-AAA) alone had no effect on FMRP reporter synthesis (Fig 1D). However, mutating all three sites (0/+1-AAA) led to a synergistic increase in FMRP at both normal and expanded repeats in rat hippocampal neurons, human SH-SY5Y neuroblastoma cells, and in RRL (Fig 1D–E, Extended Data 1E). The increase in FMRP was not due to altered mRNA expression (Fig 1F, Extended Data 1F). These findings are consistent with previous work suggesting that CGG repeats can impair FMRP translation14,22,23.
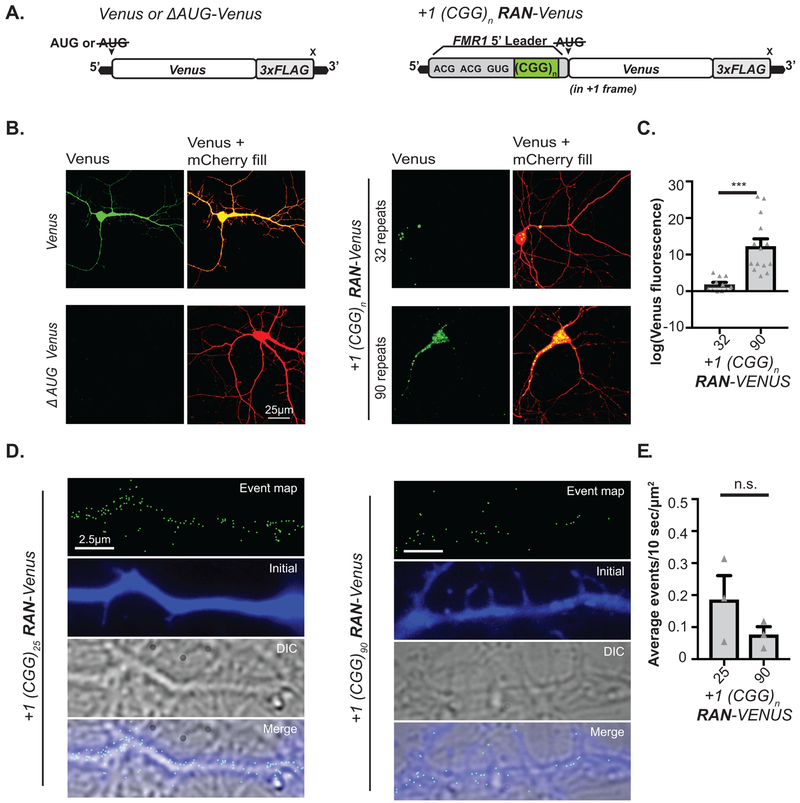
Mutation of CGG RAN initiation sites enhanced FMRP reporter expression in both neuronal soma (Fig 1G, Extended Data 1G–H) and processes (Fig 1H) at normal and expanded repeats. To determine whether RAN translation happens in neuronal processes, we first monitored +1CGG RAN translation specific reporters by immunocytochemistry (ICC). CGG RAN products accumulated in aggregates in neuronal soma and dendrites, with higher expression of reporters with larger repeats (Extended Data 2A–C). To more directly test whether RAN translation occurs in neuronal processes, we performed single-molecule imaging to detect translation events in distal dendrites. We readily detected isolated +1 CGG RAN events within dendrites at both normal and expanded repeats (Extended Data 2D–E).
The CGG repeat is required for mGluR-dependent enhancement of FMRP synthesis.
FMRP is an RNA-binding protein that interacts with and suppresses the translation of ~4% of brain mRNAs, including an enriched fraction of synaptic transcripts from genes associated with autism24. Upon activation of group I metabotropic glutamate receptors (mGluRs), FMRP is dephosphorylated and rapidly degraded, allowing for local translation of FMRP-associated transcripts (Fig 2A)25,26. FMR1 mRNA itself is translated in response to mGluR activation, creating a feedback loop that temporally limits activity-dependent translation (Fig 2A)25,27–29,. Disruption of this regulatory loop alters mGluR dependent Long Term Depression28,30,31. We therefore evaluated whether CGG repeats and CGG RAN are required for mGluR-dependent upregulation of FMRP synthesis.
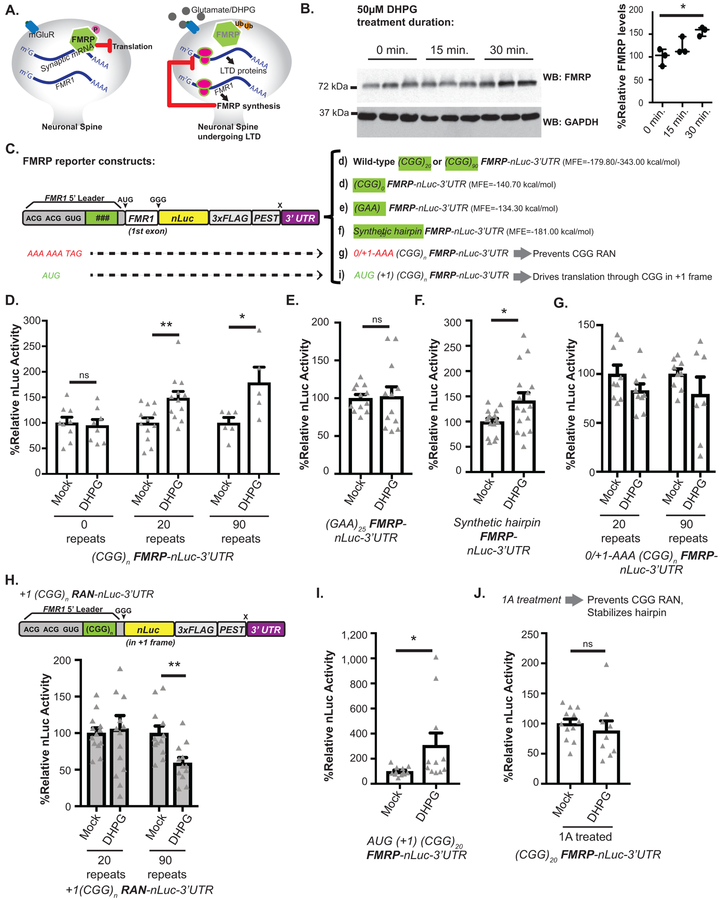
Figure 2: mGluR dependent FMRP synthesis requires RAN translation and CGG repeat.
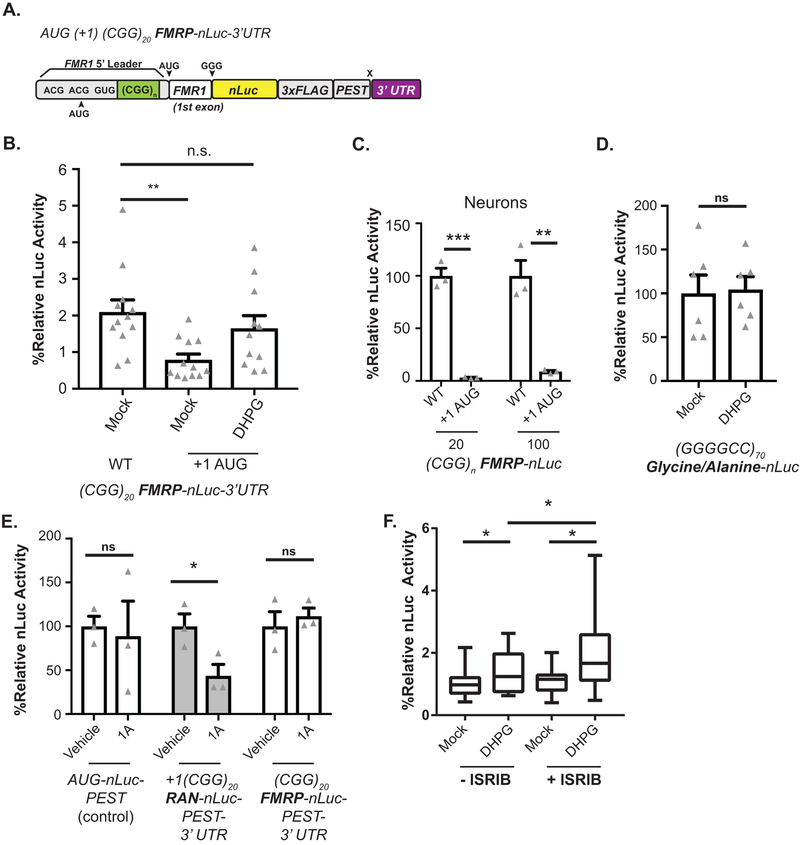
A) FMRP inhibits translation. Upon mGluR-stimulation, FMRP is ubiquitinated and degraded, which enhances translation of bound transcripts. mGluR activation also increases FMR1 mRNA translation. This new FMRP turns off local translation to temporally constrain mGluR effects. B) Left: Endogenous FMRP in rat hippocampal neurons treated with DHPG for indicated times. Right: Quantification of FMRP, normalized to GAPDH (n=3, p=0.016). C) FMRP reporters with indicated mutations to repeat (green) or RAN initiation sites. The minimum free energy (MFE) of each 5’ leader is noted. D) DHPG effect on FMRP reporters with different CGG repeats in rat hippocampal neurons (CGG0: mock n=12, DHPG n=11, p=0.34; CGG25: mock n=12, DHPG n=12, p=0.007; CGG100: mock n=5, DHPG n=6, p=0.027). E) DHPG effect on (GAA)25 FMRP-nLuc reporter expression (n=12, p=0.86). F) DHPG effect on synthetic hairpin-FMRP-nLuc reporter expression (n=16, p=0.021). G) DHPG effect on FMRP-nLuc reporters with mutated CGG RAN initiation codons (0/+1-AAA) (n=9; CGG20: p=0.16, CGG90: p=0.2). H) DHPG effect on +1 CGG RAN reporters (CGG20: mock n=13, DHPG n=14, p=0.16; CGG90: mock n=12, DHPG n=12, p=0.20). I) DHPG effect on FMRP-nLuc reporter with AUG initiation codon in place of ACG +1CGG RAN initiation codon (mock n=12, DHPG n=11, p=0.039). J) Effect of 1A pretreatment on (CGG)20 FMRP-nLuc reporter expression in neurons treated with vehicle (n=12) or DHPG (n=9, p=0.48). For all, white bars: FMRP reporter; grey bars: RAN reporter. Panel B: One-way ANOVA with correction for multiple comparisons. Panels D, E, F, G, I, J: Two sided Student t-test. Panel H: Two sided Mann Whitney U test. *p<0.05, **p<0.01. Graphs are mean +/− S.E.M.
Application of the mGluR1/5 agonist s-dihydrophenylglycine (DHPG) significantly increased endogenous FMRP expression in cultured hippocampal rat neurons (Fig 2B)25,27,29. To accurately measure DHPG-induced changes using our reporter system, we added the 3’ UTR of FMR1 (which is required for mGluR-dependent FMRP synthesis32) and included a C-terminal PEST degron sequence to stimulate reporter turnover (Fig 2C). DHPG treatment significantly enhanced expression from FMRP reporters bearing either 20 or 90 CGG repeats (Fig 2D). Surprisingly, removal of the CGG repeat ((CGG)0) prevented FMRP reporter upregulation in response to DHPG, suggesting a specific role for the repeat in FMRP translational regulation (Fig 2D).
To study this repeat-dependent effect further, we replaced the CGG repeat with a (GAA)25 repeat. This modification maintained 5’ leader length, but eliminated the stable RNA secondary structure. (GAA)25 repeat constructs also exhibited impaired upregulation of FMRP expression in response to DHPG (Fig 2E). To evaluate if any strong secondary structure could replace the CGG repeat, we replaced the CGG repeat with a synthetic non-repetitive hairpin that has perfect complementarity within its stem region (Fig 2C). This hairpin has a minimum free energy similar to that of 20 CGG repeats and suppresses basal FMRP reporter synthesis in vitro23. FMRP reporters bearing this non-repetitive significantly increased in response to DHPG (Fig 2F), suggesting that mRNA structure plays a critical role in activity-dependent FMRP synthesis.
CGG repeats may also influence FMR1 mRNA transport into dendritic processes33. We therefore tested whether repeat removal might impact FMR1 mRNA trafficking by using hybridization chain reaction (HCR) to track reporter mRNAs (Extended Data 3A). FMRP reporters containing the 3’UTR of FMR1 were transported into dendrites more efficiently than a construct lacking these sequences (AUG-nLuc, Extended Data 3B)32. However, the CGG repeat was not strictly required for dendritic localization of reporter mRNAs.
RAN translation is required for mGluR-dependent enhancement of FMRP synthesis
We next tested whether CGG RAN translation is required for DHPG dependent upregulation of FMRP. To accomplish this, we introduced 0/+1-AAA mutations into FMRP reporters bearing the PEST tag and 3’ UTR (Fig 2C). As with our earlier constructs, these mutations enhanced basal FMRP reporter expression in neurons. However, further upregulation of FMRP reporter expression in response to DHPG was lost in these reporters at both normal and expanded repeat sizes (Fig 2G).
If CGG RAN prevents ribosomes from reaching the AUG of FMRP and initiating translation there, then we would predict that processes which suppress CGG RAN would enhance FMRP synthesis. We therefore evaluated whether mGluR activation might suppress CGG RAN. In contrast to FMRP reporters, CGG RAN translation from expanded CGG repeats significantly decreased after DHPG treatment (Fig 2H). These findings suggest a dynamic and inverse relationship between CGG RAN and FMRP synthesis.
To understand how CGG RAN might be regulated by mGluR activation, we first replaced the +1CGG RAN ACG initiation codon with AUG (Extended Data 4A). As expected, this lowered steady-state neuronal FMRP reporter expression (Extended Data 4B–C). However, upon DHPG treatment, FMRP reporter signal was enhanced (almost 3-fold) (Fig 2I, Extended Data 4B). This indicates DHPG mediated activation of FMRP synthesis does not require a non-AUG uORF initiation codon. As a corollary experiment, we evaluated whether DHPG might modulate RAN translation at a different repeat expansion. We observed no response to DHPG using a RAN translation specific reporter for the C9Orf72 gene G4C2 hexanucleotide repeat in the GA dipeptide repeat reading frame (Extended Data 4D)34.
Next, we tested the impact of the small molecule compound 1A on activity-dependent FMRP synthesis35. 1A binds to non-Watson Crick G-G base pairs in CGG hairpins to stabilize the repeat structure and impair repeat RNA-RNA binding protein interactions35. Consistent with published results35,36, 1A treatment decreased neuronal +1CGG RAN reporter abundance, but had no effect on FMRP reporter expression (Extended Data 4E). Pretreatment with 1A precluded the DHPG-dependent increase in FMRP reporter synthesis (Fig 2J).
Some aspects of mGluR dependent plasticity act through phosphorylation of eIF2α and the integrated stress response37. This effect is mediated at least in part through altered translation of a uORF in OPHN137. We previously demonstrated that eIF2α phosphorylation, which classically suppresses AUG initiated translation, paradoxically enhances CGG RAN translation34. To evaluate whether eIF2α phosphorylation is involved in DHPG dependent FMRP synthesis, we treated neurons with ISRIB, which prevents phosphorylated eIF2α from impairing ternary complex turnover38, enhances memory and impairs RAN translation39. Treatment of neurons with ISRIB enhances basal FMRP reporter expression (Extended Data 4F). However, ISRIB treatment failed to impair further upregulation of the FMRP reporter in response to DHPG, suggesting that eIF2α phosphorylation dependent signaling is not required for upregulation of FMRP (Extended Data 4F).
Taken together with earlier work14,22,25,28,29, these data are most consistent with a model where downregulation of CGG RAN in response to mGluR activation enhances translation from the main FMR1 ORF, allowing for enhanced FMRP production. This regulation is dependent upon both the magnitude of basal suppression of FMRP synthesis by upstream CGG repeats and RAN translation from specific upstream near-cognate start codons.
ASOs that block CGG RAN initiation increase endogenous FMRP
The translational efficiency of ORFs on a given transcript can be enhanced by antisense oligonucleotides (ASOs) that sterically hinder initiation at upstream AUG initiated uORFs40,41. ASOs were recently also used to block in vitro RAN translation at GGGGCC repeats42. We therefore tested if CGG RAN could be impaired with an ASO. We designed and tested four different ASOs harboring 2’-O-methyl modifications and phosphorothioate backbones to singly overlap the three CGG RAN start sites. ASOs were positioned such that the near-cognate start codon was at the ASO 5’end, as this strategy was most effective for AUG uORFs41 (Extended Data 5A).
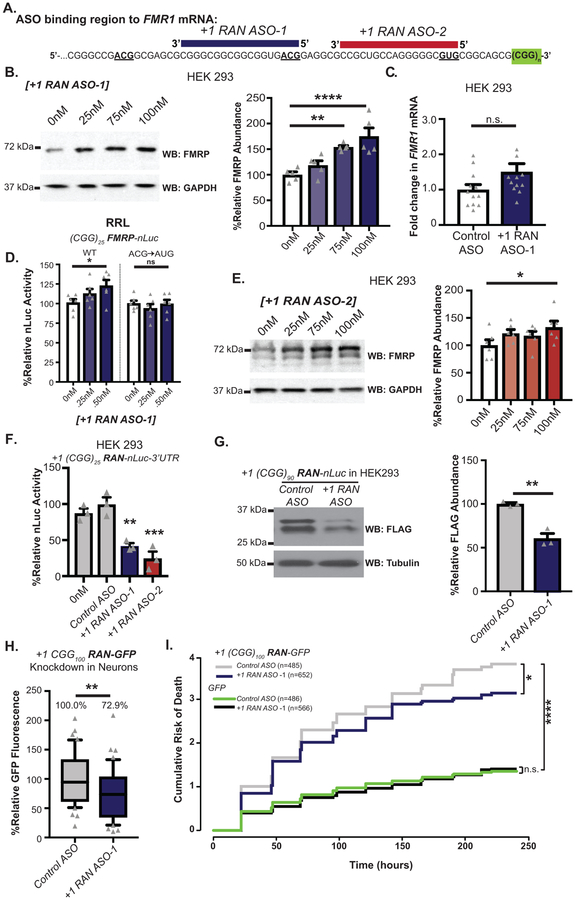
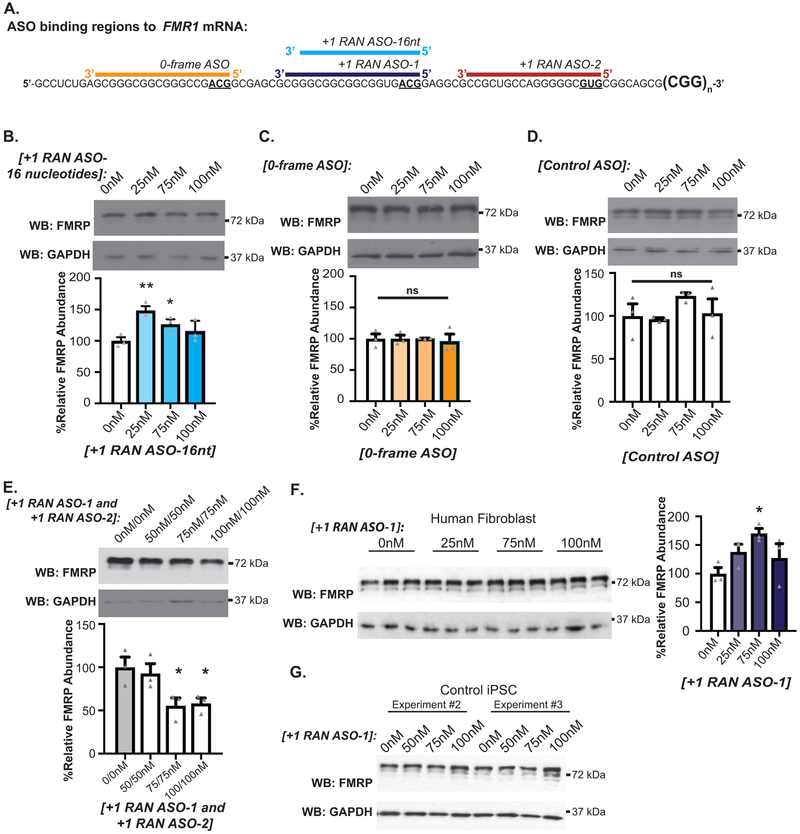
To determine whether CGG RAN inhibits endogenous FMRP synthesis, we transfected RAN ASOs into HEK293 cells, which have 23 CGG repeats in FMR1. An 18nt ASO targeting the ACG initiation codon for CGG RAN in the +1 reading frame (+1RAN ASO-1) (Fig 3A, purple) increased endogenous FMRP expression in a dose-dependent manner, peaking at 189% with 100nM ASO (Fig 3B). This occurred in the absence of significant changes in FMR1 mRNA expression (Fig 3C). To confirm this effect was mediated by enhanced translation, we mixed +1RAN ASO-1 with (CGG)25 FMRP-nLuc reporter mRNA and evaluated translation in RRL. In vitro translation of the FMRP reporter was significantly increased at 0.50nM of the ASO (Fig 3D). The same assay was performed with reporter mRNAs bearing an AUG in place of ACG, which is predicted to impair ASO binding41. This mutation eliminated the ASO effect, suggesting that complete ASO complementarity with its target and in particular the initiation codon is required to increase FMRP synthesis (Fig 3D). A 16 nucleotide ASO targeting the same initiation codon (Extended data 5A, blue) and an ASO targeting the GUG codon used in +1CGG RAN translation (+1RAN ASO-2) (Fig 3A, red) showed similar but more modest increases in endogenous FMRP (Fig 3E, Extended data 5B). Both +1RAN ASO-1 and +1RAN ASO-2 significantly decreased expression of +1CGG RAN reporters in HEK293 cells, while Control ASO had no effect relative to vehicle treated (0nM) controls (Fig 3F–G).
Figure 3: RAN targeting ASOs increase FMRP and suppress CGG repeat toxicity.
A) Non-cleaving, +1CGG RAN blocking antisense oligonucleotides (+1RAN ASO-1, purple and +1RAN ASO-2, maroon) overlap the FMR1 5’ leader and ACG start site or GUG start site, respectively. B) Left: Endogenous FMRP expression in HEK293 cells transfected with +1RAN ASO-1. Right: Quantification of 5 independent experiments, normalized to GAPDH and expressed as % of mean 0nM control (n=5; 0nM vs 25nM: p=0.2056; 0nM vs 75nM: p=0.0013; 0nM vs 100nM: p<0.001). C) RT-qPCR on endogenous FMR1 mRNA with or without +1RAN ASO-1(100nM) (n=12, p=0.015). D) In vitro (RRL) nLuc assay with WT or mutant (CGG)25 FMRP-nLuc reporter mRNAs in the presence of increasing +1RAN ASO-1. Mutant reporter has AUG in place of ACG initiation codon (n=6; WT 0nM vs 0.25nM: p=0.2092; WT 0nM vs 0.50nM: p=0.0255). E) Endogenous FMRP expression in HEK293 cells transfected with +1RAN ASO-2 (n=6; 0nM vs 25nM: p=0.1236; 0nM vs 75nM: p=0.2090; 0nM vs 100nM: p=0.0224). F) +1CGG25 RAN-nLuc signal in HEK293 cells transfected with control ASO, +1 RAN ASO-1 (100nM), or +1 RAN ASO-2 (100nM) (n=3; Control vs 0nM: p=0.5452; Control vs +1RAN ASO-1: p=0.0016; Control vs +1RAN ASO-2: p<0.001). G) Immunoblot and bar graph of +1CGG25 RAN-nLuc-Flag in HEK293 cells transfected with control ASO or +1RAN ASO-1 (100nM) (n=3; p=0.0028). H) Quantification of +1CGG RAN reporter product (+1(CGG)90 RAN-Venus) in neurons treated with +1RAN ASO-1 (n=42) or Control ASO (n=41) for 5 days (p=0.0075). I) Survival analysis by longitudinal fluorescence microscopy on rat cortical neurons transfected with +1 (CGG)100 RAN-GFP treated with 1μM +1RAN ASO-1 or Control ASO (n=485, n=552, respectively) or GFP treated with 1μM +1RAN ASO-1 or Control ASO (n=486, n=566, respectively). Panels B-H: Two sided Student’s t-test with correction for multiple comparisons. Panel I: Cox proportional hazard analysis. n.s.=not significant, *p<0.05, **p<0.01, ****p<0.001. Panel B-G: Graphs are mean +/− S.E.M. Panel H: box extends to 25th/75th percentiles with a line at mean and whiskers indicate 95% CI.
ASOs targeting the 0-frame initiation site (0-frame ASO) and a Control ASO with a non-specific nucleotide sequence had no impact on FMRP expression (Extended Data 5C–D). Interestingly, tiling both +1CGG RAN initiation sites with ASOs that singly increased FMRP synthesis actually decreased FMRP levels (Extended Data 5E), perhaps because tiling the RNA with multiple ASOs impairs ribosomal scanning40. To evaluate whether we could enhance endogenous FMRP levels in the setting of expanded CGG repeats, we transfected FXTAS patient fibroblasts with 69 repeats with +1RAN ASO-1. As at normal repeat sizes, this enhanced endogenous FMRP expression (Extended Data 5FA).
RAN targeting ASOs reduce repeat toxicity in rodent neurons.
We next used live confocal imaging of +1(CGG)100 RAN-Venus in transfected neurons to determine if RAN ASOs impact neuronal RAN translation. +1RAN ASO-1 treatment significantly decreased neuronal +1(CGG)100 RAN-Venus expression (Fig 3H). Ectopic expression of CGG repeats is toxic in neurons43. We used automated longitudinal microscopy to track survival of hundreds of individual neurons over days. Cortical Rat neurons expressing +1(CGG)100 RAN-GFP demonstrated significant toxicity compared to GFP alone (Fig 3I)43. Treatment of +1(CGG)100 RAN-GFP neurons with +1RAN ASO-1 significantly improved survival compared to Control ASO treatment, but had no effect on neurons expressing GFP alone. Together with previous studies3,14, these findings support a direct role for +1CGG RAN in CGG repeat-associated neuronal toxicity.
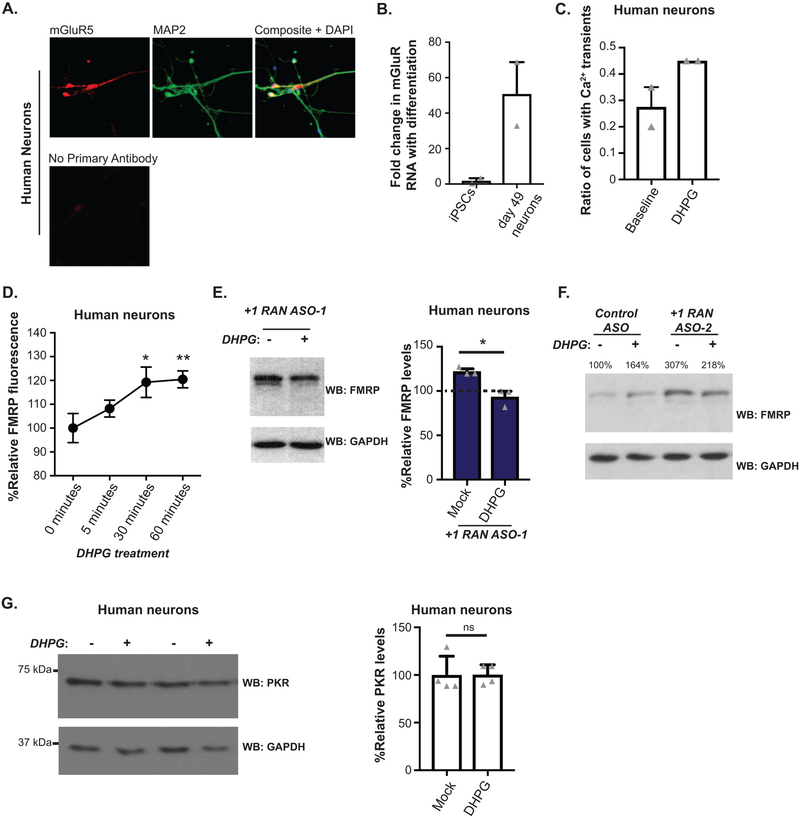
CGG RAN targeting ASOs enhance FMRP expression in human neurons
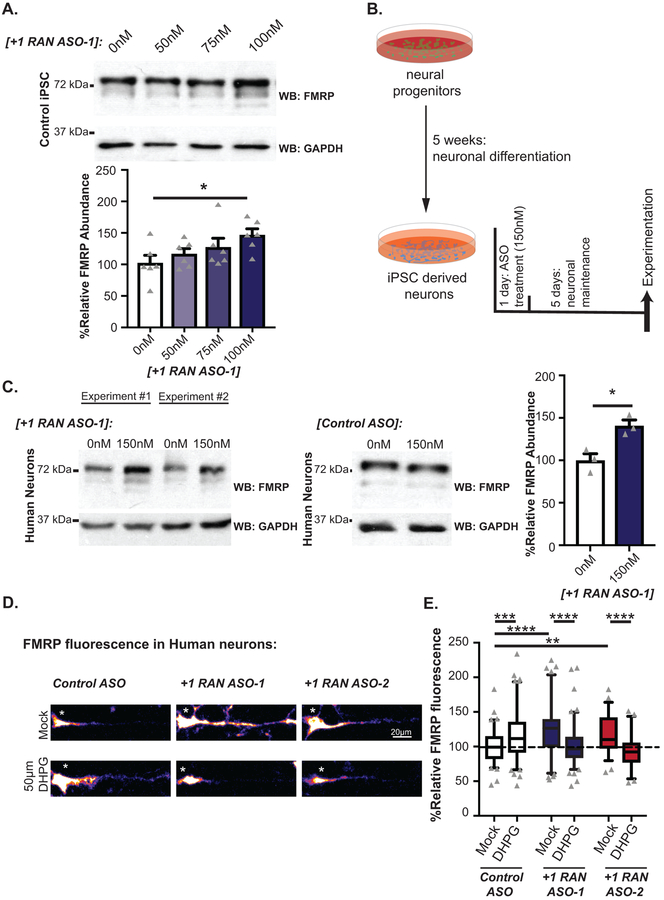
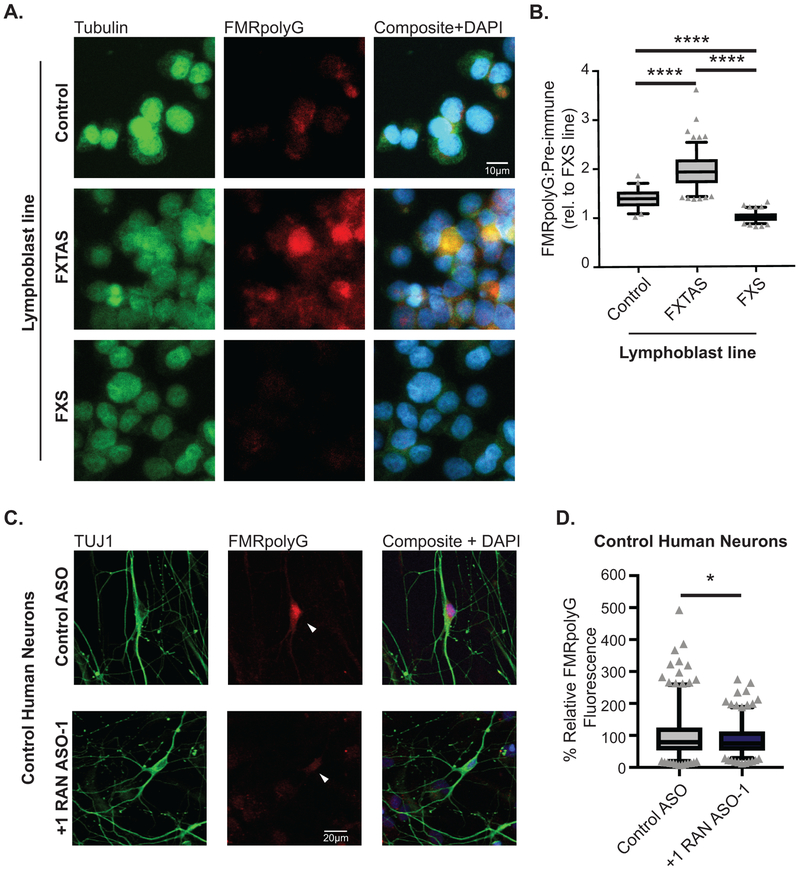
The studies above utilized either transfected human cells, non-human overexpression model systems or non-neuronal patient cell lines. To evaluate whether we could impact endogenous FMRP synthesis and its regulation in human neurons, we utilized induced pluripotent stem cells (iPSCs) harboring a normal CGG FMR1 repeat. Treatment of these cells with +1RAN ASO-1 enhanced basal endogenous FMRP abundance (Fig 4A, Extended Data 5G). These iPSCs were then differentiated into forebrain-like glutamatergic neurons (Fig 4B). Application of +1RAN ASO-1 triggered a ~41% increase in neuronal FMRP, but not in Control ASO treated neurons (Fig 4C). A similar increase in FMRP occurred after treatment with +1RAN ASO-2 targeting the GUG initiation codon (Fig 4D–E). To identify if this increase correlates with a decrease in +1RAN translation we performed ICC to detect endogenous FMRpolyG using a specific and validated antibody (NTF-1) generated against the N-terminus of the protein (Extended Data 6A–B). In control human neurons, +1RAN ASO-1 treatment led to a small but significant decrease in FMRpolyG staining compared to control ASO treated neurons (Extended Data 6C–D).
Figure 4: CGG RAN ASOs alter FMRP translation dynamics in human neurons.
A) FMRP in control iPSCs transfected with the indicated doses of +1RAN ASO-1 (n=6; 0nM vs 50nM: p=0.3857; 0nM vs 75nM: p=0.1370; 0nM vs 100nM: p=0.0126). B) Schematic of neuronal ASO treatment protocol. C)FMRP in neurons from untreated, +1RAN ASO-1 treated iPSC neurons or Control ASO treated neurons (n=3; p=0.0167). D) Representative images of control neurons treated with indicated ASOs +/− DHPG. E) Quantification of FMRP fluorescence in human neurons treated with indicated ASO (150nM), normalized to untreated control ASO performed in parallel (Control ASO/Mock n=88, Control ASO/DHPG n=101, +1RAN ASO-1/Mock n=97, +1RAN ASO-1/DHPG n=102, +1RAN ASO-2/Mock n=56, +1RAN ASO-2/DHPG n=55, where “n” is the mean CTCF signal from 5 neurons. Control ASO/Mock vs Control ASO/DHPG: p<0.001; Control ASO/Mock vs +1RAN ASO-1/Mock: p<0.001; Control ASO/Mock vs +1RAN ASO-2/Mock: p=0012; +1RAN ASO-1/Mock vs +1RAN ASO-1/DHPG: p<0.001; +1RAN ASO-2/Mock vs +1RAN ASO-2/DHPG: p<0.001). Panel A: One-way ANOVA with multiple comparisons. Panel C, E: Two sided Student’s t-tests with Bonferroni correction. *p<0.05, **p<0.01, ****p<0.0001. Panel A, C: Graphs are mean +/− S.E.M. Panel E: box extends to 25th/75th percentiles with a line at mean and whiskers indicate 95% CI.
Blocking CGG RAN abrogates human neuron mGluR-dependent upregulation of FMRP
Previous studies of mGluR-regulated FMRP synthesis have been performed solely in rodent neurons, and lack validation in human neurons. Additionally, our studies on the role of CGG RAN in regulating activity-dependent FMRP synthesis used reporter systems in rodent neurons which may not fully recapitulate the native behavior of the FMR1 transcript. To test whether CGG RAN is required for mGluR-dependent upregulation of endogenous human FMRP synthesis, we performed experiments in iPSC-derived neurons. We first confirmed these neurons expressed mGluR5 and responded to DHPG treatment (Extended Data 7A–C). As in previous studies in rodent neurons, 30 and 60 minutes of DHPG significantly increased FMRP expression, confirming that human iPSC-derived neurons display mGluR-elicited protein synthesis (Fig 4D–E, Extended Data 7D).
We next applied +1RAN ASO-1 or +1RAN ASO-2 to neurons in the presence or absence of DHPG (Fig 4D–E, Extended Data 7E–F). Both ASOs increased basal FMRP expression in the absence of DHPG. However, upon mGluR activation, neurons pretreated with RAN-targeting ASOs exhibited decreased FMRP. These DHPG-dependent effects on FMRP expression were observed in the soma and primary dendrites by ICC (Fig 4D). The decrease in FMRP is consistent with previous studies demonstrating that FMRP is rapidly degraded by the proteasome within 5 minutes of DHPG application25,26 and suggests that RAN ASOs prevent a compensatory upregulation of new FMRP synthesis. Consistent with this hypothesis, this feature was not observed in reporter assays, which lack the FMRP phosphorylation site for regulated ubiquitination26. These data support a conserved endogenous role for CGG RAN in regulating activity-driven FMRP synthesis.
To evaluate whether DHPG influences expression of other CGG repeat containing transcripts, we measured the effect of mGluR activation on endogenous double-stranded RNA activated kinase PKR, which contains 13 CGG repeats in its 5’UTR in humans. We readily detected PKR in human neurons by western blot, but we did not detect a change in PKR expression with DHPG treatment (Extended Data 7G).
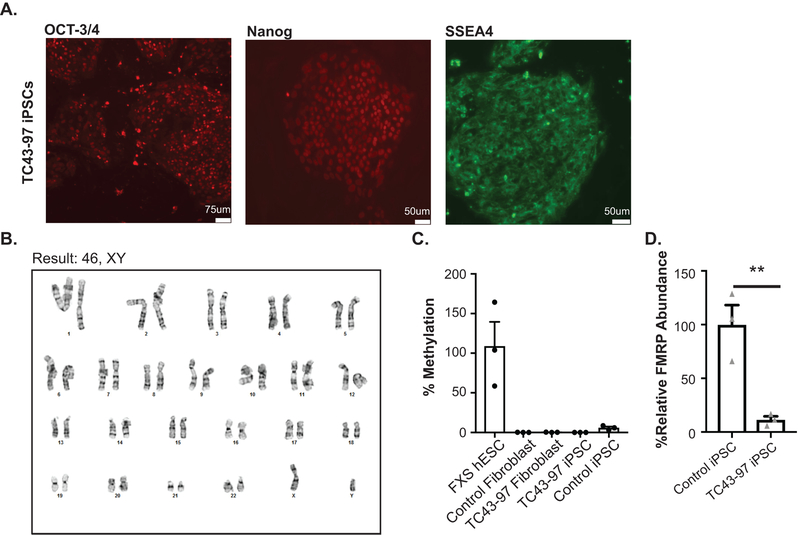
CGG RAN ASOs increase FMRP in human CGG repeat expansion neurons
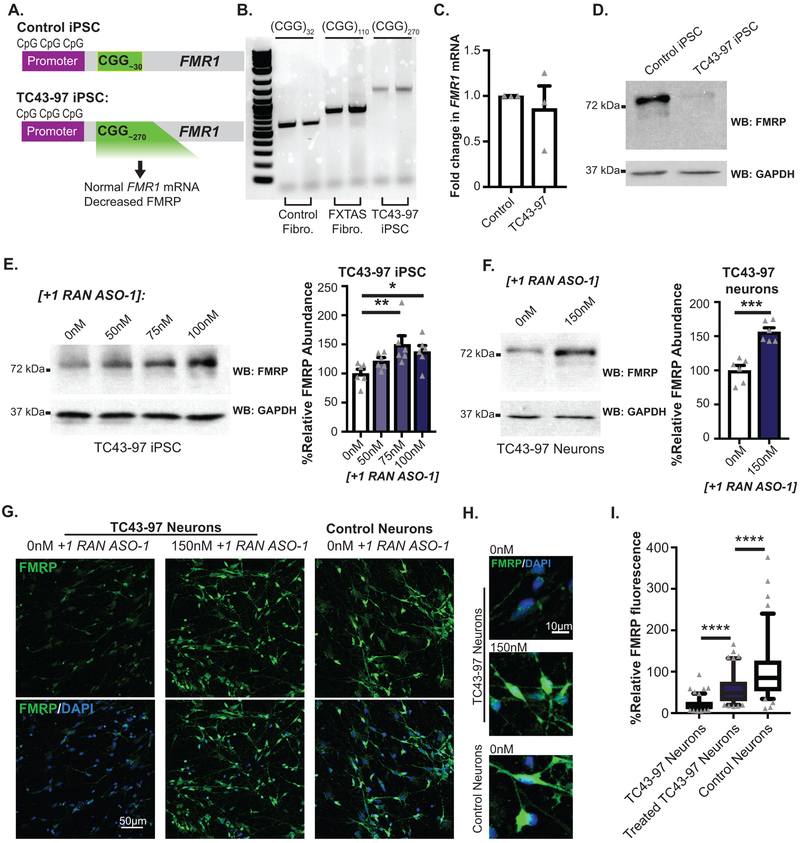
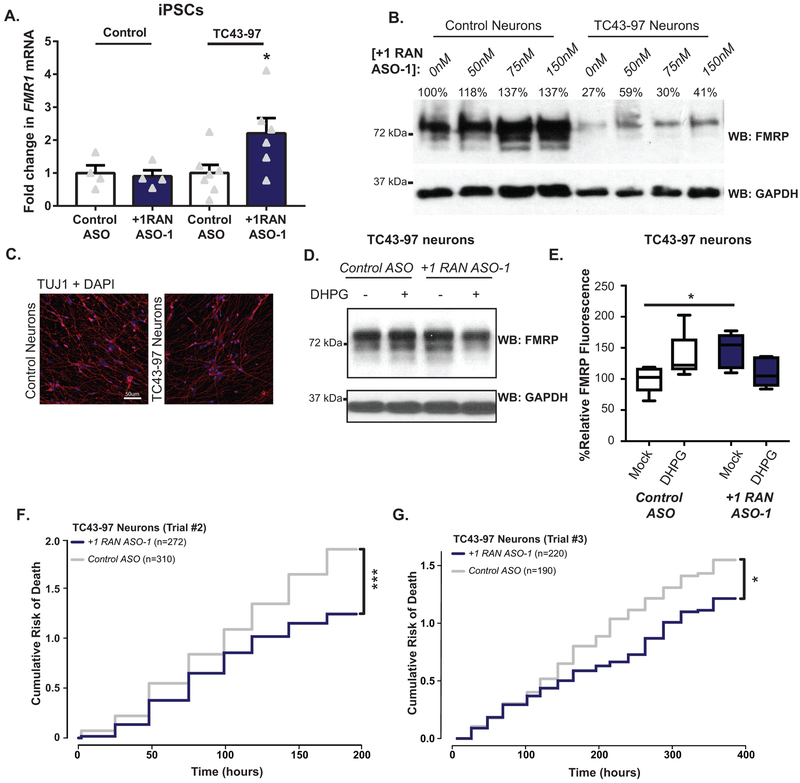
Inefficient FMRP translation contributes to some FXS cases10 and transcribed full mutations can cause neuronal inclusions and FXTAS symptoms11,12. We derived iPSCs from a previously characterized unmethylated full mutation (UFM) fibroblast line (TC43–97)44. TC43–97 UFM iPSCs (Fig 5A–D, Extended Data 8A–B) carry a predominant repeat of 270 CGGs (Fig 5B) with a range of repeats between 250–340. Bisulfite qPCR confirmed the promoter remained unmethylated after iPSC derivation (Extended Data 8C). The TC43–97 iPSCs maintained normal FMR1 mRNA levels, but exhibited significantly decreased FMRP (Fig 5C–D, Extended Data 8D).
Figure 5: RAN ASO increases FMRP in unmethylated full mutation (UFM) neurons.
A) Schematic of TC43-clone 97 (TC43–97) UFM male iPSC clones. Control iPSCs harbor 30 CGG repeats and unmethylated promoter. B) PCR amplification of 5’ leader region of FMR1 demonstrates repeat size in control fibroblasts, FXTAS fibroblasts, and TC43–97 iPSCs. C) FMR1 mRNA expression in TC43–97 and Control iPSCs (n=3; p=0.6058). D) Cropped immunoblot of FMRP expression in TC43–97 and Control iPSCs (representative of 3 independent experiments). E) FMRP expression in TC43–97 iPSCs transfected with +1RAN ASO-1, quantified on right (n=6; 0nM vs 50nM: p=0.1468; 0nM vs 75nM: p=0.0025; 0nM vs 100nM: p=0.0167). F) FMRP immunoblot in untreated and +1RAN ASO-1 treated neurons, quantified at right (n=6; p<0.001). G) FMRP (green) in TUJ1-positive TC43–97 neurons treated with +1RAN ASO-1. Blue: DAPI. 40X image. Scale bar: 50μm. H) 60x images of FMRP signal in TC43–97 or control neuronal soma and processes after indicated treatments. I) Quantification of FMRP expression in untreated TC43–97 neurons (n=120), treated TC43–97 neurons (n=100), and control neurons (n=95), where “n” is the mean CTCF signal from 5 neurons (Control vs untreated TC43–97: p<0.001; Control vs treated TC43–97: p<0.001; untreated vs treated TC43–97: p<0.001). Panel C and F: Two sided Student t-test. Panel E: One-way ANOVA with a Fisher’s LSD test. Panel I: Kruskal Wallis Test with correction for multiple comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Panel C, E, F: Graphs are mean +/− S.E.M. Panel I: box extends to 25th/75th percentiles with a line at mean and whiskers indicate 95% CI.
+1RAN ASO-1 increased FMRP in TC43–97 iPSCs in a dose-dependent manner (Fig 5E). This was accompanied by a modest increase in FMR1 mRNA expression not seen in control iPSCs (Extended Data 9A). Similarly, +1RAN ASO-1 treatment increased FMRP levels in TC43–97 derived neurons by immunoblot (Fig 5F, Extended Data 9B). FMRP increased by 2-fold in +1RAN ASO-1 treated TUJ1 positive neurons by ICC (Fig 5G–I, Extended Data 9C). As in control neurons, in TC43–97 neurons treated with +1RAN ASO-1, treatment with DHPG lowered FMRP expression (Extended Data 9D–E). Of note, the overall level of FMRP was still elevated in these cells compared to neurons treated with Control ASO alone. Thus, targeting CGG RAN in human neurons enhances neuronal FMRP expression and alters its regulation even in the setting of transcribed full mutation CGG repeats.
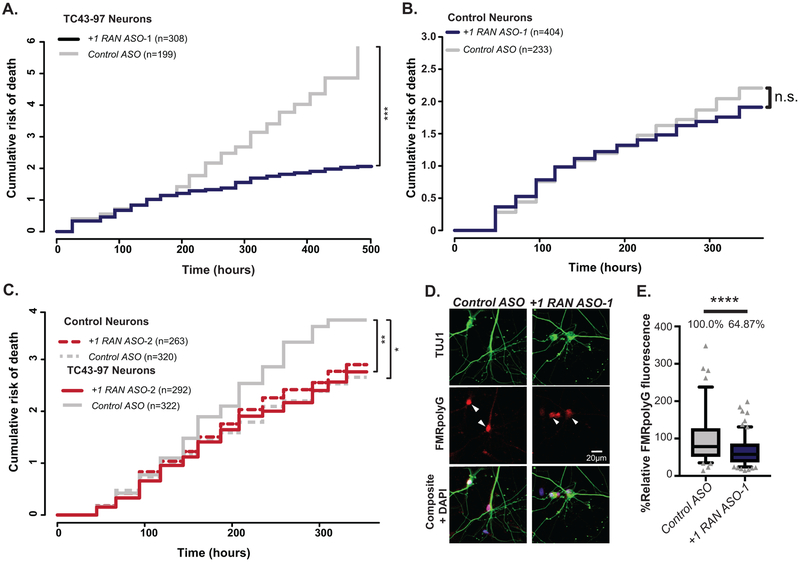
CGG RAN ASOs improve neuronal survival of human CGG repeat expansion neurons
Rescue of CGG RAN toxicity in rodent neurons relied on overexpression of CGG repeats. We therefore tested whether +1RAN ASOs could enhance survival in TC43–97 UFM neurons. Control ASO or +1RAN ASOs were applied for 24 hours and imaged beginning 24 hours after ASO removal. Using the longitudinal imaging, neurons were manually tracked over multiple days. TC43–97 neurons treated with +1RAN ASO-1 exhibited a significant decrease in their risk of death compared to neurons treated with Control ASO (Fig 6A, Extended Data 9F–G). In contrast, +1 RAN ASO-1 did not impact survival of control neurons (Fig 6B)3,13. Treatment with +1RAN ASO-2 also enhanced survival selectively in TC43–97 neurons (Fig 6C), consistent with these effects on survival resulting from an impact on RAN translation from expanded repeats. +1RAN ASO-1 decreased FMRpolyG staining in TC43–97 neurons by 30% (Fig 6D–E). These data suggest that blocking endogenous CGG RAN in human neurons with an expanded repeat enhances survival by decreasing production of FMRpolyG with a long glycine tract (Extended Data 10).
Figure 6: CGG RAN ASO enhances survival in expanded repeat human neurons.
A) Survival analysis in TC43–97 human neurons treated with 150nM +1RAN ASO-1 (n=308) or Control ASO (n=199) (p<0.001). B) Survival analysis in control human neurons treated with 150nM +1RAN ASO-1 (n=404) or Control ASO (n=233) (p=0.709). C) Survival analysis in TC43–97 (n=292, 322) and control (n=263, 320) human neurons treated with 150nM +1RAN ASO-2 or Control ASO, respectively (Control/Control ASO vs TC43–97/Control ASO: p=0.0105, TC43–97/Control ASO vs TC43=97/+1RAN ASO-2: p=0.0015). D) Immunocytochemistry to FMRpolyG (red) in mature TC43–97 neurons (TUJ1-positive (green)) treated with +1RAN ASO-1 or Control ASO treatment. E) Quantification of FMRpolyG signal (corrected total cellular fluorescence) in TC43–97 neurons treated with +1RAN ASO-1 (n=194) or Control ASO (n=93), where “n” is the mean CTCF signal from 5 neurons (p<0.001). Panel A, B, C: Cox proportional hazard analysis. Survival is plotted as cumulative risk of death. Panel E: Two sided Mann Whitney U test. Box in graph extends to the 25th and 75th percentiles of data points with a line at the mean, and whiskers indicate 95% confidence interval. n.s.=not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Discussion:
Simple tandem repeats make up a significant fraction of the human transcriptome and repeat expansions are a common cause of neurological disease1,2,16. Yet our knowledge of the native functions of transcribed repeats within the context of normal neuronal physiology is quite limited2. Here we define a physiological and conserved function for CGG repeats and non-AUG initiated translation at the FMR1 locus. CGG RAN translation acts as a regulated inhibitory upstream open reading frame that controls production of the Fragile X protein, FMRP, in neurons both basally and in response to metabotropic glutamate receptor activity. These findings provide initial evidence for physiological roles of repetitive elements in both translational control and neurobiology, and suggest a potential target for therapeutic development in Fragile X disorders.
Until now, CGG repeats were viewed largely as a static and steric hindrance to ribosomal scanning that is overcome in an unregulated fashion10,22,23. Our findings suggest that CGG repeats allow CGG RAN to act as a regulatory uORF that temporally restricts FMRP synthesis21. This is consistent with previous studies suggesting that uORF containing transcripts can undergo mGluR-dependent translation37. However, unlike uORFs in genes such as OPHN1, DHPG-dependent upregulation of FMRP did not require eIF2α phosphorylation signaling pathways based on studies using pharmacological inhibitors. In contrast, the requirement for both a CGG repeat and the native FMR1 sequence context to allow for mGluR-dependent upregulation of FMRP synthesis suggests that this sequence functions to enhance basal CGG RAN and maintain FMRP repression.
New FMRP synthesis in response to mGluR activation is thought to serve as a negative feedback loop that turns off local synaptic translation and thus constrains mGluR dependent plasticity28. Previous work in FMR1 premutation mice suggests that an inability to upregulate FMRP expression in dendrites in response to mGluR activation correlates with enhanced mGluR LTD30. As larger repeats enhance CGG RAN translation and elicit greater basal repression on downstream FMRP synthesis, their conservation in mammals and expansion in primates could reflect a selective advantage, providing an amplified gain for regulated FMRP synthesis in response to external cues17. Consistent with this, driving translation through the CGG repeat with an AUG that is out of frame with FMRP sets the basal level of FMRP translation low, facilitating a robust relative response to mGluR activity.
By mapping out a native function for CGG RAN, we identified a potential therapeutic target in fragile X disorders. CGG RAN translation products contribute to disease relevant phenotypes in FXTAS model systems, including cell based assays, Drosophila, and transgenic mice3,14. In other repeat expansion disorders, ASOs which enhance degradation of the repeat containing transcript correct disease relevant phenotypes and are moving into clinical trials45. However, eliminating disease-repeat containing transcripts is not an appealing option in FXTAS because loss of FMRP causes FXS. By targeting the major native +1CGG RAN initiation sites with non-cleaving ASOs, we simultaneously suppressed CGG repeat-elicited toxicity and enhanced endogenous FMRP synthesis in both cell-based model systems and in human neurons with large repeats. These findings suggest that approaches which target RAN translation could potentially provide a solution to two disparate problems elicited by expanded CGG repeats. In FXS, a majority of FXS patients make little or no mRNA. Recent studies suggest that transcriptional reactivation of the FMR1 locus in FXS may be feasible46,47, however this reactivation would generate mRNAs with very large CGG repeats that could impair FMRP synthesis and support translation of toxic proteins. Approaches like the ASO described here could thus be useful in combination with reactivation strategies to prevent CGG repeat-associated toxicity of the reactivated large repeat and enhance overall FMRP production.
In recent years, ASO therapies have moved from the research bench to the clinic. This was initially thought of as a strategy to target solely gain-of-function disorders. However, the recent success of nusinersen, which blocks binding of hnRNPs to SMN2 to increase production of the SMN protein in spinal muscular atrophy48, speaks to the power of these molecules to treat neurological disorders. To this end, the approaches described here represent an exciting opportunity to potentially leverage new disease insights towards effective therapeutic development.
Methods:
Plasmid Construction
Vectors expressing AUG-nLuc, +1 (CGG)n RAN-nLuc and +0 (CGG)n RAN-nLuc from pcDNA3.1(+) were described in13. All ligations were performed using Roche Rapid DNA Dephosphorylation and Ligation Kit according to manufacturer’s specification. PCR cloning was accomplished with Phusion High-Fidelity DNA Polymerase (NEB) or Platinum Taq High-Fidelity (Thermo Fisher). All site-directed mutagenesis was performed using Q5 Site-Directed Mutagenesis Kit (NEB) according to manufacturer’s protocol.
To generate RAN translation reporters bearing out-of-frame mutants, the XhoI and XbaI fragment harboring the nLuc reporter in each particular reading frame was cloned back into the desired mutant backbone plasmid containing various repeats (Kearse et al., 2016). +1 (CGG)100 GGG-nLuc-3xFLAG-PEST was generated using a two-step Q5 site directed mutagenesis strategy using +1 (CGG)0 GGG-nLuc-3xFLAG (Kearse et al., 2016) as a template. The PEST containing GGG-nLuc coding sequence was cloned into the original plasmid using EcoRV and XbaI to obtain a full (CGG)100 repeat construct.
For the (CGG)n FMRP-nLuc vector, the (CGG)100 repeat was moved from a separate pUAST vector using restriction enzymes EcoRI and XhoI. The 3’ end of the FMR1 5’ UTR and first coding exon was added by PCR cloning using XhoI and NotI. Nanoluciferase with a GGG in place of the AUG, GGG-nLuc-3xFLAG, was cloned in frame with the first FMRP exon using NotI and XbaI.
The 3’ UTR was added to the FMRP reporters to produce the (CGG)n FMRP-nLuc-3’ UTR by PCR cloning from a separate pCRII-TOPO vector bearing the sequence using restriction enzymes XmaI and XbaI. The entire nLuc-3xFLAG-PEST tag was PCR cloned out of pcDNA3.1 +1 (CGG)100 nLuc-PEST and into the (CGG)n FMRP-nLuc-3’UTR vector using NotI and XbaI. The +1 (CGG)100 nLuc-PEST vector was cut with XhoI and XbaI and ligated into the (CGG)n FMRP-nLuc-3’ UTR vectors to generate +1 (CGG)n nLuc-PEST-3’ UTR.
The Venus open reading frame was cloned into pCRII-TOPO vector bearing the C-terminal region of +1 FMRpolyG from XhoI to the first codon before the in-frame start site. This was PCR cloned in place of GFP in the +1 (CGG)100 GFP pcDNA3.1 plasmid3 using XhoI and XbaI. Repeat size in all reporters was confirmed by digestion with BlpI and XhoI digestion and gel electrophoresis on a 2% (wt/vol) agarose gel, and by Sanger sequencing specific for structured DNA at the University of Michigan Sequencing Core.
C9RAN reporters used in this study were previously described in Green et al., 2017. 5’ end of the C9orf72 intron 1 was PCR-amplified from human fibroblast DNA inserted upstream of GGG-NL-3xF in pcDNA3.1 via Nhe1.
In Vitro Transcription
pcDNA3.1(+) vectors were digested with PspOMI for linearization. pcDNA3.1(+) vectors with the 3’ UTR of FMR1 were digested with SmaI for linearization. HiScribe T7 Quick High Yield RNA Synthesis kit (NEB) was used to make capped RNA from linearized plasmids. mRNA was then polyadenylated as in Kearse et al., 2016.
In Vitro Translation and Luciferase Assay
In vitro translation reactions using the Flexi Rabbit Reticulocyte Lysate (RRL) System (Promega), were performed with nLuc reporter mRNAs and analyzed as in Kearse et al., 2016. Briefly, luciferase assays were performed using a mixture of prepared NanoGlo reagent (Promega) and in vitro reactions diluted in Glo Lysis Buffer at 25μL:25μL (Promega), and quantified using a GloMax-96 plate reader (Promega). For immunoblot analysis, 10μL RRL reactions were performed with saturating mRNA levels, mixed with 40μL of SDS sample buffer and heated at 70° for 15 minutes, and then resolved using SDS-PAGE. nLuc-3xFLAG reporter proteins were detected by immunoblot using the anti-FLAG antibody (mouse, Sigma, F1804).
Primary Hippocampal Neuron Culture and Luciferase Assay
All animal use followed NIH guidelines and was in compliance with the University of Michigan Committee on Use and Care of Animals. Hippocampi were dissected from P0–P3 Sprague-Dawley rat pups of both sexes. Cells were papain dissociated as in Sutton et al., 2006. Hippocampal neurons were plated on poly-D-Lysine coated 12-well culture plates at a density of 6.0×104 cells/plate. Hippocampal cultures were maintained for 11–12 DIV prior to transfection. On the day of transfection, cells were washed 1X with Neuron Growth Media (NGM, Neurobasal Media-A supplemented with 1X B27, 1X Glutamax) then placed in 1mL of NGM supplemented with 0.0189% kynurenic acid (NGM-KY) [wt/vol], A mixture of 510μL of Opti-MEM (Fisher), 10uL Lipofectamine 2000 (Fisher), and DNA was added to the neurons for 30 minutes. 2.5μg or 5μg nanoluciferase plasmid and pGL4.13 (firefly luciferase (FFLuc)) plasmid was added to each well for luciferase assays. 7.5μg of reporter plasmid and mCherry plasmid was transfected into cells for imaging assays. The OptiMEM/NGM-KY mixture was removed from cells followed by 2 washes in NGM-KY, and 1mL of NGM was then added. Neurons were placed back in the 37°C incubator for 24 hours.
Transfected neurons were treated for 30 minutes with 50μM (S)-3,5-dihydrophenylglycine (DHPG, Tocris) then lysed directly on plate for 10 minutes in 250μL of Glo Lysis Buffer. 1A (a gift from Matt Disney) treatment was performed 3 hours prior to DHPG treatment. ISRIB (SML0843, Sigma-Aldrich) treatment was performed 6 hours before DHPG treatment at a final concentration of 200nM. Luciferase assays were performed as described above in a 50μL:50μL prepared NanoGlo Reagent to lysate. For FFLuc quantification, 50μL of prepared ONE-Glo reagent (Promega) was used in place of NanoGlo. Nanoluciferase was normalized to firefly luciferase to control for transfection differences. Nanoluciferase assays performed in neurons were surveyed for transfection efficiency and viability (FFLuc level) and consistency (NLuc: FFLuc) due to technical variability. All runs were performed in triplicate, data points below a specified FFLuc threshold or outside a set range of variability were excluded. The same threshold was applied to all runs. Any run with >1 data point that did not meet the inclusion criteria, was eliminated from the analysis.
Single-molecule microscopy and analysis
Single-molecule imaging of Venus constructs was performed as previously described49–51. Single-molecule imaging was performed with a modified Nikon (Tokyo, Japan) N-SIM microscope equipped with a Nikon Apo TIRF 100X, NA 1.49 microscope objective, and an iXon3 EMCCD camera (Andor Technology, Belfast, UK). For single-molecule imaging of translation in live cells, Venus protein was excited with a 488nm laser line (1.5mW laser power) from a continuous wave (CW) solid-state laser (Coherent, Santa Clara, CA). The single-molecule imaging was done in epifluorescence mode. Following whole-cell photo-bleaching, single molecule time-lapse images of Venus protein were collected continuously at an exposure time of 150ms per frame. To minimize the effects of photo-damage on the cells an illumination area of ~25μm diameter was used.
Data analysis of time-lapse images was done as previously described49,50,52. Briefly, the centroid coordinates of individual molecules in each frame were determined and linked in time to construct temporal trajectories, using a standard single-particle tracking algorithm. Each Venus molecule detected was tracked from appearance (maturation) to disappearance (bleaching). Preexisting Venus fluorescent signal was used to manually define an ROI that contained neuronal dendrites, but not soma, for each dataset. From each dataset, translation events situated inside the ROI were analyzed with MATLAB (MathWorks). The analysis software is available at http://www.ccam.uchc.edu/yu.
Hybridization Chain Reaction
Mouse embryonic fibroblasts were seeded at 2.5 ×104 cells/well for 24 hours on 0.01% poly-D-lysine-coated 8-well chamber slides, then transfected with 250 ng nLuc reporters (AUG-NL-3xF, +2(CGG)100-NL-3xF, and mock) and 2:1 jetPRIME® (VWR) for 24 hours. Cells were fixed according to Molecular Instruments protocol53. Following overnight incubation with 70% ethanol, cells were rehydrated in PBS for 1 hour, permeabilized with 0.1% Triton X-100 for 5 minutes, blocked with 2% BSA for 20 minutes at room temperature, and incubated overnight with mouse anti-FLAG primary antibody in 2% BSA at 4°C. Goat anti-mouse Alexa 488 secondary antibody was applied the following morning for 1 hour at room temperature, in the dark. Following ICC, reporter RNA was detected in transfected cells using DNA probes with additional sequence complimentary to Cy5 labeled self-hybridizing hairpins53–55. Probes against the nanoluciferase sequence (Table 1) were purchased from molecularinstruments.org and applied according to manufacturer’s protocol. Coverslips were then applied to slides with ProLong Gold Antifade Mountant with DAPI. 10–20 fields per condition were imaged at 20×1.6 magnifications with Olympus IX71 fluorescent microscope and Slidebook 5.5 software.
Table 1:
Hybridization Chain Reaction Probes and DNA primer sequences used in this study.
| HCR_DNA_nLuc DNA Probe_1 | GAGGAGGGCAGCAAACGGGAAGAGTCTTCCTTTACGATATT TTGTAGCCGGCTGTCTGTCGCCAGTCCCCAACGAAATCTTCGAGTGTGAA ATATA GCATTCTTTCTTGAGGAGGGCAGCAAACGGGAAGAG |
| HCR_DNA_nLuc DNA Probe_2 | GAGGAGGGCAGCAAACGGGAAGAGTCTTCCTTTACGATATT TTGGATCGGAGTTACGGACACCCCGAGATTCTGAAACAAACTGGACACAC ATATA GCATTCTTTCTTGAGGAGGGCAGCAAACGGGAAGAG |
| HCR_DNA_nLuc DNA Probe_3 | GAGGAGGGCAGCAAACGGGAAGAGTCTTCCTTTACGATATT AAATTTTTTCGATCTGGCCCATTTGGTCGCCGCTCAGACCTTCATACGGG ATATA GCATTCTTTCTTGAGGAGGGCAGCAAACGGGAAGAG |
| HCR_DNA_nLuc DNA Probe_4 | GAGGAGGGCAGCAAACGGGAAGAGTCTTCCTTTACGATATT CGTAACCCCGTCGATTACCAGTGTGCCATAGTGCAGGATCACCTTAAAGT ATATA GCATTCTTTCTTGAGGAGGGCAGCAAACGGGAAGAG |
| Nanoluciferase qPCR primer F | CAGCCGGCTACAACCTGGAC |
| Nanoluciferase qPCR primer R | GCCCATTTTCACCGCTCAG |
| FMR1 qPCR primer F | CATGAAGATTCAATAACAGTTGC |
| FMR1 qPCR primer R | CACTTTAGCTAACCACCAACA |
| FMR1 F_up | CCCACGCCACTGAGTGCACCTCTGC |
| FMR1 R_down | AGCCCCGCACTTCCACCACCAGCTCCT |
| HPRT qPCR primer F | GGACCTCTCGAAGTGTTGGATA |
| HPRT qPCR primer R | ACGTGATTCAAATCCCTGAAGT |
| mGluR5 qPCR primer F | AATCTCCCGATGTCAAGTGGT |
| mGluR5 qPCR primer R | AGGGTTTCGGTGGTTTGTTTC |
Rat hippocampal neurons plated on Mattek dishes were maintained to DIV 5 prior to transfection and incubation with plasmid for 24-hours. HCR was performed as above. mAPPLE protein was detected using a rabbit anti-dsRed antibody (Clonetech). Confocal imaging was performed as outlined below.
RNA Quantification
Total RNA was isolated from cells using Quick-RNA Miniprep Kit (Zymo Research). DNA was eliminated using 1μL Turbo DNase (Thermo Fisher) according to the manufacturer’s protocol. For cells transfected with plasmids, this process was repeated to ensure removal of contaminating DNA. cDNA was synthesized from equal amounts of total RNA using iScript cDNA synthesis kit (Bio-Rad). qPCR reactions were performed using iQ SYBR® Green Supermix (Bio-Rad) and equal amounts of cDNA. qPCR was performed on a MyiQ thermocycler (Bio-Rad) using a two-step protocol, primers are listed in Table 1. Biological triplicates were run in technical triplicate for all experiments. Standards were run alongside samples for each primer to calculate primer efficiencies. Relative abundance was calculated for each replicate using the efficiencies of each primer and Ct values from the transcript of interest relative to the housekeeping gene.
Cell Culture and Transfection
SH-SY5Y cells (ATCC) were plated on 6-well plates for nanoluciferase and RT-qPCR assays. Cells were maintained in DMEM: F12 (Fisher) and 1% Pen/Strep. Cells were transfected with 550ng of both nanoluciferase DNA and pGL4.13 using FuGENE® HD (Promega). Cells were maintained for 24 hours post-transfection before being either lysed for RNA isolation or nanoluciferase assay.
HEK293 cells (ATCC) were maintained in DMEM with high glucose (Fisher), 10% fetal bovine serum (vol/vol), and 1% Pen/Strep (vol/vol; Fisher). 1.3×104 cells/well were plated on 96-well plates for nanoluciferase assays, 24 hours prior to transfection. For ASO treatment of reporter expressing cells, cells were transfected with 100ng nanoluciferase reporter RNA using Lipofectamine 2000. 7 hours later media was changed and 100nM ASOs were transfected with Lipofectamine RNAiMAX (Fisher) according to manufacturer’s specifications. After an additional 17 hours, cells were lysed in 200μL of Glo Lysis buffer, and reporter levels were measured by luciferase assay as detailed above. All ASO sequences are listed in Table 2. For immunoblotting of the knockdown of +1 (CGG)90 RAN-nLuc, HEK293 cells were plated on 24-well plates and transfected with 250ng of reporter DNA for 3 hours. Media was changed 3 hours later, and cells were re-transfected with 100nM Control or +1 RAN ASO-1. Cells were incubated for 48 hours with a media change after 24 hours, then lysed in 200μL of RIPA buffer. Lysates were resolved on 12% SDS-PAGE gels with subsequent immunoblotting for detection of FLAG using anti-FLAG antibody (Sigma).
Table 2:
ASOs used in this study.
| Name | Sequence | Length | Chemistry |
|---|---|---|---|
| 0-frame ASO | CGUCGGCCCGCCGCCCGC | 18 nucleotides | 2’-OMe, PS |
| +1 RAN ASO-16nt | CGUCACCGCCGCCGCC | 16 nucleotides | 2’-OMe, PS |
| +1 RAN ASO-1 | CGUCACCGCCGCCGCCCG | 18 nucleotides | 2’-OMe, PS |
| +1 RAN ASO-2 | CACGCCCCCUGGCAGCGG | 18 nucleotides | 2’-OMe, PS |
| Control ASO | CAUUGUUUUUUGUCUUCC | 18 nucleotides | 2’-OMe, PS |
For analysis of endogenous FMRP levels during ASO treatment, HEK293 cells were plated on 12-well plates, and transfected with a mixture of ASOs and 4.5μL of Lipofectamine® RNAiMAX according to manufacturer’s specifications. After 24 hours, cells were lysed in 250μL RIPA supplemented with mini complete protease inhibitors (Sigma), boiled in 6X SDS sample buffer at 90°C for 5 minutes, and then were resolved on 8% polyacrylamide gels by SDS-PAGE. FMRP was detected by subsequent immunoblotting using anti-FMRP antibody ab17722 (Abcam). FMRP levels were quantified relative to GAPDH. Rabbit anti-PKR (Abcam) antibody was used for immunoblotting of human neurons.
iPSC Reprogramming
Fibroblasts from male donors were cultured in DMEM, 10% FBS, 1X L-glutamax (Fisher), 1 mM MEM non-essential amino acids (Fisher), at 37°C and 5% CO2. For episomal reprogramming, 1× 106 fibroblasts were collected after Trypsin treatment and mixed with a set of plasmids pCXLE-hOCT3/shP53, -hSK, -hUL (Addgene), then electroporated with Neon® device (condition:1650 Volts, 10 mm width, and 3 pulses). Induced fibroblasts were plated onto 6 well plates at density of 0.5–1×104cells/well and switched 1 day later to a PSC medium mTeSR1 (StemCell Technologies). IPSC colonies appeared and were manually picked and passaged onto new matrigel coated 12-well plates, and continually grown with mTeSR1. IPSCs were passaged weekly using 0.5mM EDTA and culture medium supplemented with 10μM Y-27632 ROCK inhibitor (EMD Millipore) for 24 hours. After 5–10 passages the cells were evaluated for pluripotency by immunocytochemistry (ICC) and embryoid body differentiation. For embryoid body experiments, iPSC colonies were grown in suspension for 3–4 weeks, passaged onto 0.1% gelatin (Sigma) for another week and processed for ICC. iPSC reprogramming was confirmed by staining with antibodies against Oct-3/4, NANOG, and SSEA4 (Santa Cruz Biotech, Abcam, DSHB). IPSC lines were karyotyped by Cell Line Genetics. TC43–97 fibroblasts were generously provided by Christopher Pearson. Control fibroblasts were provided by Jack Parent.
PCR Amplification of the CGG repeat
100ng-150ng of DNA was amplified with primers “FMR1 F_up” and “FMR1 R_down” (final concentration: 500nM, Table 1) using the Expand Long Template PCR System (Sigma)56. Reactions took place in Buffer 2, and were supplemented with betaine (final concentration: 1.2M) and DMSO (5% v/v). Cycling conditions were programmed as in Saluto, et al., 2005.
iPSC Differentiation and ASO Treatment
Undifferentiated iPSCs were cultured in TeSR-E8 media (Stem Cell Technology) on MatriGel-coated plates and passaged with 1 mM EDTA every 3–4 days. Neural induction was performed using a dual-SMAD inhibition57 protocol with modifications. In brief, two wells of a 6-well plate were grown to approximately 80% confluence, dissociated with EDTA, and plated into a single well of a MatriGel-coated 6-well plate with TeSR-E8 containing 10 μM Rock Inhibitor (Y-27632). The cells were confluent the next day and neural differentiation was induced using neural maintenance media (referred here as 3N) containing 1 μM dorsomorphin and 10 μM SB431542. The cells were cultured for 12–14 days with daily media changes. Neuroepithelial sheets were then combed into large clumps, passaged, and maintained on MatriGel-coated plates in rosette media (3N containing 20 ng/ml FGF) with daily media changes until neural rosettes appeared. Rosettes were manually picked and dissociated into single cells using Accutase. Neural progenitors were plated onto MatriGel-coated plates, grown in neural expansion media (3N containing 20 ng/ml FGF and 20 ng/ml EGF) with media changes every other day, and passaged as needed using Accutase. For differentiation into neurons, neural progenitors were plated at a density of approximately 1.5 × 105 cells/mL in neural expansion media on PLO-laminin coated plates or coverslips, allowed to grow for 24 hours, and switched to neural maintenance media. Neurons were maintained for up to 6 weeks with half media changes every other day and a full media change supplemented with 1 μg/ml laminin every 10 days.
For ASO treatments, undifferentiated iPSCs were plated as small colonies on MatriGel-coated plates in TeSR-E8 containing 10 μM Rock Inhibitor and grown overnight. Media was replaced with TeSR-E8 the next day. Cells were allowed to recover for at least 4 hours and media was replaced again just prior to treatment. ASOs (0–100 nM) and RNAiMax (4.5 μl per 100 μl of prepared complexes) were diluted in Opti-MEM reduced serum media, incubated together for 5 min at room temperature, and added to cells. Cells were harvested 24 hours after treatment. For iPSC-derived neurons, ASOs (0–150 nM) were diluted in neural maintenance media and added to 5-week old (for FMRP detection assays) or 10-week old (for FMRpolyG detection assays) neurons one day after a full media change. Media was changed 24 hours after treatment. Neurons were maintained as previously stated and harvested 6 days after treatment. Immunoblotting was performed as described above. For ASO knockdown of fluorescent reporters, DIV 5 rat hippocampal cultures were transfected (+1 CGG100 RAN-Venus) and replaced with media containing 1μM of the +1 RAN ASO or the Control ASO. Neurons were placed back in the incubator, and live imaged on the 5th day.
Longitudinal microscopy of primary neurons and iPSC-derived neurons
Mixed cortical neurons were dissected from E20 Long-Evans rat pups of both sexes, as previously described58,59. Cortical neurons were cultured at 0.6 × 106 cells/mL on 96-well plates. Cultures were maintained at 37°C in neural growth media (NGM; Neurobasal A supplemented with 2% B-27 and 1% Glutamax-1 [vol/vol] (Fisher)). On DIV4, neurons were co-transfected with 0.1μg pGW1-mApple and either 0.1μg pGW1-GFP or pGW +1 (CGG)100 GFP DNA per well of a 96-well culture plate, using Lipofectamine 2000 (Invitrogen). +1 RAN ASO-1 and the Control ASO were applied to neurons at 1μM immediately following transfection, the media with ASO remained on the neurons for the entirety of the experiment. Neurons were imaged at regular 24-hour intervals beginning 24 h post-transfection using an automated fluorescence microscopy platform detailed in prior studies58,59. Image processing for each timepoint and survival analysis for automated fluorescence images were achieved by custom code written in Python or the ImageJ macro language, and cumulative hazard plots were generated using the survival package in R.
TC43–97 and wild-type NPCs were plated at a density of 4×104 cells/mL on 96-well plates. NPCs were differentiated as described above. Day 14 neurons were treated with Control ASO or +1 RAN ASOs at a final well concentration of 150nM. ASO was removed from the neurons 24 hours later. 24 hours after ASO removal, 90% of the neural maintenance media was removed from each well and replaced with base media without supplements (Neurobasal medium). iPSC-derived neuronal cultures were imaged using bright field microscopy. iPSC-derived neuronal images were automatically stitched and stacked. Neurons were manually tracked across time using custom software, Manual Survival Analysis (MASA), and survival analysis performed in R as described above.
Immunocytochemistry
Hippocampal and iPSC derived neurons were washed 2x in PBS containing 1mM MgCl2 and 0.1 mM CaCl2 (PBS-MC). Cells were fixed for 15 minutes with a solution of 4% Paraformaldehyde/4% Sucrose in PBS-MC warmed to 37°C, washed 3x in PBS-MC, and permeabilized for 5 minutes in 0.1% Triton-X in PBS-MC. Cells were blocked in 2% BSA for an hour, then incubated in primary antibody for at least 2 hours at room temperature. Reporter protein was detected with an antibody for FLAG (mouse, Sigma) and mCherry or mApple was detected with an anti-dsRed antibody (rabbit, Clonetech). A rabbit anti-mGluR5 (Abcam) antibody was used in human neurons to verify mGluR expression. FMRP was detected with a rabbit anti-FMRP antibody (Abcam), and FMRpolyG was detected using a custom developed polyclonal rabbit antibody to the N-terminal region of the protein (NTF1, NeoScientific, 1:200, epitope: EAPLPGGVRQRGGGGGGGGGG). NTF1 staining and specificity was validated in patient derived lymphoblasts with confirmed CGG repeat sizes and loss of FMR1 mRNA expression in FXS cells (Coriell, GM09237, GM06891, GM07539) with pre-immune serum staining included as a negative control15. For all staining, cells were washed 3 times, followed by incubation in secondary antibody for 1 hour. Cells were subsequently washed and placed in ProLong Gold Antifade with DAPI (Thermo Fisher).
Confocal Microscopy and Live Imaging
Fixed and live imaging was performed in an inverted Olympus FV1000 laser-scanning confocal microscope. Directly prior to live cell imaging, NGM was removed and replaced with 1mL of warmed 1x HBSS. Plates were imaged for a maximum of 1 hour to prevent imaging hyper-stressed or dying cells. For all experiments, acquisition parameters were identical between conditions within experiments. For all reporter quantification experiments, all co-transfected cells were imaged on >2 individual transfected plates. Imaging of transfected neurons was performed using a 60X objective. For human iPSC-derived neuron experiments, 20X or 40X–imaged fields were analyzed from regions of similar confluency.
Channels were imaged sequentially and optimized to eliminate bleed-through. Neurons were imaged in a series of Z-planes to resolve the entire soma and dendritic arbor. Images were analyzed in ImageJ. Average intensity composite images were derived from raw image files. For quantification of individual soma or dendrites, cell casts were made using threshold images as a guide. Dendrites were analyzed starting 20μm from soma to the most distal staining as specified by mCherry fill staining. The ROI was applied to the individual channels, and intensities were measured and normalized as specified. For all iPSC-derived neuron experiments, only TUJ1 positive cells with neuronal morphology were quantified. Corrected total cellular fluorescence (CTCF) was calculated by subtracting background fluorescence in the area of the cell from the integrated density as measured by ImageJ. CTCF was used for all immunocytochemistry experiments unless otherwise noted due to variation in cell size, in which case average fluorescence was utilized and noted in figure legend.
For imaging DHPG treated human neurons, ≥3 independent wells of wild-type human neurons were imaged. Data derived with the +1 RAN ASOs, represents multiple wells imaged from three separate rounds of differentiation with all conditions imaged and quantified in parallel. For the TC43–97 line, two independent wells were imaged. 2–4 individual frames of similar confluence were imaged. To prevent overrepresentation of individual replicates, up to 30 neurons were analyzed per frame. Each n represents the average corrected fluorescence of 5 sequentially analyzed neurons. Analysis was performed blinded. ROI’s were manually detected based on neuronal morphology and TUJ1 staining, and measured for FMRP staining. For FMRpolyG imaging, two individual wells of differentiated neurons were imaged per treatment condition. 3–5 frames of similar confluence were blindly imaged and blindly quantified per well. A minimum threshold of TUJ1 staining was implemented to eliminate non-neuronal cells. Up to the first 30 automatically measured cells in each frame were included in the analysis. Each n represents the mean value of 5 sequentially analyzed neurons. Due to the primarily somatic localization of FMRpolyG, cells were automatically detected using ImageJ. FMRpolyG staining was measured using the same parameters for each frame.
Biological replicates and statistical analyses
For all cellular nanoluciferase assays, western blots and RT-qPCR, n represents individual biological replicates derived from separate wells/plates of cells treated exactly the same. For ICC and imaging studies on transfected cells, each n represents a single cell unless otherwise specified from the specified number of plates (≥3 per condition). For in vitro assays, each n represents an individual reaction. These data analyses were not performed blind to condition.
Statistical analyses were performed in GraphPad Prism7, with the exception of the cumulative risk of death calculation, which was performed in R. For all assays, normality was confirmed using a KS normality test. For samples with normal data distributions, an unpaired Student’s t-test (two-tailed) with a 95%-CI was performed for all assays comparing two experimental conditions. A two-way ANOVA with a 95%-CI was performed for the neuron experiments comparing start site mutants to the controls run in parallel from 2–4 individual experiments run in triplicate, individually transfected wells of neurons. A one-way ANOVA with multiple comparisons was performed for all other experiments with more than two experimental conditions. A one-way ANOVA with a Fisher’s LSD test was performed on all ASO dose-response experiments, n represents individual experiments. Survival measures were calculated according to Cox proportional hazard analysis. For samples that failed the normality distribution, a Mann Whitney U test or Kruskal Wallis non-parametric ANOVA were performed dependent on the number of groups analyzed. All bar graphs show the mean +/− S.E.M. unless otherwise stated. These data analyses were performed blind to condition.
All samples were randomly assigned to experimental groups. Entire experiments were excluded only in cases of poor transfection efficiency, cell death, contamination, or poor neuronal differentiation efficiency. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications1,12,14.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Fig. 1. Impact of CGG RAN translation on FMRP reporter synthesis.
A) Left: in vitro translated (CGG)n FMRP-nluc reporter mRNAs harboring different CGG repeat sizes. ‡‡‡: FMRP-nanoluciferase-3X FLAG protein. ‡: N-terminal extension of FMRP from RAN initiation at the ACG codon in the (polyarginine) 0-frame. ‡ is detected at up to 18 repeats, but is attenuated at normal repeat sizes as previously described. ‡‡: N-terminal extension of FMRP from initiation in the 0-frame, downstream of the repeat. The ‡‡ product is only detectable in vitro, and is not detectable in cells. Right: Luciferase activity of in vitro translated FMRP-nLuc reporters (n=3; 0 vs 18: p=0.000000000000033; 18 vs 28: p=000001109; 28 vs 45: p=0.00234; 28 vs 57: p=0.0000518; 28 vs 69: p=0.0004997; 28 vs 100: p=0.001371). B) Luciferase activity from FMRP reporter in vitro after replacement of (CGG)25 repeat with unstructured (GAA)25 repeat (n=3; p=0.4940). C) Schematic of RAN translation reporters. D) Left: luciferase activity showing relative levels of FMRP, +1 RAN, and 0-frame RAN reporters at 25 and 100 repeats in HEK293 cells (n=3; CGG25: FMRP vs +1 RAN p=0.00002612, FMRP vs 0-frame RAN p=0.00001338; CGG100: FMRP vs +1 RAN p=0.0009, FMRP vs 0-frame RAN p=0.0003). E) Top: Immunoblot of (CGG)n FMRP-nLuc reporters in HEK293T cells with indicated mutations. Bottom: luciferase activity from reporters translated in vitro (RRL). +1-AUG represents insertion of AUG in place of +1 ACG RAN initiation codon. RAN initiation sites in the (CGG)n FMRP-nLuc reporters were mutated to preclude initiation in the 0-frame (0-AAA), the +1 reading frame (+1-AAA), or both (0/+1-AAA) (n=3; CGG25: WT vs 0-AAA p=0.0001, WT vs +1-AAA p=0.1977, WT vs 0/1-AAA p=0.0001; CGG100: WT vs 0-AAA p=0.0001, WT vs +1-AAA p=0.4437, WT vs 0/1-AAA p=0.0001). F) RT-qPCR to nLuc mRNA from SH-SY5Y cells expressing the indicated FMRP reporters with 100 repeats (n=3; p=0.4940). G) Flag immunocytochemistry for (CGG)100 FMRP-nLuc reporters in rat neurons co-expressing mCherry (red) to fill the cell. H) Quantification of Flag signal for the WT (CGG)100 reporter (n=23) and for the 0/+1-AAA (CGG)100 reporter (n=21), where “n” is the mean CTCF signal from 5 neurons (p=0.0258). Panel A: One-way ANOVA with multiple comparisons. Panels D, E: One-way ANOVA with multiple comparisons, within repeat groups. Panel B, F, H: two sided Student’s t-test. n.s.=not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Graphs are mean +/− S.E.M.
Extended Data Fig. 2. Live tracking of CGG RAN translation in neuronal dendrites.
A) Left: Venus with and without (ΔAUG) an AUG initiation codon serve as a positive and negative translation control. Right: Venus fluorescent proteins with the AUG deleted were inserted after the 5’ leader of FMR1, in the +1 (FMRpolyG) reading frame to serve as a reporter for +1CGG RAN. B) Live imaging of mature rat hippocampal neurons expressing indicated Venus reporters (green) and mCherry (red). C) Quantification of Venus reporter signals with 32 repeats (n=11) or 90 repeats (n=14) (p=0.0002). D) Single molecule imaging of CGG RAN translation in distal neuronal processes expressing indicated +1CGG RAN-Venus reporters after photo bleaching. Green dots represent individual translation events. E) Quantification of CGG RAN events in processes (n=3; p=0.2355). Panels C, E: Two sided Student’s t-test. n.s.=not significant, ***p<0.001. Graphs are mean +/− S.E.M.
Extended Data Fig. 3. FMR1 reporter mRNA trafficking in neuronal dendrites.
A) Mouse embryonic fibroblasts were transfected with +1 (CGG)100 RAN-nLuc reporters or mock transfected were probed for nLuc RNA by Hybridization Chain Reaction (HCR) and co-stained for FLAG. The specificity of the probes for nLuc is illustrated by the presence of signal (red) in transfected cells only. B) Left: HCR of nLuc mRNA combined with ICC to co-transfected mApple verifies dendritic export of reporter RNAs. Right: Quantification of dendritic nLuc reporter mRNA; AUG-nLuc (n=16), 0 repeat (n=16), 20 repeats (n=18), 90 repeats (n=20) (AUG vs 0: p=0.1447; AUG vs 20: p=0.0373; AUG vs 90: p=0.0044). One-way ANOVA with correction for multiple comparisons. Box extends to 25th/75th percentiles with a line at mean and whiskers indicate 95% CI.
Extended Data Fig. 4. Modifiers of mGluR dependent FMRP reporter synthesis.
A) Schematic of the AUG (+1) CGGn FMRP-nLuc-3’UTR reporter, which drives translation in the +1 RAN reading frame and reports for FMRP. B) relative nLuc values in neurons transfected with CGG25 or CGG100 FMRP-nLuc-3’UTR reporters with or without a +1 AUG mutant 5’ leader region (n=3; CGG20: p=0.0002; CGG100: p=0.0036). C) Relative nLuc values in neurons transfected with CGG25 FMRP-nLuc-3’UTR reporters: WT (n=12), +1 AUG/Mock (n=12), +1 AUG/DHPG (n=11) (WT/Mock vs +1AUG/mock: p=0.005; WT/Mock vs +1AUG/DHPG: p=0.4504). D) Rat hippocampal neurons were transfected with a nLuc construct for the C9Orf72 G4C2 hexanucleotide repeat in the Glycine/Alanine reading frame (n=6; p=0.8642). E) Hippocampal neurons were treated with 1A at 20 hours post-transfection. Quantification of nLuc expression following 3 hours of 1A treatment is represented for each indicated reporter (n=3; AUG/Vehicle vs AUG/1A: p=0.8038; RAN/Vehicle vs RAN/1A: p=0.0437; FMRP/Vehicle vs FMRP/1A: p=0.5728). F) Rat hippocampal neurons were treated with ISRIB 6 hours before treatment with vehicle or DHPG (Mock/Mock: n=18, Mock/DHPG n=18, ISRIB/Mock n=17, ISRIB/DHPG n=18; Mock/Mock vs Mock/DHPG: p=0.0342, Mock/DHPG vs ISRIB/DHPG: p=0.0678, ISRIB/Mock vs ISRIB/DHPG: p=0.0058). Panel B, C, F: One-way ANOVA with multiple comparisons. Panel D, E: Two sided Student’s t-test. n.s.=not significant, *p<0.05, **p<0.01, ***p<0.001. Graph is mean +/− S.E.M.
Extended Data Fig. 5. CGG RAN ASOs in human cell lines.
A) Schematic of other tested non-cleaving RAN blocking ASOs. Colored bars overlap the corresponding FMR1 5’ leader sequence and start sites; 0 frame ACG (orange), +1 frame ACG (+1RAN ASO-1, purple(18nt) or blue(16nt)) and +1 frame GUG (+1RAN ASO-2, maroon). B) Effect of +1RAN ASO-16 nucleotide on endogenous FMRP expression (0nM vs 25nM: p=0.0021; 0nM vs 75nM: p=0.0404; 0nM vs 100nM: p=0.2021). C) Effect of +0 RAN ASO (18nt) on endogenous FMRP expression (0nM vs 25nM: p=0.9999; 0nM vs 75nM: p=0.9997; 0nM vs 100nM: p=0.9580). D) Effect of Control ASO on endogenous FMRP expression (0nM vs 25nM: p=0.8183; 0nM vs 75nM: p=0.1780; 0nM vs 100nM: p=0.8486). E) Effect of combinatorial treatment with (+1RAN ASO-1 and +1RAN ASO-2 on endogenous FMRP expression at indicated doses (0nM vs 50nM: p=0.6231; 0nM vs 75nM: p=0.0127; 0nM vs 100nM: p=0.0171). F) Impact of +1RAN ASO-1 transfection into patient derived fibroblasts (0nM vs 50nM: p=0.2795; 0nM vs 75nM: p=0.0336; 0nM vs 100nM: p=0.5035). G) Representative immunoblot of FMRP expression after treatment with +1RAN ASO-1 (technical replicates of main figure 4a) in transfected control iPSCs. For all experiments, n=3, replicated in 3 independent experiments. For all graphs: One-way ANOVA with a Fisher’s LSD test for dose dependency. n.s.=not significant, *p<0.05, **p<0.01. Graphs are mean +/− S.E.M.
Extended Data Fig. 6. FMRpolyG expression in human cells and control human neurons.
A) Immunocytochemistry against FMRpolyG on Control ((CGG)23), FXTAS ((CGG)100–117), and FXS ((CGG)931–940, fully methylated) patient derived lymphoblasts. B) Rater-blinded quantification of FMRpolyG staining expressed as a ratio to pre-immune serum at the same concentration (μg/mL) on the same cells. Values are expressed relative to the FXS line, which does not express the FMR1 transcript (Control n=50, FXTAS n=133, FXS n=102; p=0.0000000000005). C) Immunocytochemistry to FMRpolyG (red) in mature control human neurons (TUJ1-positive (green)) treated with +1RAN ASO-1 or Control ASO treatment. D) Quantification of FMRpolyG signal with +1RAN ASO-1 (n=90) or Control ASO (n=69) treatment, where “n” is the mean CTCF signal from 5 neurons (p=0.0485). Panel B: Kruskal Wallis test with post-hoc two sided Mann Whitney U tests. Panel D: Two sided unpaired Student t-test. *p<0.05, ****p<0.0001. Box extends to 25th/75th percentiles with a line at mean and whiskers indicate 95% CI. Marked dots are only shown for values outside the 95% CI.
Extended Data Fig. 7. RAN ASOs block mGluR-dependent FMRP translation in human neurons.
A) Immunocytochemistry for mGluR5 in control human iPSC-derived neurons. B) mGluR mRNA expression as quantified by RT-PCR in Control iPSCs vs. neurons at day 49 of differentiation (n=2). C) Impact of 5 minutes of 50μM DHPG on calcium transients in human iPSC derived neurons. Graph represents number of neurons with active calcium transients compared to total number of neurons tracked over two independent neuronal cultures (n=2). D) Time course of DHPG effect on FMRP levels in human neurons (0 min: n=20, 5 min: n=35, 30 min: n=34, 60 min: n=46; 0 vs 5 min: p=0.2972, 0 vs 30 min: p=0.0158, 0 vs 60 min: p=0.0072). E) Left: Immunoblot from Control human neurons treated with +1RAN ASO-1 with or without DHPG treatment. Right: Quantification of FMRP expression in human neurons treated with DHPG after pretreatment with +1RAN ASO-1 relative to Control ASO treated neurons (n=3; p=0.0110). F) Representative western of FMRP expression after DHPG in control human neurons pretreated with the indicated ASOs and then treated with vehicle or DHPG. G) PKR has a 30-nucleotide CGG repeat in its 5’ leader. Endogenous expression of PKR was assessed after vehicle or DHPG treatment in iPSC-derived control human neurons. Left: representative immunoblot to PKR after indicated treatments. Right: Quantification of PKR expression by immunoblot in response to DHPG treatment (n=4; p=0.9696). Panel B, C, E, G: Two sided unpaired student t-test. Panel D: One-way ANOVA with multiple comparisons and post-hoc LSD. *p<0.05, **p<0.01. Graphs are mean +/− S.E.M (+/− SD for B and C).
Extended Data Fig. 8. Characterization of unmethylated Fragile X full mutation iPSC line TC43–97.
A) Detection of three pluripotency markers—OCT-3/4, Nanog, and SSEA4—in the TC43–97 iPSCs confirms successful reprogramming of the fibroblast line. B) Cytogenetic analysis of cells in metaphase revealed an apparently normal male karyotype of TC43–97 iPSCs. C) Methylation sensitive qPCR of FMR1 promoter demonstrates lack of DNA methylation in TC43–97 iPSCs. Methylation levels were calculated relative to the FX hESC condition (n=3). D) Quantification of immunoblots to FMRP in the TC43–97 iPSCs relative to Control (n=3; p=0.0088). Panel D: Two sided Student’s t-test, **p<0.01. Graphs are mean +/− S.E.M.
Extended Data Fig. 9. +1RAN ASO effects in TC43–97 neurons.
A) RT-PCR from indicated iPSC lines 24 hours after transfection with indicated ASOs: Control/Control ASO (n=4), Control/+1RAN ASO-1 (n=4), TC43–97/Control ASO (n=7), TC43–97/+1RAN ASO-1 (n=6, p=0.0522). B) Representative western blot showing relative FMRP expression in both Control and TC43–97 neurons treated with increasing doses of +1RAN ASO-1. Each lane is quantified relative to GAPDH and as a percent of the untreated control neurons. C) Soma (DAPI, blue) and processes (Tuj1, red) of differentiated neurons are TUJ1 positive in both control and TC43–97 neurons. D) Representative FMRP immunoblot from TC43–97 neurons treated with DHPG and indicated ASOs. E) Quantification of FMRP fluorescence by immunocytochemistry in human neurons treated as indicated, normalized to untreated Control ASO neurons quantified in parallel. Average fluorescence was binned for every 5 neurons consecutively analyzed to represent an individual data point. (Control ASO/Mock: n=7, Control ASO/DHPG: n=5, +1RAN ASO-1/Mock: n=6, +1RAN ASO-1/DHPG: n=5; Control ASO/Mock vs +1RAN ASO-1/Mock: p=0.0163). F) Survival analysis of TC43–97 iPSC derived neurons treated with 150nM +1RAN ASO-1 (n=272) or Control ASO (n=310), independent experiment and neuronal derivation #2 (p=0.000148). G) Survival analysis of TC43–97 iPSC derived neurons treated with 150nM +1RAN ASO-1 (n=220) or Control ASO (n=190), independent experiment and neuronal derivation #3 (p=0.027). Survival is plotted as cumulative risk of death. Panel C: Two sided Student T-test. Panel E: Two-way ANOVA with post-hoc correction for multiple comparisons. Panels F, G: Cox proportional hazard analysis. n.s.= not significant. * p<0.05. ***p<0.001. Graph is mean +/− S.E.M. Box extends to 25th/75th percentiles with a line at mean and whiskers indicate 95% CI.
Extended Data Fig. 10. Proposed Model for how RAN translation regulates FMRP synthesis.
CGG RAN regulates FMRP synthesis by limiting access of initiation complexes to the AUG initiation codon of FMRP in a repeat-dependent manner. II: mGluR activation bypasses CGG RAN, which allows for enhanced synthesis of FMRP. III: In the absence of CGG RAN or CGG repeat, steady-state FMRP synthesis increases but is decoupled from mGluR activation. IV: Non-cleaving RAN ASOs prevent CGG RAN initiation. This increases steady-state FMRP production, decreases FMRpolyG production and enhances neuronal survival.
ACKNOWLEDGEMENTS
We thank Todd and Sutton lab members and Aaron Goldstrohm for helpful discussions, commentary and technical advice. Christopher Pearson provided TC43-97 fibroblasts originally generated by Mitchell Turker44. Compound 1A was a gift from Matt Disney35. This work was funded by NIH (R01NS099280 and R01NS086810), and philanthropic donations to P.K.T. C.M.R was supported by NINDS F31 NS090883-03. M.G.K was supported by F32NS089124 and is now supported by K99GM126064. J.M.H. was funded by T32NS007222 and a post-doctoral fellowship from FRAXA. S.J.B. and B.N.F. were supported by the NIH (R01-NS097542, 1P30AG053760). S.E.W. was supported by NIH T-32-NS076401. M.I. and G.B. were supported by NINDS 5P30NS05507708. M.R.G. was supported by NIH T32NS007222. M.A.S. was supported by the Michigan Discovery Fund. F.R. and P.J.-N. were supported by Ionis Pharmaceuticals, who also funded reagent generation. IPSC work was supported by the University of Michigan Human Stem Cell and Genome Editing Core.
Footnotes
Competing interests
P.K.T. served as a consultant with Denali Therapeutics and has licensed technology through the University of Michigan to Denali. P.K.T., C.M.R, F.R., and P.J. hold a patent on ASOs targeting CGG RAN. F.R. and P.J. are paid employees of Ionis Pharmaceuticals. All other authors declare no competing interests.
References:
- 1.Paulson H in Handbook of clinical neurology Vol. 147 (eds Geschwind Daniel H., Paulson Henry L., & Klein Christine) 105–123 (Elsevier, 2018).29325606 [Google Scholar]
- 2.Hannan AJ Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet 19, 286–298, doi: 10.1038/nrg.2017.115 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Todd PK et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455, doi: 10.1016/j.neuron.2013.03.026 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zu T et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A 108, 260–265, doi: 10.1073/pnas.1013343108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verkerk AJ et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Hagerman RJ et al. Fragile X syndrome. Nat Rev Dis Primers 3, 17065, doi: 10.1038/nrdp.2017.65 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Berry-Kravis E et al. Fragile X-associated tremor/ataxia syndrome: clinical features, genetics, and testing guidelines. Movement disorders: official journal of the Movement Disorder Society 22, 2018–2030, quiz 2140, doi: 10.1002/mds.21493 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Greco CM et al. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain: a journal of neurology 125, 1760–1771 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Tassone F et al. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. American journal of medical genetics 94, 232–236 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Feng Y et al. Translational suppression by trinucleotide repeat expansion at FMR1 Science (New York, N.Y.) 268, 731–734 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Pretto D et al. Clinical and molecular implications of mosaicism in FMR1 full mutations. Frontiers in genetics 5, 318, doi: 10.3389/fgene.2014.00318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santa Maria L et al. FXTAS in an unmethylated mosaic male with fragile X syndrome from Chile. Clinical genetics 86, 378–382, doi: 10.1111/cge.12278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kearse MG et al. CGG Repeat-Associated Non-AUG Translation Utilizes a Cap-Dependent Scanning Mechanism of Initiation to Produce Toxic Proteins. Molecular cell 62, 314–322, doi: 10.1016/j.molcel.2016.02.034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sellier C et al. Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron 93, 331–347, doi: 10.1016/j.neuron.2016.12.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krans A, Skariah G, Zhang Y, Bayly B & Todd PK Neuropathology of RAN translation proteins in fragile X-associated tremor/ataxia syndrome. Acta neuropathologica communications 7, 152, doi: 10.1186/s40478-019-0782-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gemayel R, Vinces MD, Legendre M & Verstrepen KJ Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annual review of genetics 44, 445–477, doi: 10.1146/annurev-genet-072610-155046 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Eichler EE et al. Evolution of the cryptic FMR1 CGG repeat. Nature genetics 11, 301–308, doi: 10.1038/ng1195-301 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Collins SC et al. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. American journal of medical genetics. Part A 152A, 2512–2520, doi: 10.1002/ajmg.a.33626 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Lareau LF & Weissman JS Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802, doi: 10.1016/j.cell.2011.10.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingolia NT, Brar GA, Rouskin S, McGeachy AM & Weissman JS The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7, 1534–1550, doi: 10.1038/nprot.2012.086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinnebusch AG, Ivanov IP & Sonenberg N Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416, doi: 10.1126/science.aad9868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen LS, Tassone F, Sahota P & Hagerman PJ The (CGG)n repeat element within the 5’ untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum Mol Genet 12, 3067–3074, doi: 10.1093/hmg/ddg331 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Ludwig AL, Hershey JW & Hagerman PJ Initiation of translation of the FMR1 mRNA Occurs predominantly through 5’-end-dependent ribosomal scanning. J Mol Biol 407, 21–34, doi: 10.1016/j.jmb.2011.01.006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darnell JC et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261, doi: 10.1016/j.cell.2011.06.013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou L et al. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron 51, 441–454, doi: 10.1016/j.neuron.2006.07.005 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Nalavadi VC, Muddashetty RS, Gross C & Bassell GJ Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. The Journal of neuroscience 32, 2582–2587, doi: 10.1523/JNEUROSCI.5057-11.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiler IJ et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proceedings of the National Academy of Sciences of the United States of America 94, 5395–5400 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bear MF, Huber KM & Warren ST The mGluR theory of fragile X mental retardation. Trends Neurosci 27, 370–377, doi: 10.1016/j.tins.2004.04.009 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Todd PK, Mack KJ & Malter JS The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci U S A 100, 14374–14378, doi: 10.1073/pnas.2336265100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliff AJ et al. Impaired activity-dependent FMRP translation and enhanced mGluR-dependent LTD in Fragile X premutation mice. Human molecular genetics 22, 1180–1192, doi: 10.1093/hmg/dds525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber KM, Gallagher SM, Warren ST & Bear MF Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci U S A 99, 7746–7750, doi: 10.1073/pnas.122205699 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suhl JA et al. A 3’ untranslated region variant in FMR1 eliminates neuronal activity-dependent translation of FMRP by disrupting binding of the RNA-binding protein HuR. Proceedings of the National Academy of Sciences of the United States of America 112, E6553–6561, doi: 10.1073/pnas.1514260112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muslimov IA, Patel MV, Rose A & Tiedge H Spatial code recognition in neuronal RNA targeting: role of RNA-hnRNP A2 interactions. J Cell Biol 194, 441–457, doi: 10.1083/jcb.201010027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green KM et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nature communications 8, 2005, doi: 10.1038/s41467-017-02200-0 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Disney MD et al. A small molecule that targets r(CGG)(exp) and improves defects in fragile X-associated tremor ataxia syndrome. ACS chemical biology 7, 1711–1718, doi: 10.1021/cb300135h (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang WY et al. Small Molecule Recognition and Tools to Study Modulation of r(CGG)(exp) in Fragile X-Associated Tremor Ataxia Syndrome. ACS chemical biology 11, 2456–2465, doi: 10.1021/acschembio.6b00147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Prisco GV et al. Translational control of mGluR-dependent long-term depression and object-place learning by eIF2alpha. Nat Neurosci 17, 1073–1082, doi: 10.1038/nn.3754 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sidrauski C, McGeachy AM, Ingolia NT & Walter P The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. eLife 4, doi: 10.7554/eLife.05033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng W et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2alpha phosphorylation. Nature communications 9, 51, doi: 10.1038/s41467-017-02495-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang XH et al. Antisense oligonucleotides targeting translation inhibitory elements in 5’ UTRs can selectively increase protein levels. Nucleic Acids Res 45, 9528–9546, doi: 10.1093/nar/gkx632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang XH et al. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nature biotechnology 34, 875–880, doi: 10.1038/nbt.3589 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Tabet R et al. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nature communications 9, 152, doi: 10.1038/s41467-017-02643-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linsalata AE et al. DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO reports, e47498, doi: 10.15252/embr.201847498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burman RW, Popovich BW, Jacky PB & Turker MS Fully expanded FMR1 CGG repeats exhibit a length- and differentiation-dependent instability in cell hybrids that is independent of DNA methylation. Hum Mol Genet 8, 2293–2302 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Tabrizi SJ et al. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N Engl J Med 380, 2307–2316, doi: 10.1056/NEJMoa1900907 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Haenfler J et al. Targeted Reactivation of FMR1 Transcription in Fragile X Syndrome Embryonic Stem Cells. Front Mol Neurosci. 2018. August 15;11:282. doi: 10.3389/fnmol.2018.00282., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu XS et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172, 979–992 e976, doi: 10.1016/j.cell.2018.01.012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finkel RS et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 388, 3017–3026, doi: 10.1016/S0140-6736(16)31408-8 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Ifrim MF, Williams KR & Bassell GJ Single-Molecule Imaging of PSD-95 mRNA Translation in Dendrites and Its Dysregulation in a Mouse Model of Fragile X Syndrome. The Journal of neuroscience. 35, 7116–7130, doi: 10.1523/JNEUROSCI.2802-14.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatavarty V et al. Single-molecule imaging of translational output from individual RNA granules in neurons. Molecular biology of the cell 23, 918–929, doi: 10.1091/mbc.E11-07-0622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbarese E et al. Conditional knockout of tumor overexpressed gene in mouse neurons affects RNA granule assembly, granule translation, LTP and short term habituation. PloS one 8, e69989, doi: 10.1371/journal.pone.0069989 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu J, Xiao J, Ren X, Lao K & Xie XS Probing gene expression in live cells, one protein molecule at a time. Science 311, 1600–1603, doi: 10.1126/science.1119623 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Choi HM et al. Mapping a multiplexed zoo of mRNA expression. Development 143, 3632–3637, doi: 10.1242/dev.140137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huss D et al. Combinatorial analysis of mRNA expression patterns in mouse embryos using hybridization chain reaction. Cold Spring Harbor protocols 2015, 259–268, doi: 10.1101/pdb.prot083832 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Choi HM et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nature biotechnology 28, 1208–1212, doi: 10.1038/nbt.1692 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saluto A et al. An enhanced polymerase chain reaction assay to detect pre- and full mutation alleles of the fragile X mental retardation 1 gene. J Mol Diagn 7, 605–612, doi: 10.1016/S1525-1578(10)60594-6 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y, Kirwan P & Livesey FJ Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nature protocols 7, 1836–1846, doi: 10.1038/nprot.2012.116 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Barmada SJ et al. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc Natl Acad Sci U S A 112, 7821–7826, doi: 10.1073/pnas.1509744112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arrasate M, Mitra S, Schweitzer ES, Segal MR & Finkbeiner S Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810, doi: 10.1038/nature02998 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.