Supplemental Digital Content is available in the text
Keywords: compartmentalization, genitourinary tract, HIV-1, semen, urine
Abstract
HIV-1 persists indefinitely in multiple cellular reservoirs despite antiretroviral therapy. We previously demonstrated HIV-1 compartmentalization in kidney and urine. Here, we further characterized viruses in urine and when available, compared them to those present in semen from HIV-1 positive participants with detectable plasma viremia to further understand the viral dynamics in the upper and lower genitourinary tract.
Blood and urine samples were obtained from 19 HIV-1 positive participants. Simultaneous semen samples were obtained from 16 of the 19 participants. HIV-1 envelope (env) gene sequences were obtained by single-genome amplification (SGA) and neighbor-joining trees were constructed using the Kimura 2-parameter model.
HIV-1 env gene sequences were amplified from blood in 19/19 (100%) participants, urine in 18/19 (95%) participants, and semen in 12/16 (75%). In individuals from which both urine and semen samples were obtained, differences in viral shedding between the 2 sources were observed, where HIV-1 env sequences could only be amplified from either urine or semen. Longitudinal phylogenetic analysis of urine-derived env sequences from 1 participant demonstrated that urine clusters distinct from blood are maintained over time (20 weeks), consistent with viral compartmentalization and local replication. Comparison of urine and semen derived sequences demonstrated either virus compartmentalization or equilibration.
Our results demonstrate that when present, viral compartmentalization in urine persists over time. Comparison of timing of viral shedding in urine and semen samples from our cohort suggest different viral kinetics between the upper and lower genitourinary tract and sequence analysis suggests that HIV-1 populations in urine and semen can either be imported from blood or produced locally.
1. Introduction
HIV-1 can infect and persist in different organs and tissues, resulting in the generation of distinct viral populations, compartments, and reservoirs. Viral compartments restrict HIV-1 trafficking and gene flow and facilitate separate viral evolution and divergence from the peripheral blood or other anatomical sites.[1–3] Multiple viral compartments have been described in HIV-1 positive participants, including lymph nodes,[4] gut associated lymphoid tissue,[5] central nervous system,[6] kidney[7] and lower genitourinary (GU) tract.[8] Understanding the viral dynamics in these compartments is critical to informing cure strategies.
We have previously demonstrated that HIV positive participants shed virus in urine and that in some individuals those viruses are genetically distinct from the viral populations found in blood.[9] The exact source of the urine-derived viruses in the GU tract is unknown. They could originate either from the upper or lower GU tract, which is particularly relevant when analyzing urine-derived HIV-1 sequences in males where the presence of HIV-1 sequences and replication-competent viruses in semen is well documented.[8,10–13] Sequence analyses of viral RNA populations in semen suggest that viruses can originate from different mechanisms including direct import of virus from blood, compartmental evolution of the virus or clonal expansion in the seminal tract.[8] Interestingly, sequence analysis of HIV envelope (env) from urine samples suggests that virus in urine could also originate through multiple mechanisms, since in addition to virus compartmentalization and clonal amplification, we observed equilibrated viral populations between blood and urine in some study participants.[9] The differences observed between the blood and genitourinary viruses suggest that the blood does not fully reflect the genitourinary compartment in assessing the HIV-1 reservoir. Additionally, the presence of identical sequences in urine could be the result of clonal expansion of infected cells in the upper GU tract, as it has been recently described for infected T cells in the bloodstream.[14,15]
Our group has demonstrated that HIV-1 infects renal tubular epithelial (RTE) cells in vitro and in vivo and that the kidney represents a separate compartment for HIV replication.[7,16] We have shown that HIV-1 is transferred to renal epithelial cells through cell-cell contact with infected T cells[17,18] and macrophages[19] and that, once infected, renal epithelial cells can produce new viral particles that are rescued by CD4+ T cells and monocytes coming into contact with infected RTE.[17–19] Additionally, we recently demonstrated in vitro, that renal epithelial cells that acquired HIV-1 from infected macrophages, can undergo multiple rounds of cell division,[19] suggesting that proliferation of infected renal cells may contribute to HIV-1 persistence in this compartment. Since HIV can infect and persist in renal epithelial cells in vivo,[20,21] the kidney may serve as a long-term reservoir that will need to be targeted in any eradication strategy. We have recently shown in a person infected with HIV receiving a kidney from an HIV-1 positive donor that the viruses shed in urine are genetically more closely related to those present in urine-derived renal epithelial cells than to viruses present in blood,[22] supporting renal epithelial cells as 1 source of urine viruses.
Because of the close proximity and relationship between the upper and lower GU tracts in males, we have expanded our previous studies to directly compare the relationship of HIV-1 sequences from urine and semen to determine the extent of viral divergence and similarity of 1 source from the other and from the blood. Additionally, we performed viral sequence analysis on longitudinal urine samples to assess the viral evolution in this compartment over time.
2. Materials and methods
2.1. Study participants and sample processing
Ten HIV-1 positive participants (9 males and 1 female) with detectable viral load were enrolled in this study. Simultaneous blood, urine, and semen samples were obtained from 7 of the enrolled participants, while only urine and blood could be collected from the remaining 3. All participants gave informed consent, and sample collections were performed with institutional review board approval (Pro0040696). Nine additional HIV-1 positive individuals were selected from the CHAVI001 acute and established cohorts from a subset of participants from whom blood, urine, and semen samples were available.[8]
Large volumes of urine ranging from 60 to 775 ml were collected from the 10 enrolled participants while approximately 20 ml of urine was available from the CHAVI001 participants. Urine samples were spun at 400 x g for 10 minutes to separate urine supernatants from urinary cells. Supernatants were then filtered through a 0.45 μm filter unit to remove cellular debris followed by 2 hours of ultracentrifugation to pellet HIV virions as previously described.[9] Pelleted viruses were then resuspended in 400 μl of 1X phosphate-buffered saline (PBS) and either immediately subjected to RNA extraction or stored at −80°C.
Semen samples were collected in 2 ml of viral transport medium (VTM) consisting of RPMI 1640, 10% fetal bovine serum, 2 mM L-Glutamine, 200 U/ml of Nystatin, 100 U/ml Pen/Strep, and processed within 4 hours of collection. Semen was further diluted 1:2 in VTM supplemented with 100X protease inhibitor buffer and spun for 10 minutes at 800 x g to separate the seminal plasma from seminal cells. Seminal plasma was then collected and ultracentrifuged for 2 hours at 23000 rpm to pellet HIV virions. Pelleted viruses were resuspended in 200 μl of VTM and either immediately subjected to RNA extraction or stored at −80°C. The seminal cell pellet (containing sperm cells and lymphocytes) was frozen at −20°C until DNA extraction.
EDTA anticoagulated blood samples were processed within 4 hours from collection to isolate plasma and peripheral blood mononuclear cells (PBMCs) by Ficoll gradient centrifugation. Plasma was ultracentrifuged for 2 hours at 23000 rpm to pellet HIV-1 virions and then resuspended in 400 μl of 1X phosphate-buffered saline and either immediately subjected to RNA extraction or stored at −80°C. PBMCs were pelleted and stored at −20°C for DNA extraction.
2.2. Viral RNA extraction and cDNA synthesis
Viral RNA extraction and cDNA synthesis from urine and plasma was performed as previously described.[9,22] Viral RNA was also extracted from 200 μl of concentrated seminal plasma using the Roche High Pure Viral RNA Kit following the manufacturers protocol. cDNA synthesis of seminal plasma was performed as previously described.[9]
2.3. Viral DNA extraction
Viral DNA was extracted from 5 × 106 PBMC and seminal cell pellet by using the QIAamp micro kit (Qiagen) following the manufacturers instructions and eluted in 50 μl of water.
2.4. Single genome amplification
Single-genome amplification of HIV-1 env sequences was performed as previously described.[9] We performed PCR on each sample type (urine cDNA, blood plasma, PBMC, seminal plasma, and seminal cell) until the sample was exhausted or we reached > 15 sequences per sample type.
2.5. Sequencing and sequence analyses
The complete env gene amplicons were sequenced using the primer walking method as previously described.[9] Every positive reaction was inspected for double peaks using the Sequencher program 5.4.1 (Gene Codes). Any sequence with evidence of mixed bases was excluded from further analysis. All alignments were made using gene cutter (hiv.lanl.gov) and phylogenetic trees were made with MEGA6.[23] Neighbor-joining trees were constructed using the Kimura 2-parameter mode and the reliability of topologies was estimated by performing bootstrap analysis with 1000 replicates. Highlighter plots (hiv.lanl.gov) were generated to show the genetic variation of sequences in urine supernatants.
2.6. Statistical analysis
To estimate the required number of sequences the following argument was made: the expectation under the null hypothesis is that, in any 1 participant, virus sequences from urine will be found clustering with sequences from semen. Thus the frequency of urine sequences clustering with semen sequences is p0 = 0.5. If the null hypothesis is rejected because urine viruses are derived from the lower male GU tract we should find that the frequency of urine sequences clustering with semen sequences is p1 = 90%. If alpha (probability of type I error) is 0.05 the power is 0.8 with 10 sequences from urine, semen and PBMC (based on normal approximation to the binomial distribution).
3. Results
3.1. Viral compartmentalization and clonal amplification of HIV-1 in urine persist over time.
We have previously demonstrated that HIV-1 positive participants shed viruses in urine and that those viruses are compartmentalized from blood in the majority of the participants analyzed.[9] To further expand our analysis, we collected urine and blood specimens from 2 HIV-1 positive participants with detectable viremia at a single time point and from one additional participant at multiple time points post-enrollment (Table 1). We performed single-genome amplification on HIV-1 env RNA isolated from urine supernatants and blood plasma as well as on HIV-1 env DNA isolated from PBMCs. Multiple HIV-1 env sequences were amplified from each specimen in each participant (Table 1). There was no correlation between plasma viral load and the number of sequences amplified from urine (Table 1), consistent with our previous report.[9] A phylogenetic analysis of the HIV-1 env sequences amplified from urine and blood revealed 2 scenarios: urine-derived env sequences were either intermixed with blood-derived sequences (participant 40696-028 in Supplementary Fig. 1), suggesting viral equilibrium and viral exchange between urine and blood, or the urine-derived env sequences clustered either predominantly (participant 40696-049 in Supplementary Fig. 1) or exclusively separate (participant 40696-073 in Supplementary Fig. 1) from the blood-derived plasma and PBMC sequences, suggesting that in those cases the urine-derived viruses originated from a separate source.
Table 1.
Characteristics of study participants.
| Participant ID | Ethnicity | Sex | HIV diagnosis (year) | Plasma Viral Load (Log10 copies/ml) | CD4 Count | Urine #SGA Seqs | Semen (C+P) #SGA Seqs | Plasma #SGA Seqs | PBMC #SGA Seqs | |
| Urine only | ||||||||||
| 40696-028 | Black | M | 2000 | 3.8 | 27 | 19 | n/a | 19 | 8 | |
| 40696-049 | Hispanic | F | 2005 | 5.0 | 81 | 31 | n/a | 18 | 16 | |
| 40696–073∗ | Black | M | 2012 | |||||||
| Week 0 | 4.9 | 55 | 21 | 4.9 | 21 | 18 | ||||
| Week 2 | 5 | 55 | 18 | 5.0 | 21 | 22 | ||||
| Week 12 | 3.3 | 62 | 40 | 3.3 | 19 | 17 | ||||
| Week 20 | 3.3 | 62 | 19 | 3.3 | 17 | 21 | ||||
| North Carolina samples | ||||||||||
| 40696–011∗ | Black | M | 2007 | 5.3 | 60 | 23 | 0 | 15 | 19 | |
| 40696-014 | Black | M | 2010 | 3.8 | 496 | 3 | 0 | 11 | 23 | |
| 40696–031∗ | Black | M | 2001 | 4.1 | 368 | 19 | 2 | 15 | 18 | |
| 40696-047 | Asian | M | 2011 | 3.8 | 482 | 8 | 0 | 10 | 16 | |
| 40696-063 | Black | M | 2000 | 5.2 | 416 | 5 | 0 | 4 | 25 | |
| 40696-072 | Black | M | 2007 | 4.4 | 788 | 0 | 12 | 20 | 10 | |
| 40696–068∗ | Black | M | 2011 | |||||||
| Week 0 | 5.8 | 4 | 74 | 5.8 | 19 | 19 | ||||
| Week 3 | 5.8 | 4 | 7 | 5.8 | 15 | 19 | ||||
| Week 5 | 5.8 | 4 | 30 | 5.8 | n/a | n/a | ||||
| Archived samples (CHAVI 001 study) | ||||||||||
| 700011043 | Black | M | Acute | 5.8 | n/a | 1 | 0 | n/a | n/a | |
| 700011410∗ | Black | M | Chronic | 5.2 | n/a | 17 | 18 | 19 | 16 | |
| 700011161 | Black | M | Chronic | 5.7 | n/a | 14 | 32 | 19 | 18 | |
| 700010920 | Black | M | Chronic | 5.1 | n/a | 3 | 4 | n/a | n/a | |
| 700010914 | Black | M | Acute | 5.8 | n/a | 3 | 4 | n/a | n/a | |
| 700011084 | Black | M | Acute | 5.0 | n/a | 15 | 0 | 26 | 19 | |
| 700010333 | Black | M | Chronic | 4.2 | n/a | 3 | 19 | n/a | n/a | |
| 700010501 | Black | M | Chronic | 4.3 | n/a | 2 | 26 | n/a | n/a | |
| 700011145∗ | Black | M | Chronic | 4.7 | n/a | 33 | 11 | 36 | 19 |
To further understand the dynamics of viral compartmentalization in urine over time, we collected longitudinal urine and blood samples from participant 40696-073 up to week 20 post-enrollment. Phylogenetic analysis of HIV-1 env sequences amplified at each time point from week 0 to week 20 demonstrated that viral compartmentalization in urine persists over time with limited exchange of virus between urine and blood (Fig. 1a). Interestingly, we observed that as this participants viral load decreased after initiation of antiretroviral treatment (Table 1), multiple identical HIV-1 env sequences were amplified from urine. To compare sequences across all time points, we constructed a phylogenetic tree of sequences from all sample collections. As shown in Figure 1b, in addition to maintenance of viral compartmentalization between urine and blood over time, we observed 3 clusters of identical or nearly identical sequences (9/11 sequences identical, 2/11 sequences with one nucleotide mutation), containing sequences from different time points. The presence of identical sequences from different time points suggests possible clonal amplification of HIV-1 in the upper GU tract.
Figure 1.

Longitudinal phylogenetic analysis of urine-derived HIV-1 env sequences demonstrates the persistence of viral compartmentalization and clonal amplification over time. (a) Blood and urine were collected from participant 40696–073 at weeks 0, 2, 12, and 20 post-enrollment. Neighbor-joining phylogenetic trees including env sequences from each time point are shown. (b) Neighbor-joining phylogenetic tree including all the HIV-1 env sequences amplified from participant 40696–073 between weeks 0 and 20 post-enrollment. Identical HIV-1 env sequences were amplified across the different time points, supporting clonal amplification of HIV-1 in the genitourinary tract. Highlighter plots confirm the lack of sequence diversity in the selected cluster of sequences. PBMC SGA-env sequences (full red circles), plasma SGA-env sequences (open red circles) and urine SGA-env sequences (blue triangles). Bootstrap values ≥80 are shown. Genetic distance is indicated at the bottom of the figure and represents the number of nucleotide substitutions per site.
3.2. Viral shedding kinetics differs between urine and semen.
Because of the relationship between the upper and lower GU tracts in males, we next compared viral populations between urine and semen. HIV-1 env sequences were amplified from paired urine, semen, and blood samples from men with chronic HIV-1 infection in North Carolina (n = 7) or from the CHAVI 001 cohort (n = 9), 3 with acute and 6 with chronic HIV-1 infection (Table 1). Single genome amplification was performed on HIV-1 env RNA isolated from urine supernatants, blood plasma, and seminal plasma as well as on HIV-1 env DNA isolated from PBMCs and seminal cells. Interestingly, as shown in Table 1, for several participants we were able to amplify HIV-1 env sequences in urine but not in semen (n = 6), and vice versa (n = 1). The phylogenetic analysis of HIV-1 env sequences amplified from urine or semen in the participants that had at least 8 urine-derived or semen-derived sequences demonstrated compartmentalization of urine-derived sequences from blood in 3 out of 3 participants and viral equilibrium between semen and blood in one participant (Fig. 2 and Supplementary Fig. 2). The differences in viral shedding observed suggest that the viral populations found in urine and semen could originate from different cellular sources in the GU tract.
Figure 2.
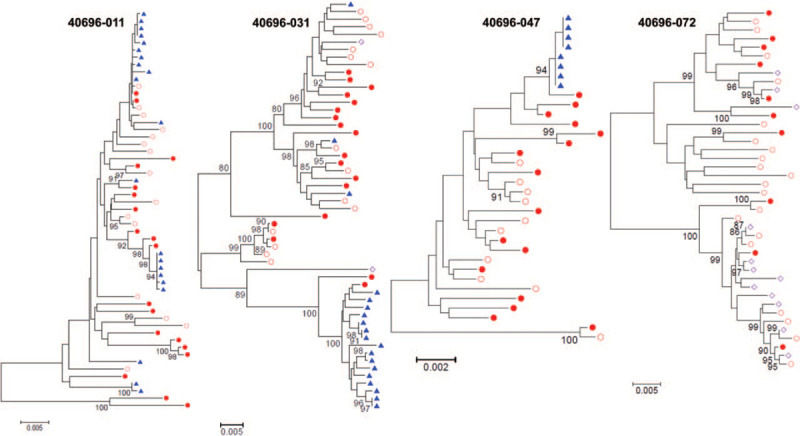
HIV-1 shedding kinetics differ between urine and semen. Neighbor-joining phylogenetic trees of HIV-1 env sequences amplified from 4 participants from which blood, urine, and semen were simultaneously collected. In 3 of the 4 participants (40696–011, 40696–031, 40696–047) multiple urine-derived HIV-1 env sequences could be amplified while no or a limited number (2 sequences for participant 40696–031) of semen-derived HIV-1 env sequences were detected. Conversely, in participant 40696–072 we amplified several HIV-1 env sequences from semen samples but not from urine. PBMC SGA-env sequences (full red circles), plasma SGA-env sequences (open red circles), urine SGA-env sequences (blue triangles), and seminal plasma SGA-env sequences (open purple diamond). Bootstrap values ≥80 are shown. Genetic distance is indicated at the bottom of the figure and represents the number of nucleotide substitutions per site.
3.3. Urine and semen-derived HIV-1 env sequences are compartmentalized in some participants and equilibrated in others.
To further our comparison of the viral populations present in urine and semen, we focused on participants from whom we were able to amplify > 10 urine and seminal sequences, one chronically infected male from the North Carolina cohort and 3 chronically infected males from the CHAVI 001 cohort (Table 1). As shown in Figure 3, we observed 2 scenarios. In 3 participants, urine-derived env sequences were intermixed with both blood and seminal-derived sequences (participants 700011161, 700011410, and 40696-068, Fig. 3a). Intermixing of these sequences suggests viral equilibrium and exchange between the blood and the GU tract, suggesting that those urine and semen-derived viruses are likely produced by infected lymphocytes trafficking from one site to the other. However, similarly to what we have observed when comparing urine and blood viral populations, viral compartmentalization of urine-derived HIV-1 env sequences from semen was observed in participant 700011145. The majority of these sequences formed independent clusters from sequences amplified from blood or semen; conversely, the majority of semen-derived sequences in this participant were intermixed with blood sequences (Fig. 3b). Notably, the independent clusters of urine-derived sequences contained several identical sequences, suggestive of clonal amplification of HIV-1 infected cells within the upper GU tract. Clonal amplification was not evident within blood or seminal samples in this participant.
Figure 3.
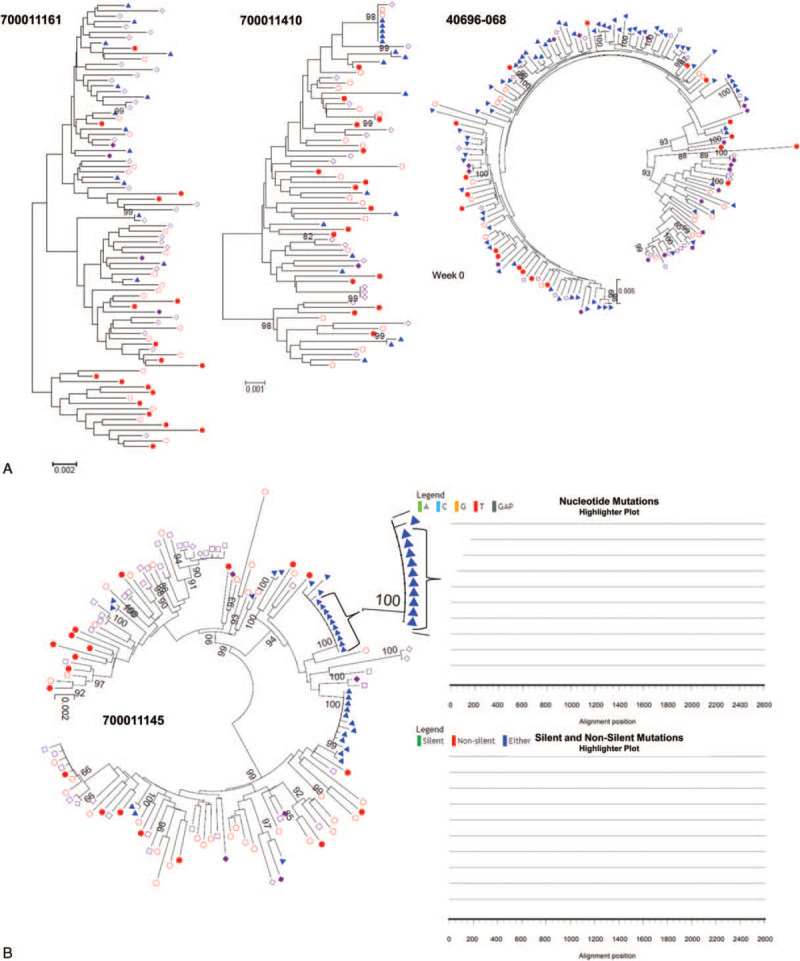
Phylogenetic analysis of urine, semen, and blood demonstrates viral equilibrium between urine and semen in some participants and compartmentalization of urine-derived sequences in others. (a) Neighbor-joining phylogenetic tree of full-length env sequences obtained via SGA from urine supernatant, blood plasma, blood PBMCs, semen supernatant, and semen cells from 4 participants. Viral equilibrium between urine and semen was observed in participants 700011161, 700011410, and 40696–068. (b) Compartmentalization of urine-derived viruses was observed in participant 700011145. PBMC SGA-env sequences (full red circles), plasma SGA-env sequences (open red circles), seminal cell SGA-env sequences (full purple diamonds), seminal plasma SGA-env sequences (open purple diamonds), and urine SGA-env sequences (blue triangles). Bootstrap values ≥80 are shown. Genetic distance is indicated at the bottom of the figure and represents the number of nucleotide substitutions per site.
To determine whether viral equilibrium between the different anatomical spaces persists overtime, we collected longitudinal blood, urine, and semen samples from participant 40696-068 up to week 5 post-enrollment. We observed that viral intermixing was maintained at each time point, suggesting that these viral dynamics are consistent for at least 5 weeks (Fig. 4). Interestingly, we observed the presence of identical sequences across sample types (PBMC, urine and semen), suggesting a common cellular source for those viruses.
Figure 4.

Longitudinal phylogenetic analysis of urine and semen-derived HIV-1 env sequences demonstrates the persistence of viral equilibrium and the presence of identical sequences across sample types. Blood, urine, and semen samples were collected from participant 40696–068 at weeks 0, 3, and 5 post-enrollment. A neighbor-joining phylogenetic tree including env sequences from all the time points is shown. Identical HIV-1 env sequences were amplified at both week 0 and 5. The highlighter plots confirm the lack of sequence diversity in the 2 selected sequence clusters. PBMC SGA-env sequences (full circles), plasma SGA-env sequences (open circles), seminal cell SGA-env sequences (full diamonds), seminal plasma SGA-env sequences (open diamonds), and urine SGA-env sequences (triangles). Symbols are colored based on time point. Bootstrap values ≥80 are shown. Genetic distance is indicated at the bottom of the figure and represents the number of nucleotide substitutions per site.
4. Discussion
Previous work has demonstrated viral compartmentalization of urine-derived HIV-1 env sequences from blood in both male and female participants,[9] suggesting the presence of a separate population of infected cells in the upper genitourinary tract. In this study we expanded our analysis to assess the viral dynamics in urine over time and compared viral populations in urine to those present in semen. Because of the anatomic relationship between the upper and lower genitourinary tracts in males, the comparison of urine, semen, and blood sequences is important to understand the GU viral dynamics and to determine the extent of viral divergence and similarity of one source from the other.
The analysis of longitudinal urine samples suggest that, when present, compartmentalization of urine-derived HIV-1 env sequences persists over time. In one study participant (40696-073), compartmentalization of urine-derived sequences was maintained up to at least 20 weeks post-enrollment, with limited evidence of viral exchange between urine and blood. These long-term viral dynamics further suggest the presence of a separate viral source in the upper GU tract. We observed that 2 HIV-1 env sequences amplified from plasma at week 12 post-enrollment clustered together with the urine-derived sequences and was separate from the other blood-derived sequences. This suggests that the GU compartment is capable of seeding the systemic circulation. Interestingly, as plasma viral load decreased in this participant, we were still able to amplify a large number of urine-derived sequences that continued to cluster independently from blood. Additionally, a higher number of identical or nearly identical sequences were amplified at later time points, supporting clonal amplification of infected cells in this compartment.
We have recently demonstrated in an HIV-1 positive kidney transplant recipient that the viruses present in urine were genetically different from the viral quasi-species found in blood but closely related to urine-derived renal epithelial cells,[22] supporting renal epithelial cells as one source of urine viruses. In addition to renal epithelial cells, some of those urine-derived viruses might originate from interstitial lymphocytes or macrophages residing in the kidney. The viral diversity observed in the urine-derived viruses could also be the result of continuous viral transfer from infected lymphocytes and macrophages to renal epithelial cells and vice versa, as previously demonstrated in vitro,[9,19] which could sustain HIV-1 replication and evolution in this compartment. Furthermore, the demonstration that some productively infected renal epithelial cells undergo multiple rounds of proliferation in vitro,[19] provides a possible mechanism for HIV-1 persistence in the kidney and may explain the amplification of several identical sequences in urine. Alternatively, these identical sequences could be the result of an activated infected cell producing identical virions. However, because identical sequences were detected across multiple time points over 20 weeks of follow-up, the presence of infected cells containing the same integrated provirus is more likely. Several studies on CD4+ T cells have demonstrated that clonal expansion of HIV-1 infected cells is a driver of HIV-1 persistence, and that those cells can produce infectious viruses.[24–28] Understanding the mechanisms sustaining HIV-1 persistence in different reservoir sites throughout the body of infected individuals is crucial to optimize HIV-1 cure strategies.
Previous studies have demonstrated that there are temporal differences in HIV-1 shedding within semen independent of blood viral dynamics.[8] We observed that viral shedding kinetics differ also between urine and semen; in a number of participants we were able to amplify HIV-1 sequences from urine but not from semen and vice versa. These data suggest that the urine and semen viruses come from different sources in the genitourinary tract.
We observed 2 different scenarios in the participants from which we obtained ≥11 sequences from both urine and semen:
-
1.
urine-derived sequences formed separate clusters from blood and semen or
-
2.
viral sequences from all the different specimens were equilibrated.
The intermixing of urine, semen, and blood sequences suggests viral exchange between these different anatomical compartments, while the compartmentalization of urine-derived sequences in some participants, indicates that those sequences originated from a separate cellular source in the upper genitourinary tract, likely renal epithelial cells or interstitial lymphocytes/macrophages present in the kidney.
Several identical sequences were amplified across multiple time points in one participant (40696–068) from which longitudinal urine, blood, and semen samples were collected. Interestingly, one cluster of identical sequences consisted of a seminal cell-derived sequence, a seminal plasma-derived sequences, and 4 urine-derived sequences. The presence of identical sequences across sample types suggest seminal cells as the source of these urine-viruses. A second cluster of identical sequences containing 3 PBMC, 5 urine, 1 seminal plasma, and 1 seminal cell-derived sequence was also detected, indicating trafficking of expanded clones of infected cells between the 3 sites.
In conclusion, our study shows that while some participants presented equilibrated viral populations in urine, semen, and blood, supporting continuous import of those viruses from blood, other participants presented compartmentalized urine viral clusters that contained several identical sequences. In this compartment, likely the kidney, the virus can undergo independent replication, as supported by the analysis of longitudinal urine samples in patient 40696–073 and previous work on kidney biopsies of HIV-1 positive participants with HIV-associated nephropathy.[7] Finally, our results demonstrate that the analysis of urine-derived HIV-1 sequences is an important non-invasive approach that can and should be used to understand this non-blood compartment and its implications for HIV-1 persistence and cure strategies.
Acknowledgments
The authors thank Mehri Mckellar and Laura Mkumba for coordinating samples procurement. HS performed experiments, analyzed the data and wrote the first draft of the manuscript. EW performed experiments, data analysis and manuscript editing. KH and JC performed experiments. FG contributed to data analysis and editing of the manuscript. MEK and MB oversaw the planning and direction of the project including analysis and interpretation of the data and editing of the manuscript. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) [RO1DK108367, P01DK056492].
Author contributions
Conceptualization: Mary Klotman, Maria Blasi.
Data curation: Hannah Stadtler, Elizabeth Wescott, Feng Gao, Maria Blasi.
Formal analysis: Hannah Stadtler, Feng Gao, Mary Klotman, Maria Blasi.
Funding acquisition: Mary Klotman.
Methodology: Hannah Stadtler, Elizabeth Wescott, Kelly Hughes, Jerry Chang.
Project administration: Maria Blasi.
Supervision: Maria Blasi.
Writing – original draft: Hannah Stadtler.
Writing – review & editing: Elizabeth Wescott, Kelly Hughes, Jerry Chang, Feng Gao, Mary Klotman, Maria Blasi.
Supplementary Material
Footnotes
Abbreviations: env = envelope, GU = genitourinary tract, PBMC = peripheral blood mononuclear cells, PBS = phosphate buffered saline, RTE = renal tubule epithelial, Seqs = sequences, SGA = single genome amplification, VTM = viral transport media.
How to cite this article: Stadtler H, Wescott E, Hughes K, Chang J, Gao F, Klotman M, Blasi M. HIV-1 diversity and compartmentalization in urine, semen, and blood. Medicine. 2020;99:46(e23063).
HS and EW contributed equally: author order was decided alphabetically.
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants number P01DK056492 and R01DK108367.
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
participants for whom phylogenetic trees are shown in Figures 1–4. Additional phylogenetic trees are shown in the Supplementary Appendix. n/a = not available, Seqs = Sequences, SGA = single genome amplification.
References
- [1].Karris MA, Smith DM. Tissue-specific HIV-1 infection: why it matters. Future Virol 2011;6:869–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nickle DC, Jensen MA, Shriner D, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol 2003;77:5540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nickle DC, Shriner D, Mittler JE, et al. Importance and detection of virus reservoirs and compartments of HIV infection. Curr Opin Microbiol 2003;6:410–6. [DOI] [PubMed] [Google Scholar]
- [4].Fletcher CV, Staskus K, Wietgrefe SW, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A 2014;111:2307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Marle G, Gill MJ, Kolodka D, et al. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology 2007;4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schnell G, Joseph S, Spudich S, et al. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog 2011;7:e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marras D, Bruggeman LA, Gao F, et al. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med 2002;8:522–6. [DOI] [PubMed] [Google Scholar]
- [8].Anderson JA, Ping LH, Dibben O, et al. HIV-1 populations in semen arise through multiple mechanisms. PLoS Pathog 2010;6:e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Blasi M, Carpenter JH, Balakumaran B, et al. Identification of HIV-1 genitourinary tract compartmentalization by analyzing the env gene sequences in urine. AIDS 2015;29:1651–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Houzet L, Matusali G, Dejucq-Rainsford N. Origins of HIV-infected leukocytes and virions in semen. J Infect Dis 2014;210: Suppl 3: S622–630. [DOI] [PubMed] [Google Scholar]
- [11].Butler DM, Delport W, Kosakovsky Pond SL, et al. The origins of sexually transmitted HIV among men who have sex with men. Sci Transl Med 2010;2:18re11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pilcher CD, Joaki G, Hoffman IF, et al. Amplified transmission of HIV-1: comparison of HIV-1 concentrations in semen and blood during acute and chronic infection. AIDS 2007;21:1723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chaillon A, Smith DM, Vanpouille C, et al. HIV Trafficking between blood and semen during early untreated HIV infection. J Acquir Immune Defic Syndr 2017;74:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maldarelli F, Wu X, Su L, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014;345:179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wagner TA, McLaughlin S, Garg K, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014;345:570–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bruggeman LA, Ross MD, Tanji N, et al. Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol 2000;11:2079–87. [DOI] [PubMed] [Google Scholar]
- [17].Blasi M, Balakumaran B, Chen P, et al. Renal epithelial cells produce and spread HIV-1 via T-cell contact. AIDS 2014;28:2345–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen P, Chen BK, Mosoian A, et al. Virological synapses allow HIV-1 uptake and gene expression in renal tubular epithelial cells. J Am Soc Nephrol 2011;22:496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hughes K, Akturk G, Gnjatic S, et al. Proliferation of HIV-infected renal epithelial cells following virus acquisition from infected macrophages. AIDS 2020;34:1581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Winston JA, Bruggeman LA, Ross MD, et al. Nephropathy and establishment of a renal reservoir of HIV type 1 during primary infection. N Engl J Med 2001;344:1979–84. [DOI] [PubMed] [Google Scholar]
- [21].Canaud G, Dejucq-Rainsford N, Avettand-Fenoel V, et al. The kidney as a reservoir for HIV-1 after renal transplantation. J Am Soc Nephrol 2014;25:407–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Blasi M, Stadtler H, Chang J, et al. Detection of Donor's HIV Strain in HIV-Positive Kidney-Transplant Recipient. N Engl J Med 2020;382:195–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Reeves DB, Duke ER, Wagner TA, et al. A majority of HIV persistence during antiretroviral therapy is due to infected cell proliferation. Nat Commun 2018;9:4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hosmane NN, Kwon KJ, Bruner KM, et al. Proliferation of latently infected CD4(+) T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J Exp Med 2017;214:959–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Simonetti FR, Sobolewski MD, Fyne E, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 2016;113:1883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Haworth KG, Schefter LE, Norgaard ZK, et al. HIV infection results in clonal expansions containing integrations within pathogenesis-related biological pathways. JCI Insight 2018;3(13.): [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mullins JI, Frenkel LM. Clonal expansion of human immunodeficiency virus-infected cells and human immunodeficiency virus persistence during antiretroviral therapy. J Infect Dis 2017;215: suppl_3: S119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


