Abstract
Nasopharyngeal carcinoma (NPC) is a malignant epithelial tumor ubiquitously associated with the Epstein‐Barr virus (EBV), which is highly prevalent in South China, Southeast Asia, and North Africa. Despite being a highly radio‐sensitive and treatable cancer, a majority of NPC patients are diagnosed in their advanced stage, and locoregional and distant relapses following definitive treatment contribute largely to cancer‐specific mortality among these patients. Given that EBV‐driven NPC is the predominant variant seen in endemic regions, various EBV detection methods have been developed and are utilized in screening, prognostication, and post‐treatment surveillance of NPC patients. While the Immunoglobulin A (IgA) serology assay is the most extensively studied EBV detection method, the detection of plasma EBV DNA released during replication or cellular apoptosis has shown superior outcomes in endemic population screening, prognostication, and detection of distant relapse. Furthermore, there is emerging evidence on the use of circulating tumor cells, microRNAs, DNA hypermethylation, and combination assays in various clinical scenarios. Herein, this paper provides a comprehensive overview of the relevant studies using various EBV detection techniques in the management of NPC. Specifically, the recent advances, clinical evidence, and challenges associated with the clinical application of EBV liquid biopsies in population screening, prognostication, and surveillance of NPC are presented.
Keywords: circulating tumor cells, EBV DNA, Epstein‐Barr virus, microRNA, nasopharyngeal carcinoma, prognosis, screening, serology, surveillance
List of abbreviations
- BARF1
BamHI‐A rightward frame 1
- BART
BamHI‐A rightward transcripts
- BHRF1
BamHI fragment H rightward open reading frame 1
- CDH4
cadherin 4
- CDKN2A
cyclin‐dependent kinase inhibitor 2A
- CK
cytokeratin
- CRT
chemoradiotherapy
- CSF
cerebrospinal fluid
- CT
chemotherapy
- CTCs
circulating tumor cells
- DAPI
4′,6‐Diamidino‐2‐Phenylindole
- DAPK
death‐associated protein kinase
- DFS
disease‐free survival
- DLEC1
Deleted in lung and esophageal cancer 1
- DMFS
distant metastasis‐free survival
- DNMT
DNA methyltransferase
- dPCR
digital polymerase chain reaction,
- EA
early antigen
- EBER
EBV‐encoded RNA
- EBER‐1
EBV‐encoded RNA‐1
- EBNA
Epstein‐barr nuclear antigen
- EBV
Epstein‐Barr virus
- ELISA
enzyme‐linked immunosorbent assay
- epCAM
epithelial cell adhesion molecules
- EU
endotoxin
- FDA
Food and Drug Administration
- IE
immunoenzymatic method
- IF
immunofluorescence
- IFA
immunofluorescence assay
- IgA
immunoglobulin A
- IgG Rta
Rta protein antibody Immunoglobulin
- IIF
Indirect immunofluorescence
- IMRT
intensity modulated radiotherapy
- LMP1
latent membrane protein 1
- LMP2
latent membrane protein 2
- LOD
limit of detection
- LRRFS
loco‐regional relapse‐free survival
- miRNA
micro‐ribonucleic acid
- NA
nuclear antigen
- NP
nasopharyngeal
- NPC
Nasopharyngeal carcinoma
- NPV
negative predictive value
- OS
overall survival
- PCR
polymerase chain reaction
- PCR
polymerase chain reaction
- PFS
progression free survival
- PPV
positive predictive value
- PPV
positive predictive value
- qPCR
quantitative polymerase chain reaction,
- QS‐RGQ
QIAsymphony Rotor‐Gene Q
- RARβ2
retinoic acid receptor beta2
- RASSF1A
RAS association domain family protein 1A
- RFS
relapse free survival
- rOD
relative optical detection
- RT
radiotherapy
- UCHL1
Ubiquitin C‐Terminal Hydrolase L1
- US
United States
- VCA
viral capsid antigen
- WIF1
WNT Inhibitory Factor
1. BACKGROUND
Nasopharyngeal carcinoma (NPC) is an endemic malignant epithelial tumor prevalent in South China, Southeast Asia, and North Africa, with an incidence of up to 30.9/100,000 person‐years among Southern Chinese males [1]. This cancer is ubiquitously associated with the Epstein‐Barr virus (EBV), and the presence of EBV‐encoded RNA (EBER) is always demonstrated in NPC [2, 3, 4]. EBV virions can be transmitted via the saliva in both asymptomatic carriers and infectious mononucleosis patients [5, 6]. During the acute infection, lytic reproduction occurs within newly infected host nasopharyngeal epithelial cells [7] which enables the propagation of EBV virions to both the surrounding host epithelial cells and B cells within the Waldeyer's ring. Subsequently, EBV infection can persist by remaining predominantly as plasmids in the latent phase [8]. These plasmids reside in memory B cells within the germinal center of the Waldeyer's ring [9], where its native predilection lies [10]. When memory B cells are signaled to terminally differentiate into plasma cells [7], viral reactivation occurs with release of virions that further infect other naïve B cells and nasopharyngeal epithelial cells [11]. During this reactivation process, EBV continues to spread and infect cells and can be identified and transmitted in the saliva [10, 12] (Figure 1).
FIGURE 1.
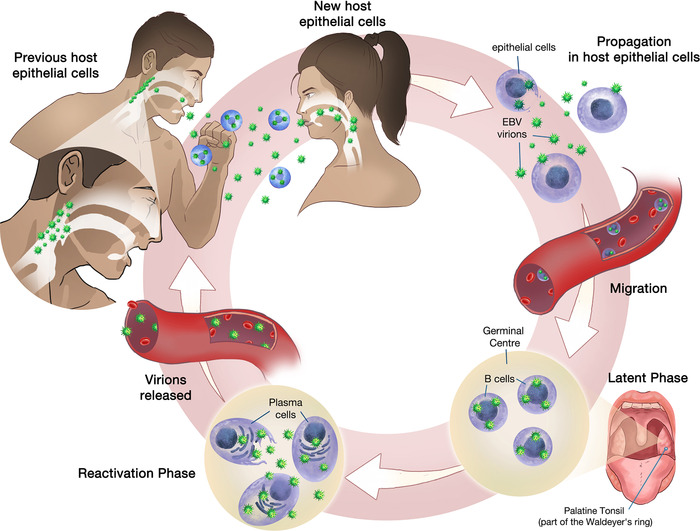
Transmission of Epstein‐Barr virus in nasopharyngeal epithelial cells
Healthy nasopharyngeal epithelial cells typically undergo growth arrest and lyse when infected by EBV virions [10]. However, nasopharyngeal mucosal inflammation can permit continued latent EBV replication to occur within the cells, mainly through molecular changes, such as p16 deletion and Cyclin D1 overexpression [13], which override the default growth inhibitory effect of cellular regulation pathways. Unregulated latent replication of EBV leads to further cellular alterations that ultimately result in malignant transformation to NPC [14].
The virions and DNA fragments released by these cancer cells during replication or cellular apoptosis offer opportunities for EBV detection in cancer screening or surveillance. Many studies have determined the feasibility of detecting EBV, ranging from serology [15, 16, 17, 18] to plasma circulating EBV or its surrogate [19, 20, 21, 22, 23, 24]. These EBV detection methods have also been validated in several clinical utilities in NPC management [25, 26, 27]. Therefore, this paper provides a comprehensive overview of the relevant studies using non‐invasive laboratory‐based EBV detection techniques in the management of NPC. Specifically, the use of these detection methods in NPC screening, prognostication, and post‐treatment surveillance are presented.
2. EBV DETECTION MARKERS
2.1. Free circulating plasma EBV DNA
In 1999, Lo et al. [28] discovered the presence of free circulating plasma EBV DNA in 55 of 57 NPC patients and only 3 of 43 healthy controls. The discovery of disproportionately high amounts of free circulating EBV DNA among NPC patients led to an explosion of new detection techniques on various possible EBV genome targets, such as BamHI‐W [28], and EBV‐encoded RNA‐1 (EBER‐1) [2]. The rationale of detecting free circulating EBV DNA in NPC management rests on the fact that NPC cells release EBV DNA during apoptosis [29, 30]. These EBV DNA fragments released by NPC are typically of shorter fragment (less than 181 base‐pairs) compared to the classical fragments released during the viral lytic phase, making them highly quantifiable using polymerase chain reaction (PCR) assays [20, 29, 31, 32, 33, 34]. Additionally, the detection of free circulating plasma EBV DNA can be applied on cerebrospinal fluid (CSF) and other samples, such as dried blood spots, nasopharyngeal swabs and aspirates, sputum and bronchial washings [34].
The detection of plasma EBV DNA in routine clinical application is limited by differences in interpretation of detection threshold and the timing of specimen collection following treatment [34]. Preiksaitis et al. [35] demonstrated significant inter‐laboratory variation in reported EBV DNA levels amongst 28 international laboratories, of which only 47.0% were within acceptable standards. Currently, no US Food and Drug Administration (FDA)‐approved assay exists, making harmonization and standardization of various assays challenging [34]. This deficiency in standardization of EBV DNA assays was addressed in the first International Standard for EBV assay released by the World Health Organization in 2011 [36]. In 2014, the Stanford Laboratory established commutability [37, 38] for two EBV PCR assays: the BamHI W assay and the commercial QIAsymphony Rotor‐Gene Q (QS‐RGQ) assay. The latest 2015 National Cancer Institute Workshop on Harmonization of EBV Testing for Nasopharyngeal Cancer further set sequential goals for test standardization [34]. The first step is to ensure standardization of sample type (plasma or serum) and DNA isolation protocol. Secondly, it recommends the development of a standardized PCR assay that targets both a single‐copy EBV gene [e.g., Epstein‐Barr virus nuclear antigen 1 (EBNA‐1)] and a multi‐copy EBV gene (e.g. BamHI‐W). This serves to maximize sensitivity of detection (via multi‐copy targets) while permitting accurate quantification of viral load (via single‐copy targets) [34]. At present, the single‐copy approach is preferred for measuring tumor burden across different patients, while the multi‐copy approach is preferred when tumor cells are present in low amounts (e.g., NPC screening of asymptomatic patients and post‐treatment monitoring for NPC recurrence). Ultimately, clinical validation of these assays across multi‐institutions is necessary in order to define test sensitivity and specificity and establish clinically relevant cut‐off values in each of these clinical applications.
Presently, most commercial assays use BamHI‐W on plasma samples because this target yields the most consistent results across various studies. Additionally, an enhancement of the detection of BamHI‐W was made by amplifying the CpG repeats on this DNA fragment, resulting in an even higher sensitivity in EBV detection among NPC patients [39]. Furthermore, Lam et al. [40] found that the positive predictive value (PPV) of detecting plasma EBV in NPC patients was improved from 11.0% to 19.6% when a sequencing‐based DNA analysis was used. This technique revealed higher amounts of plasma EBV DNA and generally longer fragment lengths of plasma viral molecules in NPC patients than in non‐NPC subjects [40].
2.2. EBV serology markers
Since NPC arises from the nasopharyngeal mucosa, a high IgA response is anticipated [33]. NPC cells produce latency‐associated EBV gene products, such as latent membrane protein 1 (LMP1), LMP2, and BamHI‐A rightward frame 1 (BARF1), but only EBNA‐1 induces strong IgG and IgA antibody responses [33]. Lytic phase EBV proteins, such as the early antigen (EA) and viral capsid antigen (VCA), are produced during the rapid viral replication phase of differentiating NPC cells. Therefore, strong IgA antibody responses against these proteins are often identified among NPC patients. However, these serological markers are not exclusively identified in NPC. IgA against EBV VCA may be raised in repeated infections of EBV or frequent reactivation of latent EBV in B cells, and can revert back to normal titers for unknown reasons without developing NPC [41]. It may also be present in systemic lupus erythematosus, possibly implicating EBV in the pathogenesis of systemic lupus erythematosus; or indicating a difficulty in suppressing the underlying latent EBV infection [42]. This relatively non‐specific nature of positive IgA results limits its accuracy in diagnosing NPC [43].
2.3. Other emerging EBV detection markers
Other potential liquid biopsy markers include circulating tumor cells (CTCs, which may comprise of micro‐metastatic tumor cells that precede distant metastasis [44]), miRNAs (small 21‐23 nucleotide non‐coding RNAs [45] that are encoded during cellular replication [46]), and DNA hypermethylation (hypermethylation of potential tumor suppressor genes [47] that occurs early in pre‐malignant cells [32, 48]). There is emerging evidence on the utility of these detection methods in NPC management. An overview of these existing and emerging detection modalities of various EBV detection markers is summarized in Table 1.
TABLE 1.
Overview of current and emerging laboratory‐based, EBV‐related NPC detection modalities
| Detection marker | Target | Method | Details |
|---|---|---|---|
|
Cell‐free DNA (EBV DNA genome fragments) |
EBNA‐1 EBNA‐2 |
dPCR, qPCR | The main issue is with non‐standardized methods and cut‐off limits, making it difficult to analyze data across papers. |
| BamHI‐W | qPCR | ||
|
Antibodies (immunoglobulins) |
IgA VCA, IgA EA, IgA EBNA‐1 | ELISA, IFA |
Serum samples are diluted at various titers, usually from 1:5 or 1:10 onwards. ELISA or IFA determines the maximum dilution level to which the antibodies can be detected. ELISA: may be automated, high throughput analysis. IFA: gold standard but technically difficult, highly operator‐dependent. |
| microRNA | Multiple (BARTs) | qRT‐PCR | Reverse transcription followed by qPCR to determine the quantity of microRNA (copy/mL). |
| CTCs | Epithelial, mesenchymal, or hybrid | Cellular capture | Size‐based or affinity‐based isolation |
| Hypermethylation of DNA | Gene alterations | PCR‐based methods | Treatment of DNA with sodium bisulfite to preserve hypermethylated islands which are detected by methylation‐independent or methylation‐specific PCR primers. |
Abbreviations: BART, BamHI‐A rightward transcript; CTC, circulating tumour cell; dPCR, digital polymerase chain reaction; EA, early antigen; EBNA, Epstein‐Barr nuclear antigen; EBV, Epstein‐Barr virus; ELISA, enzyme‐linked immunosorbent assay; IFA, immunofluorescence assay.; NPC, nasopharyngeal carcinoma; qPCR, quantitative polymerase chain reaction; VCA, viral capsid antigen.
CTCs and CTC clusters or microemboli are a rare and highly heterogenous group of cells in the peripheral blood, that exhibit different molecular features and play different roles in cancer progression [49, 50]. There are only a handful of studies on CTCs in NPC detection, and these studies differ in the method of CTC isolation, making the application of CTCs in the detection of NPC controversial. He et al. [51] reported that CTC enumeration was correlated with EBV VCA‐IgA and EBV DNA load in all NPC patients with or without distant metastasis, but Vo et al. [39] concluded that EBV DNA levels showed no correlation with CTC counts. Furthermore, You et al. [52] observed that CTCs had superior specificity (86.0% vs. 41.0%) and inferior sensitivity (42.3% vs. 81.3%) when compared with plasma EBV DNA load in diagnosing distant metastasis. It is worth noting that the first two studies used size‐based isolation systems to isolate CTCs, namely Isolation by Size of Epithelial Tumour Cells (ISET) [51] and Microsieve [39], whereas the latter study used CellSearch technology [52]. Size‐based isolation attempts to separate leukocytes from CTCs, as leukocytes are smaller, but some overlap in the size distribution of the two entities impedes accurate separation [53]. CellSearch is currently the only CTC isolation system approved by the US FDA [54]. This platform uses a marker‐based CTC enrichment technique with magnetic nanoparticles which capture antibodies that are tagged onto the epithelial cell adhesion molecules (EpCAM) expressed on CTCs. Similarly, different studies use different definitions to classify CTCs. While cells isolated by ISET method were considered CTC‐positive if they were Cytokeratin (CK)‐positive or P63‐positive, Microsieve classified CK‐positive and CD45‐negative cells as canonical CTCs, and CellSearch classified CK‐positive, EpCAM‐positive, 4′,6‐diamidino‐2‐phenylindole (DAPI)‐positive, and CD45‐negative cells as CTCs. These differences in the methods used to isolate and classify CTCs could be the primary reason behind the different conclusions reached in different studies. As CTCs are highly heterogenous, marker‐based CTC enrichment methods might miss certain sub‐populations of CTCs [55]. Hence, it is important to standardize the methods and definitions used to classify CTCs in future studies.
There are numerous reports on the role of miRNAs in the pathogenesis of NPC [56]. However, the mechanism by which deregulation of these epigenetic regulators affects cell signaling pathways in NPC is not well understood. Of the two families of EBV‐encoded miRNAthe BamHI fragment H rightward open reading frame 1 (BHRF1) and the BamHI‐A rightward transcripts (BART), the former has low expression in NPC cell lines, while the latter has been closely linked to the tumorigenesis of NPC [57]. A handful of studies have evaluated the usefulness of circulating BART miRNAs as potential biomarkers for detection of NPC [22, 46], as will be discussed later in this paper.
Hypermethylation of the CpG island of promoter region results in the silence of tumor suppressor genes, and this feature is a characteristic abnormality seen in EBV‐associated cancers [58, 59, 60]. Using a viral‐wide genome approach, Lam et al. [61] found distinct EBV DNA methylation profiles among different EBV‐associated diseases; and by analyzing methylated DNA sequences allowed differentiation between NPC and non‐NPC subjects. Aberrant methylation of tumor suppressor genes such as cadherin 4 (CDH4), cyclin‐dependent kinase inhibitor 2A (CDKN2A/p16), and RAS association domain family protein 1A (RASSF1A) are early events in NPC tumorigenesis [62, 63, 64]. This hypermethylation is mediated by EBV‐encoded LMP1 and LMP2A proteins that up‐regulates the expression of DNA methyltransferases (DNMTs), namely, DNMT1, DNMT3A, and DNMT3B [65]. The presence of early and widespread methylation changes, frequently occurring in several chromosomal locations such as 3p21.3, 9p21, and 6p21.3, led to the evaluation of these liquid biomarkers for early detection of NPC. In the first study evaluating the detection rate of hypermethylation, Chang et al. [47] compared detection rates of hypermethylated p15, p16, RASSF1A, E‐cadherin, and death‐associated protein kinase (DAPK) in tumor samples, nasopharyngeal swabs, mouth and throat rinsing fluid, plasma, and buffy coat. However, hypermethylation was found in less than 20% of peripheral blood samples as compared to 17%‐63% of nasopharyngeal swabs and 17%‐50% of mouth and throat rinsing fluids. Later on, Wong et al. [66] detected hypermethylation in at least one of the 5 genes (CDH1, DAPK1, CDKN2B, RASSF1, and CDKN2A) in 71% of plasma samples from NPC patients before treatment, with a specificity of 91%. Subsequently, the use of hypermethylation panels was heavily explored, and this will be discussed in the next section on sensitivity and specificity section.
3. SENSITIVITY & SPECIFICITY OF VARIOUS EBV DETECTION TECHNIQUES
The sensitivity and specificity of different EBV detection markers in the plasma have been studied by comparing the test positivity in known NPC patients with that in healthy controls. In 1998, Mutirangura et al. [67] demonstrated that EBV DNA was positive in NPC patients. EBV DNA has demonstrated both high sensitivity (68.8%‐100%) and high specificity (88.0%‐100%) [20, 22, 28, 68, 74] in detecting NPC. The type of sample may also contribute to the sensitivity and specificity of EBV detection. A meta‐analysis by Liu et al. [75] on 15 studies found pooled sensitivity and specificity of 91.4% and 93.2% for EBV DNA in plasma samples and of 84.4% and 76.0% for EBV DNA in serum samples. A meta‐analysis by Song et al. [76] on 27 case‐control and cohort studies demonstrated that EBV DNA detection has higher sensitivity and specificity than IgA VCA detection in the diagnosis of NPC.
The IgA serology assay is the most extensively studied EBV detection method, but no single serological marker has been shown to be optimal. IgA VCA serology yields higher sensitivity (57.4%‐98.1%) than specificity (53.7%‐94.3%) [18, 70, 74, 77, 79], while IgA EA serology demonstrates higher specificity (94.7%‐100%) than sensitivity (75.0%‐89.1%) [71, 77, 79] (Table 2). Combining various serological markers further improves the sensitivity and specificity of EBV detection by a modest extent. For instance, Fachiroh et al. [78] found that combining IgA VCA and EBNA‐1 in a joined assay yielded sensitivity of 85.4% and specificity of 90.1%, and Yu et al. [18] observed similar sensitivity of 85.1% and specificity of 90.1% when using a combination assay of IgA VCA and IgA EBNA‐1 (where the test was considered positive when either serological marker turned positive) (Table 2). A notable exception, however, was the combination of IgA VCA and IgG Rta assay, which demonstrated a higher sensitivity of 94.8% and a specificity of 98.0% [79]. More studies are required to validate if high sensitivity and specificity of such combinations may be achieved consistently across other centers.
TABLE 2.
Comparison of diagnostic potential of various EBV detection methods
| Reference | NPC patients (cases) | Controls (cases) | EBV detection modality | Sample type | Sensitivity † (%) | Specificity † (%) |
|---|---|---|---|---|---|---|
| Yu et al. (Zhongshan, 2018) [18] | 47 | 16,665 | ELISA IgA VCA > rOD value | Serum | 57.4 | 94.3 |
| ELISA IgA EBNA‐1 > rOD value | 76.6 | 96.2 | ||||
| ELISA IgA VCA and IgA EBNA‐1 > rOD value | 48.9 | 99.5 | ||||
| ELISA IgA VCA or IgA EBNA‐1 > rOD value | 85.1 | 90.1 | ||||
| Chan et al. (Hong Kong, 2017) [20] | 35 | 20,139 |
EBV DNA PCR targeting BamHI‐W, LOD > 20 EBV genomes/mL, 2 consecutive positive results 4 weeks apart |
Plasma | 97.1 | 98.6 |
| Hirai et al. (Japan, 2016) [22] | 31 | 40 | miRNA BART2‐5p | Plasma | 85 | 85 |
| miRNA BART17‐5p | Plasma | 60 | 95 | |||
| miRNA BART18‐5p | Plasma | 25 | 100 | |||
| EBV DNA BamHI‐W > 0 copies/mLl | Plasma | 100 | 100 | |||
| Wen et al. (Gaozhou, 2019) [23] | 60 | 18 | CTC > 0 | Plasma | 86.7 | ‐ ¶ |
| Mesenchymal CTC > 0 | Plasma | 50.0 | ‐ ¶ | |||
| Mao et al. (Hangzhou, 2019) [24] | 45 | 0 | CTC > 0 | Plasma | 92.6 | ‐ ¶ |
| Mesenchymal CTC > 0 | Plasma | 64.3 | ‐ ¶ | |||
| Lo et al. (Hong Kong, 1999) [28] | 57 | 43 | EBV DNA PCR targeting BamHI‐W and EBNA‐1 | Plasma | 96.5 | 93.0 |
| Sengar et al. (India, 2016) [68] | 76 | ‐ ¶ | EBV DNA PCR targeting EBNA‐1, LOD > 3.8 copies/mL | Plasma | 97.1 | ‐ ¶ |
| EBV DNA BamH1W‐76 > 7.3 copies/mL | Plasma | 96.8 | ‐ ¶ | |||
| EBV DNA BamH1w‐59 > 0.001 copies/mL | Urine | 92.9 | ‐ ¶ | |||
| EBV DNA BamH1w‐59‐Cr adjusted > 0.0005 copies/mL | Urine | 96.4 | ‐ ¶ | |||
| Yang et al. (Hong Kong, 2015) [69] | 220 | 50 | EBV DNA BamHI‐W ≥ 300 copies/mLl | Plasma | 69.1 | 88.0 |
| Hypermethylation panel of RASSF1A, WIF1, DAPK1, and RARβ2 ‡ | Plasma | 72.7 | 96.0 | |||
| Panel of RASSF1A, WIF1, DAPK1, RARβ2 and/or EBV DNA BamHI‐W ≥ 300 copies/mLl § | Plasma | 88.6 | 88.0 | |||
| 96 | 43 | Panel of RASSF1A, WIF1, DAPK1, RARβ2 ‡ | NP Brush | 95.8 | 67.4 | |
| Mai et al. (Guangzhou, 2002) [70] | 66 | 58 | EBV DNA qPCR targeting W fragment (No cutoff as nested PCR used as detection method) | Plasma/serum | 84.9 | 89.7 |
| Immunoenzymatic assay IgA‐VCA ≥1:40 | Plasma/serum | 80.3 | 89.7 | |||
| Luo et al. (Guangzhou, 2009) [71] | 160 | 76 | Immunoenzymatic assay IgA‐VCA ≥ 1:40 | Serum | 90.0 | 89.5 |
| Immunoenzymatic assay IgA‐EA ≥ 1:10 | 75.0 | 94.7 | ||||
| EBV DNA qPCR targeting BamHI‐W > 1000 copies/mL | 68.8 | 88.2 | ||||
| Leung et al. (Hong Kong, 2004) [72] | 139 | 178 | EBV DNA qPCR targeting BamHI‐W ≥ 60 copies/mL | Plasma | 95 | 98 |
| IF IgA‐VCA ≥ 1:10 | Serum | 81 | 96 | |||
| Teresa et al. (New York, 2007) [73] | 32 | 123 | IF IgA‐VCA ≥ 1:10 | Plasma | 90.6 | 53.7 |
| 22 | 86 | EBV DNA qPCR targeting BamHI‐W > 0 copies/mL | 77.3 | 91.9 | ||
| Chang et al. (Taiwan, 2008) [74] | 156 | 264 | IF IgA‐VCA ≥ 1:40 | Serum/plasma | 85.9 | 86.3 |
| ELISA IgA EA+EBNA‐1 ≥ 3.0 EU/mL | 94.2 | 82.6 | ||||
| EBV DNA qPCR targeting BamHI‐W > 0 copies/mL | 81.4 | 96.6 | ||||
| Low et al. (Singapore, 2000) [77] | 111 | 111 | IF IgA EA | Plasma | 80.1 | 100.0 |
| IF IgA VCA > 1:10 | Plasma | 88.3 | 73.9 | |||
| Fachiroh et al. (Indonesia, 2006) [78] | 151 | 254 | ELISA IgA EBNA‐1 > 0.1205 | Plasma | 88.6 | 80.1 |
| ELISA IgA VCA‐p18 > 0.2233 | Plasma | 79.8 | 70.9 | |||
| ELISA IgA EBNA‐1 with ELISA IgA VCA‐p18 > 0.3536 | Plasma | 85.4 | 90.1 | |||
| Cai et al. (Wuzhou, 2014) [79] | 211 | 203(Non‐NPC ENT patients); 210 healthy controls | Immunoenzymatic assay IgA VCA > 1:10 | Serum | 98.1 | 82.8 |
| Immunoenzymatic assay IgA EA > 1:10 | 89.1 | 98.5 | ||||
| ELISA IgG Rta > 0.49 | 90.5 | 85.2 | ||||
| ELISA IgA EBNA‐1 > 0.26 | 87.2 | 84.2 | ||||
| Immunoenzymatic assay IgA VCA with ELISA IgG Rta > 0.36 | 94.8 | 98.0 |
Sensitivity and specificity were determined by comparing test positivity in known NPC patients with that in healthy controls.
At least one gene positive.
At least one gene positive and/or EBV DNA positive.
A dash (‐) indicates that the value could not be obtained from the cited paper.
Abbreviations: BART, BamHI‐A rightward transcripts; CTC, circulating tumour cell; DAPK1, Death‐Associated Protein Kinase 1; EA, Early antigen; EBNA‐1, Epstein‐barr nuclear antigen‐1; EBV, Epstein‐barr virus; ELISA, enzyme‐linked immunosorbent assay; EU, endotoxin.; IF, Immunofluoresence; IgA, Immunoglobulin A; IgG Rta, Rta protein antibody Immunoglobulin G; LOD, limit of detection; NP, Nasopharyngeal; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction; RARβ2, retinoic acid receptor beta2; RASSF1A, Ras association domain family 1 isoform A; rOD, relative optical detection; VCA, viral capsid antigen; WIF1, WNT Inhibitory Factor 1.
Recently, emerging biomarkers have demonstrated promising outcomes. Studies on CTC are limited, but sensitivity of 50.0%‐92.6% has been demonstrated [23, 24] (Table 2). The value of EBV‐specific BART miRNAs in the detection of NPC is also being explored. Hirai et al. [22] compared the copy numbers of circulating BamHI‐W DNA and three BART miRNAs (miRNA‐BART2‐5p, miRNA‐BART17‐5p, and miRNA‐BART18‐5p) in patients’ serum samples for the initial diagnosis of NPC. They concluded that BamHI‐W DNA had a higher sensitivity and specificity for detection of NPC compared with the panel of three BART miRNAs evaluated. This study was however based on a relatively small sample of 20 patients [22] (Table 2).
In the field of hypermethylation, Tian et al. [48] found that a combination of 4 gene markers yielded a good sensitivity (95.1% when at least two genes were methylated) and specificity (85.0% when at least one gene was methylated). Individually, the gene markers CDKN2A, deleted in lung and esophageal cancer 1 (DLEC1), death‐associated protein kinase 1 (DAPK1), and ubiquitin C‐terminal hydrolase L1 (UCHL1) were methylated in 22.5%, 25.0%, 51.4%, and 64.9% of NPC patients, respectively. Hypermethylation also improves detection rates of EBV DNA. Yang et al. [69] showed that a combination assay of EBV DNA and a four‐gene hypermethylation panel [Ras association domain family 1 (RASSF1A), WNT inhibitory factor 1 (WIF1), DAPK1, retinoic acid receptor beta2 (RARβ2)] yielded sensitivity of 80.4%, 88.0%, and 96.5% for stage 1‐2, stage 3, and stage 4 NPC, respectively. Across all stages, the combination assay better predicted NPC than each individual assay. In early‐stage NPC, the hypermethylation panel demonstrated higher sensitivity (64.6% vs. 51.2%) and specificity (96.0% vs. 88.0%) than EBV DNA [69]. We postulate that the detection of early‐stage NPCs can be enhanced by the addition of hypermethylation panels to EBV DNA assays. Mechanistically, epigenetic changes occur early in NPC and are readily detectable by hypermethylation panels; in locally advanced cases, the increased tumor burden is better detected via plasma EBV DNA assays. Furthermore, through whole‐genome methylation analysis, Lam et al. [61] demonstrated that methylation profiles exhibit a disease‐associated pattern which differs for NPC, lymphoma, and infectious mononucleosis. By combining the differential methylation patterns with EBV DNA size and quantity, the study demonstrated an increased PPV of 35.1% in population screening of NPC, as compared to a PPV of 11.2% using only the conventional count‐based plasma EBV DNA detection method [61]. In the future, hypermethylation could possibly enhance EBV DNA screening abilities.
4. UTILITY OF EBV LIQUID BIOPSY IN NPC SCREENING
4.1. General population screening in a Chinese community
Historically, two large‐scale serological screening studies were performed to detect NPC in a general Southern Chinese cohort in a community setting (Table 3). Zeng et al. [15] utilized both IgA EA and VCA serological screening among 12,932 adults. The NPC detection rate was 100.5/100,000 persons, which was almost double that of the indigenous detection rate of NPC. However, in a larger study by Deng et al. [16] in 1995, a lower detection rate of 31.4/100,000 persons was identified using both IgA EA and VCA serological screening of 318,912 Chinese adults aged 30‐70 years. For screened adults aged 45‐49 years, the detection rate was 68.9/100,000 persons [16].
TABLE 3.
Screening studies on Chinese‐predominant populations
| Reference | Sample size | Age (years) | Clinical stage | EBV detecton modality | Detection rate (per 100,000 persons) | Incidence (per 100,000 person years) | PPV (%) |
|---|---|---|---|---|---|---|---|
| General population | |||||||
| Zeng et al. (1982) [15] | 12,932 | 40‐59 |
I: 69.2 II: 30.8 III: 0 IV: 0 |
IgA EA (IE) | 100.5 | 47.7 | 30 |
| IgA VCA (IE) | 1.9 | ||||||
| Deng et al. (1995) [16] | 318,912 | ‐ ¶ |
I: 25.5 II: 63.6 III: 7.3 IV: 3.6 |
IgA EA and VCA (IE) | 17.2 | ‐ ¶ | 19.16 |
|
I: 34 II: 53 III: 10.0 IV: 3.0 |
IgA VCA (IE) | 31.4 | ‐ ¶ | 1.18 | |||
| Liu et al. (2012) [17] | 28,688 | 30‐59 |
I: 14.6 II: 53.7 III: 26.8 IV: 4.9 |
IgA VCA + IgA EBNA‐1 (ELISA) | 142.9 | 20 | ‐ ¶ |
| Yu et al. (2018) [18] | 16,712 | 30‐59 | ‐ ¶ | IgA VCA + IgA EBNA‐1 (ELISA) | 149.6 | 29.0 |
Both were positive: 2.3 Either one was positive: 2.6 |
| Chan et al. (2013) [19] | 1318 | 40‐60 | ‐ ¶ | DNA (qPCR BAM‐HIW) | 227.6 |
Male: 20‐30 Female: 15‐20 |
4.3 |
| ‐ ¶ | IgA VCA (ELISA) | 75.9 | 1.2 | ||||
| Chan et al. (2017) [20] | 20,174 | 52 (40‐62) |
I: 47.1 II: 23.5 III: 23.5 IV: 5.9 |
DNA (qPCR BAM‐HIW) | 168.5 | 35 |
11.0 (NPV: 99.995) |
| High‐risk population (first‐degree relatives of NPC patients) | |||||||
| Ng et al. (2005) [83] | 929 | 42 (19‐80) | Stage I: 41 | IgA VCA and IgA NA (IIF/ELISA) |
Male: 433 Female: 499 |
Male: 24.1 Female: 9.6 |
10.7 |
| Ng et al. (2010) [84] | 1,199 | 38 (18 ‐ 78) |
Stage I: 41 Stage II: 18 |
1994‐1997: IgA VCA (IF) 1998‐2005: IgA VCA (ELISA/IF) |
1,251 |
Male: 21.6 Female: 6.8 |
8.93 (NPV: 99.7) |
EBV DNA was detected in plasma samples, and IgA in serum samples.
A dash (‐) indicates that the value could not be obtained from the cited paper.
Abbreviations: ELISA, enzyme‐linked immunosorbent assay; IE, immunoenzymatic method; IF, immunofluorescence; IIF, Indirect immunofluorescence; NA, nuclear antigen.; NPV, negative predictive value; PPV, positive predictive value.
Presently, serological detection of anti‐EBV antibodies utilizes ELISA, which is less tedious and time‐consuming and has lower intra‐observer variability [18] than traditional, indirect immunofluorescence (IF) techniques [80]. Additionally, serological detection tends to use a combination of IgA VCA and IgA EBNA‐1 [81], which has higher sensitivity and specificity than combining IgA VCA with IgA EA [18].
In this regard, two large‐scale studies using ELISA and the IgA EBNA‐1 and IgA VCA combination have been performed. Yu et al. [18] studied a population of 16,712 persons between 2009 and 2015 and obtained an increased detection rate of 149.59/100,000 persons. When both IgA EBNA‐1 and IgA VCA were seropositive, the sensitivity was 48.9%, and the specificity was 99.5%; when either of them was positive, the sensitivity was 99.5%, but the specificity dropped to 90.1% [18]. Liu et al. [17] in 2013 screened 28,688 Cantonese using the combination of IgA EBNA‐1 and IgA VCA serology, which yielded a similar detection rate of 142.9/100,000 persons.
Interestingly, Yu et al. [18] boosted NPC detection rates through implementing a longer follow‐up protocol. By stratifying the population into various risk levels based on serological titers and follow‐up duration, they eventually detected 47 NPC patients (281.24/100,000). Notably however, the compliance rate by the second year of follow‐up fell to approximately half the initial number (43.7%‐53.9%). Hence, longer‐term follow‐up protocols may be able to boost NPC detection rates, but compliance and additional expenses may limit the widespread adoption of this method.
Furthermore, despite these advancements, the PPV of serological tests remained low (Yu et al. [18]: 2.3‐2.6%) compared with that of EBV DNA (Chan et al. [20]: 11%). This implies that more test‐positive individuals may be subject to costly confirmatory tests, such as endoscopy and magnetic resonance imaging. Even though serological test retain the logistical and technical advantages in being a cheap, high‐throughput, and rapid test [81], the overall costs may still be higher than that of plasma EBV DNA detection due to the low PPV. Suggested solutions usually involve the combination of serology with alternative methods. For example, Jiang et al. [82] proposed that miRNA BART2‐5p detection has the potential to improve the PPV of mass serological screening, by distinguishing between pre‐clinical NPC cases and matched serological high‐risk controls (higher EBV VCA IgA and EBNA‐1 IgA values) with a sensitivity of 90.9% and a specificity of 54.5%.
In the recent decade, EBV DNA assays targeting BamHI‐W gene fragments have yielded much higher detection rates than earlier studies utilizing serology. Zeng et al. [15] in 1982 yielded NPC detection rate of 100.5/100,000 persons using IgA EA, followed by Deng et al. [16] in 1995 yielding detection rate of 17.2/100,000 persons using IgA EA and VCA in combination. However, when plasma EBV targeting the BAM‐HIW fragment was used to screen NPC, tChan et al. [20] in 2017 showed a higher detection rate of 168.5/100,000 persons. Similarly, a direct comparison study by Chan et al. [19] in 2013 on 1318 individuals aged 40‐60 years yielded an NPC detection rate of 227.6/100,000 using plasma EBV DNA detection, but only 75.9/100,000 when IgA VCA detection was used as the detection method.
A largescale EBV DNA study by Chan et al. [20] in 2017 screened 20,178 adults aged 40‐62 years. By using qPCR to target the BamHI‐W fragment of EBV DNA, the results surpassed earlier serological studies, with an NPC detection rate of 168.5/100,000 and PPV of 11.0%, and with high sensitivity (97.1%) and specificity (98.6%). Furthermore, the majority of patients (70.6%, 24 out of 34) had early‐stage disease (stage I or II), contrary to an unscreened cohort in the same period (19.3%, 149 of 773) [20, 33]. Earlier detection was associated with longer survival [15, 16, 19, 20], such that a 97% 3‐year progression‐free survival (PFS) rate was achieved in the landmark study [20]. Therefore, at present, EBV DNA detection targeting the Bam HI‐W fragment has demonstrated a superior detection rate and PPV as compared to serological methods of EBV detection.
4.2. NPC screening in high‐risk cohorts (first‐degree relatives of NPC patients)
There is paucity of data using EBV detection methods to screen for NPC among high‐risk cohorts. When IgA VCA and IgA EA serology were used to screen for NPC among 929 first‐degree relatives of index NPC patients, the detection rates were 433/100,000 and 499/100,000 person‐years among males and females, respectively [83]. This detection rate was more than double compared to the serological detection rates of NPC among the general population: 142.9/100,000 by Liu et al. [17] in 2012 and 149.6/100,000 by Yu et al. [18] in 2018. In 2010, Ng et al. [84] used IgA VCA to screen for NPC among 1,199 asymptomatic first‐degree relatives of index NPC patients and identified 15 patients with NPC (detection rate: 1,251/100,000 persons). The majority of these patients had early‐stage disease (Stage I: 41.2%, Stage II: 17.6%). The utility of other EBV detection methods, especially using plasma EBV DNA assay warrants further exploration to screen for NPC among high‐risk individuals.
5. UTILITY OF EBV LIQUID BIOPSY IN NPC PROGNOSTICATION
5.1. Pre‐treatment plasma EBV DNA and other EBV status detection techniques in prognostication
Lin et al. [85] in 2004 found that with a pre‐treatment plasma EBV DNA cut‐off value of 1500 copies/mL, 83.4% of those with detectable EBV DNA before treatment survived at least 2 years, as compared to 100% of those with undetectable EBV DNA (P < 0.001). Prayongrat et al. [86] in 2017 found that with a pre‐treatment plasma EBV DNA cut‐off value of 600 copies/mL, 67% of those with detectable EBV DNA before treatment survived at least 5 years, as compared to 82.6% of those with undetectable plasma EBV DNA (P < 0.05).
Across all studies using plasma EBV DNA assay, those with detectable plasma DNA before treatment had 2‐ to 5‐year overall survival (OS) rates lower than those with undetectable EBV DNA [85, 86, 87, 88, 89] (Table 4). Sengar et al. [68] found the mean OS to be 26.7 months shorter in patients with detectable pre‐treatment EBV DNA levels than in those with undetectable pre‐treatment EBV DNA levels (42.4 months vs 69.1 months). Unfortunately, cut‐off values varied across these studies, and an optimal cut‐off value is still being determined [90].
TABLE 4.
Associations of survival with pre‐treatment EBV status detected using various techniques
| Pre‐treatment EBV status | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | No. of NPC patients | EBV detection modality | Target | Cut‐off value | Detectable | Undetectable | P value | Difference ¶ | Treatment | Follow‐up (months) | Survival endpoint |
| Vo et al. (2016) [39] | 46 | EBV DNA | BamHI‐W | – | ‐ ‡ | ‐ ‡ | 0.03 | ‐ ‡ |
Stage I‐II: RT Stage III‐IV: CTRT |
18.7 | OS |
| EBNA‐1 | ‐ ‡ | ‐ ‡ | 0.02 | ‐ ‡ | OS | OS | |||||
| CTC | Canonical | ‐ ‡ | ‐ ‡ | 0.66 | ‐ ‡ | OS | |||||
| Potential | ‐ ‡ | ‐ ‡ | 0.13 | ‐ ‡ | OS | ||||||
| Sengar et al. (2016) [68] | 76 | EBV DNA | EBNA‐1 | 3.8 copies/mL | 42.4 mo nths (mean) | 69.1 mo nths (mean) | 0.023 | 26.7 mo nths | CT and CTRT | 38.8 | OS |
| BamH1W‐76 | 7.3 copies/mL | 42.4 months (mean) | 69.1 months (mean) | 0.023 | 26.7 months | OS | |||||
| Lin et al. (2004) [85] | 99 | EBV DNA | BamHI‐W | 1500 copies/mL | 83.4% | 100% | <0.001 | 16.6% | CTRT | 30 | 2‐year OS rate |
| 66.4% | 88.8% | 0.02 | 22.4% | 2‐year RFS rate | |||||||
| Prayongrat et al. (2017) [86] † | 204 | EBV DNA | BamHI‐W | 600 copies/mL | 66.6% | 86.6% | 0.005 | 20.0% | IMRT | 35.1 | 3‐year PFS rate |
| 64.9% | 74.5% | 0.005 | 9.6% | 5‐year PFS rate | |||||||
| 80.8% | 94% | 0.028 | 13.2% | 3‐year OS rate | |||||||
| 67.0% | 82.6% | 0.028 | 15.6% | 5‐year OS rate | |||||||
| Peng et al. (2016) [87] | 584 | EBV DNA | BamHI‐W | 2010 copies/mL | 78.1% | 93.6% | <0.001 | 15.5% | IMRT | 38.2 (4.6‐58.6) | 3‐year DFS rate |
| 92.3% | 98.9% | <0.001 | 6.6% | 3‐year OS rate | |||||||
| 90.9% | 96.9% | 0.004 | 6.0% | 3‐year LRRFS rate | |||||||
| 85.5% | 96.6% | <.001 | 11.1% | 3‐year DMFS rate | |||||||
| Twu et al. (2007) [88] | 114 s | EBV DNA | BamHI‐W | 1500 copies/mL | 60.3% | 93.1% | <0.0001 | 32.8% | CTRT | 46 | 4‐year OS rate |
| 54.4% | 77.9% | 0.0009 | 23.5% | 4‐year RFS rate | |||||||
| 43.1% | 17.5% | 0.0027 | 25.6% | Relapse | |||||||
| Serology | VCA IgA | 1:80 | 29.9% | 27.9% | 0.754 | 2.0% | Relapse | ||||
| VCA IgG | 1:2560 | 31.8% | 27.8% | 0.697 | 4.0% | Relapse | |||||
| Ling et al. (2009) [89] | 317 | Serology | IgA EA/VCA | ≥1:160 | 43% | 65% | 0.01 | 22.0% | RT | 110 | 5‐year Survival rate |
| Chan et al. (2002) [95] | 170 | EBV DNA | BamHI‐W | 4000 copies/mL | ‐ ‡ | ‐ ‡ | 0.023 | ‐ ‡ | RT ± CT | 29 | 1‐year PFS rate |
| ‐ ‡ | ‐ ‡ | <0.001 | ‐ ‡ | OS | |||||||
| ‐ ‡ | ‐ ‡ | 0.005 | ‐ ‡ | Time to local recurrence | |||||||
| ‐ ‡ | ‐ ‡ | <0.001 | ‐ ‡ | Time to distant recurrence | |||||||
| Chen et al. (2018) [111] | 68 | CTC | Affinity‐based | 3 | 34.1% | 11.5% | 0.047 | 22.6% | IMRT+/‐ CT | 12 | 1Y disease progression |
Interpreting the table using Prayongrat et al. (2017) [86] as an example: using a cut‐off value of 600 copies/mL, 66.6% of those with detectable EBV DNA levels survived 3 years, whereas 86.6% of those with undetectable levels survived 3 years.
Difference in survival endpoint for those with detectable versus undetectable pre‐treatment values for the respective EBV detection modalities.
A dash (‐) indicates that the value could not be obtained from the cited paper.
Abbreviations: CT, chemotherapy; CTC, circulating tumour cells.; DFS, disease‐free survival; DMFS, distant metastasis‐free survival; EA, early antigen; EBV, epstein‐barr virus; Ig, Immunoglobulin; IMRT, intensity modulated radiotherapy; LRRFS, loco‐regional relapse‐free survival; OS, overall survival, PFS, progression free survival; RFS, relapse free survival; RT, radiotherapy; VCA, viral capsid antigen.
Additionally, pre‐treatment plasma EBV DNA may be incorporated into existing staging systems because it is highly predictive of survival. Leung et al. [91] found that stage II NPC patients could be further stratified into high‐risk (with a survival rate similar to stage I patients: about 90%) and low‐risk groups (with a survival rate similar to stage III patients: approximately 63%) according to the pre‐treatment plasma EBV DNA level, although the cut‐off value was unreported. Kitpanit et al. [92] found that integration of pre‐treatment plasma EBV DNA levels (with a cut‐off value of 2300 copies/mL) into the 8th edition of the American Joint Committee on Cancer (AJCC) TNM staging system allows better survival prediction than using the TNM staging system alone (Harrell's C‐concordance index increased from 0.690 to 0.760 for OS and from 0.650 to 0.744 for PFS). This improved staging system using pre‐treatment plasma EBV DNA may possibly allow a more accurate selection of high‐risk patients who might benefit from treatment intensification [92].
On the other hand, IgA EBV serology has not been shown to be consistent in prognosticating NPC patients [88, 89]. Twu et al. [88] showed that while the presence of plasma EBV DNA before treatment was associated with survival, pre‐treatment IgA and IgG VCA titers did not demonstrate any association with survival or risk of cancer relapse. In a separate NPC cohort, Ling et al. [89] found that those with undetectable IgA EA/VCA before treatment had higher 5‐year survival rate than those with detectable values. In this study, there was no end‐to‐end comparison with plasma EBV DNA in prognosticating this cohort of NPC patients.
CTCs have been explored to be possibly linked to survival [111], although a head‐to‐head comparison by Vo et al. [39] showed that pre‐treatment EBV DNA was better associated to survival than pre‐treatment CTC.
5.2. Post‐treatment plasma EBV DNA and other EBV status detection techniques in prognostication
While 80%‐99% of NPC patients would achieve complete remission after treatment [94, 95], 30%‐40% of patients with locally advanced (stage III‐IV) NPC would eventually recur [95]. Lo et al. [96] observed that patients who relapsed within the first year had higher median levels of plasma EBV DNA t than those without relapse (median: 41,756 vs. 5807 copies/mL). Furthermore, post‐treatment plasma EBV DNA was found to be a prognostic marker of NPC independent of the tumor stage (P = 0.003) [96]. Using a cut‐off of 500 copies/mL, Chan et al. [95] showed that the 1‐year PFS and OS rates were higher in patients with undetectable post‐treatment EBV DNA levels than in those with detectable levels (PFS: 93% vs. 48%; OS: 97% vs. 76%). Hence,post‐treatment plasma EBV DNA levels can be used to identify patients at risk of developing recurrence.
Survival is consistently poorer among NPC patients with detectable post‐treatment plasma EBV DNA levels than among those with undetectable plasma EBV DNA levels [85, 88, 93, 95] (Table 5). The difference in OS rate between patients with detectable and undetectable plasma EBV DNA levels appears to be more stark using post‐treatment plasma EBV DNA (21.0%‐76.1%) [85‐88, 93, 95] than using pre‐treatment EBV DNA (6.6%‐32.8%) [85, 86, 87, 88, 89] (Tables 4 and 5). It was hypothesized that patients with persistently elevated plasma EBV DNA levels after definitive intensity‐modulated radiotherapy may have minimal residual disease [95] and hence, may benefit from adjuvant chemotherapy [97]. However, a randomized phase III study led by Chan et al. [93] did not show any prolonged recurrence free survival with adjuvant chemotherapy (cisplatin and gemcitabine) following definitive treatment among patients with detectable post‐treatment plasma EBV DNA. Nevertheless, post‐treatment plasma EBV DNA level was associated with the risks of locoregional failure, distant metastasis, and death. More recently, Hui et al. [25] validated that integrating post‐treatment plasma EBV DNA levels with the TNM staging system improved risk stratification of NPC. Additionally, low‐risk NPC patients were identified using recursive partition model analysis with the view that they could potentially be spared from adjuvant chemotherapy.
TABLE 5.
Associations of survival with post‐treatment EBVstatus detected using various techniques
| Post‐treatment EBV status (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | No. of NPC patients | Treatment | Timing of sample collection after treatment (months) | Modality | Target | Cut‐off value | Detectable | Undetectable | P value | Difference ¶ | Median follow‐up (months) | Survival endpoint |
| Lin et al. (2004) [85] | 99 | CTRT |
RT: 0.25 CT: 1,2 |
EBV DNA | BamHI‐W | 1500 copies/mL | 56.3 | 96.7 | <0.001 | 40.4 | 30.0 | 2‐year OS rate |
| 28.6 | 84.2 | <0.001 | 55.6 | 2‐year RFS rate | ||||||||
| Prayongrat et al. (2017) [86] | 204 | IMRT | 3 | EBV DNA | BamHI‐W | 600 copies/mL | 0 | 79.2 | < 0.001 | 79.2 | 35.1 | 3‐year PFS rate |
| 0 | 72.1 | 72.1 | 5‐year PFS rate | |||||||||
| 18.8 | 89.7 | <0.001 | 70.9 | 3‐year OS rate | ||||||||
| 0 | 76.1 | 76.1 | 5‐year OS rate | |||||||||
| Peng et al. (2016) [87] | 584 | IMRT | <1 | EBV DNA | BamHI‐W | 0 copies/mL | 49.9 | 88.5 | <0.001 | 38.6 | 38.2 | DFS |
| 0 copies/mL | 72.1 | 97.5 | <0.001 | 25.4 | OS | |||||||
| 86.6 | 94.3 | 0.019 | 7.7 | LRRFS | ||||||||
| 60.5 | 93.3 | <0.001 | 32.8 | DMFS | ||||||||
| Twu et al. (2007) [88] | 114 s | CTRT | 0.25 |
EBV DNA |
BamHI‐W | 0 copies/mL | 30.8 | 84.6 | <0.0001 | 53.8 | 46 | 4‐year OS rate |
| 15.4 | 74 | <0.0001 | 58.6 | 4‐year RFS rate | ||||||||
| 84.6 | 21.8 | 0.0002 | 62.8 | Relapse | ||||||||
| Chan et al. (2018) [93] | 789 | RT/CRT | ‐ ‡ | EBV DNA | BamHI‐W | ‐ ‡ | 51.3 | 82.8 | <0.001 | 31.5 | 79.2 | 3‐year RFS rate |
| 45.4 | 77.1 | 31.7 | 5‐year RFS rate | |||||||||
| 70.8 | 93.3 | <0.001 | 22.5 | 3‐year OS rate | ||||||||
| 60.2 | 87.3 | 27.1 | 5‐year OS rate | |||||||||
| 58.3 | 86.7 | <0.001 | 28.4 | 3‐year LRFS rate | ||||||||
| 51.5 | 80.3 | 28.8 | 5‐year LRFS rate | |||||||||
| 61.6 | 88.5 | <0.001 | 26.9 | 3‐year DMFS rate | ||||||||
| 54.2 | 83.1 | 28.9 | 5‐year DMFS rate | |||||||||
| Chan et al. (2002) [95] | 170 | RT ± CT | 1.5‐2 | EBV DNA | BamHI‐W | 500 copies/mL | 48 | 93 | <0.001 | 45 | 29 | 1‐year PFS rate |
| 76 | 97 | <0.001 | 21 | 1‐year OS rate | ||||||||
| ‐ ‡ | ‐ ‡ | 0.572 | ‐ ‡ | Time to local recurrence | ||||||||
| ‐ ‡ | ‐ ‡ | <0.001 | ‐ ‡ | Time to distant recurrence | ||||||||
Difference in percentage of subjects achieving the respective survival endpoint for those with detectable vs undetectable post‐treatment EBV DNA levels.
A dash (‐) indicates that the value could not be obtained from the cited paper.
Abbreviations: CRT, chemoradiotherapy; CT, chemotherapy; DFS, disease‐free survival; DMFS, distant metastasis‐free survival.; EBV, epstein‐barr virus; IMRT, intensity modulated radiotherapy; LRRFS, loco‐regional relapse‐free survival; OS, overall survival, PFS, progression free survival; RFS, relapse free survival; RT, radiotherapy.
The role of real‐time monitoring of EBV DNA levels during chemotherapy was also found to be useful. Wang et al. [27] found that during the first month after chemotherapy, a slower EBV DNA clearance rate, determined by longer half‐life (T1/2) value, was associated with poorer survival rates. More recently, Lv et al. [26] also suggested that trajectory of plasma EBV clearance following induction chemotherapy can be used to modulate intensification or de‐intensification of subsequent treatment. They measured and tracked plasma EBV DNA levels after each cycle of induction chemotherapy and clustered these NPC patients into early responders, intermediate responders, late responders, and treatment‐resistant subjects. Significant differences in disease‐free survival were observed between these groups. On the other hand, unlike EBV DNA, post‐treatment IgA and IgG VCA titers demonstrated no association with survival or the risk of cancer relapse [88].
5.3. Prognostic value of plasma EBV DNA and other EBV status detection techniques in cancer staging
Across studies, plasma EBV DNA levels have consistently been shown to be associated with NPC stage [28, 39, 86] (Table 6), suggesting a direct correlation between plasma EBV DNA titers and tumor burden. EBV IgA serology [89] and miRNA titers [21, 46] have also been shown to significantly associate with stage, but conflicting reports exist regarding the prognostic role of CTC [39, 98]. Si et al. [98] demonstrated a significant association with stage for epithelial and mesenchymal CTCs (P < 0.05), but Vo et al. [39] found no significant association with stage for canonical and potential CTCs.
TABLE 6.
Associations of plasma EBV DNA levels with TNM stage
| Reference | Modality | Target | Cut‐off value | No. of NPC patients | Median follow‐up (months) | TNM stage | P value |
|---|---|---|---|---|---|---|---|
| Lo et al. (1999) [28] | EBV DNA | BamHI‐W and EBNA‐1 | 5 copies/mL | 57 | ‐ ‡ | Stage (I‐II vs. III‐IV) | <0.001 |
| Vo et al. (2016) [39] | EBV DNA | BamHI‐W | ‐ ‡ | 46 | 18.7 | Stage (I vs. II‐III vs. IV) | 0.0002 |
| EBNA‐1 | Stage (I vs. II‐III vs. IV) | 0.001 | |||||
| CTC | Canonical | Stage (I vs. II‐III vs. IV) | 0.825 | ||||
| Potential | Stage (I vs. II‐III vs. IV) | 0.300 | |||||
| Zhang et al. (2015) [46] | miRNA | BART7 | ‐ ‡ | 89 | ‐ ‡ | N status | <0.001 |
| Stage | <0.001 | ||||||
| BART13 | ‐ ‡ | ‐ ‡ | N status | 0.003 | |||
| Stage | 0.001 | ||||||
| Prayongrat et al. (2017) [86] | EBV DNA | BamHI‐W | 600 copies/mL | 204 | 35.1 | T status (T1‐T2 vs. T3‐T4) | 0.007 |
| N status (N0‐N1 vs. N2‐N3) | 0.051 | ||||||
| Stage | 0.036 | ||||||
| Ling et al. (2009) [89] | Serology | IgA EA/VCA | 1:160 | 317 | ‐ ‡ | Stage (I‐II vs. III‐IV) | 0.01 |
| Si et al. (2016) [98] | CTC |
Epithelial Mesenchymal Hybrid |
‐ ‡ | 81 | ‐ ‡ | Stage | <0.05 (total, mesenchymal, hybrid † ) |
| Lymph node spread | <0.05 (mesenchymal, hybrid) | ||||||
| Cumulative distal and lymph node spread | <0.05 (hybrid) |
Hybrid CTC numbers are inversely correlated with stage.
A dash (‐) indicates that the value could not be obtained from the cited paper.
Abbreviations: BART, BamHI‐A rightward transcripts.; CTC, circulating tumour cells; EA, early antigen; EBNA‐1, Epstein‐barr nuclear antigen‐1; EBV, epstein‐barr virus; IgA, immunoglobulin; M, metastasis; miRNA, micro‐ribonucleic acid; N, node; NP, nasopharyngeal; NP, nasopharyngeal; T, tumour; VCA, viral capsid antigen.
6. UTILITY OF LIQUID BIOPSY IN POST‐TREATMENT NPC SURVEILLANCE
Local recurrence (around 10‐15%) [99] and distant metastasis (approximately 20%) [100] have contributed largely to treatment failure and death of NPC. Hence, there is an urgent need for an effective surveillance method beyond routine physical examination and cross‐sectional imaging modalities so as to detect recurrence early. As a proof of concept of using plasma EBV DNA, Fan et al. [101] found that 85% (11 out of 13) of patients with relapse had detectable EBV DNA levels.
Additionally, plasma EBV DNA levels tend to be much higher in patients with distant metastasis than in those with locoregional recurrence. For example, Hsu et al. [102] found that the median EBV‐DNA level amongst patients with distant metastases was 1965 copies/mL (range, 8‐20,762,916 copies/mL), compared to 264 copies/mL (range 13‐12,707 copies/mL) amongst patients with locoregional recurrence. EBV DNA is positive in nearly all patients with distant metastasis (65.1%‐100%); whereas only 20.0%‐100% of patients with locoregional recurrence had detectable EBV DNA levels [101, 102, 103, 104, 105, 106, 107, 108] (Table 7). This difference in EBV DNA levels between NPC patients with distant metastasis and those with locoregional recurrence may be due to a higher disease burden among metastatic NPC patients [108]. An alternative hypothesis is that recurrent NPC cells emerge from irradiated tumor sites, and face difficulty of efflux into the plasma due to irradiation‐associated stromal fibrosis and decreased vascularity [108]. Additionally, EBV DNA is not entirely specific because 12%‐32% of disease‐free patients have detectable EBV DNA levels, which are usually transient but sometimes may remain persistently elevated [102, 108]. The persistence of elevated plasma EBV DNA levels could have resulted from lytic reactivation (e.g. triggered by oxidative cell stress from irradiation), persistent latent EBV infection, other EBV‐related tumors (e.g., lymphoma) [108], and even unrelated diseases such as lung cryptococcal infection [102]. It is understandable that to attain optimal cost‐efficiency, a balance between sensitivity and specificity can be optimized by altering the cut‐off value. For instance, by using a very high cut‐off value of 1,000,000 copies/mL, Cao et al. [109] obtained 100% specificity in the detection of both distant metastases and local recurrence, but at a significantly low sensitivity (27.3% for distant metastases and 0% for local recurrence).
TABLE 7.
Utility of EBV status detection techniques in surveillance of treated nasopharyngeal carcinoma
| Endpoint (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference | Sample size | Stage at diagnosis | Treatment | Surveillance regime (months) | Median follow‐up (months) | Method | Cut‐off (copies/ml) | Relapse | Metastasis | Locoregional recurrence | Disease free |
| Fan et al. (2004) [101] | 124 | I to IV | ‐ § | ‐ § | ‐ § | EBV DNA BamH I‐W and EBER‐1 | ‐ § | 84.6 | ‐ § | ‐ § | 0.0 |
| IgA EA | 87.5 | ‐ § | ‐ § | 44.4 | |||||||
| Hsu et al. (2013) [102] | 389 | I‐IV (mostly II‐IV) | RT ± CT | Interval: 3‐6 | 48 (24‐78) | EBV DNA BamH I‐W | 400 | 46.3 | 65.1 | 20.0 |
6.3 (< 6 M) † 1.1 (> 6 M) † |
| Ferrari et al. (2012) [103] | 36 | IIb to IVb | CT and RT |
First: 0 Interval: 2‐6 |
36 (IQR 24‐48) | EBV DNA BamH I‐W | 350 | 71.4 | 100.0 | 50.0 | 0.0 |
| Leung et al. (2003) [104] | 189 | I to IV | RT | ‐ § | ‐ § | EBV DNA BamH I‐W | ‐ § | 79.6 | 96.0 |
62.5 (Stage I‐II: 41.6; Stage III‐IV: 83.3) |
‐ § |
| Hong et al. (2004) [105] | 372 | III to IV | RT ± CT |
First: 3 Interval: 3‐4 |
‐ § | EBV DNA EBNA‐1 | ‐ § | 81.5 | 96.4 | 65.4 | ‐ § |
| Wang et al. (2011) [106] | 245 | I to IV | RT ± CT |
First: 1‐2 Interval: 1‐6 |
60 (45‐74 | EBV DNA BamH I‐W | 0 | 100.0 | 100.0 | 100.0 | 0.0 |
| Chan et al. (2016) [107] | 31 | III, IV | RT + CT |
First: 1‐1.5 Interval: 2‐6 |
33.7 (7‐39.3) | EBV DNA BamH I‐W | 500 | 88.9 | 100 | 66.7 | 0.0 |
| Li et al. (2017) [108] | 385 | ‐ § |
Stage I: IMRT Stage II‐IVb: IMRT and CT |
First: 3 Interval: 3‐6 |
52.8 (9.3‐ 73.8) |
EBV DNA BamH I‐W | 0 | 73.1 | 93.9 | 56.4 ‡ | 12.8 |
In the disease‐free group, 6.3% of patients had transient EBV DNA values above cut‐off, whereas 1.1% had detectable EBV DNA levels after 6 months.
The patients who developed locoregional recurrence with distant metastases were excluded.
A dash (‐) indicates that the value could not be obtained from the cited paper.
There are several advantages of using free circulating EBV DNA as a liquid biomarker for cancer surveillance. First, a temporal relationship has been confirmed between EBV DNA level and the onset of cancer relapse [105, 110]. Furthermore, EBV DNA could turn positive even before the onset of distant metastases or locoregional recurrence. In one study by Hong et al. [105], during post‐therapy surveillance, the majority of NPC patients (81%) had detectable EBV DNA (cut‐off value: 100 copies/mL) prior to or at confirmation of recurrence. However, 35% of the NPC patients with biopsy‐proven locoregional recurrence had undetectable EBV DNA either prior to and at confirmation of recurrence. Therefore, EBV DNA is more useful in detecting distant metastasis than locoregional recurrence in the surveillance scenario.The presence of detectable plasma EBV DNA will prompt physicians to investigate for possible recurrence in the surveillance scenario. In another study of 245 previously treated NPC patients, 5 with initially perceived clinical recurrence but with undetectable EBV levels were eventually confirmed on further investigation to be disease‐free [106]. Thus, EBV DNA may also aid as supporting evidence in addressing equivocal cases of recurrence.
7. CONCLUSIONS
Numerous EBV detection methods have been used to screen, prognosticate, and detect early cancer relapses in NPC patients. Presently, the detection of plasma EBV DNA exhibits a higher sensitivity and specificity than serological methods in population screening of NPC in endemic areas. Additionally, targeting the BamHI‐W fragment of EBV DNA yields superior sensitivity over other EBV targets. There is limited data on the utility of plasma EBV DNA in screening for NPC amongst high‐risk cohorts. However, testing for the presence of plasma EBV DNA after treatment has shown promise in the prognostication of NPC. Emerging data also suggest that incorporating pre‐treatment detection of plasma EBV into the AJCC staging system improves prognostication of NPC patients. In the surveillance scenario, detectable plasma EBV DNA following initial undetectable EBV DNA level is useful for detecting distant relapse and less so for locoregional recurrence. Emerging laboratory‐based NPC detection techniques via miRNAs, hypermethylation gene panels, and combinations may aid in advancing the field of liquid biopsy of NPC management.
DECLARATIONS
AUTHORS’ CONTRIBUTIONS
Development of research topic: R Tan, SKA Phua, YL Soong, LLE Oon, KS Chan, SS Lucky, J Mong, MH Tan, CM Lim
Data accrual and analysis: R Tan, SKA Phua, YL Soong, LLE Oon, KS Chan, SS Lucky, J Mong MH Tan, CM Lim
Manuscript writing, vetting and final approval: R Tan, SKA Phua, YL Soong, LLE Oon, KS Chan, SS Lucky, J Mong, MH Tan, CM Lim.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST STATEMENT
Minhan Tan is the CEO of Lucence Diagnostics. The rest of the authors declare no conflict of interest.
FUNDING
Not applicable.
ACKNOWLEDGEMENTS
We acknowledge the Head Foundation Singapore for their generous support to fund the inaugural Singapore NPC Workshop held in January 2019. During the workshop, the thematic discussion of liquid biopsy in NPC management was presented and summarized.
Tan R, Phua SKA, Soong YL, et al. Clinical utility of Epstein‐Barr virus DNA and other liquid biopsy markers in nasopharyngeal carcinoma. Cancer Commun. 2020;40:564–585. 10.1002/cac2.12100
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Cao S‐M, Simons MJ, Qian C‐N. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer. 2011;30(2):114‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsai S‐T, Jin Y‐T, Su I‐J. Expression of EBER1 in primary and metastatic nasopharyngeal carcinoma tissues using in situ hybridization: A correlation with WHO histologic subtypes. Cancer. 1996;77(2):231‐6. [DOI] [PubMed] [Google Scholar]
- 3. Young LS, Dawson CW. Epstein‐Barr virus and nasopharyngeal carcinoma. Chin J Cancer. 2014;33(12):581‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakanishi Y, Wakisaka N, Kondo S, Endo K, Sugimoto H, Hatano M, et al. Progression of understanding for the role of Epstein‐Barr virus and management of nasopharyngeal carcinoma. Cancer Metastasis Rev. 2017;36(3):435‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hadinoto V, Shapiro M, Greenough TC, Sullivan JL, Luzuriaga K, Thorley‐Lawson DA. On the dynamics of acute EBV infection and the pathogenesis of infectious mononucleosis. Blood. 2008;111(3):1420‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luzuriaga K, Sullivan JL. Infectious Mononucleosis. N Engl J Med. 2010;362(21):1993‐2000. [DOI] [PubMed] [Google Scholar]
- 7. Thorley‐Lawson DA. EBV Persistence–Introducing the Virus. Curr Top Microbiol Immunol. 2015;390(Pt 1):151‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lupton S, Levine AJ. Mapping genetic elements of Epstein‐Barr virus that facilitate extrachromosomal persistence of Epstein‐Barr virus‐derived plasmids in human cells. Mol Cell Biol. 1985;5(10):2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laichalk LL, Hochberg D, Babcock GJ, Freeman RB, Thorley‐Lawson DA. The Dispersal of Mucosal Memory B Cells: Evidence from Persistent EBV Infection. Immunity. 2002;16(5):745‐54. [DOI] [PubMed] [Google Scholar]
- 10. Tsang CM, Deng W, Yip YL, Zeng M‐S, Lo KW, Tsao SW. Epstein‐Barr virus infection and persistence in nasopharyngeal epithelial cells. Chin J Cancer. 2014;33(11):549‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hammerschmidt W. The Epigenetic Life Cycle of Epstein–Barr Virus In: Münz C, editor. Epstein Barr Virus Volume 1: One Herpes Virus: Many Diseases. Cham: Springer International Publishing; 2015. p. 103‐17. [DOI] [PubMed] [Google Scholar]
- 12. Tao Q, Young LS, Woodman CB, Murray PG. Epstein‐Barr virus (EBV) and its associated human cancers–genetics, epigenetics, pathobiology and novel therapeutics. Front Biosci. 2006;11(2):2672‐713. [DOI] [PubMed] [Google Scholar]
- 13. Niedobitek G. Epstein‐Barr virus infection in the pathogenesis of nasopharyngeal carcinoma. Molecular Pathology : MP. 2000;53(5):248‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsao SW, Tsang CM, Lo KW. Epstein‐Barr virus infection and nasopharyngeal carcinoma. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2017;372(1732):20160270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeng Y, Jan MG, Zhang Q, Zhang LG, Li HY, Wu YC, et al. Serological mass survey for early detection of nasopharyngeal carcinoma in wuzhou city, china. Int J Cancer. 1982;29(2):139‐41. [DOI] [PubMed] [Google Scholar]
- 16. Deng H, Zeng Y, Lei Y, Zhao Z, Wang P, Li B, et al. Serological survey of nasopharyngeal carcinoma in 21 cities of south China. Chin Med J. 1995;108(4):300‐3. [PubMed] [Google Scholar]
- 17. Liu Z, Ji M‐F, Huang Q‐H, Fang F, Liu Q, Jia W‐H, et al. Two Epstein‐Barr Virus–Related Serologic Antibody Tests in Nasopharyngeal Carcinoma Screening: Results From the Initial Phase of a Cluster Randomized Controlled Trial in Southern China. Am J Epidemiol. 2012;177(3):242‐50. [DOI] [PubMed] [Google Scholar]
- 18. Yu X, Ji M, Cheng W, Wu B, Du Y, Cao S. Assessment of the Long‐term Diagnostic Performance of a New Serological Screening Scheme in Large‐scale Nasopharyngeal Carcinoma Screening. J Cancer. 2018;9:2093‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan KCA, Hung ECW, Woo JKS, Chan PKS, Leung S‐F, Lai FPT, et al. Early detection of nasopharyngeal carcinoma by plasma Epstein‐Barr virus DNA analysis in a surveillance program. Cancer. 2013;119(10):1838‐44. [DOI] [PubMed] [Google Scholar]
- 20. Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, et al. Analysis of Plasma Epstein–Barr Virus DNA to Screen for Nasopharyngeal Cancer. N Engl J Med. 2017;377(6):513‐22. [DOI] [PubMed] [Google Scholar]
- 21. Zheng X‐H, Lu L‐X, Cui C, Chen M‐Y, Li X‐Z, Jia W‐H. Epstein‐Barr virus mir‐bart1‐5p detection via nasopharyngeal brush sampling is effective for diagnosing nasopharyngeal carcinoma. Oncotarget. 2015;7(4):4972‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirai N, Wakisaka N, Kondo S, Aga M, Moriyama‐Kita M, Ueno T, et al. Potential Interest in Circulating miR‐BART17‐5p As a Post‐Treatment Biomarker for Prediction of Recurrence in Epstein‐Barr Virus‐Related Nasopharyngeal Carcinoma. PLoS One. 2016;11(9):e0163609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wen Z, Li Z, Yong P, Liang D, Xie D, Chen H, et al. Detection and clinical significance of circulating tumor cells in patients with nasopharyngeal carcinoma. Oncol Lett. 2019;18(3):2537‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yanjiao Mao WY, Chen Jiangtian, Deng Qinghua, Wu Shixiu. Cell counts and clinical significance of circulating tumor cells in patients diagnosed with nasopharyngeal carcinoma. Int J Clin Exp Med. 2019;12(7):9167‐74. [Google Scholar]
- 25. Hui E, Li W, Ma B, Lam W, Chan K, Mo F, et al. Integrating post‐radiotherapy plasma Epstein‐Barr virus DNA and TNM stage for risk stratification of nasopharyngeal carcinoma to adjuvant therapy. Ann Oncol. 2020. [DOI] [PubMed] [Google Scholar]
- 26. Lv J, Chen Y, Zhou G, Qi Z, Tan KRL, Wang H, et al. Liquid biopsy tracking during sequential chemo‐radiotherapy identifies distinct prognostic phenotypes in nasopharyngeal carcinoma. Nat Commun. 2019;10(1):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W‐Y, Twu C‐W, Chen H‐H, Jan J‐S, Jiang R‐S, Chao JYC, et al. Plasma EBV DNA clearance rate as a novel prognostic marker for metastatic/recurrent nasopharyngeal carcinoma. Clin Cancer Res. 2010;16(3):1016‐24. [DOI] [PubMed] [Google Scholar]
- 28. Lo YMD, Chan LYS, Lo K‐W, Leung S‐F, Zhang J, Chan ATC, et al. Quantitative Analysis of Cell‐free Epstein‐Barr Virus DNA in Plasma of Patients with Nasopharyngeal Carcinoma. Cancer Res. 1999;59(6):1188. [PubMed] [Google Scholar]
- 29. Chan KCA, Zhang J, Chan ATC, Lei KIK, Leung S‐F, Chan LYS, et al. Molecular Characterization of Circulating EBV DNA in the Plasma of Nasopharyngeal Carcinoma and Lymphoma Patients. Cancer Res. 2003;63(9):2028. [PubMed] [Google Scholar]
- 30. Lam WKJ, Chan KCA, Lo YMD. Plasma Epstein–Barr virus DNA as an archetypal circulating tumour DNA marker. J Pathol. 2019;247(5):641‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch R‐D, et al. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin from Apoptotic and Necrotic Cells. Cancer Res. 2001;61(4):1659. [PubMed] [Google Scholar]
- 32. Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. PNAS. 2005;102(45):16368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Y, Sann KN, Chua M. The evolution of Epstein‐Barr virus detection in nasopharyngeal carcinoma. Cancer Biology and Medicine. 2018;15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim KY, Le Q‐T, Yom SS, Pinsky BA, Bratman SV, Ng RHW, et al. Current State of PCR‐Based Epstein‐Barr Virus DNA Testing for Nasopharyngeal Cancer. J Natl Cancer Inst. 2017;109(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Preiksaitis J, Pang X, Fox J, Fenton J, Caliendo A, Miller G, et al. Interlaboratory Comparison of Epstein‐Barr Virus Viral Load Assays. Am J Transplant. 2009;9:269‐79. [DOI] [PubMed] [Google Scholar]
- 36. Fryer JF, Heath A, Wilkinson DE, Minor PD. World Health Organization. Biologicals U, Standardization WHOECoB. Collaborative study to evaluate the proposed 1st WHO international standards for Epstein‐Barr Virus (EBV) for Nucleic Acid Amplification Technology (NAT)‐Based Assays / by Fryer Jacqueline F., Heath Alan B., Wilkinson Dianna E., D Philip. Minor and the Collaborative Study Group. Geneva: World Health Organization; 2011. [Google Scholar]
- 37. Abeynayake J, Johnson R, Libiran P, Sahoo MK, Cao H, Bowen R, et al. Commutability of the Epstein‐Barr Virus WHO International Standard across Two Quantitative PCR Methods. J Clin Microbiol. 2014;52(10):3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vesper HW, Miller WG, Myers GL. Reference materials and commutability. The Clinical Biochemist Reviews. 2007;28(4):139‐47. [PMC free article] [PubMed] [Google Scholar]
- 39. Vo JH, Nei WL, Hu M, Phyo WM, Wang F, Fong KW, et al. Comparison of Circulating Tumour Cells and Circulating Cell‐Free Epstein‐Barr Virus DNA in Patients with Nasopharyngeal Carcinoma Undergoing Radiotherapy. Sci Rep. 2016;6(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lam WKJ, Jiang P, Chan KCA, Cheng SH, Zhang H, Peng W, et al. Sequencing‐based counting and size profiling of plasma Epstein–Barr virus DNA enhance population screening of nasopharyngeal carcinoma. Proc Natl Acad Sci. 2018;115(22):E5115‐E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lo S, Ho W‐K, Wei WI. Outcome of Patients With Positive Epstein‐Barr Virus Serologic Status in the Absence of Nasopharyngeal Carcinoma in Hong Kong. Arch Otolaryngol Head Neck Surg. 2004;130(6):770‐2. [DOI] [PubMed] [Google Scholar]
- 42. Draborg A, Jørgensen J, Müller H, Nielsen C, Jacobsen S, Iversen L, et al. Epstein‐Barr virus early antigen diffuse (EBV‐EA/D)‐directed immunoglobulin A antibodies in systemic lupus erythematosus patients. Scand J Rheumatol. 2012;41:280‐9. [DOI] [PubMed] [Google Scholar]
- 43. Pickard A, Chen C‐J, Diehl SR, Liu M‐Y, Cheng Y‐J, Hsu W‐L, et al. Epstein‐Barr virus seroreactivity among unaffected individuals within high‐risk nasopharyngeal carcinoma families in Taiwan. Int J Cancer. 2004;111(1):117‐23. [DOI] [PubMed] [Google Scholar]
- 44. Li F, Liu J, Song D, Zhang Q, Ding N, He X. Circulating tumor cells in the blood of poorly differentiated nasal squamous cell carcinoma patients: correlation with treatment response. Acta Otolaryngol. 2016;136(11):1164‐7. [DOI] [PubMed] [Google Scholar]
- 45. Wang M, Yu F, Wu W, Wang Y, Ding H, Qian L. Epstein‐Barr virus‐encoded microRNAs as regulators in host immune responses. International Journal of Biological Sciences. 2018;14:565‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang G, Zong J, Lin S, Verhoeven RJA, Tong S, Chen Y, et al. Circulating Epstein–Barr virus microRNAs miR‐BART7 and miR‐BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer. 2015;136(5):E301‐E12. [DOI] [PubMed] [Google Scholar]
- 47. Chang HW, Chan A, Kwong DLW, Wei WI, Sham JST, Yuen APW. Evaluation of hypermethylated tumor suppressor genes as tumor markers in mouth and throat rinsing fluid, nasopharyngeal swab and peripheral blood of nasopharygeal carcinoma patient. Int J Cancer. 2003;105(6):851‐5. [DOI] [PubMed] [Google Scholar]
- 48. Tian F, Yip SP, Kwong DLW, Lin Z, Yang Z, Wu VWC. Promoter hypermethylation of tumor suppressor genes in serum as potential biomarker for the diagnosis of nasopharyngeal carcinoma. Cancer Epidemiol. 2013;37(5):708‐13. [DOI] [PubMed] [Google Scholar]
- 49. He Y, Zhang L, Cheng G, Yuan R, Zhuang Y, Zhang D, et al. Upregulation of circulating miR‐21 is associated with poor prognosis of nasopharyngeal carcinoma. International Journal of Clinical and Experimental Pathology. 2017;10:7362‐8. [PMC free article] [PubMed] [Google Scholar]
- 50. Pantel K, Alix‐Panabieres C. Real‐time liquid biopsy in cancer patients: fact or fiction? Cancer Res. 2013;73(21):6384‐8. [DOI] [PubMed] [Google Scholar]
- 51. He C, Huang X, Su X, Tang T, Zhang X, Ma J, et al. The association between circulating tumor cells and Epstein‐Barr virus activation in patients with nasopharyngeal carcinoma. Cancer Biol Ther. 2017;18(11):888‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. You R, Liu Y‐P, Lin M, Huang P‐Y, Tang L‐Q, Zhang Y‐N, et al. Relationship of circulating tumor cells and Epstein–Barr virus DNA to progression‐free survival and overall survival in metastatic nasopharyngeal carcinoma patients. Int J Cancer. 2019;145(10):2873‐83. [DOI] [PubMed] [Google Scholar]
- 53. Xu L, Mao X, Imrali A, Syed F, Mutsvangwa K, Berney D, et al. Optimization and evaluation of a novel size based circulating tumor cell isolation system. PLoS One. 2015;10(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tamminga M, Andree KC, Hiltermann TJN, Jayat M, Schuuring E, van den Bos H, et al. Detection of Circulating Tumor Cells in the Diagnostic Leukapheresis Product of Non‐Small‐Cell Lung Cancer Patients Comparing CellSearch® and ISET. Cancers. 2020;12(4):896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mong J, Tan MH. Size‐Based Enrichment Technologies for Non‐cancerous Tumor‐Derived Cells in Blood. Trends Biotechnol. 2018;36(5):511‐22. [DOI] [PubMed] [Google Scholar]
- 56. Lee KT‐W, Tan J‐K, Lam AK‐y, Gan S‐Y. MicroRNAs serving as potential biomarkers and therapeutic targets in nasopharyngeal carcinoma: A critical review. Crit Rev Oncol Hematol. 2016;103:1‐9. [DOI] [PubMed] [Google Scholar]
- 57. Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y, et al. The emerging role of Epstein‐Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer. 2018;9(16):2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69(7):2766‐74. [DOI] [PubMed] [Google Scholar]
- 59. Jiang W, Liu N, Chen X‐Z, Sun Y, Li B, Ren X‐Y, et al. Genome‐Wide Identification of a Methylation Gene Panel as a Prognostic Biomarker in Nasopharyngeal Carcinoma. Mol Cancer Ther. 2015;14(12):2864. [DOI] [PubMed] [Google Scholar]
- 60. Li L, Zhang Y, Fan Y, Sun K, Su X, Du Z, et al. Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes. Epigenomics. 2015;7(2):155‐73. [DOI] [PubMed] [Google Scholar]
- 61. Lam WKJ, Jiang P, Chan KCA, Peng W, Shang H, Heung MMS, et al. Methylation analysis of plasma DNA informs etiologies of Epstein‐Barr virus‐associated diseases. Nat Commun. 2019;10(1):3256‐. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kwong J, Lo KW, To KF, Teo PM, Johnson PJ, Huang DP. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin Cancer Res. 2002;8(1):131‐7. [PubMed] [Google Scholar]
- 63. Shao Y, Jiang H, Wu X, Luo Y, Tang W. p16 promoter hypermethylation is associated with increased risk of nasopharyngeal carcinoma. Mol Clin Oncol. 2014;2(6):1121‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Du C, Huang T, Sun D, Mo Y, Feng H, Zhou X, et al. CDH4 as a novel putative tumor suppressor gene epigenetically silenced by promoter hypermethylation in nasopharyngeal carcinoma. Cancer Lett. 2011;309(1):54‐61. [DOI] [PubMed] [Google Scholar]
- 65. Niller HH, Wolf H, Minarovits J. Viral hit and run‐oncogenesis: Genetic and epigenetic scenarios. Cancer Lett. 2011;305(2):200‐17. [DOI] [PubMed] [Google Scholar]
- 66. Wong T‐S, Kwong DL‐W, Sham JS‐T, Wei WI, Kwong Y‐L, Yuen AP‐W. Quantitative Plasma Hypermethylated DNA Markers of Undifferentiated Nasopharyngeal Carcinoma. Clin Cancer Res. 2004;10(7):2401. [DOI] [PubMed] [Google Scholar]
- 67. Mutirangura A, Pornthanakasem W, Theamboonlers A, Sriuranpong V, Lertsanguansinchi P, Yenrudi S, et al. Epstein‐Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res. 1998;4(3):665‐9. [PubMed] [Google Scholar]
- 68. Sengar M, Chorghe S, Jadhav K, Singh S, Laskar SG, Pai P, et al. Cell‐free Epstein–Barr virus‐DNA in patients with nasopharyngeal carcinoma: Plasma versus urine. Head Neck. 2016;38(S1):E1666‐E73. [DOI] [PubMed] [Google Scholar]
- 69. Yang X, Dai W, Kwong DL‐w, Szeto CYY, Wong EH‐w, Ng WT, et al. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation‐sensitive high resolution melting. Int J Cancer. 2015;136(4):E127‐E35. [DOI] [PubMed] [Google Scholar]
- 70. Mai S‐J, Zong Y, Zhang M, Zhong B, Lin S. Detection of Epstein‐Barr virus DNA in plasma/serum: a useful serological indicator for diagnosis of nasopharyngeal carcinoma. Chin Med J. 2002;115(12):1895‐7. [PubMed] [Google Scholar]
- 71. Luo Y‐L, Ou G‐P, Chi P‐D, Liang Y‐N, Liu Y‐H, Huang M‐Y. Combined determination of Epstein‐Barr virus‐related antibodies and antigens for diagnosis of nasopharyngeal carcinoma. Ai Zheng. 2009;28(1):76‐8. [PubMed] [Google Scholar]
- 72. Leung S‐f, Tam JS, Chan AT, Zee B, Chan LY, Huang DP, et al. Improved accuracy of detection of nasopharyngeal carcinoma by combined application of circulating Epstein–Barr virus DNA and anti‐Epstein–Barr viral capsid antigen IgA antibody. Clin Chem. 2004;50(2):339‐45. [DOI] [PubMed] [Google Scholar]
- 73. Teresa M, Yu G, Hu K, Li JC. Plasma Epstein‐Barr virus immunoglobulin A and DNA for nasopharyngeal carcinoma screening in the United States. Otolaryngology—Head and Neck Surgery. 2007;136(6):992‐7. [DOI] [PubMed] [Google Scholar]
- 74. Chang K‐P, Hsu C‐L, Chang Y‐L, Tsang N‐M, Chen C‐K, Lee T‐J, et al. Complementary serum test of antibodies to Epstein‐Barr virus nuclear antigen‐1 and early antigen: a possible alternative for primary screening of nasopharyngeal carcinoma. Oral Oncol. 2008;44(8):784‐92. [DOI] [PubMed] [Google Scholar]
- 75. Liu Y, Fang Z, Liu L, Yang S, Zhang L. Detection of Epstein‐Barr virus DNA in serum or plasma for nasopharyngeal cancer: a meta‐analysis. Genetic testing and molecular biomarkers. 2011;15(7‐8):495‐502. [DOI] [PubMed] [Google Scholar]
- 76. Song C, Yang S. A meta‐analysis on the EBV DNA and VCA‐IgA in diagnosis of Nasopharyngeal Carcinoma. Pakistan Journal of Medical Sciences. 2013;29(3):885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Low W‐K, Leong J‐L, Goh Y‐H, Fong K‐W. Diagnostic Value of Epstein‐Barr Viral Serology in Nasopharyngeal Carcinoma. Otolaryngol Head Neck Surg. 2000;123(4):505‐7. [DOI] [PubMed] [Google Scholar]
- 78. Fachiroh J, Paramita DK, Hariwiyanto B, Harijadi A, Dahlia HL, Indrasari SR, et al. Single‐Assay Combination of Epstein‐Barr Virus (EBV) EBNA1‐ and Viral Capsid Antigen‐p18‐Derived Synthetic Peptides for Measuring Anti‐EBV Immunoglobulin G (IgG) and IgA Antibody Levels in Sera from Nasopharyngeal Carcinoma Patients: Options for Field Screening. J Clin Microbiol. 2006;44(4):1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cai Y‐L, Li J, Lu A‐Y, Zheng Y‐M, Zhong W‐M, Wang W, et al. Diagnostic significance of combined detection of Epstein‐Barr virus antibodies, VCA/IgA, EA/IgA, Rta/IgG and EBNA1/IgA for nasopharyngeal carcinoma. Asian Pac J Cancer Prev. 2014;15(5):2001‐6. [DOI] [PubMed] [Google Scholar]
- 80. Chen Y, Zhao W, Lin L, Xiao X, Zhou X, Ming H, et al. Nasopharyngeal Epstein‐Barr Virus Load: An Efficient Supplementary Method for Population‐Based Nasopharyngeal Carcinoma Screening. PLoS One. 2015;10(7):e0132669‐e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li T, Guo X, Ji M, Li F, Wang H, Cheng W, et al. Establishment and validation of a two‐step screening scheme for improved performance of serological screening of nasopharyngeal carcinoma. Cancer Med. 2018;7(4):1458‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jiang C, Chen J, Xie S, Zhang L, Xiang Y, Lung M, et al. Evaluation of circulating EBV microRNA BART2‐5p in facilitating early detection and screening of nasopharyngeal carcinoma. Int J Cancer. 2018;143(12):3209‐17. [DOI] [PubMed] [Google Scholar]
- 83. Ng W‐T, Yau T‐K, Yung RWH, Sze W‐M, Tsang AHL, Law ALY, et al. Screening for family members of patients with nasopharyngeal carcinoma. Int J Cancer. 2005;113(6):998‐1001. [DOI] [PubMed] [Google Scholar]
- 84. Ng WT, Choi CW, Lee MC, Law LY, Yau TK, Lee AW. Outcomes of nasopharyngeal carcinoma screening for high risk family members in Hong Kong. Fam Cancer. 2010;9(2):221‐8. [DOI] [PubMed] [Google Scholar]
- 85. Lin J‐C, Wang W‐Y, Chen KY, Wei Y‐H, Liang W‐M, Jan J‐S, et al. Quantification of Plasma Epstein–Barr Virus DNA in Patients with Advanced Nasopharyngeal Carcinoma. N Engl J Med. 2004;350(24):2461‐70. [DOI] [PubMed] [Google Scholar]
- 86. Prayongrat A, Chakkabat C, Kannarunimit D, Hansasuta P, Lertbutsayanukul C. Prevalence and significance of plasma Epstein‐Barr Virus DNA level in nasopharyngeal carcinoma. J Radiat Res. 2017;58(4):509‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Peng H, Guo R, Chen L, Zhang Y, Li W‐F, Mao Y‐P, et al. Prognostic impact of plasma Epstein‐Barr virus DNA in patients with nasopharyngeal carcinoma treated using intensity‐modulated radiation therapy. Sci Rep. 2016;6:22000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Twu C‐W, Wang W‐Y, Liang W‐M, Jan J‐S, Jiang R‐S, Chao J, et al. Comparison of the prognostic impact of serum anti‐EBV antibody and plasma EBV DNA assays in nasopharyngeal carcinoma. International Journal of Radiation Oncology • Biology • Physics. 2007;67(1):130‐7. [DOI] [PubMed] [Google Scholar]
- 89. Ling W, Cao S‐M, Huang Q‐H, Li Y‐H, Deng M‐Q. Prognostic implication of pretreatment titer of serum immunoglobulin A against Epstein‐Barr virus capsid antigen in nasopharyngeal carcinoma patients in Sihui, Guangdong. Ai Zheng. 2009;28(1):57‐9. [PubMed] [Google Scholar]
- 90. Zhang J, Shu C, Song Y, Li Q, Huang J, Ma X. Epstein–Barr virus DNA level as a novel prognostic factor in nasopharyngeal carcinoma: A meta‐analysis. Medicine (Baltimore). 2016;95(40). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Leung S‐f, Zee B, Ma BB, Hui EP, Mo F, Lai M, et al. Plasma Epstein‐Barr Viral Deoxyribonucleic Acid Quantitation Complements Tumor‐Node‐Metastasis Staging Prognostication in Nasopharyngeal Carcinoma. J Clin Oncol. 2006;24(34):5414‐8. [DOI] [PubMed] [Google Scholar]
- 92. Kitpanit S, Jittapiromsak N, Sriyook A, Prayongrat A, Kannarunimit D, Chakkabat C, et al. Comparison between the seventh and eighth edition of the AJCC/UICC staging system for nasopharyngeal cancer integrated with pretreatment plasma Epstein–Barr virus DNA level in a non‐Chinese population: secondary analysis from a prospective randomized trial. Jpn J Clin Oncol. 2019;49(12):1100‐13. [DOI] [PubMed] [Google Scholar]
- 93. Chan ATC, Hui EP, Ngan RKC, Tung SY, Cheng ACK, Ng WT, et al. Analysis of Plasma Epstein‐Barr Virus DNA in Nasopharyngeal Cancer After Chemoradiation to Identify High‐Risk Patients for Adjuvant Chemotherapy: A Randomized Controlled Trial. J Clin Oncol. 2018;36(31):3091‐100. [DOI] [PubMed] [Google Scholar]
- 94. Lee AWM, Poon YF, Foo W, Law SCK, Cheung FK, Chan DKK, et al. Retrospective analysis of 5037 patients with nasopharyngeal carcinoma treated during 1976‐1985: Overall survival and patterns of failure. International Journal of Radiation Oncology*Biology*Physics. 1992;23(2):261‐70. [DOI] [PubMed] [Google Scholar]
- 95. Chan ATC, Lo YMD, Zee B, Chan LYS, Ma BBY, Leung S‐F, et al. Plasma Epstein‐Barr Virus DNA and Residual Disease After Radiotherapy for Undifferentiated Nasopharyngeal Carcinoma. J Natl Cancer Inst. 2002;94(21):1614‐9. [DOI] [PubMed] [Google Scholar]
- 96. Lo YMD, Chan ATC, Chan LYS, Leung S‐F, Lam C‐W, Huang DP, et al. Molecular Prognostication of Nasopharyngeal Carcinoma by Quantitative Analysis of Circulating Epstein‐Barr Virus DNA. Cancer Res. 2000;60(24):6878. [PubMed] [Google Scholar]
- 97. Bhattacharyya T, Babu G, Kainickal CT. Current role of chemotherapy in nonmetastatic nasopharyngeal cancer. J Oncol. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Si Y, Lan G, Deng Z, Wang Y, Lu Y, Qin Y, et al. Distribution and clinical significance of circulating tumor cells in nasopharyngeal carcinoma. Jpn J Clin Oncol. 2016;46(7):622‐30. [DOI] [PubMed] [Google Scholar]
- 99. Wang L, Guo Y, Xu J, Chen Z, Jiang X, Zhang L, et al. Clinical Analysis of Recurrence Patterns in Patients With Nasopharyngeal Carcinoma Treated With Intensity‐Modulated Radiotherapy. Annals of Otology, Rhinology & Laryngology. 2017;126(12):789‐97. [DOI] [PubMed] [Google Scholar]
- 100. Farias TP, Dias FL, Lima RA, Kligerman J, de Sá GM, Barbosa MM, et al. Prognostic Factors and Outcome for Nasopharyngeal Carcinoma. Arch Otolaryngol Head Neck Surg. 2003;129(7):794‐9. [DOI] [PubMed] [Google Scholar]
- 101. Fan H, Nicholls J, Chua D, Chan KH, Sham J, Lee S, et al. Laboratory markers of tumor burden in nasopharyngeal carcinoma: A comparison of viral load and serologic tests for Epstein‐Barr virus. Int J Cancer. 2004;112(6):1036‐41. [DOI] [PubMed] [Google Scholar]
- 102. Hsu C‐L, Chan S‐C, Chang K‐P, Lin T‐L, Lin C‐Y, Hsieh C‐H, et al. Clinical scenario of EBV DNA follow‐up in patients of treated localized nasopharyngeal carcinoma. Oral Oncol. 2013;49(6):620‐5. [DOI] [PubMed] [Google Scholar]
- 103. Ferrari D, Codecà C, Bertuzzi C, Broggio F, Crepaldi F, Luciani A, et al. Role of plasma EBV DNA levels in predicting recurrence of nasopharyngeal carcinoma in a western population. BMC Cancer. 2012;12(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Leung S‐f, Lo YMD, Chan ATC, To K‐f, To E, Chan LYS, et al. Disparity of Sensitivities in Detection of Radiation‐Naïve and Postirradiation Recurrent Nasopharyngeal Carcinoma of the Undifferentiated Type by Quantitative Analysis of Circulating Epstein‐Barr Virus DNA. Clin Cancer Res. 2003;9(9):3431. [PubMed] [Google Scholar]
- 105. Hong R‐L, Lin C‐Y, Ting L‐L, Ko J‐Y, Hsu M‐M. Comparison of clinical and molecular surveillance in patients with advanced nasopharyngeal carcinoma after primary therapy. Cancer. 2004;100(7):1429‐37. [DOI] [PubMed] [Google Scholar]
- 106. Wang W‐Y, Twu C‐W, Lin W‐Y, Jiang R‐S, Liang K‐L, Chen K‐W, et al. Plasma Epstein‐Barr virus DNA screening followed by 18F‐fluoro‐2‐deoxy‐D‐glucose positron emission tomography in detecting posttreatment failures of nasopharyngeal carcinoma. Cancer. 2011;117(19):4452‐9. [DOI] [PubMed] [Google Scholar]
- 107. Chan ATC, Ma BBY, Lo YMD, Leung SF, Kwan WH, Hui EP, et al. Phase II Study of Neoadjuvant Carboplatin and Paclitaxel Followed by Radiotherapy and Concurrent Cisplatin in Patients With Locoregionally Advanced Nasopharyngeal Carcinoma: Therapeutic Monitoring With Plasma Epstein‐Barr Virus DNA. J Clin Oncol. 2016;22(15):3053‐60. [DOI] [PubMed] [Google Scholar]
- 108. Li W‐F, Zhang Y, Huang X‐B, Du X‐J, Tang L‐L, Chen L, et al. Prognostic value of plasma Epstein–Barr virus DNA level during posttreatment follow‐up in the patients with nasopharyngeal carcinoma having undergone intensity‐modulated radiotherapy. Chin J Cancer. 2017;36(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cao S, Min H, Gao J, Hong M, Xiao X, Zhang C, et al. Significance of cell‐free epstein‐barr virus DNA in monitoring prognosis of nasopharyngeal carcinoma. Chinese Journal of Clinical Oncology. 2004;1(3):190‐5. [PubMed] [Google Scholar]
- 110. Dennis Lo YM, Chan LYS, Chan ATC, Leung S‐F, Lo K‐W, Zhang J, et al. Quantitative and Temporal Correlation between Circulating Cell‐Free Epstein‐Barr Virus DNA and Tumor Recurrence in Nasopharyngeal Carcinoma. Cancer Res. 1999;59(21):5452. [PubMed] [Google Scholar]
- 111. Chen Z, Xu L, Xu X, Yuan C. The clinical value of detecting circulating tumour cells in the peripheral blood of nasopharyngeal carcinoma patients. Oncol Lett. 2018;15(5):6283‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


