Abstract
Background:
Several studies have explored the prognostic value of MicroRNA-153 (miR-153) in various cancers, but obtained inconsistent results. Thus, we conducted a meta-analysis to assess the prognostic significance of miR-153 for patients with cancer.
Methods:
Eligible studies were identified by searching the online databases Pubmed, Embase, Web of Science, Medline,and the China National Knowledge Infrastructure (CNKI) up to March 2020. Hazard ratios (HRs) with 95% CIs and were calculated to clarify the correlation between miR-153 expression and prognosis of different cancers. Odds ratios (ORs) with 95% CI were selected to appraise the correlation between miR-153 with clinicopathological characteristics of cancer patients.
Results:
In total, 933 patients from 11 articles were enrolled in our meta-analysis. The results revealed that low miR-153 expression was significantly correlated with poor overall survival (OS) (HR = 2.45, 95% CI = 1.66–3.63, P < .001), but not with disease-free survival (DFS) (HR = 1.67, 95% CI = 0.45–6.19, P = .442). Subgroup analysis found that low miR-153 expression was associated with worse OS in the reported directly from articles group (HR = 2.67, 95% CI: 1.32–5.37, P = .006), survival curves group (HR = 2.10, 95% CI: 1.56–2.84, P < .001), digestive system tumor (HR = 2.76, 95% CI: 1.73–4.41, P < .001), and breast cancer (HR = 4.01, 95% CI: 1.46–11.04, P = .007).
Moreover, cancer patients with low miR-153 expression were prone to poor tumor differentiation(poor vs well+moderate, OR = 2.41, 95% CI = 1.52–3.82, P < .001), earlier lymph node metastasis (present vs absent, OR = 2.19, 95% CI = 1.12–4.25, P = .021) and earlier distant metastasis (present vs absent,OR = 8.24, 95% CI = 2.93–23.21, P < .001), but not associated with age,gender and TNM stage.
Conclusions:
This meta-analysis indicated that low miR-153 expression is associated with poor prognosis. miR-153 may serve as an effective predictive biomarker for tumor prognosis, especially for digestive system tumor and breast cancer.
Keywords: cancer, meta-analysis, microRNA-153, prognosis
1. Introduction
Cancer is a multi-factorial disease and is one of the leading causes of death worldwide. According to American Cancer Society estimates, the projected numbers of newly diagnosed cases and deaths are 17.6 and 6.0 million, respectively, in the United States in 2019.[1] Though there have been substantial advances in diagnosis and treatment methods, the 5-year survival for a majority of malignancies still remains low in general.[2] Therefore, many scientists have made tremendous efforts to search for the new biomarkers for determining or predicting the prognosis for cancer.
MicroRNAs (miRNAs), a kind of endogenous non-coding single-stranded RNAs, (18–25 nucleotides in length), regulate protein translation at the post-transcriptional level by pairing with complementary sequences in the 3’-untranslated region (3’-UTR) of target mRNAs.[3] miRNAs play a critical role in the regulation of cell cycle, apoptosis, proliferation and differentiation.[4,5] MiR-153, as a novel tumor-related miRNA, was initially found in seven miRNAs specifically expressed in human and mouse brain tissues.[6] MiR-153 was downregulated in various cancers, including glioblastoma,[7] oral cancer,[8] breast cancer,[9] and ovarian cancer.[10]
Many studies investigated the prognostic value of circulating miR-153 in various cancers. Some studies found that the downregulation of circulating miR-153 was associated with worse outcome in cancer patients.[11–18] However, some other studies showed insignificant or opposite results.[19,20] We have therefore performed a meta-analysis to assess whether miR-153 expression was associated with prognosis and clinicopathological factors in cancer patients.
2. Methods
2.1. Search strategies
We searched Pubmed, Embase, Web of Science, Medline, and the China National Knowledge Infrastructure (CNKI) up to March 2020 to identify relevant studies. The key search terms were used as follows with multiple combinations: (“MicroRNA 153” OR “miR-153”) AND (“carcinoma” OR “cancer” OR “tumor” OR “neoplasm” OR “malignancy”) AND (“prognosis” OR “prognostic”). Further manual inspection was performed to improve the integrity of the eligible papers by going through the title and abstract. Moreover, references in relevant publications were also browsed. The present study was meta-analysis and did not involve the collection of samples. Therefore, ethical approval was not required.
2.2. Inclusion and exclusion criteria
The studies were included in our meta-analysis if they met the following inclusion criteria:
-
1.
miR-153 expression evaluated in the human tissues;
-
2.
Tumors should be confirmed by histological or pathological examinations;
-
3.
The main outcome of interest focus on prognostic factors;
-
4.
Sufficient information provided to calculate the odds ratios (ORs) or hazard ratio (HRs) estimates and their 95% confidence intervals (95%CIs).
The exclusion criteria were as follow:
-
1.
letters, case reports, reviews, and conference abstracts without original data;
-
2.
duplicate publications;
-
3.
articles from which the relevant data could not be extracted.
2.3. Data extraction and quality assessment
The studies information of this meta-analysis were retrieved by the reporting checklists of Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) guidelines.[21] Data extraction was conducted independently by 2 investigators (HMQ and LCF) from identified research in agreement with prescribed standards, during which disagreements were resolved by reaching a consensus on all contents. The extracted data elements mainly included the following information: first author, publication year, country, age, cancer type, total number of patients, outcome measure, method, recruitment time, HR obtain method and NOS scores. Clinicopathological factors included age, gender, tumor differentiation, lymph node metastasis, TNM stage, and distant metastasis. When HRs and their 95% CIs were given in the articles, these data were extracted directly. If the prognosis was plotted as Kaplan–Meier survival curve, the data were digitized by the software Engauge Digitizer version 4.1 and calculated as described.[22,23] The Newcastle–Ottawa Scale (NOS) was used to assess the quality of included studies.[24] Three aspects were considered in the NOS criteria:
-
1.
subject selection, 0 to 4;
-
2.
comparability of subject, 0 to 2;
-
3.
clinical outcome: 0 to 3.
The range of NOS scores is from 0 to 9; and a score ≥6 means a good quality.
2.4. Statistical analysis
HRs with 95%CIs were calculated the association between miR-153 expression and the overall survival (OS) and disease-free survival (DFS) of cancer patients. ORs with 95% CIs were used to assess the association of miR-153 expression with clinicopathological characteristics. The evaluation of statistical heterogeneity was finished by using the Cochrans Q statistic and I2 tests.[25] If the heterogeneity was significant between studies (I2 > 50% or P < .10), the random-effects model was used; otherwise, the fixed-effects model was used.[26] Both Beggs test and Eggers test were used to evaluate the potential publication bias.[27] The statistical analyses were performed using STATA version 12.0 software (Stata Corporation, Collage Station, Texas, USA). All P values were two-sided and P < .05 was considered statistically significant.
3. Results
3.1. Literature search and study characteristics
The literature screening process is illustrated in Figure 1. A total of 162 articles from the 5 databases and two articles from a manual reference search were initially selected. After removing duplicates, 83 studies remained. After reading the titles and abstracts, 55 irrelevant studies were excluded. Of the remaining studies, 17 articles were excluded for 10 articles without focusing on this topic and 7 articles without sufficient data. Finally, altogether 11 articles including 933 patients were selected for the meta-analysis.[11–20,28] Among the articles, 10 reported the correlation of miR-153 expression with OS, whereas only 4 studies reported the correlation between mir-153 expression and DFS. Among the studies, 1 evaluated oral squamous cell carcinoma, 1 evaluated colorectal cancer, 2 evaluated gastric cancer, 1 evaluated pancreatic ductal adenocarcinoma, 1 evaluated lung cancer, 1 evaluated prostate cancer, 1 evaluated glioma and 2 evaluated breast cancer. All of the included studies were published from 2013 to 2019. The basic characteristics of the involved studies are presented in Table 1.
Figure 1.
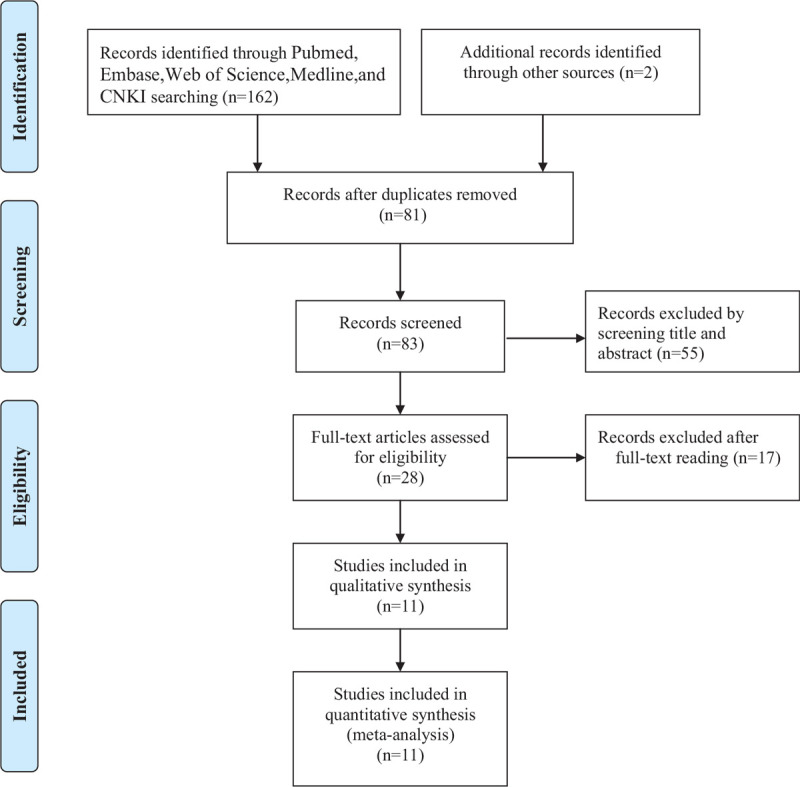
Flow diagram of study selection in present meta-analysis.
Table 1.
Characteristics of the included studies.
| First author (year) | N | Cancer type | Country | Age(year) | target gene | Recruitment time | method | Outcome measure | HR obtain method | NOS score |
| Xu, (2013) | e | OSCC | China | NR | SNAI1 and ZEB2 | 2006-2007 | qRT-PCR | OS | survival curves | 6 |
| Zhang, (2013) | 60 | CRC | UK | >60, 91.67% | MMP-9 | NA | qRT-PCR | DFS | reported directly | 7 |
| Zhang, (2015) | 80 | Gastric cancer | China | ≥65, 46.25% | EMT. | 2006.1–2008.12 | qRT-PCR | OS,DFS | reported directly | 8 |
| Bai, (2015) | 80 | PDAC | China | >60, 65% | SNAI1 | 2005.1–2010.12 | qRT-PCR | OS | reported directly | 8 |
| Chen, (2015) | 137 | Lung cancer | China | ≥60, 61.31% | NR | 2007.1–2013.4 | qRT-PCR | OS | reported directly | 7 |
| Liu, (2018) | 93 | Cervical cancer | China | >50, 48.39% | NR | 2008.3–2011.9 | qRT-PCR | OS | reported directly | 7 |
| Ouyang, (2018) | 83 | Gastric cancer | China | median 64.7 (47–82) | Kruppel-like factor 5 | 2011.3–2012.4 | qRT-PCR | OS | survival curves | 8 |
| Bi (2019) | 143 | Prostate cancer | China | ≥60, 69.23% | NR | 2014.4–2018.3 | qRT-PCR | OS | reported directly | 7 |
| Shi, (2019) | 60 | Breast cancer | China | median 51.5 | EMT | 2012.1–2013.12 | qRT-PCR | OS,DFS | reported directly | 7 |
| Zhaol, (2019) | 55 | Glioma | China | ≥50, 52.73% | SNAI1 | 2015.1–2017.7 | qRT-PCR | OS,DFS | survival curves | 8 |
| Zuo, (2019) | 67 | Breast cancer | China | mean52.5 (31–69) | RUNX2 | 2010.9–2012.3 | qRT-PCR | OS | survival curves | 8 |
3.2. Meta-analysis results
The main results of this meta-analysis are listed in Table 2. Our analysis showed that low miR-153 expression predicted poor survival in cancer patients (HR = 2.45, 95% CI = 1.66–3.63, P < .001) for heterogeneity (I2 = 67.0%, P = .001) (Fig. 2).
Table 2.
Main meta-analysis results of miR-153 expression in cancer patients.
| Heterogeneity | ||||||
| Analysis | Numbers of studies | HR(95%CI) | P value | χ2 | I2 (%) | P value |
| OS | 10 | 2.45 (1.66–3.63) | <.001 | 27.28 | 67.0 | .001 |
| HR obtain method | ||||||
| survival curves | 4 | 2.10 (1.56–2.84) | <.001 | 0.50 | 0.0 | .919 |
| reported directly | 6 | 2.67 (1.32–5.37) | .006 | 23.14 | 78.4 | <.001 |
| Cancer type | ||||||
| Digestive system tumor | 4 | 2.76 (1.73–4.41) | <.001 | 8.61 | 65.2 | .035 |
| Breast cancer | 2 | 4.01 (1.46–11.04) | .007 | 1.56 | 35.9 | .212 |
| Other cancer | 4 | 1.73 (0.74–4.04) | .206 | 12.82 | 76.6 | .005 |
| DFS | 4 | 1.67 (0.45–6.19) | .442 | 21.35 | 86.0 | <.001 |
| HR obtain method | ||||||
| reported directly | 3 | 1.57 (0.19–13.13) | .676 | 20.18 | 90.1 | <.001 |
| survival curves | 1 | 2.08 (1.13–3.83) | ||||
| Ethnicity | ||||||
| Asian | 3 | 2.69 (1.64–4.42) | <.001 | 21.35 | 1.0 | .364 |
| Caucasian | 1 | 0.21 (0.08–0.58) | ||||
| Cancer type | ||||||
| Digestive system tumor | 2 | 0.96 (0.05–18.80) | .976 | 16.17 | 93.8 | <.001 |
| Other cancer | 2 | 2.35 (1.34–4.12) | <.003 | 1.00 | 0.1 | .317 |
Figure 2.
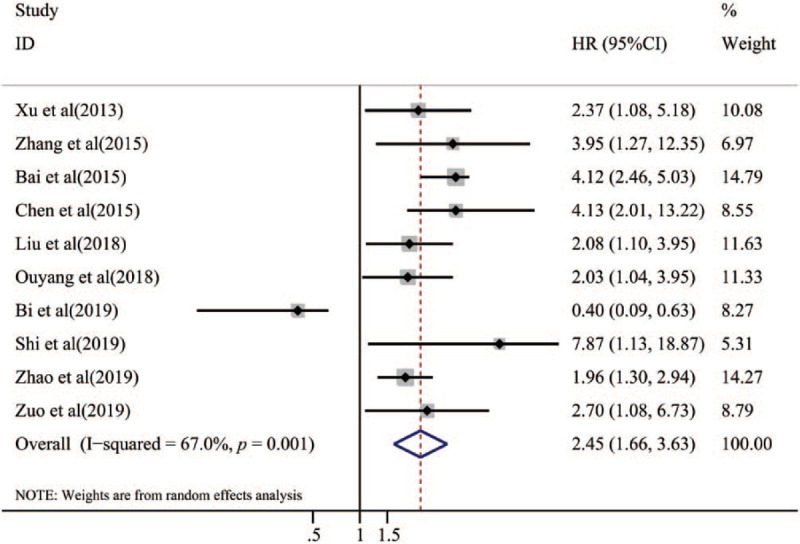
Forest plot of the relationship between miR-153 expression and overall survival.
To lessen the impact of heterogeneity, subgroup analyses were performed for HR obtain method, and cancer type (Table 2). Subgroup analysis based on the HR obtain method suggested that low expression of miR-153 predicted poor OS for both the reported directly from articles group (HR = 2.67, 95% CI: 1.32–5.37, P = .006) and, survival curves group (HR = 2.10, 95% CI: 1.56–2.84, P < .001).Furthermore, the subgroup analyses classified by cancer type validated that low expression of miR-153 was a unfavorable prognostic factor in patients with digestive system tumor (HR = 2.76, 95% CI: 1.73–4.41,P < .001) and breast cancer (HR = 4.01, 95% CI: 1.46–11.04,P = .007). Nevertheless, there was no significant association between miR-153 expression and DFS in patients with cancer (HR = 1.67, 95% CI = 0.45–6.19, P = .442; Fig. 3). The subgroup analysis of DFS was also performed according to HR obtain method, ethnicity and cancer type. The subgroup analyses classified by HR obtain method suggested that there was no significant association between miR-153 expression and DFS in patients with the reported directly from articles group (HR = 0.1.57, 95% CI: 0.19–13.13, P = .676). Subgroup analysis by ethnicity suggested that patients with low expression of miR-153 predicted poor prognosis in Asian (HR = 2.69, 95%CI: 1.64–4.42, P < .001). Furthermore, the subgroup analyses classified by cancer type validated that there was no significant association between low expression and DFS in patients with digestive system tumor (HR = 0.96, 95% CI: 0.05–18.80, P = .976).
Figure 3.
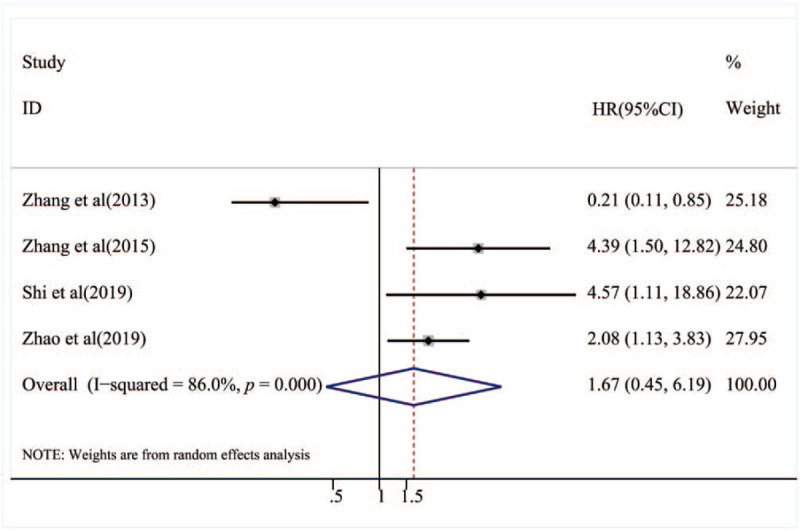
Forest plot of the relationship between miR-153 expression and disease-free survival.
3.3. Association between miR-153 expression and clinicopathological characteristics
Meta-analysis of the relationship between miR-153 expression and clinicopathological characteristics failed to show a significant association of low mir-153 expression with age (OR = 1.33, 95% CI: 0.90–2.56, P = .201), gender (OR = 1.08, 95% CI: 0.74–1.58, P = .687), or TNM stage (OR = 2.28, 95% CI: 0.92–5.63, P = .075) (Table 3). In contrast, low miR-153 expression was significantly related to poor tumor differentiation (poor vs well+moderate, OR = 2.41, 95%CI = 1.52–3.82, P < .001), earlier lymph node metastasis (present vs absent, OR = 2.19, 95% CI = 1.12–4.25, P = .021) and earlier distant metastasis (present vs absent,OR = 8.24, 95% CI = 2.93–23.21, P < .001) (Table 3).
Table 3.
Results of the association of miR-153 expression with clinicopathological features.
| Clinicopathological parameter | N | OR (95% CI) | P value | Heterogeneity test (Q, I2, P value) |
| Age (<60 vs ≥ 60 years) | 3 | 1.33 (0.86–2.06) | .201 | 0.25, 0, .884 |
| Gender (male vs female) | 6 | 1.08 (0.74–1.58) | .687 | 1.41, 0.0,.923 |
| Tumor differentiation (poor vs well+moderate) | 4 | 2.41 (1.52–3.82) | <.001 | 0.17, 0.0, .982 |
| Lymph node metastasis (Present vs Absent) | 9 | 2.19 (1.12–4.25) | .021 | 35.31, 77.3, <.001 |
| TNM stage (IV-III vs I-II) | 10 | 2.28 (0.92–5.63) | .075 | 73.62, 87.8, <.001 |
| Distant metastasis (Present vs Absent) | 3 | 8.24 (2.93–23.21) | <.001 | 1.08, 0.0, .582 |
3.4. Publication bias
In this meta-analysis, both Beggs test and Eggers test were used to check the potential publication bias. No publication bias was found in the meta-analysis with OS (P = .325) and DFS (P = .992) when tested by Beggs test(Figs. 4 and 5). In addition, no publication bias was found in the meta-analysis with OS (P = .664) and DFS(P = .497) when tested by Eggers test.
Figure 4.
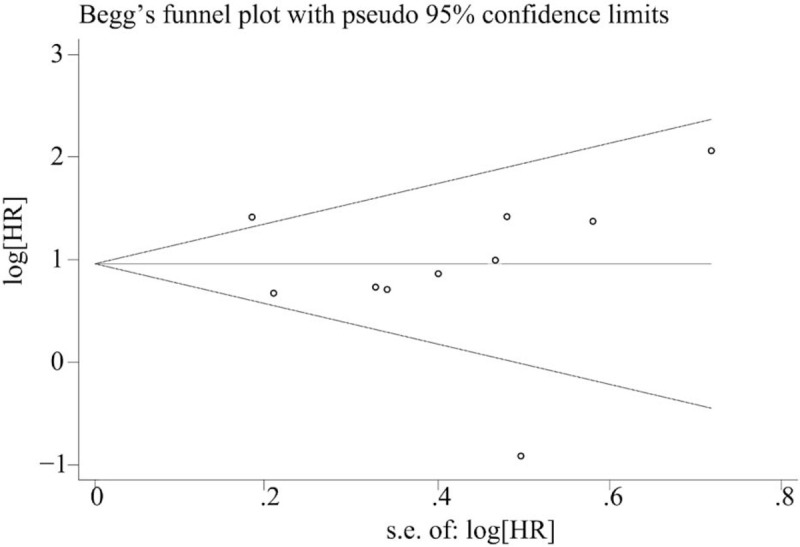
Funnel plot of publication bias on the relationship between miR-153 expression and overall survival.
Figure 5.
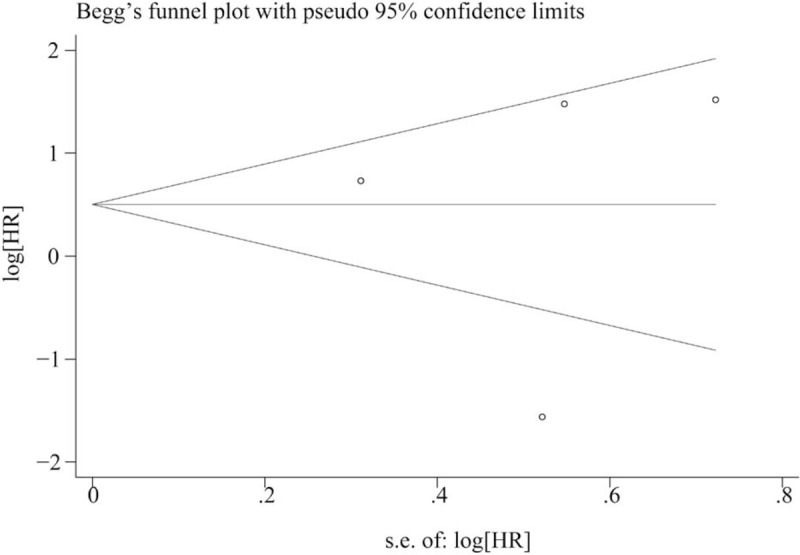
Funnel plot of publication bias on the relationship between miR-153 expression and disease-free survival.
4. Discussion
Cancer remains a severe threat to human health and cancer incidence shows an increasing trend recently.[1] Deregulation of miR-153 has recently been observed in several common human cancer, and previous studies have shown that miR-153 is involved in the regulation of various cancer cells through different pathways. For instance, Zhang et al [19] has reported that miR-153 was downregulated in gastric cancer, which promoted the migration and invasion of the SGC-7901 cells by suppressing SNAI1-induced EMT, and loss of miR-153 expression was associated with poor gastric cancer prognosis. Wang et al[29] also suggested that miR-153 suppressed Snail protein translation and subsequently decreased cell metastasis. In addition, miR-153 promotes the oncogenesis and development of hepatocellular carcinoma via activating the Wnt/β-catenin signal pathway.[30] Downregulation of miR-153 expression was associated with tumor progression and metastasis as well as poor prognosis in patients with breast cancer.[18]
However, Wu et al[31] reported that miR-153 was up-regulated in prostate cancer and miR-153 expression promoted tumor cell proliferation and migration by inhibiting the expression of PTEN. Zhang et al[28] showed that miR-153 upregulation promoted colorectal cancer invasiveness by inducing matrix metalloprotease enzyme 9 production and enhanced platinum-based chemotherapy resistance directly regulating FOXO3a. Several studies have identified a significant relationship between miR-153 expression levels and the prognosis of various malignant cancers. However, the results are not consistent. Therefore, we conducted this meta-analysis to explore the association between miR-153 expression and the survival prognosis and clinicopathological features in cancer patients.
The current study presented the first meta-analysis to comprehensively evaluate the relationship between miR-153 expression and prognosis and clinicopathological characteristics of tumors. A total of 11 eligible studies containing 933 patients were enrolled in this meta-analysis. The pooled results revealed that low miR-153 expression was significantly associated with poor OS in patients with cancers. These results give us some enlightenment that low miR-153 expression is predictive of poor cancer outcome and may represent a most promising target for anti-cancer therapy. Furthermore, the subsequent pooled results also demonstrated that low expression of miR-153 was associated with tumor differentiation, lymph node metastasis and distant metastasis.
This meta-analysis also has some limitations, and the results should be interpreted with caution. First, data presented in the current meta-analysis were not applicable to all countries worldwide, because most of them were derived from China. Second, part of the HR value was calculated using a survival curve, which may lead to some error. Third, because of the relatively small sample size, we were unable to aggregate results based on a single type of tumor.
In conclusion, our meta-analysis demonstrated that low expression of miR-153 was significantly correlated with poor OS and may serve as an effective predictive biomarker for tumor prognosis, especially for digestive system tumor and breast cancer. Future larger scale prospective and standard investigations should be conducted to confirm our results.
Author contributions
Conceptualization: Lixia Hu.
Data curation: Mengqin Huang, Chengfa Li.
Formal analysis: Mengqin Huang, Chengfa Li, Fanliang Kong.
Methodology: Lixia Hu, Yan Wu, Qianqian Yuan.
Project administration: Mengqin Huang, Lixia Hu.
Software: Qianqian Yuan, Lixia Hu.
Writing – original draft: Mengqin Huang, Yan Wu, Qianqian Yuan.
Writing – review & editing: Lixia Hu.
Footnotes
Abbreviations: CI = confidence interval, CNKI = China National Knowledge Infrastructure, DFS = disease-free survival, HR = hazard ratio, NOS = Newcastle–Ottawa Scale, OR = odds ratios, OS = overall survival, PRISMA = Preferred Reporting Items for Systematic Reviews and Meta-analysis.
How to cite this article: Huang M, Li C, Kong F, Wu Y, Yuan Q, Hu L. Prognostic and clinicopathological significance of MicroRNA-153 in human cancers: a meta-analysis. Medicine. 2020;99:46(e22833).
MH and CL contributed equally to this work and should be considered as co-first authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
CRC = colorectal cancer, DFS = disease-free survival, HR = hazard ratio, N = number of patients, NOS = Newcastle-Ottawa Scale, NR = not reported, OS = overall survival, OSCC = oral squamous cell carcinoma, PDAC = pancreatic ductal adenocarcinoma, qRT-PCR = quantitative real-time reverse transcription polymerase chain reaction.
CI = confidence interval, DFS = disease-free survival, HR = hazard ratio, OS = overall survival.
CI = confidence interval, N = numbers of studies, OR = odds ratio.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- [2].Ellis L, Woods LM, Esteve J, et al. Cancer incidence, survival and mortality: explaining the concepts. Int J Cancer 2014;135:1774–82. [DOI] [PubMed] [Google Scholar]
- [3].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene 2014;33:135–47. [DOI] [PubMed] [Google Scholar]
- [5].Visone R, Croce CM. MiRNAs and cancer. Am J Pathol 2009;174:1131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sempere LF, Freemantle S, Pitha-Rowe I, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 2004;5:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xu J, Liao X, Wong C. Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer 2010;126:1029–35. [DOI] [PubMed] [Google Scholar]
- [8].Xu Q, Sun Q, Zhang J, et al. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis 2013;34:539–49. [DOI] [PubMed] [Google Scholar]
- [9].Anaya-Ruiz M, Cebada J, Delgado-Lopez G, et al. Perez-Santos JL. miR-153 silencing induces apoptosis in the MDA-MB-231 breast cancer cell line. Asian Pac J Cancer Prev 2013;14:2983–6. [PubMed] [Google Scholar]
- [10].Kim TH, Kim YK, Kwon Y, et al. Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology 2010;57:734–43. [DOI] [PubMed] [Google Scholar]
- [11].Xu Q, Sun Q, Zhang JJ, et al. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis 2013;34:539–49. [DOI] [PubMed] [Google Scholar]
- [12].Bai ZH, Sun JL, Wang XB, et al. MicroRNA-153 is a prognostic marker and inhibits cell migration and invasion by targeting SNAI1 in human pancreatic ductal adenocarcinoma. Oncol Rep 2015;34:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [13].Chen WJ, Zhang EN, Zhong ZK, et al. MicroRNA-153 expression and prognosis in non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:8671–5. [PMC free article] [PubMed] [Google Scholar]
- [14].Liu L, Lai XJ, Yuan CJ, et al. Aberrant expression of miR-153 is associated with the poor prognosis of cervical cancer. Oncol Lett 2018;15:9183–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ouyang YL, Yuan WJ, Qiu SN. MicroRNA-153 functions as a tumor suppressor in gastric cancer via targeting Kruppel-like factor 5. Exp Ther Med 2018;16:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [16].Shi DL, Li Y, Fan LQ, et al. Upregulation Of miR-153 inhibits triple-negative breast cancer progression by targeting ZEB2-Mediated EMT and contributes to better prognosis. Onco Targets Ther 2019;12:9611–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhao W, Yin CY, Jiang J, et al. MicroRNA-153 suppresses cell invasion by targeting SNAI1 and predicts patient prognosis in glioma. Oncol Lett 2019;17:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zuo Z, Ye F, Liu Z, et al. MicroRNA-153 inhibits cell proliferation, migrationinvasion and epithelial-mesenchymal transition in breast cancer via direct targeting of RUNX2. Exp Therapeutic Med 2019;17:4693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang ZL, Sun JL, Bai ZH, et al. MicroRNA-153 acts as a prognostic marker in gastric cancer and its role in cell migration and invasion. Oncotargets Therapy 2015;8:357–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bi CW, Zhang GY, Bai Y, et al. Increased expression of miR-153 predicts poor prognosis for patients with prostate cancer. Medicine (Baltimore) 2019;98:e16705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 2009;339:b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815–34. [DOI] [PubMed] [Google Scholar]
- [23].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [25].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [27].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang L, Pickard K, Jenei V, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res 2013;73:6435–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Z, Liu C. MiR-153 regulates metastases of gastric cancer through Snail. Tumor Biology 2016;37:15509–15. [DOI] [PubMed] [Google Scholar]
- [30].Hua HW, Jiang F, Huang Q, et al. MicroRNA-153 promotes Wnt/β-catenin activation in hepatocellular carcinoma through suppression of WWOX. Oncotarget 2015;6:3840–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu Z, He B, He J, et al. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate 2013;73:596–604. [DOI] [PubMed] [Google Scholar]


