List of Abbreviations
- APEX1
apurinic/apyrimidinic endonuclease
- BER
base excision repair
- CI
confidence interval
- CNS
central nerve system
- eQTL
expression quantitative trait loci
- FADS1
fatty acid desaturase 1
- FADS2
fatty acid desaturase 2
- FEN1
flap endonuclease 1
- FPRP
false‐positive report probability
- GTEx
Genotype‐Tissue Expression
- GWAS
genome‐wide association study
- HWE
Hardy‐Weinberg equilibrium
- LIG3
DNA ligase III
- N/A
not applicable
- OGG1
8‐oxoguanine DNA glycosylase 1
- OR
odds ratio
- PARP1
poly(ADP)ribose polymerase 1
- SNP
single nucleotide polymorphism
- TMEM258
transmembrane protein 258
- XRCC1
x‐ray repair cross‐complementing group 1
Dear Editor,
Neuroblastoma is the most common non‐central nerve system (CNS) solid tumor in pediatrics [1]. Neuroblastoma accounts for approximately 8% of all pediatric cancers but disproportionally causes a high cancer mortality (15%) in children [2]. Pediatric patients with low‐risk neuroblastoma witness a 5‐year overall survival rate > 90%, whereas the 5‐year overall survival rate in high‐risk neuroblastoma pediatric patients is < 40% [3].
Genetic susceptibility to neuroblastoma is a promising area of research and needs to be fully investigated. For sporadic neuroblastoma, genome‐wide association studies (GWASs) have identified over a dozen causal genetic loci. Studies of candidate genes also reported a decent number of variants predisposing to neuroblastoma. However, the known genetic alternations still could not unveil the full genetic underpinnings of neuroblastoma.
The base excision repair (BER) pathway, one of the DNA repair systems, is responsible for repairing numerous oxidized and alkylated bases by recognizing and excising damaged bases [4]. Many core proteins are involved in the BER pathway, including poly(ADP)ribose polymerase 1 (PARP1), human 8‐oxoguanine DNA glycosylase (OGG1), flap endonuclease 1 (FEN1), apurinic/apyrimidinic endonuclease 1 (APEX1), DNA ligase III (LIG3), and x‐ray repair cross‐complementing group 1 (XRCC1). OGG1 is a bifunctional enzyme (DNA glycosylase and AP lyase) that incises at abasic sites via an AP lyase activity, leaving a single‐strand DNA break intermediate. APEX1 initiates the repair of abasic sites in DNA by cleaving the phosphodiester backbone 5′ to an AP site, creating a nick in the DNA backbone. FEN1 participates in the penultimate steps of Okazaki fragment maturation and 5′‐flap removal during long‐patch BER. LIG3 catalyzes the last stage of BER by sealing the gap. XRCC1 and PARP1 serve as the scaffold protein. Intensive evidence suggests that aberrant BER pathway proteins result in a variety of diseases, especially cancers [4]. Single nucleotide polymorphisms (SNPs) of the BER pathway genes are associated with the risk of various cancer types. Functional analysis revealed that SNPs in the BER pathway genes may modify the kinetics of BER proteins and the DNA repair capacity of the BER system, ultimately affecting carcinogenesis [4]. However, evidence regarding the role of BER pathway gene SNPs in the risk of neuroblastoma waits to be added. To identify more neuroblastoma susceptibility variations in the BER pathway genes, we performed a case‐control study in children at three center hospitals in East China.
This study was conducted in Children's Hospital of Nanjing Medical University (Nanjing, Jiangsu), Anhui Provincial Children's Hospital (Hefei, Anhui), and Yuying Children's Hospital of Wenzhou Medical University (Wenzhou, Zhejiang) in East China. A total of 313 neuroblastoma pediatric patients and 762 cancer‐free children were recruited in this study. The characteristics of the study subjects are summarized in Supplementary Table S1. Age (P = 0.823) and gender (P = 0.610) were distributed equivalently between the two groups. The study design and participant recruitment were described in our previous work [5].
We successfully genotyped 20 SNPs from 6 BER pathway genes in 313 neuroblastoma pediatric patients and 762 control children (Table 1). Specifically, 3 PARP1, 3 OGG1, 2 FEN1, 3 APEX1, 3 LIG3, and 6 XRCC1 SNPs were genotyped. The genotypic distributions of all candidate SNPs were in Hardy‐Weinberg equilibrium (P ≥ 0.05) in the controls. The rs174538 of the FEN1 gene was associated with decreased neuroblastoma risk under the dominant model (adjusted odd ratio [OR] = 0.71, 95% confidence interval [CI] = 0.54‐0.93, P = 0.012). However, no significant associations with neuroblastoma risk were found for the remaining SNPs in the single‐locus analysis (all P ≥ 0.05; Supplementary Figure S1).
TABLE 1.
Association between the SNPs in BER pathway genes and neuroblastoma susceptibility in eastern Chinese children
| Allele | Neuroblastoma patients | Cancer‐free controls | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | A | B | AA | AB | BB | Undetectable | AA | AB | BB | Undetectable | Adjusted OR (95% CI) a | P value a | Adjusted OR (95% CI) b | P value b | Adjusted OR (95% CI) c | P value c | HWE |
| PARP1 | rs1136410 | A | G | 102 | 151 | 59 | 1 | 240 | 387 | 135 | 0 | 1.03 (0.70‐1.51) | 0.893 | 0.95 (0.72‐1.26) | 0.714 | 1.08 (0.77‐1.52) | 0.660 | 0.328 |
| rs2666428 | T | C | 198 | 97 | 17 | 1 | 461 | 262 | 39 | 0 | 1.01 (0.56‐1.83) | 0.982 | 0.88 (0.67‐1.15) | 0.343 | 1.06 (0.59‐1.91) | 0.840 | 0.822 | |
| rs8679 | A | G | 269 | 40 | 3 | 1 | 651 | 107 | 4 | 0 | 1.77 (0.39‐7.96) | 0.459 | 0.92 (0.62‐1.35) | 0.672 | 1.79 (0.40‐8.08) | 0.447 | 0.860 | |
| OGG1 | ||||||||||||||||||
| rs1052133 | G | C | 128 | 139 | 45 | 1 | 284 | 368 | 110 | 0 | 0.91 (0.60‐1.36) | 0.634 | 0.85 (0.65‐1.11) | 0.241 | 1.00 (0.69‐1.46) | 1.000 | 0.600 | |
| rs159153 | T | C | 260 | 50 | 2 | 1 | 614 | 142 | 6 | 0 | 0.79 (0.16‐3.94) | 0.770 | 0.84 (0.59‐1.19) | 0.330 | 0.81 (0.16‐4.06) | 0.799 | 0.478 | |
| rs293795 | A | G | 275 | 36 | 1 | 1 | 691 | 69 | 2 | 0 | 1.17 (0.11‐13.01) | 0.897 | 1.31 (0.86‐1.99) | 0.216 | 1.14 (0.10‐12.63) | 0.915 | 0.842 | |
| FEN1 | ||||||||||||||||||
| rs174538 | A | G | 142 | 131 | 40 | 0 | 281 | 362 | 119 | 0 | 0.67 (0.45‐1.02) | 0.059 | 0.71 (0.54‐0.93) | 0.012 | 0.80 (0.54‐1.17) | 0.248 | 0.893 | |
| rs4246215 | T | G | 126 | 140 | 47 | 0 | 278 | 365 | 119 | 0 | 0.89 (0.60‐1.33) | 0.568 | 0.87 (0.66‐1.13) | 0.294 | 0.97 (0.67‐1.40) | 0.868 | 0.964 | |
| APEX1 | ||||||||||||||||||
| rs1130409 | T | G | 109 | 143 | 61 | 0 | 271 | 346 | 145 | 0 | 1.03 (0.71‐1.50) | 0.870 | 1.02 (0.78‐1.35) | 0.874 | 1.02 (0.73‐1.43) | 0.904 | 0.067 | |
| rs1760944 | T | G | 101 | 163 | 49 | 0 | 250 | 372 | 140 | 0 | 0.87 (0.58‐1.29) | 0.486 | 1.02 (0.77‐1.35) | 0.899 | 0.83 (0.58‐1.19) | 0.307 | 0.937 | |
| rs3136817 | T | C | 250 | 60 | 3 | 0 | 630 | 124 | 8 | 0 | 0.98 (0.26‐3.74) | 0.977 | 1.21 (0.86‐1.69) | 0.273 | 0.95 (0.25‐3.60) | 0.935 | 0.496 | |
| LIG3 | ||||||||||||||||||
| rs1052536 | C | T | 156 | 136 | 21 | 0 | 381 | 316 | 65 | 0 | 0.79 (0.46‐1.33) | 0.369 | 1.01 (0.77‐1.31) | 0.963 | 0.77 (0.46‐1.28) | 0.310 | 0.964 | |
| rs3744356 | C | T | 307 | 6 | 0 | 0 | 737 | 25 | 0 | 0 | N/A | N/A | 0.59 (0.24‐1.45) | 0.251 | N/A | N/A | 0.645 | |
| rs4796030 | A | C | 101 | 147 | 65 | 0 | 255 | 371 | 136 | 0 | 1.19 (0.82‐1.73) | 0.367 | 1.05 (0.79‐1.39) | 0.750 | 1.19 (0.86‐1.66) | 0.297 | 0.958 | |
| XRCC1 | ||||||||||||||||||
| rs1799782 | G | A | 152 | 136 | 25 | 0 | 347 | 336 | 79 | 0 | 0.73 (0.45‐1.20) | 0.216 | 0.90 (0.69‐1.17) | 0.424 | 0.76 (0.47‐1.21) | 0.249 | 0.860 | |
| rs25487 | C | T | 177 | 115 | 21 | 0 | 420 | 302 | 40 | 0 | 1.21 (0.69‐2.11) | 0.510 | 0.93 (0.71‐1.21) | 0.574 | 1.27 (0.73‐2.19) | 0.397 | 0.129 | |
| rs25489 | C | T | 250 | 60 | 3 | 0 | 603 | 153 | 6 | 0 | 1.24 (0.31‐5.00) | 0.765 | 0.94 (0.68‐1.31) | 0.723 | 1.26 (0.31‐5.06) | 0.750 | 0.271 | |
| rs2682585 | G | A | 245 | 61 | 7 | 0 | 600 | 155 | 7 | 0 | 2.51 (0.87‐7.25) | 0.089 | 1.03 (0.75‐1.42) | 0.869 | 2.53 (0.88‐7.30) | 0.085 | 0.384 | |
| rs3810378 | G | C | 168 | 122 | 23 | 0 | 398 | 320 | 44 | 0 | 1.22 (0.71‐2.08) | 0.476 | 0.94 (0.72‐1.22) | 0.618 | 1.28 (0.76‐2.15) | 0.363 | 0.050 | |
| rs915927 | T | C | 239 | 73 | 1 | 0 | 613 | 142 | 7 | 0 | 0.37 (0.05‐3.06) | 0.360 | 1.28 (0.93‐1.76) | 0.128 | 0.35 (0.04‐2.89) | 0.332 | 0.698 | |
Adjusted for age and gender for homozygous model (BB vs. AA).
Adjusted for age and gender for dominant model (BB/AB vs. AA).
Adjusted for age and gender for recessive model (AA/AB vs. BB).
Abbreviations: BER, base excision repair; SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; HWE, Hardy‐Weinberg equilibrium; N/A, not applicable; PARP1, poly(ADP)ribose polymerase 1; OGG1, human 8‐oxoguanine DNA glycosylase; FEN1, flap endonuclease 1; APEX1, apurinic/apyrimidinic endonuclease; LIG3, DNA ligase III; XRCC1, x‐ray repair cross‐complementing group 1.
We conducted the stratified analyses (Supplementary Table S2) to eliminate potential influences of FEN1 genotypes on neuroblastoma susceptibility by adjusting confounding factors (age, gender, and site of tumor origin). The protective role of rs174538 AG/GG in decreasing neuroblastoma risk was found in subgroups of age ≤18 months (adjusted OR = 0.60, 95% CI = 0.40‐0.89, P = 0.011), females (adjusted OR = 0.59, 95% CI = 0.40‐0.87, P = 0.009), and tumors arising from the mediastinum (adjusted OR = 0.53, 95% CI = 0.35‐0.81, P = 0.003). Combined analysis stated that the 2 protective genotypes (rs174538 AG/GG and rs4246215 TG/GG genotypes) also decreased neuroblastoma risk in the following subgroups: age ≤ 18 months (adjusted OR = 0.62, 95% CI = 0.42‐0.93, P = 0.019), females (adjusted OR = 0.61, 95% CI = 0.41‐0.91, P = 0.015), and tumors originated from the mediastinum (adjusted OR = 0.54, 95% CI = 0.36‐0.83, P = 0.005).
We carried out false‐positive report probability (FPRP) analysis to validate significant associations (Supplementary Table S3). The threshold for FPRP was preset as 0.2. At the prior probability level of 0.1, significant associations with FEN1 rs174538 A > G (GG/AG vs. AA) remained noteworthy in all subjects (FPRP = 0.121) as well as in the subgroups of females (FPRP = 0.185) and tumors originating from the mediastinum (FPRP = 0.160). In the combined analysis, significant findings for 2 vs. 0‐1 protective genotypes (FPRP = 0.166) and its subgroup tumors originated from the mediastinum (FPRP = 0.183) could be called noteworthy.
We further explored the biological effects of FEN1 rs174538 A > G on the neighboring gene expression by using released data from Genotype‐Tissue Expression (GTEx) Portal (https://www.gtexportal.org/). We observed that rs174538 A allele was significantly associated with increased mRNA expression levels of fatty acid desaturase 2 (FADS2) and transmembrane protein 258 (TMEM258) in the whole blood, nerve‐tibial, and cell‐cultured fibroblasts (Figure 1A). The rs174538 A allele was also associated with increased expression of fatty acid desaturase 1 (FADS1) mRNA in the whole blood, but with decreased expression of FADS1 mRNA in the nerve‐tibial (Figure 1B).
FIGURE 1.
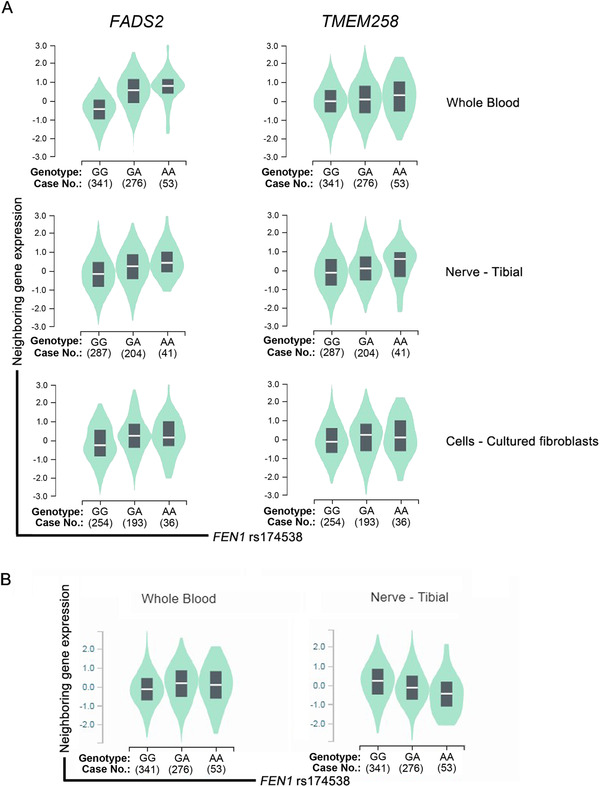
eQTL analysis of the neuroblastoma risk factor FEN1 rs174538 A > G. A. FADS2 and TMEM258 levels in the whole blood, nerve‐tibial, and cell‐cultured fibroblasts; B. FADS1 level in the whole blood and nerve‐tibial.
Abbreviations: eQTL, expression quantitative loci; FEN1, flap endonuclease 1; FADS2, fatty acid desaturase 2; TMEM258, transmembrane protein 258
The implication of the BER pathway gene SNPs in cancer susceptibility has been highly documented. Plenty of SNPs within the BER pathway genes were found to predispose to various types of cancer. Our group previously carried out a study on BER gene polymorphisms and Wilms tumor susceptibility [6]. Significant associations with Wilms tumor susceptibility were shown for the OGG1 rs1052133, FEN1 rs174538, and FEN1 rs4246215 polymorphisms. Regarding the association of the BER pathway gene SNPs with neuroblastoma risk, only 3 studies were available by far; and all of them were performed by our research group. In these studies, we found that, none of the studied APEX1 polymorphisms were associated with neuroblastoma risk [5]. Such a negative association was also observed between neuroblastoma risk and polymorphisms in the OGG1 [7] and LIG3 genes [8]. However, all these studies were conducted to analyze a single gene in the BER pathway, and the results need to be validated in another independent study. Thus, here we attempted to validate the previous studies by adopting a systematical analysis of potentially functional SNPs in 6 core genes in the BER pathway. In the current study, no significant relationships were detected between neuroblastoma risk and the SNPs in PARP1, OGG1, APEX1, LIG3, and XRCC1 genes. Such results strengthen the previous findings that these variations may be too weak to impact neuroblastoma risk. To be noted, significant conferring roles of the same BER SNPs to the risk of other cancer types have been detected, such as PARP1 rs1136410 and thyroid cancer [9], OGG1 rs1052133 and Wilms tumor [6], FEN1 rs4246215 and Wilms tumor [6], APEX1 rs1130409 and renal cell carcinoma [10], LIG3 rs1052536 and lung cancer [11]. The different roles of these SNPs in specific cancer types indicated that specific cancer types should be set before interpreting the role of SNPs.
Excitedly, we demonstrated that the rs174538 of the FEN1 gene could protect from neuroblastoma. FEN1 is a structure‐specific nuclease involved in the removal of 5′‐flap during long‐patch BER and the maturation of Okazaki fragments in DNA replication. Moreover, FEN1 is also characterized as a 5′ exonuclease and a gap‐dependent endonuclease, which mediates apoptotic DNA degradation during apoptosis. The FEN1 gene is mapped to chromosome 11 (11q12.2). Yang et al. [12] identified that the rs174538 A allele of the FEN1 gene decreased risk for lung cancer by decreasing FEN1 expression. Moreover, they detected that coke oven workers who carried the AA genotype have significantly lower DNA damage level than those with GG or GA genotypes. In a meta‐analysis conducted for the overall cancer, the results suggested that the subjects with FEN1 rs174538 A allele have a decreased susceptibility to cancer in Chinese populations [13]. We further performed online expression quantitative trait loci (eQTL) analysis to interpret the possible mechanism of how rs174538 impacts neuroblastoma risk. eQTL evidence suggested that the A allele in rs174538 was significantly associated with the increased mRNA expression levels of FADS2 and TMEM258. Further functional experiments conducted in neuroblastoma cells are needed to show how the FEN1 rs174538 A allele can be associated with altered expressions of these genes. FADS2 was found to function as a potential oncogene in some types of cancer [14]. TMEM258 is a central mediator of endoplasmic reticulum quality control and intestinal homeostasis, yet its role in cancer remains unknown [15]. The exact relationship of FADS2 and TMEM258 with neuroblastoma risk waits to be elucidated. Taken together, the significant role of rs174538 A allele in cancer deserves more attention for further exploration. Although at the preliminary stage, our findings represent a novel mechanism by which rs174538 may modulate the expression of multiple nearby genes, thereby impacting the risk of neuroblastoma.
Our study has several limitations. First, the sample sizes were small in some stratification analyses. Second, the number of analyzed SNPs was limited. Another limitation was the lack of incorporating analysis on environment factors and genetic‐environmental factors. The fourth limitation was that the current study only focused on the subjects of the Han population. Replication of these findings in additional individuals of non‐Chinese descent should be helpful to validate our findings.
In conclusion, we showed a robust association of genetic variants in the FEN1 gene with neuroblastoma risk in a relatively large sample size of pediatric patients in East China. Intensive future research is warranted to extend the role of FEN1 gene loci in neuroblastoma susceptibility in individuals of non‐Chinese ancestries.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, and was approved by the Ethics Committees of Children's Hospital of Nanjing Medical University, Anhui Provincial Children's Hospital, and the Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University. Each participant signed an informed consent before participating to this study.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
All data generated or analyzed during this study are included in this published article and its additional files.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDINGS
This work was supported by the grants from Natural Science Foundation of Guangdong Province (2019A1515010360) and Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease (2019B030301004).
AUTHORS’ CONTRIBUTIONS
Z.Z., J.Z., H.L., J.H., and Y.W. designed the study, performed the experiments and wrote the manuscript. C.Z., Y.F., H.Z., H.W., and Y.W. collected the clinical samples and information. Z.Z. and J.H. analyzed the data and prepared all the tables and figures. Z.Z., J.H., and Y.W. coordinated the study. All authors reviewed and approved the final manuscript.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
Not applicable.
Contributor Information
Yizhen Wang, Email: hejing198374@gmail.com.
Jing He, Email: ywff018@163.com.
REFERENCES
- 1. Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. 2019;39(1):22 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362(23):2202‐11. 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Newman EA, Abdessalam S, Aldrink JH, Austin M, Heaton TE, Bruny J et al. Update on neuroblastoma. J Pediatr Surg. 2019;54(3):383‐9. 10.1016/j.jpedsurg.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 4. Tudek B. Base excision repair modulation as a risk factor for human cancers. Mol Aspects Med. 2007;28(3‐4):258‐75. 10.1016/j.mam.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 5. Liu J, Jia W, Hua RX, Zhu J, Zhang J, Yang T, et al. APEX1 Polymorphisms and Neuroblastoma Risk in Chinese Children: A Three‐Center Case‐Control Study. Oxidative Medicine and Cellular Longevity. 2019;2019:5736175 10.1155/2019/5736175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu J, Jia W, Wu C, Fu W, Xia H, Liu G, et al. Base Excision Repair Gene Polymorphisms and Wilms Tumor Susceptibility. EBioMedicine. 2018;33:88‐93. 10.1016/j.ebiom.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang YZ, Zhuo ZJ, Fang Y, Li L, Zhang J, He J, et al. Functional Polymorphisms in hOGG1 Gene and Neuroblastoma Risk in Chinese Children. J Cancer. 2018;9(23):4521‐6. 10.7150/jca.27983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng J, Wei K, Xin Y, Zhao P, Zhang J, Jia W, et al. Lack of associations between LIG3 gene polymorphisms and neuroblastoma susceptibility in Chinese children. J Cancer. 2019;10(23):5722‐6. 10.7150/jca.33605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bashir K, Sarwar R, Saeed S, Mahjabeen I, Kayani MA. Interaction among susceptibility genotypes of PARP1 SNPs in thyroid carcinoma. PLoS One. 2018;13(9):e0199007 10.1371/journal.pone.0199007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao Q, Qin C, Meng X, Ju X, Ding Q, Wang M, et al. Genetic polymorphisms in APE1 are associated with renal cell carcinoma risk in a Chinese population. Mol Carcinog. 2011;50(11):863‐70. 10.1002/mc.20791. [DOI] [PubMed] [Google Scholar]
- 11. Landi S, Gemignani F, Canzian F, Gaborieau V, Barale R, Landi D, et al. DNA repair and cell cycle control genes and the risk of young‐onset lung cancer. Cancer Res. 2006;66(22):11062‐9. 10.1158/0008-5472.CAN-06-1039. [DOI] [PubMed] [Google Scholar]
- 12. Yang M, Guo H, Wu C, He Y, Yu D, Zhou L, et al. Functional FEN1 polymorphisms are associated with DNA damage levels and lung cancer risk. Hum Mutat. 2009;30(9):1320‐8. 10.1002/humu.21060. [DOI] [PubMed] [Google Scholar]
- 13. Gao XR, Zhang SL, Yang YF, Han GR. FEN1 ‐69G>A and 4150G>T polymorphisms and cancer risk in Chinese population. Sci Rep. 2014;4:6183 10.1038/srep06183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vriens K, Christen S, Parik S, Broekaert D, Yoshinaga K, Talebi A, et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566(7744):403‐6. 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graham DB, Lefkovith A, Deelen P, de Klein N, Varma M, Boroughs A, et al. TMEM258 Is a Component of the Oligosaccharyltransferase Complex Controlling ER Stress and Intestinal Inflammation. Cell Rep. 2016;17(11):2955‐65. 10.1016/j.celrep.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files.


