Abstract
The search for effective coronavirus disease (COVID-19) therapy has attracted a great deal of scientific interest due to its unprecedented health care system overload worldwide. We have carried out a study to investigate the in silico effects of the most abundant pomegranate peel extract constituents on the multi-step process of serious acute respiratory syndrome coronavirus 2 (SARS-CoV-2) internalization in the host cells. Binding affinities and interactions of ellagic acid, gallic acid, punicalagin and punicalin were studied on four selected protein targets with a significant and confirmed role in the process of the entry of virus into a host cell. The protein targets used in this study were: SARS-CoV-2 spike glycoprotein, angiotensin-converting enzyme 2, furin and transmembrane serine protease 2. The results showed that the constituents of pomegranate peel extracts, namely punicalagin and punicalin had very promising potential for significant interactions with the selected protein targets and were therefore deemed good candidates for further in vitro and in vivo evaluation.
Keywords: COVID-19, SARS-CoV-2, Molecular docking, Pomegranate peel extract, Punicalagin, Punicalin
Introduction
Coronaviruses are members of the Coronaviridae family and can cause respiratory tract infections in humans which are usually mild, although some beta coronaviruses, including the Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV) and novel coronavirus SARS-CoV-2 may induce severe symptoms [1, 2]. Differently from the former two, the SARS-CoV-2 virus has spread rapidly with a high human-to-human transmission rate, affecting the respiratory system and causing serious clinical impairment with extremely high mortality worldwide, putting the World Health Organization in a position to declare pandemic on March 11, 2020 [3]. The structure of SARS-CoV-2 virus is well determined and the virus itself is composed of four main structural proteins such as spike (S) glycoprotein, small envelope (E) glycoprotein, membrane (M) glycoprotein and nucleocapsid (N) protein, as well as several accessory proteins [4]. The S glycoprotein, which is incorporated into the viral envelope, is crucial for virus binding to a specific receptor and fusion with target host cells. It has already been well established that SARS-CoV-2 virus enters the cells of the respiratory system by interacting with angiotensin-converting enzyme 2 (ACE2) as its specific receptor [5]. The ACE2 receptors are highly expressed throughout the respiratory tract cells such as nasal epithelial cells, goblet/secretory cells and type II pneumocytes of the lung [6]. The virus-receptor attachment occurs at the binding domain presented at 331 and 524 residues of the virus S glycoprotein [7]. The S glycoprotein comprises of two functional subunits known as S1, which is an amino (N)-terminal subunit and S2, a carboxyl (C)-terminal subunit. The surface (S1) subunit binds to the ACE2 receptor, while the transmembrane subunit (S2) interacts with the host cell membrane to facilitate the fusion of the viral cell membrane with the host cell membrane, and thus initiate the process of endocytosis. The process of membrane fusion depends on host cell proteases that are responsible for S glycoprotein cleavage at S1/S2 and S2′ site [8, 9]. One of these is the enzyme transmembrane serine protease 2 (TMPRSS2) which is, together with ACE2, required for SARS-CoV-2 cell entry [10]. Both of these enzymes are highly expressed on the surface of the epithelial cells of the respiratory system, and after the fusion the TMPRSS2 will cleave the S glycoprotein. However, it has become evident that the S glycoprotein is activated by a two-step process requiring another enzyme, furin. Furin is a membrane-bound member of the proprotein convertases family which processes latent precursor proteins into their biologically active products. During the viral entry into the host cell, the SARS-CoV-2 S- glycoprotein is first pre-cleaved by furin at the S1/S2 site and then subsequently cleaved by the TMPRS2 enzyme at S2′ site [11, 12]. Some recent studies have shown that cleavage of the S glycoprotein by furin might increase the binding affinity of the SARS-CoV-2 virus to ACE2 as its receptor [13]. It is clear that in addition to the ACE2, the furin/TMPRSS2 coexistence is a crucial human host factor for the SARS-CoV-2 infection, and these proteases are potential therapeutic targets for the coronavirus infection. Different substances, like camostat mesylate as a TMPRSS2 inhibitor, as well as several furin inhibitors, could potentially attenuate or block the virus entry process [14].
However, there are currently no medications proven to be effective for the prevention or treatment of the SARS-CoV-2 infection, and researchers are desperately trying to discover molecules with efficient antiviral activity. Including natural products from herbal medicine [15]. One of those is pomegranate (Punica granatum L., Punicaceae family) which is a well-known fruit consumed worldwide. Pomegranate has been traditionally used in the folk medicine due to its beneficial health properties and treatment of different chronic diseases like diabetes type 2 [16, 17], atherosclerosis [18, 19], cardiovascular diseases [20], inflammatory diseases [21] or cancer [22]. Pomegranate peel extracts are particularly rich with phytobiotics such as hydrolysable tannins (ellagitannin, punicalagin, punicalin, gallic and ellagic acid), flavonoids, anthocyanins and other phenols. These polyphenols possess a wide range of biologic properties including anti-inflammatory, antioxidant, hypoglycemic, lipid-lowering, antihypertensive or antimicrobial effects [17, 23, 24].
Several studies have reported the inhibitory effect of pomegranate extracts on the influenza virus, herpes virus, poxviruses and human immunodeficiency virus [25, 26]. In a computational study the pomegranate ligand molecules, namely the punicalagin, punicalin and ellagic acid, showed a strong interaction with the catalytic and substrate binding residues of the hepatitis C virus (HCV) NS3/4A protease. In an additional experiment, these polyphenols specifically blocked the NS3/4A protease in vitro [27]. Recent in vitro studies have shown that the antiviral effect of pomegranate peel extracts (PoPEx) on influenza virus is associated with the inhibition of viral absorption and RNA transcription [28, 29].
Our review of the available literature has shown that a possible anti-SARS-CoV-2 properties of PoPEx have not been studied so far. Based on the existing data, we hypothesized that polyphenols derived from PoPEx have potential interactions with SARS-CoV-2 S-glycoprotein, as well as ACE2, furin and TMPRSS2 enzymes.
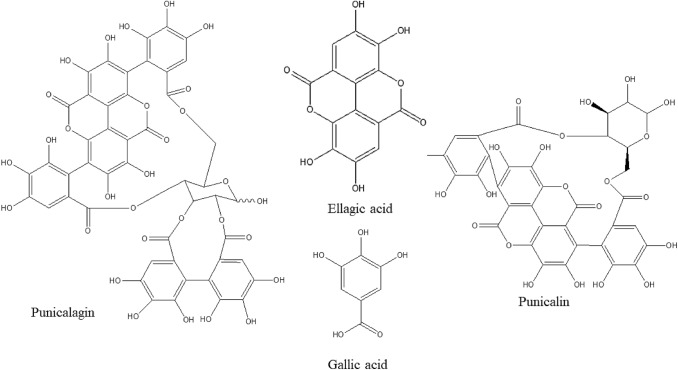
Therefore, the aim of this study was to explore the anti SARS-CoV-2 properties of four major ellagitannin members derived from PoPEx i.e. punicalagin, punicalin, ellagic acid and gallic acid using in silico tools (Fig. 1).
Fig. 1.
Chemical structure of the most abundant components of the pomegranate peel extract
Materials and methods
In silico study
Protein active sites, protein-ligand interactions, binding affinities of the selected protein targets with umifenovir, lopinavir, camostat, and selected PoPEx constituents (punicalagin, punicalin, ellagic acid and gallic acid) were determined through the following protocol steps.
Proteins
In the present study, the S glycoprotein, ACE2, furin and TMPRSS2 were selected as known key proteins for SARS-CoV-2 virus internalization and as relevant therapy targets. The crystal structures of furin and ACE2 in a complex with the S glycoprotein used for docking studies were downloaded from the Protein Data Bank (PDB; www.pdb.org, PDB ID:5MIM and www.pdb.org, PDB ID:6LZG, respectively). The protein structures of TMPRSS2 built by homology modelling were also used for a molecular docking analysis. All protein structures were prepared for the docking analysis using Yasara Structure (http://www.yasara.org/) [30, 31]. This procedure included deletion of solvents from the PDB files, adding hydrogens and charges to the structure, and process of energy minimization.
Protein active site prediction
Selected protein targets were analysed for druggable regions using the online DoGSiteScorer tool (https://proteins.plus/#dogsite) provided by the University of Hamburg – Center for Bioinformatics [32, 33]. Pocket sites with a favourable volume, surface area and drug score were further evaluated regarding constitutive amino acid residues. The grid box cube which included all pocket site amino acid residues was then used for the docking analysis.
Ligands
The 3D structures of molecules of ellagic acid, gallic acid, umifenovir, lopinavir and camostat were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) whereas Chemspider (http://www.chemspider.com/) was used to download the punicalagin and punicalin molecules. All ligands were further prepared for the docking study using the biocomputing software Yasara Structure (v. 20.4.24) energy minimization process.
Molecular docking
By predicting druggable active sites, working cavities were created with the grid box generated around amino acid descriptors within a distance of 1 Å. Then docking procedure was conducted through the Yasara Structure software based on the AutoDock Vina algorithm and AMBER03 force field [34]. Output files of the best binding affinity docking positions were further analysed with visualization software (Discovery Studio v.20.1.0.19295).
The results obtained employing the DogSiteScorer online tool were expressed through a tabular presentation of shape, size, drug score and apolar amino acid ratio values. The highest drug score value suggested an energetically favourable site for interaction with ligands. Furthermore, the selected pocket sites were additionally characterized with enclosure parameters and constitutive amino acid residues. Based on these results, input data for docking calculation were defined. The molecular docking results were outlined through tabular free binding energy values and 2D illustrations of interactions for the most stable protein-ligand complexes.
Results
Protein active site prediction
The DoGSiteScorer tool uses a Difference of Gaussian filter to detect potential binding pockets - entirely based on the 3D structure of the protein. Energetically favourable pockets were uncovered for all analysed targets. The most druggable pockets for the S glycoprotein (Drug score: 0.79), ACE2 (Drug score: 0.83), furin (Drug score: 0.73) and TMPRSS2 (Drug score: 0.84) were selected for performing further molecular docking studies (Table 1). A high apolar amino acid ratio implies a promising drug target and 0.63 as the highest value was detected for the S glycoprotein. The other selected pockets apolar amino acid ratio values for ACE2, furin and TMPRSS2 were 0.38, 0.26 and 0.40, respectively.
Table 1.
DoGSiteScorer descriptors results for predicted pockets of S glycoprotein, ACE2, furin and TMPRSS2 proteins
| Proteins | Volume | Surface | Drug score | Simple score | Apolar amino acid ratio | |
|---|---|---|---|---|---|---|
| S glycoprotein | P_0 | 404.61 | 682.33 | 0.79 | 0.31 | 0.63 |
| P_1 | 281.86 | 515.54 | 0.5 | 0.14 | 0.29 | |
| P_2 | 215.87 | 460.4 | 0.59 | 0.01 | 0.45 | |
| ACE2 | P_0 | 876.74 | 1068.47 | 0.83 | 0.56 | 0.38 |
| P_1 | 639.94 | 896.33 | 0.73 | 0.42 | 0.35 | |
| Furin | P_0 | 545.34 | 635.62 | 0.73 | 0.29 | 0.26 |
| P_1 | 541.25 | 943.85 | 0.68 | 0.37 | 0.37 | |
| TMPRSS2 | P_0 | 525.12 | 813.49 | 0.84 | 0.27 | 0.40 |
| P_1 | 350.59 | 628.49 | 0.69 | 0.24 | 0.52 |
ACE2- angiotensin converting enzyme 2; TMPRSS2 – transmembrane serine protease 2
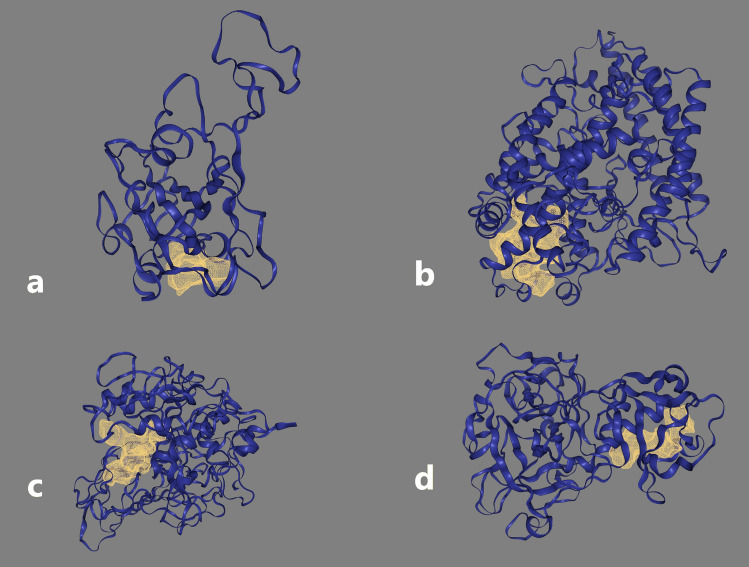
Positions of the selected pocket sites for different protein targets of interest are shown in Fig. 2.
Fig. 2.
Selected active sites of analysed protein targets; (a) S glycoprotein, (b) ACE2, (c) Furin and (d) TMPRSS2
Amino acid residues that define the predicted pocket site and are involved in interactions with potential ligands are shown in Table 2. A pockets comparison revealed that the number of residues of the integral amino acid varied from 19 to 43. The highest number of amino acids was found at furin predicted active site.
Table 2.
Amino acid descriptors of selected active pockets for S glycoprotein, ACE2, furin and TMPRSS2 proteins
| S glycoprotein | ACE2 | Furin | TMPRSS2 |
|---|---|---|---|
| Leu_335_E | Tyr_279_A | Ile_151_A | Val_76_A |
| Cys_336_E | Val_283_A | Leu_152_A | Cys_77_A |
| Pro_337_E | Phe_285_A | Asp_153_A | Gln_78_A |
| Phe_338_E | Gly_286_A | Asp_154_A | Asp_79_A |
| Gly_339_E | Gln_287_A | Asp_191_A | Trp_81_A |
| Phe_342_E | Lys_288_A | Asn_192_A | Asn_82_A |
| Asn_343_E | Pro_289_A | His_194_A | Asn_84_A |
| Ala_344_E | Asn_290_A | Arg_197_A | Tyr_85_A |
| Val_362_E | Ile_291_A | Cys_198_A | Ala_88_A |
| Ala_363_E | Met_366_A | Leu_227_A | Arg_91_A |
| Asp_364_E | Leu_370_A | Asp_228_A | Asp_92_A |
| Val_367_E | Glu_406_A | Ser_251_A | Phe_112_A_B |
| Leu_368_E | Ser_409_A | Ala_252_A | Met_119_A |
| Ser_371_E | Leu_410_A | Ser_253_A | Lys_120_A |
| Ser_373_E | Ser_411_A | Trp_254_A | Leu_121_A |
| Phe_374_E | Ala_413_A | Gly_255_A | Asn_122_A |
| Trp_436_E | Thr_414_A | Pro_256_A | Ala_125_A |
| Leu_441_E | Pro_415_A | Glu_257_A | Val_128_A |
| Arg_509_E | Lys_416_A | Asp_258_A | Asp_129_A |
| Leu_418_A | Asp_259_A | Ile_130_A | |
| Phe_428_A | Gly_260_A | Tyr_131_A | |
| Gln_429_A | Lys_261_A | Leu_134_A | |
| Glu_430_A | Thr_262_A | Tyr_135_A | |
| Asp_431_A | Val_263_A | Val_146_A | |
| Thr_434_A | Asp_264_A | Pro_281_A | |
| Glu_435_A | Trp_291_A | Thr_397_A | |
| Asn_437_A | Ala_292_A | Asp_398_A | |
| Phe_438_A | Ser_293_A | Tyr_401_A | |
| Lys_441_A | Gly_294_A | Arg_402_A | |
| Gln_442_A | Asn_295_A | Arg_405_A | |
| Ile_446_A | Asp_301_A | ||
| Phe_523_A | Ser_302_A | ||
| Gln_526_A | Cys_303_A | ||
| Cys_530_A | Cys_305_A | ||
| Leu_539_A | Asp_306_A | ||
| His_540_A | Thr_309_A | ||
| Lys_541_A | Glu_331_A | ||
| Cys_542_A | His_364_A | ||
| Tyr_587_A | Thr_367_A | ||
| Ser_368_A | |||
| Ala_371_A | |||
| Pro_372_A | |||
| Arg_519_A | |||
ACE2- angiotensin converting enzyme 2; TMPRSS2 – transmembrane serine protease 2
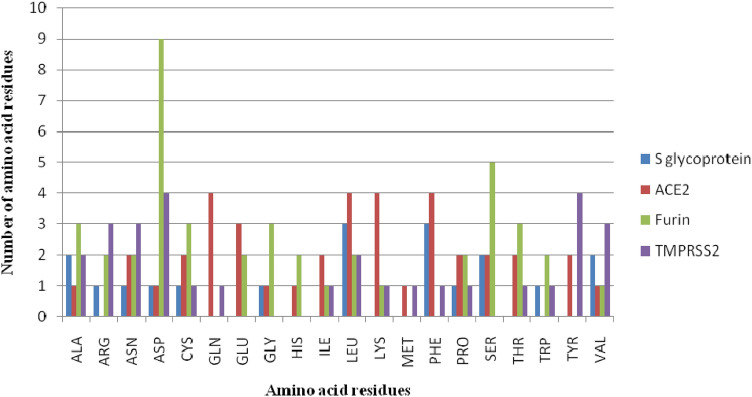
Amino acid descriptor analysis revealed the occurrence of 20 amino acids at the predicted active sites. The frequencies of these residues were found to be different and markedly depended on the analysed protein targets. The most frequent amino acids were Asp and Ser, which were constituents of the furin predicted active site. The most abundant amino acids for the S glycoprotein were Leu and Phe with 3 repetitions. The same amino acids together with Glu and Leu were also noticed as the building blocks of the ACE2 active site. Asp and Tyr were also found to be the most significant amino acids at the TMPRSS2 active site with 4 repetitions (Fig. 3).
Fig. 3.
Amino acid frequencies at targets predicted active sites of S glycoprotein, ACE2, furin and TMPRSS2 proteins (ACE2- angiotensin converting enzyme 2; TMPRSS2 – transmembrane serine protease 2)
The best drug score values for analysed targets varied between 0.73 and 0.84. Beside the drug scores, pocket sites from all selected targets were characterized through the size, shape and amino acid descriptors. The size and shape descriptors together with a drug score value for every predicted active site of the selected targets are summarized in Table 3. The selected pocket′s volumes for the S glycoprotein, ACE2, furin and TMPRSS2 proteins were 404.6, 876.74, 545.34 and 525.12 Å3, respectively. The biggest volume, surface and depth of the selected pocket were detected for ACE2. The results revealed equable enclosure parameters for ACE2, furin and TMPRSS2 (0.10–0.11), while the highest value for this parameter was determined for the S glycoprotein (0.24).
Table 3.
Summarized size and shape descriptors and drug score values for the selected active sites of the analysed protein targets
| Protein target | Size and shape descriptors | Drug score | |||
|---|---|---|---|---|---|
| Volume (Å3) | Surface (Å2) | Depth (Å) | Enclosure | ||
| S glycoprotein | 404.61 | 682.33 | 19.30 | 0.24 | 0.79 |
| ACE2 | 876.74 | 1068.47 | 23.22 | 0.11 | 0.83 |
| Furin | 545.34 | 635.62 | 15.99 | 0.11 | 0.73 |
| TMPRSS2 | 525.12 | 813.49 | 21.77 | 0.10 | 0.84 |
Å – (ångström) is a metric non-SI unit of length equal to 10−10 m; ACE2- angiotensin converting enzyme 2; TMPRSS2 – transmembrane serine protease 2
Binding energy
The results showed that some PoPEx compounds, especially punicalagin and punicalin formed more stable complexes than positive controls with amino acid residues at the active sites of the selected protein targets (Table 4). The complex of lopinavir with ACE2 had a higher binding energy value (−7.512 kcal/mol) than ACE2 with any of the investigated PopEx constituents. Nevertheless, the binding affinity of punicalin and punicalagin was significant even for this protein target (−7.353 and − 7.144 kcal/mol, respectively).
Table 4.
Summary binding energy results of the selected PoPEx constituents on the selected protein targets
| Protein target | Compound | Binding energy [kcal/mol] | Interacting amino acid residues |
|---|---|---|---|
| S glycoprotein | Punicalin | −7.406 | Ser 371, Leu 368, Val 367 |
| Punicalagin | −7.312 | Asn 343, Asn 370, Ser 371, Leu 335, Leu 368, Phe 342 | |
| Lopinavir | −6.508 | Ala 363, Asp 364, Leu 335, Leu 368, Phe 374, Val 362 | |
| Ellagic acid | −6.114 | Ala 372, Phe 377, Tyr 369 | |
| Umifenovir | −5.821 | Cys 336, Leu 368, Phe 338, Phe 342, Val 362, Val 367 | |
| Gallic acid | −4.808 | Phe 374, Tyr 369 | |
| ACE2 | Lopinavir | −7.512 | Asn 290, Asp 367, Glu 406, Ile 291, Leu 370, Lys 441, Met 366, Thr 276, Thr 445 |
| Punicalin | −7.353 | Asn 290, Asp 292, Asp 367, Lys 441, Ser 280 | |
| Punicalagin | −7.144 | Asp 292, Lys 441, Phe 428, Pro 289 | |
| Ellagic acid | −6.854 | Asp 292, Asp 367, Gln 442, Lys 441 | |
| Umifenovir | −6.558 | Ala 413, Asp 367, Glu 406, Leu 370, Leu 410, Lys 441, Ser 409 | |
| Gallic acid | −5.244 | Gln 442, Ile 291, Lys 441, Thr 445 | |
| Furin | Punicalin | −9.725 | Asp 258, Glu 257, Gly 255, His 194, Pro 256, Ser 368, Thr 365 |
| Punicalagin | −9.385 | Arg 298, Asp 259, Gly 366, His 194 | |
| Ellagic acid | −7.801 | Ala 532, Arg 490, Gly 307, Ser 311, Trp 531 | |
| Gallic acid | −7.486 | Asp 258, Asp 301, Asp 306, Cys 303, Glu 257, Pro 256, Thr 262 | |
| Sulconazole | −6.923 | Ala 532, Phe 528, Trp 531, Val 263 | |
| TMPRSS2 | Punicalin | −8.168 | Arg 87, Arg 91, Arg 405, Asn 97, Met 404 |
| Punicalagin | −7.358 | Arg 87, Arg 91, Arg 405, Asn 97, Gly 408, Met 404 | |
| Camostat | −7.069 | Arg 87, Arg 405, Asn 97, Met 404, Phe 99 | |
| Ellagic acid | −6.829 | Arg 87, Arg 405 | |
| Gallic acid | 5.709 | Ala 88, Arg 91, Asp 92, Asp 129, Tyr 401 |
ACE2- angiotensin converting enzyme 2; TMPRSS2 - transmembrane serine protease 2
Analysis of ligand-receptor interactions
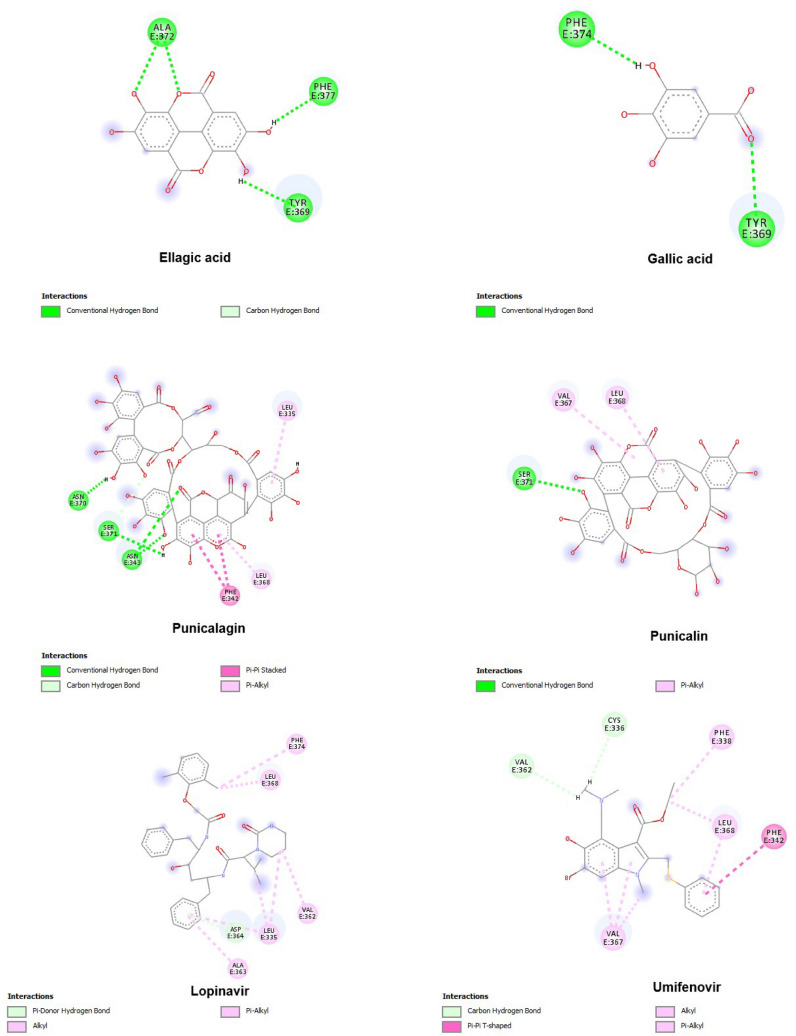
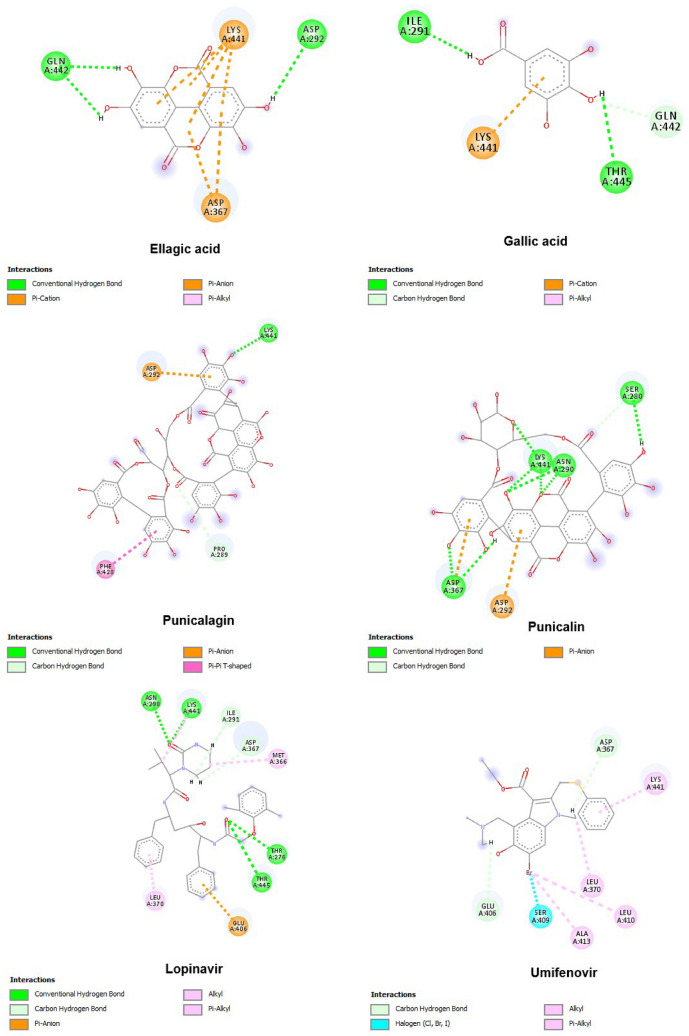
Docking analysis was further extended to the ligand-receptor type of interactions analysis. Amino acid residues were colour-coded according to the nature of interactions they were involved in (hydrogen bonding, ionic bonding, cationic bond, Pi-Pi bonding). Molecular docking results showed that all PoPEx ligands interacted through hydrogen bonds with amino acid residues at the S glycoprotein predicted active site. Among all the analysed ligands, punicalin demonstrated the strongest interaction with the S glycoprotein with free binding energy of −7.406 kcal/mol and formation of a hydrogen bond with the Ser 371 amino acid. The molecule also showed two Pi-alkyl interactions with Val 367 and Leu 368 residues at the predicted active site. Punicalagin was another PoPEX constituent with more stable binding conformation than reference substance lopinavir at the predicted active site. Although having slightly higher energy (−7.312 kcal/mol) than punicalin, this ligand-protein complex was stabilized through three hydrogen bonds (Asn 343, Asn 370 and Ser 371). Furthermore, this complex was additionally stabilized through Pi-Pi stacked (Phe 342) and Pi-alkyl (Leu 335 and Leu 368) interactions at the predicted active site (Fig. 4). Umifenovir, as the second positive control was a lower affinity binder for protein target than lopinavir.
Fig. 4.
S glycoprotein amino acids involved in interactions with selected PoPEx ligands and positive controls
Of all analysed targets, ACE2 was the only one that formed the most stable complex with lopinavir (−7.512 kcal/mol) as a positive control. The binding energies for ellagic acid, gallic acid, punicalagin and punicalin were found to be −6.854, −5.244, −7.144 and −7.353 kcal/mol, respectively. Nevertheless, all tested PoPEx ligands showed significant binding affinity at the ACE2 predicted active site with all ligand-protein complexes stabilized through hydrogen bonds. Amino acid residues Lys 441 involved in hydrogen bond interactions in the most stable lopinavir-ACE2 complex was also found to be important for stabilization of complexes punicalin-ACE2 and punicalagin-ACE2. Moreover, Asn 290 and Thr 445 key amino acid residues for stabilization of lopinavir conformation with minimum energy were also found to be significant in stabilization of punicalin and gallic acid, respectively (Fig. 5).
Fig. 5.
ACE2 amino acids involved in interactions with selected PoPEx ligands and positive controls
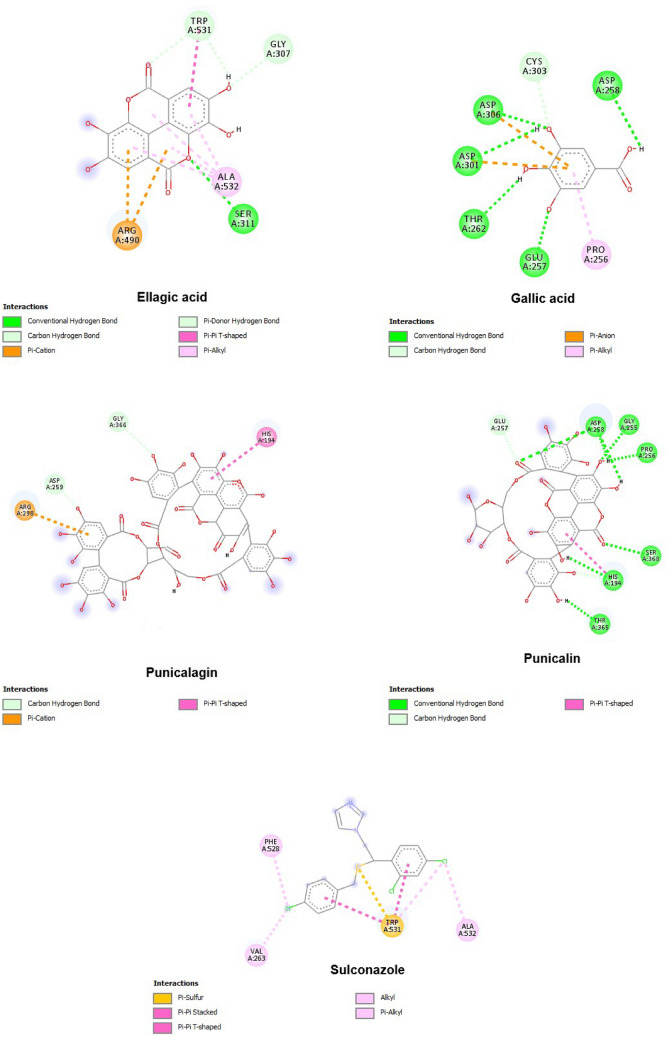
The outlined results revealed that all PoPEx constituents formed the most stable complexes with furin. This was the only protein target analysed in this study that formed lower energy complexes with all investigated ligands compared to the positive control. The structure of the furin complex with ellagic, gallic acid and punicalin emphasized influence of amino acids Ser 311, Asp 258, Asp 301, Asp 306, Glu 257, Thr 262, Asp 258, Gly 255, His 194, Pro 256, Ser 368 and Thr 365 in polar interactions (Fig. 6).
Fig. 6.
Furin amino acids involved in interactions with selected PoPEx ligands and sulconazole
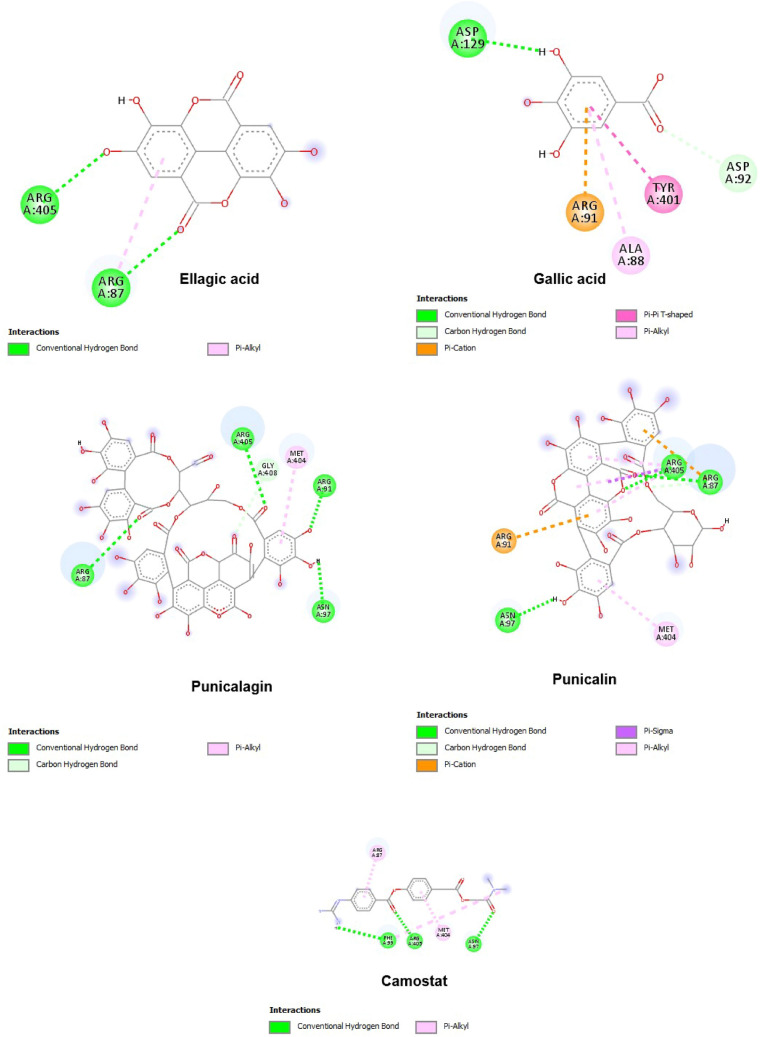
Punicalagin and punicalin showed intensive interactions with TMPRSS2 amino acid residues at the predicted active site with binding energy values of −7.358 and −8.168 kcal/mol, and with four and three hydrogen bonds, respectively. These binding energy values revealed higher affinity for the analysed target than camostat as a positive control (−7.069 kcal/mol). Polar interactions with Asn 97 and Arg 405 residues were essential for stabilization of TMPRSS2 complexes with punicalagin, punicalin as well as camostat (Fig. 7).
Fig. 7.
TMPRSS2 amino acids involved in interactions with selected PoPEx ligands and camostat
Discussion
A viral infection begins after initial entry of the virus into a host cell where the replication process starts. The quantitative measure of virus production through the process of replication is called viral load. Although it has not been definitively established whether high values of viral load correspond to more severe symptoms, Liu et al. [35] showed that mild clinical manifestation of SARS-CoV-2 infections could be associated with lower virus quantities detected in the patients [35]. Therefore, the virus entry process represents a crucial step in the SARS-CoV-2 infection and research activities seek to identify specific binders that would be able to block the viral entry proteins.
In the protein structure, sites with certain physicochemical and spatial characteristics are a prerequisite for a stable interaction between proteins and ligands. Non-polar amino acid ratios for selected ACE2, furin and TMPRSS2 pockets were lower than for the second ranged predicted pockets. However, drug score value was generally considered to be a more important druggability indicator. Drug score values obtained for all four proteins predicted pockets were found to exceed 0.5. Therefore, all the predicted active sites were deemed druggable binding pocket sites [33]. The best drug score (0.84) was revealed for TMPRSS2. Recent studies confirmed the DogSiteScorer capability to identify pocket active sites at the newly determined SARS-CoV-2 crystal structures [36, 37].
The results of this study showed that protein active site’s volume results ranged from 404.61 to 876.74 Å3. According to Nayal and Honig [38] non-drug binding sites’ volume values were lower than 328 Å3 hence all analysed pockets were considered as drug binding regarding the volume. The largest pocket volume and surface detected were for ACE2. The binding site depth above 4 Å is considered satisfactory, with average values of 6.8–11.4 Å for drug-binding sites. Depth values of the analysed protein targets ranged between 15.99 Å (furin) and 23.22 Å (ACE2) which was above the statistical average according to the literature [38]. Low values for enclosure parameters for all targets showed that pocket sites were not entirely on the surface of the proteins, but partly buried inside the structure. Cavities with hydrophobic amino acid residues such as Phe and Leu were found to exhibit high binding affinity. A possible explanation was that they did not go through a desolvation process before their interaction with ligands [39].
The most frequent amino acids at the selected druggable active sites were Leu and Phe for the S glycoprotein, Gln, Leu, Lys and Phe for ACE2, Asp for furin and Asp and Tyr for TMPRSS2. Asp was the most frequent single amino acid with 9 repetitions at the furin active site. This amino acid residue was mainly involved in the interactions as a hydrogen bond acceptor where polar interactions were especially favourable for complex protein-ligand stability [40]. Asp was also an acidic part of the so-called “classical” catalytic triad, three coordinated amino acids (together with Ser and His) which generated a charge-relay system for activating a nucleophilic attack on enzyme substrate. They were usually found close to each other at active sites of important enzymes such as proteases [41].
In our docking study at the predicted active sites on the S glycoprotein and ACE2 two molecules were used as positive controls, i.e. lopinavir and umifenovir. Some molecules such as chloroquine and hydrochloroquine strongly inhibited the SARS-CoV-2 infection in vitro, whereas others, like remdesivir have been recently conditionally approved for the treatment of hospitalized patients with a severe COVID-19 infection [42, 43]. Our selection of positive controls was based on their affinity for binding on the S glycoprotein (lopinavir) as well as their mechanism of action (umifenovir). Results from similar docking studies showed that lopinavir exhibited good binding affinity for the S glycoprotein while hydrochloroquine was found to be a poor binder due to a relatively small molecule -hypothetically [44]. Additionally, umifenovir, as an in vitro proven broad-spectrum antiviral drug [45] showed a propensity to reduce viral (SARS-CoV-2) load in a clinical pilot trial conducted in China in February 2020 [46]. Our results showed better binding affinity of lopinavir than umifenovir at the predicted active site of the S glycoprotein with a binding energy value of −6.508 kcal/mol which corresponded with previously published results from Ozbil and Cubuk [44].
Results from the present study showed that punicalin and punicalagin exhibited higher affinity for the predicted druggable active site on the S glycoprotein than the positive controls. Both of the analysed tannins formed complexes at the active site which were stabilized with hydrogen bonds interaction between Asn 343, Asn 370 and Ser 371 amino acid residues for punicalagin and only Ser 371 for punicalin. Furthermore, the complexes were additionally stabilized with hydrophobic interactions, such as Pi-alkyl and Pi-Pi interactions. Interestingly, the Phe 342 residue which stabilized the best pose of punicalagin in a complex with the S glycoprotein through a Pi-pi stacked interaction was found to be the central residue for allosteric disruption of the S glycoprotein (homology modelled structure) interaction with ACE2 [47].
Docking results showed that none of the PoPEx complexes were more stable than the one of lopinavir with ACE2. Even though punicalagin, punicalin and ellagic acid showed lower affinity for this target, (as compared to the lopinavir), they were all found to be better binders than umifenovir (binding energy = −6.558 kcal/mol). Although blocking ACE2 and preventing it from an interaction with the S glycoprotein has been definitely recognized as a novel therapy approach for SARS-CoV-2 infections, ACE2 also acts as one of the important homeostasis regulators via the renin-angiotensin system [48]. Hence, careful consideration of various potential side effects that ACE2 inhibition could cause is of paramount importance, especially concerning the reported increase in pulmonary inflammation [49]. Hopefully, the anti-inflammatory activity of PoPEx constituents (especially punicalagin) have already been reported by different investigations and could provide supplementary beneficial effects in COVID-19 patients therapy [50, 51]. Yet, considering all abovementioned, a more adequate approach would be to use a soluble form of ACE2 as a virus trap rather than a membrane protein [52].
Two proteases with a crucial role in the process of S glycoprotein cleavage in two biologically active subunits are furin and TMPRSS2. Sheybani et al. [12] have conducted a study to evaluate inhibitory effects of the water-soluble vitamin B folic acid (folate) on furin activity. They suggested folic acid as inexpensive and safe compound for the prevention or early stages treatment of respiratory infections associated with SARS-CoV-2 [12]. The results of our study confirmed that punicalin and punicalagin interacted with furin active site amino acid residues forming more stable complexes than sulconazole as a positive control. Sulconazole, a broad-spectrum anti-fungal agent, was found to be the most promising candidate with anti-furin activity from the collection of about 8000 molecules [53]. Contrary to the less significant results of binding energies achieved in docking analysis with the S glycoprotein and ACE2, ellagic and gallic acids formed stable complexes with furin revealing binding energies to be −7.801 and − 7.486 kcal/mol, respectively.
The best binding affinity for all active sites was detected for punicalin. The ligand conformation in the most stable complex was stabilized through hydrogen bonds interactions with Asp 258, Gly 255, His 194, Pro 256, Ser 368 and Thr 365 amino acid residues from the active site. The His 194 amino acid residue was also involved in the stabilization of the punicalagin-furin complex, but through a Pi-Pi interaction.
Although molecular docking analysis is critically dependent on the crystallized structures of the analysed targets, recent advances in the field of homology modelling enabled interaction predictions even for non-crystallized structures. Molecular docking study of PoPEx constituents at the predicted TMPRSS2 active site revealed that all analysed ligands interacted through hydrogen bonds with this enzyme target. Yet, only punicalin and punicalagin were better binders than camostat a positive control. In a recently conducted study this clinically proven serine protease inhibitor significantly decreased the SARS-CoV-2 entry and consequently the viral infection [10]. Amino acid residues involved in the interactions with punicalagin and punicalin were Arg 405, Arg 87, Asn 97 and Arg 91 additionally for punicalagin. Arg 405 and Asn 97 were also the key residues involved in the stabilization of the camostat-TMPRSS2 complex through conventional hydrogen bonds, while Arg 87 additionally stabilized the structure through a Pi-alkyl interaction.
Although, several studies have evaluated the activity of natural products as anti-SARS-CoV-2 agents, they were primarily focused on single targets with confirmed role in virus internalization. In this study naturally derived polyphenols were simultaneously studied against four the most important anti-SARS-CoV-2 targets. This is the first study so far that analysed the effect of PoPEx constituents (punicalagin, punicalin, ellagic acid and gallic acid) on major multiple targets responsible for virus internalization.
Based on the available literature data, punicalagin after oral administration in rats yields plasma concentration of 30 μg/mL, which proves its absorption from digestive tract and its availability in biologically relevant concentration in body fluids [54]. These findings encourage us to continue with in vitro and bioavailability studies in humans, which would enable us to evaluate the clinical potential of PoPEx constituents for treatment or prevention of COVID-19.
Conclusion
The results described in the present study highlight the potential inhibitory activity of PoPEx polyphenols against the SARS-CoV-2 internalization process. Furthermore, the presented molecular docking study demonstrated that PoPEx constituents, i.e. punicalagin and punicalin are promising candidates for further anti-SARS-CoV-2 in vitro studies. Being the ingredients of a natural product that is used as food, these candidates also have a confirmed safety profile which is their additional and important advantage in the disease treatment.
Compliance with ethical standards
Conflict of interest
The author(s) declare that there is no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of V The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aćimović J, Jandrić L, Đakovic-Dević J, Bojanić J, Subotić B, Radojčić T, Rodić-Vukmir N, Zeljković B. Epidemiological characteristics of COVID-19 infection in the Republic of Srpska: a hundred days survey. Scr Med. 2020;51:74–80. doi: 10.5937/scriptamed51-27298. [DOI] [Google Scholar]
- 4.Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabi FA, Al Zoubi MS, Kasasbeh GA, Salameh DM, Al-Nasser AD (2020) SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens 9. 10.3390/pathogens9030231 [DOI] [PMC free article] [PubMed]
- 6.Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, Talavera-Lopez C, Maatz H, Reichart D, Sampaziotis F, Worlock KB, Yoshida M, Barnes JL, Network HCALB. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M, Hofmann-Winkler H, Pöhlmann S (2018) Priming time: how cellular proteases arm coronavirus spike proteins:71–98. 10.1007/978-3-319-75474-1_4
- 9.Hoffmann M, Kleine-Weber H, Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleine-Weber H, Elzayat MT, Hoffmann M, Pohlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci Rep. 2018;8:16597. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheybani Z, Dokoohaki MH, Negahdaripour M, Dehdashti M, Zolghadr H, Moghadami M, Masoompour SM, Zolghadr AR (2020) The role of folic acid in the management of respiratory disease caused by COVID-19. ChemRxiv. Preprint. 10.26434/chemrxiv.12034980.v1
- 13.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, Hennig BP, Kreuter M, Conrad C, Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janković S. Current status and future perspective of coronavirus disease 2019: a review. Scr Med. 2020;51:101–109. doi: 10.5937/scriptamed51-27183. [DOI] [Google Scholar]
- 16.Banihani S, Swedan S, Alguraan Z. Pomegranate and type 2 diabetes. Nutr Res. 2013;33:341–348. doi: 10.1016/j.nutres.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Grabež M, Škrbić R, Stojiljković MP, Rudić-Grujić V, Paunović M, Arsić A, Petrović S, Vučić V, Mirjanić-Azarić B, Šavikin K, Menković N, Janković T, Vasiljević N. Beneficial effects of pomegranate peel extract on plasma lipid profile, fatty acids levels and blood pressure in patients with diabetes mellitus type-2: a randomized, double-blind, placebo-controlled study. J Funct Foods. 2020;64:103692. doi: 10.1016/j.jff.2019.103692. [DOI] [Google Scholar]
- 18.Al-Jarallah A, Igdoura F, Zhang Y, Tenedero CB, White EJ, MacDonald ME, Igdoura SA, Trigatti BL. The effect of pomegranate extract on coronary artery atherosclerosis in SR-BI/APOE double knockout mice. Atherosclerosis. 2013;228:80–89. doi: 10.1016/j.atherosclerosis.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 19.Akhtar S, Ismail T, Fraternale D, Sestili P. Pomegranate peel and peel extracts: chemistry and food features. Food Chem. 2015;174:417–425. doi: 10.1016/j.foodchem.2014.11.035. [DOI] [PubMed] [Google Scholar]
- 20.Sohrab G, Roshan H, Ebrahimof S, Nikpayam O, Sotoudeh G, Siasi F. Effects of pomegranate juice consumption on blood pressure and lipid profile in patients with type 2 diabetes: a single-blind randomized clinical trial. Clin Nutr ESPEN. 2019;29:30–35. doi: 10.1016/j.clnesp.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Danesi F, Ferguson LR (2017) Could pomegranate juice help in the control of inflammatory diseases? Nutrients 9. 10.3390/nu9090958 [DOI] [PMC free article] [PubMed]
- 22.Orgil O, Schwartz E, Baruch L, Matityahu I, Mahajna J, Amir R. The antioxidative and anti-proliferative potential of non-edible organs of the pomegranate fruit and tree. LWT Food Sci Technol. 2014;58:571–577. doi: 10.1016/j.lwt.2014.03.030. [DOI] [Google Scholar]
- 23.Saeed M, Naveed M, BiBi J, Kamboh AA, Arain MA, Shah QA, Alagawany M, El-Hack MEA, Abdel-Latif MA, Yatoo MI, Tiwari R, Chakraborty S, Dhama K. The promising pharmacological effects and therapeutic/medicinal applications of Punica Granatum L. (pomegranate) as a functional food in humans and animals. Recent Patents Inflamm Allergy Drug Discov. 2018;12:24–38. doi: 10.2174/1872213X12666180221154713. [DOI] [PubMed] [Google Scholar]
- 24.Lansky EP, Newman RA. Punica granatum (pomegranate) and its potential for prevention and treatment of inflammation and cancer. J Ethnopharmacol. 2007;109:177–206. doi: 10.1016/j.jep.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Haidari M, Ali M, Ward Casscells S, 3rd, Madjid M. Pomegranate (Punica granatum) purified polyphenol extract inhibits influenza virus and has a synergistic effect with oseltamivir. Phytomedicine. 2009;16:1127–1136. doi: 10.1016/j.phymed.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Howell AB, D’Souza DH. The pomegranate: effects on bacteria and viruses that influence human health. Evid Based Complement Alternat Med. 2013;2013:606212. doi: 10.1155/2013/606212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy BU, Mullick R, Kumar A, Sudha G, Srinivasan N, Das S. Small molecule inhibitors of HCV replication from pomegranate. Sci Rep. 2014;4:5411. doi: 10.1038/srep05411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moradi M-T, Karimi A, Shahrani M, Hashemi L, Ghaffari-Goosheh M-S. Anti-influenza virus activity and phenolic content of pomegranate (Punica granatum L.) Peel extract and fractions. Avicenna J Med Biotechnol. 2019;11:285. [PMC free article] [PubMed] [Google Scholar]
- 29.Moradi M-T, Karimi A, Rafieian-kopaei M, Rabiei-Faradonbeh M, Momtaz H. Pomegranate peel extract inhibits internalization and replication of the influenza virus: an in vitro study. Avicenna J Phytomedicine. 2020;10:143. [PMC free article] [PubMed] [Google Scholar]
- 30.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H (2020) Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed]
- 31.Krieger E, Geretti E, Brandner B, Goger B, Wells TN, Kungl AJ. A structural and dynamic model for the interaction of interleukin-8 and glycosaminoglycans: support from isothermal fluorescence titrations. Proteins. 2004;54:768–775. doi: 10.1002/prot.10590. [DOI] [PubMed] [Google Scholar]
- 32.Volkamer A, Griewel A, Grombacher T, Rarey M. Analyzing the topology of active sites: on the prediction of pockets and subpockets. J Chem Inf Model. 2010;50:2041–2052. doi: 10.1021/ci100241y. [DOI] [PubMed] [Google Scholar]
- 33.Volkamer A, Kuhn D, Grombacher T, Rippmann F, Rarey M. Combining global and local measures for structure-based druggability predictions. J Chem Inf Model. 2012;52:360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 34.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Yan L-M, Wan L, Xiang T-X, Le A, Liu J-M, Peiris M, Poon LLM, Zhang W. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–657. doi: 10.1016/s1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odhar HA, Ahjel SW, Albeer A, Hashim AF, Rayshan AM, Humadi SS. Molecular docking and dynamics simulation of FDA approved drugs with the main protease from 2019 novel coronavirus. Bioinformation. 2020;16:236–244. doi: 10.6026/97320630016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X, He S, Zhou Z, Zhou Z, Chen Q, Yan Y, Zhang C, Shan H, Chen S (2020) Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 10.1016/j.apsb.2020.04.009 [DOI] [PMC free article] [PubMed]
- 38.Nayal M, Honig B. On the nature of cavities on protein surfaces: application to the identification of drug-binding sites. Proteins. 2006;63:892–906. doi: 10.1002/prot.20897. [DOI] [PubMed] [Google Scholar]
- 39.Young T, Hua L, Huang X, Abel R, Friesner R, Berne BJ. Dewetting transitions in protein cavities. Proteins. 2010;78:1856–1869. doi: 10.1002/prot.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saberi Fathi SM, Tuszynski JA. A simple method for finding a protein’s ligand-binding pockets. BMC Struct Biol. 2014;14:18. doi: 10.1186/1472-6807-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ekici OD, Paetzel M, Dalbey RE. Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 2008;17:2023–2037. doi: 10.1110/ps.035436.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, Zhan S, Lu R, Li H, Tan W, Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grein J, Ohmagari N, Shin D, Diaz G, Asperges E, Castagna A, Feldt T, Green G, Green ML, Lescure FX, Nicastri E, Oda R, Yo K, Quiros-Roldan E, Studemeister A, Redinski J, Ahmed S, Bernett J, Chelliah D, Chen D, Chihara S, Cohen SH, Cunningham J, D’Arminio Monforte A, Ismail S, Kato H, Lapadula G, L’Her E, Maeno T, Majumder S, Massari M, Mora-Rillo M, Mutoh Y, Nguyen D, Verweij E, Zoufaly A, Osinusi AO, DeZure A, Zhao Y, Zhong L, Chokkalingam A, Elboudwarej E, Telep L, Timbs L, Henne I, Sellers S, Cao H, Tan SK, Winterbourne L, Desai P, Mera R, Gaggar A, Myers RP, Brainard DM, Childs R, Flanigan T. Compassionate use of Remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozbil M, Cubuk H (2020) Comparison of clinically approved molecules on SARS-CoV-2 drug target proteins: a molecular docking study. ChemRxiv. Preprint. 10.26434/chemrxiv.12090828.v1 [DOI] [PMC free article] [PubMed]
- 45.Pecheur EI, Borisevich V, Halfmann P, Morrey JD, Smee DF, Prichard M, Mire CE, Kawaoka Y, Geisbert TW, Polyak SJ. The synthetic antiviral drug arbidol inhibits globally prevalent pathogenic viruses. J Virol. 2016;90:3086–3092. doi: 10.1128/JVI.02077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall DC, Jr, Ji HF. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med Infect Dis. 2020;35:101646. doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roshanravan N, Ghaffari S, Hedayati M. Angiotensin converting enzyme-2 as therapeutic target in COVID-19. Diabetes Metab Syndr. 2020;14:637–639. doi: 10.1016/j.dsx.2020.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.BenSaad LA, Kim KH, Quah CC, Kim WR, Shahimi M. Anti-inflammatory potential of ellagic acid, gallic acid and punicalagin A&B isolated from Punica granatum. BMC Complement Altern Med. 2017;17:47. doi: 10.1186/s12906-017-1555-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X, Yin P, Wan C, Chong X, Liu M, Cheng P, Chen J, Liu F, Xu J. Punicalagin inhibits inflammation in LPS-induced RAW264.7 macrophages via the suppression of TLR4-mediated MAPKs and NF-kappaB activation. Inflammation. 2014;37:956–965. doi: 10.1007/s10753-014-9816-2. [DOI] [PubMed] [Google Scholar]
- 52.Alhenc-Gelas F, Drueke TB. Blockade of SARS-CoV-2 infection by recombinant soluble ACE2. Kidney Int. 2020;97:1091–1093. doi: 10.1016/j.kint.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villoutreix BO, Creemers J, Léger Y, Siegfried G, Decroly E, Evrard S, Khatib A-M (2020) Targeting furin activity through in silico and in vitro drug repurposing strategy for SARS-CoV-2 spike glycoprotein cleavage repression. Pharmacodynamics. Preprint. 10.21203/rs.3.rs-25856/v1
- 54.Cerda B, Llorach R, Ceron JJ, Espin JC, Tomas-Barberan FA. Evaluation of the bioavailability and metabolism in the rat of punicalagin, an antioxidant polyphenol from pomegranate juice. Eur J Nutr. 2003;42:18–28. doi: 10.1007/s00394-003-0396-4. [DOI] [PubMed] [Google Scholar]