Abstract
Strategies for drug discovery and repositioning are an urgent need with respect to COVID-19. We developed “REDIAL-2020”, a suite of machine learning models for estimating small molecule activity from molecular structure, for a range of SARS-CoV-2 related assays. Each classifier is based on three distinct types of descriptors (fingerprint, physicochemical, and pharmacophore) for parallel model development. These models were trained using high throughput screening data from the NCATS COVID19 portal (https://opendata.ncats.nih.gov/covid19/index.html), with multiple categorical machine learning algorithms. The “best models” are combined in an ensemble consensus predictor that outperforms single models where external validation is available. This suite of machine learning models is available through the DrugCentral web portal (http://drugcentral.org/Redial). Acceptable input formats are: drug name, PubChem CID, or SMILES; the output is an estimate of anti-SARS-CoV-2 activities. The web application reports estimated activity across three areas (viral entry, viral replication, and live virus infectivity) spanning six independent models, followed by a similarity search that displays the most similar molecules to the query among experimentally determined data. The ML models have 60% to 74% external predictivity, based on three separate datasets. Complementing the NCATS COVID19 portal, REDIAL-2020 can serve as a rapid online tool for identifying active molecules for COVID-19 treatment. The source code and specific models are available through Github (https://github.com/sirimullalab/redial-2020), or via Docker Hub (https://hub.docker.com/r/sirimullalab/redial-2020) for users preferring a containerized version.
Introduction
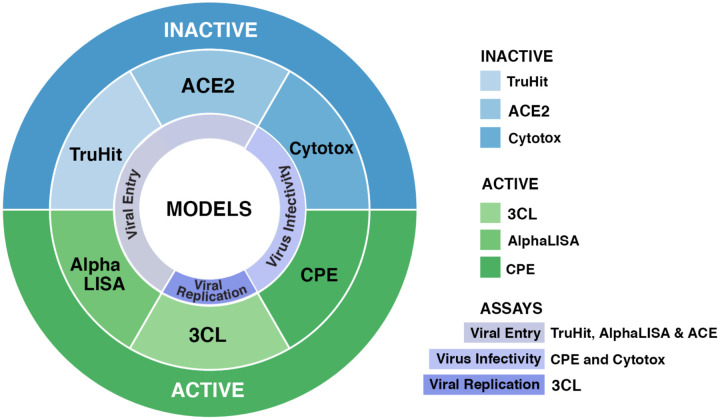
Currently, there is an urgent need to find drugs and effective treatment options for coronavirus disease 2019 (COVID-19). Here, we present a suite of machine learning (ML) models termed “REDIAL-2020” that forecast activities for live viral infectivity, viral entry, and viral replication, specifically for SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2). This application can serve the scientific community when prioritizing compounds for in vitro screening and may ultimately accelerate the identification of novel drug candidates for the COVID-19 treatment. REDIAL-2020 currently consists of six independently trained ML models and includes a similarity/substructure search module that queries the underlying experimental dataset for similar compounds. These ML models were trained using experimental data generated by the following assays: the SARS-CoV-2 cytopathic effect (CPE) assay and its host cell cytotoxicity counterscreen; the Spike-ACE2 protein-protein interaction (AlphaLISA) assay and its TruHit counterscreen, as well as an angiotensin- converting enzyme 2 (ACE2) enzymatic activity assay; and 3C-like (3CL) proteinase enzymatic activity assay. The assays represent three distinct categories: i) viral entry (CPE1 and host cell cytotoxicity counterscreen2); ii) viral replication (3CL enzymatic activity); and iii) live virus infectivity (AlphaLISA, TruHit counterscreen, and ACE2 enzymatic activity),3 as described in the National Center for Advancing Translational Sciences (NCATS) COVID-19 portal.4 We retrieved these datasets from the NCATS COVID19 portal.5 The NCATS team is committed to performing a range of COVID19-related viral and host target assays, as well as analyzing the results.6
These ML models are integrated into a user-friendly web portal that allows input using three different formats: i) drug name, both as International Nonproprietary Name, INNs (e.g., hydroxychloroquine) or as trade name (e.g., Plaquenil); ii) PubChem CID,7 i.e., PubChem Compound ID number (e.g., 3652 for hydroxychloroquine); or iii) using the chemical structure encoded in the SMILES (Simplified Molecular-Input Line-Entry System) format,8 respectively. The workflow and output, regardless of input format, is identical and described below. Drug repositioning requires computational support,9 and data-driven decision making offers a pragmatic approach to identifying optimal candidates while minimizing the risk of failure. Since molecular properties and bioactivities can be described as a function of chemical structure, cheminformatics-based predictive models are becoming increasingly useful in drug discovery and repositioning research. Specifically, anti-SARS-CoV-2 models based on high throughput data could be used as a prioritization step when planning experiments, particularly for large molecular libraries, thus decreasing the number of experiments and reducing downstream costs. REDIAL-2020 could serve such a purpose and help the scientific community reduce the number of molecules before experimental tests for anti-SARS-CoV-2 activity. This suite of ML models can also be used via the command line for large scale virtual screening. As more reliable data sets become available in the public domain, we plan to tune the ML models further, add additional models based on SARS-CoV-2 assays, and make these models available in future releases of REDIAL-2020.
Live Virus Infectivity Assays
The SARS-CoV-2 cytopathic effect (CPE) assay measures the ability of a compound to reverse the cytopathic effect induced by the virus in Vero E6 host cells. As cell viability is reduced by viral infection, the CPE assay measures the compound’s ability to restore cell function (cytoprotection). While this assay does not provide any information concerning the mechanism of action, it can be used to screen for antiviral activity in a high-throughput manner. However, there is the possibility that the compound itself may exhibit a certain degree of cytotoxicity, which could also reduce cell viability. Since this confounds the interpretation of CPE assay results, masking the cyto-protective activity, a counter-screen to measure host (Vero E6) cell cytotoxicity is used to detect such compounds. Thus, a net, positive result from the combined CPE assays consists of a compound showing a protective effect but no cytotoxicity.
Viral Entry Assays
The Spike-ACE2 protein-protein interaction (AlphaLISA) assay measures a compound’s ability to disrupt the interaction between the viral Spike protein and its human receptor protein, ACE2 (angiotensin-converting enzyme type 2).10 The surface of the ACE2 protein is the primary host factor recognized and targeted by SARS-CoV-2 virions.11 This binding event between the SARS-CoV-2 Spike protein and the host ACE2 protein initiates binding of the viral capsid and leads to viral entry into host cells. Thus, disrupting the Spike-ACE2 interaction is likely to reduce the ability of SARS-CoV-2 virions to infect host cells. This assay has two counterscreens, as follows. The TruHit counterscreen is used to determine false positives, i.e., compounds that interfere with the AlphaLISA readout in a non-specific manner, or with assay signal generation and/or detection. It uses the biotin-streptavidin interaction (one of the strongest known non-covalent drug-protein interactions) because other compounds are unlikely to disturb it. Consequently, any compound showing interference with this interaction is most likely a false positive. Common interfering agents are oxygen scavengers or molecules with spectral properties sensitive to the 600–700 nm wavelengths used in AlphaLISA. The second counterscreen is an enzymatic assay that measures human ACE2 inhibition to identify compounds that could potentially disrupt endogenous enzyme function. ACE2 lowers blood pressure by catalyzing the hydrolysis of angiotensin II (a vasoconstrictor peptide) into the vasodilator angiotensin (1–7).12 While blocking the Spike-ACE2 interaction may stop viral entry, drugs effective in this manner could cause unwanted side-effects by blocking the endogenous vasodilating function of ACE2. Thus, the ACE2 assay serves to detect such eventualities and to de-risk such off-target events.
Viral Replication Assays
Following entry into the host cell, the main SARS-CoV-2 replication enzyme is 3C-like proteinase (3CL), also called “main protease” or Mpro,13 which cleaves the two SARS-CoV-2 polyproteins into various proteins (e.g., RNA polymerases, helicases, and methyltransferases, etc.), which are essential to the viral life cycle. Since inhibiting the 3CL protein disrupts the viral replication process, this makes 3CL an attractive drug target.14 The SARS-CoV-2 3CL biochemical assay measures compounds’ ability to inhibit recombinant 3CL cleavage of a fluorescently labeled peptide substrate.
Note on assays and models terminology.
Throughout this paper, we refer to assay and model names as follows: “CPE” for SARS-CoV-2 cytopathic effect, “cytotox” for host cell cytotoxicity counterscreen, “AlphaLISA” for Spike-ACE2 protein-protein interaction, “TruHit” for Spike-ACE2 protein-protein interaction counterscreen, “ACE2” for ACE2 enzymatic activity and “3CL” for 3CL enzymatic activity.
Results and Discussion
Preliminary Data Analysis
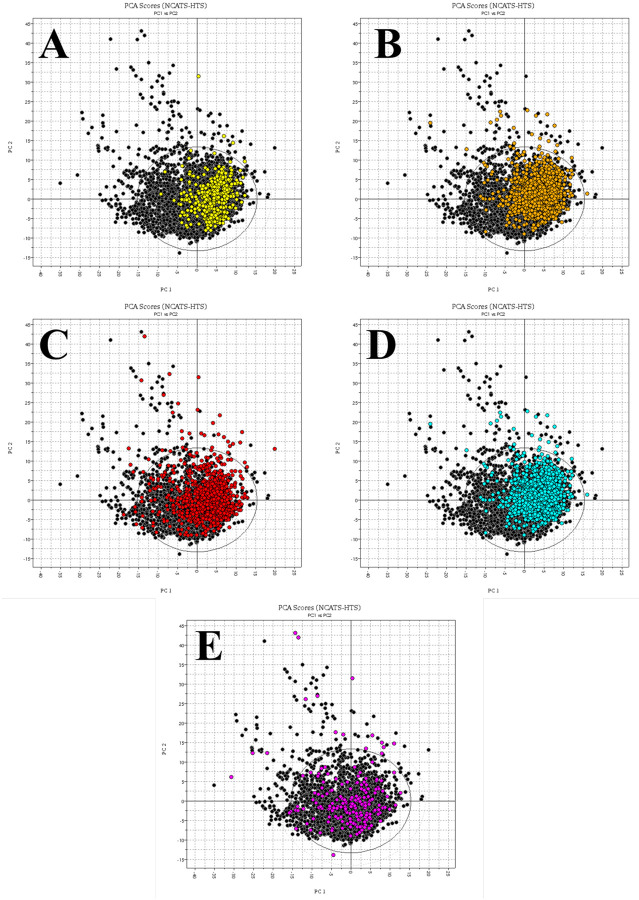
Prior to developing ML models, unsupervised learning should be used to evaluate the data and seek patterns that might guide successive steps. Hence, upon definition of the experimental categories (see Methods for details), we inspected the data using principal component analysis (PCA)15 and applied it using VolSurf+16 molecular descriptors. For both CPE and cytotox, clusters emerge along the first principal component (PC1; Figure 1). For CPE data, the majority of compounds showing high to moderate CPE activity are grouped in the right-hand of Figure 1A. At the same time, compounds with high to moderate cytotoxicity are grouped in the right-hand region of Figure 1B. By inspecting the loading score plot for VolSurf+ descriptors that are likely to contribute to these patterns, we identified membrane permeability (estimated using logP, the logarithm of the octanol/water partition coefficient) and water solubility (estimated using logS, the logarithm of the thermodynamic aqueous solubility) as major contributors to the first latent variable (see Supporting Information Figure SI1). Compounds with low logP/high logS, clustered in the left-hand region of the score plot, are less likely to be active in the CPE assay and more likely to be non-cytotoxic.
Figure 1:
PCA scores plot of the molecules tested in NCATS SARS-CoV-2 experiments based on VolSurf+ descriptors. A) CPE compounds colored by CPE categories: high/moderate activity in yellow and low activity in black; B) cytotoxic compounds colored by cytotoxicity categories: high/moderate cytotoxic in orange and low (not) cytotoxic in black. C) AlphaLISA compounds colored by Spike-ACE2 interaction blockers categories: high/moderate (strong) blockers in red and low (weak) blockers in black. D) TruHit compounds, colored by AlphaLISA readout interfering categories: high/moderate interfering in cyan and low interfering in black. E) ACE2 compounds, colored by ACE2 inhibition categories: high/moderate (strong) inhibitors in magenta and low (weak) inhibitors in black.
The distribution of actives was also visualized for AlphaLISA and TruHit compounds in Figures 1C and 1D, respectively (see also Table 1). For the AlphaLISA assay, although clustering is less pronounced respect to CPE (Figure 1A), the right-hand part of the plot does capture most of the high/moderate activity compounds. Such distribution of actives in the right-hand region was not observed for ACE2 actives (Figure 1E). Thus, permeability and solubility are not the major determinants of this ACE2 inhibition assay.
Table 1.
Number of compounds excluded for each model upon filtering with the logP and logS criteria.
| Assay | Excluded Actives (relative percentage) | Excluded Inactives (relative percentage) |
|---|---|---|
| CPE | 21 (4%) | 1417 (32%) |
| citotox | 83 (8%) | 1331 (34%) |
| AlphaLISA | 117 (15%) | 1104 (48%) |
| TruHit | 118 (15%) | 1100 (48%) |
| ACE2 | 62 (34%) | 1159 (40%) |
| 3CL | 65 (30%) | 2467 (37%) |
The results of this preliminary analysis can be used to filter input data prior to machine learning. For example, the majority of the compounds placed on the left side of the Figure 1 PCA plot are inactive (exception for ACE2). Therefore, prior to developing the ML models, we applied cutoff filters based on compounds calculated logP and logS using ALOGPS17 to every dataset except for ACE2. These filters help narrow the focus of ML model development on features derived only from compounds for which simple property criteria (e.g., logP and logS) cannot be used to distinguish actives from inactives -- specifically, the right-hand regions in Figure 1. The initial number of compounds, after data wrangling, was 4,954. Upon use of the logS and logP filters, each dataset was reduced in size (Table 1). However, the fraction of active compounds excluded from the ACE2 dataset was quite high (34%). Hence, logP and logS filters were not applied for ACE2 inhibition, and the complete dataset was used for model development. For 3CL enzymatic activity, data from NCATS was retrieved separately. The initial set contained 12,263 compounds. However, data wrangling identified ~1,850 duplicate and ~3,000 “inconclusive” entries, which were discarded. Additional entries were removed during the desalting and physicochemical feature generation as VolSurf descriptors could not be computed for some of the compounds. The final 3CL dataset contains 6,961 entries, with 222 active and 6,739 inactive compounds. Given that the fraction of active 3CL compounds filtered would have been 30%, the logP/logS filters were not applied. There were no significant activity clusters detected in the 3CL dataset via PCA-VolSurf+ (see Supporting Information Figure SI2).
With respect to actives vs. inactives, the six NCATS assays are highly unbalanced, with a disproportionate ratio of active (few) compounds compared to inactive (many) compounds. For example, there were ~9 times more inactives than actives and ~3 times more non-cytotoxic compounds than cytotoxic compounds for the CPE and cytotoxicity assays, respectively. Thus, in order to avoid over-training for the dominant category, each model was derived using random selection wherein compounds from the majority class were selected in equal proportion to those of the minority class. Our balanced dataset numbers were as follows: 736 for CPE, 1,662 for cytotox, 1,260 for AlphaLISA, 1,668 for TruHit, 206 for ACE2 and 442 for 3CL.
Models Comparison and Selection
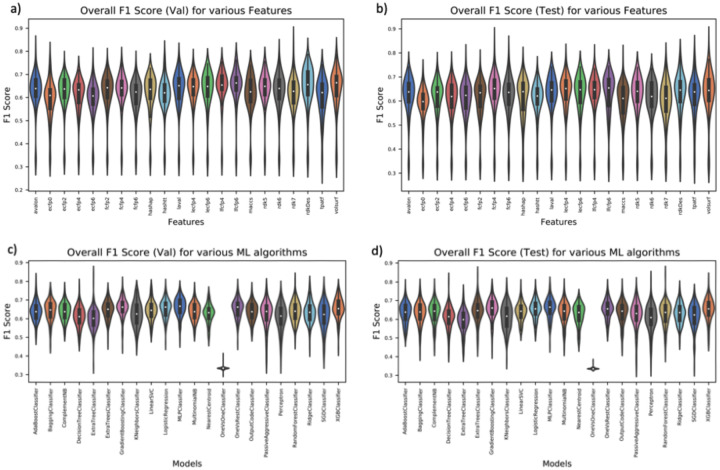
To evaluate anti-SARS-CoV-2 activities of novel chemicals, we implemented six predictive models based on consensus methods. Of the two consensus methods (voting-based and probability score-based) evaluated, the voting-based consensus model showed better performance (Figures SI13-SI16 in supporting information). Thus, the voting-based method is implemented in the REDIAL-2020 app. Consensus models were generated based on the top three performing models trained on fingerprint, pharmacophore, and physicochemical descriptors (see Methods section for details). First, we selected a fingerprint model from an initial evaluation of 19 different fingerprint descriptor methods. This was combined with a Topological Pharmacophore Atom Triplets Fingerprints (TPATF) model. Finally, the rdkit or VolSurf+ descriptor-based model provided a third model, based on physicochemical properties. All these models were trained with 22 different classifiers available in scikit-learn.18 Figure 2 (a–d) summarizes the comparison between various features and ML algorithms. Figures 2a and 2b compare the performance of each feature across 22 ML algorithms (classifiers) and 6 assays. Figures 2c and 2d compare the performance of each classifier across 22 features and 6 assays. For example, the violin plot for the avalon feature (Figure 2a) summarizes F1-scores from all 6 assays (and 22 classifiers). Among descriptors, VolSurf+ and lfcfp6 outperformed others, whereas the gradient boost classifier and the MLP (multilayer perceptron) classifier performed better among ML algorithms. See Supporting Information Figures SI9-SI20 for more detailed comparisons across different features and ML algorithms with respect to individual models.
Figure 2:
Comparison between the following, across the 6 assays: a) features for the validation set; b) features for the test set; c) ML algorithms for validation; and d) ML algorithms for the test set, respectively.
Two options for the consensus model were considered, based on the potential overlap between VolSurf+ and rdkit descriptors: fingerprint+TPATF+rdkit, and fingerprint+TPATF+VolSurf, respectively. VolSurf+ descriptors outperformed rdkit in CPE and TruHit, whilst rdkit outperformed VolSurf+ in cytotox, AlphaLISA, ACE2, and 3CL based on the tested evaluation metrics such as Accuracy, F1-score, and AUC (see Supporting Information Figures SI13-SI18). However, the situation changed when considering consensus models. Inclusion of VolSurf+ yielded better models for the AlphaLISA, TruHit, and ACE2 voting-based consensus models, whereas including rdkit yielded better consensus models for the CPE, cytotox, and 3CL consensus assays. Supporting Information Figures SI3-SI8 show a comparison of the best models from each feature category. Concerning the web portal, we implemented consensus model predictions based on the rdkit descriptors, since RDKit is open-source software that can be ported and dockerized without restrictions. Out of the six ML models, four (CPE, cytotox, TruHit, and 3CL) were implemented as consensus models with the rdkit descriptors, with the remaining two (AlphaLISA and ACE2) implemented as rdkit descriptor-based only. Tables 1 and 2 summarize the evaluation scores and the confusion matrices, respectively, for all models implemented in REDIAL-2020.
Table 2.
Summary of the results of the best models.
| Model | Validation set results | Test set results | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC | F1 | SEN | PREC | AUC | ACC | F1 SEN PREC | AUC | |||
| CPE | 0.794 | 0.794 | 0.794 | 0.795 | 0.794 | 0.725 | 0.725 | 0.726 | 0.727 | 0.725 |
| cytotox | 0.771 | 0.771 | 0.771 | 0.772 | 0.771 | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 |
| AlphaLISA | 0.788 | 0.788 | 0.788 | 0.79 | 0.789 | 0.762 | 0.762 | 0.762 | 0.762 | 0.762 |
| TruHit | 0.762 | 0.762 | 0.762 | 0.762 | 0.762 | 0.727 | 0.727 | 0.727 | 0.727 | 0.727 |
| ACE2 | 0.806 | 0.805 | 0.804 | 0.812 | 0.804 | 0.452 | 0.452 | 0.453 | 0.453 | 0.452 |
| 3CL | 0.803 | 0.802 | 0.803 | 0.81 | 0.803 | 0.672 | 0.671 | 0.671 | 0.672 | 0.671 |
ACC, Accuracy; F1, F1 score; SEN, sensitivity; PREC, precision; AUC, area under the receiver operating characteristic curve.
Submission Webpage
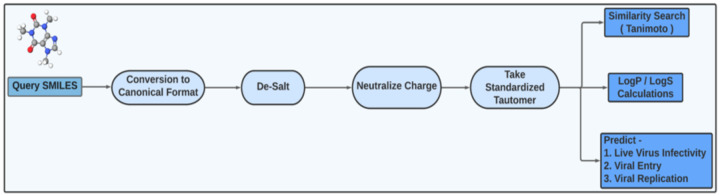
By accessing REDIAL-2020 (http://drugcentral.org/Redial) from any web browser, including mobile devices, the submission page is displayed (Figure 4). The web server accepts SMILES, drug names, or PubChem CIDs as input. The User Interface (UI) at the top of the page allows users to navigate various options (Figure 4). The UI provides summary information about the six models, such as model type, which descriptor categories were used for training, and the evaluation scores. The UI further depicts the processes of cleaning the chemical structures (encoded as SMILES) prior to training the ML models. Input queries such as drug name and PubChem CID are converted to SMILES prior to processing. Each SMILES string input is subject to four different steps, namely, converting the SMILES into canonical SMILES,19 removing salts (if included), neutralizing formal charges (except permanent ones), and standardizing tautomers. REDIAL-2020 predicts the CPE, cytotox, AlphaLISA, TruHit, ACE2, and 3CL of the given compounds. The workflow of operations performed on the submitted query SMILES through the redial webapp are summarized in Figure 3.
Figure 4:
REDIAL-2020 submission page.
Figure 3:
REDIAL-2020 prediction workflow.
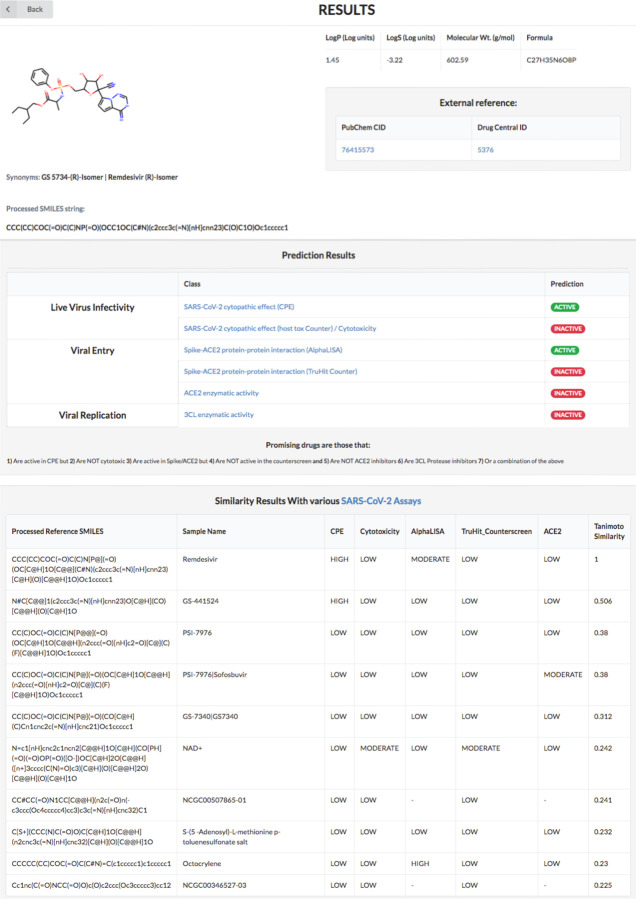
Figure 5 shows an output panel example, which is loaded on the same web page. REDIAL-2020 links directly to DrugCentral20,21 for approved drugs, and to PubChem for chemicals (where available), enabling easy access to additional information about the query molecule. Using REDIAL-2020 estimates, promising anti-SARS-CoV-2 compounds would be as follows: a) active in the CPE assay and b) inactive in the cytotox assay; or c) active in the AlphaLISA assay but d) inactive in the TruHit assay while e) not blocking ACE2; or f) active in the 3CL assay; or any combination of the above three. A schematic representation of the “best profile” that can be defined for a molecule, after running all the prediction models, is depicted in Figure 6.
Figure 5:
Screenshot of the webpage displaying ML estimates and similarity results for a query molecule.
Figure 6:
Schematic representation of the most desirable profile for anti-SARS-CoV-2 activities that can be observed via REDIAL-2020 predictions, based on the six assays of interest.
Similarity Search
For each query molecule, a fingerprint-based Tanomoto22 similarity search is conducted against molecules included in the model input datasets. For reference, we have used datasets from the NCATS COVID19 portal5 for the similarity search. The top 10 similar molecules to that of the query molecule, based on Tanimoto coefficient23 scores, are displayed (see Figure 5).
External Predictivity
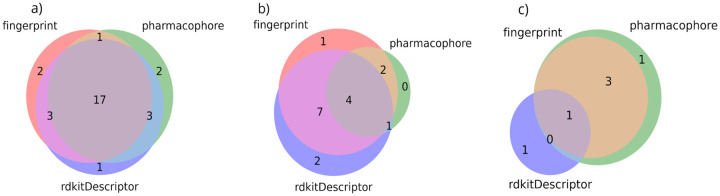
To confirm the utility of our models, we collected three additional datasets from the literature and submitted these molecules (external to our training/test sets) as input for prediction. First, we used a recently developed database for COVID-19 experiments24 to explore and download published in vitro COVID-19 bioactivity data for approved drugs which was reported in various recent papers.13,25–32 After removing drugs already included in the NCATS experiments, we identified 39 external drugs active in anti-SARS-CoV-2 CPE assays (Supporting Information Table SI1). Out of 39 drugs, 24 were predicted as active by at least two models and by the consensus model (Figure 7), and 15 drugs were predicted as inactive. Among those predicted to be inactive, independent experiments suggest they are weakly active. Specifically, bortezomib (S. Bradfute, personal communication), methotrexate, omeprazole, ouabain, and phenazopyridine (C. Jonsson, personal communication) show below 30% cell survival when tested at 10 μM. An additional set of six drugs from this list are undergoing testing in the Jonsson lab.
Figure 7:
Venn diagrams showing the distribution of external set predictions according to the ML models for a) CPE, 24 actives; b) CPE, 14 actives; c) 3CL, 6 actives. See the text for details.
The second external CPE set was collected from a recently published screen that included 21 compounds from the ReFRAME library,33 validated in dose response across multiple cell lines.32 Out of 21 most potent compounds, 19 were identified as an external set to our CPE model (Supporting Information Table SI2). Among these 19 compounds, 14 (~74%) were correctly predicted by consensus models (17 by at least one model) as active. The third dataset of 3CL (Mpro) inhibitors, identified13 6 inhibitors: ebselen(0.67 μM), disulfiram(9.35 μM), tideglusib(1.55 μM), carmofur(1.82 μM), shikonin(15.75 μM) and PX-12(21.39μM), respectively. Among these 6 inhibitors, our consensus 3CL model predicted 4 of them as actives, and all 6 of them as actives by at least one of the three 3CL ML models. The consensus model predicts all potent (activity less than 2μM) compounds correctly inhibitors, namely ebselen, tideglusib, and carmofur, respectively.
Thus, the REDIAL-2020 suite of models correctly predicts ~60% of the external compounds for the CPE assay, 74% of the external compounds for the ReFRAME library,33 and 66.67% of the external compounds for 3CL inhibitors13, respectively. Although these values appear to underestimate previous model performance in the validation and external sets (see Table 3), it has been noted that CPE experiments are affected by significant intra- and inter-experiment variability.24 Hence, we cannot exclude the possibility that some of the experiments performed by other laboratories are not directly comparable with results from the NCATS COVID19 portal.5
Table 3.
Confusion matrix values for each “best” model
| Model | Validation set results | Test set results | ||||||
|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | TP | TN | FP | FN | |
| CPE | 39 | 42 | 9 | 12 | 35 | 39 | 12 | 16 |
| cytotox | 95 | 97 | 27 | 30 | 85 | 90 | 35 | 40 |
| AlphaLISA | 78 | 71 | 24 | 16 | 71 | 73 | 21 | 24 |
| TruHit | 93 | 96 | 28 | 31 | 92 | 89 | 35 | 33 |
| ACE2 | 11 | 14 | 2 | 4 | 7 | 7 | 8 | 9 |
| 3CL | 29 | 24 | 9 | 4 | 24 | 21 | 12 | 10 |
TP, true positives; TN, true negatives; FP, false positives; FN, false negatives
Conclusion
Here, we described “REDIAL-2020”, an open-source, open-access, open-science application for estimating anti-SARS-CoV-2 activities from molecular structure. By leveraging the recently available data from NCATS, we developed six categorical ML models: CPE, cytotox, AlphaLISA, TruHit, ACE2, and 3CL. Input data from NCATS were used to train and validate multiple ML models using a variety of algorithms and features. The best performing models, in terms of F1 scores and test set predictions, were exposed on the REDIAL-2020 portal. Furthermore, a similarity search against the input data is conducted for every submitted molecule. The top 10 most similar molecules to the query molecule from the existing COVID-19 databases, together with associated experimental data, are displayed. This allows users to evaluate the confidence of the ML predictions.
REDIAL-2020 provides a fast and reliable way to screen novel compounds for anti-SARS-CoV-2 activities. These would be compounds that block live virus infectivity (CPE active but not cytotoxic); or compounds that block viral entry (blocking the Spike/ACE2 interaction while not interfering with the assay and lacking ACE2 inhibition properties); or compounds that block viral replication, by inhibiting 3CL (Mpro) protease; or a combination of the above, respectively. REDIAL-2020 is available on Github and Dockerhub as well, and the command-line version supports large scale virtual screening purposes. Future developments of REDIAL-2020 could include additional ML models based on, e.g., newly released TMPRSS2 inhibition assay34 data from the NCATS COVID19 portal, and perhaps other assay data as they become available in the public domain. We will continue to update and enhance the ML models, and make these models available in future releases of REDIAL-2020.
Methods
Data
Data for the SARS-CoV-2 cytopathic effect (CPE), the Vero E6 (host cell) cytotoxicity counterscreen, the Spike-ACE2 protein-protein interaction (AlphaLISA), TruHit (AlphaLISA counterscreen), ACE2 enzymatic inhibition conterscreen, and 3CL enzymatic inhibition were obtained from the NCATS COVID19 portal.4,5 Assay data were mapped to DrugCentral2020 to retrieve DrugCentral IDs, SMILES strings, and drug names. The experimental results from NCATS were mined to define activity and significance classes. The procedure for data extraction, wrangling, and post-processing are detailed as follows.
Mining NCATS COVID19 Data
All operations described below were performed using the Knime35 platform. NCATS data associated with the aforementioned assays was downloaded from the COVID19 portal.4,5 The files contained over 23,000 compounds generated by high-throughput screening (HTS) experiments. When possible, each compound was cross-linked to drugs annotated in DrugCentral, to separate approved drugs from the rest of the tested compounds. Matching NCATS compounds to DrugCentral was conducted in three sequential steps: by InChI (International Chemical Identifier),36 by synonym (name), and by matching CAS (Chemical Abstracts Service) Registry Numbers. First, NCATS molecules were matched by InChI. Molecules that did not match were then queried by drug name and associated synonyms, as annotated in DrugCentral. Finally, if not matched by either InChI or name, molecules were matched by CAS number. If none of the above steps resulted in a match, then the molecule in question was not classified as an approved drug. At the end of this process, 4,954 unique molecules (2,273 approved drugs and 2,681 chemicals) were stored. Whenever possible, for approved drugs and other chemicals, SMILES were retrieved from DrugCentral and from PubChem, respectively. Otherwise, the original SMILES strings were retained. Bioactivity data was mined according to the “curve class” and “maximum response” parameters.37 The “curve class” evaluates the quality of the dose-response curve and, consequently, the quality of the measurement. The “maximum response” is the maximum response value detected during the experiment. The “ACTIVITY CLASS” and a “SIGNIFICANCE CLASS” were defined using criteria described in Table 4 and Table 5, respectively. As a final data wrangling step, all compounds were categorized, and assay data grouped to have a unique record per molecule for each assay. When more than one assay was measured for the same molecule, only the datapoint with the best curve class was retained.
Table 4:
Criteria applied to each experimental dataset that define the various activity categories. Depending on the assay type, MAX RESPONSE could have either negative or positive values. In any case, since a greater (absolute) value always represents high activity, the absolute value of MAX RESPONSE is used in the definition of activity classes.
| Cutoff | ACTIVITY CLASS |
|---|---|
| MAX RESPONSE > 66 | HIGH |
| 33 ≥ MAX RESPONSE ≤ 66 | MODERATE |
| MAX RESPONSE < 33 | LOW |
Table 5:
The significance classes represent the quality of the measurement and are defined in accordance with the different shape of the dose-response curve (CURVE CLASS2).37
| CURVE CLASS2 | SIGNIFICANCE CLASS |
|---|---|
| 1.1 1.2 2.1 2.2 | HIGH |
| 1.3 1.4 2.3 3.4 | MODERATE |
| 3 | LOW |
| 4 | INACTIVEa |
| 5 | INCONCLUSIVE |
A type 4 curve is a flat curve, which indicates that the compound does not show any activity.
Data Filtering
For each assay, the data was labeled as positive and negative. The compounds with “LOW” activity class were treated as negative, whereas “HIGH” and “MODERATE” were treated as positive compounds. Molecular structures were standardized using five different filters using RDKit38 features: (i) SMILES strings were converted into canonical SMILES strings. Some of the SMILES were not converted SMILES, were discarded. (ii) RDKit Salt Stripper was implemented to obtain the salt stripped molecules. The “donotRemoveEverything” feature helps by leaving the last salt when the entire canonical SMILES string contains only the salts, (iii) The RDKit “Uncharger” feature neutralizes the molecules by adding or removing hydrogen atoms, (iv) The canonical tautomer was used obtained from RDKit, and (v) logP and logS filters were defined as follows: (logP < 1 and logP > 9) and (logS > −3 and logS < −7.5). Such thresholds were arbitrarily defined to maximize the number of negative data excluded while minimizing the positive data excluded. However, for the 3CL dataset, logP and logS filters were not implemented. The final task was the removal of duplicate compounds resulting from desalting, neutralizing, and standardizing tautomers.
Molecular Descriptors
A total of 22 features of three distinct types (fingerprints-based, pharmacophore-based, and physicochemical descriptors based) were implemented. Fingerprints were converted into a bit vector of either 1,024 or 16,384 lengths. Pharmacophore type was also a bit vector of size 2,692, whereas RDKit and VolSurf+ descriptors were of length 200 and 128, respectively.
Fingerprints-based:
This includes the circular, path-based, and substructure keys.39,40 Circular fingerprints include the extended-connectivity fingerprints (ECFPx) and feature-connectivity fingerprints (FCFPx), where x is 0, 2, 4, and 6 are the bond length or diameter for each circular atom environment. ECFP consists of the element, number of heavy atoms, isotope, number of hydrogen atoms, and ring information, whereas FCFP consists of pharmacophore features.
Substructure keys Avalon and the public Molecular ACCess System (MACCS) are two distinct types of fingerprints that are substructure keys. The Avalon fingerprint, used here, is a bit vector of size 1,024. It includes feature classes such as atom count, atom symbol path, augmented atom, and augmented symbol path, etc. MACCS structural keys are 166-bit structural key descriptors. Each bit here is associated with a SMARTS pattern and belongs to the dictionary-based fingerprint class. Path-based fingerprints include RDKx (where x is 5, 6, 7), topological torsion (TT), HashTT, atom pair (AP), and HashAP. The size of each fingerprint is 1024 for all of them.
A longer version of the fingerprint, of 16384 bits, was also used for comparison. This longer version is represented by the prefix” L”: LAvalon, LECFP6, LECFP4, LFCFP6 and LFCFP4.
Pharmacophore-based:
Topological pharmacophore atomic triplets fingerprints (TPATF) were obtained using maychemtools.41 TPATF describes the ligand sites that are necessary for molecular recognition of a macromolecule or a ligand, and passes that information to the ML model to be trained. Ligand SMILES strings were passed through a Perl script to generate TPATF. The basis sets of atomic triplets were generated using two different constraints (i) triangle rule, i.e., the length of each side of a triangle cannot exceed the sum of the lengths of the other two sides; and (ii) elimination of redundant pharmacophores related by symmetry. The default pharmacophore atomic types Hydrogen Bond Donor (HBD), hydrogen bond acceptor (HBA), positively ionizable (PI), negatively ionizable (NI), H (hydrophobic), and Ar (aromatic) were used during generation of TPATF.42
Physicochemical descriptors:
This includes the RDKit molecular descriptors and VolSurf+ descriptors. For RDKit descriptors, a set of 200 descriptors were used, which were obtained from RDKit.38 They are either experimental properties or theoretical descriptors, which are e.g. molar refractivity, logP, heavy atom counts, bond counts, molecular weight, topological polar surface area.
A total of 128 descriptors were obtained using VolSurf+ software. VolSurf+ is a computational approach aimed at describing the structural, physicochemical and pharmacokinetic features of a molecule starting from a 3D map of the interaction energies between the molecule and chemical probes (GRID-based molecular interaction fields, or MIFs).43 VolSurf+ compresses the information present in MIFs into numerical descriptors, which are simple to use and interpret.16
Model Development
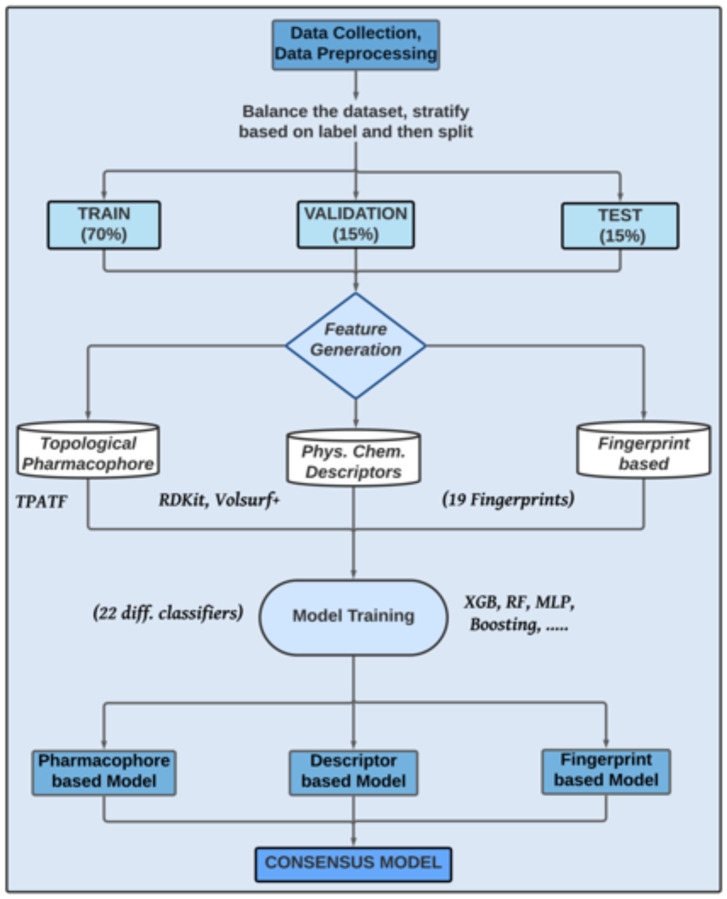
For each classifier, several machine learning models were developed, employing three categories of features and 22 distinct machine learning algorithms from the scikit-learn package.18 Figure 8 briefly shows the workflow of the model generation. The three different categories of features employed were 1) chemical fingerprints, 2) physicochemical descriptors, and 3) topological pharmacophore descriptors. For fingerprint-based descriptors, 19 different RDKit fingerprints were tested. For physicochemical descriptors, Volsurf+ and RDKit descriptors were employed. For Topological pharmacophore descriptors, TPATF fingerprints from Mayachemtools were employed. For each model, input data was split into a 70% training set, 15% validation set, and 15% test set using a stratified sampling. Table 6 depicts the number of compounds used in training, validation, and test sets for each model. Compounds discussed in the External Predictivity section (Figure 7) were not part of this workflow and are not included in Table 6.
Figure 8.
ML model development workflow.
Table 6:
Summary of datasets used for each model.
| Type | Count | Total Actives | Training count | Validation count | Test count | Training Actives | Validation Actives | Test Actives |
|---|---|---|---|---|---|---|---|---|
| CPE | 736 | 368 | 532 | 102 | 102 | 266 | 51 | 51 |
| cytotox | 1662 | 877 | 1163 | 249 | 250 | 581 | 125 | 125 |
| AlphaLisa | 1260 | 630 | 882 | 189 | 189 | 441 | 94 | 95 |
| TruHit | 1658 | 829 | 1161 | 248 | 249 | 580 | 124 | 125 |
| ACE2 | 206 | 103 | 144 | 31 | 31 | 72 | 15 | 16 |
| 3CL | 442 | 221 | 309 | 66 | 67 | 154 | 33 | 34 |
All of the models were trained leveraging 22 different classifiers available in scikit-learn. Initially, 968 different models were trained using 22 classifiers (Complement Naive Bayes, Extreme Gradient Boosting, KNeighbors, Gradient Boosting, Perceptron, One Vs Rest , Extra-Tree, Ridge, One Vs One, Bagging, Random Forest, Output Code, Passive Aggressive, Linear SVC, Stochastic Gradient Descent, Logistic Regression, Extra Trees, Multinomial Naive Bayes, Ada Boost, Decision Tree, Nearest Centroid, Multi-layer perceptron) and 22 distinct features (ECFP0, ECFP2, ECFP4, LECFP4, ECFP6, LECFP6, FCFP2, FCFP4, LFCFP4, FCFP6, LFCFP6, RDK5, RDK6, RDK7, Avalon, LAvalon, MACCS, HashTT, HashAP, VolSurf+, TPATF, and RDKit descriptors) based on the default configurations of the classifiers. Finally, the best suited combination of classifiers and features were selected for hyperparameter tuning.
Similarity Search
We used an ECFP4 bit vector fingerprint with 1024 bits, and TC calculations, for the fingerprints present in the database along with that of a query molecule, are computed on the fly. The Tanimoto coefficient represents the overlap of features between molecules as the ratio of the number of common features to the total number of features in each fingerprint. The coefficient ranges from 0 to 1, with 1 corresponding to identical fingerprints.
Hyperparameter Optimization
The best performing default models based on validation sets were selected for hyperparameter optimization. These models were optimized using a grid search method. Scikit-learn provides the package for grid search hyperparameter optimization using cross validation. However, it is slow and does not offer a grid search with a separate validation set. Thus, we used a freely available software “hypopt” for the hyperparameter tuning of each model.44 The models were optimized and selected based on the validation F1 score. The outperforming models were saved and used for the evaluation of external datasets.
Evaluation Metrics
The model performances were evaluated by five distinct evaluation metrics available in scikit-learn: accuracy, recall, precision, F1 score, and area under the receiver operating characteristic (ROC) curve (AUC). In general, the accuracy measures the total number of correct predictions among the total numbers of instances evaluated. The recall was used to measure the fraction of total positives that are correctly classified, whereas precision estimates the fraction of total positives that are correctly predicted from the total predicted positives. F1 score is the harmonic mean of recall and precision. The score ranges from 0 to 1 for each of the metrics, where 1 is a perfect score. Moreover, AUC was computed for each of the classifiers. A perfect classifier yields an AUC of 1, whereas a random model has an expected AUC of 0.5.
Acknowledgement
We thank the High-Performance Computing support staff (Marc T. Hertlein and Leopoldo A. Hernandez) at The University of Texas at El Paso for assistance in using the Chanti cluster. We also acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper. URL: http://www.tacc.utexas.edu. Access to unpublished SARS-CoV-2 experimental data from Dr. Colleen Jonsson (University of Tennessee Health Sciences Center) and Dr. Steven Bradfute (University of New Mexico Health Sciences Center) is gratefully acknowledged.
Funding
Dr. Sirimulla acknowledges support from the National Science Foundation through NSF-PREM grant #DMR-1827745. The DrugCentral component of this work is funded by NIH Common Fund U24 CA224370.
Footnotes
Web Portal
REDIAL-2020 is available online at http://drugcentral.org/Redial.
Code Availability
All of the codes and the trained models are available at https://github.com/sirimullalab/redial-2020.
Conflicts of Interest
T.I.O. has received honoraria or consulted for Abbott, AstraZeneca, Chiron, Genentech, Infinity Pharmaceuticals, Merz Pharmaceuticals, Merck Darmstadt, Mitsubishi Tanabe, Novartis, Ono Pharmaceuticals, Pfizer, Roche, Sanofi and Wyeth. He is on the Scientific Advisory Board of ChemDiv Inc. and InSilico Medicine.
References
- 1.Gorshkov K. et al. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv (2020) doi: 10.1101/2020.05.16.091520 [DOI] [Google Scholar]
- 2.Sun H., Wang Y., Cheff D. M., Hall M. D. & Shen M. Predictive models for estimating cytotoxicity on the basis of chemical structures. Bioorg. Med. Chem. 28, 115422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson Q. M. et al. Targeting ACE2-RBD interaction as a platform for COVID19 therapeutics: Development and drug repurposing screen of an AlphaLISA proximity assay. bioRxiv (2020) doi: 10.1101/2020.06.16.154708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brimacombe K. R. et al. An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv (2020) doi: 10.1101/2020.06.04.135046 [DOI] [Google Scholar]
- 5.NCATS OpenData COVID-19. https://opendata.ncats.nih.gov/covid19/assays.
- 6.Huang R. et al. Massive-scale biological activity-based modeling identifies novel antiviral leads against SARS-CoV-2. bioRxiv (2020) doi: 10.1101/2020.07.27.223578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S. et al. PubChem Substance and Compound databases. Nucleic Acids Res. 44, D1202–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988). [Google Scholar]
- 9.Oprea T. I. et al. Associating drugs, targets and clinical outcomes into an integrated network affords a new platform for computer-aided drug repurposing. Mol. Inform. 30, 100–111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M. et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millet J. K. & Whittaker G. R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 517, 3–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keidar S., Kaplan M. & Gamliel-Lazarovich A. ACE2 of the heart: from angiotensin I to angiotensin (1--7). Cardiovasc. Res. 73, 463–469 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Jin Z. et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y. & Jung S.-H. An Overview of Severe Acute Respiratory Syndrome–Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem. 59, 6595–6628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carey R. N., Wold S. & Westgard J. O. Principal component analysis. Alternative to referee methods in method comparison studies. Anal. Chem. 47, 1824–1829 (1975). [DOI] [PubMed] [Google Scholar]
- 16.Cruciani G., Pastor M. & Guba W. VolSurf: a new tool for the pharmacokinetic optimization of lead compounds. Eur. J. Pharm. Sci. 11 Suppl 2, S29–39 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Tetko I. V. et al. Virtual computational chemistry laboratory--design and description. J. Comput. Aided Mol. Des. 19, 453–463 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Pedregosa F. et al. Scikit-learn: Machine learning in Python. the Journal of machine Learning research 12, 2825–2830 (2011). [Google Scholar]
- 19.Weininger D., Weininger A. & Weininger J. L. SMILES. 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Comput. Sci. 29, 97–101 (1989). [Google Scholar]
- 20.Ursu O. et al. DrugCentral 2018: an update. Nucleic Acids Res. 47, D963–D970 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ursu O. et al. DrugCentral: online drug compendium. Nucleic Acids Res. 45, D932–D939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers D. J. & Tanimoto T. T. A Computer Program for Classifying Plants. Science 132, 1115–1118 (1960). [DOI] [PubMed] [Google Scholar]
- 23.Whittle M., Gillet V. J., Willett P., Alex A. & Loesel J. Enhancing the effectiveness of virtual screening by fusing nearest neighbor lists: a comparison of similarity coefficients. J. Chem. Inf. Comput. Sci. 44, 1840–1848 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Kuleshov M. V. et al. The COVID-19 Gene and Drug Set Library. Res Sq (2020) doi: 10.21203/rs.3.rs-28582/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon S. et al. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs. Antimicrob. Agents Chemother. 64, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weston S., Haupt R., Logue J., Matthews K. & Frieman M. B. FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2 in vitro. 2020.03.25.008482 (2020) doi: 10.1101/2020.03.25.008482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Touret F. et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 10, 13093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing J. et al. Reversal of Infected Host Gene Expression Identifies Repurposed Drug Candidates for COVID-19. bioRxiv (2020) doi: 10.1101/2020.04.07.030734 [DOI] [Google Scholar]
- 29.Riva L. et al. A Large-scale Drug Repositioning Survey for SARS-CoV-2 Antivirals. bioRxiv (2020) doi: 10.1101/2020.04.16.044016 [DOI] [Google Scholar]
- 30.Choy K.-T. et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 178, 104786 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirabelli C. et al. Morphological Cell Profiling of SARS-CoV-2 Infection Identifies Drug Repurposing Candidates for COVID-19. bioRxiv (2020) doi: 10.1101/2020.05.27.117184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riva L. et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature (2020) doi: 10.1038/s41586-020-2577-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janes J. et al. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc. Natl. Acad. Sci. U. S. A. 115, 10750–10755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrimp J. H. et al. An Enzymatic TMPRSS2 Assay for Assessment of Clinical Candidates and Discovery of Inhibitors as Potential Treatment of COVID-19. bioRxiv (2020) doi: 10.1101/2020.06.23.167544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berthold Michael R., Cebron N., Dill F. & Others. KNIME: the Konstanz Information Miner Studies in Classification, Data Analysis, and Knowledge Organization. Springer. ISSN; 1431–8814 (2007). [Google Scholar]
- 36.Heller S. R., McNaught A., Pletnev I., Stein S. & Tchekhovskoi D. InChI, the IUPAC International Chemical Identifier. J. Cheminform. 7, 23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seethala R. & Zhang L. Handbook of Drug Screening. (CRC Press, 2016). [Google Scholar]
- 38.Landrum G. & Others RDKit: Open-source cheminformatics. (2006). [Google Scholar]
- 39.Riniker S. & Landrum G. A. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminform. 5, 26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers D. & Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Sud M. MayaChemTools: An Open Source Package for Computational Drug Discovery. J. Chem. Inf. Model. 56, 2292–2297 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Bonachéra F., Parent B., Barbosa F., Froloff N. & Horvath D. Fuzzy tricentric pharmacophore fingerprints. 1. Topological fuzzy pharmacophore triplets and adapted molecular similarity scoring schemes. J. Chem. Inf. Model. 46, 2457–2477 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Goodford P. J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 28, 849–857 (1985). [DOI] [PubMed] [Google Scholar]
- 44.Hypopt. https://github.com/cgnorthcutt/hypopt.