Abstract
Corona Virus Disease 19 (COVID-19) pandemic has created an alarming situation across the globe. Varieties of diagnostic protocols are being developed for the diagnosis of COVID-19. Many of these diagnostic protocols however, have limitations such as for example unacceptable no of false-positive and false-negative cases, particularly during the early stages of infection. At present, the real-time (quantitative) reverse transcriptase-polymerase chain reaction (RT-PCR) is considered the gold standard for COVID-19 diagnosis. However, RT-PCR based tests are complex, expensive, time consuming and involve pre-processing of samples. A swift, sensitive, inexpensive protocol for mass screening is urgently needed to contain this pandemic. There is urgent need to harness new powerful technologies for accurate detection not only of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) but also combating the emergence of pandemics of new viruses as well. To overcome the current challenges, the authors propose a diagnostic protocol based on Surface-enhanced Raman Spectroscopy (SERS) coupled with microfluidic devices containing integrated microchannels functionalized either with vertically aligned Au/Ag coated carbon nanotubes or with disposable electrospun micro/nano-filter membranes. These devices have the potential to successfully trap viruses from diverse biological fluids/secretions including saliva, nasopharyngeal, tear etc. These can thus enrich the viral titre and enable accurate identification of the viruses from their respective Raman signatures. If the device is successfully developed and proven to detect target viruses, it would facilitate rapid screening of symptomatic as well as asymptomatic individuals of COVID-19. This would be a valuable diagnostic tool not only for mass screening of current COVID-19 pandemic but also in viral pandemic outbreaks of future.
Keywords: Raman Spectroscopy, SARS-CoV-2, Microfluidic Platforms
Background of hypothesis
Rapid spread of the COVID-19 pandemic has created alarming situation all over the world. Originating in China, it has rapidly spread like a wild fire all over the world, thus ravaging the healthcare systems and economies of several countries. This pandemic has far reaching consequences even beyond its physical spread as it has severely dented global economic structure. This unprecedented situation underscores the urgent need for novel rapid diagnostic protocols that could be used to screen symptomatic as well as asymptomatic persons who could be potential spreaders. It would be highly desirable to develop long lasting, reliable diagnostic assays for detection of not only the current pathogens but also those virus variants that might emerge in future.
Corona viruses have a larger genome made up of ribonucleic acids (RNA) of size ranging between 26.4 and 32.7 kilo bases (kb) [1]. These viruses are generally spherical in shape with the diameter of ~150–160 nm (nm) [2]. These Corona viruses are members of Severe Acute Respiratory Syndrome (SARS) virus family, contains specific proteins responsible for membrane (M), nucleocapsid (N), envelope (E), and spike (S). Some strains of Sever Acute Respiratory Syndrome Corona Virus (SARS-CoV) encode a special enzyme called hemagglutinin esterase (HE) protein [2]. The binding of SARS-CoV-2 with angiotensin-converting enzyme 2 (ACE2) through S protein enables the entry of it into the host cells. The activation of viruses with the help of host cellular proteases called type II transmembrane serine protease (TMPRSS2) is suggested to result in human-to human transmission [3].
Currently several diagnostic test assays are available for detection and confirmation of SARS-CoV-2 infections. These diagnostic tests mainly fall into two categories i.e. molecular assay, and serological and immunological assays [4]. Most molecular assays require expensive equipment, highly skilled personnel to detect assay signals. The immuno assays can vary in sensitivity and specificity, and are limited by the delay in time between initial virus infection and anti-viral antibody production, which can result in detection of only a subset of SARS-CoV-2 infections giving positive results at the time of testing. In addition, assay times for existing genomic profile and serological tests can take three hours to as long as five days. Most of the existing diagnostic techniques such as RT-PCR technique, and enzyme-linked immunosorbent assay (ELISA) are quite expensive.
In this paper, we present our hypothesis for development of novel prototype of microchannel integrated platforms that are coupled either with Ag-functionalized cellulose based surface-enhanced Raman scattering strips or with Au/Ag functionalized carbon nanotubes for subsequent detection of SARS-CoV-2 in the body fluids such as tears, saliva, nasal and throat swabs by using Surface-enhanced Raman Spectroscopy. Such platforms, in our opinion, would highly be rapid and robust device that can capture SARS- CoV-2 based on its size and facilitate detection of virus in real time directly from the samples. No pre-processing or labeling of the sample would be necessary prior to detection, thus making it a simple and rapid protocol. It will have high sensitivity of detection because of two features: trapping of viruses will help concentrating the sample at detection area. Use of plasmonic material such as Au/Ag would enhance “Raman Signal” (RS) intensity. Because of the high sensitivity of the assay, the protocol has the potential to improve the range of detection of minute traces of SARS- CoV-2 in nasal, throat, saliva and conjunctival swabs. As asymptomatic individuals could be potential spreaders, reducing the threshold of detection could greatly help to combat the rapid spread of COVID-19.
To the best of our knowledge, this will be the first and a novel approach to develop a diagnostic technique based on the Surface-enhanced Raman Spectroscopy coupled with microfluidic platform for COVID-19 diagnosis by using tear, saliva, nasal and throat swabs.
Statement to hypothesis
The authors of the collaborative study propose a diagnostic tool based on SERS coupled with microfluidic devices containing integrated microchannels functionalized either with disposable electrospun micro/nano-filter membranes or with vertically aligned Au/Ag coated carbon nanotubes for the detection of SARS-CoV-2.
SERS is a very selective and highly sensitive technique which can be employed in pathogen identification. SERS enhances the Raman effect, using plasmonic properties of metallic nanostructures [5]. At the early stage of a disease, when the biomarkers are significantly low in quantity, they are brought in the proximity of metallic nanostructures under optimum conditions, inherent Raman scattering of the particles is enhanced. The vibration signatures produced by SERS provide information on molecular and structural details of the biological samples [5]. Earlier reports suggest that viruses can be captured between aligned carbon nanotubes, when their sizes match the intertubular distance [6], [7]. This can be achieved without any specific labeling while keeping the virus viable and its structure intact, obtaining 106–109 times signal amplification. Carbon nanotube captured virus is characterized for its surface antigens using Raman spectra for comparison with various pathogens [6], [7].
Therefore, upon coupling with plasmonic materials such as gold nanoparticles, SERS has the ability to detect a single molecule with the highest level of sensitivity, and applicability to detect the live cells without staining or labeling even at lower concentration of virus in the sample. A study reported by Li and co-workers showed the presence of few electrostatic hotspots for the SARS-CoV that can be detected by Raman Spectroscopy [8]. Therefore, the authors believe that the detection of SARS-CoV-2 can be performed based on the electrostatic signals generated via its antigen interactions. Thus, successful identification of one of the sites of characteristic electrostatic interactions can lead to a reliable and highly sensitive rapid detection method for SARS-CoV-2.
Yeh and co-workers demonstrated the use of carbon nanotube arrays for the effective trapping (based on size) and label free detection of virus using respective Raman signature [6], [7]. Thus, the authors believe that trapping of SARS-CoV-2 viruses by using similar, and appropriately modified microfluidic devices coupled with SERS will be useful in detecting COVID-19 viruses from clinical samples. Further, it is speculated that such a protocol might attract significant diagnostic value and clinical potential.
A few previous studies reported that once SARS-CoV-2 virus gets established in the intra-ocular region, it might then spread to the respiratory tract via the nasolacrimal duct - a duct connecting the ocular space to the nasal passages. It has also been observed that the COVID-19 virus can spread from the respiratory tract to the intra-ocular region [9]. Most of respiratory viruses have been documented to possess ocular tropism, causing ocular complications in infected individuals and establishing a respiratory infection following ocular exposure [10]. The novel coronavirus RNA was also detected in tears and conjunctival samples from infected individuals [11]. The conjunctival epithelium and conducting airways appear to be potential portals of infection for SARS-CoV-2 [12]. The presence of ACE2 and TMPRSS2 in conjunctival and corneal epithelial cells suggest the ocular surface as a secondary site of infection following respiratory tract, or possibly even as the initial portal of entry to an individual [13]. Therefore, the authors believe that it is important to check the presence of SARS-CoV-2 in the tear sample along with other systemic fluids. A recent report from Tata Memorial Center (TMC) group showed the presence of RNA in saliva by using Raman Spectroscopy [14]. Thus, it is worth collecting and analyzing tear and saliva samples along with the nasal and throat swabs for the detection of SARS-CoV-2. The authors believe that the proposed technique is expected to have an ability to detect mild and asymptomatic cases via SERS. It would thus be helpful in early diagnosis and contact tracing, which will be an essential step in preventing the silent spread of the virus.
The aim is to develop the prototype which facilitates size dependent trapping of SARS-CoV-2 from the body fluid samples in microfluidic device and identification of the virus by using Surface-enhanced Raman Spectroscopy. This procedure has certain advantages. The device permits increasing concentration of the virus in the sample (without ultra centrifugation) to facilitate detection. Raman spectroscopic detection takes very short time (~few mins) and needs lesser operating cost than any other current tools. Yeh et al have shown that viruses can be accurately distinguished on the basis of their Raman signature [6], [7]. Recent paper has shown this to be true for SARS-CoV-2 [14]. At present RT PCR is considered as a gold standard protocol, therefore validation of our prototype with the existing tool is necessary to determine the specificity and sensitivity of the newly proposed tool. The proposed method can be applicable for detection of SARS-CoV-2 as well as its variants (mutated forms), as it is based on physical properties of the virus which are less likely to be altered by mutations. At present this prototype will be developed for COVID-19 diagnosis, with appropriate alterations and functionalization of the sensing materials, carbon nanotubes porosity and geometry. It would be useful in detection of any other similar viruses as well as bacteria in future. Therefore, the authors propose to use two models; one with gold and other with silver nanoparticles as plasmonic material.
Proposed prototype
The two following proposed models are based on the unique concept of SERS coupled with microfluidic device for the detection of COVID-19 from biological fluids.
Model 1
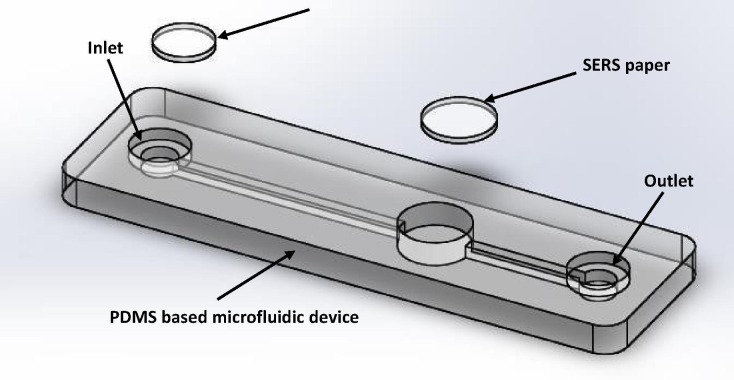
A cost-effective Polydimethylsiloxane (PDMS) based microfluidic device with integrated disposable type micro/nanofilter and SERS strips is proposed for rapid screening of asymptomatic COVID-19 patients. Fig. 1 depicts the basic design of disposable type PDMS microfluidic device. In the microfluidic platform, the microchannels will be integrated with disposable electrospun micro/nano-filter membrane and the detection well can be functionalized with a disposable Ag functionalized cellulose paper based SERS strip. The overall PDMS microfluidic device will be fabricated using 3D printing process to facilitate batch production. Initial trials will be tested using simulated body fluids.
Fig. 1.
Proposed microfluidic platform for virus detection.
Model 2
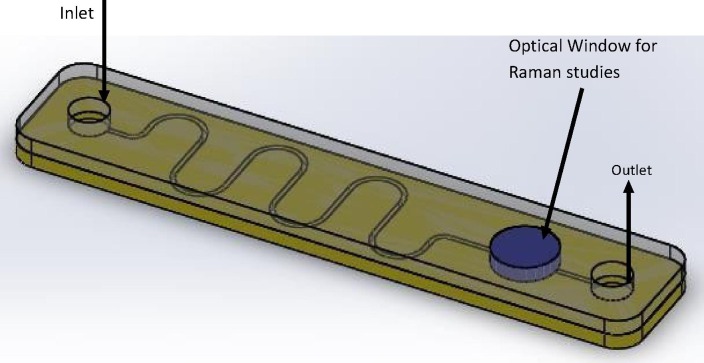
Integrated microfluidic platforms with vertically aligned carbon nanotubes (VACNTs) functionalized microchannel, which can be used for high-risk patients with COVID-19 symptoms. Fig. 2 depicts the basic design of VACNTs integrated microfluidic device.
Fig. 2.
Proposed VACNTs functionalized microfluidic platform.
Apart from this application, the proposed devices would be a universal detection platform, and can be extended for various types of biosensor applications with suitable functionalization of the sensing materials.
Application of Raman spectroscopy for the detection of SARS-CoV-2
The Ag/Au nanoparticles present in the detection well will enhance the Raman signal. Raman signal will be analyzed via machine learning algorithms that can be trained on the signals of known virus molecules. Spectrum of test sample will be matched with previously recorded standard reference spectrum of SARS-CoV-2.
The device essentially traps viruses in nanotubes. Au/Ag-VACNTs not only help to segregate but also concentrate the viruses from samples (on repeated applications). This localized increase in the titre helps in accurate identification and enhances the sensitivity of the assay. Presence of plasmonic AU-NPs further enhances the RS. The trapped viruses can be eluted and characterized either by sequencing or by other molecular analyses, if necessary, to confirm the specificity of the target virus. Importantly, this assay does not involve labeling of virus particles and processing of the samples prior to introduction into the device. As consequent detection of SERS takes only a few minutes the proposed tool may become a robust, reliable, highly sensitive and very rapid detection method of choice across the globe. Hence, we believe that this assay would provide a viable, long term alternative for reliable diagnosis of viral identity. Further, positive outcome of this tool will facilitate designing and manufacturing of versatile, portable platforms to counteract future outbreak of viral epidemics with steep reduction in the response time.
Conclusion
It is believed that this technique would provide a viable, long term alternative for reliable diagnosis of SARS-CoV-2 detection. Importantly, positive outcome of any of these two prototypes will facilitate the design and manufacture of versatile, portable platforms to contain future outbreaks of viral epidemics with steep reduction in the response time.
Limitations of concept
Validation of the prototypes is needed in the initial stage in determining the specificity and sensitivity of the proposed prototypes as against gold standard methods such as RT-PCR.
Conflict of interest
All authors declare no conflict of interest
Funding sources
No funding was availed to disclose
Author contribution
The authors of the study jointly proposed this diagnostic tool for the detection of SARS-CoV, and are planning to submit this concept as a multicentric proposal for seeking research grant to Indian Government funding agencies. Instead of individual authors, the role of each organization is listed as follows;
-
1.Aditya Jyot Foundation For Twinkling Little Eyes
-
•Conceptualization of prototype
-
•Literature survey
-
•Preparation of manuscript
-
•Development of proposal
-
•
-
2.PSG Institute of Advanced Studies
-
•Designing and Development of microfluidic device
-
•Preparation of manuscript
-
•Provision of Technical support
-
•
-
3.PSG Institute of Technology and Applied Research
-
•Designing and Development of microfluidic device
-
•Preparation of manuscript
-
•Provision of Technical support
-
•
-
4.Advanced Centre for Treatment Research and Education in Cancer
-
•Preparation of manuscript
-
•Provision of Technical support for Raman Spectroscopy
-
•
References
- 1.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J Microbiol Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. Available from: http://www.sciencedirect.com/science/article/pii/S1684118220300827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kannan S, Ali PSS, Sheeza A, Hemalatha K. COVID-19 (Novel Coronavirus) - Recent trends. Eur Rev Med Pharmacol Sci. 2020;24:2006–11. [DOI] [PubMed]
- 3.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a C. [DOI] [PMC free article] [PubMed]
- 4.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J. Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci [Internet] 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. Available from: https://doi.org/10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore T.J., Moody A.S., Payne T.D., Sarabia G.M., Daniel A.R., Sharma B. In vitro and in vivo SERS biosensing for disease diagnosis. Biosensors. 2018;8(2) doi: 10.3390/bios8020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh Y.-T., Tang Y.i., Sebastian A., Dasgupta A., Perea-Lopez N., Albert I. Tunable and label-free virus enrichment for ultrasensitive virus detection using carbon nanotube arrays. Sci Adv. 2016;2(10):e1601026. doi: 10.1126/sciadv.1601026. Available from: https://advances.sciencemag.org/lookup/doi/10.1126/sciadv.1601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeh Y.-T., Gulino K., Zhang Y., Sabestien A., Chou T.-W., Zhou B. A rapid and label-free platform for virus capture and identification from clinical samples. Proc Natl Acad Sci U S A. 2020;117(2):895–901. doi: 10.1073/pnas.1910113117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W. Structural Identification of the Electrostatic Hot Spots for Severe Acute Respiratory Syndrome Coronavirus Spike Protein to Be Complexed with Its Receptor ACE2 and Its Neutralizing Antibodies. 2020 Feb 18 [cited 2020 Sep 28]; Available from: https://www.preprints.org/manuscript/202002.0265/v1.
- 9.DeBroff B.M. COVID-19: ocular manifestations, ocular secretions, and ocular portal of entry. Adv Ophthalmol Vis Syst [Internet] 2020;10(2):48–49. Available from: http://medcraveonline.com. [Google Scholar]
- 10.Belser J.A., Rota P.A., Tumpey T.M. Ocular tropism of respiratory viruses. Microbiol Mol Biol Rev. 2013;77(1):144–156. doi: 10.1128/MMBR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia J., Tong J., Liu M., Shen Y.e., Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS‐CoV‐2 infection. J Med Virol. 2020;92(6):589–594. doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hui K.P.Y., Cheung M.-C., Perera R.A.P.M., Ng K.-C., Bui C.H.T., Ho J.C.W. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med [Internet] 2020;8(7):687–695. doi: 10.1016/S2213-2600(20)30193-4. Available from: https://doi.org/10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L., Xu Z., Castiglione G.M., Soiberman U.S., Eberhart C.G., Duh E.J. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul Surf [Internet] 2020;18(4):537–544. doi: 10.1016/j.jtos.2020.06.007. Available from: https://pubmed.ncbi.nlm.nih.gov/32544566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai S., Vatsa Mishra S., Joshi A., Sarkar D., Hole A., Mishra R. Raman Spectroscopy based detection of RNA viruses in saliva: a preliminary report. J Biophotonics [Internet] 2020 doi: 10.1002/jbio.202000189. Available from: https://europepmc.org/articles/PMC7361326. [DOI] [PMC free article] [PubMed] [Google Scholar]