Abstract
White sponge nevus (WSN) is a benign autosomal dominant disorder characterized by the formation of white spongy plaques in the oral mucosa. Keratin (KRT) 13 is highly expressed in the mucosa, and mutations in this gene have been commonly associated with WSN patients. However, it remains unknown whether there is a causal relationship between KRT13 mutations and WSN and what the underlying mechanisms might be. Here, we use mouse genetic models to demonstrate that Krt13 is crucial for the maintenance of epithelial integrity. Krt13 knockout mice show a WSN-like phenotype in several tissues, including the tongue, buccal mucosa, and esophagus. Transcriptome analyses uncover that Krt13 regulates a cohort of gene networks in tongue epithelial cells, including epithelial differentiation, immune responses, stress-activated kinase signaling, and metabolic processes. We also provide evidence that epithelial cells without Krt13 are susceptible to mechanical stresses experienced during postnatal life, resulting in unbalanced cell proliferation and differentiation. These data demonstrate that Krt13 is essential for maintaining epithelial homeostasis and loss of Krt13 causes the WSN-like phenotype in mice.
Keywords: Keratin, mucosa, epithelial cell, white sponge nevus, mouse model
INTRODUCTION
White sponge nevus (WSN) is a condition characterized by the formation of white thickened patches on the non-keratinized stratified squamous epithelium. Lesions are most commonly found in oral mucosa, including the buccal mucosa, tongue, floor of the mouth, and alveolar mucosa. They are less frequently found in extra-oral sites, such as the esophagus and urogenital mucosa (Frithiof and Bánóczy, 1976; Jorgenson and Levin, 1981; Cai et al., 2015). WSN is a benign overgrowth of the epithelial cells and generally asymptomatic, but the folds in the tissue can promote bacterial/fungal growth and lead to infection and pain (Sadeghi and Witkop, 1979; Marrelli et al., 2012). Also, the change in physical appearance, especially in the oral mucosa, can be troublesome to some patients. Available treatments, such as antibiotics or antifungal mouth rinse, are limited to treat the infection but do not affect the course of the disease (Otobe et al., 2007; Dufrasne et al., 2011; Satriano et al., 2012). There is a great need for better understanding of the disease mechanisms so that effective treatments can be developed.
The pathogenic changes of WSN show typical characteristics of epidermal keratin disorders. Indeed, studies in human patients and animal models over the past two decades have identified a keratin pair, keratin (KRT) 4 and 13, as central components in the WSN pathogenesis. A 3-bp deletion in KRT4 that removes an asparagine residue and a point mutation in KRT13 that causes a leucine-to-proline substitution were first reported in families affected by WSN (Richard et al., 1995; Rugg et al., 1995). Since then, many missense mutations in the KRT4 and KRT13 genes have been reported in WSN patients (Cai et al., 2015). The causal relationship between KRT4 and WSN has been demonstrated by mouse genetic studies as mice with the targeted deletion of Krt4 or the ENU-induced point mutation in Krt4 show WSN-type of lesions in lingual mucosa and other epithelial tissues (Ness et al., 1998; McGowan et al., 2007). However, there is no evidence demonstrating a causal relationship between KRT13 and WSN. Also, how defects in KRT4/KRT13 intermediate filaments might lead to the development of WSN is unknown.
To determine the role of KRT13 in WSN, we have used CRISPR/Cas9 to generate a novel mouse line carrying a Krt13 allele with a premature stop codon inserted in exon 1. Our analyses, including DNA sequencing, qRT-PCR, western blotting, and immunostaining on tissue sections, confirm that the mutated allele of Krt13 introduced by CRISPR/Cas9 is a knockout (KO) allele. We find that Krt13 KO mice recapitulate the phenotypes of WSN patients in multiple tissues, including the tongue, buccal mucosa, and esophagus. We provide evidence that epithelial cells without Krt13 fail to tolerate mechanical stresses experienced in postnatal life, resulting in an imbalance between cell differentiation and proliferation. Furthermore, our unbiased transcriptome analyses uncover novel regulatory networks of Krt13 in tongue epithelial cells. These studies show a causal link between Krt13 and WSN and unveil foundational mechanisms governing Krt13 function in epithelia.
RESULTS
Krt13 is expressed in both basal and differentiated epithelial cells in the mouse mucosa.
To investigate the possible role of Krt13 in mucosa and WSN, we first examined the expression pattern of Krt13 in mice at various developmental stages [embryonic day (E) 17.5, newborn, and postnatal day (P) 20] by immunostaining. We focused on the tongue since it is the most commonly affected tissue in human patients. At E17.5, expression of Krt13 was observed in mucosa covering the entire mouth, including the palate, dorsal tongue, ventral tongue, and the floor of the mouth (Fig. 1A). At P0, Krt13 expression was limited to the interpapillary cell column on the dorsal surface of the tongue. On the ventral surface, all epithelial cells were positive for Krt13 (Fig. 1B, C). This expression pattern persists as development proceeds. On the dorsal tongue epithelia at P20, a strong Krt13 staining signal was observed in cells in the interpapillary region and the keratinized layer, but not in filiform papillae. The expression of Krt13 remained ubiquitous in the ventral tongue epithelium (Fig. 1D, E). Although the staining intensity of Krt13 in the basal cells was weaker than the suprabasal layers, we observed consistent staining of Krt13 in the basal cells of the tongue epithelia.
Figure 1. Krt13 expression in developing mouse oral mucosa.
(A) Left, diagram showing the sagittal section view of E17.5 mouse head and the oral cavity. Right, E17.5 head sagittal sections stained with Krt13. Krt13 is expressed in mucosa covering the palate and dorsal tongue (a), ventral tongue and floor of the mouth (b). Scale bar, 100 μm.
(B-E) Immunohistochemical staining showing Krt13 expression in tongue epithelia at P0 and P20. fp, filiform papilla; ip, interpapillary region. Scale bar, 50 μm.
(F-I) Co-immunostaining of Krt13 with Sox2 showing Krt13 expression in both basal cells (arrows) and cells in the suprabasal layers. No Krt13 expression is found in the filiform papilla (asterisks). Scale bar, 25 μm in F, G, I, and 50 μm in H.
To determine whether Krt13 is indeed expressed in both differentiated cells and basal cells, we performed Krt13 co-immunostaining experiments with antibodies against Sox2. Sox2 regulates the commitment of the epithelium to squamous cell fate during development and is enriched in basally situated progenitor cells in epithelial tissues (Okubo et al., 2009; Arnold et al., 2011). In both P0 and P20 tongues, we found that Krt13 and Sox2 were coexpressed in the basal cells (Fig. 1F-I). Sox2-negative and Krt13-positive cells were observed in the suprabasal layers, indicating that Sox2 is turned off when epithelial cells undergo differentiation and move towards the upper layers. These data suggest that Krt13 is expressed in both basal cells and differentiated keratinocytes in the mouse oral mucosa.
Krt13 deletion leads to the WSN-like phenotype in the mouse mucosa.
To determine if the loss of Krt13 causes a WSN-like phenotype in mice, we used CRISPR/Cas9 to generate mice carrying a Krt13 allele with a premature stop codon inserted in exon 1 (referred to as Krt13cr). PCR genotyping followed by sequencing confirmed the correct mutation introduced by CRISPR/Cas9 (Fig. 2A-C). At P0, immunostaining revealed no expression of Krt13 in the tongue of Krt13cr/cr mice, demonstrating that the Krt13cr allele is a null allele (Fig. 2D, therefore referred to as Krt13 KO). Heterozygous intercrosses showed that the KO allele segregates in a Mendelian fashion and there is no significant difference in body weight between wild type (WT) and Krt13 KO mice, suggesting that Krt13 is not required for normal development.
Figure 2. Generation of Krt13 KO mice using CRISPR/Cas9.
(A) Diagram of the Krt13 WT and Krt13cr alleles.
(B) PCR genotyping of Krt13 mice.
(C) Sequencing confirmation of the Krt13 WT and Krt13cr alleles.
(D) Sagittal sections of dissected P0 tongues stained with Krt13. No Krt13 expression is detected in Krt13cr/cr tongue sections. Scale bar, 50 μm.
At three weeks of age, the tongue from Krt13 KO mice appeared white and wrinkled, especially on the ventral surface (Fig. 3A-D). In WT mice, two big lingual vessel branches were evident at the ventral side of the tongue. However, they were invisible in the Krt13 KO mice because of the opaque and wrinkled appearance of the tongue. Histological analysis revealed a variety of changes in the Krt13 KO tongue (Fig. 3E-H). In the WT tongue, column-like basal cells formed a single innermost layer. Differentiated epithelial cells became flattened in the suprabasal layer. Cells in the granular layer contained many keratohyalin granules. A compact keratin layer was found to cover both the dorsal and ventral surfaces of the tongue. In the Krt13 KO tongue, basal cells appeared jumbled together. Increased nuclear atypia and vacuolization were found in both basal and suprabasal layers. Cells remained cuboidal shaped in the suprabasal layers and keratohyalin granules were missing. The outermost keratin layer was disorganized and appeared foamy. All these histological changes were found in both dorsal and ventral epithelia of the tongue, although the epithelial folds were most prominent in the ventral side, especially close to the root.
Figure 3. WSN-like phenotype in Krt13 KO tongues.
(A-D) Gross morphology showing white and wrinkled appearance of Krt13 KO tongues at P20. Scale bar, 1 mm.
(E-H) H&E staining on tongue sagittal sections showing severe abnormalities in Krt13 KO mice, including thickened epithelium, immature suprabasal cells, loss of keratohyalin granules, and disorganized keratin layer. Scale bar, 100 μm.
(I-P) TEM analysis showing ultrastructural abnormalities in Krt13 KO tongue epithelia, including disrupted layer structure (I, J) and massive cytoplasmic vacuolization (J, asterisk in P), increased intercellular gaps (K, L), broken desmosomes (arrowheads in M, N), loss of keratohyalin granules and accumulation of big lipid droplets in the granular cells (arrows in O and arrowheads in Q, respectively). BL, basal layer; Sp, spinous layer; Gr, granular layer; SC, stratum corneum; Dm, desmosome.
Scale bar, 10 μm in I, J; 2 μm in K, L, O, P; 100 nm in M, N.
To examine the cellular morphology in detail, we performed transmission electron microscopy analysis on the tongue (Fig. 3I-P). Similar to what we found in H&E staining, the typical layer structure (columnar basal cells and flattened squamous suprabasal cells) was lost in Krt13 KO tongue epithelia. Most cells remained abnormally round or cuboidal as they moved to the upper layers. Intercellular gaps seemed to increase, but desmosomes still formed. Interestingly, desmosomes were often found broken and the intermediate filaments running from the desmosomes to the cytoplasm were missing or reduced in Krt13 KO tissues. Massive cytoplasmic vacuolization was found in all cell layers. Keratohyalin granules were completely missing and instead many big lipid droplets were found in the granular cells in the Krt13 KO tongue epithelia.
To determine whether Krt13 KO mice show defects in other tissues, we performed morphological and histological analyses on buccal mucosa and esophagus, where Krt13 is also highly expressed (Fig. S1). Similar to the tongue, Krt13 staining was found in both basal cells and differentiated epithelial cells in these tissues. Severe abnormalities including thickened epithelium, immature suprabasal cells, loss of keratohyalin granules, and disorganized keratin layer were found in the buccal mucosa and esophagus of P20 Krt13 KO mice. These experiments indicate a major role of Krt13 in maintaining the integrity of multiple epithelia, presumably through maintaining the balance of epithelial proliferation and differentiation. The phenotypes we observed in Krt13 KO mice mimic the pathological changes in WSN patients.
Loss of Krt13 leads to imbalanced proliferation and differentiation in epithelial cells.
The morphological changes in Krt13 KO mice indicate that loss of Krt13 likely affects epithelial cell proliferation and differentiation. To determine whether a disruption in cell differentiation indeed had occurred, we analyzed P20 Krt13 KO ventral tongues for various markers. The transcription factor p63 is highly expressed in the basal layer of the epidermis and the stratified squamous epithelium. It functions to maintain stem-cell character or proliferative capacity of the basal cells (Fuchs and Raghavan, 2002; Zhang et al., 2017). In WT tongues, p63 expression was limited to the basal layer. A few cells in the spinous layer immediately next to the basal layer were weakly positive for p63. In Krt13 KO mice, we observed p63 positive cells expanded to upper layers in the tongue epithelium (Fig. 4A, B). Krt5 is another marker for basal progenitor cells (Iwasaki et al., 2006; Okubo et al., 2009). Its expression was significantly reduced in the spinous and granular layers in WT tongues. In Krt13 KO mice, we observed that Krt5 positive cells spanned to the spinous and granular layers of the epithelium (Fig. 4C, D). Loricrin is a marker for differentiated epithelial cells. In WT tongues, its expression was found in the spinous and granular layers, but not in the terminally differentiated stratum corneum layer. In Krt13 KO tongue epithelium, we observed normal expression of loricrin in the spinous and granular layers. Interestingly, many loricrin positive cells were found in the thickened stratum corneum layer (Fig. 4E, F).
Figure 4. Disrupted proliferation and differentiation in Krt13 KO tongue epithelial cells.
(A-F) Krt13 deletion leads to disrupted epithelial differentiation in P20 mouse tongues. Sagittal sections of P20 ventral tongue stained with basal cell marker p63 (A-B) and Krt5 (C-D), or a differentiated epithelial marker loricrin (E-F). Note disrupted layer structure (B, D, F) and ectopic p63+ cells in the suprabasal layers (arrows in B’) in Krt13 KO tongues.
(G-K) Krt13 deletion leads to epithelial cell overproliferation in P20 mouse tongues. Sagittal sections of P20 ventral tongue stained with proliferation markers PCNA (G-H) and PH3 (I-J). Note proliferating cells are no longer limited to the basal layer, but expand to the upper layers in Krt13 KO tongues (arrows in H’, J’). Numbers of PH3(+) cells from WT and Krt13 KO sections were compared using the Student’s t-test (K). n= 14 (**, P < 0.01).
BL, basal layer; Sp, spinous layer; Gr, granular layer; SC, stratum corneum. Dotted lines represent the epithelial surface. Scale bar, 50 μm.
The folded and thickened epithelium in Krt13 KO tongues suggests that the loss of Krt13 leads to hyperplasia. We tested this idea by staining tissue sections for proliferating cell nuclear antigen (PCNA), a marker for cell proliferation. In WT tongues, proliferating cells were found in the basal layer with occasional staining of isolated cells in the spinous layer. In Krt13 KO epithelium, we observed proliferating cells expanded to the upper layers of the epithelium (Fig. 4G, H). Similar results were observed using another mitosis-specific marker, phosphohistone H3 (PH3) (Fig. 4I-K).
Our data show that Krt13 expression in the tongue starts at embryonic stages, and loss of Krt13 causes epithelial proliferation and differentiation defects at P20. To determine whether the phenotype that we observed is due to the direct loss of Krt13 or secondary to the postnatal stresses (e.g., suckling and biting), we collected tongues at P0 and performed histological analyses. We did not observe any abnormalities in the gross morphology, structural architecture, or epithelial cell proliferation (Fig. S2). These data indicate that loss of Krt13 alone does not cause morphological changes of epithelial cells, but rather impairs the maintenance of tissue homeostasis in postnatal life.
Krt13 regulates a transcriptional program to maintain tissue homeostasis.
To precisely define the molecular mechanism and identify downstream mediators of the phenotype in Krt13 KO mice, we performed unbiased whole-transcriptome profiling using deep RNA sequencing (RNA-Seq). Total RNA was extracted from the whole tongue at P0 (prior to the appearance of any noticeable Krt13-induced changes) or the ventral tongue epithelium at P20 (when the phenotype in Krt13 KO mice is prominent). Transcriptomic differences between Krt13 KO and control tongues were compared to reveal global changes in gene expression. At P0, we found that 125 genes exhibited a >2-fold change with a P-value <0.05 (referred to as differentially expressed genes), and only 10 out of 125 genes were downregulated (Fig. 5A and Table S1). As expected, Krt13 was one of them. We next performed Gene Ontology (GO) analysis on differentially expressed genes between WT and Krt13 KO tongues. We found that the most enriched GO term was keratinization. Proinflammatory responses, such as genes involved in immune cell chemotaxis pathway, positive regulation of interleukin-6 production, and positive regulator of NF-κB signaling, were also enriched in Krt13 KO tongues. Other enriched genes were involved in stress-activated protein kinase signaling and threonine/lipid metabolic processes (Fig. 5B and Table S1).
Figure 5. Downstream targets regulated by Krt13 in tongue epithelial cells.
(A) Transcriptome changes in response to the loss of Krt13 at P0, revealed by RNA-Seq. Lists of top ten genes with the highest fold change. Red, down-regulated; Blue, up-regulated expression.
(B) Enriched GO terms for the differentially expressed genes between WT and Krt13 KO tongues at P0.
(C) Enriched GO terms for the upregulated genes in P20 Krt13 KO tongues compared to WT.
(D) Heatmap from P20 RNA-Seq datasets showing 33 genes related to mitotic nuclear division and cell cycle regulation that are upregulated in Krt13 KO tongues.
(E) qRT-PCR showing that mechanic stress can directly induce the expression of Cyclin E family of genes in Krt13 KO tongues while WT tongues are more resistant to such treatment. All data represent mean ± SEM of three biological replicates. GAPDH was used as a control. Quantification of data was compared using ANOVA followed by Turkey’s multiple comparison test (*, P < 0.05; **, P < 0.01; n.s., not significant).
At P20, the number of differentially expressed genes increased to 2907, reflecting increasing severity in phenotype over time. GO analysis of the 1076 upregulated genes revealed that a distinct set of cell cycle control genes that were upregulated, which is consistent with our histological findings that epithelial cells in Krt13 KO tissues are hyperproliferative (Fig. 5C and Table S1). Epidermal growth factor receptor (EGFR) signaling pathway is known to control cell proliferation, and activating mutations have been associated with many cancers (Wee and Wang, 2017). We observed an upregulation of several EGFR signaling genes in the Krt13 KO samples. Sphingolipids are a major class of the membrane lipids and they provide the support structure for all cell membranes (Bartke and Hannun, 2009). Asymmetric distribution of sphingolipids and other lipids also plays an important role in polarizing epithelial cells into apical and basolateral domains, which is essential for normal epithelial cell function (van IJzendoorn et al., 2019). We found enrichment of genes for the sphingolipid biosynthetic process, which might contribute to the disrupted epithelial layer structure and big lipid droplets that we observed in the Krt13 KO epithelial cells. In addition, many changes that we observed at P0 persisted, such as proinflammatory responses and stress-activated protein kinase signaling. Among the downregulated 1831 genes, we found enrichment of actin filament binding, metallopeptidase activity, and calcium ion binding genes (Table S1).
Since the GO analysis between Krt13 KO and WT tissues at P20 revealed that mitotic cell division was the most represented term, we chose to investigate the cell cycle control genes in more detail. Heatmap derived from the RNA-Seq data associated with the GO terms “mitotic nuclear division” and “regulation of cell cycle” exhibited upregulation of 33 genes in Krt13 KO samples (Fig. 5D). We validated the expression changes of all 33 DEGs in the P20 Krt13 KO tongue by qRT-PCR. Except for 7 genes with very low expression (> 35 cycles), all the other 26 genes showed significant upregulation compared to WT (Fig. S3). Cyclin D1 controls the G1 to S phase transition. Its ability to activate the cyclin dependent kinases (CDKs) is the most extensively studied mechanism for cell cycle control and oncogenic actions (Sherr, 1994; Sicinski et al., 1995). Transgenic overexpression of Cyclin D1 in the epithelium of the tongue, esophagus, and forestomach tissues leads to tissue overgrowth (Nakagawa et al., 1997). We did not observe upregulation of Cyclin D1. However, other G1/S specific cyclins, Cyclin E1 and E2, were significantly upregulated in Krt13 KO samples.
To directly test the idea that postnatal stress contributes to the overproliferation phenotype of Krt13 KO mice, we performed an in vitro culture experiment (Fig. 5E). We dissected and cultured P0 tongues on a flat silicone surface with ventral side up. Insect pins were used to hold the cultured tongues in place so that mechanical stresses can be applied to the ventral surface. We gently scraped the tongue surface with cotton end tips, 30 times per session, three sessions an hour, to mimic the mechanic stresses that mice might encounter in postnatal life. Total RNA was then collected to investigate changes in gene expression induced by the scrapings. In WT tongues, the treatment did not cause changes to the expression of Cyclin E1 and E2. However, Krt13 KO tongues showed a striking increase in the expression of these two genes. This finding indicates that Krt13 is directly coupled with cell cycle control and loss of Krt13 makes epithelial cells susceptible to stress-induced overproliferation.
DISCUSSION
The experiments described here establish an essential role of Krt13 in maintaining the homeostasis of mucosa in mice. In particular, we report that (1) Krt13 is expressed in both progenitor cells in the basal layer and differentiated cells in the suprabasal layers; (2) loss of Krt13 leads to disrupted epithelial cell differentiation and overproliferation in mouse tongues, which resembles pathological changes seen in human WSN patients; (3) Krt13 is expressed in several epithelia other than tongue, including buccal mucosa and esophagus, and loss of Krt13 leads to disrupted epithelial integrity in these tissues as well; and (4) Krt13 knockout causes significant transcriptome changes and Cyclin E family might play a critical role in contributing to the phenotype development.
Model of Krt13 function in epithelial cells
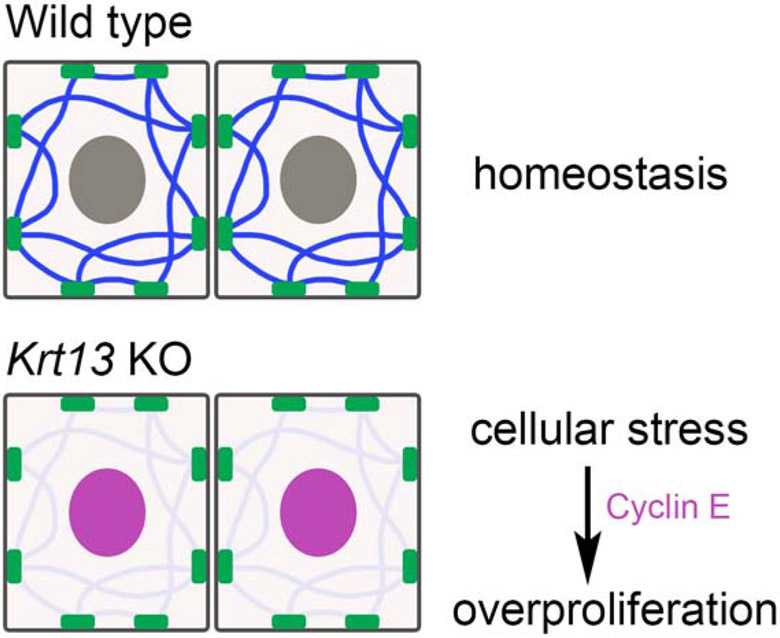
One of the primary roles of keratin filaments in epithelial cells is to provide structural support (Coulombe et al., 1991; Jacob et al., 2018). The expression of Krt13 in the tongue begins at embryonic stages, and we did not observe any abnormalities in the gross morphology, structural architecture, or epithelial cell proliferation in newborn mice. This age-dependent appearance of the tongue phenotype leads us to propose the model that loss of Krt13 does not directly cause the overproliferation of epithelial cells but instead affects the maintenance of tissue homeostasis (Fig. 6). We hypothesize that mechanical stresses during postnatal life, e.g., sucking milk and chewing solid food, contribute to the development of the tongue phenotype in the Krt13 KO mice, possibly through the regulation of cell cycle. Our in vivo transcriptome analysis and our in vitro culture experiments highlight the contribution of cell overproliferation to the pathological changes in postnatal Krt13 KO tongues.
Figure 6. Model of Krt13 function in epithelia.
(Top) Krt13 is crucial for maintaining the homeostasis of epithelial tissues.
(Bottom) Disruption of this mechanism results in increased cellular stress and unbalanced cell proliferation/differentiation, possibly through the Cyclin E family and/or other targets.
Cyclin E controls the G1 to S phase transition during normal cell cycle control and tumorigenesis by activating CDK2 (Donnellan and Chetty, 1999; Siu et al., 2012). In this study, we found that the expression of both members of Cyclin E, Cyclin E1 and Cyclin E2, were upregulated in P20 tongues of Krt13 KO mice. Also, we showed that mechanical stress can directly induce the expression of these genes in cultured Krt13 KO tongues. It is tempting to hypothesize that Krt13 functions through the regulation of Cyclin E1/E2. Cyclin E1 and E2 KO mice develop to term normally. Other than some Cyclin E2 KO males showing reduced fertility and testicular atrophy, there are no observed abnormalities in these mutant mice (Geng et al., 2003). We can attempt to rescue the Krt13 KO tongue phenotype with the knockout of Cyclin E1/E2 to determine to what extent Cyclin E causes the Krt13 KO phenotype.
Krt13 mutations and potential partners in epithelial cells
Although we demonstrated that Krt13 KO mice develop a WSN-like phenotype in multiple epithelial tissues, it is important to point out that these mutant mice completely lack the expression of Krt13, whereas many of WSN patients are associated with point mutations in the Krt13 gene. It is reasonable to predict that any defects that alter Krt13-mediated intermediate filament assembly, either loss of Krt13 protein or expressing mutant forms of Krt13, will result in the same mechanical instability to the epithelial cells. However, to draw a definite conclusion that both genetic modifications cause identical pathogenesis would require the generation of mice carrying the dominant point mutations seen in WSN patients.
Type I and II keratins are strictly interdependent for the formation of coiled-coil heterodimers to initiate the assembly of intermediate filaments (Kim and Coulombe, 2007; Jacob et al., 2018). Krt13 is a type I keratin, and Krt4 is believed to be its type II partner. Krt4 KO mice have been previously generated and mutant mice also show WSN-type lesions in lingual mucosa and other epithelial tissues (Ness et al., 1998). Interestingly, Krt4 KO mice did not exhibit any phenotypes in the tongue until five months of age. The less severe phenotype of Krt4 KO mice might be explained by the residual protective function of Krt13 paired with another type II keratin in the absence of Krt4. Based on our RNA-Seq data, many of the 26 type II keratins are expressed in mouse tongue epithelia, which includes Krt6a, Krt6b, and Krt78. They might serve as partners of Krt13 in the mouse mucosa.
MATERIALS AND METHODS
Mouse lines and husbandry
The Krt13 knockout mouse allele was generated using CRISPR/Cas9. A sgRNA (GACTTCGGAGGTGTCGATGG) targeting exon 1 of Krt13 was selected and synthesized, as described previously (Pelletier et al., 2015). Briefly, a dsDNA template containing the sequences for a T7 promoter, the sgRNA, and a tracrRNA scaffold was generated by tandem PCR amplification and purification on a QIAGEN column (QIAGEN 28106). In vitro transcription of this dsDNA template was performed using the MEGAshortscript T7 Kit (Invitrogen AM1354), and the products were purified using the MEGAclear Transcription Clean-Up Kit (Invitrogen AM1908). A ssODN HDR template containing a premature stop codon was synthesized by IDT (GAGGTTTTGGCGGTGGCTTTGGTGACTTCGGAGGTGTCGATAAaTAAaTAAGAATTC CCTCCTCTCTGGCAACGAGAAGATCACCATGCAGAACCTC). The sgRNA, Cas9 protein, and HDR template were injected into C57BL/6 embryos using the service provided by the Genome Editing and Animal Models Core (GEAM) at the University of Wisconsin-Madison. Correctly targeted founders were backcrossed to C57BL/6 mice for a total of four generations. Primers used for genotyping the Krt13 knockout allele were Krt13-F: CTGCTCATCTCGGTTTGTCA and Krt13-R: TCCACACCTGGTCGTCTGTA. All mice were handled and housed according to the approved Institutional Animal Care and Use Committee (IACUC) protocol M005675 of the University of Wisconsin-Madison.
Tissue Processing and Immunostaining
For the immunostaining of embryonic tissues, embryos were collected and processed as described previously (Dong et al., 2018). Briefly, embryos were embedded in OCT, fresh frozen, cryosectioned at 14 μm thickness, and slides were stored at −80°C before immunostaining. Cryosections were thawed and fixed for 10 minutes in 4% formaldehyde and then washed three times with PBST (0.1% Triton in PBS). Sections were blocked for one hour with 5% normal donkey serum in PBST. Slides were incubated overnight at 4°C in primary antibodies in a humid chamber. Next, slides were rinsed three times in PBST and incubated for one hour at room temperature in secondary antibodies. 1:500 dilutions of Alexa Fluor 488- and/or 594-conjugated donkey anti-mouse, rat, or rabbit IgG antibodies (Thermo Fisher Scientific, A-21202, A-21209, and A-21206) were applied and DAPI was included in the secondary antibody solution at a 1:1000 concentration. After incubation with secondary antibodies, slides were rinsed three times in PBS and coverslipped with Fluoromount-G (SouthernBiotech, 0100-01). Confocal images were acquired using a Nikon A1R-Si+ Confocal Microscope with NIS elements software.
Postnatal tissues were collected and fixed in 4% formaldehyde overnight, embedded in paraffin wax and sectioned at 4 μm. After de-waxing, the sections were stained with Hematoxylin and Eosin or immunostained for histological analyses. Immunohistochemical analysis was carried out using the VECTASTAIN® ABC-AP Staining KIT (Alkaline Phosphatase, Rabbit IgG) as recommended by the manufacturer. Images for H&E staining and immunohistochemistry were captured at 40x using an EVOS® XL Core Cell Imaging System (Thermo Fisher Scientific). Immunofluorescence staining on postnatal paraffin tissues was carried similarly as frozen sections after de-waxing and antigen retrieval.
The following primary antibodies were used: Rabbit anti-Krt13 (ab92551; Abcam; 1:1000), Rat anti-SOX2 (14-9811-82; Thermo Fisher Scientific; 1:100), Rabbit anti-p63 antibody (ab124762; Abcam; 1:2000), Rabbit anti-Krt5 (905501; BioLegend; 1:2000), Rabbit anti-Loricrin (ab85679; Abcam; 1:200), mouse monoclonal anti-PCNA (2586, Cell Signaling Technology; 1:2000), Rabbit anti-PH3 (9701; Cell Signaling Technology; 1:200). Tissue sections from control and mutant mice were immunostained in parallel to insure identical processing.
Transmission electron microscope (TEM)
TEM analysis on mouse tongues was performed using room temperature chemical fixation on samples that were fixed with 2.0% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M Phosphate buffer, post-fixed in 1.0% osmium tetroxide, dehydrated in a graded series of acetone, and embedded in Durcupan ACM (Fluka). Ultrathin sections (90-100 nm) were post-stained with uranyl acetate and lead citrate. Samples were viewed with a Philips CM120 transmission electron microscope.
RNA isolation, RNA-Seq, and quantitative real-time PCR (qRT-PCR)
The whole P0 mouse tongues or ventral side of P20 tongues were dissected from WT and Krt13−/− mice. RNA was extracted using Trizol (Invitrogen) and RNeasy (Qiagen) kits. Four or five biologically independent samples were collected for each genotype. The quality of RNA were determined using Agilent 2100 Bioanalyzer. cDNA was generated using the Invitrogen SuperScript IV reverse transcription kit followed by second-strand cDNA synthesis and then subjected to standard Illumina adaptor ligation and sequencing. Gene ontology analysis was performed using the Gene Ontology Consortium online tool (http://geneontology.org/). Quantitative real-time RT-PCR was performed in triplicate in 20 μl reactions with SYBR Premix Ex Taq II ROX plus (Takara, RR82LR) with 40 ng first-strand cDNA and 0.2 μg each forward and reverse primers. Samples were cycled once at 95 °C for 2 minutes, followed by 40 cycles of 95, 58, and 72 °C for 30 seconds each. Relative mRNA level was calculated using the ΔΔCT method with GAPDH as an endogenous control. Primers used for qRT-PCR (Table S2) were selected from the PrimerBank database (Spandidos et al., 2010).
Quantification and statistical analysis
For RNA-Seq analysis, ENCODE guidelines and best practices for RNA-Seq were followed. Briefly, adapter-trimmed paired-end 150 bp strand-specific Illumina reads were aligned to the Mus musculus GRCm38.p5 genome (assembly accession GCA_00001635.7) with Skewer v0.1.123 and STAR v2.5.3a softwares (Dobin et al., 2013; Jiang et al., 2014). Expression estimation was conducted using RSEM v1.3.0 (Li and Dewey, 2011). To test for differential expression among individual group contrasts, expected read counts were used as input into edgeR v3.16.5 (Robinson et al., 2010). Significance of the negative-binomial test was adjusted with a Benjamini-Hochberg FDR correction at the 5% level (Reiner et al., 2003). Prior to statistical analysis with edgeR, independent filtering was performed, requiring a threshold of least 2 reads per million in at least 3 samples. The validity of the Benjamini-Hochberg FDR multiple testing procedure was evalated by inspection of the uncorrected p-value distribution.
Enrichment analysis of differentially expressed genes was performed with Kyoto Encyclopedia of Genes and Genomes (KEGG) curated pathways and geneSCF (Kanehisa et al., 2002; Subhash and Kanduri, 2016).
For quantification of mRNA by qRT-PCR, statistical analyses were performed with two-tailed unpaired Student’s t-test between two experimental groups and one-way analysis of variance (ANOVA) for more than two experimental groups followed by Tukey’s test for multiple comparisons using the GraphPad Prism 8 software. P-values less than 0.05 were considered significant.
Supplementary Material
Figure S1. WSN-like phenotype in buccal mucosa and esophagus of Krt13 KO mice.
(A-C) Histological analysis on mouse buccal mucosa. Co-immunostaining of Krt13 with Sox2 showing Krt13 expression in both basal cells and differentiated cells in WT mouse buccal mucosa (A). H&E staining showing severe abnormalities in buccal mucosa of Krt13 KO mice, including thickened epithelium, immature suprabasal cells, loss of keratohyalin granules, and disorganized keratin layer (B-C). (D-F) Histological analysis on mouse esophagus showing similar WSN-like characteristics in Krt13 KO mice. Scale bar, 50 μ m.
Figure S2. No obvious phenotype in Krt13 KO tongues at P0.
(A-D) Gross morphology and H&E staining showing no obvious defects in Krt13 KO tongues at P0 (ventral side shown). (E-H) Krt13 deletion does not lead to significant differentiation/proliferation defects in mouse tongues at P0. Sagittal sections of P0 ventral tongue stained with a basal cell marker Krt5 (E-F) or a cell proliferation marker PCNA (G-H). Dotted lines represent the epithelial surface. Scale bar, 1 mm in A-B, 200 μm in C-D, 50 μm in E-H.
Figure S3. Validation of the RNA-Seq data by qRT-PCR using RNA extracted from P20 WT and Krt13 KO tongues.
26 genes related to the mitotic nuclear division and cell cycle regulation show an increased expression in the P20 Krt13 KO tongue. All data represent mean ± SEM of three biological replicates. GAPDH was used as a control. Quantification of data between two groups was compared using the Student’s t-test. (**, P < 0.01).
Table S1. Differential gene expression between the WT and Krt13 KO tongues and Gene Ontology analysis, related to Figure 5.
Table S2. List of primers used for qRT-PCR analysis, related to Figure 5.
Highlights:
Krt13 is expressed in both progenitor and differentiated cells in the mouse tongue epithelium;
Krt13 knockout leads to disrupted epithelial integrity, which resembles pathological changes seen in human WSN patients;
Cell overproliferation controlled by the Cyclin E family might contribute to the Krt13 knockout phenotype.
ACKNOWLEDGMENTS
The authors would like to thank Ruth Sullivan and Gary Wood for their feedbacks. We acknowledge Kathy Krentz and the Genome Editing and Animal Models Core (GEAM) at the UW Biotechnology Center for mouse pronuclear injections, Beth Gray and the Comparative Pathology Laboratory at the UW Research Animal Resources Center for paraffin sectioning and H&E staining, Sandra Splinter BonDurant and Mark Berres and the UW Gene Expression Center for RNA-Seq service, the UW SMPH EM Facility for TEM service, and the UW Biochemistry Optical Core for providing the confocal microscope.
FUNDING
The work was supported by grants from the National Institutes of Health (the Skin Diseases Research Center Core Grant P30AR066524, the University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520, and R01GM129259 to H.C.). BJL is supported by the Department of Veterans Affairs VA Merit Review Award I01BX003315. HC was also supported in part by the Gary S. Wood Dermatology Research Bascom Endowed Professorship.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AVAILABILITY
The RN-Seq datasets have been deposited in Gene Expression Omnibus (GEO) under accession number GEO: GSE145215.
REFERENCES:
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N and Hochedlinger K (2011) 'Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice', Cell Stem Cell 9(4): 317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke N and Hannun YA (2009) 'Bioactive sphingolipids: metabolism and function', J Lipid Res 50 Suppl: S91–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Jiang B, Yu F, Yang J, Chen Z, Liu J, Wei R, Zhao S, Wang X and Liu S (2015) 'Current approaches to the diagnosis and treatment of white sponge nevus', Expert Rev Mol Med 17: e9. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Hutton ME, Vassar R and Fuchs E (1991) 'A function for keratins and a common thread among different types of epidermolysis bullosa simplex diseases', J Cell Biol 115(6): 1661–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR (2013) 'STAR: ultrafast universal RNA-seq aligner', Bioinformatics 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Vold S, Olvera-Jaramillo C and Chang H (2018) 'Functional redundancy of frizzled 3 and frizzled 6 in planar cell polarity control of mouse hair follicles', Development 145(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnellan R and Chetty R (1999) 'Cyclin E in human cancers', FASEB J 13(8): 773–80. [DOI] [PubMed] [Google Scholar]
- Dufrasne L, Magremanne M, Parent D and Evrard L (2011) '[Current therapeutic approach of the white sponge naevus of the oral cavity]', Bull Group Int Rech Sci Stomatol Odontol 50(1): 1–5. [PubMed] [Google Scholar]
- Frithiof L and Bánóczy J (1976) 'White sponge nevus (leukoedema exfoliativum mucosae oris): ultrastructural observations', Oral Surg Oral Med Oral Pathol 41(5): 607–22. [DOI] [PubMed] [Google Scholar]
- Fuchs E and Raghavan S (2002) 'Getting under the skin of epidermal morphogenesis', Nat Rev Genet 3(3): 199–209. [DOI] [PubMed] [Google Scholar]
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H and Sicinski P (2003) 'Cyclin E ablation in the mouse', Cell 114(4): 431–43. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Yoshizawa H and Aoyagi H (2006) 'Immunohistochemical expression of keratins 13 and 14 in the lingual epithelium of rats during the morphogenesis of filiform papillae', Arch Oral Biol 51(5): 416–26. [DOI] [PubMed] [Google Scholar]
- Jacob JT, Coulombe PA, Kwan R and Omary MB (2018) 'Types I and II Keratin Intermediate Filaments', Cold Spring Harb Perspect Biol 10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW and Zhu S (2014) 'Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads', BMC Bioinformatics 15: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson RJ and Levin S (1981) 'White sponge nevus', Arch Dermatol 117(2): 73–6. [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S and Nakaya A (2002) 'The KEGG databases at GenomeNet', Nucleic Acids Res 30(1): 42–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S and Coulombe PA (2007) 'Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm', Genes Dev 21(13): 1581–97. [DOI] [PubMed] [Google Scholar]
- Li B and Dewey CN (2011) 'RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome', BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrelli M, Tatullo M, Dipalma G and Inchingolo F (2012) 'Oral infection by Staphylococcus aureus in patients affected by White Sponge Nevus: a description of two cases occurred in the same family', Int J Med Sci 9(1): 47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan KA, Fuchs H, Hrabé de Angelis M and Barsh GS (2007) 'Identification of a Keratin 4 mutation in a chemically induced mouse mutant that models white sponge nevus', J Invest Dermatol 127(1): 60–4. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Wang TC, Zukerberg L, Odze R, Togawa K, May GH, Wilson J and Rustgi AK (1997) 'The targeting of the cyclin D1 oncogene by an Epstein-Barr virus promoter in transgenic mice causes dysplasia in the tongue, esophagus and forestomach', Oncogene 14(10): 1185–90. [DOI] [PubMed] [Google Scholar]
- Ness SL, Edelmann W, Jenkins TD, Liedtke W, Rustgi AK and Kucherlapati R (1998) 'Mouse keratin 4 is necessary for internal epithelial integrity', J Biol Chem 273(37): 23904–11. [DOI] [PubMed] [Google Scholar]
- Okubo T, Clark C and Hogan BL (2009) 'Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate', Stem Cells 27(2): 442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otobe IF, de Sousa SO, Matthews RW and Migliari DA (2007) 'White sponge naevus: improvement with tetracycline mouth rinse: report of four cases', Clin Exp Dermatol 32(6): 749–51. [DOI] [PubMed] [Google Scholar]
- Pelletier S, Gingras S and Green DR (2015) 'Mouse genome engineering via CRISPR-Cas9 for study of immune function', Immunity 42(1): 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D and Benjamini Y (2003) 'Identifying differentially expressed genes using false discovery rate controlling procedures', Bioinformatics 19(3): 368–75. [DOI] [PubMed] [Google Scholar]
- Richard G, De Laurenzi V, Didona B, Bale SJ and Compton JG (1995) 'Keratin 13 point mutation underlies the hereditary mucosal epithelial disorder white sponge nevus', Nat Genet 11(4): 453–5. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ and Smyth GK (2010) 'edgeR: a Bioconductor package for differential expression analysis of digital gene expression data', Bioinformatics 26(1): 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg EL, McLean WH, Allison WE, Lunny DP, Macleod RI, Felix DH, Lane EB and Munro CS (1995) 'A mutation in the mucosal keratin K4 is associated with oral white sponge nevus', Nat Genet 11(4): 450–2. [DOI] [PubMed] [Google Scholar]
- Sadeghi EM and Witkop CJ (1979) 'The presence of Candida albicans in hereditary benign intraepithelial dyskeratosis. An ultrastructural observation', Oral Surg Oral Med Oral Pathol 48(4): 342–6. [DOI] [PubMed] [Google Scholar]
- Satriano RA, Errichetti E and Baroni A (2012) 'White sponge nevus treated with chlorhexidine', J Dermatol 39(8): 742–3. [DOI] [PubMed] [Google Scholar]
- Sherr CJ (1994) 'G1 phase progression: cycling on cue', Cell 79(4): 551–5. [DOI] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, Haslam SZ, Bronson RT, Elledge SJ and Weinberg RA (1995) 'Cyclin D1 provides a link between development and oncogenesis in the retina and breast', Cell 82(4): 621–30. [DOI] [PubMed] [Google Scholar]
- Siu KT, Rosner MR and Minella AC (2012) 'An integrated view of cyclin E function and regulation', Cell Cycle 11(1): 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H and Seed B (2010) 'PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification', Nucleic Acids Res 38(Database issue): D792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhash S and Kanduri C (2016) 'GeneSCF: a real-time based functional enrichment tool with support for multiple organisms', BMC Bioinformatics 17(1): 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van IJzendoorn SCD, Agnetti J and Gassama-Diagne A (2019) 'Mechanisms behind the polarized distribution of lipids in epithelial cells', Biochim Biophys Acta Biomembr 1862(2): 183145. [DOI] [PubMed] [Google Scholar]
- Wee P and Wang Z (2017) 'Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways', Cancers (Basel) 9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jiang M, Kim E, Lin S, Liu K, Lan X and Que J (2017) 'Development and stem cells of the esophagus', Semin Cell Dev Biol 66: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. WSN-like phenotype in buccal mucosa and esophagus of Krt13 KO mice.
(A-C) Histological analysis on mouse buccal mucosa. Co-immunostaining of Krt13 with Sox2 showing Krt13 expression in both basal cells and differentiated cells in WT mouse buccal mucosa (A). H&E staining showing severe abnormalities in buccal mucosa of Krt13 KO mice, including thickened epithelium, immature suprabasal cells, loss of keratohyalin granules, and disorganized keratin layer (B-C). (D-F) Histological analysis on mouse esophagus showing similar WSN-like characteristics in Krt13 KO mice. Scale bar, 50 μ m.
Figure S2. No obvious phenotype in Krt13 KO tongues at P0.
(A-D) Gross morphology and H&E staining showing no obvious defects in Krt13 KO tongues at P0 (ventral side shown). (E-H) Krt13 deletion does not lead to significant differentiation/proliferation defects in mouse tongues at P0. Sagittal sections of P0 ventral tongue stained with a basal cell marker Krt5 (E-F) or a cell proliferation marker PCNA (G-H). Dotted lines represent the epithelial surface. Scale bar, 1 mm in A-B, 200 μm in C-D, 50 μm in E-H.
Figure S3. Validation of the RNA-Seq data by qRT-PCR using RNA extracted from P20 WT and Krt13 KO tongues.
26 genes related to the mitotic nuclear division and cell cycle regulation show an increased expression in the P20 Krt13 KO tongue. All data represent mean ± SEM of three biological replicates. GAPDH was used as a control. Quantification of data between two groups was compared using the Student’s t-test. (**, P < 0.01).
Table S1. Differential gene expression between the WT and Krt13 KO tongues and Gene Ontology analysis, related to Figure 5.
Table S2. List of primers used for qRT-PCR analysis, related to Figure 5.
Data Availability Statement
The RN-Seq datasets have been deposited in Gene Expression Omnibus (GEO) under accession number GEO: GSE145215.