Abstract
Radiation therapy (RT) is highly effective due to its ability to physically focus the treatment to target the tumor while sparing normal tissue and its ability to be combined with systemic therapy. This systemic therapy can be utilized before RT as an adjuvant or induction treatment, during RT as a radiation “sensitizer,” or following RT as a part of combined modality therapy. As part of a unique concept of using radiation as “focused biology” we investigated how tumors and normal tissues adapt to clinically relevant multi-fraction (MF) and single-dose (SD) radiation to observe whether the adaptations can induce susceptibility to cell killing by available drugs or by immune enhancement. We identified an adaptation occurring after MF (3 × 2 Gy) that induced cell killing when AKT-mTOR inhibitors were delivered following cessation of RT. Additionally, we identified inducible changes in integrin expression 2 months following cessation of RT that differ between MF (1 Gy x 10) and SD (10 Gy) that remain targetable compared to pre-RT. Adaptation is reflected across different “omics” studies, and thus the range of possible molecular targets is not only broad but also time, dose, and schedule dependent. While much remains to be studied about the radiation adaptive response, radiation should be characterized by its molecular perturbations in addition to physical dose. Consideration of the adaptive effects should result in the design of a tailored radiotherapy treatment plan that accounts for specific molecular changes to be targeted as part of precision multi-modality cancer treatment.
Introduction
The sophistication of radiation therapy (RT) technology has increased markedly in the last two decades, revolutionizing tumor targeting and normal tissue sparing. New dose and fraction sizes, along with the application of stereotaxis, allow safer delivery of a large single dose or a few larger fraction (hypofractionation) treatments. Additionally, the use of charged particle therapy and high dose-rate brachytherapy limits normal tissue injury and potentially results in novel radiation biology. The basic tenets of radiation biology for tumor control are traditionally the 4 R’s: repopulation, redistribution, repair, and reoxygenation (1). With an increased understanding of tumor response, additional “R’s” have been suggested, including “radiosensitivity” and immunological “rejection” (2–5). The mechanisms behind these concepts are known to be complicated and interact with one another during treatment (e.g. DNA damage and immune response, stem cell repopulation and radioresistance), and understanding these responses, which are likely operational for all cancer therapeutics, offers novel precision-medicine treatments.
Recognizing that data from experimental doses used in the laboratory were often not clinically relevant, we examined the impact of clinically relevant RT doses and schedules along with nonsteroidal anti-inflammatory drugs (NSAIDs) on tumors and normal tissues (6–8). Very early in the current era of immuno-oncology and check-point inhibitors, we demonstrated that fractionated radiation triggers canonical immune response pathways in both tumors and normal tissues (9). These observations led to the novel hypothesis that multi-fraction (MF) radiation induces cellular adaptations in tumors that can be targeted with drugs to which the tumors had little to no sensitivity before RT. As described in this review, the data demonstrated that single dose- (SD) and MF-induced changes depended on the underlying cell type (brain, breast and prostate tumor and endothelial cells) and occurred even at doses with little cell killing (7). Additionally, cancer stem cell repopulation can lead to acquired radioresistance (10), demonstrating another type of radioadaptive response. Thus, RT adaptation is relevant to the tumor as a whole, individual tumor cells, and surrounding normal tissue beyond the high-dose field margin.
Previous publications from our laboratories demonstrated that MF radiation (e.g. 1 Gy x 10 over 5 days) induced more differential mRNA and microRNA gene expression compared to SD radiation (e.g. 10 Gy x 1) (11,12). Herein we discuss new findings that support the applicability of radiation-induced adaptations for new therapeutic combinations (13). These adaptations can accompany the advances in physical and spatial focusing that are part of “Accurate, Precision Radiation Medicine” (14). A new paradigm established in a recent NCI workshop (13) emphasizes the importance of describing radiation in both the physical dose, Gy, and in biological perturbations. This includes the adaptations discussed in this review to potentially exploit RT-inducible targets for cancer care including developing radio-mitigators and protectants of normal tissue injury.
In essence, we consider radiation “as a drug” where the pharmacokinetics and pharmacodynamics (PK/PD) can be used in unique ways to impact both local therapy and distant metastases. The technical capability of RT allows the treatment to be focused in a wide range of targets, doses, and schedules, a concept we labeled “focused biology” (15). While we recognize the limitations of laboratory models and that there is much work to be done for these adaptations to be clinically applicable, this report provides insight into the current state of the science.
I. Different adaptations occur between MF and SD radiation.
The range and depth of “omics” analyses continually expand with studies on coding and non-coding RNAs, metabolomics, and proteomics. Our initial work on fractionation was performed in vitro and in vivo in prostate, breast, and brain tumor cell lines, and demonstrated that the tumor microenvironment influences gene expression patterns after both SD and MF (7). The pattern of gene expression differed for the different cell lines with most changes occurring in the immune response pathways (6,9,16). Both the extent and stability of changes in gene expression were greater following MF compared to SD. Additionally, findings from both our laboratory and others demonstrated that gene expression changes can vary extensively depending on whether the dose is delivered as SD or MF (7,9,17). These studies clearly demonstrated that MF exposure was shown to alter several genes, selectively providing the opportunity to explore molecular target-directed interventions to enhance the tumor response to radiation.
microRNA expression as a form of RT adaptation: dose/fractionation response
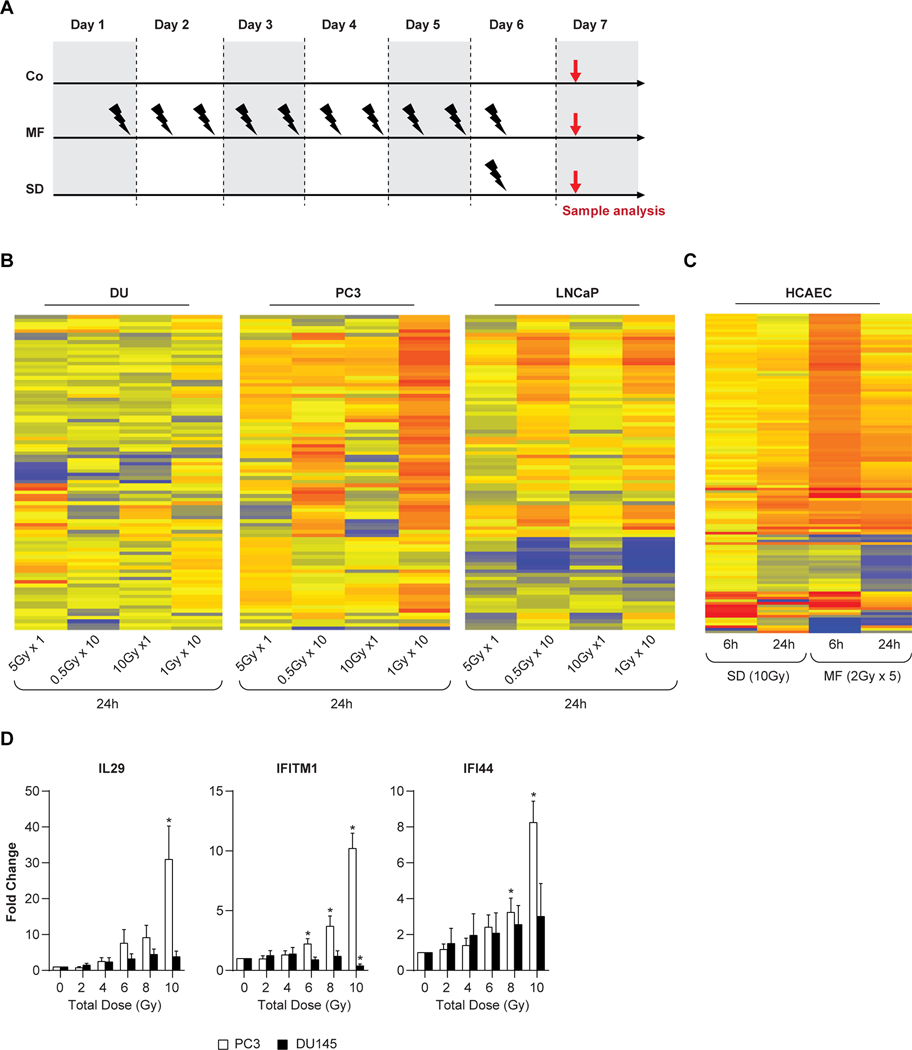
To further develop the efficacy of molecular targeted therapy following RT adaptation, we studied microRNA (miRNA) expression patterns after SD ranging from 5 to 10 Gy and MF of 10 fractions ranging from 0.5 to 1 Gy (Fig. 1A), using 3 human prostate tumor cell lines with different p53 status: LNCaP (p53 wildtype), PC3 (p53-null) and DU145 (p53-mutated). The RT-induced changes in miRNA expression pattern depend on dose size and fractionation (Fig. 1B). Despite significant changes in miRNA expression, the surviving fraction following 0.5 Gy x 10 was approximately 85%. Significant differences in the miRNA profiles of breast cancer cell line MDA-MB-361 after SD versus MF exposure have also been reported (18), signifying a dependency on dose fractionation and the presence of a radioadaptive miRNA response. We also studied the differential mRNA and miRNA expression pattern after SD (10 Gy) and MF (2Gy x 5) in normal human coronary artery endothelial cells (HCAEC) (19). The miRNA expression pattern in HCAEC was significantly altered between SD and MF at both 6h and 24h after the final RT dose (Fig. 1C). There were only 17 miRNAs altered after SD, in contrast to 103 differentially altered miRNAs detectable after MF. Among the altered miRNAs, only 5 were common to SD and MF, pointing towards the importance of dose delivery in post RT-druggable targets. Another study using a mouse model with Lewis lung carcinoma cells (LLC1) reported that gene and miRNA expression profiles are dependent upon whether the cells received SD or MF radiation (17), further demonstrating the importance of the type of dose delivery in a preclinical model. These dose delivery-dependent miRNA adaptations are important for continuation of radiotherapy and cancer treatment but could be strengthened with development of predictive miRNA biomarkers. A pretreatment signature that measures radiosensitivity of head and neck squamous cell carcinoma to SD exposure shows the potential of predictive miRNA biomarkers (20); however, more studies are needed to determine the feasibility of using such biomarkers for predicting response to MF radiation.
Figure 1: Adaptation and inflection point.
Schematic representation of experimental set up for Controls, along with single dose and fractionated radiation for PC3 and DU145 cells (A). Heat map of differentially expressed genes in prostate carcinoma cells (PC3and DU145) after SD (5 Gy, 10 Gy) and MF (0.5 Gy and 1 Gy x 10) radiation at 24 hours (B) Heat map of differentially expressed genes in HCAEC at 6 and 24 hours after an SD (10Gy) and 6 and 24 hours after the final dose of MF (2Gyx5) irradiation (C). Yellow to red, upregulated; blue, downregulated genes. Inflection point kinetics of immune genes for interleukin 29 (IL29), interferon induced transmembrane protein 1 (IFITM1), and interferon induced protein 44 (IFI44) in multifractionated treated PC3 and DU145 cells as assessed by real time RT-PCR. Treatment was 1 Gy, 3x/day with at least 6 hours between fractions (D).
Early inflection point
Using prostate cancer and HCAEC cell lines, we demonstrated that cells subjected to MF promote a pro-immunogenic molecular signature, among other changes (6,9,21). To investigate the time course of the adaptation, DU145 and PC3 cells were irradiated with 1 Gy every 6 hours, 2–3 times per day, for total doses from 2–10 Gy. The data demonstrate an inflection point starting from the 6th 1Gy fraction (Fig. 1D) such that the timing and number of fractions is relevant for the induction of a pro-immunogenic molecular signature.
Adaptation to immune checkpoint blockade
Early radiation-induced adaptations can be exploited for immunotherapy and molecular-targeted therapy (16,22–24). Dewan and Vanpouille-Box demonstrated that certain radiation doses and fractions, in combination with immune checkpoint inhibitors, were capable of inducing an immune response that produced an abscopal effect (23,25). Cytosolic DNA that accumulates as a consequence of radiation activates the cGAS/STING pathway with downstream induction of interferon type I (IFN-I) and IFN-stimulated genes. This response can be antagonized by the DNA exonuclease Trex1. IFN-I is produced during acute viral infections and plays a key role in the activation of cytotoxic CD8 T-cells that eliminate infected cells. In tumors, the acute IFN-I production triggered by radiation elicits anti-tumor CD8 T-cells (23,26); however, chronic IFN-I stimulation has been associated with therapeutic resistance (27,28), underscoring the importance of the adaptation as an evolving response that influences the type of tumor microenvironment that develops.
Given that radiation can generate anti-tumor T-cells, another type of early adaptation observed in response to fractionated radiation is the upregulation by tumor cells of immune checkpoint ligand PDL-1, which protected tumor cells from immune-mediated rejection. In this case, concomitant blockade of PDL-1 improved responses by enabling T-cells to reject the tumor, providing an example of a targetable adaptation that sensitizes the tumor to immunotherapy. PDL-1 upregulation was also implicated in radiation-induced tumor equilibrium as a chronic adaptation that led to a standstill between the tumor and the anti-tumor T-cells (29). We have recently shown that upregulation of the ectonucleotidase CD73 on breast cancer cells represents another example of early adaptation to radiation (30). CD73 generates adenosine, a pleotropic immunosuppressive mediator, preventing the infiltration of the tumor by antigen-presenting cells while increasing regulatory T-cells. The details of CD73 signaling pathways and metabolism in tumors have been previously reviewed in depth (31,32). In our recent study, antibody-mediated blockade of CD73 improved tumor response to radiation significantly.
These above findings shed light on how radiation might be used in combination therapy by modifying treatment based on biological adaptation. Effective treatment should strategically exploit adaptations rather than empirically following an initial regimen and examining changes only at the time of disease progression. Our previous work demonstrated that change in expression of a gene is not predictable based upon its initial expression (9).
II. Adaptations can be targeted and lead to cell killing
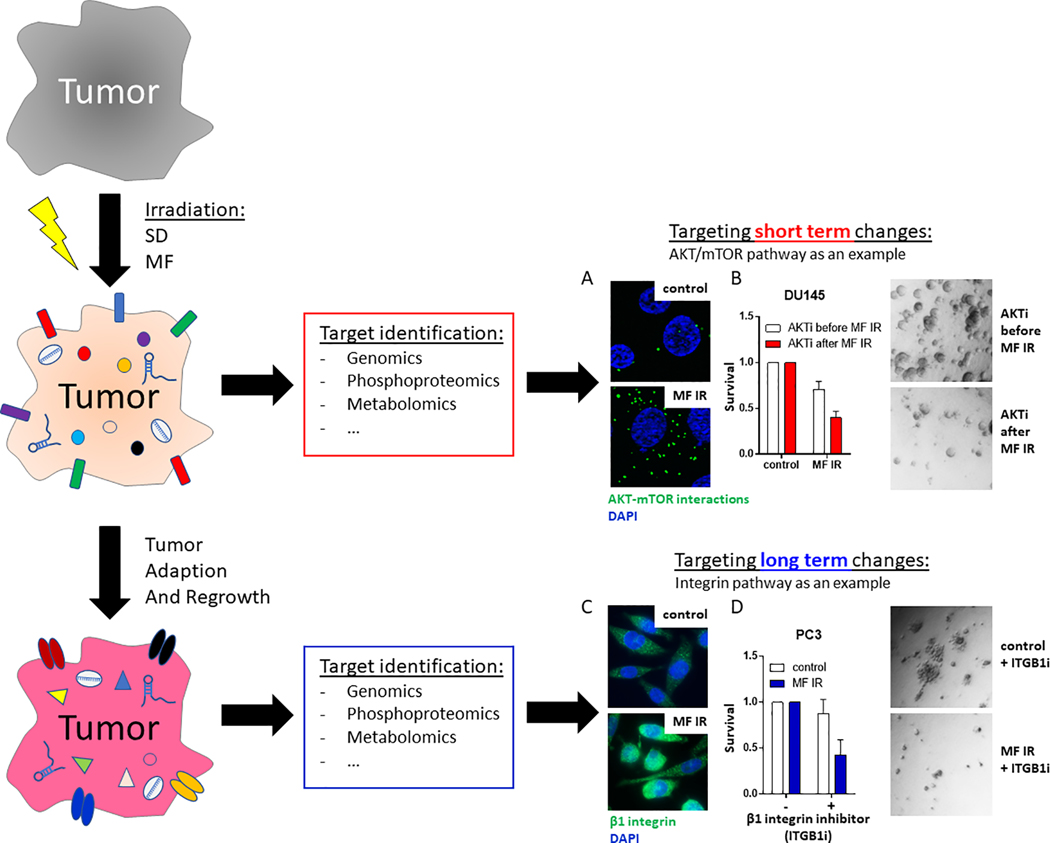
There are a range of changes that occur to enable molecular target definition (33). In proof-of-principle experiments we measured phospho-protein changes that occurred 2 hours after the last radiation dose. Using a more physiological 3-dimensional cell culture system and with the goal of targeting inducible changes post-radiation, we demonstrated targetable activation of the AKT/mTOR (Fig. 2) (34). To test the hypothesis of dose and fractionation dependence of adaptation, a regimen of three daily 2 Gy fractions was compared to a single dose of 6 Gy. Phospho-proteomic upregulation of AKT and mTOR and increased protein-protein interaction were observed in DU145 cells at 2 h after multifractionated RT (MF, 3 × 2 Gy) but not after a single dose of 6 Gy (Fig. 2A). When drug treatment was given before and during fractionated RT there was no enhancement in cell killing (Fig. 2B); however, when given 2 hours following completion of MF radiation—at which point the activated AKT/mTOR signaling was observed—the efficacy of the AKT inhibitor was significantly increased.
Figure 2: Target identification and inhibition for short-term and long-term cultures.
Tumors were irradiated with single dose (SD) or multifractionated (MF) irradiation. Within the first 24 h, short-term changes in mRNA, protein phosphorylation and metabolism were examined. (A) Activated AKT-mTOR signaling was (B) targeted with a small molecule AKT inhibitor (AKTi) Additionally, long-term changes in target expression were evaluated after tumor regrowth. (C) At 2 months after irradiation, β1 integrin was overexpressed in prostate cancer cells. (D) Inhibition of β1 integrin (ITGB1i) resulted in decreased clonogenic survival of MF-irradiated cells.
Early work by Aryankalayil, indicated that adaptation can persist up to 72 hours, the latest time point tested (9). To examine the duration of the adaptation, Eke treated PC3 cells with MF (1 Gy x 10) and SD (10 Gy x 1) and cultured cells for 2 months (34). Initial cell growth was significantly slower but resumed at pre-radiation growth rate after approximately 6 weeks. Based on a long-standing interest in integrin biology and radiation enhancement with integrin-targeted therapy (35–37), the expression of integrins and the ability to target them was studied in the cells that survived SD and MF at 2 months following their last radiation treatment (Fig. 2C, D). Integrin β1 and β4 were upregulated after SD and MF compared to the mock treated controls. The ability of antibodies against β1 (AIIB2) to kill cells was enhanced following long-term adaptation after radiation when compared to unirradiated control cultures. These experiments demonstrate that post-RT adaptation persists in the surviving cells, and can be targeted long after radiation treatment (38).
Our ongoing observations from the inducible changes demonstrate there are more changes early after radiation from MF compared to SD (12,16,21,38); however, at 2 months the pattern is reversed with cells that survived SD showing more changes. These data will be the subject of future reports. As noted below, understanding and exploiting this adaptation is a key emphasis of improved molecular-targeted therapy.
III. Metabolic adaptations after radiation
Alterations in tumor metabolism, with a focus on glucose utilization, have been studied after radiation injury with the goal of more effectively destroying cancer cells (39–41). However, these studies do not necessarily take into account the speed with which these changes may occur, differences in SD versus MF schedules and to what extent these changes continue post radiation.
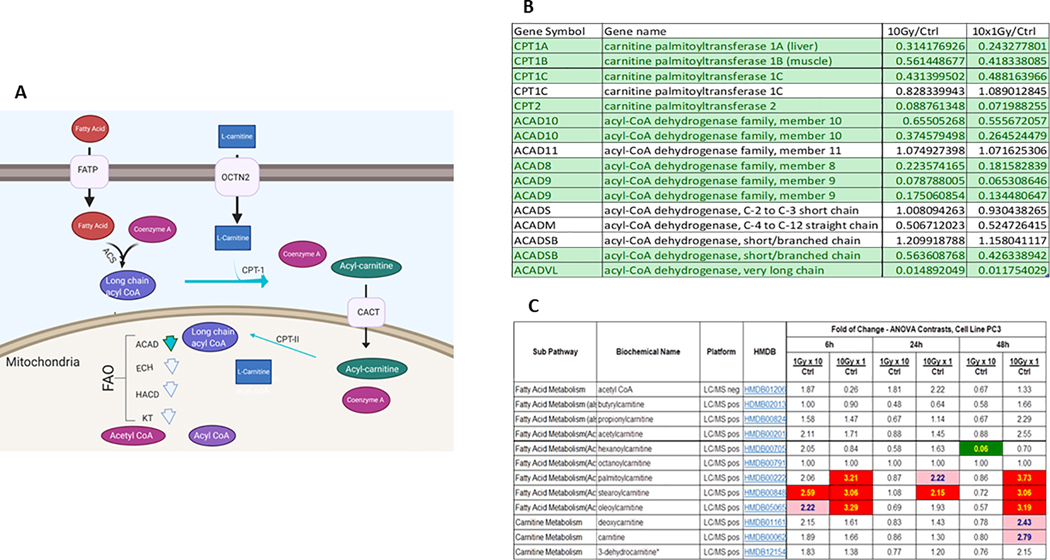
Preliminary metabolomic data from our laboratory indicates dynamic metabolic changes assessed by both gene expression and metabolite content at early time points between 6h, 24h and 48h after both MF (1 Gy x 10) and SD (10 Gy x 1) radiation in PC3, DU145, and LNCaP cells (Supplemental Table S1). Of interest were the changes in fatty acid oxidation after radiation treatment. Long chain fatty acids enter the cell primarily through a protein mediated system (42). Once in the cell they are bound to Coenzyme A via acyl-CoA synthases (43). Fatty acids then bind to L-carnitine via Carnitine palmitoyltransferase 1 (CPT-1), producing an acylcarnitine which is then ferried into the inner mitochondrial membrane for fuel in fatty acid oxidation (FAO) (Fig. 3A) (44). Microarray data from 24h post 10 Gy or 1 Gy x 10 radiation demonstrated downregulation of genes which regulate the long chain FAO pathway (Fig. 3B). IPA analysis (data not shown) indicates that these changes in FAO pathway gene expression lead to significant perturbations of the FAO pathway (Fig. 3C). This was consistent with observed alterations in certain acylcarnitines in PC3 cells after MF and SD radiation compared to controls. Increases in acylcarnitines are associated with impaired FAO (45,46). Information on acylcarnitine expression is routinely obtained using blood and serum samples (47) which might serve as a biomarker of effect. Differential carnitine and acylcarnitine expression has recently been proposed as a biomarker in hepatocellular carcinoma (48).
Figure 3: Perturbations in lipid metabolism after SD and MF radiation.
Figure 3A. Schematic representation of long chain fatty acid transport across cell membrane and into the mitochondria for FAO. Fatty acids utilize a protein mediated system to cross the cell membrane, for simplicity this is marked as fatty acid transport protein (FATP). Fatty acids are converted to acyl-CoAs to activate them. To enter the mitochondria, acyl-CoAs must be bound to carnitine. CPT1 converts acyl-Coas to acylcarnitines which allows the fatty acid to enter the inner mitochondrial membrane through CACT. CPTII converts acylcarnitines to acyl-CoAs. The acyl-CoA undergoes FAO. Chain length specific acyl-CoA dehydrogenase (ACAD) perform the initial step of FAO. The following three steps are catalyzed by enoyl-CoA hydratase (ECH), 3-Hydroxyacyl-CoA Dehydrogenases (HACD), and 3-Ketothiolases (KT). This produces one acetyl-CoA, one shortened acyl-CoA, NADH and FADH2, which can be fed into the TCA cycle. The shortened acyl-COA may undergo another round of FAO. Green arrows indicate decreased expression of enzymes in PC3 cells 24h post radiation as indicated in figure 3B.
Figure 3B. Microarray data from human prostate cancer cell PC3 24h after the end of SD or MF radiation exposure. Results are presented as irradiated divided by control. Green indicates that genes were statistically, significantly down regulated by paired T-test. CPT1 is considered the rate limiting enzyme for long chain fatty acid entry into the mitochondria for FAO. CPTII activates acyl-carnitines for FAO. Various members of dehydrogenase family each display specificity for fatty acid chains and are the first step of FAO.
Figure 3C. Metabolic data from PC3 cells 6h, 24h, and 48h after single dose (SD) or multifractionated (MF) radiation. Results indicate significant buildup of specific acylcarnitines at 6 and 48h after single dose(SD) radiation. Asterisk (*) indicates compounds identified based on the mass spectrometry data, but that do not currently have a reference standard to verify the identity.
Fatty acid metabolism is notably altered in cancer cells (49). This has recently spurred an interest in inhibitors of lipid metabolism, particularly fatty acid oxidation as novel treatments. This work is complex due to the multifaceted cell survival roles played by constituents of lipid metabolism. Some studies suggest L-carnitine itself has anti-tumorigenic effects due to its function as an HDAC inhibitor(50). Etomoxir, a CPT-1 inhibitor, has been tested as an anti-cancer agent (51). Another compound of interest, Mildronate, decreases L-carnitine entry into the cell by blocking organic cation transporter 2 (OCTN2) and inhibits endogenous L-carnitine production (52). Mildronate has been shown to decrease tumor growth in a rodent model of glioblastoma (53). Avocatin B, derived from avocados, has also been tested as a novel therapeutic in leukemia cells (54). It prevents FAO, potentially through competitive inhibition of fatty acids, accumulates in the mitochondria and increases ROS accumulation which triggers apoptosis (55). A more thorough understanding of the roles of short and medium chain fatty acids, acylcarnitines and fatty acid oxidation on cancer survival is necessary to develop effective combination therapies.
Ongoing work to characterize and utilize the potential tumor adaptation includes detailed studies of phospho-proteomics, metabolomics, DNA repair, and epigenetic changes, as well as further in vivo studies (16,34,56,57). Long-term studies of tumors post irradiation conducted with Citrin at NCI demonstrate long-term up regulation of integrins in PC3 tumors (38). Mitchell recently demonstrated an improvement in tumor growth delay when using a relatively standard combined chemo-radiation treatment (58). Extending drug treatment for 2 weeks post-radiation had a significantly greater effect than using the drug for 1 week only. The second week of drug treatment alone inhibited radiation-induced tumor vasculogenesis and thus delayed recurrent tumor growth. This finding is consistent with studies done by Brown et al where induction of SDF-1 is used to monitor radiation-induced vasculogenesis (59). Detailed analysis is now in progress; notably at week 1, many more transcriptional changes were observed with combined drug and radiation compared to drug or radiation alone suggesting perhaps the possibility of identifying novel targets for treatment.
While the adaptive paradigm needs more in vivo confirmation, the data demonstrate a) RT can induce an adaptive response that depends on dose and fractionation; b) adaptive changes occur early in treatment (days) and at the end of a week of treatment, and can persist for months, with the time interval to be studied; c) exploiting the adaptation by either taking advantage of it or interfering with it have potential for novel treatment opportunities; and d) the dose fraction size can impact immunotherapy with check-point inhibitors.
IV. Normal tissue adaptations can be exploited as biomarkers of injury and exposure
Acute and delayed normal tissue radiation damage is a dose-limiting factor for radiotherapy and a major concern of accidental radiation exposure. Our previous studies with HCAEC demonstrated that normal tissues also undergo radiation-induced molecular adaptations (6,7) (Fig. 1C) that could be exploited as biomarkers to predict and avoid or mitigate normal tissue injury, thereby improving patient outcomes. The potential of using RNA as a biomarker of tissue-specific injury and of general radiation exposure is increasing with the description of non-coding RNAs that are relatively stable in the blood, are organized in complex regulatory networks, and provide information on tissue-specific changes and identify pathways for injury mitigation.
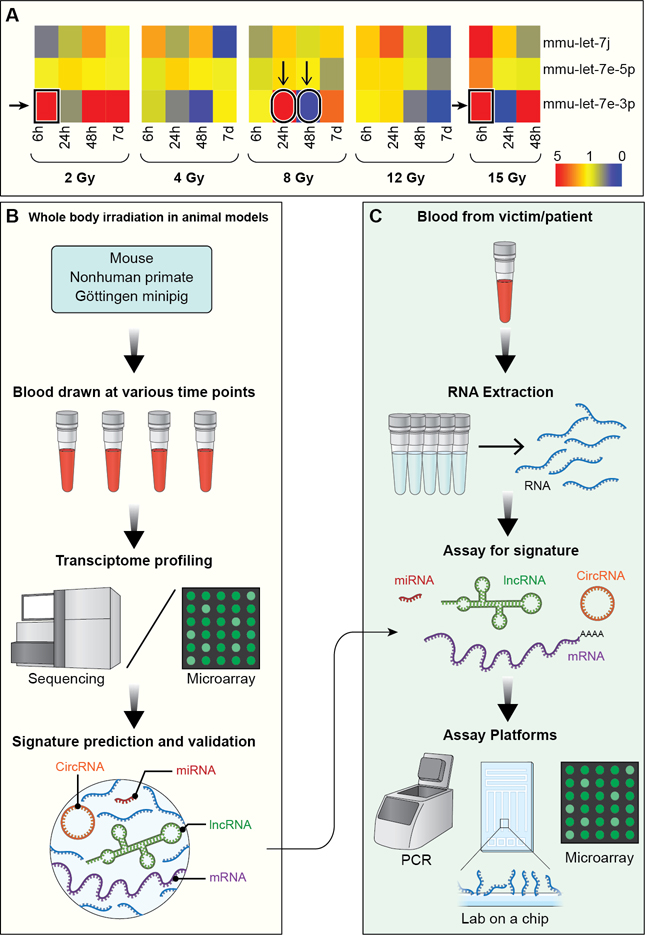
The implementation of biomarkers for radiation exposure and damaged normal tissue requires an analytical solution similar to the time- and dose-oriented changes for cancer treatment. Such a solution will require complex analyses using various models (e.g. Random Decision Forest, Support Vector Machine) and ultimately a time- and dose-oriented Markov decision tree (60). Critical to this development is identification of stable, reproducible and readily assayable markers that have a high specificity and sensitivity for detection in blood or other body fluids. To this end, Aryankalayil used a whole-body irradiated mouse model to identify significant alterations in the expression patterns of long non-coding RNAs (lncRNAs) and of miRNAs and target mRNAs at different timepoints after various levels of exposure (61,62). Importantly, these studies demonstrated that to triage victims of a radiological incident, multiple RNA biomarkers are needed to differentiate dose at different timepoints following exposure. For example, let-7e-3p may have utility within set of biomarkers, but the varying up- and down-regulation across doses and timepoints make it insufficient as a single marker (Fig. 4A). Additionally, several other groups reported plasma- and serum-based miRNA signatures that distinguish dose, including high vs. low and lethal vs. sub-lethal doses (63–65), further validating the potential of circulating RNA biomarkers for radiation biodosimetry and the importance of multiple biomarkers.
Figure 4: Predicting radiation exposure and normal tissue injury using an integrated RNA biomarker approach.
A) Heatmap displays fold change values of 3 different miRNAs of let-7 family, which share a seed sequence indicating shared targets, expressed in the blood of mice collected at various timepoints after whole body irradiation with various doses. Red signifies upregulated expression and blue signifies downregulated expression in comparison to the sham-irradiated controls. B) Schematic shows the methodology of utilizing animal models to predict dose-differentiating, RNA-based signatures from approximately 300–500μL of blood. C) Intended workflow, starting with a 500μL blood draw and using different high throughput diagnostic platforms, shows application of predicted signatures for triaging victims of accidental radiation exposure and assessment of normal tissue injury after radiotherapy.
Ongoing work continues to identify circulating RNA biomarkers for a biodosimetry decision tree using mouse, mini-pig, and non-human primate models (Fig. 4B). Tissue-specific injury markers are also being identified, which could be most useful for discerning tumor and normal tissue adaptations during radiotherapy. Additionally, studies of the microenvironment immune response to local tumor irradiation have demonstrated the role of normal tissue immune regulation on tumor recurrence, metastasis, and response to therapy (66–68). Investigation into molecular biomarkers of this immune response could therefore be informative in guiding more effective treatment regimens. Ultimately, molecular signatures for each application (e.g. biodosimetry, stromal response to radiotherapy) can be implemented into a molecular diagnostic workflow (Fig. 4C). While blood-based RNA is currently the primary molecule of interest, other sources of circulating markers, such as exosome-derived RNA, are also being evaluated.
V. Gaps to fill toward clinical implementation
Clinical application requires a more thorough understanding of the underlying biology and the development of biomarkers or imaging strategies to assess the tumor and normal tissue. Clinical trials could then be designed with biomarkers to validate the preclinical data. Table 1 includes ongoing studies and considerations to further develop clinical targeting of RT-adaptation.
Table 1.
Gaps and work-in-progress
| Topic | Ongoing and future work | Details and hypotheses |
|---|---|---|
| Broadening of cell lines | Expand study of cell lines with germline mutations, with initial focus on lung cancer | Lung cancer has a dual potential for immunotherapy and molecular-targeted therapies |
| Increase in spectrum of prostate cancer cell lines | Prostate cancer often treated with primary radiation, so this will enable adaptive effects to be studied | |
| Development of a platform to study the response to radiation in patient-derived xenografts, cell lines, and organoids, ideally from consecutive biopsies | Personalized medicine will enable more accurate precision medicine | |
| Target determination | Identification of biomarkers of adaptation from an array of “omics” combined with proteomics or phosphor-proteomics | Likely to help define specific targets, including non-coding RNAs, e.g. miRNA (69) as potential targets |
| Use of immunohistochemistry and profiling of subclones over different time points to study the proportion of cells that adapt, in addition to examining the cells that survive the post-exposure “drugging” | Adaptations are likely to be heterogeneous, possibly transient, and tumor adaptation to drugs need to be determined | |
| Timing and transition of adaptation | Study additional tumor types and additional time points between the end of radiation and 2 months, as well as beyond 2 months | There are at least 2 general adaptation points: 1) starts during therapy, within days, whereby MF produces more changes than SD; and 2) starts by 2 months or later and SD predominates. This time course needs to be better defined |
| Mechanism of adaptation | Epigenetic adaptations | Preliminary data suggest that there are epigenetic changes and, if so, when does this occur and for how long does it persist? |
| Ongoing in vivo studies to examine growth delay and related biological changes for radiation, drugs, and the combination | Potential collaborations with laboratories studying charged particle therapy | |
| Normal tissue changes | Expansion of the whole-body and organ-specific radiation biomarkers | Provide biomarkers for radiation biodosimetry and normal tissue adaptations from therapy |
| Pursuant to our study of endothelial changes (19) further study of radiation inducible endothelial changes from in vitro cultures, organ-on-a-chip and in vivo experiments | Understand the role of endothelial damage in radiation injury. (This is supported in a number of laboratories by NIAIDa and BARDAb programs). | |
| Clinical applicability | Through collaboration, obtain clinical samples for normal tissue biomarkers with groups studying radiation biodosimetry | Clinical samples are limited by underlying medical conditions, dose delivered, and volume of tissue irradiated |
| Investigate the use of “radiation as a drug,” as part of an overall approach from the “Shades of Gy” workshop (13) toward “accurate, precision radiation medicine” (14) | Prospective intervention trials will depend on preclinical data. Some pre- and post-RT sampling may be done including pre- operative radiotherapy, intraoperative radiotherapy and brachytherapy |
NIAID Radiation and Nuclear Countermeasures Program. Available at: https://www.niaid.nih.gov/research/radiation-nuclear-countermeasures-program. Accessed April 24, 2020
BARDA Radiological/nuclear medical countermeasures. Availalble at: https://www.medicalcountermeasures.gov/barda/cbrn/radiological-and-nuclear-countermeasures/ Accessed April 24, 2020
VI. Summary
Evolution in the technological delivery of RT is accompanied by rapid progress in cancer genetics and biology to decipher key molecular signaling and survival pathways and identify druggable targets. In this setting, as described in this review, the biological perturbations induced by radiation could potentially be exploited to enhance precision oncology. This includes pathways that are induced immediately after treatment and that may be sustained throughout radiation fractionation and beyond, for weeks/months after the end of radiotherapy, as a result of what we have described as RT-induced adaptation. Using a range of “omics” it is possible to interrogate the time and type of adaptation radiation induces in cancer, when compared to the pre-treatment phenotype. We speculate that different pre-treatment genotypes will have specified types of adaptation (e.g. p53 normal vs abnormal) that will help guide the initial treatment as is being studied in the match-type trials (70). A correct interpretation of these results enables rational design of multimodality therapies. Some adaptations take place within the first 5–10 or so fractions (2–4 days) and may persist for a number of days; long-term adaptations occur up to 2 months after treatment completion, and possibly longer. Early adaptation was traditionally called a “stress response” and can now be interrogated on how it is changing the tumor biological phenotype. In each model, the exact timing and how different forms of adaptation occur and how long they persist remain to be defined. Likewise, adaptation may be influenced by the irradiated tumor microenvironment, and the crosstalk between the tumor and the host. Growing evidence shows that the early changes induced by radiation in surviving cancer cells can improve the recognition of the tumor by the adaptive immune system and generate targets for immunotherapy (22,71).
While little information exists about the mechanisms of late adaptation on the tumor interaction with the immune system, some clinical papers suggest that pretreatment with radiation of chemoradiation may enhance effects of immune checkpoint blockade (72). Research in this area is warranted, particularly since novel immunotherapies are becoming rapidly available.
The PK/PD of radiation does depend on dose and fractionation and on type of radiation (e.g., photons versus particles), such that radiation can and should be considered “like a drug”, and function as a key partner within the precision medicine paradigm. In multiple models we found that the initial gene expression pattern did not predict radiation-induced gene response, indicating that investigation of adaptation mechanisms and pathways requires sequential interrogation. As outlined in Table 1, much remains to be done. Nonetheless recognizing that adaptation occurs offers the opportunity for unique approaches to cancer treatment. It is clear that post-radiation, the tumor and normal tissue “know” they have been irradiated and display persistent changes that depend on the type of radiation, dose and fractionation and whether radiation was given alone or with other modifiers, as in combination with chemotherapy, molecular-targeted therapy or immunotherapy.
Defining an individual tumor’s adaptation to a radiation regimen permits testing treatments against newly emerged targets. Most importantly, radiotherapy can be used multiple times to enable testing different permutations of combinations. Indeed, drugs and/or immunological agents may become more effective after the adaptation, potentially inducing susceptibility to a drug based on cellular adaptation rather than a mutation. This could have major impact on return on investment for drug development as a drug deemed unlikely to work based on the initial tumor profile may become effective after adaptation. Complex analysis of biomarkers of both tumor and normal tissue response are warranted. The ability to utilize the focused biology (12) from RT is a potentially unique enhancement of both local and systemic therapy adding a new dimension to accurate, precision cancer care.
Supplementary Material
Supplementary Table S1. Metabolic data from PC3 cells 6h, 24h, and 48h after SD or MF radiation. Samples were sent to Metabolon for processing. Results indicate decrease in glycolysis intermediates in SD sample at 6h. No significant changes to TCA cycle intermediates. No significant changes to ATP at any time points measured.
Acknowledgments
This study was supported by the NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research (grant ZIA BC 010670 to C.N.C).
For research collaboration: Murali Cherukuri, Radiation Biology Branch; Deborah Citrin, Radiation Oncology Branch; James Hodge, Laboratory of Tumor Immunology and Biology; Mansoor Ahmed, Radiation Research Program; Laurel MacMillan and Rocco Casagrande, Gryphon Scientific, Takoma Park, MD; and for support of the projects, Kevin Camphausen, Chief Radiation Oncology Branch.
Footnotes
Disclosure of potential conflicts of interest:
The authors report no conflicting interests.
REFERENCES
- 1.Pajonk F, Vlashi E, McBride WH. Radiation resistance of cancer stem cells: The 4 R’s of radiobiology revisited. Stem Cells. NIH Public Access; 2010. page 639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golden EB, Formenti SC. Is tumor (R)ejection by the immune system the “5th R” of radiobiology? Oncoimmunology. Landes Bioscience; 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crowther AJ, Ocasio JK, Fang F, Meidinger J, Wu J, Deal AM, et al. Radiation sensitivity in a preclinical mouse model of medulloblastoma relies on the function of the intrinsic apoptotic pathway. Cancer Res. American Association for Cancer Research Inc.; 2016;76:3211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todorovic V, Prevc A, Zakelj MN, Savarin M, Brozic A, Groselj B, et al. Mechanisms of different response to ionizing irradiation in isogenic head and neck cancer cell lines. Radiat Oncol. BioMed Central Ltd.; 2019;14:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steel GG, Mcmillan TJ, Peacock JH. The 5rs of radiobiology. Int. J. Radiat. Biol. Informa Healthcare; 1989. page 1045–8. [DOI] [PubMed] [Google Scholar]
- 6.Palayoor ST, John-Aryankalayil M, Makinde AY, Falduto MT, Magnuson SR, Coleman CN. Differential expression of stress and immune response pathway transcripts and miRNAs in normal human endothelial cells subjected to fractionated or single-dose radiation. Mol Cancer Res. American Association for Cancer Research Inc.; 2014;12:1002–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai M-H, Cook JA, Chandramouli GVR, DeGraff W, Yan H, Zhao S, et al. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res. 2007;67:3845–52. [DOI] [PubMed] [Google Scholar]
- 8.John-Aryankalayil M, Palayoor ST, Cerna D, Falduto MT, Magnuson SR, Coleman CN. NS-398, ibuprofen, and cyclooxygenase-2 RNA interference produce significantly different gene expression profiles in prostate cancer cells. Mol Cancer Ther. 2009;8:261–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.John-Aryankalayil M, Palayoor ST, Cerna D, Simone CB, Falduto MT, Magnuson SR, et al. Fractionated radiation therapy can induce a molecular profile for therapeutic targeting. Radiat Res. 2010;174:446–58. [DOI] [PubMed] [Google Scholar]
- 10.Kim JJ, Tannock IF. Repopulation of cancer cells during therapy: An important cause of treatment failure. Nat. Rev. Cancer. Nat Rev Cancer; 2005. page 516–25. [DOI] [PubMed] [Google Scholar]
- 11.Makinde AY, John-Aryankalayil M, Palayoor ST, Cerna D, Coleman CN. Radiation survivors: understanding and exploiting the phenotype following fractionated radiation therapy. Mol Cancer Res. 2013;11:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makinde AY, Eke I, Aryankalayil MJ, Ahmed MM, Coleman CN. Exploiting Gene Expression Kinetics in Conventional Radiotherapy, Hyperfractionation, and Hypofractionation for Targeted Therapy. Semin Radiat Oncol. 2016;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed MM, Coleman CN, Mendonca M, Bentzen S, Vikram B, Seltzer SM, et al. Workshop Report for Cancer Research: Defining the Shades of Gy: Utilizing the Biological Consequences of Radiotherapy in the Development of New Treatment Approaches-Meeting Viewpoint. Cancer Res. 2018;78:2166–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman CN, Prasanna PGS, Bernhard EJ, Buchsbaum JC, Ahmed MM, Capala J, et al. Accurate, Precision Radiation Medicine: A Meta-Strategy for Impacting Cancer Care, Global Health, and Nuclear Policy and Mitigating Radiation Injury From Necessary Medical Use, Space Exploration, and Potential Terrorism. Int J Radiat Oncol. 2018;101:250–3. [DOI] [PubMed] [Google Scholar]
- 15.Coleman CN. Linking radiation oncology and imaging through molecular biology (or now that therapy and diagnosis have separated, it’s time to get together again!). Radiology. Radiological Society of North America; 2003;228:29–35. [DOI] [PubMed] [Google Scholar]
- 16.Simone CB, John-Aryankalayil M, Palayoor ST, Makinde AY, Cerna D, Falduto MT, et al. mRNA Expression profiles for prostate cancer following fractionated irradiation are influenced by p53 status. Transl Oncol. 2013;6:573–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stankevicius V, Kuodyte K, Schveigert D, Bulotiene D, Paulauskas T, Daniunaite K, et al. Gene and miRNA expression profiles of mouse Lewis lung carcinoma LLC1 cells following single or fractionated dose irradiation. Oncol Lett. Spandidos Publications; 2017;13:4190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung CM, Chen TW, Li SC, Ho MR, Hu LY, Liu WS, et al. MicroRNA expression profiles in human breast cancer cells after multifraction and single-dose radiation treatment. Oncol Rep. Spandidos Publications; 2014;31:2147–56. [DOI] [PubMed] [Google Scholar]
- 19.Palayoor ST, J-Aryankalayil M, Makinde AY, Cerna D, Falduto MT, Magnuson SR, et al. Gene expression profile of coronary artery cells treated with nonsteroidal anti-inflammatory drugs reveals off-target effects. J Cardiovasc Pharmacol. 2012;59:487–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Jong MC, Ten Hoeve JJ, Grénman R, Wessels LF, Kerkhoven R, Te Riele H, et al. Pretreatment microRNA expression impacting on epithelial-to-mesenchymal transition predicts intrinsic radiosensitivity in head and neck cancer cell lines and patients. Clin Cancer Res. American Association for Cancer Research Inc.; 2015;21:5630–8. [DOI] [PubMed] [Google Scholar]
- 21.Aryankalayil MJ, Makinde AY, Gameiro SR, Hodge JW, Rivera-Solis PP, Palayoor ST, et al. Defining molecular signature of pro-immunogenic radiotherapy targets in human prostate cancer cells. Radiat Res. 2014;182:139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demaria S, Coleman CN, Formenti SC. Radiotherapy: Changing the Game in Immunotherapy. Trends in Cancer. 2016;2:286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, Schneider RJ, et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun. NLM (Medline); 2017;8:15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed MM, Hodge JW, Guha C, Bernhard EJ, Vikram B, Coleman CN. Harnessing the potential of radiation-induced immune modulation for cancer therapy. Cancer Immunol Res. 2013;1:280–4. [DOI] [PubMed] [Google Scholar]
- 25.Dewan MZ, Galloway AE, Kawashima N, Dewyngaert JK, Babb JS, Formenti SC, et al. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti-CTLA-4 antibody. Clin Cancer Res. Clin Cancer Res; 2009;15:5379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burnette BC, Liang H, Lee Y, Chlewicki L, Khodarev NN, Weichselbaum RR, et al. The efficacy of radiotherapy relies upon induction of type I interferon-dependent innate and adaptive immunity. Cancer Res. Cancer Res; 2011;71:2488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. Cell Press; 2016;167:1540–1554.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DSA, Baker SW, Khodarev N, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. Proc Natl Acad Sci U S A; 2008;105:18490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang H, Deng L, Chmura S, Burnette B, Liadis N, Darga T, et al. Radiation-Induced Equilibrium Is a Balance between Tumor Cell Proliferation and T Cell–Mediated Killing. J Immunol. The American Association of Immunologists; 2013;190:5874–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wennerberg E, Spada S, Rudqvist N-P, Lhuillier C, Gruber S, Chen Q, et al. CD73 blockade promotes dendritic cell infiltration of irradiated tumors and tumor rejection [published online ahead of print, 2020 Feb 11]. Cancer Immunol Res. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B CD73: A novel target for cancer immunotherapy. Cancer Res. NIH Public Access; 2010. page 6407–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Leve S, Wirsdörfer F, Jendrossek V. Targeting the immunomodulatory CD73/adenosine system to improve the therapeutic gain of radiotherapy. Front. Immunol. Frontiers Media S.A.; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eke I, Makinde AY, Aryankalayil MJ, Ahmed MM, Coleman CN. Comprehensive molecular tumor profiling in radiation oncology: How it could be used for precision medicine. Cancer Lett. 2016;382:118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eke I, Makinde AY, Aryankalayil MJ, Sandfort V, Palayoor ST, Rath BH, et al. Exploiting radiation-induced signaling to increase the susceptibility of resistant cancer cells to targeted drugs: AKT and mTOR inhibitors as an example. Mol Cancer Ther. 2018;17:355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eke I, Deuse Y, Hehlgans S, Gurtner K, Krause M, Baumann M, et al. β 1 Integrin/FAK/cortactin signaling is essential for human head and neck cancer resistance to radiotherapy. J Clin Invest. 2012;122:1529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eke I, Zscheppang K, Dickreuter E, Hickmann L, Mazzeo E, Unger K, et al. Simultaneous β1 integrin-EGFR targeting and radiosensitization of human head and neck cancer. J Natl Cancer Inst. 2015;107. [DOI] [PubMed] [Google Scholar]
- 37.Eke I, Dickreuter E, Cordes N. Enhanced radiosensitivity of head and neck squamous cell carcinoma cells by β1 integrin inhibition. Radiother Oncol. 2012;104. [DOI] [PubMed] [Google Scholar]
- 38.Eke I, Makinde AY, Aryankalayil MJ, Reedy JL, Citrin DE, Chopra S, et al. Long-term Tumor Adaptation after Radiotherapy: Therapeutic Implications for Targeting Integrins in Prostate Cancer. Mol Cancer Res. 2018;16:1855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nylander V, Ingerslev LR, Andersen E, Fabre O, Garde C, Rasmussen M, et al. Ionizing radiation potentiates high-fat diet-induced insulin resistance and reprograms skeletal muscle and adipose progenitor cells. Diabetes. American Diabetes Association Inc.; 2016;65:3573–84. [DOI] [PubMed] [Google Scholar]
- 40.Kumari N, Das A, Bhatt AN. Interleukin-6 confers radio-resistance by inducing Akt-mediated glycolysis and reducing mitochondrial damage in cells. J Biochem. Oxford Academic; 2019;167:259–65. [DOI] [PubMed] [Google Scholar]
- 41.Shen H, Hau E, Joshi S, Dilda PJ, McDonald KL. Sensitization of glioblastoma cells to irradiation by modulating the glucose metabolism. Mol Cancer Ther. American Association for Cancer Research Inc.; 2015;14:1794–804. [DOI] [PubMed] [Google Scholar]
- 42.Schwenk RW, Holloway GP, Luiken JJFP, Bonen A, Glatz JFC. Fatty acid transport across the cell membrane: Regulation by fatty acid transporters Prostaglandins Leukot Essent Fat Acids Churchill Livingstone; 2010;82:149–54. [DOI] [PubMed] [Google Scholar]
- 43.Knottnerus SJG, Bleeker JC, Wüst RCI, Ferdinandusse S, IJlst L, Wijburg FA, et al. Disorders of mitochondrial long-chain fatty acid oxidation and the carnitine shuttle Rev. Endocr. Metab. Disord. Springer New York LLC; 2018. page 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo CY, Ann DK. When fats commit crimes: Fatty acid metabolism, cancer stemness and therapeutic resistance. Cancer Commun. BioMed Central Ltd.; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE, et al. Plasma Acylcarnitine Profiles Suggest Incomplete Long-Chain Fatty Acid β-Oxidation and Altered Tricarboxylic Acid Cycle Activity in Type 2 Diabetic African-American Women. J Nutr. Oxford University Press (OUP); 2009;139:1073–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saiki S, Hatano T, Fujimaki M, Ishikawa KI, Mori A, Oji Y, et al. Decreased long-chain acylcarnitines from insufficient β-oxidation as potential early diagnostic markers for Parkinson’s disease. Sci Rep. Nature Publishing Group; 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rinaldo P, Cowan TM, Matern D. Acylcarnitine profile analysis. Genet Med. 2008;10:156. [DOI] [PubMed] [Google Scholar]
- 48.Li S, Gao D, Jiang Y. Function, detection and alteration of acylcarnitine metabolism in hepatocellular carcinoma. Metabolites [Internet]. MDPI AG; 2019. [cited 2020 Feb 13];9:36 Available from: http://www.mdpi.com/2218-1989/9/2/36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer Br. J. Cancer. Springer Nature; 2020. page 4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, Liu N, Guo H, Liao S, Li X, Yang C, et al. L-Carnitine Is an Endogenous HDAC Inhibitor Selectively Inhibiting Cancer Cell Growth In Vivo and In Vitro. PLoS One. 2012;7:e49062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dheeraj A, Agarwal C, Schlaepfer IR, Raben D, Singh R, Agarwal R, et al. A novel approach to target hypoxic cancer cells via combining β-oxidation inhibitor etomoxir with radiation. Hypoxia (Auckland, NZ). 2018;6:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liepinsh E, Makarova E, Sevostjanovs E, Hartmane D, Cirule H, Zharkova-Malkova O, et al. Carnitine and γ-Butyrobetaine Stimulate Elimination of Meldonium due to Competition for OCTN2-mediated Transport. Basic Clin Pharmacol Toxicol. 2017;120:450–6. [DOI] [PubMed] [Google Scholar]
- 53.Fink MA, Paland H, Herzog S, Grube M, Vogelgesang S, Weitmann K, et al. L-carnitine–mediated tumor cell protection and poor patient survival associated with OCTN2 overexpression in glioblastoma multiforme. Clin Cancer Res. American Association for Cancer Research Inc.; 2019;25:2874–86. [DOI] [PubMed] [Google Scholar]
- 54.Lee EA, Angka L, Rota SG, Hanlon T, Mitchell A, Hurren R, et al. Targeting mitochondria with avocatin B induces selective leukemia cell death. Cancer Res. American Association for Cancer Research Inc.; 2015;75:2478–88. [DOI] [PubMed] [Google Scholar]
- 55.Tcheng M, Samudio I, Lee EA, Minden MD, Spagnuolo PA. The mitochondria target drug avocatin B synergizes with induction chemotherapeutics to induce leukemia cell death Leuk. Lymphoma. Taylor and Francis Ltd; 2017. page 986–8. [DOI] [PubMed] [Google Scholar]
- 56.Makinde AY, Eke I, Aryankalayil MJ, Ahmed MM, Coleman CN. Exploiting Gene Expression Kinetics in Conventional Radiotherapy, Hyperfractionation, and Hypofractionation for Targeted Therapy. Semin Radiat Oncol. 2016;26:254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eke I, Zong D, Aryankalayil MJ, Sandfort V, Bylicky MA, Rath BH, et al. 53BP1/RIF1 signaling promotes cell survival after multifractionated radiotherapy. Nucleic Acids Res. 2020;48:1314–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naz S, Sowers A, Choudhuri R, Wissler M, Gamson J, Mathias A, et al. Abemaciclib, a selective CDK4/6 inhibitor, enhances the radiosensitivity of non–small cell lung cancer in vitro and in vivo. Clin Cancer Res. American Association for Cancer Research Inc.; 2018;24:3994–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown JM, Thomas R, Nagpal S, Recht L. Macrophage exclusion after radiation therapy (MERT): A new and effective way to increase the therapeutic ratio of radiotherapy Radiother. Oncol. Elsevier Ireland Ltd; 2020. page 159–64. [DOI] [PubMed] [Google Scholar]
- 60.Sonnenberg FA, Beck JR. Markov Models in Medical Decision Making. Med Decis Mak. 1993;13:322–38. [DOI] [PubMed] [Google Scholar]
- 61.Aryankalayil MJ, Chopra S, Levin J, Eke I, Makinde A, Das S, et al. Radiation-Induced Long Noncoding RNAs in a Mouse Model after Whole-Body Irradiation. Radiat Res. Radiation Research Society; 2018;189:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aryankalayil MJ, Chopra S, Makinde A, Eke I, Levin J, Shankavaram U, et al. Microarray analysis of miRNA expression profiles following whole body irradiation in a mouse model Biomarkers. Taylor and Francis Ltd; 2018;23:689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao F, Liu P, Narayanan J, Yang M, Fish BL, Liu Y, et al. Changes in miRNA in the lung and whole blood after whole thorax irradiation in rats. Sci Rep. Nature Publishing Group; 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacob NK, Cooley JV., Yee TN, Jacob J, Alder H, Wickramasinghe P, et al. Identification of Sensitive Serum microRNA Biomarkers for Radiation Biodosimetry. PLoS One. PLoS One; 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Acharya SS, Fendler W, Watson J, Hamilton A, Pan Y, Gaudiano E, et al. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci Transl Med. 2015;7:287ra69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rafat M, Aguilera TA, Vilalta M, Bronsart LL, Soto LA, Von Eyben R, et al. Macrophages promote circulating tumor cell-mediated local recurrence following radiotherapy in immunosuppressed patients. Cancer Res. American Association for Cancer Research Inc.; 2018;78:4241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heinonen M, Milliat F, Benadjaoud MA, François A, Buard V, Tarlet G, et al. Temporal clustering analysis of endothelial cell gene expression following exposure to a conventional radiotherapy dose fraction using Gaussian process clustering. PLoS One. Public Library of Science; 2018;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guipaud O, Jaillet C, Clément-Colmou K, François A, Supiot S, Milliat F. The importance of the vascular endothelial barrier in the immune-inflammatory response induced by radiotherapy. Br. J. Radiol. British Institute of Radiology; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. Nature Publishing Group; 2017. page 203–21. [DOI] [PubMed] [Google Scholar]
- 70.Flaherty KT, Gray R, Chen A, Li S, Patton D, Hamilton SR, et al. THE MOLECULAR ANALYSIS FOR THERAPY CHOICE (NCI-MATCH) TRIAL: LESSONS for GENOMIC TRIAL DESIGN. JNCI J Natl Cancer Inst. Oxford University Press (OUP); 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Formenti SC, Rudqvist NP, Golden E, Cooper B, Wennerberg E, Lhuillier C, et al. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nat. Med. Nature Publishing Group; 2018. page 1845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaverdian N, Lisberg AE, Bornazyan K, Veruttipong D, Goldman JW, Formenti SC, et al. Previous radiotherapy and the clinical activity and toxicity of pembrolizumab in the treatment of non-small-cell lung cancer: a secondary analysis of the KEYNOTE-001 phase 1 trial Lancet Oncol. Lancet Publishing Group; 2017;18:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Metabolic data from PC3 cells 6h, 24h, and 48h after SD or MF radiation. Samples were sent to Metabolon for processing. Results indicate decrease in glycolysis intermediates in SD sample at 6h. No significant changes to TCA cycle intermediates. No significant changes to ATP at any time points measured.