Abstract
The importance of skeletal muscle for rib development and patterning in the mouse embryo has not been resolved, largely because different experimental approaches have yielded disparate results. In this study, we utilize both gene knockouts and muscle cell ablation approaches to re-visit the extent to which rib growth and patterning are dependent on developing musculature. Consistent with previous studies, we show that rib formation is highly dependent on the MYOD family of myogenic regulatory factors (MRFs), and demonstrate that the extent of rib formation is gene-, allele-, and dosage-dependent. In the absence of Myf5 and MyoD, one allele of Mrf4 is sufficient for extensive rib growth, although patterning is abnormal. Under conditions of limiting MRF dosage, MyoD is identified as a positive regulator of rib patterning, presumably due to improved intercostal muscle development. In contrast to previous muscle ablation studies, we show that diphtheria toxin A (DTA)-mediated ablation of muscle progenitors or differentiated muscle, using MyoDiCre or HSA-Cre drivers, respectively, profoundly disrupts rib growth and patterning. Further, a comparison of three independently derived Rosa26-based DTA knockin alleles demonstrates that the degree of rib perturbations in MyoDiCre/DTA embryos is markedly dependent on the DTA allele used, and may in part explain discrepancies with previous findings. The results support the conclusion that the extent and quality of rib formation is largely dependent on the dosage of Myf5 and Mrf4, and that both early myotome-sclerotome interactions, as well as later muscle-rib interactions, are important for proper rib growth and patterning.
Keywords: rib development, sclerotome, skeletal muscle, MyoD, Myf5, Mrf4, Myf6, myogenin, diphtheria toxin, DTA, cell ablation, musculoskeletal, myotome, myoblast, myogenesis, mouse embryo, knockout, Cre recombinase
Introduction
In vertebrate embryos, somites give rise to the musculoskeletal system of the trunk, dermis of the back, and limb skeletal muscles. Soon after their formation by sequential segmentation of the paraxial mesoderm, somites are transformed into three-layered structures comprised of the epithelial dermomyotome dorsally, the mesenchymal sclerotome ventrally, and the myotome − the earliest differentiating skeletal muscle of the embryo, derived from the dermomyotome − interposed between the sclerotome and dermomyotome. The ribs are comprised of a small proximal portion (costal head, neck and tubercles) associated with each corresponding thoracic vertebra, and a distal portion, which constitutes most of the length of each rib and either attaches to the sternum or “floats,” depending on anteroposterior position. Proximal and distal ribs originate from progenitors of the medial and lateral sclerotome of thoracic somites, respectively (Evans, 2003; Huang et al., 2000), and their development is controlled by distinct genetic mechanisms. Thus, mutations in Pax1 (Dietrich and Gruss, 1995; Koseki et al., 1993; Wallin et al., 1994) and Uncx4.1 (Evans, 2003; Mansouri et al., 2000) selectively affect proximal rib development, whereas somatopleural signals—notably, members of the BMP family—are required for specification of the lateral somite (Pourquié et al., 1996; Tonegawa et al., 1997) and for proper growth of the distal rib into the somatopleure (Sudo et al., 2001). Further, distal rib development and patterning is specifically disrupted by perturbations of skeletal muscle development (Braun et al., 1992; Braun and Arnold, 1995; Dickman et al., 1999; Hasty et al., 1993; Henderson et al., 1999; Huang et al., 2003; Tallquist et al., 2000; Tremblay et al., 1998).
In the mouse, all four members of the myogenic regulatory gene family (MyoD, Myf5, Mrf4 and myogenin) are activated over a narrow developmental window (E9.5 to E9.75) in the ventrolateral (hypaxial) myotome/dermomyotome of thoracic somites (Chen et al., 2002; Chen and Goldhamer, 2004; Kassar-Duchossoy et al., 2004; Summerbell et al., 2002, 2000; Tajbakhsh et al., 1997; Vinagre et al., 2010) in close proximity to sclerotomal progenitors of the distal ribs (Scaal, 2016). The consequences on rib development of knocking out members of this gene family are highly variable and are both gene- and allele-specific. Notably, the severity of rib defects is correlated with the degree to which myotome formation or function is disrupted. Thus, the original knockout allele of Myf5 (Myf5m1) causes a delay of approximately 2 days in myotome formation and mice exhibit severe truncations of the distal ribs (Braun et al., 1992). Embryos carrying other Myf5 null alleles show varying degrees of rib defects from no effect to severe (Kassar-Duchossoy et al., 2004; Kaul et al., 2000; Tajbakhsh et al., 1996; Tallquist et al., 2000), a range of phenotypes that likely reflects the degree to which expression of the closely linked Mrf4 gene is also affected in cis, which effectively produces double knockout mice in severe cases (Floss et al., 1996; Kassar-Duchossoy et al., 2004; Tallquist et al., 2000; Yoon et al., 1997). Mild to severe rib defects were also described in three independently derived Mrf4 knockout lines (Braun and Arnold, 1995; Patapoutian et al., 1995; Zhang et al., 1995), with severe rib truncations being restricted to the Mrf4 allele that causes a substantial delay in myotome formation and loss of Myf5 expression in the early somite (Braun and Arnold, 1995; Floss et al., 1996; Yoon et al., 1997). Collectively, these data support the conclusion that either Myf5 or Mrf4 is required for the timely formation of the myotome, and substantially delayed myotome formation results in severe rib developmental defects. Myotome formation is normal in mice carrying null mutations in either MyoD and myogenin, and these mice exhibit no rib defects or comparatively modest rib patterning defects, respectively (Hasty et al., 1993; Rudnicki et al., 1992). Muscle differentiation is severely impaired in myogenin KO mice (Hasty et al., 1993; Nabeshima et al., 1993), and rib patterning defects in these mice likely reflect a structural and biomechanical role of intercostal muscles in rib morphogenesis (Hasty et al., 1993).
Muscle ablation studies have yielded results that are seemingly at odds with the considerable evidence supporting the importance of skeletal muscle in rib development. DTA-mediated ablation of all differentiated skeletal muscle using a myogenin-Cre driver (Li et al., 2005) did not result in rib abnormalities (Gensch et al., 2008). Further, despite the early expression of Mrf4 (also known as Myf6) in the hypaxial dermomyotome/myotome (Kassar-Duchossoy et al., 2004; Summerbell et al., 2002; Vinagre et al., 2010), and the eventual loss of all differentiated skeletal muscle in Myf6-Cre/DTA mice, rib development was unaffected (Haldar et al., 2008). Finally, whereas variable and sometimes severe rib defects were observed in two independent Myf5-Cre/DTA models (Gensch et al., 2008; Haldar et al., 2008) cells of rib primordia were lineage labeled in Myf5-Cre embryos, likely as a consequence of transient activity of the Myf5 locus in the paraxial mesoderm (Gensch et al., 2008), thereby complicating interpretation of rib abnormalities in Myf5-Cre/DTA mice. In fact, investigators concluded that these rib defects were the result of direct, cell-autonomous, killing of skeletogenic cells (Gensch et al., 2008; Haldar et al., 2008), rather than the consequence of perturbations of myotome or intercostal muscle development.
Here, we used both DTA-mediated conditional cell ablation and MRF knockout mice to re-investigate the requirement for skeletal muscles in rib development. We show that a single allele of Mrf4 is sufficient for substantial rib development in the absence of Myf5 and MyoD, and that MyoD is a positive effector of rib patterning under conditions of limiting MRF gene dosage. Ablation of either MyoD-expressing progenitors or differentiated muscle profoundly disrupted rib development, phenocopying features of severely affected knockouts of Myf5 and Mrf4. Further, the data support the conclusion that ongoing muscle-rib interactions, beyond somite stages, are required for normal rib growth and patterning. Finally, the severity of rib defects was highly dependent on the DTA allele employed, emphasizing the impact of choice of DTA allele on biological outcomes and their interpretations.
Materials and Methods
Mice and Genotyping
Animal procedures were reviewed and approved by the University of Connecticut’s Institutional Animal Care and Use Committee. Knockin and transgenic mice used in this study include the following: MyoDiCre (Kanisicak et al., 2009; JAX #014140, MyoDm1 (Rudnicki et al., 1992; JAX #002523, Myf5m1 (Braun et al., 1992; JAX #002522), Myf5loxP (Kassar-Duchossoy et al., 2004), ROSA26-eGFP-DTA (Ivanova et al., 2005; JAX #006331), ROSA-DTA (Voehringer et al., 2008; JAX # 009669), Rosa26-DTA176 (Wu et al., 2006; JAX #010527), HSA-Cre79 (Miniou et al., 1999; JAX #006139), and R26NZG and R26NG (Yamamoto et al., 2009; JAX# 012429). MyoDiCre expresses Improved Cre (iCre), which was modified from native P1 Cre such that codon usage was optimized for mammalian expression, an optimal Kozak consensus sequence was used, CpG content was minimized to reduce the likelihood of epigenetic silencing, and putative cryptic splice sites were eliminated (Shimshek et al., 2002). Experimental animals were maintained on an enriched FVB background. PCR-based genotyping of DNA isolated from tail biopsies or yolk sacs was done as previously described (Wood et al., 2013). Primers and PCR conditions for genotyping are listed in the Supplementary Table.
Mouse Crosses
Crosses to generate experimental embryos for DTA ablation and lineage labeling introduced the Cre allele through the male germline, as introduction through the female can occasionally result in Cre deposition in the egg and consequent global recombination (Wood et al., 2013; Yamamoto et al., 2009). Embryos with different combinations of MRF alleles were typically generated by crossing heterozygous parents, although in some cases, homozygous Myf5loxP/loxP mice were used to increase the efficiency of generating embryos of the desired genotype. Control embryos were either wild-type, or carried the Cre driver or DTA allele. No phenotypic differences among these genotypes were observed. Embryos were collected between E10.5 and E17.5, with noon on the day of the vaginal plug considered E0.5.
Alcian Blue and Alizarin Red Staining
Embryos were dissected into 1X PBS (20 mM sodium phosphate, 0.10 M NaCl, pH 7.4), after which they were fixed for 1 week in 95% ethanol, and then transferred to 100% acetone for another 1 week. During incubations in ethanol and acetone, solutions were changed two times. Following fixation and fat removal, embryos were transferred to Alcian Blue 8GX (Acros Organics) and Alizarin Red S (Sigma) staining solution (1 ml 0.14% AB in 70% ethanol, 1 ml 0.12% AR in 95% ethanol, adjusted to a final volume of 10 ml with 70% ethanol after adjusting to pH 3.5 with glacial acetic acid). After 3 days of incubation at 37°C with intermittent agitation, embryos were washed in tap water four times for 10 min each and then cleared in 1% potassium hydroxide (KOH) for 1–3 days at room temperature (RT), depending on embryonic stage. Embryos were then transferred to 20% glycerol in 1% KOH at RT. Embryos were incubated in this solution, with periodic changing of solution when needed, until completely cleared. Embryos were then transferred to 50% glycerol in 1% KOH, 80% glycerol in 1% KOH, and 100% glycerol, each for 1 day at RT. Embryos were stored in 100% glycerol.
Paraffin Embedding and Immunofluorescence
Embryos destined for paraffin embedding were dissected into 1X PBS, after which they were fixed in 2% paraformaldehyde (PFA) in 1X PBS for 2 hr at 4°C. Following fixation, embryos were washed three times for 10 min in 1X PBS. Embryos were then dehydrated by two, 10 min washes each at 35%, 50%, and 70% ethanol in 1X PBS. After storing overnight (O/N) in 70% ethanol in dH2O, embryos were incubated four times for 10 min each in 100% ethanol, after which they were transferred to a 1:1 ethanol/xylene solution for 5 min. Embryos were washed three times for 5 min in 60°C xylene, and then transferred to a 1:1 xylene and paraffin solution for 10 min at 60°C. Following four, 10 min incubations in 60°C paraffin, embryos were positioned in paraffin blocks and allowed to cool O/N at RT. Paraffin embedded embryos were stored at 4°C.
Paraffin embedded embryos were serial sectioned at 10 μm, collected on Superfrost Plus slides (Fisher), dried O/N at 37°C, and stored at 4°C. Antigen retrieval was used for detection of all proteins. Sections were removed from 4°C and allowed to warm to RT. Sections then underwent deparaffinization consisting of two, 5 min washes in xylene, followed by 5 min in 100% ethanol. Sections were then rehydrated in a stepwise manner in 95%, 85%, 70% and 50% ethanol each for 3 min, after which sections were washed 3 times for 5 min in 1X PBS. For antigen retrieval, slides were incubated for 6 min in −20°C methanol at −20°C, and washed in 1X PBS as above. Slides were then placed in 10 mM sodium citrate buffer, pH 6.0, which had been heated to 90°C in a water bath. Slides were incubated for 20 min at 90°C, after which they were rinsed in 1X PBS as before.
The following antibodies and working dilutions were used for immunofluorescence. SOX9: rabbit polyclonal antiserum (1:100 dilution; catalog #AB5535, Millipore), and MyHC: mAb MF20 (1:1 dilution of hybridoma supernatant; Developmental Studies Hybridoma Bank). For detection of SOX9, sections were blocked O/N at 4°C in PBSMBT (1X PBS, 1.5% Non-fat dried milk, 1.5% BSA, 0.1% Triton X-100). Sections were then incubated in primary antibody for 1 hr at RT. After three, 5 min washes in 1X PBS, sections were incubated in a 1:250 dilution of goat anti-rabbit Alexa Fluor 568 (Life Technologies) in PBSMBT for 1 hr at RT, washed in 1X PBS as above, and the signal amplified by incubation for 1 hr at RT in a 1:250 dilution of donkey anti-goat Alexa Fluor 568 (Life Technologies) in PBSMBT. Slides were washed in 1X PBS, stained with DAPI (0.1 μg/ml in 1X PBS) and coverslipped using Fluoro-Gel (Electron Microscopy Sciences). For the detection of MyHC, paraffin sections were rehydrated as stated above, and then processed for antigen retrieval and staining as previously described (Wood et al., 2013).
Whole Mount In Situ Hybridization and X-gal Staining
Whole mount in situ hybridization with digoxigenin-labeled probes followed published protocols (Henrique et al., 1995; Yamamoto et al., 2007), with minor modifications (Wood et al., 2013). Fgf4 (Hebert et al., 1990; Vinagre et al., 2010), and Pdgfa (IMAGE clone number 3495629,(Vinagre et al., 2010) probes have been previously described. Whole mount X-gal staining for detection of β-galactosidase (β-gal) was performed as previously described (Yamamoto et al., 2007).
RT-qPCR
Embryos were dissected into 1X PBS, heads were removed and embryos were frozen in liquid nitrogen, and stored at −80°C. For RNA extraction, samples were removed from −80°C and dissociated in RTL buffer by trituration using a pipette. RNA extraction was performed using the Qiagen mini kit (Cat #74104, Qiagen) and the DNase I kit (Cat #79254, Qiagen) according to the manufacturer’s protocol, and quantified using a Nanodrop (Thermo Fisher Scientific). First strand synthesis was performed using the iScript Advanced cDNA synthesis kit (BioRad) using 2.8 to 6 μg of RNA, depending on RNA yields. For qPCR, all samples were run using TaqMan Fast Advanced Master Mix (#4444964, Life Technologies) in 96-well plates on a BioRad CFX96 qPCR Real Time System machine. Each time point represents three biological replicates and each biological replicate was run in triplicate for each gene. Each biological replicate represents one embryo. Samples were analyzed using CFX Manager software. To minimize variability, the average Ct of the reference gene Gapdh was used in ΔCt calculations. Either available gene-specific assays, or primer/probe combinations that were designed for this study by the manufacturer (Life Technologies) were used as follows: MyoD (Mm00440387_m1), Myf5 (Mm00435125_m1), Mrf4 (Mm00435126_m1), myogenin (Mm00446195_g1), Gapdh (Mm99999915_g1), and Rosa26 exon1 (For. 5’-CAGAGAGCCTCGGCTAGGTA-3’, Rev. 5’-CTCCACCACGCTCGGA-3’, Probe: 5’-TCCCCGCAAACGCA-3’).
Detection of Apoptotic Cells by TUNEL Staining
The DeadEnd fluorometric TUNEL system (Promega) was used to detect apoptotic cells. Paraffin embedded embryos were processed as above. After rehydration, sections were fixed in 4% PFA in 1X PBS for 15 min at RT, then rinsed two times in 1X PBS. Sections were then incubated in 20 μg/ml proteinase K for 10 min, washed in 1X PBS, then refixed in 4% PFA in 1X PBS for 5 min at RT. After incubating in Equilibration Buffer for 10 min at RT, sections were incubated at 37°C for 1 hr in TUNEL reaction cocktail. Upon completion, sections were placed in 2X SSC for 15 min to halt the reaction, washed in 1X PBS, counterstained with DAPI and coverslipped using Fluoro-Gel. Note that GFP fluorescence from the R26GDTA allele is destroyed during tissue processing and does not interfere with fluorometric detection of apoptotic cells.
Microscopy and Image Collection
Imaging of whole mount E11.5 and E12.5 embryos was performed on a Leica MZFLIII fluorescence stereomicroscope equipped with a Spot RT3 camera and Spot Advanced image capture software (Diagnostic Instruments). ABAR stained fetuses were imaged using either the Leica MZFLIII stereomicroscope or a Pentax K-30 equipped with a SMC Pentax-D FA Macro 100mm F2.8 WR lens. Slides were imaged using a Nikon E600 upright microscope equipped with a Spot RT3 camera, or a Leica SP8 confocal microscope. All images were processed and assembled in Photoshop with only minor adjustments made to image brightness and contrast, when necessary.
Results
MRF dosage and positive effector functions of MyoD
To directly compare the consequences of muscle ablation and loss of MRF gene expression on rib formation, we produced mice with varying allelic combinations of Myf5, Mrf4 and MyoD, and assessed rib development at E17.5 by Alcian Blue and Alizarin Red (ABAR) staining. Three knockout alleles were used for this analysis: MyoDm1 (Rudnicki et al., 1992; the original Myf5 allele, Myf5m1 (Braun et al., 1992), which also abrogates early expression of the closely linked Mrf4 gene (Yoon et al., 1997); and Myf5loxP, a null allele of Myf5 that does not significantly affect myotomal expression of Mrf4 (Kassar-Duchossoy et al., 2004). As expected, Myf5m1/m1 mice, regardless of the status of the MyoD gene, exhibited profound truncations of the distal ribs (Fig. 1A, A’, B, B’; Table 1), with only the proximal ribs and the proximal-most portion of the distal ribs retained, as described (Braun et al., 1992). In contrast, MyoDm1/m1;Myf5loxP/+ mice, which harbor one wild-type allele of Myf5 and two functional alleles of Mrf4 (Table 1) showed a complete rescue of rib development (Fig. 1H, H’). Reducing the MRF dosage to one functional allele of both Myf5 and Mrf4 (MyoDm1/m1;Myf5m1/+ mice; Table 1) resulted in relatively modest rib defects, including imperfect spacing of anterior ribs and irregular spacing and morphology of more posterior ribs (Fig. 1F). Despite considerable patterning defects, MyoDm1/m1;Myf5loxP/loxP mice, which have two functional alleles of Mrf4 and are null for both Myf5 and MyoD (Table 1), were able to support formation of the rib cage (Fig. 1D). Notably, even a single functional allele of Mrf4 (MyoDm1/m1;Myf5m1/loxP mice; Table 1) supported rib development, with the extent of rib growth and the types and severity of patterning defects resembling mice with two functional Mrf4 alleles (Fig. 1C, D). Rib cage abnormalities in mice with one or two functional Mrf4 alleles included exaggeration of the aforementioned defects, anteroposterior compression of anterior ribs (likely owing to poor intercostal muscle development), and inconsistent connection of the non-floating false ribs to their more anterior neighbors distally (Fig. 1C). Finally, the ossified portion of vertebral rib segments showed pronounced regional thickenings or dilations that tended to be at a similar position along the proximodistal rib axis (Fig. 1C’, D’).
Figure 1.

Severity of rib defects in MRF knockout mice is gene-, allele-, and dosage-dependent. E17.5 fetuses of the genotypes shown were ABAR-stained for cartilage and bone. (A-H) Fetuses are ordered from most to least severe rib phenotypes. In some cases, differences in phenotypes between mice in adjacent panels were modest or inconsistent (C,D and E,F). Mice lacking both Myf5 and Mrf4 (A, B) showed the most severe phenotype, regardless of the presence of MyoD (B). One or two alleles of Mrf4 was sufficient for substantial rib development, although patterning was disrupted (C, D). Note that the number of active Mrf4 alleles was dictated by the Myf5 allele used (see Table 1 and text). MyoD substantially improved rib patterning in mice carrying one or two active Mrf4 alleles (C, D, vs E, G). (A’-H’) Dorsal views of fetuses corresponding to panels A-H. Rib rudiments in the most severely affected fetuses (A’, B’) typically terminated with a dilated knob-like structure (arrows). In C’, D’, E’, and G’, arrows show areas of abnormal rib thickenings. Forelimbs were removed to better visualize the ribs. Images shown do not necessarily reflect the same magnification. Fetuses were magnified to fit individual panels. Poor staining of posterior vertebrae in A is a staining artifact and not a consistent feature of this genotype.
Table 1.
MyoD and Myf5 null alleles used in this study, with resulting gene dosages of MyoD, Myf5 and Mrf4. The Myf5m1 allele (Braun et al., 1992) also abrogates expression of the closely linked Mrf4 gene in the embryo (Yoon et al., 1997), whereas myotomal Mrf4 expression is not appreciably affected in Myf5loxP/loxP embryos (Kassar-Duchossoy et al., 2004).
| (A) MyoDm1/m1;Myf5m1/m1 | 0 | 0 | 0 | 0 |
| (B) MyoDm1/+;Myf5m1/m1 | 1 | 0 | 0 | 1 |
| (C) MyoDm1/m1;Myf5m1/loxP | 0 | 0 | 1 | 1 |
| (D) MyoDm1/m1;Myf5bxP/loxP | 0 | 0 | 2 | 2 |
| (E) MyoDm1/+;Myf5m1/loxP | 1 | 0 | 1 | 2 |
| (F) Myo Dm1/m1;Myf5m1/+ | 0 | 1 | 1 | 2 |
| (G) MyoDm1/+;Myf5loxP/loxP | 1 | 0 | 2 | 3 |
| (H) MyoDm1/m1;Myf5loxP/+ | 0 | 1 | 2 | 3 |
Although MyoD-null mice do not exhibit rib defects (Rudnicki et al., 1992) and MyoD has not been implicated in rib development, MyoD had a substantial, positive effect on rib patterning under conditions of limiting MRF dosage. Thus, in mice carrying either one or two Mrf4 alleles in the absence of Myf5, a single dose of MyoD dramatically improved rib patterning (compare Fig. 1C and E; Fig. 1D and G; Table 1). The positive effector function of MyoD is most likely due to improved development of intercostal muscles, as Mrf4 alone can drive substantial, but only transient (at least through E12.5), muscle development in embryos lacking MyoD and Myf5 (Kassar-Duchossoy et al., 2004).
Ablation of developing skeletal muscle results in severe rib defects
To reassess the consequences of muscle ablation on rib development, we conditionally expressed wild-type DTA from the Rosa26eGFP-DTA allele (Ivanova et al., 2005; referred to here as R26GDTA) using the MyoDiCre driver (Kanisicak et al., 2009; Yamamoto et al., 2009), which is expressed in essentially all muscle progenitors (Wood et al., 2013). MyoDiCre-dependent expression of R26GDTA is highly effective at permanently ablating all skeletal muscle progenitors and differentiated muscle, as indicated by the complete loss of detectable PAX7, MYF5, MYOD, and myosin heavy chain (MyHC) proteins by E12.5, and MyoD and Myf5 mRNA by E13.5 (Wood et al., 2013). Further, unlike Myf5-Cre (Gensch et al., 2008; Haldar et al., 2008), MyoDiCre activity has not been detected in skeletogenic progenitors (Kanisicak et al., 2009; Wood et al., 2013), thereby avoiding the complication of direct ablation of rib progenitors. ABAR staining of MyoDiCre/+;R26GDTA/+ E17.5 fetuses revealed severe truncations of the distal ribs and other rib malformations (Fig. 2). The bony, dorsal, portion of most distal rib segments was greatly shortened and misshapen, had irregular contours, and exhibited a regional thickening characteristic of MRF knockout mice (Fig. 1C’, D’; Fig. 2B’). The cartilaginous portions of the distal ribs showed greater variability in their extent of development. Whereas most ribs either lacked cartilaginous elements entirely or cartilage only “capped” truncated bony segments (Fig. 2C, C’, D, D’), rib cartilage segments occasionally extended to the sternum. In fact, rib 2 typically was small but complete, with connections at both vertebral and sternal ends (Fig. 2B, D’). Cartilage growth of more posterior ribs showed greater variability, often manifested as one or more highly irregular and bifurcated cartilaginous structures with no obvious physical connection to dorsal bony elements (Fig. 2B, C, C’, D, D’). Close examination of dissected tissues did not reveal any obvious structural continuity between dorsal bony elements and these amorphous cartilaginous elements or evidence of rib breakage (data not shown), arguing against their introduction during handling or staining artifacts as causes of this apparent discontinuity.
Figure 2.
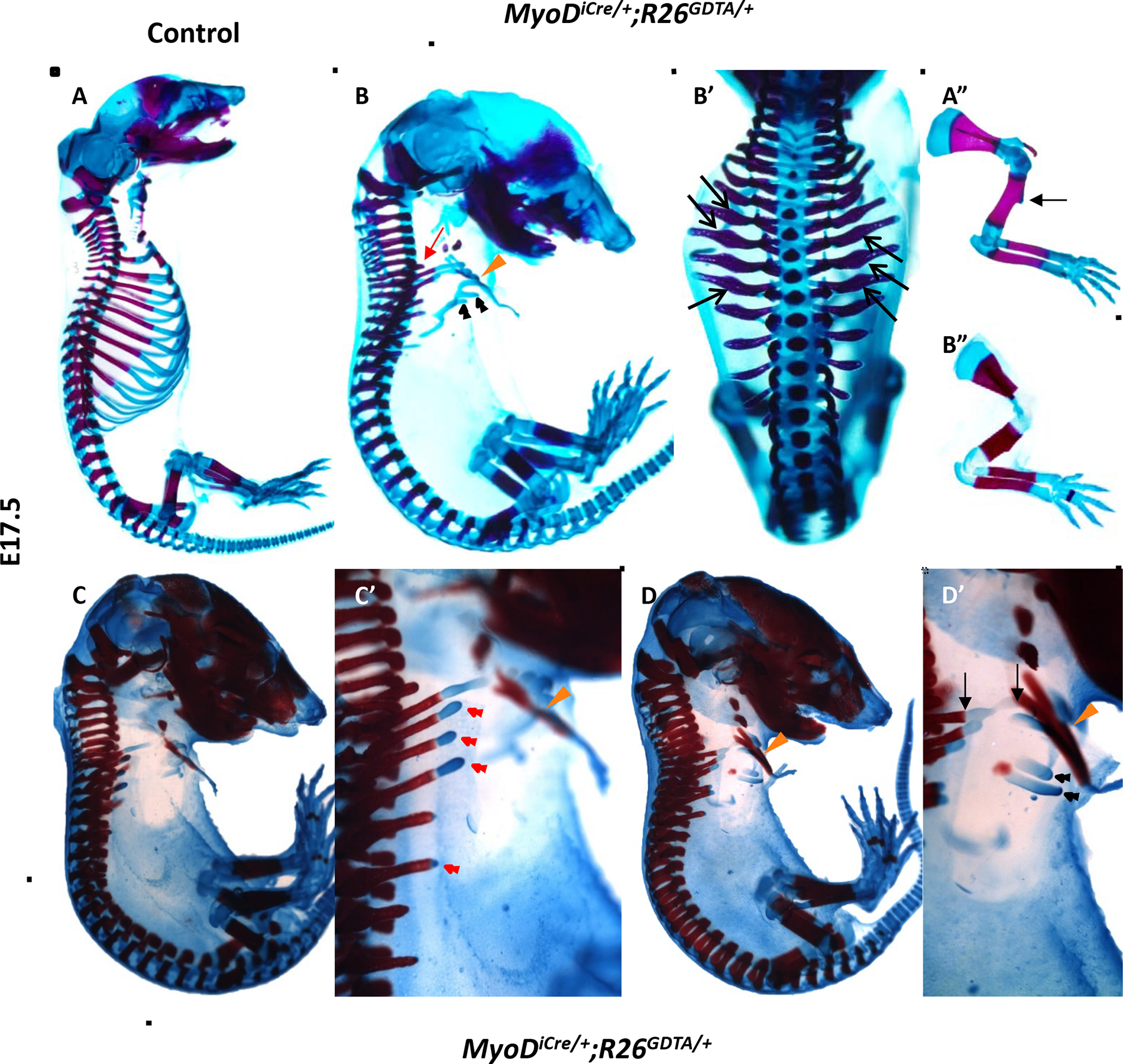
Ablation of MyoD+ progenitors markedly disrupts rib development, as shown by whole mount ABAR staining of E17.5 fetuses. (A, A”) Whole mount (A) and forelimb (A”) of control fetus. The deltoid tuberosity of the humerus (arrow) is shown (A”). (B, B’) Lateral (B) and dorsal (B’) view of a MyoDiCre/+;R26GDTA/+ fetus. The first rib is marked with a red arrow. All ribs were severely truncated, except for the second rib, which typically approximated, and sometimes attached, to the sternum (orange arrowhead). In some fetuses, including the one shown, the third rib was less severely affected than the first rib or more posterior ribs. As in MRF knockout mice, ribs were irregularly shaped and often had local thickenings (B’, black arrows). Amorphous “floating” cartilaginous rib structures (black arrowheads), with no obvious connection to more dorsal bony elements, were often observed (B). (B”) The forelimb of the MyoDiCre/+;R26GDTA/+ fetus shown in B and B’. The humerus is shorter than in control mice, the size of the deltoid tuberosity is greatly reduced, and the radius and ulna are essentially touching. (C, C’, D, D’) Two additional examples of the rib phenotypes in MyoDiCre/+;R26GDTA/+ fetuses. (C’ and D’) are higher magnification images of C and D. These fetuses share all major features with the fetus shown in B and B’. Truncated ribs are often capped with cartilage (C’, red arrowheads). In the fetus shown in D and D’, the second rib approximates the sternum (orange arrowhead). At higher magnification, continuity between the portion of the second rib delimited by arrows is evident. Black arrowheads in D’ indicate floating cartilaginous rib structures. Forelimbs were removed to better visualize the ribs.
In addition to rib defects, MyoDiCre/+;R26GDTA/+ fetuses exhibited defects of the appendicular skeleton, phenocopying several of the features of MyoDm1/m1;Myf5m1/m1 amyogenic fetuses (Rot-Nikcevic et al., 2005; Rudnicki et al., 1993). The most prominent limb skeletal anomalies in MyoDiCre/+;R26GDTA/+ mice included loss or severe reduction of the deltoid tuberosity of the humerus, decreased separation of the zeuogopodal bones (radius/ulna and tibia/fibula), shortening and thickening of the humerus, and reduced length of the primary diaphyseal ossification center of the humerus (Fig. 2A”, B”; data not shown).
Pdgfa and several Fgfs have been implicated in mediating myotome-sclerotome interactions important for rib development (Grass et al., 1996; Huang et al., 2003; Patapoutian et al., 1995; Tallquist et al., 2000; Vinagre et al., 2010). Consistent with the effects of abrogating Myf5 and Mrf4 expression in the mouse, targeting expression of DTA to MyoD+ progenitors disrupted Pdgfa and Fgf4 expression. E10.5 MyoDiCre/+;R26GDTA/+ embryos exhibited a diminution of myotomal MyHC expression, particularly in the hypaxial domain of interlimb somites (Supplementary Fig. 1; Wood et al., 2013) and, correspondingly, expression of Fgf4 was differentially lost in hypaxial myotomes at this stage (Fig. 3A, B). This differential effect on expression of Fgf4 in the hypaxial myotome phenocopies the effect of Dll1-Hoxa10 expression in transgenic mice, which showed severe rib truncations (Carapuço et al., 2005; Vinagre et al., 2010) that resemble those of Myf5m1/m1 embryos (Braun et al., 1992). Myotomal expression of both Pdgfa and Fgf4 was reduced at E11.5 (Fig. 3C–F), with preferential mRNA loss in hypaxial myotomal domains, although there was also an apparent reduction in mRNA abundance in epaxial somite domains, most evident for Fgf4 in cervical somites (Fig. 3C, D). Reduced and delayed effects on gene expression in epaxial domains of thoracic somites likely reflect the lower levels and delayed onset of MyoD expression (and therefore MyoDiCre expression) in epaxial myotomes (Chen et al., 2002; Chen and Goldhamer, 2004), and the correspondingly slower kinetics of DTA-mediated ablation.
Figure 3.
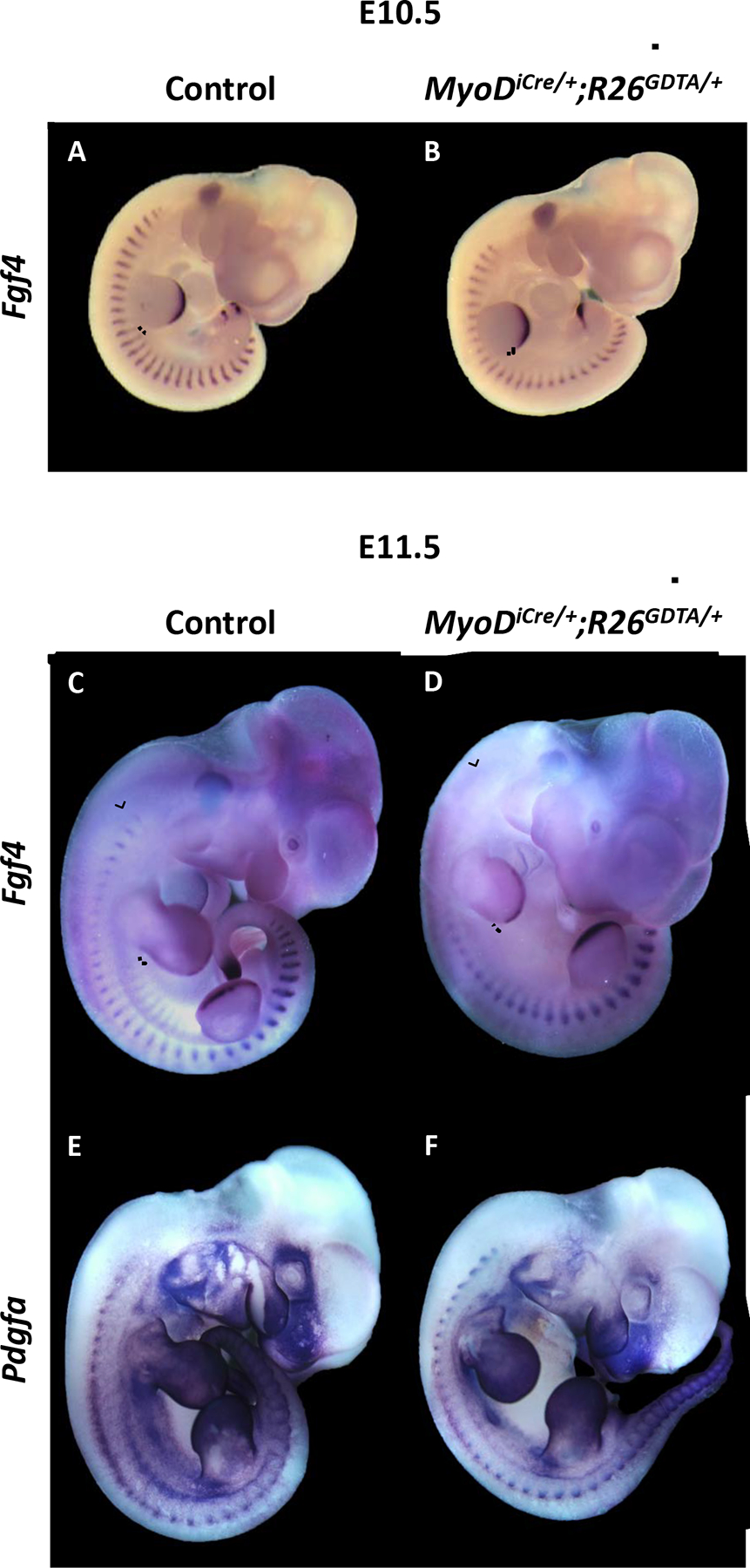
Whole mount in situ hybridization for Fgf4 and Pdgfa transcripts in control and MyoDiCre/+;R26GDTA/+ embryos. (A-D) Hypaxial expression of Fgf4 is lost by E10.5 in the interlimb somites of MyoDiCre/+;R26GDTA/+ embryos (A, B). The differential effect of DTA expression on Fgf4 expression in both hypaxial and epaxial somite domains is also evident at E11.5 (C, D). The ventrolateral extent of Fgf4 expression in control embryos, and the corresponding position in DTA-expressing embryos, is shown by a dotted line. Epaxial loss of Fgf4 caused by DTA expression and the corresponding area in the control is shown by brackets. (E, F) Diminution of myotomal Pdgfa expression in an E11.5 MyoDiCre/+;R26GDTA/+ embryo (F) compared to a control (E).
Selectively targeting differentiated muscle for ablation also causes severe rib truncations
As ablation of MyoD+ cells eliminates both myogenic progenitors and differentiated skeletal muscle (Wood et al., 2013), analysis of MyoDiCre/+;R26GDTA/+ mice cannot distinguish whether muscle progenitors, differentiated muscle, or both, are required for normal rib development. Therefore, we specifically evaluated the requirement for differentiated muscle using HSA-Cre79 (HSA-Cre; also known as ACTA1-Cre) transgenic mice, in which Cre expression is directed by the α-skeletal actin promoter (Miniou et al., 1999). Cre-dependent reporter gene expression was evident in the myotomes of anterior thoracic somites by E9.5 (Fig. 4B), and MyHC-positive muscle was undetectable by E11.5 in HSA-Cre;R26GDTA/+ embryos (Wood et al., 2013). Notably, HSA-Cre;R26GDTA/+ embryos exhibited severe rib malformations (Fig. 4D–F). Rib growth and patterning defects were distinct from those of MyoDiCre/+;R26GDTA/+ embryos (Fig. 2B, C, D), which likely represents differences in the timing and pattern of expression of HSA-Cre and MyoDiCre drivers (Kanisicak et al., 2009; Miniou et al., 1999; Wood et al., 2013; Yamamoto et al., 2009). For example, unlike MyoDiCre/+;R26GDTA/+ fetuses, HSA-Cre;R26GDTA/+ fetuses lacked all but the most proximal portions of the distal ribs of thoracic ribs 1–3, but showed greater growth of more posterior ribs—albeit to varying degrees—compared to MyoDiCre/+;R26GDTA/+ mice (Fig. 2 and 4). Gross rib malformations in HSA-Cre;R26GDTA/+ mice suggest that muscle progenitors alone can provide limited trophic support, but are not sufficient to provide the appropriate signaling environment or structural and biomechanical influences required for normal rib development. We note, however, that muscle progenitor numbers were noticeably reduced between E11.5 and E12.5 in HSA-Cre;R26GDTA/+ embryos (Wood et al., 2013), presumably because the kinetics of ablation as cells underwent differentiation and expressed DTA outpaced progenitor cell expansion by proliferation. Thus, data presented here may under-represent the capacity of muscle progenitors to provide trophic support for rib development.
Figure 4.

DTA-mediated ablation of differentiated skeletal muscles causes marked rib truncations. (A-C) GFP lineage labeling in HSA-Cre;R26NG/+ embryos. GFP expression was detected in the myotomes of cervical somites at E9.0 (A), in anterior thoracic somites at E9.5 (B), and in somites along most of the body axis by E10.5 (C). White arrowheads delimit the anteroposterior extent of somitic GFP expression at each stage, and blue arrowheads identify GFP expression in the branchial arches. The asterisks in A and B show GFP expression in the heart (Miniou et al., 1999). Background fluorescence is visible in the head and surface of the embryos. (D-F) ABAR staining of three E17.5 HSA-Cre;R26GDTA/+ fetuses showing the consistency of rib malformations. (F’) Dorsal views of embryos shown in F. HSA-Cre;R26GDTA/+ fetuses show more severe truncations of anterior ribs compared to posterior ribs. Growth and patterning defects are distinct from those of MyoDiCre/+;R26GDTA/+ fetuses. Forelimbs were removed to better visualize the ribs. The first rib in each panel is designated with a red arrow.
Severity of rib abnormalities is dependent on choice of DTA allele
The dramatic effect of muscle ablation on rib formation shown here contrasts with previous studies in which targeting muscle progenitors and differentiated muscle (Myf6-Cre; Haldar et al., 2008) or differentiated muscle alone (myogenin-Cre; Gensch et al., 2008), had no effect on rib development. As these studies utilized different Cre drivers and Cre-dependent DTA alleles, we specifically tested the impact of choice of DTA allele on the kinetics of muscle ablation and extent of rib development. For this analysis, we crossed MyoDiCre mice with mice carrying Rosa26-DTA176 (Wu et al., 2006; referred to as R26DTA176), which encodes the attenuated form of DTA used by Haldar et al. (2008), as well as to an independently derived DTA line (ROSA-DTA; referred to as R26DTA) in which wild-type DTA was knocked into the Rosa26 locus (Voehringer et al., 2008). While muscle-specific nlacZ expression was undetectable in most MyoDiCre/+;R26NZG/GDTA embryos between E11.5 and E13.5 (Fig. 5B, F) (Wood et al., 2013), reporter gene expression in MyoDiCre/+;R26NZG/DTA176 embryos was substantially reduced, but readily detected (Fig 5C, G; Wood et al., 2013), a result consistent with the reduced activity of the attenuated form of DTA (Wu et al., 2006). Correspondingly, whereas MyHC staining was dramatically reduced at E11.5 and not detected at E12.5 in MyoDiCre/+;R26GDTA/+ mice, residual MyHC staining persisted in MyoDiCre/+;R26DTA176/+ fetuses until at least E16.5 (Wood et al., 2013). Unexpectedly, MyoDiCre/+;R26DTA/+ mice, despite expressing wild-type DTA, also showed slower kinetics of muscle ablation, as indicated by persistent β-gal staining at E11.5 and E12.5 (Fig. 5D, H). As an additional read-out of ablation kinetics, we also quantified MRF gene expression in the bodies of MyoDiCre/DTA mice by RT-qPCR. Notably, mRNA levels of MyoD, Myf5 and myogenin were significantly lower in E13.5 R26GDTA embryos compared to R26DTA176/+ and R26DTA/+ embryos (Fig. 6). Mrf4 abundance trended lower in R26GDTA embryos but did not reach statistical significance. Our previous (Wood et al., 2013) and present analysis is consistent with the work of Comai et al. (2014), who showed that loss of skeletal muscle in Myf5-Cre/DTA fetuses was severe when the R26GDTA allele was employed, in contrast to findings when other DTA alleles were used (Gensch et al., 2008; Haldar et al., 2008).
Figure 5.

Comparison of MyoDiCre-dependent nlacZ expression in three DTA mouse lines, as revealed by X-gal staining for β-gal. Representative images are shown. (A, E) Control embryos lacking a DTA allele. All myogenic regions of the embryo stain intensely for nlacZ. (B, F) nlacZ was typically undetectable in GDTA embryos. Occasionally, a barely detectable signal was observed in somites (e.g. B, arrow). (C, G) In embryos expressing DTA176, β-gal staining was substantially diminished, but remained evident in interlimb somites, limb buds, and in cervical regions of the embryos. (D, H) MyoDiCre/+ embryos carrying the independently derived wild-type DTA allele R26DTA showed β-gal staining that was comparable to, or greater than, that of MyoDiCre/+;R26DTA176/+ embryos (C, G). In C, D, G, and H, brackets and arrows show staining in interlimb somites and limb buds, respectively. Staining in cervical regions is not labeled.
Figure 6.

RT-qPCR analysis of MRF gene expression in DTA embryos between E11.5 and E13.5. Heads were removed prior to processing tissues for RNA extraction. Controls carried one of the three DTA alleles but lacked MyoDiCre. ΔCt values were calculated using Gapdh as the internal control. Data is represented as the mean of three biological replicates ± standard error. Each biological replicate is the mean of three technical replicates. Two-way ANOVA with Tukey’s multiple comparison statistical test was performed. * = p≤0.05, ** = p≤0.01, *** = p≤0.001, and **** = p≤0.0001. Statistical analyses were not performed between stages or between MRFs.
RT-qPCR analyses also showed that E11.5 to E13.5 R26GDTA/+ embryos had significantly higher Rosa26 transcript levels than either R26DTA176/+ or R26DTA/+ embryos (Supplementary Fig. 2), which may explain, at least in part, the more rapid ablation observed by conditional expression of the R26GDTA allele. The exon 1 primers used do not distinguish between wild-type and knocked in Rosa26 alleles; therefore, the actual differences in expression between DTA alleles may be greater than indicated by this analysis, assuming no differences between lines in expression levels from the unmanipulated allele. Although recombination efficiencies were not quantified, differences in kinetics of cell ablation could also reflect differences in the relative susceptibility of each DTA allele to Cre-mediated recombination (Liu et al., 2013).
Associated with the slower cell ablation kinetics, MyoDiCre/+;R26DTA176/+ and MyoDiCre/+;R26DTA/+ mice exhibited a dramatic increase in rib development and partial normalization of rib morphology (Fig. 7), with the extent of rib development resembling knockout mice carrying one or two functional alleles of Mrf4 (see Fig. 1). These data suggest that differences in the extent and kinetics of muscle ablation during critical periods of rib development contributed to the wide differences in rib growth and patterning observed between the present and previous studies (Gensch et al., 2008; Haldar et al., 2008).
Figure 7.

Partial normalization of rib development in MyoDiCre mice when cell ablation is mediated by R26DTA176 or R26DTA alleles. E17.5 fetuses were ABAR-stained for cartilage and bone. (A-C) Lateral views of MyoDiCre/+;R26DTA176/+ fetuses reflecting the range of phenotypes observed. (D-F) MyoDiCre/+;R26DTA/+ fetuses show a comparable or greater degree of rib development compared to MyoDiCre/+;R26DTA176/+ embryos. Forelimbs were removed to better visualize the ribs.
Perturbation of SOX9 expression and pronounced apoptosis in muscle-ablated embryos
A priori, development of ribs from sclerotomal progenitors could depend on interactions with skeletal muscle at any of several levels, including specification, proliferation, survival, condensation, or differentiation of cartilage or bone. To define the developmental status of presumptive skeletogenic progenitors in MyoDiCre/+;R26GDTA/+ embryos, sections of control and DTA-expressing embryos were immunofluorescently stained for SOX9 at E11.5 and E12.5. SOX9 is an obligatory transcription factor for cartilage development, and marks cells from early pre-cartilage mesenchymal condensations through mature cartilage stages (Akiyama et al., 2002; Bi et al., 1999; Wright et al., 1995). In control embryos, the entire length of pre-cartilage rib condensations at both stages stained strongly for SOX9 (Fig. 8A, G), as well as with the lectin peanut agglutinin (PNA; data not shown). In MyoDiCre/+;R26GDTA/+ embryos, pre-cartilage condensations of the distal rib rudiments typically included only the proximal-most portion, likely corresponding in length to the extent of rib outgrowth observed at E17.5. These condensations stained for SOX9 (Fig. 8D, J) and were PNA+ (data not shown), although staining intensity was weaker than in wild-type embryos. Importantly, areas of SOX9 expression in E11.5 MyoDiCre/+;R26GDTA/+ embryos were associated with MyHC+ skeletal muscle, whereas MyHC staining was absent adjacent to more ventral mesenchymal tissue that was negative for SOX9 (Fig. 8C, F) and PNA (data not shown). Interestingly, however, although SOX9+ condensations were absent from all but the most proximodorsal anatomical locations, DAPI-stained sections of E11.5 and E12.5 embryos showed apparent accumulations of mesenchyme organized into thin rod-like structures at ventral, somatopleural regions (Fig. 8F, L). Such accumulations apparently formed independent of SOX9 function, although we cannot rule out the possibility of transient SOX9 expression prior to E11.5 (see Wright et al., 1995). Collectively, these results indicate that skeletal muscle is required, directly or indirectly, for SOX9 expression, and in the absence of SOX9, mesenchymal rib progenitors are unable to condense into definitive, pre-cartilage rib anlagen in embryonic regions where ribs are ultimately absent in DTA embryos.
Figure 8.

Lack of SOX9 and MyHC staining is associated with areas of apoptosis in MyoDiCre/+;R26GDTA/+ embryos. (A-L) Each set of three images for each genotype and stage represent adjacent transverse 10 μm paraffin sections stained for either SOX9, TUNEL, or MyHC, and counterstained with DAPI. (A-C) At E11.5, pre-cartilage rib condensations of control embryos stained intensively for SOX9 (A), and little or no TUNEL signal was detected (B). Developing intercostal muscles are MyHC-positive (C). Note that a typical section plane may not capture the entire proximodistal extent of a given rib. (D-F) In MyoDiCre/+;R26GDTA/+ embryos, the ventral portions of ribs (brackets) lack SOX9 expression (D), are surrounded by MyHC-negative tissue (F) and are associated with intense TUNEL activity (E). Although the TUNEL signal co-localizes with apparent cell aggregations, apoptotic cells cannot be definitively assigned to a particular cell type due to the lack of markers for these aggregates and for DTA-expressing muscle cells. In proximodorsal regions of the distal rib shown, SOX9 expression is weak (D, arrow) and is associated with muscle that is still MyHC-positive (F, orange arrow). The corresponding TUNEL signal (E, orange arrow) likely represents dying muscle. (E’-E’”) Confocal images corresponding to the bracketed region in E showing co-localization of the DAPI-stained cell aggregate and the fluorescent TUNEL signal. Images are of a 1 μm optical section. The dotted line in E” represents the approximate boundary of the cell aggregate. The scale bar represents 100 μm. (G-I) Transverse section of E12.5 control embryo through the proximal aspect of a distal rib dorsally, and through developing intercostal muscles ventrally. As at E11.5, the SOX9 (G) and MyHC (I) signals are intense and the TUNEL signal (H) is low or at background. (J-L) At E12.5, the SOX9 signal persists dorsoproximally (J, arrow) and TUNEL staining is associated with cell aggregates that lack SOX9 staining (K, bracket). MyHC is now absent both in ventral and dorsal embryonic regions (L). (M, N) Merged images of three adjacent 10 μm frontal sections through the ventral body wall of E11.5 control and MyoDiCre/+;R26GDTA/+ embryos stained for SOX9, TUNEL, or MyHC. In the control embryo (M), six cross-sectional profiles of SOX9+ rib primordia, alternating with MyHC-positive developing intercostal muscles, are shown. The TUNEL signal is at background or very low. The apparent overlap of SOX9 and MyHC signals on the right side of the image is a consequence of imperfect alignment of sections that are separated by up to 30 μm. A merged image of a MyoDiCre/+;R26GDTA/+ embryo (N) essentially shows only TUNEL staining because of the lack of SOX9 and MyHC signals in the ventral body wall. The near continuous TUNEL signal along the anteroposterior axis may represent apoptosis of both the musculature and rib progenitors. Scale bars represent 200 μm.
As SOX9 is required for survival of limb bud mesenchymal cells (Akiyama et al., 2002), we sought to address whether SOX9-negative mesenchyme in MyoDiCre/+;R26GDTA/+ embryos undergoes apoptosis. Notably, TUNEL staining was associated with ventral mesenchymal accumulations at both E11.5 and E12.5 (Fig. 8E, K). Confocal imaging confirmed that the TUNEL signal co-localized with the DAPI-stained rod-like structures (Fig. 8E’–E’”). We note that it was not possible to definitively establish that these TUNEL-positive aggregates represent presumptive rib structures because of the lack of cell-specific markers for these aggregates or for closely associated DTA-targeted muscle at this stage. However, frontal sections of E11.5 embryos through somatopleural regions, which reveal alternating cross-sections of rib and intercostal elements in control embryos (Fig. 8M), showed a near continuous region of apoptotic cells along the antero-posterior axis of MyoDiCre/+;R26GDTA/+ embryos (Fig. 8N), consistent with the possibility that the TUNEL signal represents both dying skeletal muscle and adjacent mesenchyme. More proximodorsally, TUNEL staining was reduced or absent in SOX9+ aggregates and apoptosis was largely confined to skeletal muscles targeted for DTA-mediated ablation (Fig. 8F). The weaker TUNEL signal and persistent MyHC staining more dorsally at E11.5 is consistent with the delayed kinetics of muscle ablation in the epaxial domains of interlimb somites. These data demonstrate that interactions between the early myotome and sclerotome are not sufficient for normal rib development, and point to the importance of sustained interactions with developing musculature for the continued growth and morphogenesis of the ribs.
Discussion
The dependence of distal rib development on skeletal muscle has been suggested by MRF knockout studies from many labs. Yet, the high degree of allele-specific variability in rib defects, from none to severe (Braun et al., 1992; Braun and Arnold, 1995; Kaul et al., 2000; Patapoutian et al., 1995; Tajbakhsh et al., 1996; Tallquist et al., 2000; Zhang et al., 1995), together with heretofore unexplained discordance between gene loss-of-function analyses and muscle cell ablation studies, has led to uncertainties as to the functions of the MRFs, and skeletal muscle more generally, in rib development. Here, we re-visited the role of skeletal muscle in rib development using a combination of genetic and cell ablation approaches.
Severity of rib defects is dependent on MRF dosage and is gene- and allele-specific
Most early studies attributed rib defects in Mrf4 knockout mice to a reduction or loss of Myf5 expression (Braun and Arnold, 1995; Floss et al., 1996; Patapoutian et al., 1995; Yoon et al., 1997). However, together with Mrf4 knockout phenotypes, the variability in rib phenotypes exhibited by the collection of Myf5 knockout alleles (Braun et al., 1992; Kaul et al., 2000; Tajbakhsh et al., 1996; Tallquist et al., 2000) and the demonstration that levels of myotomal Mrf4 expression are strongly influenced by the targeting design of the linked Myf5 allele (Kassar-Duchossoy et al., 2004), indicate that both Myf5 and Mrf4 play a role in rib development. Although all allelic combinations of Myf5 and Mrf4 could not be tested, the present results directly support the conclusion that the combined gene dosage of Mrf4 and Myf5 determine the quality of rib growth and patterning. The results also point to gene-specific effects, as mice carrying one allele each of Myf5 and Mrf4 showed better patterning than mice with two Mrf4 alleles, when MyoD was also knocked out. That either Myf5 or Mrf4 is required to support rib development was also demonstrated by the studies of Vinagre et al. (2010), who showed that the loss of ribs in Dll-Hoxa10 transgenic embryos was preceded by loss of expression of both Mrf4 and Myf5 in the hypaxial domain of interlimb somites and that forced expression of Mrf4 in these mice rescued rib development. A priori, a requirement for Myf5 or Mrf4 in rib development could reflect unique transcriptional activities of these MRFs, although this possibility seems unlikely, as rib development was normal when MyoD (Tallquist et al., 2000) or myogenin (Wang et al., 1996) was knocked into the Myf5 locus. However, the possibility of transcriptional targets shared by MYF5 and MRF4, but not the other MRFs, cannot be formally ruled out by these knockin studies, since Mrf4 can support rib development in the absence of Myf5, and the status of Mrf4 expression was not noted in these studies (Tallquist et al., 2000; Wang et al., 1996; Wang and Jaenisch, 1997).
In light of the collective data indicating that the extent and quality of rib development can largely be explained by gene dosage of Myf5 and Mrf4, parsimony would argue against the hypothesis that cis-acting effects on the expression of one or more unknown rib-determining genes (Kaul et al., 2000) are responsible for the rib phenotype in Myf5m1 mice. Although there is no definitive explanation for the observation that myotome formation is reportedly delayed in Myf5-null mice that have a normal rib cage (Myf5ΔloxP/ΔloxP mice; Kaul et al., 2000), Vinagre et al. (2010) showed that Mrf4 expression in the hypaxial myotome is retained in Myf5ΔloxP/ΔloxP embryos, consistent with the evidence that either Myf5 or Mrf4 is required for rib development.
Interestingly, the MRF genetic hierarchy has not been conserved across vertebrates. In the mouse, Myf5 and Mrf4 are expressed earlier than MyoD in somites (Chen et al., 2002; Chen and Goldhamer, 2004; Kassar-Duchossoy et al., 2004; Summerbell et al., 2002, 2000; Tajbakhsh et al., 1997; Vinagre et al., 2010) and function genetically upstream of MyoD in trunk myogenesis (Kassar-Duchossoy et al., 2004). Notably, Mrf4 is sufficient to program myogenic progenitors and drive early myogenesis (Kassar-Duchossoy et al., 2004), and this capacity can explain the substantial rib development in embryos lacking both Myf5 and MyoD. In contrast, Mrf4 is expressed after Myf5 and MyoD in the frog, zebrafish, and chick, and, like myogenin, its expression is initiated at the onset of differentiation (Berti et al., 2015; Gaspera et al., 2006; Hinits et al., 2009, 2007; Mok et al., 2015; Schnapp et al., 2009). Further, mrf4 is not expressed and muscle development is prevented in zebrafish embryos lacking both myf5 and myod, placing mrf4 genetically downstream of these MRFs (Hinits et al., 2011, 2009, 2007; Schnapp et al., 2009). Formal proof that developing musculature is necessary for rib development in zebrafish is lacking, as myf5/myod double mutant fish die before the onset of trunk skeletogenesis (Hinits et al., 2011). It is reasonable to hypothesize, however, that in fish and perhaps other non-mammalian vertebrates, MyoD and Myf5 are key muscle determinants of rib development, with MyoD subserving the rib functions of mouse Mrf4. In this regard, it will be important to determine whether zebrafish myod is sufficient to support rib development in embryos lacking Myf5 and Mrf4.
The presence and severity of rib defects is dictated by muscle ablation kinetics
Using DTA-mediated cell ablation approaches, we confirmed that developing skeletal muscle is indeed essential for growth and patterning of the distal ribs. Gross rib malformations were observed when muscle progenitors and differentiating muscle were ablated in MyoDiCre mice (Kanisicak et al., 2009; Wood et al., 2013), as well as when differentiating muscle was specifically targeted in HSA-Cre mice (Miniou et al., 1999). These results contrast with previous studies in which rib development was unaffected in Myf6-Cre/DTA (Haldar et al., 2008) and myogenin-Cre/DTA (Gensch et al., 2008) mice. Mrf4 is expressed in muscle progenitors of the hypaxial somite, slightly earlier than MyoD (E9.5 versus E9.75; (Chen et al., 2002; Chen and Goldhamer, 2004; Kassar-Duchossoy et al., 2004; Summerbell et al., 2002, 2000; Tajbakhsh et al., 1997; Vinagre et al., 2010). In contrast to MyoD, however, Mrf4-expressing progenitors constitute only a small fraction of myogenic progenitors, and ablation of the Mrf4 lineage did not significantly affect early myogenesis (Haldar et al., 2008). While this expression/ablation profile alone would predict that the rib cage would be unaffected in Myf6-Cre/DTA mice, the absence of rib defects is perhaps more surprising when considering that Mrf4 is broadly expressed in differentiated muscle and Myf6-Cre/DTA mice lack skeletal muscle at birth (Haldar et al., 2008). Similarly, differentiated muscle was lost by E18.5 in myogenin-Cre/DTA fetuses, yet rib abnormalities were not observed (Gensch et al., 2008). Of note, however, significant muscle ablation was not observed until E14.5 or later in Myf6-Cre/DTA (Haldar et a., 2008) and myogenin-Cre/DTA (Gensch et al., 2008) mice. This is in contrast to ablation kinetics in HSA-Cre;R26GDTA/+ mice reported here, in which MyHC-positive muscle was undetectable by E11.5. As ossifying cartilage models of the ribs are already present by E14.5 (Kaufman and Bard, 1999), the comparatively protracted kinetics of muscle ablation relative to rib morphogenesis in these previous studies likely explains the absence of rib malformations.
Many factors could influence muscle ablation kinetics, including the strength and spatiotemporal specificity of the promoter driving Cre, levels of Cre protein, recombination efficiency and promoter strength of the DTA allele, and the use of wild-type or attenuated DTA proteins. In this latter regard, the impact of choice of DTA allele was directly shown here by comparing muscle ablation kinetics and the severity of rib malformations in MyoDiCre embryos carrying one of three distinct DTA alleles, all of which are knockin alleles at the Rosa26 locus. Notably, compared to R26GDTA mice, muscle ablation was slower and rib development was substantially improved in mice harboring R26DTA176, which expresses the attenuated DTA variant (Wu et al., 2006) used by Haldar et al. (2008). Comai et al. (2014) also observed more efficient muscle ablation with the R26GDTA allele. Surprisingly, muscle cell ablation was also protracted, and rib development improved, with an independent wild-type DTA allele (R26DTA; Voehringer et al., 2008), relative to R26GDTA. Whether the greater efficiency of R26GDTA-mediated ablation is related to an apparently higher transcriptional output from this allele, as suggested by RT-qPCR analyses, or to allele-specific differences in efficiencies of Cre-dependent recombination, is unclear. Regardless of mechanism, it is reasonable to suggest that use of distinct DTA alleles in the present and previous studies influenced muscle ablation kinetics and rib phenotypes.
Interactions with skeletal muscle beyond somite stages are required for normal rib development
In E11.5 and E12.5 MyoDiCre/+;R26GDTA/+ embryos, rod-like mesenchymal aggregates were evident in ventrolateral somatopleural regions that lacked definitive rib structures at later stages. These aggregates did not stain for SOX9, an obligate cartilage transcription factor (Akiyama et al., 2002; Bi et al., 1999; Wright et al., 1995) required for survival of pre-cartilage progenitors (Akiyama et al., 2002), and the surrounding tissue was devoid of MyHC+ skeletal muscle. Although it was not possible to definitively establish that these mesenchymal aggregates represented rib precursors, because of the lack of available markers, the aggregates appeared to be continuous with more proximodorsal structures that stained positively for SOX9 and were associated with MyHC+ muscle (prior to its eventual loss). Notably, SOX9-negative aggregates showed robust apoptotic activity at E11.5 and E12.5, whereas TUNEL staining was not observed in SOX9-positive mesenchymal accumulations. Taken together, these data suggest a model in which skeletal muscle associated with presumptive rib progenitors directly or indirectly is required for SOX9 expression, and failure to express SOX9 results in apoptosis of presumptive rib progenitors. In this model, development of proximodorsal aspects of the distal ribs can be explained by the relative perdurance of skeletal muscle in these regions, ultimately dictated by the spatiotemporal pattern of MyoDiCre expression and DTA activity.
The correlation between delayed myotome formation and severe rib truncations reported by many studies (Braun et al., 1992; Braun and Arnold, 1995; Floss et al., 1996; Tajbakhsh et al., 1996; Tallquist et al., 2000; Wang and Jaenisch, 1997; Yoon et al., 1997) points to the importance of early myotome-sclerotome interactions in rib development. Indeed, these early studies led to the identification of myotomally expressed PDGFA and FGFs as likely mediators of such interactions (Floss et al., 1996; Grass et al., 1996; Huang et al., 2003; Patapoutian et al., 1995; Tallquist et al., 2000; Vinagre et al., 2010). That muscle is also required at late somite stages and beyond is suggested by the present data, as effects on myotomal gene expression in MyoDiCre/+;R26GDTA/+ embryos were first evident at E10.5 [(approximately 1 to 2 days after formation of the thoracic somites, depending on anteroposterior position; Theiler, 1989)], and the lack of SOX9 expression and increased apoptosis in apparent rib mesenchyme at E11.5 and E12.5 were restricted to anatomical regions that lacked skeletal muscle. The disruption of rib development and patterning in HSA-Cre/DTA embryos, and the corrective action of MyoD on rib patterning under conditions of limiting MRF dosage, also are consistent with the sustained influence of skeletal muscle on rib development. Collectively, available data indicate that early myotome-sclerotome interactions are necessary, but not sufficient, for normal rib development, and that sustained interactions are required for the continued growth and patterning of the ribs. Precisely defining the developmental window during which muscle interactions are required will require Cre drivers that are muscle specific, highly efficient, and allow temporal control of Cre activity.
Supplementary Material
Highlights.
MyoDiCre/DTA-dependent skeletal muscle ablation profoundly disrupts rib development
One allele of Mrf4 drives substantial rib development in the absence of Myf5 and MyoD
Specifically ablating differentiated muscle causes gross rib malformations
The severity of rib defects is highly dependent on choice of DTA allele
Muscle interactions beyond somite stages are required for normal rib development
Acknowledgements
We thank Drs. Michael Rudnicki and Shahragim Tajbakhsh for providing MyoDm1 and Myf5loxP mice, respectively, and Dr. Moisés Mallo for supplying the Fgf4 and Pdgfa cDNAs. We also thank Dr. Masakazu Yamamoto, Anthony Patelunas, and Cory Jubinville for critical review of the manuscript. This work was supported, in part, by NIH grant R01AR052777 to D.J.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Akiyama H, Chaboissier M-C, Martin JF, Schedl A, Crombrugghe B. de, 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16, 2813–2828. 10.1101/gad.1017802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti F, Nogueira JM, Wöhrle S, Sobreira DR, Hawrot K, Dietrich S, 2015. Time course and side-by-side analysis of mesodermal, pre-myogenic, myogenic and differentiated cell markers in the chicken model for skeletal muscle formation. J Anat 227, 361–82. 10.1111/joa.12353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, Crombrugghe B. de, 1999. Sox9 is required for cartilage formation. Nature Genetics 22, 85–89. 10.1038/8792 [DOI] [PubMed] [Google Scholar]
- Braun T, Arnold HH, 1995. Inactivation of Myf-6 and Myf-5 genes in mice leads to alterations in skeletal muscle development. EMBO J. 14, 1176–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Rudnicki MA, Arnold HH, Jaenisch R, 1992. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell 71, 369–382. [DOI] [PubMed] [Google Scholar]
- Carapuço M, Nóvoa A, Bobola N, Mallo M, 2005. Hox genes specify vertebral types in the presomitic mesoderm. Genes Dev 19, 2116–2121. 10.1101/gad.338705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JCJ, Goldhamer DJ, 2004. The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Developmental Biology 265, 502–512. [DOI] [PubMed] [Google Scholar]
- Chen JCJ, Ramachandran R, Goldhamer DJ, 2002. Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Developmental Biology 245, 213–223. 10.1006/dbio.2002.0638 [DOI] [PubMed] [Google Scholar]
- Dickman ED, Rogers R, Conway SJ, 1999. Abnormal skeletogenesis occurs coincident with increased apoptosis in the Splotch (Sp2H) mutant: Putative roles for Pax3 and PDGFRα in rib patterning. Anatomical Rec 255, 353–361. [DOI] [PubMed] [Google Scholar]
- Dietrich S, Gruss P, 1995. undulated Phenotypes Suggest a Role of Pax-1 for the Development of Vertebral and Extravertebral Structures. Dev Biol 167, 529–548. 10.1006/dbio.1995.1047 [DOI] [PubMed] [Google Scholar]
- Evans DJR, 2003. Contribution of somitic cells to the avian ribs. Dev Biol 256, 115–127. 10.1016/s0012-1606(02)00117-3 [DOI] [PubMed] [Google Scholar]
- Floss T, Arnold HH, Braun T, 1996. Myf-5(m1)/Myf-6(m1) compound heterozygous mouse mutants down-regulate Myf-5 expression and exert rib defects: evidence for long-range cis effects on Myf-5 transcription. Dev Biol 174, 140–7. 10.1006/dbio.1996.0058 [DOI] [PubMed] [Google Scholar]
- Gaspera BD, Sequeira I, Charbonnier F, Becker C, Shi D-L, Chanoine C, 2006. Spatio-temporal expression of MRF4 transcripts and protein duringXenopus laevis embryogenesis. Dev Dynam 235, 524–529. 10.1002/dvdy.20628 [DOI] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T, 2008. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development 135, 1597–1604. 10.1242/dev.019331 [DOI] [PubMed] [Google Scholar]
- Grass S, Arnold HH, Braun T, 1996. Alterations in somite patterning of Myf-5-deficient mice: a possible role for FGF-4 and FGF-6. Development 122, 141–150. [DOI] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR, 2008. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Developmental cell 14, 437–445. 10.1016/j.devcel.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH, 1993. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364, 501–506. 10.1038/364501a0 [DOI] [PubMed] [Google Scholar]
- Hebert JM, Basilico C, Goldfarb M, Haub O, Martin GR, 1990. Isolation of cDNAs encoding four mouse FGF family members and characterization of their expression patterns during embryogenesis. Developmental Biology 138, 454–463. [DOI] [PubMed] [Google Scholar]
- Henderson DJ, Conway SJ, Copp AJ, 1999. Rib truncations and fusions in the Sp2H mouse reveal a role for Pax3 in specification of the ventro-lateral and posterior parts of the somite. Developmental Biology 209, 143–158. 10.1006/dbio.1999.9215 [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D, 1995. Expression of a Delta homologue in prospective neurons in the chick. Nature 375, 787–790. 10.1038/375787a0 [DOI] [PubMed] [Google Scholar]
- Hinits Y, Osborn DPS, Carvajal JJ, Rigby PWJ, Hughes SM, 2007. Mrf4 (myf6) is dynamically expressed in differentiated zebrafish skeletal muscle. Gene Expr Patterns 7, 738–745. 10.1016/j.modgep.2007.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Osborn DPS, Hughes SM, 2009. Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Dev Camb Engl 136, 403–14. 10.1242/dev.028019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Williams VC, Sweetman D, Donn TM, Ma TP, Moens CB, Hughes SM, 2011. Defective cranial skeletal development, larval lethality and haploinsufficiency in Myod mutant zebrafish. Dev Biol 358, 102–12. 10.1016/j.ydbio.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Stolte D, Kurz H, Ehehalt F, Cann GM, Stockdale FE, Patel K, Christ B, 2003. Ventral axial organs regulate expression of myotomal Fgf-8 that influences rib development. Dev Biol 255, 30–47. 10.1016/s0012-1606(02)00051-9 [DOI] [PubMed] [Google Scholar]
- Huang R, Zhi Q, Schmidt C, Wilting J, Brand-Saberi B, Christ B, 2000. Sclerotomal origin of the ribs. Development 127, 527–532.10631173 [Google Scholar]
- Ivanova A, Signore M, Caro N, Greene NDE, Copp AJ, Martinez-Barbera JP, 2005. In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis (New York, N.Y. : 2000) 43, 129–135. 10.1002/gene.20162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ, 2009. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Developmental biology 332, 131–141. 10.1016/j.ydbio.2009.05.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S, 2004. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431, 466–471. 10.1038/nature02876 [DOI] [PubMed] [Google Scholar]
- Kaufman MH, Bard JBL, 1999. The Anatomical Basis of Mouse Development 51–59. 10.1016/b978-012402060-3/50030-1 [DOI] [Google Scholar]
- Kaul A, Koster M, Neuhaus H, Braun T, 2000. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell 102, 17–19. 10.1016/s0092-8674(00)00006-4 [DOI] [PubMed] [Google Scholar]
- Koseki H, Wallin J, Wilting J, Mizutani Y, Kispert A, Ebensperger C, Herrmann BG, Christ B, Balling R, 1993. A role for Pax-1 as a mediator of notochordal signals during the dorsoventral specification of vertebrae. Development 119, 649–660. [DOI] [PubMed] [Google Scholar]
- Li S, Czubryt MP, McAnally J, Bassel-Duby R, Richardson JA, Wiebel FF, Nordheim A, Olson EN, 2005. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc National Acad Sci 102, 1082–1087. 10.1073/pnas.0409103102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Willet SG, Bankaitis ED, Xu Y, Wright CVE, Gu G, 2013. Non-parallel recombination limits Cre-LoxP-based reporters as precise indicators of conditional genetic manipulation. Genesis New York N Y 2000 51, 436–42. 10.1002/dvg.22384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Voss AK, Thomas T, Yokota Y, Gruss P, 2000. Uncx4.1 is required for the formation of the pedicles and proximal ribs and acts upstream of Pax9. Development 127, 2251–2258. [DOI] [PubMed] [Google Scholar]
- Miniou P, Tiziano D, Frugier T, Roblot N, Meur ML, Melki J, 1999. Gene targeting restricted to mouse striated muscle lineage. Nucleic acids research 27, e27–27. 10.1093/nar/27.19.e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok GF, Mohammed RH, Sweetman D, 2015. Expression of myogenic regulatory factors in chicken embryos during somite and limb development. J Anat 227, 352–60. 10.1111/joa.12340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y, 1993. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364, 532–535. 10.1038/364532a0 [DOI] [PubMed] [Google Scholar]
- Patapoutian A, Yoon JK, Miner JH, Wang S, Stark K, Wold B, 1995. Disruption of the mouse MRF4 gene identifies multiple waves of myogenesis in the myotome. Development 121, 3347–3358. [DOI] [PubMed] [Google Scholar]
- Pourquié O, Fan C-M, Coltey M, Hirsinger E, Watanabe Y, Bréant C, Francis-West P, Brickell P, Tessier-Lavigne M, Douarin NML, 1996. Lateral and Axial Signals Involved in Avian Somite Patterning: A Role for BMP4. Cell 84, 461–471. 10.1016/s0092-8674(00)81291-x [DOI] [PubMed] [Google Scholar]
- Rot-Nikcevic I, Reddy T, Downing KJ, Belliveau AC, Hallgrímsson B, Hall BK, Kablar B, 2005. Myf5 −/− :MyoD −/− amyogenic fetuses reveal the importance of early contraction and static loading by striated muscle in mouse skeletogenesis. Dev Genes Evol 216, 1–9. 10.1007/s00427-005-0024-9 [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R, 1992. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71, 383–390. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PNJ, Stead RH, Braun T, Arnold H-H, Jaenisch R, 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351–1359. 10.1016/0092-8674(93)90621-v [DOI] [PubMed] [Google Scholar]
- Scaal M, 2016. Early development of the vertebral column. Semin Cell Dev Biol 49, 83–91. 10.1016/j.semcdb.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Schnapp E, Pistocchi AS, Karampetsou E, Foglia E, Lamia CL, Cotelli F, Cossu G, 2009. Induced early expression of mrf4 but not myog rescues myogenesis in the myod/myf5 double-morphant zebrafish embryo. J Cell Sci 122, 481–8. 10.1242/jcs.038356 [DOI] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hübner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R, 2002. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32, 19–26. 10.1002/gene.10023 [DOI] [PubMed] [Google Scholar]
- Sudo H, Takahashi Y, Tonegawa A, Arase Y, Aoyama H, Mizutani-Koseki Y, Moriya H, Wilting J, Christ B, Koseki H, 2001. Inductive Signals from the Somatopleure Mediated by Bone Morphogenetic Proteins Are Essential for the Formation of the Sternal Component of Avian Ribs. Dev Biol 232, 284–300. 10.1006/dbio.2001.0198 [DOI] [PubMed] [Google Scholar]
- Summerbell D, Ashby PR, Coutelle O, Cox D, Yee S, Rigby PW, 2000. The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Dev Camb Engl 127, 3745–57. [DOI] [PubMed] [Google Scholar]
- Summerbell D, Halai C, Rigby PWJ, 2002. Expression of the myogenic regulatory factor Mrf4 precedes or is contemporaneous with that of Myf5 in the somitic bud. Mech Develop 117, 331–335. 10.1016/s0925-4773(02)00208-3 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Buckingham M, 1996. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature 384, 266–270. 10.1038/384266a0 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M, 1997. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89, 127–138. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellström M, Soriano P, 2000. Early myotome specification regulates PDGFA expression and axial skeleton development. Dev Camb Engl 127, 5059–70. [DOI] [PubMed] [Google Scholar]
- Tonegawa A, Funayama N, Ueno N, Takahashi Y, 1997. Mesodermal subdivision along the mediolateral axis in chicken controlled by different concentrations of BMP-4. Dev Camb Engl 124, 1975–84. [DOI] [PubMed] [Google Scholar]
- Tremblay P, Dietrich S, Mericskay M, Schubert FR, Li Z, Paulin D, 1998. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Dev Biol 203, 49–61. 10.1006/dbio.1998.9041 [DOI] [PubMed] [Google Scholar]
- Vinagre T, Moncaut N, Carapuço M, Nóvoa A, Bom J, Mallo M, 2010. Evidence for a myotomal Hox/Myf cascade governing nonautonomous control of rib specification within global vertebral domains. Developmental cell 18, 655–661. 10.1016/j.devcel.2010.02.011 [DOI] [PubMed] [Google Scholar]
- Voehringer D, Liang H-E, Locksley RM, 2008. Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J. Immunol 180, 4742–4753. 10.4049/jimmunol.180.7.4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin J, Wilting J, Koseki H, Fritsch R, Christ B, Balling R, 1994. The role of Pax-1 in axial skeleton development. Dev Camb Engl 120, 1109–21. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jaenisch R, 1997. Myogenin can substitute for Myf5 in promoting myogenesis but less efficiently. Development 124, 2507–2513. [DOI] [PubMed] [Google Scholar]
- Wang Y, Schnegelsberg PNJ, Dausman J, Jaenisch R, 1996. Functional redundancy of the muscle-specific transcription factors Myf5 and myogenin. Nature 379, 823–825. 10.1038/379823a0 [DOI] [PubMed] [Google Scholar]
- Wood WM, Etemad S, Yamamoto M, Goldhamer DJ, 2013. MyoD-expressing progenitors are essential for skeletal myogenesis and satellite cell development. Developmental biology 384, 114–127. 10.1016/j.ydbio.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P, 1995. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nature Genetics 9, 15–20. 10.1038/ng0195-15 [DOI] [PubMed] [Google Scholar]
- Wu S, Wu Y, Capecchi MR, 2006. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development 133, 581–590. 10.1242/dev.02236 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ, 2009. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis (New York, N.Y. : 2000) 47, 107–114. 10.1002/dvg.20474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Watt CD, Schmidt RJ, Kuscuoglu U, Miesfeld RL, Goldhamer DJ, 2007. Cloning and characterization of a novel MyoD enhancer-binding factor. Mechanisms of development 124, 715–728. 10.1016/j.mod.2007.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JK, Olson EN, Arnold HH, Wold BJ, 1997. Different MRF4 knockout alleles differentially disrupt Myf-5 expression: cis-regulatory interactions at the MRF4/Myf-5 locus. Developmental Biology 188, 349–362. 10.1006/dbio.1997.8670 [DOI] [PubMed] [Google Scholar]
- Zhang W, Behringer RR, Olson EN, 1995. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev 9, 1388–1399. 10.1101/gad.9.11.1388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


