Abstract
Background.
Alcohol-associated liver disease accounts for half of cirrhosis-related deaths worldwide. The spectrum of disease varies from simple steatosis to fibrosis, cirrhosis, and ultimately hepatocellular carcinoma. Understanding the disease on a molecular level helps us to develop therapeutic targets.
Aim.
We performed transcriptomic analysis in liver and ileum from chronic plus binge ethanol-fed mice, and we assessed the role of selected differentially expressed genes and their association with serum bile acids and gut microbiota.
Methods.
Wild-type mice were subjected to a chronic Lieber-DeCarli diet model for 8 weeks followed by one binge of ethanol. RNA-seq analysis was performed on liver and ileum samples. Associations between selected differentially regulated genes and serum bile acid profile or fecal bacterial profiling (16S rDNA sequencing) were investigated.
Results.
We provide a comprehensive transcriptomic analysis to identify differentially expressed genes, KEGG pathways, and gene ontology functions in liver and ileum from chronic plus binge ethanol-fed mice. In liver, we identified solute carrier organic anion transporter family, member 1a1 (Slco1a1; encoding for organic anion transporting polypeptides (OATP) 1A1), as the most down-regulated mRNA, and it negatively correlated with serum cholic acid level. Prokineticin 2 (Prok2) mRNA, a cytokine-like molecule associated with gastrointestinal tract inflammation, was significantly down-regulated in ethanol-fed mice. Prok2 mRNA expression was negatively correlated with abundance of Allobaculum (genus), Coprococcus (genus), Lachnospiraceae (family), Lactococcus (genus), and Cobriobacteriaceae (family), while it positively correlated with Bacteroides (genus).
Conclusions.
RNA-seq analysis revealed unique transcriptomic signatures in the liver and intestine following chronic plus binge ethanol feeding.
Keywords: gene expression, microbiome, 16S rDNA sequencing, bile acids, metabolome
Introduction
Alcohol-associated liver disease is a leading cause of death accounting for 0.9% of deaths globally and 48% of liver cirrhosis-associated deaths in United States [1]. Patients with severe alcoholic hepatitis present a 30-day mortality as high as 50% without treatment [2]. For these patients, corticosteroids remain the only therapy with a short-term advantage over placebo, but liver transplantation is the only cure [3]. Recent work on the communication between gut-liver axis has allowed a better understanding of the pathogenesis of alcohol-associated liver disease. Using 16S rDNA sequencing, the association between gut microbiome and alcohol-associated liver disease in human and rodents has been reported by several studies [4–6]. As essential metabolites of gut microbiota, serum bile acid profiles have been characterized in patients with alcohol-associated liver disease [7]. Total and conjugated serum bile acids were significantly increased in patients with alcoholic hepatitis compared with controls; total and conjugated bile acids were correlated positively with disease severity (MELD) [7]. To enhance our understanding of the mechanisms of alcohol-associated liver disease at transcript level, we applied RNA-Seq analysis using liver and intestine samples from control- and chronic plus binge ethanol-fed mice. By combining the transcriptomic analysis with metabolomics on serum bile acid profile and 16S metagenomics, we identified top dysregulated genes solute carrier organic anion transporter family, member 1a1 (Slco1a1) in liver and Prokineticin 2 (Prok2) in ileum. Slco1a1 expression was negatively correlated with serum cholic acid, and Prok2 expression was correlated with abundance of several bacterial genera, such as Allobaculum. We, for the first time, provide a detailed intestinal and hepatic transcriptomic analysis in a model of chronic plus binge ethanol feeding in mice.
Methods
Mice.
C57BL/6 female mice purchased from Charles River at the age of 9–10 weeks were subjected to a chronic Lieber-DeCarli diet for 8 weeks as described previously [8, 9]. Mice were gavaged with a single dose of ethanol (5g/kg bodyweight) at the end of treatment and euthanized 9 hours later. Control mice were fed with an isocaloric amount of dextrose. All animal studies were approved by Institutional Animal Care and Use Committee of the University of California, San Diego.
RNA isolation and RNA sequencing.
Total RNA from liver and ileum were extracted using RNAeasy Mini Kit as previously described [8]. RNA-seq libraries were sequenced on Illumina NovaSeq 6000 platform. Sequence reads were aligned to mouse genome (NCBI GRCm38.p6) using Salmon version 0.14.1 [10]. Differentially expressed genes (DEGs) were identified using DESeq2 package [11]. Gene expression fold change ≥1.5 and adjusted P value < 0.05 were set as the threshold values for KEGG over-representation analysis and gene ontology (GO) over-representation analysis with R package clusterProfiler [12]. Sequence data were registered at NCBI under BioProject PRJNA597350. Sequence reads are available at NCBI under the following consecutive BioSample IDs: SAMN13673201-SAMN13673357 as previously described [8].
Real-time qPCR.
cDNA was generated from extracted total RNA, and real-time qPCR was performed as previously described [6]. Primer sequences for Slco1a1 are forward 5’-TAATCGGGCCAATCTTC −3’ and reverse 5’-ACTCCCATAATGCCCTTGG −3’. Primer sequences for Prok2 are forward 5’-CGGAGGATGCACCACACCT-3’ and reverse 5’-TTTCCGGGCCAAGCAAATAAACCG-3’. Real-time qPCR was performed using SYBR Premix (Biorad) on ABI StepOnePlus instrument. Data were expressed as fold change over control-treated animals. 18S was used as the housekeeping gene.
Serum bile acid measurement.
Serum bile acids from control- and ethanol-fed mice were analyzed by LC-MS as described earlier [13, 8].
DNA extraction and 16S rDNA sequencing.
DNA was extracted from mouse feces with QIAmp Fast DNA Stool Mini kit (Qiagen) and concentrated with Zymo gDNA Clean and Concentration kit (Zymo) as previously described [8]. Briefly, V3-V4 hypervariable segment of the 16S rDNA gene was amplified by PCR reactions and subsequently sequenced on the Illumina Miseq platform. Sequence reads were analyzed with Quantitative Insights Into Microbial Ecology 2 (QIIME2; available at https://qiime2.org) [14]. Representative OTUs from each set were chosen at a minimum sequence identity of 97% which uses the Greengenes database [15]. Sequence data were registered at NCBI under BioProject PRJNA597350. Sequence reads are available at NCBI under the following consecutive BioSample IDs:SAMN13673358-SAMN13673442 [8].
Statistical analysis.
All data are expressed as mean ± s.e.m. except when stated otherwise. Comparisons between two groups were performed by Mann-Whitney-Wilcoxon test. Spearman’s correlation was conducted for correlation analysis. Statistical analysis was performed using R statistical software, R version V.3.6.2, 2019 (R Foundation for Statistical Computing) and GraphPad Prism (V.8.4.2). A P value <0.05 was considered statistically significant.
Results
Liver transcriptomic profile changes in response to chronic plus binge ethanol feeding in mice
Wild-type C57BL/6 mice were subjected to chronic plus binge ethanol feeding for 8 weeks. To visualize differentially expressed genes (DEGs) in liver after ethanol treatment, we plotted significantly altered genes in a volcano plot (Figure 1A). In total, we identified 1244 up-regulated genes and 993 down-regulated genes when comparing ethanol treatment with controls (Figure 1B). Top ten up- and down-regulated genes are listed in Table 1. Interestingly, many of them were noncoding RNAs (Gm40489, Gm36841, Gm33519, Gm40784, Gm34982, 4933401D09Rik, LOC115488496, Gm32468, Gm28548, Gm33730, LOC115487163, Gm32463, Gm36180). We further performed functional analyses of DEGs using KEGG (Figure 1C) and GO biological process (Figure 2A) databases. KEGG pathway analysis revealed potentially important roles of MAPK signaling, cytochrome P450, steroid hormone biosynthesis, and glutathione metabolism (Figure 1C). Top GO terms of DEGs indicated altered functions of: response to interferon-beta, leukocyte migration and chemotaxis, and alcohol metabolic process (Figure 2A). Next, we evaluated selected genes, GO terms, and KEGG pathways in our dataset that are differentially regulated following chronic plus binge ethanol feeding [1]. We show genes significantly up- and down-regulated involved in immune response, inflammation, fatty acid oxidation, glucose metabolism, triglyceride metabolism, bile acid metabolism, apoptosis, cell cycle, and growth factors (Table 2). Selected GO biological process analysis revealed several differentially regulated pathways in liver including lipid, triglyceride, cholesterol, and alcohol metabolism (Figure 2B). In addition to the top 20 GO terms, cholesterol-associated GO terms were also predicted to be significantly enriched after ethanol feeding (Figure 2C).
Figure 1. RNA-seq analysis in liver in response to chronic plus binge ethanol: differentially expressed genes (DEGs) and KEGG pathway analysis.
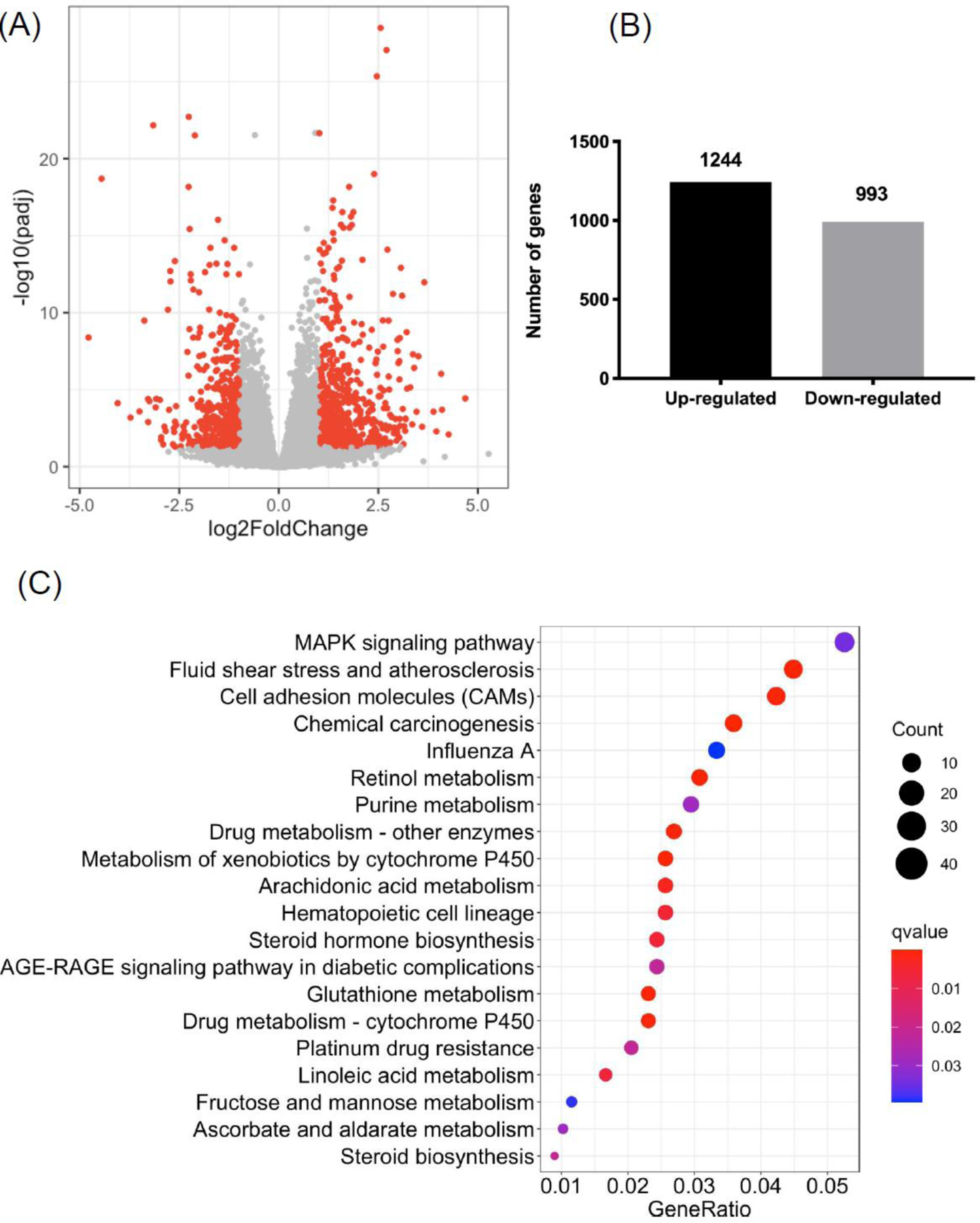
(A) Volcano plot illustrating DEGs in liver between ethanol-fed and control mice (absolute fold change > 2; adjusted P value < 0.05). (B) Number of up- and down-regulated genes in liver in response of ethanol compared with control. (C) Top 20 significant KEGG pathways in ethanol-fed mice compared with controls (q value < 0.05). Control group: n=10; Ethanol group: n=19. For figure 1(B) and 1(C), cutoff for DEGs is absolute fold change > 1.5, adjusted P value < 0.05.
Table 1.
List of top 10 up- and down-regulated genes in the liver.
| Gene symbol | Fold change | P value | Adjusted P |
|---|---|---|---|
| Up-regulated genes | |||
| Gm40489 | 25.69 | 1.22E-06 | 3.68E-05 |
| Rplp2-ps1 | 19.24 | 9.41E-04 | 8.07E-03 |
| Gm36841 | 17.12 | 9.69E-06 | 2.01E-04 |
| Gm33519 | 16.90 | 1.60E-08 | 9.31E-07 |
| Nrcam | 15.54 | 5.35E-04 | 5.13E-03 |
| Gm40784 | 14.77 | 1.38E-05 | 2.67E-04 |
| Cyp2b10 | 12.59 | 3.27E-15 | 1.08E-12 |
| Gm34982 | 12.12 | 2.26E-04 | 2.57E-03 |
| 4933401 D09Rik | 11.35 | 7.82E-10 | 6.84E-08 |
| LOC115488496 | 11.00 | 1.40E-05 | 2.70E-04 |
| Down-regulated genes | |||
| Slco1a1 | −27.47 | 2.96E-11 | 4.10E-09 |
| Gm32468 | −21.89 | 1.08E-22 | 2.02E-19 |
| Gm28548 | −16.52 | 2.93E-06 | 7.57E-05 |
| Gm33730 | −13.23 | 4.09E-05 | 6.52E-04 |
| LOC115487163 | −11.32 | 1.33E-05 | 2.60E-04 |
| Akr1c18 | −10.40 | 1.72E-12 | 3.29E-10 |
| Gm32463 | −9.73 | 1.37E-06 | 4.04E-05 |
| Klrg1 | −9.71 | 9.30E-05 | 1.26E-03 |
| Serpina9 | −9.51 | 2.03E-06 | 5.62E-05 |
| Gm36180 | −8.90 | 1.68E-26 | 6.90E-23 |
Figure 2. Gene ontology (GO) biological processes and gene-concept networks of cholesterol-associated GO terms in liver in response to chronic plus binge ethanol.
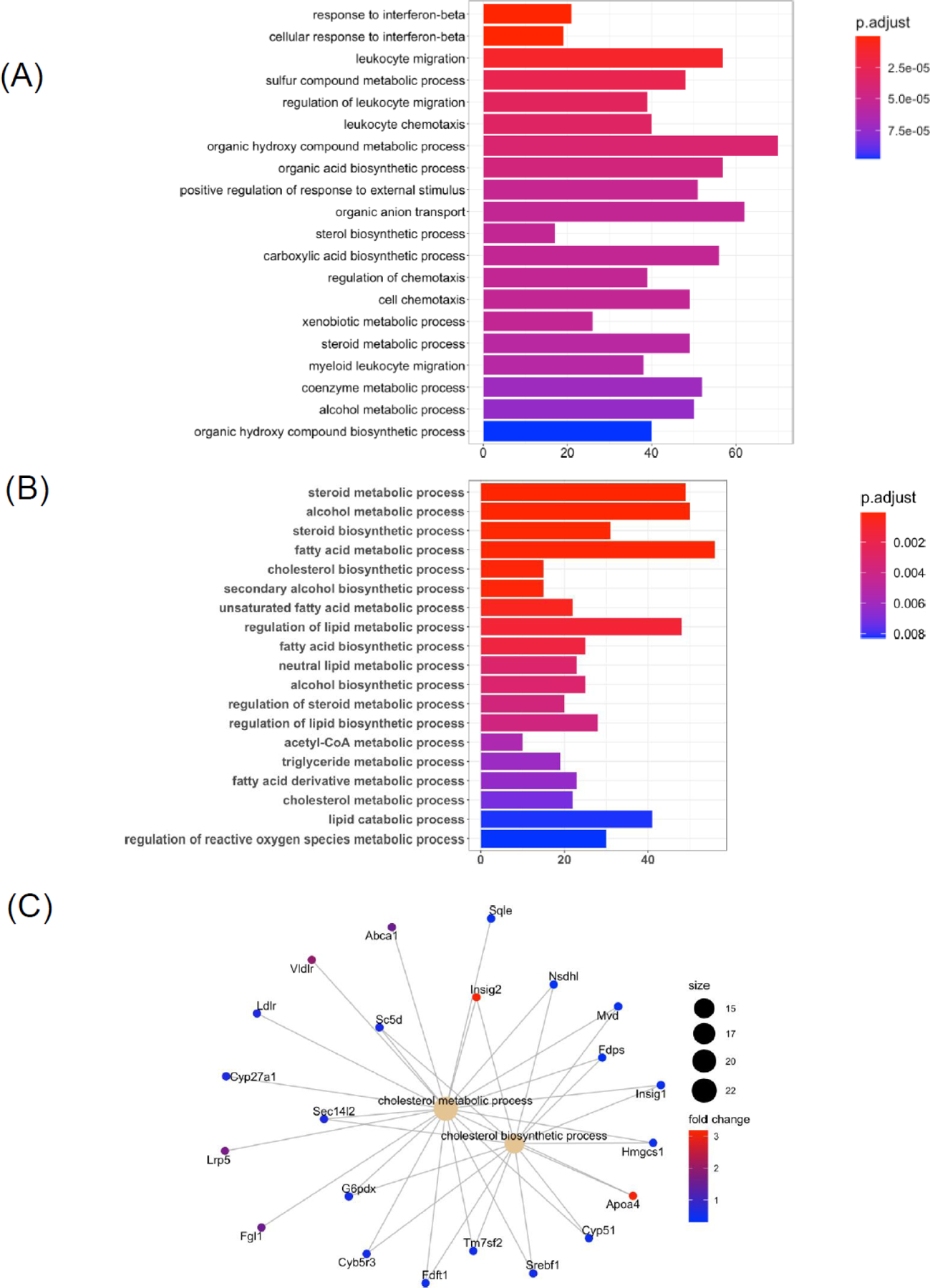
(A) Bar chart of top 20 significant GO terms of liver differentially expressed genes (DEGs) following ethanol treatment compared with controls. (B) Bar chart of selected and significantly different GO terms following chronic plus binge ethanol feeding. (C) Gene-concept networks of cholesterol metabolic process and cholesterol biosynthetic process. Control group: n=10; Ethanol group: n=19. Significant GO terms cutoff: adjusted P value < 0.05.
Table 2.
Selected genes in liver differentially regulated following chronic plus binge ethanol feeding in mice.
| Gene symbol | Fold change | Adjusted P | Function |
|---|---|---|---|
| Tlr3 | −1.38 | 1.13E-02 | Immune response |
| Tlr5 | 1.33 | 2.17E-02 | Immune response |
| Tlr7 | −1.35 | 7.09E-02 | Immune response |
| Tlr12 | 1.48 | 8.91E-02 | Immune response |
| ll1a | 1.53 | 4.16E-02 | Inflammation |
| ll1b | 1.61 | 9.92E-02 | Inflammation |
| Pdk4 | 1.48 | 3.31E-01 | Fatty acid oxidation |
| Gpx3 | 1.93 | 4.86E-05 | Oxidative stress |
| Nqo1 | 1.44 | 1.88E-02 | Oxidative stress |
| Ppp1r3b | −1.50 | 1.46E-02 | Glucose metabolism |
| Thrsp | −4.44 | 4.15E-09 | Triglyceride biosynthesis |
| Fasn | −2.08 | 7.27E-03 | Triglyceride biosynthesis |
| Abcb11 | −1.62 | 9.20E-06 | Bile acid metabolism |
| Cyp27a1 | −1.56 | 7.38E-04 | Bile acid metabolism |
| Bcl6 | −2.08 | 1.01E-02 | Apoptosis |
| Casp3 | −1.36 | 1.28E-02 | Apoptosis |
| Casp9 | −1.37 | 1.52E-02 | Apoptosis |
| Foxm1 | −1.98 | 7.23E-04 | Cell cycle |
| Cdkn1a | 5.04 | 1.25E-03 | Cell cycle |
| Hgf | 1.92 | 1.45E-05 | Growth factor |
| Vegfa | −1.56 | 1.60E-03 | Growth factor |
| Vegfb | −1.65 | 2.74E-06 | Growth factor |
Correlation of hepatic Slco1a1 expression with serum bile acids
Among the top 10 down-regulated genes in liver, we found that gene Slco1a1 was down-regulated over 27 folds in response to ethanol (Table 1). We validated the expression of Slco1a1 by real-time PCR as shown in Figure 3A. Consistent with sequencing results, Slco1a1 expression was significantly reduced in ethanol-fed mice (Figure 3A). As one of the three abundant organic anion transporting polypeptide (OATP) hepatic isoforms, Slco1a1 encodes for protein OATP1A1. OATP1 as sodium-independent hepatic bile acid uptake transporter is involved in the enterohepatic circulation of bile acids [16]. To investigate if downregulation of Slco1a1 was associated with changes in serum bile acid profile, we correlated hepatic Slco1a1 expression with selected highly abundant bile acids in mice (Figure 3B). Serum cholic acid level was negatively correlated with Slco1a1 expression (R=−0.45, Figure 3C).
Figure 3. Correlation between hepatic Slco1a1 expression and serum bile acid profiling.
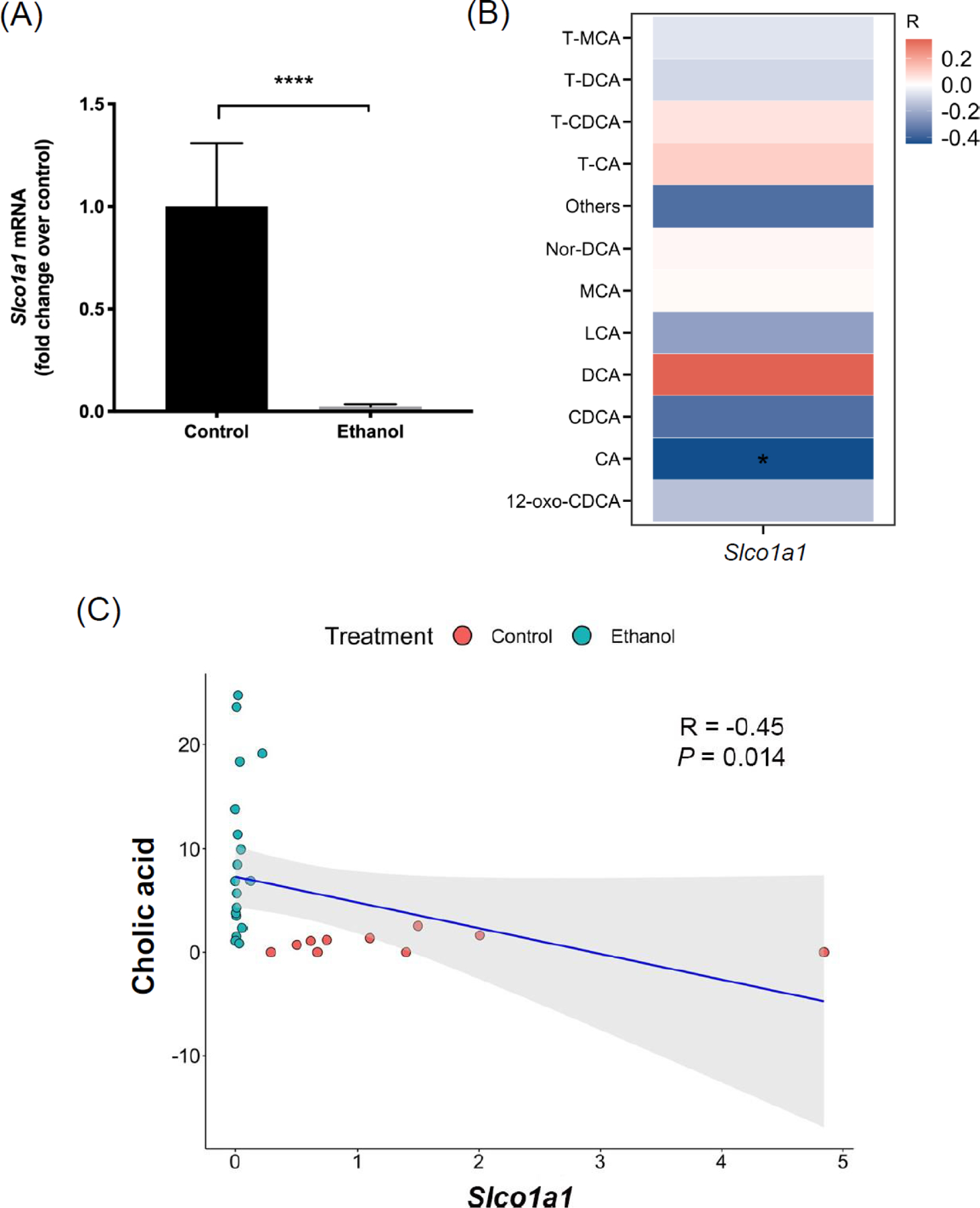
(A) Hepatic mRNA level of Slco1a1 in control and ethanol groups (real-time qPCR). (B) Heatmap of Spearman correlation between hepatic Slco1a1 mRNA and serum bile acids. Red color indicates positive correlation, blue color indicates negative correlation. (C) Spearman correlation between cholic acid and Slco1a1. Control group: n=10; Ethanol group: n=19. *P < 0.05. ****P < 0.0001. Slco1a1, Solute carrier organic anion transporter family, member 1a1; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; MCA, muricholic acid; T-CA, taurocholic acid; T-CDCA, taurochenodeoxycholic acid; T-DCA, taurodeoxycholic acid; T-MCA, tauromuricholic acid.
Intestine transcriptomic profile changes in response to chronic plus binge ethanol feeding in mice
To reveal how chronic plus binge ethanol feeding affects intestinal gene expression, we focused on DEGs in ileum from mice treated with ethanol compared to controls (Figure 4A). The terminal ileum is known to actively take up bile acids from the intestinal luminal and secrete them into the portal vein, where they reach the liver and are transported into hepatocytes [17]. In total, we found 931 genes were up-regulated and 891 genes were down-regulated in response to ethanol treatment (Figure 4B). Top 10 dysregulated genes are listed in Table 3. Interestingly, we found that Prok2, an important cytokine-like molecule in gut inflammation (encoding for protein prokineticin 2) [18], was significantly down-regulated after ethanol treatment (Table 3). Top 20 significant KEGG pathways are indicated in Figure 4C. The pathway analysis identified a subset of pathways such as cell cycle, steroid hormone biosynthesis, PPAR signaling pathway, IL-17 signaling pathway, and cytochrome p450-associated pathways (Figure 4C). The GO biological process analysis highlighted functions such as chromosome segregation, nuclear division, nuclear chromosome segregation, organelle fission, and sister chromatid segregation (Figure 5A). Next, we evaluated selected intestinal genes that were previously reported [19, 20] to be associated with intestinal damage such as tight junction, apoptosis, and inflammation following chronic plus binge ethanol feeding (Table 4). Selected GO terms revealed significantly changed cell cycle and fatty acid metabolism functions (Figure 5B), and KEGG pathway analysis identified several inflammation-associated pathways altered by chronic plus binge ethanol in mice (Figure 5C). We also found triglyceride metabolic process and catabolic process were predicted to be altered in ethanol-fed mice (Fgf21, G6pc, Apoa1, Pck1, Pnpla3, and Scd1, Figure 5D).
Figure 4. RNA-seq analysis in ileum in response to chronic plus binge ethanol: differentially expressed genes (DEGs) and KEGG pathway analysis.

(A) Volcano plot illustrating DEGs in ileum between ethanol-fed and control mice (absolute fold change > 2; adjusted P value < 0.05). (B) Number of up- and down-regulated genes in ileum in response to ethanol compared with control. (C) Top 20 significant KEGG pathways in ethanol-fed mice compared with controls (q value < 0.05). Control group: n=10; Ethanol group: n=19. For figure 1(B) and 1(C), cutoff for DEGs is absolute fold change > 1.5, adjusted P value <0.05.
Table 3.
List of top 10 up- and down-regulated genes in the ileum.
| Gene symbol | Fold change | P value | Adjusted P |
|---|---|---|---|
| Up-regulated genes | |||
| Amy2a5 | 57.25 | 3.21E-04 | 2.69E-03 |
| Psca | 35.84 | 8.67E-07 | 1.73E-05 |
| Meltf | 15.97 | 1.33E-18 | 7.92E-16 |
| Ankrd1 | 12.55 | 7.03E-05 | 7.59E-04 |
| Cyp2d9 | 12.29 | 5.93E-30 | 4.48E-26 |
| Gm33148 | 11.11 | 6.63E-05 | 7.23E-04 |
| Inhbe | 9.84 | 4.09E-03 | 2.14E-02 |
| Gm16596 | 9.56 | 3.93E-16 | 1.44E-13 |
| Smok2a | 8.43 | 1.71E-03 | 1.06E-02 |
| Gm36505 | 8.36 | 4.27E-04 | 3.41E-03 |
| Down-regulated genes | |||
| Gm41145 | −20.05 | 1.26E-11 | 1.15E-09 |
| Hephl1 | −19.46 | 6.86E-23 | 1.20E-19 |
| G6pc2 | −11.90 | 2.16E-07 | 5.28E-06 |
| Gm29932 | −10.21 | 2.68E-16 | 1.04E-13 |
| Adcy10 | −9.26 | 1.96E-07 | 4.84E-06 |
| Gm40449 | −8.69 | 2.96E-10 | 1.65E-08 |
| Gm16104 | −8.33 | 2.47E-06 | 4.30E-05 |
| Prok2 | −7.57 | 3.52E-05 | 4.24E-04 |
| 1700016G22Rik | −7.28 | 2.43E-06 | 4.24E-05 |
| Gm39181 | −7.11 | 2.58E-05 | 3.26E-04 |
Figure 5. Gene ontology (GO) biological processes and gene-concept networks of triglyceride-associated GO terms in ileum in response to chronic plus binge ethanol.
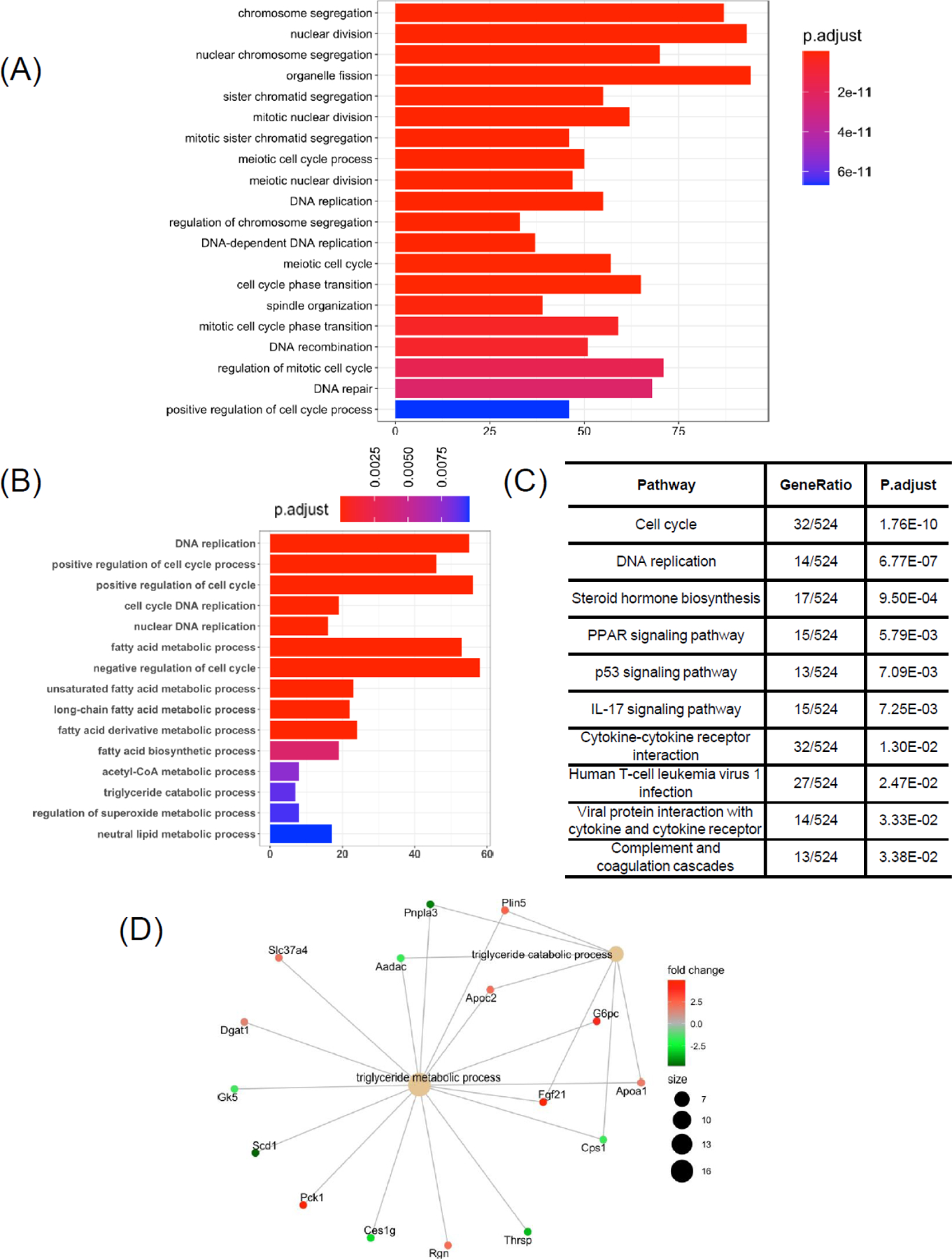
(A) Bar chart of top 20 significant GO terms of ileum differentially expressed genes (DEGs) following ethanol treatment compared with controls. (B) Bar chart of selected and significantly different GO terms following chronic plus binge ethanol feeding. (C) Selected and significantly changed KEGG pathways following chronic plus binge ethanol feeding. (D) Gene-concept networks of triglyceride metabolic process and triglyceride biosynthetic process. Control group: n=10; Ethanol group: n=19. Significant GO terms cutoff: adjusted P value < 0.05.
Table 4.
Genes in ileum that are differentially regulated following chronic plus binge ethanol feeding in mice.
| Gene symbol | Fold change | Adjusted P | Function |
|---|---|---|---|
| Cldn8 | −3.54 | 1.86E-09 | Tight junction |
| Cldn11 | −1.65 | 4.77E-02 | Tight junction |
| Cldn14 | 2.28 | 5.73E-05 | Tight junction |
| Casp2 | −1.32 | 6.95E-06 | Apoptosis |
| Casp4 | 1.93 | 7.06E-08 | Apoptosis |
| Casp6 | −1.16 | 4.42E-03 | Apoptosis |
| Tnf | 2.06 | 5.66E-03 | Inflammation |
| ll1a | 1.80 | 1.38E-02 | Inflammation |
| ll1b | 1.50 | 7.57E-02 | Inflammation |
Correlation of intestinal Prok2 expression with abundance of gut bacteria
We validated the expression of Prok2 in ileum by real-time PCR. Consistent with RNA sequencing, the expression of Prok2 in ethanol-fed mice was significantly decreased compared to control group (Figure 6B). To explore if Prok2 mRNA expression is affected by the abundance of gut bacteria from 16S sequencing, we performed Spearman correlation analysis (Figure 6A). Interestingly, ileal Prok2 expression was significantly correlated with Allobaculum (genus), Coprococcus (genus), Lachnospiraceae (family), Lactococcus (genus), Bacteroides (genus), and Coriobacteriaceae (Family) (Figure 5A). The Spearman correlation graph between Prok2 expression and Allobaculum abundance is shown in Figure 6C (R = −0.52, P = 0.0037).
Figure 6. Correlation between ileal Prok2 expression and metagenomic (16S rDNA sequencing) data.
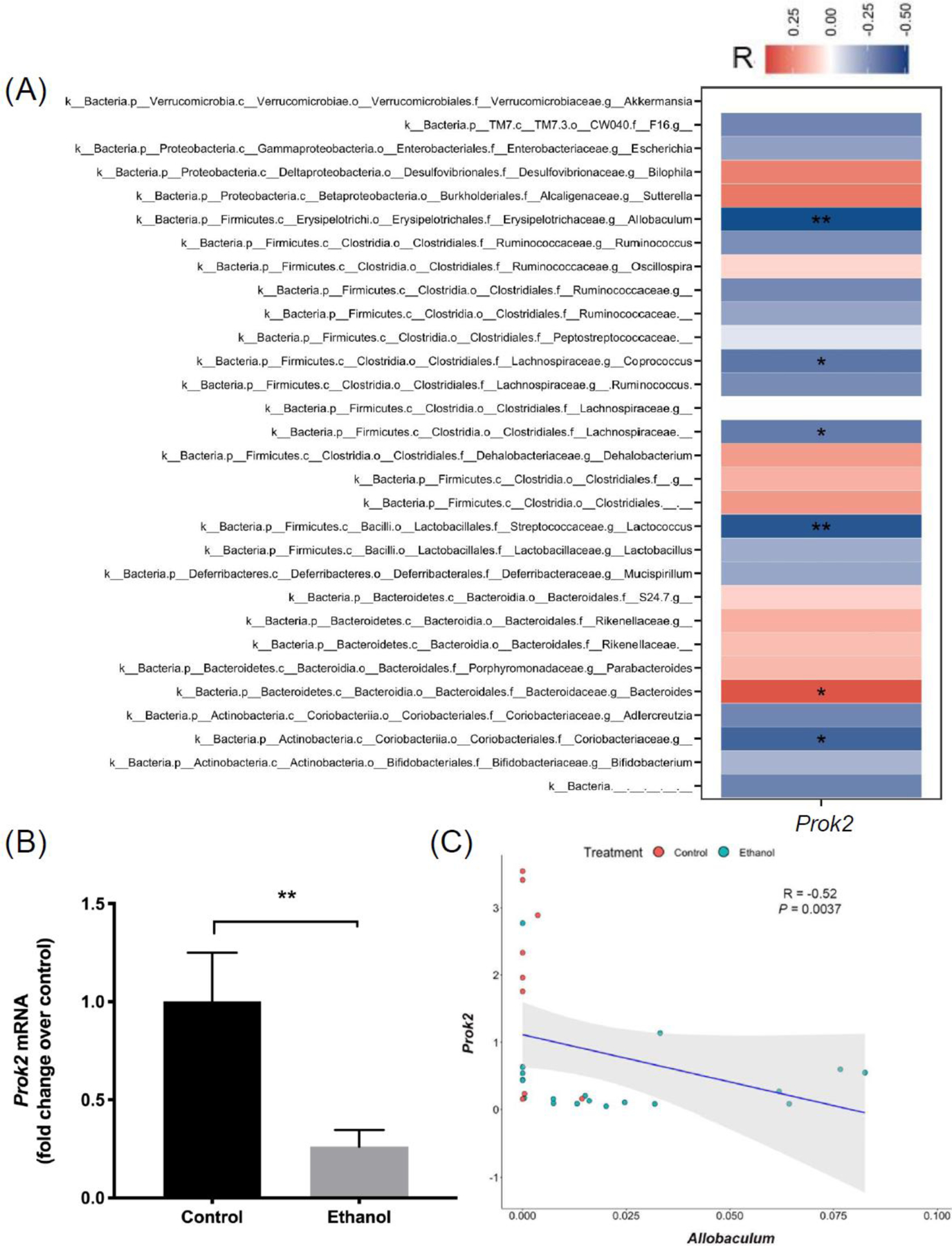
(A) mRNA level of ileal Prok2 in control and ethanol groups (real-time qPCR). (B) Heatmap of Spearman correlation between ileal Prok2 mRNA and the 30 most abundant bacteria at genus level, determined from 16S rDNA gene sequencing. Red color indicates positive correlation, blue color indicates negative correlation. (C) Spearman correlation between ileal Prok2 mRNA and Allobaculum abundance. Control group: n=10; Ethanol group: n=19. *P < 0.05, ****P < 0.0001.
Discussion
In this study, we performed RNA-seq analysis to comprehensively understand the liver and intestine transcriptomic signatures in mice following chronic plus binge ethanol feeding. By investigating the relationship between hepatic gene expression and serum bile acid profile in the context of ethanol-induced liver disease, we identified a negative correlation between suppressed Slco1a1 mRNA and serum cholic acid. Further, we found that ethanol-fed mice showed a significant reduction in ileum Prok2 expression, which was negatively correlated with Allobaculum abundance.
The liver transcriptome and associated functional annotations in alcoholic-associated liver disease have been reported both in animal studies and patients with alcoholic hepatitis [13, 21–23]. In our study, we observed a strong activation of mitogen-activated protein kinase (MAPK) pathway, which plays important roles in regulating insulin sensitivity and secretion [24]. Previous data suggest an LPS-dependent enhanced activation of extracellular receptor-activated kinases 1/2 (ERK1/2) contributes to increased tumor necrosis factor (TNF-α production after chronic ethanol feeding. Inhibition of ERK1/2 decreased LPS-dependent production of TNF-α mRNA [25]. Therefore, MAPK signaling could be an important component of inflammatory response in alcoholic-associated liver disease. In line with previous study [13], GO biological process analysis revealed activation of several functions including triglyceride metabolic process, lipid catabolic process, and interleukin-1 production in response to ethanol. Among all DEGs, we reported Slco1a1, encoding for OATP1A1, as the top down-regulated gene in liver after ethanol treatment. The role of OATP1 as sodium-independent hepatic bile acid uptake transporter has not been thoroughly studied in the pathogenesis of alcoholic-associated liver disease. Previous studies suggested that OATP family are essential for efficient uptake of unconjugated bile acids. Slco1a/1b knockout mice showed elevated levels of unconjugated bile acids in plasma [26]. Moreover, OATPs were shown to contribute to hepatic uptake of conjugated bile acids in coordination with sodium-dependent taurocholate cotransporting polypeptide (NTCP) as the predominant mediator [16]. In our study, we discovered that Slco1a1 expression was negatively correlated with serum cholic acid level. The significant reduction of Slco1a1 expression in liver may constitute the increase of serum cholic acid in response to ethanol or vice versa, but whether this association accounts for pathogenesis of the disease remains to be studied.
RNA-seq analysis in ileum identified Prok2, encoding for prokineticin 2, as one of the top 10 down-regulated genes in ethanol-fed mice. As an inflammatory cytokine-like molecule expressed by macrophages and neutrophils at the sites of injuries, Prok2 expression was up-regulated over 10 folds in colonic mucosal biopsy samples from patients with active ulcerative colitis compared with control samples, and increased Prok2 was positively correlated with Il1b expression [18]. Since Prok2 primary transcript has two alternative splice variants, the decreased expression in our study might be a different variant or it may play different roles in alcoholic-associated liver disease. Considering the connection between Prok2 and gut inflammation [18], we investigated the relationship between Prok2 and gut bacteria signature by integrating the ileum transcriptomic analysis with 16S metagenomic analysis. We found a negative correlation between Prok2 and Allobaculum abundance following chronic plus binge ethanol feeding. Previous study showed an increased fecal abundance of Allobbaculum in mice fed with ethanol compared with control groups [27]. Further, mice fed with high-fat diet also showed increased Allobaculum abundance in fecal samples [28]. This opens possibilities of targeting Prok2 expression as a way to either modulate intestinal inflammation or gut microbial composition.
To our knowledge, we present the first comprehensive analysis by integrating transcriptomic analysis in liver and gut with metabolomic analysis and metagenomic analysis in a model of chronic plus binge ethanol-induced liver disease in mice. Binge drinking might be a significant risk factor for progression of liver disease in patients with alcohol use disorder [29]. Future studies should be performed to validate these results in human patients, and mechanistic studies are needed to reveal whether the correlation we saw in our study are causative effects or associations.
Acknowledgements
This study was supported in part by NIH grants R01 AA24726, R01 AA020703, U01 AA026939, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to B.S.). The study was also supported by the NIDDK-funded San Diego Digestive Diseases Research Center (P30 DK120515) and by the NIAAA-funded Southern California Research Center for ALPD and Cirrhosis (P50 AA011999).
Abbreviations:
- DEG
differentially expressed gene
- GO
gene ontology
- Slco1a1
solute carrier organic anion transporter family, member 1a1
- OATP
organic anion transporting polypeptides
- NTCP
sodium-dependent taurocholate cotransporting polypeptide
- TNF
tumor necrosis factor
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Disclosures: B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics and Patara Pharmaceuticals. B.S.’s institution UC San Diego has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company and Axial Biotherapeutics.
References
- 1.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141(5):1572–85. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramond MJ, Poynard T, Rueff B, Mathurin P, Theodore C, Chaput JC et al. A randomized trial of prednisolone in patients with severe alcoholic hepatitis. N Engl J Med. 1992;326(8):507–12. doi: 10.1056/NEJM199202203260802. [DOI] [PubMed] [Google Scholar]
- 3.Saberi B, Dadabhai AS, Jang YY, Gurakar A, Mezey E. Current Management of Alcoholic Hepatitis and Future Therapies. J Clin Transl Hepatol. 2016;4(2):113–22. doi: 10.14218/JCTH.2016.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan AW, Fouts DE, Brandl J, Starkel P, Torralba M, Schott E et al. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53(1):96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA et al. Colonic microbiome is altered in alcoholism. Am J Physiol-Gastr L. 2012;302(9):G966–G78. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575(7783):505–511. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandl K, Hartmann P, Jih LJ, Pizzo DP, Argemi J, Ventura-Cots M et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J Hepatol. 2018;69(2):396–405. doi: 10.1016/j.jhep.2018.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu H, Jiang L, Gao B, Gautam N, Alamoudi JA, Lang S et al. The selective PPAR-delta agonist seladelpar reduces ethanol-induced liver disease by restoring gut barrier function and bile acid homeostasis in mice. Translational Research. 2020. doi: 10.1016/j.trsl.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang AM, Inamine T, Hochrath K, Chen P, Wang L, Llorente C et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127(7):2829–41. doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–9. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, Llorente C et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology. 2018;67(6):2150–66. doi: 10.1002/hep.29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slijepcevic D, Abbing RLPR, Katafuchi T, Blank A, Donkers JM, van Hoppe S et al. Hepatic Uptake of Conjugated Bile Acids Is Mediated by Both Sodium Taurocholate Cotransporting Polypeptide and Organic Anion Transporting Polypeptides and Modulated by Intestinal Sensing of Plasma Bile Acid Levels in Mice. Hepatology. 2017;66(5):1631–43. doi: 10.1002/hep.29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Augustin O, Sanchez de Medina F. Intestinal bile acid physiology and pathophysiology. World J Gastroenterol. 2008;14(37):5630–40. doi: 10.3748/wjg.14.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson RP, Lilley E, Panesar M, Bhalay G, Langridge S, Tian SS et al. Increased prokineticin 2 expression in gut inflammation: role in visceral pain and intestinal ion transport. Neurogastroenterol Motil. 2012;24(1):65–75, e12. doi: 10.1111/j.1365-2982.2011.01804.x. [DOI] [PubMed] [Google Scholar]
- 19.Pradhan-Sundd T, Vats R, Russell JO, Singh S, Michael AA, Molina L et al. Dysregulated Bile Transporters and Impaired Tight Junctions During Chronic Liver Injury in Mice. Gastroenterology. 2018;155(4):1218–32 e24. doi: 10.1053/j.gastro.2018.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyongyosi B, Cho Y, Lowe P, Calenda CD, Iracheta-Vellve A, Satishchandran A et al. Alcohol-induced IL-17A production in Paneth cells amplifies endoplasmic reticulum stress, apoptosis, and inflammasome-IL-18 activation in the proximal small intestine in mice. Mucosal Immunol. 2019;12(4):930–44. doi: 10.1038/s41385-019-0170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Gong M, French BA, Liao G, Li J, Tillman B et al. Aberrant modulation of the BRCA1 and G1/S cell cycle pathways in alcoholic hepatitis patients with Mallory Denk Bodies revealed by RNA sequencing. Oncotarget. 2015;6(40):42491–503. doi: 10.18632/oncotarget.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khanova E, Wu R, Wang W, Yan R, Chen Y, French SW et al. Pyroptosis by caspase11/4-gasdermin-D pathway in alcoholic hepatitis in mice and patients. Hepatology. 2018;67(5):1737–53. doi: 10.1002/hep.29645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, French BA, Li J, Tillman B, French SW. Altered regulation of miR-34a and miR-483–3p in alcoholic hepatitis and DDC fed mice. Exp Mol Pathol. 2015;99(3):552–7. doi: 10.1016/j.yexmp.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Thompson BJ, Hietakangas V, Cohen SM. MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet. 2011;7(12):e1002429. doi: 10.1371/journal.pgen.1002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishore R, Hill JR, McMullen MR, Frenkel J, Nagy LE. ERK1/2 and Egr-1 contribute to increased TNF-alpha production in rat Kupffer cells after chronic ethanol feeding. Am J Physiol Gastrointest Liver Physiol. 2002;282(1):G6–15. doi: 10.1152/ajpgi.00328.2001. [DOI] [PubMed] [Google Scholar]
- 26.van de Steeg E, Wagenaar E, van der Kruijssen CM, Burggraaff JE, de Waart DR, Elferink RP et al. Organic anion transporting polypeptide 1a/1b-knockout mice provide insights into hepatic handling of bilirubin, bile acids, and drugs. J Clin Invest. 2010;120(8):2942–52. doi: 10.1172/jci42168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8(1):e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62(12):1787–94. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 29.Seitz HK, Bataller R, Cortez-Pinto H, Gao B, Gual A, Lackner C et al. Alcoholic liver disease. Nat Rev Dis Primers. 2018;4(1):16. doi: 10.1038/s41572-018-0014-7. [DOI] [PubMed] [Google Scholar]


