Abstract
DNA methylation on cytosine in CpG islands generates 5-methylcytosine (5mC), and further modification of 5mC can result in the oxidized variants 5-hydroxymethyl (5hmC), 5-formyl (5fC), and 5-carboxy (5caC). Base excision repair (BER) is crucial for both genome maintenance and active DNA demethylation of modified cytosine products and involves substrate-product channeling from nucleotide insertion by DNA polymerase (pol) β to the subsequent ligation step. Here, we report that, in contrast to the pol β mismatch insertion products (dCTP, dATP, and dTTP), the nicked products after pol β dGTP insertion can be ligated by DNA ligase I or DNA ligase III/XRCC1 complex when a 5mC oxidation modification is present opposite in the template position in vitro. A Pol β K280A mutation, which perturbates the stabilization of these base modifications within the active site, hinders the BER ligases. Moreover, the nicked repair intermediates that mimic pol β mismatch insertion products, i.e., with 3’-preinserted dGMP or dTMP opposite templating 5hmC, 5fC or 5caC, can be efficiently ligated, whereas preinserted 3’-dAMP or dCMP mismatches result in failed ligation reactions. These findings herein contribute to our understanding of the insertion tendencies of pol β opposite different cytosine base forms, the ligation properties of DNA ligase I and DNA ligase III/XRCC1 complex in the context of gapped and nicked damage-containing repair intermediates, and the efficiency and fidelity of substrate channeling during the final steps of BER in situations involving oxidative 5mC base modifications in the template strand.
Keywords: Base excision repair, DNA polymerase β, DNA ligase I, DNA ligase III, CpG islands, DNA methylation, 5-methylcytosine
1. INTRODUCTION
The CpG dinucleotides in genomic DNA are hotspots for mutations, including G to T transitions in the human p53 gene, and epigenetic modifications play a fundamental role in embryonic development, transcriptional regulation, and epigenetic integrity (1–3). The CpG sequences are subject to many chemical modifications that vary from no structural impact on the DNA helix to certain helix-distorting lesions (4,5). The variety of modifications includes methylation of cytosine in the context of CpG islands, where the methyl group from S-adenosyl methionine is transferred to cytosine by DNA methyltransferases (DNMT1–3) (6–8). This DNA methylation generates 5-methylcytosine (5mC), an alteration that is used as an epigenetic mark in the regulation of gene expression and has an important role in loss of genomic imprinting, such as the allelic methylation differences at imprinted genes (9–11). Further chemical modifications of 5mC can occur via oxidation by Fe2+- and α-ketoglutarate-dependent dioxygenases of the Ten Eleven Translocation (TET) family of proteins, leading to the formation of progressively oxidized 5mC variants: 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and finally 5-carboxylcytosine (5caC) (12–14). The significance of 5mC in diagnosis and therapy for many types of cancer has been extensively demonstrated (15–18). Moreover, in recently reported studies, the contribution of 5hmC and DNA damage-induced changes in the levels of TET enzymes to the mechanisms underlying neurodevelopmental abnormalities and neurodegenerative disorders has been shown (19–22). Nevertheless, little is known about the functions of 5hmC in mammalian biology.
The elimination of the oxidative 5mC derivatives is mediated by active DNA demethylation through base excision repair (BER), a process important for maintaining the methylation status of CpG sites (23–27). However, we lack understanding of the mechanism by which the repair proteins contribute to the potential mutational effect of oxidative 5hmC modifications in vitro. Biochemical studies have indicated the presence of substrate channeling in BER that involves coordination of the substrate-product hand off between the different enzymes (28–30). This mechanism prevents the accumulation of toxic and mutagenic repair intermediates that could promote harmful nuclease activities and cytotoxicity, as well as trigger cell cycle arrest and apoptosis (30–32). The BER pathway is initiated by a DNA damage-specific DNA glycosylase, which removes a single-base lesion via hydrolysis of the N-glycosidic bond (33). Thymine DNA glycosylase (TDG) catalyzes the removal of 5fC and 5caC, which leaves an abasic (AP) site in double-stranded DNA (34,35). AP endonuclease 1 (APE1) then cleaves the phosphodiester backbone at the AP site, resulting in the formation of a single-strand break (one nucleotide gap) with 3’-hydroxyl (3’-OH) and 5’-deoxyribosephosphate (5’-dRP) termini (36). Pol β then removes the 5’-dRP group and conducts template-directed gap filling DNA synthesis (37). Pol β nucleotide insertion products generate a nicked substrate for the subsequent and final DNA ligation step of the pathway (38). BER DNA ligases complete repair, which requires high fidelity DNA synthesis by pol β, a polymerase that discriminates against the incorporation of incorrect nucleotides to varying degrees on the basis of base pairing (39,40). At the final step of the process, DNA ligase I or DNA ligase III/X-ray repair cross-complementing protein 1 (XRCC1) complex catalyze phosphodiester bond formation between the adjacent 3’-OH and 5’-P termini of the nicked repair intermediate (41,42). Notably, a physical interaction between pol β and DNA ligase I during the repair of single-base DNA lesions has been reported, and pol β is also known to physically interact with XRCC1 (43–46). We previously demonstrated that pol β 8-oxodGTP insertion leads to DNA ligase failure, and that pol β mismatch insertion products, excluding Watson-Crick like dGTP:T base-pairs, cannot be channeled to the ligation step in vitro (47–50).
Recently, it has been shown that pol β can incorporate dGTP opposite 5hmC or 5caC, and that the enzyme active site can accommodate these epigenetic base modifications on the template DNA strand (51). Moreover, the ternary complex structure of pol β with a non-hydrolysable dGTP analog revealed an altered conformation in which the 5’-phosphate backbone of the template base modification is repositioned and stabilized by the Lysine 280 (K280) side chain. However, how these structural adjustments that pol β undergoes during bypass of an epigenetic base damage located on template strand might impact downstream steps of BER and whether the repair products of pol β (e.g., mismatch nucleotide insertions opposite the oxidative 5mC modifications) can be channeled to the final DNA ligation step remains undetermined. Successful ligation relies on the formation of a Watson-Crick base pair between 3’-OH and 5’-P termini of the nicked repair product. Thus, DNA ligases may fail in the presence of modified or damaged DNA ends, resulting in the formation of ligation intermediates harboring an adenylate (AMP) block at the 5’-end (38,42).
In the present study, we examined coordinated repair at the downstream steps of BER pathway when oxidatively modified 5mC products (5hmC, 5fC, and 5caC) were placed in the template strand. For this purpose, using one nucleotide gapped DNA substrates, we investigated the effect of pol β mismatch nucleotide insertion opposite 5hmC, 5fC, and 5caC on the channeling of the resulting nicked intermediate to the BER ligases (DNA ligase I and DNA ligase III/XRCC1 complex) in vitro. Moreover, in this study, using the nicked DNA substrates that mimic pol β mismatch nucleotide insertion products, we evaluated the substrate specificity of the BER ligases for their ability to join DNA ends in the presence of templating 5hmC, 5fC, or 5caC in vitro. The results presented herein offer novel insight into the importance of an interplay between key repair proteins, pol β and the BER DNA ligases, on the fidelity of the substrate-product channeling mechanism when the repair machinery encounters an epigenetically important cytosine base modification in the template strand.
2. Materials and methods
2.1. Preparation of DNA substrates
Oligodeoxyribonucleotides with and without a 6-carboxyfluorescein (FAM) label were obtained from Integrated DNA Technologies (IDT; Coralville, IA). The template oligonucleotides (34-mer) with 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), or 5-carboxylcytosine (5caC) were from the Midland Reagent Company Inc (Midland, Texas). The DNA substrates were prepared as described previously (47–50,52–55). Briefly, the upstream (17-mer) and downstream (16-mer) primers were annealed in the presence of the complementary oligonucleotide (34-mer) to prepare the one nucleotide gapped DNA substrates. The upstream (18-mer) primer with a 3’-preinserted mismatch (dAMP, dTMP, dGMP, or dCMP) and the downstream (16-mer) oligonucleotide were similarly annealed in the presence of the 34-mer to prepare the nicked DNA substrates. The sequence information for the double-stranded DNA substrates is presented in Supplementary Table 1.
2.2. Protein purifications
Recombinant full-length human DNA ligase I was purified as previously described (47–50,52–55). Briefly, the protein was overexpressed in Rosetta2 (DE3) cells at 37 °C, and the cells were grown overnight at 16 °C. After cell lysis by sonication at 4 °C in the lysis buffer containing 40 mM HEPES (pH 7.5), 200 mM NaCl, 10% glycerol, and cOmplete Protease Inhibitor Cocktail (Roche), and clarification by centrifugation, the His-tagged protein was loaded onto a HisTrap HP column (GE Healthcare) and purified with an increasing imidazole gradient (0–500 mM) elution at 4 °C, and then subsequently loaded onto a HiTrap Q HP column (GE Healthcare) and eluted with a linear gradient of NaCl up to 1 M. Finally, the collected fractions were separated through a Superdex 200 Increase 10/300 column (GE Healthcare). The resulting pure fractions of DNA ligase I were combined, concentrated with a centrifugal filter unit, and stored in aliquots at −80 °C. The N- and C-terminal domains of DNA ligase I were purified as described before (50). Recombinant (GST-tagged pGEX4T3) wild-type full-length human DNA polymerase β was purified as previously described (47–50,52–55). Briefly, the protein was overexpressed in One Shot BL21(DE3)pLysS E. coli cells (Invitrogen) and grown at 37 °C with IPTG induction. The cells were then grown overnight at 16 °C. After centrifugation, the cells were lysed at 4 °C by sonication in lysis buffer containing 25 mM HEPES (pH 7.5), 500 mM NaCl, 0.1% NP40, and a protease inhibitor cocktail. The lysate was pelleted at 10,444 × g for 1 h and then clarified by filtration. The pol β supernatant was loaded onto a GSTrap HP column (GE Health Sciences) and purified with elution buffer containing 50 mM Tris-HCl (pH 8.0) and 10 mM reduced glutathione. The collected fractions were subsequently passed through a HiTrap Desalting HP column in a buffer containing 150 mM NaCl and 20 mM NaH2PO4 (pH 7.0), and then further purified by Superdex 200 Increase 10/300 chromatography (GE Healthcare). Pol β K280A mutant was constructed using site-directed mutagenesis with synthetic primers, confirmed by sequencing of the coding region, and purified as described for the wild-type protein above. Recombinant full-length human DNA polymerase λ was overexpressed and purified as reported (49,54,55). Briefly, the clarified lysate was first passed through a Q-Sepharose column, and then the enzyme was purified using a nickel column at 4 °C. The protein was eluted with an imidazole gradient, and pol λ was further purified using Mono S column chromatography with NaCl gradient elution at 4 °C. All proteins used in this study were dialyzed against storage buffer (25 mM TrisHCl, pH 7.4, 100 mM KCl, 1 mM TCEP, and 10% glycerol), concentrated, frozen in liquid nitrogen, and stored at −80 °C. The purified proteins used in this study are presented in Supplementary Figure 1A.
2.3. Ligation assay
The ligation assays were performed under steady-state conditions using the nicked DNA substrates (Supplementary Table 1) as described previously (47–50,52–55). The reaction was performed in a mixture containing 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 100 μg ml−1 BSA, 10% glycerol, and the DNA substrate (500 nM) in a final volume of 10 μl. The ligation assays were initiated with the addition of DNA ligase (10 nM). The reaction mixture was incubated at 37 °C and stopped at the indicated time points within the figure legends. The reactions were then mixed with an equal amount of gel loading buffer (95% formamide, 20 mM EDTA, 0.02% bromophenol blue, and 0.02% xylene cyanol) and separated by electrophoresis on an 18% polyacrylamide gel as described previously (47–50,52–55). The resulting gels were scanned with a Typhoon PhosphorImager (Amersham Typhoon RGB), and the data were analyzed using ImageQuant software.
2.4. Pol β nucleotide insertion and insertion coupled to DNA ligation assays
The nucleotide insertion and insertion/ligation coupled assays were performed under steady-state conditions using the one nucleotide gapped DNA substrates (Supplementary Table 1) as described previously (47–50). Briefly, the reactions were performed in a mixture containing 50 mM TrisHCl, pH 7.4, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 1 mM ATP, 10% glycerol, and 0.1 mg/ml BSA at 37 °C. Reactions were initiated by mixing a solution containing 100 nM DNA and 100 μM dNTP (dATP, dTTP, dGTP, or dCTP) with pol β alone (50 nM) or the mixture of pol β and a DNA ligase (100 nM). Pol β nucleotide insertion and coupled insertion/ligation assays were performed similarly in the presence of the pol β mutant (K280A) or the N- and C-terminal domains of DNA ligase I. Reactions were quenched by addition of 100 mM EDTA, mixed with gel loading buffer, and the reaction products were separated, and the data were analyzed as described above.
3. Results
3.1. The ligation of pol β dGTP or mismatch insertions opposite oxidized forms of 5mC
In order to examine the ligation efficiency of pol β nucleotide insertions into one nucleotide gapped DNA substrate with oxidized forms of 5mC (5hmC, 5fC, or 5caC) in the template position (Supplementary Table 1), we performed coupled reactions to measure pol β insertion and DNA ligation simultaneously in vitro. These reaction mixtures included pol β, DNA ligase (DNA ligase I or DNA ligase III/XRCC1 complex), and dNTP (Figure 1A). Our results revealed ~2-fold less amount of ligation products after pol β dGTP insertion opposite 5hmC, 5fC, and 5caC by DNA ligase I (Figure 1B, lanes 7–10, 12–15, and 17–20, respectively), in comparison with ligation of the repair intermediates after pol β dGTP:C correct insertion (Figures 1B, lanes 2–5). We obtained similar results in the coupled reactions containing pol β and DNA ligase III/XRCC1 complex (Supplementary Figure 1B). In both cases, the ligation products increased over time (Figures 1C and Supplementary Figure 1C), and there was no significant difference between BER ligases (Supplementary Figure 2).
Figure 1.

The ligation of pol β dGTP insertions opposite oxidized forms of 5mC. (A) Illustration of the one nucleotide gapped DNA substrate and reaction products observed in the coupled assays including pol β and DNA ligase. (B) Lanes 1, 6, 11, and 16 are the negative enzyme controls of the one nucleotide gapped DNA substrates with template C, 5hmC, 5fC, and 5caC, respectively. Lanes 2–5, 7–10, 12–15, and 17–20 are the ligation products after pol β dGTP insertion opposite C, 5hmC, 5fC, and 5caC, respectively, and correspond to time points 10, 30, 45, and 60 s. (C) The graph shows time-dependent changes in the ligation products. The data represent the average of three independent experiments ± SD.
We next evaluated the ligation of pol β insertion mismatches opposite the 5mC modifications as described above (Figure 1A). In contrast to the results obtained with pol β dGTP insertions, the products in the presence of pol β mismatches (dATP, dTTP, and dCTP) were self-ligation products, i.e., direct ligation of the one nucleotide gapped DNA with a template 5hmC, 5fC, or 5caC, by DNA ligase I or the DNA ligase III/XRCC1 complex. More specifically, the coupled reaction that includes pol β, DNA ligase I and dATP (Figure 2, lanes 2–4) yielded DNA products of similar size, i.e., without nucleotide insertion, to those observed in the reactions including DNA ligase I alone (Figure 2, lanes 5–19). We obtained similar results with the DNA ligase III/XRCC1 complex (Supplementary Figure 3). We previously reported this “self-ligation” of gapped DNA with a template unmodified C to which pol β inserts mismatches dCTP, dATP, and dTTP (50). For the other two reactions (i.e., pol β with dTTP or dCTP in the presence of substrates with 5hmC, 5fC, or 5caC), there were also only self-ligation products in the coupled reactions (Supplementary Figures 4 and 5). This outcome could be due to low nucleotide insertion efficiency of pol β as reported previously (51). Consistent with this idea, we observed a very low amount of pol β dATP insertions opposite 5hmC in the insertion reaction (Supplementary Figure 6, lanes 11–15) and of corresponding self-ligation products in the coupled reaction (Supplementary Figure6, lanes 16–20), when compared to pol β dATP:T insertions and subsequent complete ligation (Supplementary Figure 6, lanes 1–10).
Figure 2.

The ligation of pol β mismatch insertions opposite oxidized forms of 5mC by DNA ligase I. Lanes 1–4 are the ligation products after pol β dGTP:C, dATP:5hmC, dATP:5fC, and dATP:5caC insertions, respectively. Lanes 5, 10, and 15 are the negative enzyme controls of the one nucleotide gapped DNA substrates with template 5hmC, 5fC, and 5caC, respectively. Lanes 6–9, 11–14, and 16–19 are the self-ligation products of the one nucleotide gapped DNA with template 5hmC, 5fC, and 5caC, respectively, and correspond to time points 10, 30, 45, and 60 s. The gel is a representative of three independent experiments.
We hypothesize that this self-ligation product could be in a transient intermediate conformation at the active site in which the template and incoming nucleotide bases do not form hydrogen bonds with each other but instead form a staggered conformation. Indeed, this conformation has been previously shown in the pol β structure with primer-template T-C and A-C mismatches in which the template strand shifts upstream and the bases of the mismatched pair partially overlap where the nascent base pair-binding domain (N-subdomain) of the protein is in an intermediate position between an opened or a closed state (56).
3.2. Effects of pol β active site mutants on the ligation of mismatch insertions opposite oxidative 5mC modifications
In light of the results described above, we next examined Lys280 of pol β, a residue that plays an important role in the structural repositioning within the enzyme active site to accommodate the oxidative variants of 5mC in the template position (51). Similar to the wild-type enzyme, in the presence of the K280A mutant, we obtained pol β dGTP insertion products and their complete ligation for template C (Figure 3A, lanes 2–5 and 7–10), 5hmC (Figure 3A, lanes 12–15 and 17–20), 5fC (Figure 4A, lanes 2–5 and 7–10), and 5caC (Figure 4A, lanes 12–15 and 17–20). However, in all cases, the amount of ligation products was ~ 2 to 4-fold lower after dGTP insertion by K280A relative to wild-type pol β (Figures 3B and 4B). In the presence of dATP, pol β insertions opposite 5hmC, 5fC, or 5caC yielded the self-ligation products in the coupled reactions that included pol β K280A and DNA ligase I (Supplementary Figure 7A), similar to the results with wild-type pol β (Figure 2). We then tested a pol β active site mutant at position Asp256, which plays a critical role in the activation of the primer O3′ nucleophile and coordination of the catalytic metal ion required for the nucleotidyl transferase reaction (57). In the presence of either correct dGTP:C or mismatch nucleotide insertions (dATP opposite 5hmC, 5fC, or 5caC), in the reactions including pol β D256A mutant and DNA ligase I, we only obtained self-ligation products of the one nucleotide gapped DNA (Supplementary Figure 7B).
Figure 3.
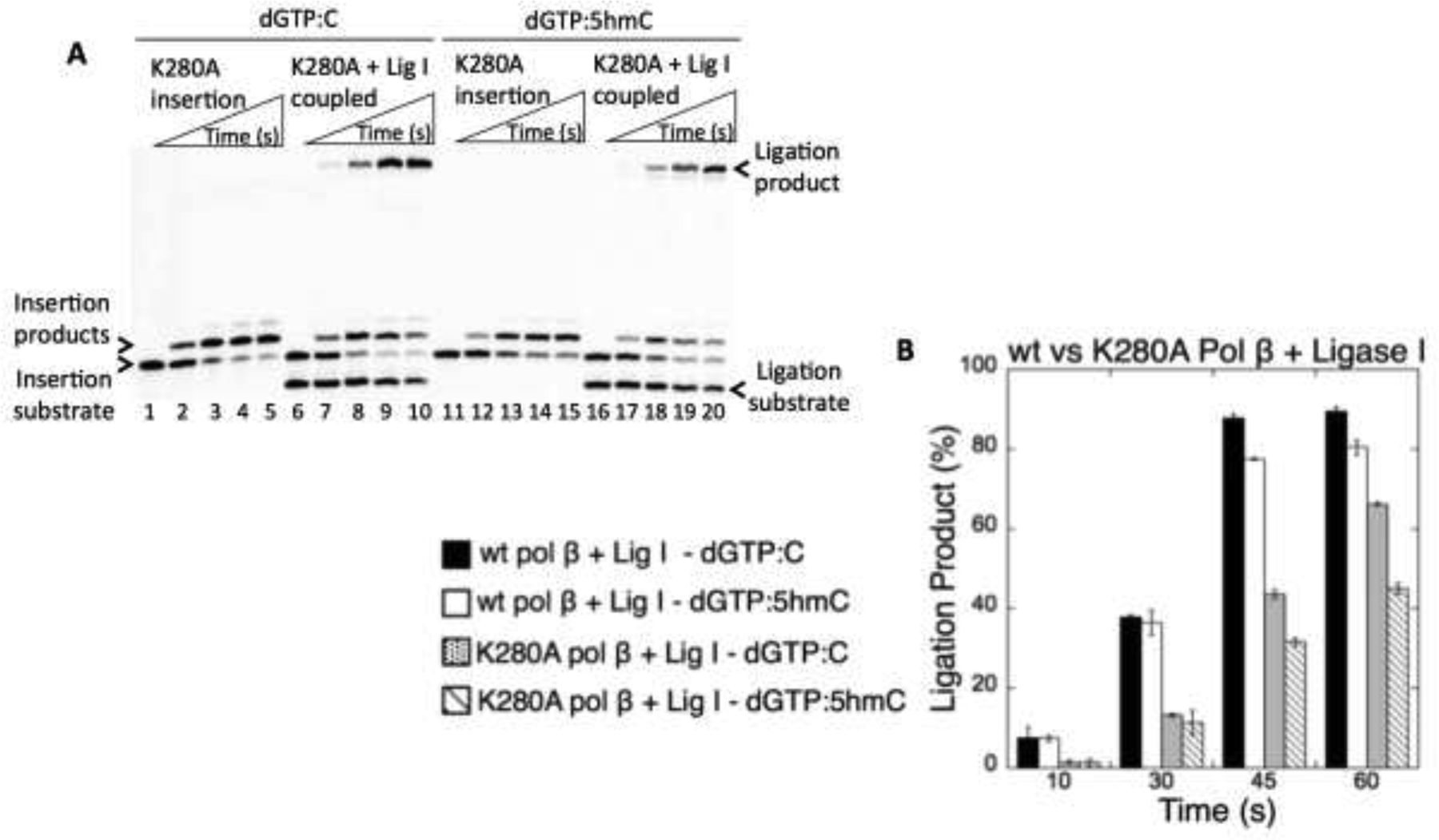
Effect of pol β K280 mutation on the ligation of dGTP insertions opposite C and 5hmC by DNA ligase I. (A) Lanes 1 and 11 (insertion) and lanes 6 and 16 (coupled) are the negative enzyme controls of the one nucleotide gapped DNA substrates with template C and 5hmC. Lanes 2–5 and 12–15 are pol β dGTP insertion products, and lanes 7–10 and 17–20 are pol β dGTP insertion coupled to ligation products, and correspond to time points of 10, 30, 45, and 60 s. (B) The graph shows time-dependent changes in the ligation products in coupled reactions including wild-type pol β and DNA ligase I vs pol β K280A and DNA ligase I. The data represent the average of three independent experiments ± SD.
Figure 4.

Effect of pol β K280 mutation on the ligation of dGTP insertions opposite 5fC and 5caC by DNA ligase I. (A) Lanes 1 and 11 (insertion) and lanes 6 and 16 (coupled) are the negative enzyme controls of the one nucleotide gapped DNA substrates with template 5fC and 5caC. Lanes 2–5 and 12–15 are pol β dGTP insertion products, and lanes 7–10 and 17–20 are pol β dGTP insertion coupled to ligation products, and correspond to time points of 10, 30, 45, and 60 s. (B) The graph shows time-dependent changes in the ligation products in coupled reactions including wild-type pol β and DNA ligase I vs pol β K280A and DNA ligase I. The data represent the average of three independent experiments ± SD.
3.3. Impact of interaction between pol β and DNA ligase I on the ligation of 5mC template modifications
The N-terminal domain of DNA ligase I participates in a protein-protein interaction with pol β (43,44) and governs the channeling of repair intermediates from pol β nucleotide insertion to ligation during BER (50). We examined the effect of this non-catalytic domain of DNA ligase I in the ligation of pol β dGTP insertions opposite 5hmC, 5fC, and 5caC using either full-length or a C-terminal fragment of DNA ligase I. Similar to the results with the full-length protein (Figure 1), we obtained ligation products after pol β dGTP insertion opposite 5hmC or 5caC (Figure 5A, lanes 2–5 and 11–14, respectively) with the C-terminal domain of DNA ligase I. However, the amount of the ligation product was significantly lower in comparison to the full-length protein that harbors both C- and N-terminal domains, indicating an importance of the physical interaction (Figure 5B). As expected, there was no ligation products in the coupled reaction including only the N-terminal domain of DNA ligase I, which lacks the catalytic core (Figure 5A, lanes 6–9 and 15–18). For pol β dATP mismatch insertions, there were self-ligation or no ligation products in the presence of either the C-terminal or N-terminal domain of ligase I, respectively (Supplementary Figure 8).
Figure 5.

Effect of pol β and DNA ligase I interaction on the ligation of dGTP insertions opposite oxidized forms of 5mC. (A) Lanes 1 and 10 are the negative enzyme controls of the one nucleotide gapped DNA substrates with template 5hmC and 5caC, respectively. Lanes 2–5 and 6–9 are pol β dGTP:5hmC insertion coupled to ligation products in the presence of C-terminal and N-terminal domains of DNA ligase I, respectively, and correspond to time points of 10, 30, 45, and 60 s. Lanes 11–14 and 15–18 are pol β dGTP:5caC insertion coupled to ligation products in the presence of C-terminal and N-terminal domains of DNA ligase I, respectively, and correspond to time points of 10, 30, 45, and 60 s. (B) The graph shows time-dependent changes in the ligation products in the coupled reactions including pol β and full-length DNA ligase I vs pol β and C-terminal domain of DNA ligase I. The data represent the average of three independent experiments ± SD.
3.4. Comparison of ligation efficiency after pol λ dGTP insertion opposite oxidized forms of 5mC
Pol λ, exhibiting both dRP lyase and gap-filling activities, is considered a back-up polymerase for pol β and participates in BER of lesions generated by monofunctional alkylating agents and oxidative stressors (58,59). We compared the roles of X-family DNA polymerases, pol β vs pol λ, by investigating the efficiency of DNA ligation coupled to pol λ dGTP insertion in the presence of the oxidized forms of 5mC using a coupled assay as described above. Interestingly, our results showed that pol λ dGTP insertions (Figure 6A) opposite 5hmC (lanes 5–8), 5fC (lanes 9–12), or 5caC (lanes 13–16) result in the formation of both products (full ligation and self-ligation), which accumulated simultaneously at the same time points in the coupled reaction (Figure 6B). The amount of complete ligation products by pol λ was ~4-fold lower when compared with those of pol β and DNA ligase I, suggesting a bias towards self-ligation in reactions containing pol λ (Supplementary Figure 9A). We note that we obtained similar levels of complete ligation products after pol λ dGTP insertions opposite unmodified C in comparison to reactions with pol β (Supplementary Figure 9B). These observations suggest that pol λ may undergo a distinct conformational change to accommodate the carboxy modification on the template DNA strand with an incoming dGTP. This could be due to the unique structural features of pol λ, such as the presence of loop 1 that is absent in pol β and plays role in mismatch discrimination in the enzyme active site (60). Future structure analysis of pol λ in complex with epigenetic 5mC modifications is required to fully interpret our results here.
Figure 6.

Ligation of pol λ dGTP insertions opposite oxidized forms of 5mC by DNA ligase I. (A) Lanes 1–3 are the negative enzyme controls of the one nucleotide gapped DNA substrates with template 5hmC, 5fC, and 5caC, respectively. Lane 4 is the insertion coupled to ligation products for dGTP:C, and lanes 5–8, 9–12, and 13–16 are the insertion coupled to ligation products for dGTP:5hmC, dGTP:5fC, and dGTP:5caC, respectively, and correspond to time points 10, 30, 45, and 60 s. (B) The graph shows time-dependent changes in the ligation products in coupled reactions including pol β and DNA ligase I vs pol λ and DNA ligase I. The data represent the average of three independent experiments ± SD.
3.5. Ligation of nicked repair intermediates with 5mC modifications
To understand the specificity of the BER DNA ligases (DNA ligase I and the DNA ligase III/XRCC1 complex) for repair intermediates that mimic pol β mismatch insertion products with the oxidized forms of 5mC in the template position, we measured ligation in reactions containing DNA ligase alone and a nicked DNA substrate with a 3’-preinserted mismatch (dAMP, dTMP, dGMP, or dCMP) opposite template 5hmC, 5fC, or 5caC (Figure 7A).
Figure 7.

Ligation of the nicked repair intermediates with preinserted 3’-dGMP opposite oxidized forms of 5mC by DNA ligase I. (A) Illustration of the nicked DNA substrate and reaction products observed in the ligation assays including DNA ligase. (B) Lanes 1, 8, and 15 are the negative enzyme controls of the nicked DNA substrates with template 5hmC, 5fC, and 5caC, respectively. Lanes 2–7, 9–14, and 16–21 are the ligation products and correspond to time points of 0.5, 1, 3, 5, 8, and 10 min.
DNA ligase I was able to ligate the nicked DNA substrates with preinserted 3’-dGMP opposite 5hmC, 5fC, and 5caC (Figure 7B, lanes 2–7, 9–14, and 16–21, respectively), with an increase in the amount of ligation products over the time course (Figure 8A). We obtained similar results in ligation reactions including the DNA ligase III/XRCC1 complex, indicating the mutagenic potential of misinsertion by pol β (Figure 10A and Supplementary Figure 12A). In contrast to the results observed in the coupled reactions (Figure 2), we obtained both complete and failed (5’-AMP containing) ligation products for nicked DNA with 3’-preinserted mismatches by either DNA ligase I (Figures 8–9) or DNA ligase III/XRCC1 complex (Figures 10–11). For the nicked DNA substrates with preinserted 3’-dTMP opposite 5hmC, 5fC, or 5caC, DNA ligase I (Figure 8B) and DNA ligase III/XRCC1 complex (Figure 10B) yielded very efficient ligation (Supplementary Figures 10 and 12B). For the nicked DNA substrates with preinserted 3’-dAMP and 3’-dCMP mismatches opposite 5hmC, 5fC, and 5caC, we observed a comparatively lower amount of ligation products by DNA ligase I (Figure 9) or the DNA ligase III/XRCC1 complex (Figure 11), with the complete products accumulating simultaneously with ligation failure products (Supplementary Figures 11 and 13). In control ligation reactions with the nicked DNA substrates harboring 3’-dGMP opposite unmodified C, we confirmed efficient ligation of undamaged DNA ends by both BER DNA ligases and its comparison with those of template 5mC modifications (Supplementary Figure 14). Overall, results here reveal that the BER DNA ligases exhibit similar mismatch specificity for ligation of templating oxidized 5mC modifications. However, they show different fidelity depending on the identity of the mismatch and the type of template base (5hmC, 5fC, or 5caC) within the nicked repair intermediate (Supplementary Figure 15), and their activities can also be significantly influenced by the presence of pol β.
Figure 8.

Substrate specificity of DNA ligase I for the ligation of the repair intermediates with preinserted 3’-dGMP and 3’-dTMP opposite oxidized forms of 5mC. The graphs show time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data represent the average of three independent experiments ± SD. The gel images are presented in Figure 7B and Supplementary Figure 10.
Figure 10.

Substrate specificity of DNA ligase III/XRCC1 complex for the ligation of the repair intermediates with preinserted 3’-dGMP and 3’-dTMP opposite oxidized forms of 5mC. The graphs show time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data represent the average of three independent experiments ± SD. The gel images are presented in Supplementary Figure 12.
Figure 9.

Substrate specificity of DNA ligase I for the ligation of the repair intermediates with preinserted 3’-dAMP and 3’-dCMP opposite oxidized forms of 5mC. The graphs show time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data represent the average of three independent experiments ± SD. The gel images are presented in Supplementary Figure 11.
Figure 11.

Substrate specificity of DNA ligase III/XRCC1 complex for the ligation of the repair intermediates with preinserted 3’-dAMP and 3’-dCMP opposite oxidized forms of 5mC. The graphs show time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data represent the average of three independent experiments ± SD. The gel images are presented in Supplementary Figure 13.
4. Discussion
The methylation of cytosine bases in DNA generates an epigenetic mark known as 5mC, and the successive conversion of 5mC by TET protein-mediated oxidation results in 5hmC, 5fC, and 5caC (9,11,13,14). Although the roles of 5mC in regulation of gene expression are well established, the effect of the epigenetic 5mC oxidative modifications on DNA replication and repair remains poorly understood. BER, a multi-step enzymatic pathway involving substrate-product hand off between repair proteins, plays an important role in genome maintenance and active DNA demethylation (23,27–32). Moreover, it has been reported that TET enzymes interact with many BER proteins, such as TDG, DNA ligase III, and XRCC1 (61).
The oxidative modifications of 5mC in DNA favor Watson-Crick base pairing with an unmodified guanine in vitro (62,63). Nonetheless, the 5mC derivatives induce mutations with a broad spectrum in E. coli. 5fC, in particular, can result in C-T transitions and one nucleotide deletions when they are bypassed in the template DNA strand by translesion DNA polymerases (pol η and κ) during DNA replication, and can also form DNA-protein conjugates with histone proteins in mammalian cells in vivo (64–66). Moreover, pol β, the major BER DNA polymerase, can accommodate 5hmC and 5fC as a template base that coordinates base pairing with an incoming dGTP at the active site (51). To date, the potential mutagenic impact of DNA polymerase-mediated nucleotide insertions opposite the oxidative 5mC base modifications during DNA repair has remained unknown. Furthermore, DNA ligases, which catalyze the ultimate step at the end of many DNA repair responses following gap filling, along with DNA polymerases, remain prime candidates for genome instability (38). Our study demonstrates the range of potential repair outcomes during the downstream substrate-product channeling steps (e.g., DNA ligation) between central BER proteins, namely pol β and DNA ligase I or DNA ligase III, in the context of oxidatively modified and epigenetically important base modifications of 5mC.
Our results, in particular, reveal that the products of pol β dGTP insertion opposite oxidative 5mC base modifications can be efficiently channeled to the BER ligases and ligated in similar way with those of pol β dGTP insertion opposite unmodified C. We previously reported the same efficient substrate-product hand off between pol β and the DNA ligases for Watson-Crick like dGTP opposite a template T (50). Such a scenario would lead to the formation of a mutagenic repair intermediate, which we hypothesize could be in and of itself a very serious form of damage and potentially be converted into a persistent DNA strand break that blocks transcription or results in a double-strand break upon DNA replication. In this study, and in our previous reports (50), we found that all other possible pol β mismatch insertions (dATP, dCTP, dTTP) opposite either a 5mC modification or unmodified C can also yield self-ligation products (i.e., potential single nucleotide deletion mutagenesis products), indicating a lack of effective substrate channeling and incomplete repair outcomes. We suggest that these situations could provide an opportunity for a proofreading enzyme such as APE1 to remove a mismatched base at the 3’-end through its exonuclease activity (67). In addition, we have previously observed that BER ligases can fail and generate abortive ligation products with a 5’-adenylate in the presence of an incoming 8-oxodGTP inserted by pol β opposite template A or C (47–49). Moreover, our prior studies have also demonstrated that the BER ligases can fail on the APE1 incised gap filled nicked product (i.e., a 3’-OH and 5’-dRP nick), resulting in the formation of a 5’-adenylated-dRP-containing ligation damage intermediate (52–54). Interestingly, in this study, we did not observe ligation failure for pol β repair reactions in the presence of a template 5mC base modification. In contrast, when comparing the ligation efficiency after dGTP insertion opposite a 5mC base modification between reactions with both pol β and DNA ligase I (Figure 1) and reactions with DNA ligase I alone (Figures 8A and 10A), we found that the presence of pol β significantly facilitated ligation of the repair intermediate (e.g., ~80% in coupled reaction vs ~10% in ligation reaction for 60 sec). These findings are supported by our results that reveal a lack of coordination between pol β and DNA ligase I when using the K280A mutant that destabilizes the template 5mC base modifications (Figures 3 and 4) or C-terminal domain of DNA ligase I that is devoid of pol β interaction N-terminal domain (Figure 5). Collectively, the results indicate that the fidelity of substrate channeling from pol β mismatch insertion to ligation by the BER DNA ligases during the final steps of the repair pathway in situations involving oxidative 5mC base modifications in the template strand could be an important determinant of the repair efficiency (Figure 12A).
Figure 12.

The models show the overall findings of this study and in our previous reports for pol β gap filling, DNA ligation and substrate-product channeling during base excision repair. (A) Substrate channeling from pol β correct vs mismatch insertion to ligation by the BER DNA ligases during the final steps of the repair pathway in situations involving oxidative 5mC base modifications in the template strand. (B) Substrate specificity of the BER ligases for joining of 3’-preinserted correct vs mismatches when the oxidative 5mC modifications are present in the template position.
In this study, we also investigated the substrate specificity and 3’-end surveillance mechanism of the BER ligases for joining of 3’-preinserted mismatches opposite 5mC base modifications. These nicked ligase substrates that mimic products of mismatch insertions by an error-prone DNA polymerase could be mutagenic DNA intermediates formed during DNA repair or replication processes. Our results reveal that ligation of the nicked DNA with preinserted 3’-dGMP or 3’-dTMP, when the oxidative 5mC modifications are present in the template position, are efficient. We note that we previously reported efficient ligation of 3’-dGMP opposite T and 3’-dTMP opposite unmodified C (50). Moreover, in this study and our previous work, we showed that BER ligase failure (i.e., the formation of an abortive ligation product with 5’-adenylate) occurs during joining of DNA termini with either a 3’-dAMP or a 3’-dCMP opposite either a 5mC modification or unmodified C. Overall our findings suggest that BER ligases show distinct efficiency depending on the architecture of the base pairing at the ends of DNA and favor sealing of G-T and T-C non-canonical mispairs (Figure 12B).
With regard to the biological importance of the study, we envision two scenarios in which BER might take place opposite a 5mC or alternate oxidative modification. In the first scenario, there may be situations where simultaneous oxidation of 5mC and the template G takes place, possibly leading to classic OGG1-mediated BER of 8-oxoG and the generation of a one nucleotide gap intermediate opposite the oxidized 5mC modifications. Obviously, such an event would involve the gap being filled by pol β and subsequent hand off to one of the BER ligases to complete the BER response. In the second scenario, one that is likely much more rare, any situation involving a gap generated opposite a 5mC base modification could end with a single-nucleotide gap opposite the cytosine oxidative product that might serve as a substrate for pol β and the BER ligases. We suggest that recruitment of DNA glycosylases (e.g., OGG1) to CpG sites could be facilitated through protein-protein interactions that are coordinated by TET enzymes and their BER interacting partners (e.g. XRCC1 that interacts with multiple DNA glycosylases and stimulates OGG1 activity), perhaps setting off one of the scenarios described above (61,68,69). In our previous study (23), we in fact showed an inefficient first step of the active demethylation process that involves TDG-initiated BER when guanine is oxidized in the CpG dinucleotide. However, it currently remains unknown whether the enzymatic activity of OGG1 is affected by structural abnormalities (e.g., simultaneously oxidized 5mC and G) in a CpG dinucleotide. Further biochemical and biological studies are required to elucidate how the multiprotein BER complex functions to balance genome maintenance repair reactions with precise regulation of chemical modifications within epigenetically important sites (70,71).
Supplementary Material
Highlights.
Base excision repair is crucial for DNA demethylation of modified cytosine products
DNA ligase I and DNA ligase III/XRCC1 complex ligate pol β dGTP insertion products
Substrate channeling of pol β mismatches opposite oxidative 5mC modifications
Distinct ligation efficiency of nicked repair intermediates with 5mC modifications
Acknowledgements
The expression plasmid vector for human DNA ligase I and the purified protein of DNA ligase III/XRCC1 complex are generous gifts from Dr. Alan E. Tomkinson (University of New Mexico). The author thanks Samuel H. Wilson (NIEHS/NIH) for providing the expression vector of human DNA polymerases β and λ as well as the purified protein of pol β D256A mutant. The author thanks David M Wilson III (Boost Scientific) for providing editing services and helpful discussions.
Funding
This work was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health grant R00ES026191.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
None
References
- 1.Cooper DN, Youssoufian H, The CpG dinucleotide and human genetic disease, Hum. Genet 78 (1988) 151–155. [DOI] [PubMed] [Google Scholar]
- 2.Pfeifer GP, Tang M, Denissenko MF, Mutation hotspots and DNA methylation, Curr. Top. Microbiol. Immunol 249 (2000) 1–19. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB, The fundamental role of epigenetic events in cancer, Nat. Rev. Genet 3 (2002) 415–428. [DOI] [PubMed] [Google Scholar]
- 4.Liyanage VR, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, Davie JR, DNA modifications: function and applications in normal and disease states, Biology 3 (2014) 670–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito S, Kuraoka I, Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword, DNA Repair 32 (2015) 52–57. [DOI] [PubMed] [Google Scholar]
- 6.Bird A, The essentials of DNA methylation, Cell 70 (1992) 5–8. [DOI] [PubMed] [Google Scholar]
- 7.Siegfried Z, Cedar H, DNA methylation: a molecular lock, Curr. Biol 7 (1997) R305–R307. [DOI] [PubMed] [Google Scholar]
- 8.Hermann A, Gowher H, Jeltsch A, Biochemistry and biology of mammalian DNA methyltransferases, Cell Mol. Life Sci 61 (2004) 2571–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branco MR, Ficz G, Reik W, Uncovering the role of 5-hydroxymethylcytosine in the epigenome, Nat. Rev. Genet 13 (2011) 7–13. [DOI] [PubMed] [Google Scholar]
- 10.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, Gao J, Liu P, Li L, Xu GL, Jin P, He C, Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming, Cell 153 (2013) 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalgo ML, Jones PA, Mutagenic and epigenetic effects of DNA methylation, Mutat. Res 386 (1997) 107–118. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen KD, Helin K, Role of TET enzymes in DNA methylation, development, and cancer, Genes. Dev 30 (2016) 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y, Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine, Science 333 (2011) 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A, Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1, Science 324 (2009) 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye C, Li L, 5-hydroxymethylcytosine: a new insight into epigenetics in cancer. Cancer Biol. Ther 15 (2014) 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan L, Shi YG, Tet family proteins and 5-hydroxymethylcytosine in development and disease, Development 139 (2012) 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinney SR, Pradhan S, Ten eleven translocation enzymes and 5-hydroxymethylcytosine in mammalian development and cancer, Adv. Exp. Med. Biol 754 (2013) 57–79. [DOI] [PubMed] [Google Scholar]
- 18.Vasanthakumar A, Godley LA, 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy, Cancer Genet. 208 (2015) 167–177. [DOI] [PubMed] [Google Scholar]
- 19.Wen L, Tang F, Genomic distribution and possible functions of DNA hydroxymethylation in the brain, Genomics 104 (2014) 341–346. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Bernstein A, Chen D, Jin P, 5-hydroxymethylcytosine: A new player in brain disorders? Exp. Neurol 268 (2015) 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Mahdawi S, Virmouni SA, Pook MA, The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases, Front. Neurosci 8 (2014) 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang D, Zhang Y, Hart RP, Chen J, Herrup K, Li J, Alteration in 5-hydroxymethylcytosine-mediated epigenetic regulation leads to Purkinje cell vulnerability in ATM deficiency, Brain 138 (2015) 3520–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sassa A, Caglayan M, Dyrkheeva NS, Beard WA, Wilson SH, Base excision repair of tandem modifications in a methylated CpG dinucleotide, J. Biol. Chem 289 (2014) 13996–14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A, Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair, Cell 146 (2011) 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL, Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair, Nuc. Acids Res 35 (2007) 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohli RM, Zhang Y, TET enzymes TDG and the dynamics of DNA demethylation, Nature 502 (2013) 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellacosa A, Drohat AC, Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites, DNA Repair 32 (2015) 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson SH, Kunkel TA, Passing the baton in base excision repair, Nat. Struct. Biol 7 (2000) 176–178. [DOI] [PubMed] [Google Scholar]
- 29.Prasad R, Shock DD, Beard WA, Wilson SH, Substrate channeling in mammalian base excision repair pathways: passing the baton, J. Biol. Chem 285 (2010) 40479–40488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parikh SS, Mol CD, Hosfield DJ, Tainer JA, Envisioning the molecular choreography of DNA base excision repair, Curr. Opin. Struct. Biol 9 (1999) 37–47. [DOI] [PubMed] [Google Scholar]
- 31.Prasad R, Beard WA, Batra VK, Liu Y, Shock DD, Wilson SH, A review of recent experiments on step-to-step “hand-off” of the DNA intermediates in mammalian base excision repair pathways, Mol. Biol 45 (2011) 586–600. [PMC free article] [PubMed] [Google Scholar]
- 32.Prasad R, Williams JG, Hou EW, Wilson SH, Pol β associated complex and base excision repair factors in mouse fibroblasts, Nuc. Acids Res 40 (2012) 11571–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J-K, Active DNA demethylation mediated by DNA Glycosylases, Annu. Rev. Genet 43 (2009) 143–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL, Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA, Science 333 (2011) 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maiti A, Drohat AC, Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites, J. Biol. Chem 286 (2011) 35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitzgerald ME, Drohat AC, Coordinating the initial steps of base excision repair. Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex, J. Biol. Chem 283 (2008) 32680–32690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beard WA, Wilson SH, Structure and mechanism of DNA polymerase β, Biochemistry 53 (2014) 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Çağlayan M, Interplay between DNA polymerases and DNA ligases: Influence on substrate channeling and the fidelity of DNA ligation, J. Mol. Biol 19 (2019) 30235–30239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beard WA, Shock DD, Vande Berg BJ, Wilson SH, Efficiency of correct nucleotide insertion governs DNA polymerase fidelity, J. Biol. Chem 277 (2002) 47393–47398. [DOI] [PubMed] [Google Scholar]
- 40.Freudenthal BD, Beard WA, Shock DD, Wilson SH, Observing a DNA polymerase choose right from wrong, Cell 154 (2013) 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sleeth KM, Robson RL, Dianov GL, Exchangeability of mammalian DNA ligases between base excision repair pathways, Biochemistry 43 (2004) 12924–12930. [DOI] [PubMed] [Google Scholar]
- 42.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T, DNA ligases: structure, reaction mechanism, and function, Chem. Rev 106 (2006) 687–699. [DOI] [PubMed] [Google Scholar]
- 43.Prasad R, Singhal RK, Srivastava DD, Molina JT, Tomkinson AE, Wilson SH, Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis, J. Biol. Chem 271 (1996) 16000–16007. [DOI] [PubMed] [Google Scholar]
- 44.Dimitriadi EK, Prasad R, Vaske MK, Chen L, Tomkinson AE, Lewis MS, Wilson SH, Thermodynamics of human DNA ligase I trimerization and association with DNA polymerase β, J. Biol. Chem 273 (1998) 20540–20550. [DOI] [PubMed] [Google Scholar]
- 45.Marintchev A, Gryk MR, Mullen GP, Site-directed mutagenesis analysis of the structural interaction of the single-strand-break protein, X-ray crosscomplementing group 1, with DNA polymerase β, Nuc. Acids Res 31 (2003) 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marintchev A, Robertson A, Dimitriadis EK, Prasad R, Wilson SH, Mullen GP, Domain specific interaction in the XRCC1-DNA polymerase β complex, Nuc. Acids Res 28 (2000) 2049–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Çağlayan M, Wilson SH, Oxidant and environmental toxicant-induced effects compromise DNA ligation during base excision DNA repair, DNA Repair 35 (2015) 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Çağlayan M, Wilson SH, Role of DNA polymerase β oxidized nucleotide insertion in DNA ligation failure, J. Radiat. Res 58 (2017) 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Çağlayan M, Horton JK, Dai DP, Stefanick DF, Wilson SH, Oxidized nucleotide insertion by pol β confounds ligation during base excision repair, Nat. Commun 8 (2017) 14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Çağlayan M, The ligation of pol β mismatch insertion products governs the formation of promutagenic base excision DNA repair intermediates, Nuc. Acids Res 48 (2020) 3708–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howard M, Foley G, Shock D, Batra VK, Wilson SH, Molecular basis for the faithful replication of 5-methylcytosine and its oxidized forms by DNA polymerase β, J. Biol. Chem 294 (2019) 7194–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cağlayan M, Batra VK, Sassa A, Prasad R, Wilson SH, Role of polymerase β in complementing aprataxin deficiency during abasic-site base excision repair, Nat. Struct. Mol. Biol 21 (2014) 497–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Çağlayan M, Horton JK, Prasad R, Wilson SH, Complementation of aprataxin deficiency by base excision repair enzymes, Nuc. Acids Res 43 (2015) 2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Çağlayan M, Prasad R, Krasich R, Longley MJ, Kadoda K, Tsuda M, Sasanuma H, Takeda S, Tano K, Copeland WC, Wilson SH, Complementation of aprataxin deficiency by base excision repair enzymes in mitochondrial extracts, Nuc. Acids Res 17 (2017) 10079–10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Çağlayan M, Wilson SH, Pol μ dGTP mismatch insertion opposite T coupled with ligation reveals a promutagenic DNA intermediate during double strand break repair, Nat. Commun 9 (2018) 4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krahn JM, Beard WA, Wilson SH, Structural insights into DNA polymerase β deterrents for misincorporation support an induced-fit mechanism for fidelity, Structure 12 (2004) 1823–1832. [DOI] [PubMed] [Google Scholar]
- 57.Batra VK, Perera L, Lin P, Shock DD, Beard WA, Pedersen LC, Pedersen LG, Wilson SH, Amino acid substitution in the active site of DNA polymerase β explains the energy barrier of the nucleotidyl transfer reaction, J. Am. Chem. Soc 135 (2013) 8078–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Braithwaite EK, Kedar PS, Stumpo DJ, Bertocci B, Freedman JH, Samson LD, Wilson SH, DNA polymerases β and λ mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts, Plos One 5 (2010) e12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown JA, Pack LR, Sanman LE, Suo Z, Efficiency and fidelity of human DNA polymerases λ and β during gap-filling DNA synthesis, DNA Repair 10 (2011) 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bebenek K, Pedersen LC, Kunkel TA, Structure-function studies of DNA polymerase λ, Biochemistry 53 (2014) 2781–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muller U, Bauer C, Siegl M, Rottach A, Leonhardt H, TET-mediated oxidation of methylcytosine causes TDG or NEIL glycosylase dependent gene reactivation, Nuc. Acids Res 42 (2014) 8592–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szulik MW, Pallan PS, Nocek B, Voehler M, Banerjee S, Brooks S, Joachimiak A, Egli M, Eichman BF, Stone MP, Differential stabilities and sequence-dependent base pair opening dynamics of Watson-Crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine, Biochemistry 54 (2015) 1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karino N, Ueno Y, Matsuda A, Synthesis and properties of oligonucleotides containing 5-formyl-2’-deoxycytidine: in vitro DNA polymerase reactions on DNA templates containing 5-formyl-2’-deoxycytidine, Nuc. Acids Res 29 (2001) 2456–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xing XW, Liu YL, Vargas M, Wang Y, Feng YQ, Zhou X, Yuan BF, Mutagenic and cytotoxic properties of oxidation products of 5-methylcytosine revealed by nextgeneration sequencing, PLoS One 8 (2013) e72993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamiya H, Tsuchiya H, Karino N, Ueno Y, Matsuda A, Harashima H, Mutagenicity of 5-formylcytosine, an oxidation product of 5-methylcytosine, in DNA in mammalian cells, J. Biochem 132 (2002) 551–555. [DOI] [PubMed] [Google Scholar]
- 66.Shibutani T, Ito S, Toda M, Kanao R, Collins LB, Shibata M, Urabe M, Koseki H, Masuda Y, Swenberg JA, Masutani C, Hanaoka F, Iwai S, Kuraoka I, Guanine-5-carboxylcytosine base pairs mimic mismatches during DNA replication, Sci. Rep 4 (2014) 5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitaker AM, Freudenthal BD, APE1: A skilled nucleic acid surgeon, DNA Repair 71 (2018) 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campalans A, Marsin S, Nakabeppu Y, O’connor TR, Boiteux S, Radicella JP, XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair, DNA Repair 4 (2005) 826–835. [DOI] [PubMed] [Google Scholar]
- 69.Marsin S, Vidal AE, Sossou M, Murcia JM. Page F. Le, Boiteux S, Murcia G. de, Radicella JP, Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1, J. Biol. Chem 278 (2003) 44068–44074. [DOI] [PubMed] [Google Scholar]
- 70.Putiri EL, Robertson KD, Epigenetic mechanisms and genome stability, Clin. Epigen 2 (2011) 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dabin J, Fortuny A, Polo SE, Epigenome maintenance in response to DNA damage, Mol. Cell 62 (2016) 712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


