Abstract
We screened cell line and plasma-derived exosomes for molecules that localize to mitochondria or that reflect mitochondrial integrity. SH-SY5Y cell-derived exosomes contained humanin, citrate synthase, and fibroblast growth factor 21 protein, and plasma-derived exosomes contained humanin, voltage-dependent anion-selective channel 1, and transcription factor A protein. Nuclear mitochondrial (NUMT) DNA complicated analyses of mitochondrial DNA (mtDNA), which otherwise suggested exosomes contain at most very low amounts of extended mtDNA sequences but likely contain degraded pieces of mtDNA. Cell and plasma-derived exosomes contained several mtDNA-derived mRNA sequences, including those for ND2, CO2, and humanin. These results can guide exosome-focused, mitochondria-pertinent biomarker development.
Keywords: biomarker, exosome, mitochondria, mitochondrial DNA, NUMT
1. Introduction
Exosomes are 30–150 nm vesicles initially formed through the invagination of early endosome membranes [1–3]. An accumulation of exosome vesicles within a parent endosome defines its transition to a unique structure, the multivesicular body (MVB), which continues to generate and collect exosomes that at this stage are also referred to as intra-luminal vesicles (ILVs). While proteins contained within endosome membrane carry forward into exosome membrane, during these endocytosis-like events invaginating endosome/MVB membrane also comes to enclose adjacent cytosol that can contain cytosolic protein, lipid, RNA, and surprisingly even DNA. MVBs can go on to fuse with the plasma membrane, with the subsequent release of their exosome cargo to the extracellular space [4]. This release may serve various functions, including elimination of intracellular waste, transfer of materials between cells, inter-cellular signaling, or even inter-tissue signaling [4–6].
Extracellular exosomes transfer from the tissues they arise from into fluid tissues, including blood. It is possible to concentrate exosomes from blood and quantify their protein, lipid, RNA, and DNA contents. Selective harvesting of tissue-specific exosomes from blood also appears feasible [7]. For example, studies report the isolation of potentially brain-generated exosomes from blood [8–11]. For these reasons there is considerable interest in using exosomes as biomarkers of human health and physiology, including brain diseases, disease progression, and drug target engagement [12].
Although the small size of exosomes (less than 150 nm diameter) should preclude the incorporation of intact mitochondria (over 500 nm), exosomes reportedly contain molecules that commonly associate with mitochondria, including cardiolipin, mitochondrial DNA (mtDNA), and mitochondrial proteins [8, 13–16]. Not all studies are positive, though, and some investigators suggest exosome proteomes may lack mitochondrial proteins [17]. Exosome origin may also influence whether and what mitochondrial contents are present.
Mitochondrial dysfunction is increasingly implicated in human diseases, including neurodegenerative diseases, and increasingly mitochondria are targeted by therapeutic interventions [18, 19]. Because exosomes are practical to obtain, reportedly contain mitochondrial components, and may represent accessible derivatives of otherwise difficult to access tissues including the brain, they could potentially serve as clinically practical biomarkers of mitochondrial status and perhaps function. For this reason, we sought to better characterize mitochondria-derived or mitochondria-relevant exosome contents that could inform the development of clinically useful mitochondrial biomarkers.
2. Materials and Methods
2.1. Cell culture and plasma
Exosome-free fetal bovine serum (FBS) was generated from standard FBS (Cat. No. PS-FB1, Peak Serum) by ultracentrifugation at 120,000 g overnight at 4°C. We did not dilute the FBS in phosphate buffered saline (PBS) prior to the ultracentrifugation, a step followed by many investigators, as a prior study reported ultracentrifugation of diluted versus undiluted FBS yielded comparable levels of vesicle depletion with the extended centrifugation protocol we utilized [20]. The supernatant was filtered through a 0.22 micron filter (Ultrafree-Centrifugal Filter Unit, Millipore), aliquoted into 50 ml tubes, and stored at −20°C prior to use.
Human SH-SY5Y and mtDNA-depleted SH-SY5Y ρ0 neuroblastoma cells were grown in high glucose Dulbecco’s modified Eagle’s medium (DMEM) (Cat. No. VWRL0100-0500, VWR International) supplemented with 10% exosome-free FBS, 1% of a penicillin-streptomycin stock solution (Cat. No. 30-001-CI, Fisher Scientific), pyruvate (100 mg/ml), and uridine (50 mg/ml). Human NT2 and mtDNA-depleted NT2 ρ0 teratocarcinoma cells were cultured in DMEM supplemented with 10% exosome-free FBS, 1% penicillin-streptomycin, pyruvate (200 mg/ml), and uridine (150 mg/ml).
Plasma used in these analyses derived from human subjects enrolled in clinical studies at the University of Kansas Alzheimer’s Disease Center (KUADC). The KUADC maintains phlebotomy and biospecimen protocols approved by the University of Kansas Medical Center’s institutional review board and all individuals participating in KUADC studies provide informed consent prior to phlebotomy. As part of these KUADC studies blood is collected in tubes containing acid-citrate-dextrose anticoagulant. Within two hours of phlebotomy a routine centrifugation process is used to separate the plasma, which is stored frozen at −800C pending utilization.
2.2. Exosome harvesting
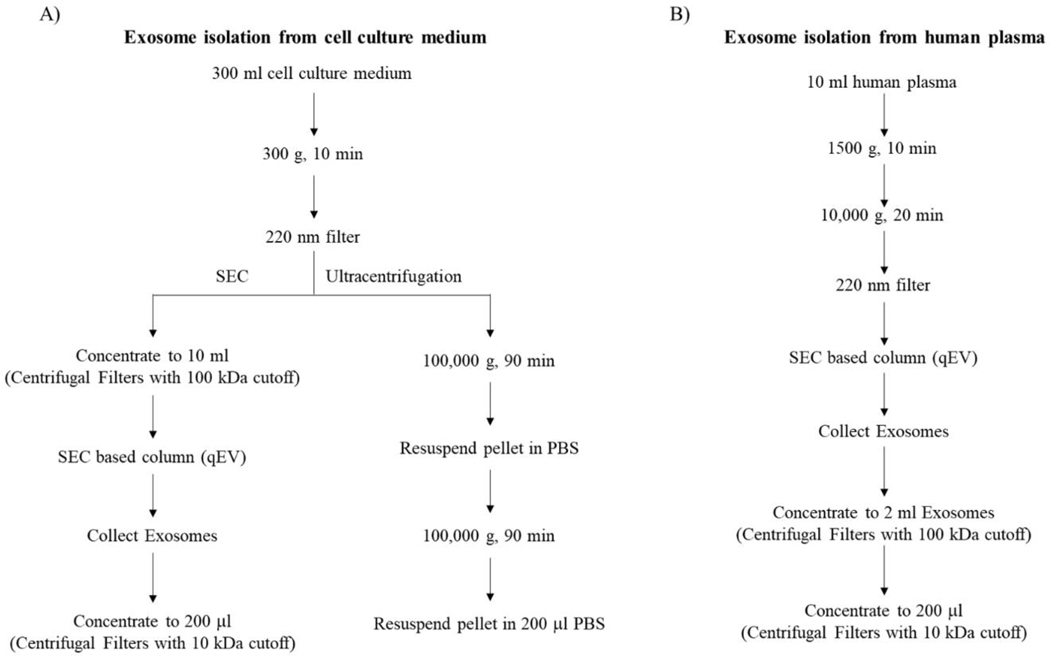
A previously described ultracentrifugation method was used to harvest exosomes from SH-SY5Y cells [21]. Briefly, cells were maintained in medium supplemented with exosome-free FBS for 24 hours. 300 ml of conditioned medium was collected and centrifuged at 300 g for 10 minutes to remove dead cells and cell debris. The supernatant was then passed through a 0.22 micron filter and centrifuged at 100,000 g for 90 minutes at 4°C to pellet the exosomes. The exosome pellet was resuspended in PBS and centrifuged a second time at 100,000 g for 90 minutes at 4°C. The exosome pellet was resuspended in 200 μl of PBS and frozen at −80°C until use. Particle concentrations and size distributions were analyzed with a Nanosight system (Figure 1A).
Figure 1. Exosome isolation workflows.
(A) Exosome isolation from cell culture medium using SEC qEV size exclusion columns or ultracentrifugation. (B) Exosome isolation from plasma using SEC qEV size exclusion columns.
For size exclusion chromatography (SEC)-based harvesting of SH-SY5Y, SH-SY5Y ρ0, NT2, and NT2 ρ0 cell exosomes we followed the instructions that accompanied a commercial SEC kit (Cat. No. qEV10/70nm, Izon). Specifically, we collected 300 ml of 24 hour-conditioned medium from ~75% confluent culture flasks, centrifuged the medium to pellet whole cells (300 g for 10 minutes at 4°C), passed the entire supernatant through a 0.22 micron filter to further remove structures larger than exosomes, concentrated the filtered medium with a 100,000 nominal molecular weight limit (NMWL) column (Centricon Plus-70 Centrifugal Filter with a 100 kDa cutoff, Millipore) at 3,500 g and 4°C, and resuspended the exosomes captured in the filter into a 10 ml volume. The suspension was added to a qEV10 column (Izon) and eluted with PBS into successive collection tubes using an Izon Automatic Fraction Collector (AFC), with each tube receiving 5 ml of eluent. The eluents from tubes 5–8 were combined, and the resulting 20 ml volume was reduced to 200 μl using a 10,000 NMWL column (Amicon Ultra-15 Centrifugal Filter with a 10 kDa cutoff, Millipore) (Figure 1A).
To harvest plasma exosomes, we centrifuged 10 ml of plasma to pellet whole cells/platelets (1,500 g for 10 minutes, then 10,000 g for 20 minutes, both at 4°C) and passed the supernatant through a 0.22 micron filter before adding it to a qEV10 column. As was the case with the cell culture collection procedure, we collected 5 ml aliquots and pooled fractions 5–8. The sample was then added to a 100,000 NMWL column, and the exosomes were retrieved from the filter and suspended within a 1–2 ml volume. That suspension was added to a 10,000 NMWL column, and the exosomes were retrieved from the filter and suspended within a final 200 μl volume (Figure 1B).
2.3. Nanoparticle tracking analysis and transmission electron microscopy
Particle concentrations and size distributions were analyzed by Nanoparticle tracking analysis (NTA) using a NanoSight LM10 instrument (Malvern Panalytical). Exosomes from human plasma and cell culture medium were suspended in 200 ul PBS. The mixture was further diluted as needed with PBS to obtain a concentration of 108 particles per ml. The laser sample chamber was loaded with approximately 500 ul of exosome solution using a 1 ml disposable syringe and the camera level was set to 14. Three 1-minute videos were recorded for each sample. Videos were analyzed with NTA 2.3 software (NanoSight).
For transmission electron microscopy (TEM) we covered 30 μl of purified exosomes with a carbon coated grid. Grid washing was accomplished by touching the grid to six drops of distilled water. Excess water was removed, and a drop of 1% uranyl acetate aqueous solution was added for 5 seconds. The grid was air dried at room temperature, and the sample was imaged using a JEM-1400 Transmission Electron Microscope (JEOL).
2.4. Immunochemistry
Exosome samples were boiled in a loading buffer containing sodium dodecyl sulfate (SDS) and β-mercaptoethanol at 95°C for 10 minutes. The lysates were resolved through SDS-polyacrylamide gel electrophoresis (PAGE) using 4–15% Criterion TGX Tris-glycine polyacrylamide gels (Bio-Rad). Gel proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Cat. No. 10061–492, GE Healthcare). Membranes were blocked in 5% bovine serum albumin (BSA) with PBS-Tween 20 (PBST) at room temperature. The membranes were then probed with primary antibodies to annexin V (1:1000, Cell Signaling, Cat. No. 8555), CD9 (1:1000, Cell Signaling, Cat. No. 13174), CD63 (1:1000, Invitrogen, Cat. No. 10628D), CD81 (1:1000, Invitrogen, Cat. No. 10630D), β-integrin (1:1000, Cell Signaling, Cat. No. 9699), Tsg101 (1:1000, Abcam, Cat. No. ab83), calnexin (1:1000, Cell Signaling, Cat. No. 2679), GM130 (1:1000, Cell Signaling, Cat. No. 12480), citrate synthase (1:1000, Cell Signaling, Cat. No. 14309), SOD2 (1:1000, Cell Signaling, Cat. No. 13141), CO2 (1:2000, Abcam, Cat. No. ab79393), cytochrome C (1:400, Abcam, Cat. No. ab13575), VDAC1 (1:1000, Abcam, Cat. No. ab15895), TFAM (1:2000, Abcam Cat. No. ab131607), and FGF 21 (1:2000, Invitrogen, Cat. No. MA5–25558) in 5% BSA with PBST overnight at 4°C. Membranes were washed 3 times with PBST and placed in secondary antibody at 1:4000 dilutions in 5% non-fat milk with PBST for 1 hour at room temperature. Membranes were washed 3 times with PBST and incubated with Super Signal West Femto Chemiluminesence Reagent (Life Technologies, Cat. No. 34095). Images were captured using a Chemidoc imaging station (Bio-Rad).
A human putative humanin peptide (MT-RNR2) enzyme-linked immunosorbent assay (ELISA) kit (Biomatik) was used to measure humanin levels in SH-SY5Y cell and plasma-derived exosomes. As part of this assay, exosome preparations were subjected to two freeze-thaw cycles to disrupt membranes.
Where indicated, prior to generating protein lysates exosome or post-exosome fractions were treated with trypsin to remove free or external protein contamination. To accomplish this, we pre-warmed trypsin aliquots to 37°C and added trypsin to the exosome or post-exosome fractions to a final concentration of 2 mg/ml, for 10 minutes, at room temperature.
2.5. Vmax Assays
We measured cytochrome oxidase, NADH:ubiquinone oxidoreductase, and citrate synthase Vmax activities in exosomes isolated from 300 ml of SH-SY5Y cell medium or 10 ml of human plasma. The Vmax assays used were previously described [22].
2.6. DNA Analyses
Genomic DNA from cultured cells, cell culture exosomes, or plasma exosomes was extracted using a standard phenol-chloroform-isoamyl alcohol protocol [23]. To verify the efficacy of a DNA digestion protocol, 1 g of cell culture DNA was directly treated with 1 μl of DNase I (DNA-free™ DNA Removal Kit, Invitrogen, Cat. No. AM1906) for 30 minutes at 370C, and the DNase I was removed according to the kit instructions. When specified, to address potential contamination of exosome DNA by free DNA, prior to lysing the exosomes we incubated 200 ul exosomes with 1 μl of DNase I for 30 minutes at 37oC. The DNase I was then removed and the DNA remaining within the exosomes was extracted.
Standard polymerase chain reaction (PCR) was performed using AmpliTaq™ DNA Polymerase (Applied Biosystems, Cat. No. N8080152). Amplifications exceeding 5000 base pairs used the Expand Long Range dNTP Pack (Roche, Cat. No. 04829034001). Amplifications were performed using primers that anneal ND2, CO2, humanin, β-actin, the ends of the mtDNA pseudogene described by Herrnstadt et al (NUMT-inclusive) [24], and just beyond the ends of that pseudogene (NUMT-exclusive). Table 1 provides these primer sequences, product sizes, and cycling parameters. PCR products were electrophoresed in 2% (for short amplifications) or 1% (for long amplifications) agarose gels and visualized using a Biorad Imager.
Table 1.
Primers used for PCR, RT-PCR amplification.
| Primer Name | Primer Sequence | Product Size | Tm or cycling parameters | |
|---|---|---|---|---|
| mt 262–388 (control region) | Forward Reverse |
5’-CACTTTCCACACAGACATCA-3’ 5’-TGGTTAGGCTGGTGTTAGGG-3’ |
127 | One cycle of 5 min at 95°C followed by 40 cycles of 10 sec at 95°C, 30 sec at 60°C |
| β-globin | Forward Reverse |
5’-GTGCACCTGACTCCTGAGGAGA-3’ 5’-CCTTGATACCAACCTGCCCAG-3’ |
101 | One cycle of 5 min at 95°C followed by 40 cycles of 10 sec at 95°C, 30 sec at 60°C |
| mt-tRNALeu | Forward Reverse |
S’-CACCCAAGAACAGGGTTTGT-3’ 5’-TGGCCATGGGTATGTTGTTA-3’ |
107 | One cycle of 5 min at 95°C followed by 40 cycles of 10 sec at 95°C, 30 sec at 60°C |
| ND1 | Forward Reverse |
5’-CCACCTCTAGCCTAGCCGTTT-3’ 5’-TGTTTGGGCTACTGCTGGC-3’ |
97 | One cycle of 5 min at 95°C followed by 40 cycles of 10 sec at 95°C, 30 sec at 60°C |
| ND2 (DNA primer) | Forward Reverse |
5’-TCCCCACCATCATAGCCA-3’ 5’-GGGTTTTGCAGTCCTTAG-3’ |
302 | 50.8 |
| ND2 (cDNA primer) | Forward Reverse |
5’-TAAAACTAGGAATAGCCCCC-3’ 5’-TTGAGTAGTAGGAATGCGGT-3’ |
349 | 50 |
| Numt-exclusive | Forward Reverse |
5’-TGATTTATCTCTCCACACTAGCAGAGACCAAC-3’ 5’-GCTACAAAAAATGTTGAGCCGTAGATG--3’ |
5947 | 57.3 |
| Numt-inclusive | Forward Reverse |
5’-GAGTCCGAACJAGJCTCAGGCT-3’ 5’-CTCGAAGTAClCTGAGGCTTGTAGGAGG-3’ |
5840 | 57.3 |
| CO2 | Forward Reverse |
5’GCTATTCCCCTATCATAGAAG-3’ 5’-GGGAATTAATTCTAGGACGATG-3’ |
612 | 53.6 |
| Humanin | Forward Reverse |
5’-CCGCGGTACCCTAACCGTGC-3’ 5’-ACGGGGGAAGGCGCTTTGTG-3’ |
600 | 60 |
| β-actin | Forward Reverse |
5’-TTAATAGTCATTCCAAATATGA-3’ 5’-GGGACAAAAAAGGGGGAAGG-3’ |
246 | 50 |
For experiments requiring quantitative PCR (qPCR), 10 ng of genomic DNA was amplified with primers directed to the mtDNA control region, mt-tRNALeu, ND1, or β-globin using a PowerUp SYBR Green Master Mix (Life Technologies, Cat. No. A25741). Table 1 provides the primer sequences and cycling parameters. Reactions were performed in triplicate. For the control region and β-globin products serial amplicon dilution was used to generate standard curves, which were used to assign sample copy numbers. The cycle-threshold (CT)-based ΔΔCT method was used to determine the relative amount of tRNALeu and ND1 mtDNA.
To distinguish between amplification of authentic mtDNA and mtDNA nuclear pseudogenes, also referred to as nuclear mtDNA (NUMT) sequences, we used genomic DNA from NT2 and NT2 ρ0 cells or their exosomes to take advantage of their ND2 G5460A polymorphism. For some experiments we used standard PCR with primers to the ND2 gene to directly amplify genomic DNA. For other experiments, the genomic DNA was first subjected to amplification by primers that anneal the ends of a previously described 5840 nucleotide NUMT that includes ND2 sequence (NUMT-inclusive primers), or to mtDNA sequences that reside just beyond the ends of that NUMT (NUMT-exclusive primers) [24, 25]. The PCR products from these long PCR amplifications were purified with a GeneJET PCR Purification kit (Cat. No. K0702, Thermo Fisher) and used for nested PCR with the ND2 primers. ND2 amplicons were purified with the GeneJET PCR Purification kit and subjected to digestion by the Hph I restriction enzyme (New England Biolabs, Cat. No. R0158S).
2.7. RNA analyses
We used an RNeasy Plus Mini Kit (Qiagen, Cat. No. 74134) and TRIzol LS Reagent (Life Technologies, Cat. No. 15596018) to extract total RNA. Where indicated, to remove DNA contamination following RNA extraction but prior to cDNA synthesis we treated RNA samples with DNase I (DNA-free DNA Removal Kit, Invitrogen, Cat. No. AM1906). DNase I was subsequently removed per kit instructions. Samples were analyzed for quality using an Agilent Bioanalyzer 2100 RNA Pico Chip.
To perform reverse transcription, we used an iScript RT qPCR master mix (Bio-Rad, Cat. No. 1708840). Reverse transcription PCR was performed using a MyCycler thermal cycler (BioRad). cDNA amplifications utilized the ND2, CO2, humanin, and β-actin primer sets specified in Table 1. RT-PCR products were electrophoresed through 1% agarose gels containing ethidium bromide and visualized under ultraviolet (UV) light.
2.8. Statistics
Group comparisons were by unpaired Student’s t-test. Analyses were conducted with α=0.05 to protect against Type I error. When indicated, variation is identified as standard deviation (SD) or standard error of the mean (SEM).
3. Results
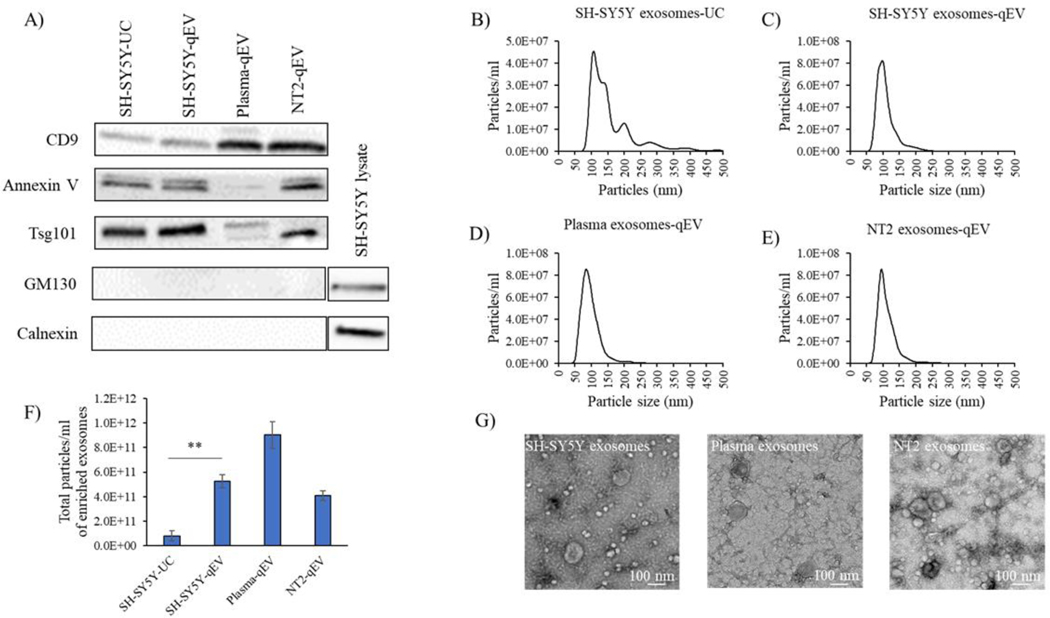
We considered two approaches for generating exosomes, ultracentrifugation and SEC. Exosome purification was performed from 300 ml of cell culture medium or 10 ml of human plasma. Ultracentrifugation and SEC exosome purification protocols are summarized in Figure 1. Figure 2 shows quality assessments of exosomes derived from human SH-SY5Y cells, NT2 cells, and plasma. Immunochemical screening found both approaches generated exosome fractions containing robust amounts of exosome proteins (CD9, annexin V, and Tsg101) without apparent Golgi (GM130) or endoplasmic reticulum (calnexin) protein contamination (Figure 2A). NTA indicated SEC exosome fractions contained fewer particles that exceeded the expected 30–150 nm distribution than ultracentrifugation (Figure 2B–E). Consistent with previous reports [26, 27], qEV columns generated better particle yields (Figure 2F) that on TEM showed appropriate size and morphology (Figure 2G). Based partly on comparisons between ultracentrifugation and SEC-generated exosomes from SH-SY5Y cell conditioned medium, and partly on the greater ease of the SEC approach, for the rest of our studies we used the SEC-based qEV column method to prepare exosomes.
Figure 2. Characterization and quantification of plasma and cell-derived exosomes.
(A) Western blot analysis of exosome markers CD-9, Annexin V, Tsg101; the Golgi marker GM130; and the endoplasmic reticulum marker calnexin in exosomes isolated from SH-SY5Y cell culture medium, NT2 cell culture medium, and human plasma. For Western blots without clear exosome staining we used whole cell lysates to demonstrate antibody integrity. Data shown are representative of three experiments; UC=ultracentrifugation. (B-E) Exosome concentration and size distribution as determined by Nanosight analysis. The y-axis shows particle number and the x-axis shows particle diameter (nm). Exosomes from SH-SY5Y cells, NT2 cells, or plasma were derived using the technique indicated in each panel. (F) Isolation technique-specific particle yields of exosome fractions isolated from human plasma, SH-SY5Y cell culture medium, and NT2 cell culture medium via qEV column or ultracentrifugation. Data shown as mean ± SE, n=3 experiments; ** p < 0.01 SH-SY5Y qEV versus SH-SY5Y-UC. (G) TEM verification of plasma, SH-SY5Y cell, and NT2 cell-derived exosome fractions.
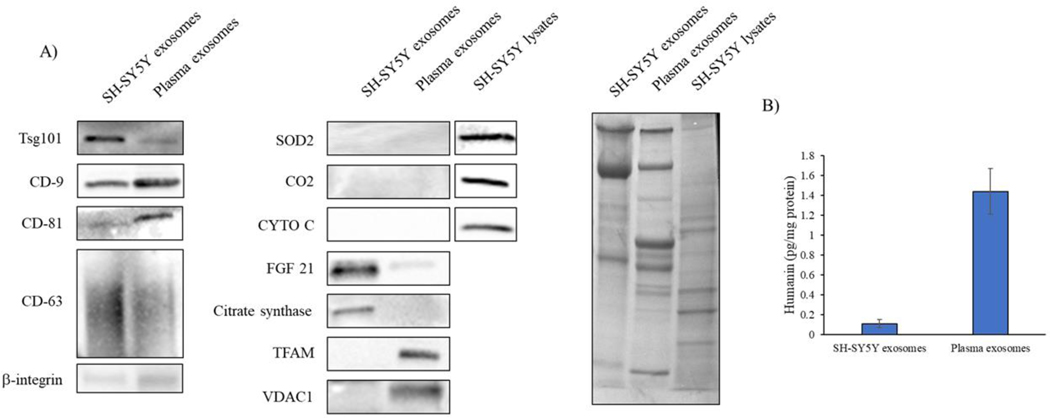
We assessed the quality of SH-SY5Y cell and plasma exosomes from SEC based qEV10 column exosome fractions 5–8. The validation panel showed that relative to SH-SY5Y cell-derived exosomes, plasma-derived exosomes contain less Tsg101 and CD63, but more CD9 and CD81. We did not resolve percentages of positively vs negatively stained exosomes. β-integrin is used as a marker of both exosomes and microvesicles [28, 29]; the presence of β-integrin in our exosome preparations emphasizes we cannot absolutely rule out a small degree of microvesicle contamination. With this in mind we screened for the presence of selected proteins that localize to mitochondria, including superoxide dismutase 2 (SOD2), cytochrome C oxidase subunit 2 (CO2), cytochrome C (CYTO C), citrate synthase, transcription factor A (TFAM), and voltage-dependent anion-selective channel 1 (VDAC1); for the presence of FGF21, whose levels reportedly reflect mitochondrial functional integrity; and humanin, whose levels reportedly reflect mitochondrial functional integrity and which may reside within both mitochondria and the cytoplasm. Figure 3 shows FGF21, citrate synthase, and humanin were present with SH-SY5Y cell exosomes, and TFAM, VDAC1, and humanin were present with plasma exosomes. The humanin signal appeared far more robust in the plasma exosomes than it did in the SH-SY5Y exosomes.
Figure 3. Characterization of mitochondrial and mitochondrial-related proteins in plasma and SH-SY5Y-derived exosomes.
Exosomes were isolated from SH-SY5Y-conditioned cell culture medium and human plasma using qEV SEC. (A) We used immunochemistry to Tsg101, CD9, CD81, CD63, and β-integrin to validate exosome content. Levels of superoxide dismutase 2 (SOD2), cytochrome C oxidase subunit 2 (CO2), cytochrome C (CYTO C), fibroblast growth factor21 (FGF21), citrate synthase, transcription factor A (TFAM), and voltage-dependent anion-selective channel 1 (VDAC1) were measured by Western blot. For Western blots without clear exosome staining we used whole cell lysates to demonstrate antibody integrity. Equal protein amounts were loaded into each lane of an SDS-PAGE gel, and amido black staining was used to further illustrate protein loading. Data shown are representative of at least three experiments. (B) Humanin levels in the SH-SY5Y cell-derived and plasma exosomes as measured through ELISA. Data are mean ± SEM; n = 3 experiments.
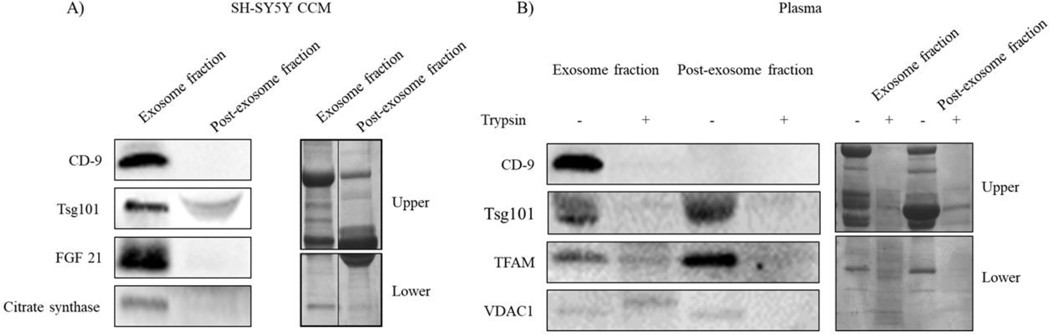
To assess whether the mitochondrial-associated proteins detected in the exosome fractions were truly present within the exosomes and not contaminants, we compared exosome and post-exosome fraction protein content. Western blots showed the CD9 exosome marker was present in the exosome fraction but not the post-exosome fraction (Figure 4A). NTA revealed the post-exosome fraction lacked particles (data not shown). FGF21 and citrate synthase were present in the SH-SY5Y cell culture medium exosome but not post-exosome fractions (Figure 4A), which suggests these proteins were present within the exosomes, and not a consequence of free protein contamination. Humanin protein in the SH-SY5Y cell and plasma post-exosome fractions was below the limits of ELISA detection (data not shown).
Figure 4. Status of exosome-derived proteins in the post-exosome SEC fractions and following trypsin digestion.
(A) Western blots for CD-9, TSG101, FGF21, and citrate synthase proteins in exosome and post-exosome fractions generated from SH-SY5Y cell culture medium (CCM). Amido black staining illustrates the degree of overall protein loading. Note the amido black blots were cut horizontally during the course of data generation, thereby generating upper and lower sections; we present the amido black data in a way that clearly acknowledges this. (B) Western blots for CD-9, TSG101, TFAM, and VDAC1 proteins in the exosome and post-exosome fractions generated from plasma, with and without exposure of those fractions to trypsin. Amido black staining illustrates the degree of overall protein loading. Note the amido black blots were cut horizontally during the course of data generation, thereby generating upper and lower sections; we present the amido black data in a way that clearly acknowledges this. Data shown are representative of two or three experiments.
In contrast, TFAM and VDAC1 were found in both the plasma exosome and post-exosome fractions (Figure 4B). We therefore treated these samples with trypsin to eliminate free protein contamination. The trypsin treatment completely or almost completely removed TFAM and VDAC1 from the post-exosome fraction and CD-9, a membrane protein, from the exosome fraction but did not appreciably reduce VDAC1 and only partly reduced TFAM from the exosome fraction (Figure 4B). This suggests at least some of the VDAC1 and TFAM proteins present in the plasma exosome fraction were contained within the exosomes themselves and not simply free protein contaminants.
To further pursue the finding that exosomes contain mitochondria-pertinent peptides, we assayed SH-SY5Y and plasma exosome cytochrome oxidase, NADH:ubiquinone oxidoreductase, and citrate synthase Vmax activities. None of these enzymes showed a measurable activity (data not shown).
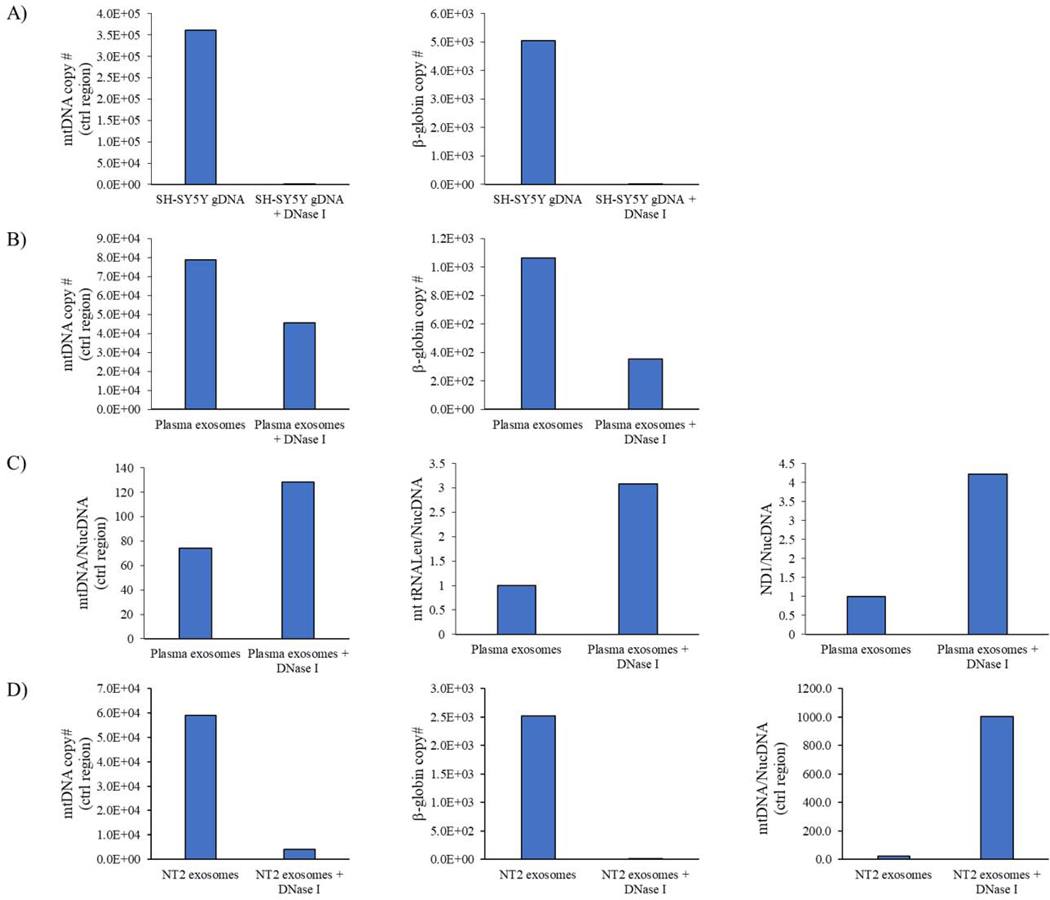
Next, we tested whether exosomes from cell culture medium or plasma contained mtDNA. To minimize the presence of sample contamination by free DNA, before extracting exosome DNA we treated our exosome samples with DNase I, at an amount that was found to eliminate DNA from an SH-SY5Y cell genomic DNA sample (Figure 5A). With the exosomes, even with the DNase I treatment primers directed to the mtDNA control region, mt-tRNALeu, and ND1 generated a detectable product (Figure 5B–D). The DNase I treatment did, however, affect mtDNA target and nuclear DNA target cycle thresholds and ratios. DNase I-treated samples showed higher ratios of mtDNA to nuclear DNA-directed amplification (Figure 5B–D), which could indicate free DNA contamination predominantly consists of nuclear DNA.
Figure 5. Effect of DNase I on exosome fraction mtDNA and nuclear DNA content.
(A) Quantitative PCR shows the DNase I treatment we used was stringent enough to eliminate both mtDNA and nuclear DNA from SH-SY5Y cell-generated genomic DNA samples. (B) Quantitative PCR shows DNase I treatment of the plasma exosome fraction does not fully eliminate mtDNA or nuclear DNA from the fraction. (C) DNase I treatment consistently increases the plasma exosome fraction mtDNA:nuclear DNA copy number ratio. For the control region amplification, the ratio derives from absolute copy numbers. For the mt-tRNALeu and ND1 amplifications, the ratios derive from relative copy numbers. (D) In an NT2 cell-derived exosome fraction, DNase I treatment does not eliminate all mtDNA amplification, eliminates most nuclear DNA amplification, and increases the mtDNA:nuclear DNA ratio. Ctrl=control.
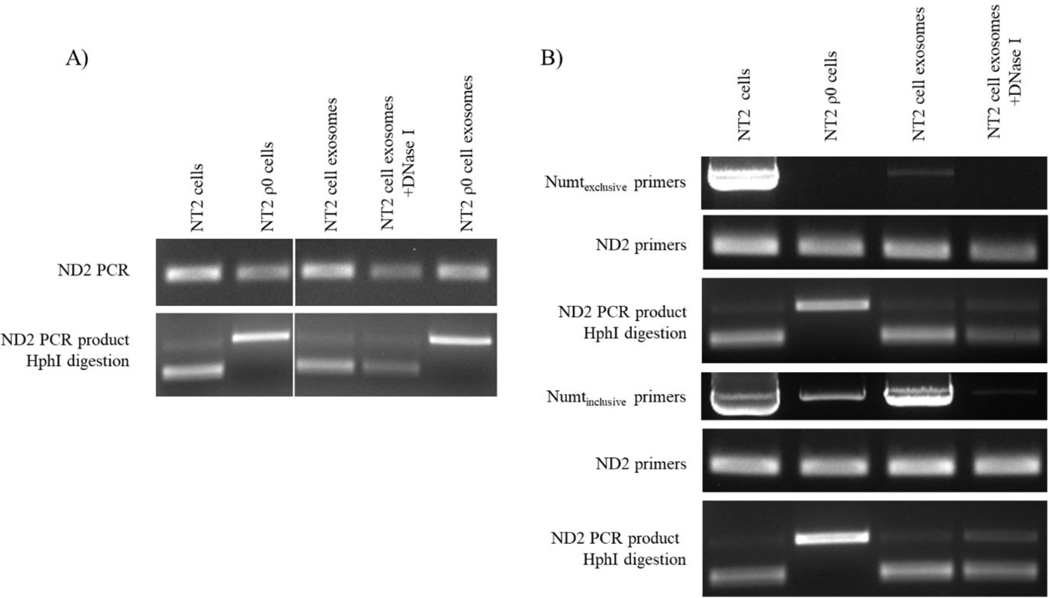
While the data in Figure 5B–D indicate mtDNA-directed primers amplify DNA carried specifically within exosomes, primers directed to small stretches of the mtDNA can also amplify nucleus-localized mtDNA pseudogenes, otherwise known as NUMT sequences. For this reason, we specifically tested whether amplification by an mtDNA-directed primer set arose from templating of authentic mtDNA or NUMT sequence. To accomplish this, we used DNA extracted from NT2 cells, NT2 ρ0 cells, and exosomes from these cell lines. NT2 cell mtDNA has an ND2 G to A transition at nucleotide 5460, which creates an Hph I restriction site; these cells also contain ND2 sequence within a 5840 base pair NUMT whose 5460-equivalent nucleotide is G [24]. We found primers directed to ND2 amplified DNA from both NT2 and NT2 ρ0 cell-derived exosomes (Figure 6A). The ND2 product generated from NT2 ρ0 exosome DNA showed no Hph I digestion, which indicates it arose via amplification of NUMT DNA. The ND2 product generated from NT2 cell exosome-derived, DNase I-treated exosome DNA was digested by Hph I, which indicates these exosomes contained some authentic mtDNA.
Figure 6. Interrogation and characterization of exosome fraction DNA using mtDNA and NUMT-directed primers.
(A) DNA isolated from NT2 cells, NT2 ρ0 cells, NT2 cell-derived exosomes, and NT2 ρ0 cell-derived exosomes, with and without DNase I-treatment, was amplified with primers to the mtDNA ND2 gene. The products of this PCR reaction were then digested with Hph I. Restriction of the ND2 band indicates 5460A, which is found in authentic mtDNA. Absence of ND2 band restriction indicates 5460G, which is not present in NT2 mtDNA and is found in at least one ND2 NUMT. (B) DNA isolated from NT2 cells, NT2 ρ0 cells, and NT2 cell-derived exosomes treated or not treated with DNase I was first amplified with primers targeted to just beyond the borders of the Herrnstadt et al. NUMT (Numtexclusive primers), or just within the boundaries of the NUMT (Numtinclusive primers). Following these PCR amplifications aliquots were taken and subjected to PCR amplification with primers to ND2. The ND2 amplicons were then digested with Hph I. Data shown are representative of three experiments.
To further assess if the ND2 primers templated authentic mtDNA carried within NT2 cell-derived exosomes or from the 5840 base pair NUMT sequence carried within the exosomes, we amplified NT2 cell exosome DNA with primers that annealed the ends of the NUMT (Numtinclusive), or else fell just beyond the borders of the NUMT (Numtexclusive). The primers that annealed the NUMT ends generated a robust PCR product, while the primers that fell beyond the NUMT borders produced a very weak band (Figure 6B). Using DNA isolated from DNase I-treated exosomes, both Numtinclusive and Numtexclusive primers failed to produce a visible band. These results indicate if exosomes do contain the target ~6000 base pair DNA sequences, the amount resides below the limit of clear PCR detection. Both exosome-derived long PCR amplifications were purified and used to template a nested reaction using the ND2 primers. Each reaction produced a visible ND2 band, and in both cases Hph I digested the band. This further suggests authentic mtDNA present within exosomes was amplified to produce the result shown in Figure 6A. Collectively these findings argue exosomes contain short, possibly degraded pieces of authentic mtDNA whose copy numbers exceed that of extended authentic mtDNA sequences.
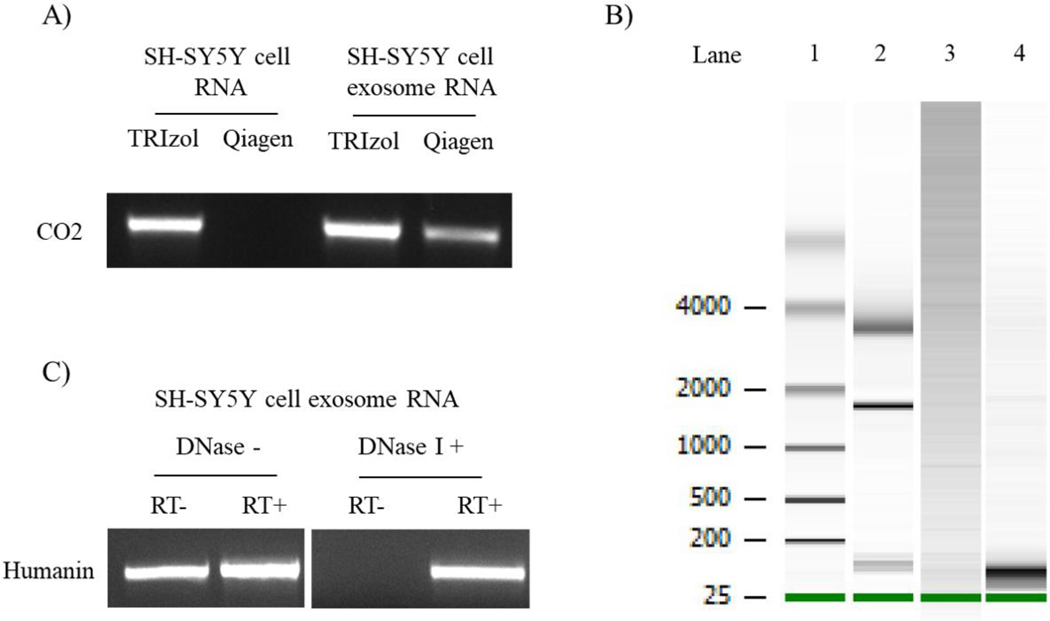
The presence of NUMT or fragmented mtDNA within exosomes could confound detection or quantification of mtDNA-derived exosome RNA. We therefore considered how different RNA extraction techniques and DNase I treatment affected exosome RNA sample quality. Figure 7A demonstrates RNA extracted from SH-SY5Y cells using an RNeasy Plus Mini Kit (Qiagen), when subjected to PCR in the absence of reverse transcription, did not generate a CO2 amplicon. This suggests sample DNA contamination in this case is below the level of PCR detection. However, a clear CO2 band was apparent when exosome RNA was used to template the CO2 PCR (Figure 7A). This indicates the RNeasy Plus Mini Kit does not fully eliminate mtDNA contamination of exosome RNA. An Agilent Bioanalyzer analysis showed exosome RNA isolated with the Qiagen kit contained bands over 4000 base pairs long, which further suggests DNA contamination (Figure 7B). TRIzol-based RNA extraction followed by DNase I treatment yielded more RNA and minimized the presence of these larger bands (Figure 7B). For TRIzol-extracted SH-SY5Y exosome RNA, without DNase I primers directed to humanin amplified a product in the absence of reverse transcriptase, but after DNase I humanin amplification occurred only when reverse transcriptase was present (Figure 7C). DNase I, therefore, appeared to effectively remove DNA contamination from our exosome RNA samples.
Figure 7. Characterization of exosome fraction RNA.
(A) Total RNA derived from SH-SY5Y cells or SH-SY5Y cell-derived exosomes using TRIzol or RNeasy Plus Mini Kit columns was subjected to PCR amplification using primers to CO2. The column-based method eliminated genomic DNA contamination from the SH-SY5Y cell RNA sample, but not from the SH-SY5Y cell-derived exosome fraction. (B) The panel shows an Agilent Bioanalyzer 2100 RNA Pico Chip analysis of SH-SY5Y cell exosome fraction RNA. Lane 1 is an RNA ladder. Lane 2 is an RNA standard that shows 5S (120 nt), 18S (1900 nt) and 28S (4700nt) rRNA bands. Lane 3 represents RNA isolated from SH-SY5Y cell-derived exosome fractions using an RNeasy Plus Mini Kit. Lane 4 represents RNA isolated from the SH-SY5Y cell-derived exosome fraction using TRIzol; as part of this standard protocol following its extraction this TRIzol-generated RNA was treated with DNase I, and the RNA from this exosome fraction predominantly consisted of small RNAs. (C) TRIzol-generated RNA from SH-SY5Y cell-derived exosome fractions, with or without subsequent DNase I treatment, was subjected to PCR using primers to humanin following reverse transcription to cDNA, or in the absence of reverse transcription to cDNA. RT+, following reverse transcriptase; RT-, no reverse transcription.
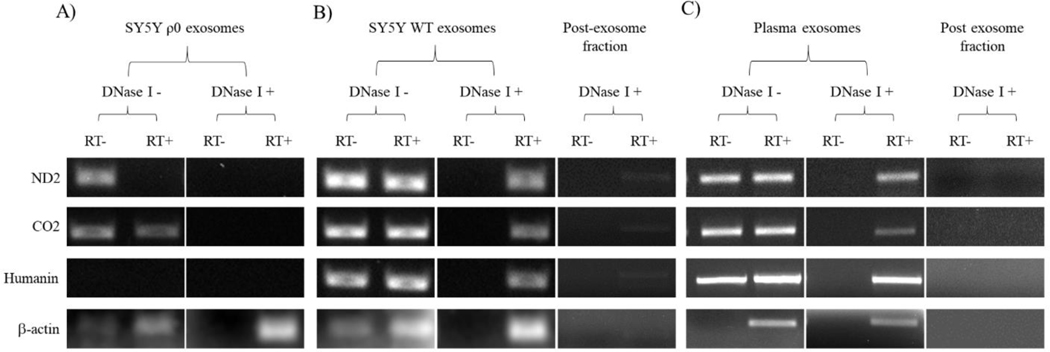
We next screened TRIzol-extracted, DNase I-treated and untreated RNA samples from SH-SY5Y ρ0 cell exosomes, SH-SY5Y cell exosomes, and plasma exosomes for mtDNA-derived mRNA transcripts. With the SH-SY5Y ρ0 cell exosome RNA, prior to DNase I treatment the ND2 and CO2 primers produced bands in the absence of reverse transcriptase (Figure 8A). In the presence of reverse transcriptase, the ND2 band was no longer seen, which could potentially reflect dilution of DNA contaminants by reverse transcription-generated cDNAs. Following treatment with DNase I, neither the ND2 nor CO2 primers produced a visible band, either with or without reverse transcriptase, which suggests DNA NUMT or mtDNA fragment contamination was eliminated and no mtDNA-derived RNA was present. Humanin primers failed to generate a band regardless of whether DNase I or reverse transcriptase was added, indicating that in addition to an absence of humanin mtDNA or mtDNA transcription in SH-SY5Y ρ0 cells, humanin-containing NUMT amplification is less robust than amplification of ND2 and CO2-containing NUMTs.
Figure 8. Interrogation of exosome fraction RNA samples for mtDNA-derived mRNA transcripts.
(A) RNA from SH-SY5Y ρ0 cell-derived exosome fractions, with or without post-isolation DNase I treatment, was subjected to amplification with primers to mtDNA-derived mRNA transcripts (ND2, CO2, and humanin). Primers to β-actin, a nuclear gene-derived transcript, were also utilized. The PCR reactions were performed without prior reverse transcription of the RNA to cDNA (RT-) and following reverse transcription of the RNA to cDNA (RT+), and the resulting PCR products were analyzed using gel electrophoresis. (B) An equivalent analysis is shown for RNA from SH-SY5Y cell-derived exosome fractions. In addition, we analyzed RNA isolated from the post-exosome fractions. (C) An equivalent analysis is shown for RNA from plasma-derived exosome and post-exosome fractions. For all cases the data shown are representative of three experiments.
For the SH-SY5Y cell exosome RNA, prior to the addition of DNase I primers to ND2, CO2, and humanin produced visible PCR bands even in the absence of reverse transcriptase (Figure 8B). After the addition of DNase I, ND2, CO2, and humanin bands were still produced, but only in the presence of reverse transcriptase. The plasma exosome RNA produced an amplification pattern that exactly recapitulated the SH-SY5Y cell exosome RNA amplification pattern (Figure 8C). DNase I-treated and untreated post-exosome fractions generated very little to no ND2, CO2, humanin, or β-actin amplicons during reverse-transcription PCR, which indicates SH-SY5Y cell culture medium and plasma contamination by free mRNA was below a meaningful limit of detection (Figure 8B and C). This suggests reverse-transcription PCR-mediated amplification of the exosome mRNA fraction was not a consequence of free mRNA sample contamination.
4. Discussion
We tested whether exosomes contain within them proteins that localize to mitochondria, proteins whose levels reflect mitochondrial function, mtDNA, or mtDNA-transcribed RNA. We took steps to minimize confounding by external contaminants as well as other internal components. Our results show that mitochondria-related proteins are present within exosomes. In addition to containing DNA, they likely also contain at least degraded pieces of authentic mtDNA. MtDNA-transcribed RNA can also reside within exosomes.
SEC segregates sample contents according to constituent size. When using qEV columns to perform SEC, fractions 5 through 8 primarily contain exosomes, while free proteins mostly reside in the post-exosome 12 through 16 fractions. Fractions 9 through 11 contain both exosomes and free protein, so some mixing can occur. qEV SEC columns reportedly separate exosomes and free protein more effectively than other methods including ultracentrifugation, density gradient purification, and precipitation-based techniques [21, 27, 30]. Here, we found that humanin, FGF 21, and citrate synthase were present in the SH-SY5Y cell-derived exosome fractions. Further experiments are needed to investigate whether these mitochondria-related proteins reside within exosomes, adhere to their surface, or both.
Plasma exosomes appeared to contain at least some humanin, VDAC1, and TFAM protein, but we cannot rule out concomitant sample contamination especially by free TFAM or TFAM that was on rather than in exosomes. In general, it is only useful to interpret our TFAM analysis from a qualitative rather than quantitative perspective for two reasons. The complete elimination of CD9 protein, an exosome marker in which only 40% of the protein extends external to the vesicle membrane [31], following trypsin treatment could reflect an overly-stringent trypsin protocol. Freeze-thawing of frozen plasma could also potentially compromise membrane integrity [32]. Exosome membrane disruption could minimize its ability to protect internal constituents from interventions intended to eliminate external contaminants.
While other studies report exosomes contain mtDNA [8, 33–35], these studies did not account for potential confounding by NUMT DNA sequences. The presence of NUMT DNA could give the false impression that mtDNA is present within exosomes when it is not, or skew quantification of truly internalized authentic mtDNA. These are not abstract concerns, as we found exosomes may contain NUMT DNA. We nevertheless were able to conclude NT2 cell-derived exosomes do contain authentic mtDNA because the NT2 ND2 gene, but not its major NUMTs, carries a unique G5460A transition-dependent HphI restriction site and NT2 cell-derived exosome ND2 DNA contained this restriction site. Beyond demonstrating NT2 cell-derived exosomes contain at least some authentic mtDNA fragments, we could not clearly establish the presence of extended mtDNA sequences.
As is the case with nuclear DNA [35], it is worth considering why and how mtDNA enters exosomes. The general assumption for nuclear DNA is that exosome internalization facilitates disposal of DNA waste, which may be used to signal other cells. This does not address the question of whether endosomes directly access the nucleus, or if nuclei first transfer pieces of DNA to the cytoplasm. The issue of endosome access to mtDNA seems more straightforward, because mitochondria and, therefore, mtDNA already reside in the cytoplasm. The presence of mtDNA within exosomes could reflect incorporation of mtDNA that is no longer surrounded by mitochondrial membrane, or of mtDNA residing within mitochondrial vesicles created through mitochondrial fission or mitophagy. Also, while our NUMT amplification experiments indicate exosomes contain only or mostly degraded mtDNA, our data do not address whether its degradation occurs before or after it enters an endosome or exosome. In either case, what drives its degradation is unclear, although a recent report adds mtDNA polymerase γ (POLG) to a list of known mitochondrial exonucleases and suggests in addition to mtDNA synthesis it plays a critical role in mtDNA degradation [36].
Similar questions pertain to RNA. We do not know if mtDNA-transcribed RNA enters exosomes independently or as constituents of membrane-bound, fission-formed mitochondrial vesicles. Unlike mtDNA, precedent for RNA export from the generating organelle to the surrounding cytosol at least exists [37]. Like mtDNA, RNA-based biomarker applications will need to account for potential confounding by DNA, including NUMT DNA.
Hashimoto et al. discovered the humanin transcript in 2001 [38]. It matches 16S mtDNA gene sequence and presumably arises from an open reading frame within the 16S gene, although it remains possible NUMT sequence supports expression [39, 40]. Whether humanin translation occurs at mitochondrial or cytosolic ribosomes, or both, remains uncertain but humanin RNA clearly resides in cytoplasm. Humanin expression and protein levels are sensitive to cell stress, and the protein reportedly participates in cytosolic stress pathways and responses. Cells further secrete humanin protein, which suggests it also functions to communicate stress between cells and perhaps tissues. We believe these characteristics, in conjunction with our new data, justify further development of humanin-based exosome biomarkers.
We did not attempt to measure ATP or respiration, which is a limitation as some report the presence of functional mitochondria in extracellular vesicles [41–43]. Our focus, rather, was on the simple presence versus absence of protein, mRNA, or mtDNA components in exosomes rather than mitochondrial functional integrity. Other limitations of this study relate to assay sensitivity and specificity. Our failure to detect some mitochondrial proteins, mitochondrial enzyme Vmax activities, or definitive evidence of extended mtDNA sequences within exosomes could reflect a true absence, or that values simply fell below the threshold for unequivocal detection.
We considered that endpoints we detected could reflect non-exosome contaminants, and although we took steps to minimize this concern, we cannot completely exclude this possibility. If secreted, mitochondrial-derived vesicles (MDVs) could represent one such confounder, as their size and exosome size overlap [44, 45]. We also cannot rule out the possibility that our exosome preparations contained a limited amount of contamination by small microvesicles that populate the tail of the microvesicle size spectrum. Assuming our results reflect the true status of the exosomes we studied we do not know whether the contents of these exosomes reflect the contents of exosomes from other origins. We focused strictly on exosomes themselves, and our findings may not extrapolate well or at all to a family of usually larger extracellular vesicles, including microvesicles, that are often analyzed in conjunction with exosomes [2, 3].
We did not determine the percentage of exosomes that contain mitochondrial proteins or resolve our exosome fractions by tetraspanin distribution patterns. Although we evaluated for the presence of mitochondrial components in cell lines and plasma, our goal was mostly to demonstrate presence or absence, not to compare relative amounts.
Despite these limitations, our current studies provide insight that can help guide the development of exosome-derived mitochondrial biomarkers. We provide evidence that mitochondria-related proteins reside within plasma and cell-derived exosomes. Our data indicate potential confounding by NUMT sequences as an issue to consider, and that exosome mtDNA may primarily exist as fragments. When considering mtDNA as an endpoint, treating exosomes with DNase prior to DNA extraction and selecting primers that minimize NUMT sequence amplification will help avoid confounding by free DNA contamination and nuclear pseudogenes. When considering RNA as an endpoint, DNase treatment following RNA extraction will help avoid confounding by NUMT DNA and mtDNA fragments.
Proving a “negative” is a challenging scientific endeavor. Here, one imperative negative was to establish our measured targets were not contaminants. In situations where the need to prove a negative apply, we also narrowly interpret our data or minimize interpretations. Experimental replication will no doubt inform the accuracy of our findings and conclusions.
5. Conclusions
Exosomes contain mitochondrial components and cell components whose levels depend on or reflect mitochondrial function. Our study suggests exosomes could serve as “liquid biopsies” that inform the status of mitochondria within difficult to obtain tissues. Our study further identifies confounds that could adversely affect the precision and accuracy of exosome-derived mitochondrial biomarkers. Proper endpoint selection and awareness of confounds will facilitate further development.
Highlights.
Exosomes contain mitochondria-pertinent proteins
Exosomes contain at least degraded authentic mtDNA
Exosomes contain mtDNA-encoded mRNA
Acknowledgements
This work was supported by the University of Kansas Alzheimer’s Disease Center (P30AG035982). XW was supported by a Mabel Woodyard Fellowship award. The University of Kansas Medical Center Electron Microscope Research Laboratory is supported, in part, by NIH/NIGMS COBRE grant P20GM104936. The JEOL JEM-1400 transmission electron microscope was purchased with funds from NIH grant 1S10RR027564.
Abbreviations:
- AFC
automatic fraction collector
- BSA
bovine serum albumin
- CCM
cell culture medium
- cDNA
complementary DNA
- CO2
cytochrome oxidase subunit 2
- CT
cycle threshold
- Ctrl
control
- Cyto C
cytochrome C
- DMEM
Dulbecco’s modified Eagle medium
- ELISA
enzyme linked immunosorbent assay
- FBS
fetal bovine serum
- FGF
fibroblast growth factor
- ILV
intra-luminal vesicle
- KUADC
University of Kansas Alzheimer’s Disease Center
- MT
mitochondrial
- MDVs
mitochondrial-derived vesicles
- mtDNA
mitochondrial DNA
- MVB
multivesicular body
- ND1
NADH dehydrogenase subunit 1
- ND2
NADH dehydrogenase subunit 2
- NMWL
nominal molecular weight limit
- nt
nucleotide
- NTA
nanoparticle tracking analysis
- NUMT
nuclear mitochondrial DNA
- PBS
phosphate buffered saline
- PBST
phosphate buffered saline with Tween 20
- PCR
polymerase chain reaction
- POLG
mtDNA polymerase γ
- PVDF
polyvinylidene difluoride
- qPCR
quantitative PCR
- RT
reverse transcriptase
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SEC
size exclusion chromatography
- SEM
standard error of the mean
- SOD2
superoxide dismutase 2
- SD
standard deviation
- TEM
transmission electron microscopy
- TFAM
transcription factor A of the mitochondria
- UC
ultracentrifugation
- UV
ultraviolet
- VDAC1
voltage-dependent anion-selective channel 1
Footnotes
Financial Disclosures: The authors report no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Meldolesi J (2018) Exosomes and Ectosomes in Intercellular Communication. Curr Biol 28, R435–R444. [DOI] [PubMed] [Google Scholar]
- [2].Shah R, Patel T, Freedman JE (2018) Circulating Extracellular Vesicles in Human Disease. N Engl J Med 379, 958–966. [DOI] [PubMed] [Google Scholar]
- [3].Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Urbanelli L, Magini A, Buratta S, Brozzi A, Sagini K, et al. (2013) Signaling pathways in exosomes biogenesis, secretion and fate. Genes (Basel) 4, 152–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ha D, Yang N, Nadithe V (2016) Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B 6, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Conigliaro A, Cicchini C (2018) Exosome-Mediated Signaling in Epithelial to Mesenchymal Transition and Tumor Progression. J Clin Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kapogiannis D, Mustapic M, Shardell MD, Berkowitz ST, Diehl TC, et al. (2019) Association of Extracellular Vesicle Biomarkers With Alzheimer Disease in the Baltimore Longitudinal Study of Aging. JAMA Neurol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guescini M, Genedani S, Stocchi V, Agnati LF (2010) Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm (Vienna) 117, 1–4. [DOI] [PubMed] [Google Scholar]
- [9].Athauda D, Gulyani S, Karnati HK, Li Y, Tweedie D, et al. (2019) Utility of Neuronal-Derived Exosomes to Examine Molecular Mechanisms That Affect Motor Function in Patients With Parkinson Disease: A Secondary Analysis of the Exenatide-PD Trial. JAMA Neurol 76, 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mustapic M, Eitan E, Werner JK Jr., Berkowitz ST, Lazaropoulos MP, et al. (2017) Plasma Extracellular Vesicles Enriched for Neuronal Origin: A Potential Window into Brain Pathologic Processes. Front Neurosci 11, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cumba Garcia LM, Peterson TE, Cepeda MA, Johnson AJ, Parney IF (2019) Isolation and Analysis of Plasma-Derived Exosomes in Patients With Glioma. Front Oncol 9, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Boukouris S, Mathivanan S (2015) Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl 9, 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dang VD, Jella KK, Ragheb RRT, Denslow ND, Alli AA (2017) Lipidomic and proteomic analysis of exosomes from mouse cortical collecting duct cells. Faseb j 31, 5399–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Haraszti RA, Didiot MC, Sapp E, Leszyk J, Shaffer SA, et al. (2016) High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracell Vesicles 5, 32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Choi DS, Kim DK, Kim YK, Gho YS (2013) Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 13, 1554–1571. [DOI] [PubMed] [Google Scholar]
- [16].Burke M, Choksawangkarn W, Edwards N, Ostrand-Rosenberg S, Fenselau C (2014) Exosomes from myeloid-derived suppressor cells carry biologically active proteins. J Proteome Res 13, 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mears R, Craven RA, Hanrahan S, Totty N, Upton C, et al. (2004) Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics 4, 4019–4031. [DOI] [PubMed] [Google Scholar]
- [18].Swerdlow RH (2007) Treating neurodegeneration by modifying mitochondria: potential solutions to a “complex” problem. Antioxid Redox Signal 9, 1591–1603. [DOI] [PubMed] [Google Scholar]
- [19].Swerdlow RH (2009) Mitochondrial medicine and the neurodegenerative mitochondriopathies. Pharmaceuticals 2, 150–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shelke GV, Lässer C, Gho YS, Lötvall J (2014) Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, et al. (2015) Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles 4, 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ghosh S, Patel N, Rahn D, McAllister J, Sadeghi S, et al. (2007) The thiazolidinedione pioglitazone alters mitochondrial function in human neuron-like cells. Mol Pharmacol 71, 1695–1702. [DOI] [PubMed] [Google Scholar]
- [23].Clarke JD (2009) Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc 2009, pdb.prot5177. [DOI] [PubMed] [Google Scholar]
- [24].Herrnstadt C, Clevenger W, Ghosh SS, Anderson C, Fahy E, et al. (1999) A novel mitochondrial DNA-like sequence in the human nuclear genome. Genomics 60, 67–77. [DOI] [PubMed] [Google Scholar]
- [25].Swerdlow RH, Redpath GT, Binder DR, Davis JN 2nd, VandenBerg SR (2006) Mitochondrial DNA depletion analysis by pseudogene ratioing. J Neurosci Methods 150, 265–271. [DOI] [PubMed] [Google Scholar]
- [26].Patel GK, Khan MA, Zubair H, Srivastava SK, Khushman M, et al. (2019) Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci Rep 9, 5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Buschmann D, Kirchner B, Hermann S, Marte M, Wurmser C, et al. (2018) Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J Extracell Vesicles 7, 1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chulpanova DS, Kitaeva KV, James V, Rizvanov AA, Solovyeva VV (2018) Therapeutic Prospects of Extracellular Vesicles in Cancer Treatment. Front Immunol 9, 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Paolillo M, Schinelli S (2017) Integrins and Exosomes, a Dangerous Liaison in Cancer Progression. Cancers (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vaswani K, Koh YQ, Almughlliq FB, Peiris HN, Mitchell MD (2017) A method for the isolation and enrichment of purified bovine milk exosomes. Reprod Biol 17, 341–348. [DOI] [PubMed] [Google Scholar]
- [31].Tang X, Chang C, Guo J, Lincoln V, Liang C, et al. (2019) Tumour-Secreted Hsp90alpha on External Surface of Exosomes Mediates Tumour - Stromal Cell Communication via Autocrine and Paracrine Mechanisms. Sci Rep 9, 15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cheng Y, Zeng Q, Han Q, Xia W (2019) Effect of pH, temperature and freezing-thawing on quantity changes and cellular uptake of exosomes. Protein Cell 10, 295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Soltesz B, Urbancsek R, Pos O, Hajas O, Forgacs IN, et al. (2019) Quantification of peripheral whole blood, cell-free plasma and exosome encapsulated mitochondrial DNA copy numbers in patients with atrial fibrillation. J Biotechnol 299, 66–71. [DOI] [PubMed] [Google Scholar]
- [34].Tsilioni I, Theoharides TC (2018) Extracellular vesicles are increased in the serum of children with autism spectrum disorder, contain mitochondrial DNA, and stimulate human microglia to secrete IL-1beta. J Neuroinflammation 15, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, et al. (2017) Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A 114, E9066–e9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Medeiros TC, Thomas RL, Ghillebert R, Graef M (2018) Autophagy balances mtDNA synthesis and degradation by DNA polymerase POLG during starvation. J Cell Biol 217, 1601–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, et al. (2015) The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 21, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hashimoto Y, Ito Y, Niikura T, Shao Z, Hata M, et al. (2001) Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein. Biochem Biophys Res Commun 283, 460–468. [DOI] [PubMed] [Google Scholar]
- [39].Niikura T, Chiba T, Aiso S, Matsuoka M, Nishimoto I (2004) Humanin: after the discovery. Mol Neurobiol 30, 327–340. [DOI] [PubMed] [Google Scholar]
- [40].Gong Z, Tas E, Muzumdar R (2014) Humanin and age-related diseases: a new link? Front Endocrinol (Lausanne) 5, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hough KP, Trevor JL, Strenkowski JG, Wang Y, Chacko BK, et al. (2018) Exosomal transfer of mitochondria from airway myeloid-derived regulatory cells to T cells. Redox Biol 18, 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hill BG, Shiva S, Ballinger S, Zhang J, Darley-Usmar VM (2019) Bioenergetics and translational metabolism: implications for genetics, physiology and precision medicine. Biol Chem 401, 3–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Morrison TJ, Jackson MV, Cunningham EK, Kissenpfennig A, McAuley DF, et al. (2017) Mesenchymal Stromal Cells Modulate Macrophages in Clinically Relevant Lung Injury Models by Extracellular Vesicle Mitochondrial Transfer. Am J Respir Crit Care Med 196, 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sugiura A, McLelland GL, Fon EA, McBride HM (2014) A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. Embo j 33, 2142–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bozi LH, Bechara LR, Dos Santos AF, Campos JC (2016) Mitochondrial-derived vesicles: a new player in cardiac mitochondrial quality control. J Physiol 594, 6077–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]