Abstract
Several antiepileptic drugs (AEDs) have US Food and Drug Administration (FDA) approval for use as mood stabilizers in bipolar disorder (BD), but not all BD patients respond to these AED mood stabilizers (AED‐MSs). To identify genetic polymorphisms that contribute to the variability in AED‐MS response, we performed a discovery genome‐wide association study (GWAS) of 199 BD patients from the Mayo Clinic Bipolar Disorder Biobank. Most of these patients had been treated with the AED‐MS valproate/divalproex and/or lamotrigine. AED‐MS response was assessed using the Alda scale, which quantifies clinical improvement while accounting for potential confounding factors. We identified two genome‐wide significant single‐nucleotide polymorphism (SNP) signals that mapped to the THSD7A (rs78835388, P = 7.1E‐09) and SLC35F3 (rs114872993, P = 3.2E‐08) genes. We also identified two genes with statistically significant gene‐level associations: ABCC1 (P = 6.7E‐07; top SNP rs875740, P = 2.0E‐6), and DISP1 (P = 8.9E‐07; top SNP rs34701716, P = 8.9E‐07). THSD7A SNPs were previously found to be associated with risk for several psychiatric disorders, including BD. Both THSD7A and SLC35F3 are expressed in excitatory/glutamatergic and inhibitory/γ‐aminobutyric acidergic (GABAergic) neurons, which are targets of AED‐MSs. ABCC1 is involved in the transport of valproate and lamotrigine metabolites, and the SNPs in ABCC1 and DISP1 with the strongest evidence of association in our GWAS are strong splicing quantitative trait loci in the human gut, suggesting a possible influence on drug absorption. In conclusion, our pharmacogenomic study identified novel genetic loci that appear to contribute to AED‐MS treatment response, and may facilitate precision medicine in BD.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Antiepileptic mood stabilizers (AED‐MSs) are the second most important pharmacotherapy for bipolar disorder (BD) after lithium. AED‐MS treatment response varies widely among BD patients. No genome‐wide association study for AED‐MS response in BD has been reported.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ This study asked what genetic polymorphisms might be associated with variation in response to AED‐MS therapy in the treatment of BD patients, and what the underlying mechanisms might be.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ We identified novel loci that mapped to the THSD7A, SLC35F, ABCC1, and DISP1 genes that were associated with variation in AED‐MS response in BD. THSD7A and SLC35F are expressed in excitatory and inhibitory neurons where AED‐MSs are known to act. ABCC1 may be involved in the transport of AED‐MS metabolites.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ This study discovered genetic markers that may help predict AED‐MS treatment response in BD and it provided initial insight into potential mechanisms underlying the pharmacodynamics and pharmacokinetics of AED‐MS response in BD.
Bipolar disorder (BD) is a chronic debilitating mental illness characterized by recurrent episodes of mania/hypomania and depression. The disease negatively impacts daily functioning and poses an increased risk of comorbid medical conditions and suicide. Although lithium is the gold standard pharmacotherapy for BD, a substantial proportion of patients fail to respond optimally to lithium and/or are unable to adhere to lithium treatment due to side effects and toxicity (> 40%). 1 Several antiepileptic drugs (AED) are US Food and Drug Administration (FDA) approved for BD treatment. These AEDs are categorized as predominantly antimanic (e.g., divalproex sodium and carbamazepine) or predominantly antidepressant (e.g., lamotrigine) mood stabilizers (AED‐MSs). 2
The proposed mechanisms of AEDs for prevention of epileptic seizures are blockade of voltage‐gated sodium and calcium channels, enhancement of synaptic inhibition mediated by γ‐aminobutyric acid type A receptors, and/or inhibition of synaptic excitation mediated by ionotropic glutamate receptors. 3 , 4 Whether these mechanisms of action are also responsible for the mood‐stabilizing properties of some AEDs in BD remains unclear. Nevertheless, despite potential mechanistic differences, all AED‐MSs are believed to indirectly dampen excessive intracellular and intercellular signaling, which may be central to BD pathophysiology. 5
As is true for lithium, not all BD patients respond to AED‐MSs, and genetic polymorphisms may contribute to the observed variation in clinical response to these medications. Thus, the study of the pharmacogenomics of members of this class of medications in the context of BD treatment is warranted. Although genome‐wide association studies (GWASs) have been applied to study the pharmacogenomics of lithium response in BD, 6 prior pharmacogenomic studies of AED‐MSs in BD have been limited to a small set of candidate genes 7 , 8 and, to our knowledge, no GWAS of AED‐MS treatment response in BD has been conducted. In contrast to candidate gene studies, GWASs may reveal unknown genes that contribute to the mood‐stabilizing properties of AED‐MSs, thereby identifying genetic predictors of their treatment response to facilitate greater precision and individualization of treatment selection for BD patients.
In this study, we performed a discovery GWAS for AED‐MSs (valproate/divalproex, lamotrigine, carbamazepine, and oxcarbazepine) treatment response in BD patients using the Alda score as the phenotype, 9 and identified several novel genome‐wide significant loci. Recognizing the complications of studying a class of drugs with different pharmacokinetics, this study was designed primarily to identify genes related to their—presumed—common pharmacodynamics, and we interpret our findings accordingly. For genomic regions with statistically significant associations, we further performed drug‐stratified analyses to explore whether the observed associations were driven specifically by a genetic influence on the response to valproate/divalproex and/or lamotrigine.
METHODS
Subjects and pharmacotherapy
This study included 199 European American subjects from the Mayo Clinic Bipolar Disorder Biobank. 10 The biobank was approved by the Mayo Clinic Institutional Review Board, and recruited patients aged 18–80 years with a Diagnostic and Statistical Manual of Mental Disorders 4th Edition Text Revision (DSM‐IV‐TR) or DSM‐5 diagnosis of bipolar I, bipolar II, bipolar not otherwise specified, or schizoaffective bipolar type. All subjects provided written informed consent for use of their data and biospecimens in future studies. Exclusion criteria included inability or unwillingness to provide written informed consent, active suicidal ideation, or active psychosis. Clinical phenotypes were confirmed by the Structured Clinical Interview for DSM‐IV. 11 Further clinical data were collected using the Bipolar Biobank Clinical Questionnaire, which included a retrospective evaluation of long‐term psychotropic medication treatment response. The 199 European American subjects included in this study had been treated with one or more AED‐MSs (valproate/divalproex, lamotrigine, carbamazepine, and/or oxcarbazepine), had treatment response quantified using the Alda scale, and had been previously genotyped on a genome‐wide platform.
Measurement of AED‐MS response by Alda scale
AED‐MS response was measured by the Alda scale, which was developed to retrospectively evaluate lithium treatment response in BD by assessing the degree of clinical improvement while weighting clinical factors relevant to the observed improvement. 9 The Alda scale was adopted by the Consortium on Lithium Genetics (ConLiGen) for a large multisite GWAS. 6 This scale has two components: the A score and the B score (both range from 0 to 10). The A score is a composite measure of clinical improvement in frequency, duration, and severity of illness episodes. A higher A score represents a greater improvement. The B score quantifies confidence in the A score by assessing potential confounding factors that may influence treatment response. A smaller B score means less influence of confounding factors on the clinical improvement measured by the A score, and thus greater confidence in the A score. 9 The total Alda score, calculated by subtracting the B score from the A score, was used to quantify treatment response. A larger total Alda score represents a better treatment outcome. Moderate‐to‐good inter‐rater reliability for A, B, and total Alda scores has been reported (intraclass correlation = 0.66, 0.59, and 0.74, respectively), as well as a moderate‐to‐good test‐retest reliability across an average of 17 months (intraclass correlation = 0.62, 0.62, and 0.60, respectively). 9 , 12 The investigator who oversaw the Alda scoring of patients in this study (M.A.F.) took part in the original studies of the inter‐rater agreement and reliability of the Alda scale. 9
In this study, we defined AED‐MS treatment outcomes using the total Alda score as a continuous phenotype, which made it possible to include all subjects and preserve statistical power while accounting for the degree of confidence in the treatment response assessment. For subjects with Alda scores for more than one AED‐MS (valproate/divalproex, lamotrigine, carbamazepine, and/or oxcarbazepine), the Alda score corresponding to the treatment episode with the lowest B score (and thus higher confidence in the A score) was used. If two or more medications had tied lowest B scores, the A scores were averaged and then used to calculate the total score. Approximately 90% of Alda scores included in this study represent a response to a medication taken for more than one year.
Genotyping and GWAS analysis
DNA samples from subjects included in this study were previously genotyped using Illumina HumanOmniExpress BeadChips (Illumina Inc., San Diego, CA). 13 Genotyping, quality control, and imputation procedures have been described elsewhere. 13 After quality control and imputation, a total of 7,368,687 SNPs were included in the GWAS analysis. GWAS analyses were performed in PLINK v1.90b3v, 14 with additional regression analyses performed using R‐3.2.3. 15 Prior to performing genetic analyses, we used linear regression to test whether sex, bipolar type, and rapid cycling (four or more depressive or manic episodes in a year) were associated with treatment outcomes defined by the total Alda score. For the genome‐wide analyses, we used linear regression to assess the association between each SNP (in terms of allele dosage) and total Alda score as a continuous variable. Although the analysis was limited to subjects of European ancestry and principal components were not associated with Alda scores, the first principal component was included as a covariate in the model to avoid bias from population stratification. GWAS results were annotated by human genome assembly GRCh37 for genomic location and nearby genes. SNP associations with P < 5E‐08 were considered statistically significant.
Gene‐level and pathway‐level association analysis
Gene‐level and pathway‐level association tests were performed in the GWAS functional mapping and annotation platform FUMA, 16 which implements MAGMA gene‐and‐pathway based analyses using GWAS summary statistics. MAGMA first annotates SNPs onto genes, and then uses the P values of the SNPs to compute a single P value for each gene, 17 while accounting for linkage disequilibrium across the region; this gene‐level P value represents the overall statistical evidence that genetic variation in or near the gene is associated with the phenotype. By substantially reducing the multiple testing burden and combining evidence of association across SNPs that map to the same gene, gene‐level tests can be much more powerful than SNP‐level tests. 18 Statistical significance for gene‐level association was set at the Bonferroni‐corrected threshold P < 2.9E‐06 (corrected for the 17,049 analyzed protein‐coding genes). We also used MAGMA to perform gene‐set (i.e., pathway) enrichment analysis including pathways defined in KEGG, GO processes, and Biocarta. 17 Statistical significance for pathway‐level association was set at the Bonferroni‐corrected threshold for the number of pathways used.
For genes with genome‐wide significant evidence of association at the gene level, we also used PrediXcan 19 to estimate gene expression in whole blood for each patient in our study based on prediction models trained on the Depression Genes and Networks (DGN) transcriptome data set, and tested for association of the estimated gene expression with Alda scores. 20 This analysis allowed us to infer the direction of observed gene‐level effects, in terms of gene expression level association with AED‐MS response.
Drug‐stratified analyses
For most subjects in our GWAS, the total Alda score represented response to either valproate/divalproex (n = 87) or lamotrigine (n = 76). To determine whether the significant associations in our GWAS were driven by a specific AED‐MS medication (valproate/divalproex or lamotrigine), we additionally performed analysis of Alda scores for the two medications separately and generated regional association plots for regions encompassing the significant genes ± 10 kilobase (kb). The –log10(P) values for associations in each treatment group were then overlaid with the corresponding results from the original GWAS of the full sample, to provide a qualitative comparison. More significant association (i.e., lower P values) in a given treatment group (valproate/divalproex vs. lamotrigine) might suggest that association of the genetic variants with response to this particular treatment contributed more to the observed genome‐wide association in the full sample of 199 patients.
Functional annotations of genome‐wide significant signals
The Genotype‐Tissue Expression (GTEx) project offers a comprehensive data resource for the systemic study of genetic variation and gene expression regulation in multiple human tissues collected from nearly 1,000 individuals. 21 We performed functional annotation of the top SNPs and genes using the GTEx Portal V8 Release, 22 which provides information on gene expression across tissue types, expression quantitative trait locus (eQTL), and splicing quantitative trait locus (sQTL) analyses. For cell‐type‐specific gene expression patterns in the human brain, we conducted searches in the Allen Brain Map Transcriptomics Explorer, 23 which contains transcriptomic data of sorted single‐cell nuclei from human cortical regions. 24 , 25 We also searched for potential methylation quantitative trait loci among our top SNPs using QTLbase. 26
RESULTS
Subject characteristics and Alda scores
Patient characteristics, including sex, age, race, and clinical measurements and outcomes, are listed in Table 1 . The majority of participants had bipolar I disorder (73%; n = 146). Most Alda scores were based on valproate/divalproex (44%; n = 87) or lamotrigine (38%; n = 76), and about 11% (n = 21) were based on multiple medications. The distributions of the total Alda score and its components are shown in Figure S1 . Women had slightly higher Alda scores than men (P = 0.03). Neither BD type nor history of rapid cycling (four or more depressive or manic episodes in a year) was significantly associated with total Alda score.
Table 1.
Subject characteristics (N = 199)
| Female, n (%) | 113 (56.8%) |
| Age at interview, mean (SD) | 42.1 (14.4) |
| Race (European Americans), n (%) | 199 (100.0%) |
| Bipolar type | |
| Bipolar I, n (%) | 146 (73.4%) |
| Bipolar II, n (%) | 53 (26.6%) |
| Rapid cycling, n (%) | 134 (67.3%) |
| Sources of total Alda score | |
| Valproate/Divalproex (V), n (%) | 86 (43.7%) |
| Lamotrigine (L), n (%) | 76 (38.2%) |
| Carbamazepine (C), n (%) | 8 (4.0%) |
| Oxcarbazepine (O), n (%) | 7 (3.5%) |
| V and L, n (%) | 15 (7.5%) |
| V and O, n (%) | 1 (0.5%) |
| C and O, n (%) | 1 (0.5%) |
| V, L, and C, n (%) | 1 (0.5%) |
| L, C, and O, n (%) | 3 (1.5%) |
| Alda scale scores | |
| A score, mean (SD) | 5.9 (2.4) |
| B score, mean (SD) | 2.5 (1.2) |
| Total score, mean (SD) | 3.4 (2.7) |
| Alda B score on which the total Alda score was based | |
| ≤4, n (%) | 188 (94.5%) |
| >5, n (%) | 11 (5.5%) |
GWAS of AED‐MS treatment response
The Manhattan plot and Q‐Q plot showing SNP associations with AED‐MS treatment response are presented in Figure 1a and Figure S2 a, respectively. Table 2 lists details of the SNPs with the lowest P values at each of the top five loci. SNPs at two independent loci showed genome‐wide significant association with AED‐MS response (P < 5E‐08): rs78835388 (P = 7.1E‐09), which mapped to intron 1 of THSD7A, and rs114872993 (P = 3.2E‐08), which mapped to an intergenic region between SLC35F3 and COA6, approximately 22.4 kb 3’‐downstream of SLC35F3 and 26.6 kb 5’‐upstream of COA6. Although the minor allele frequencies (MAFs) of these SNPs are low (Table 2 ), the genes to which they mapped may be functionally relevant for the associated phenotype as described subsequently. Other suggestive associations (P < 5E‐06) included SNPs that map to the DISP1, VAV3, and ABCC1 genes. The minor allele of the DISP1 SNP, rs34701716, was associated with higher Alda scores corresponding to better treatment response (a positive β in the linear regression model), while the minor alleles of the other four SNPs were associated with lower Alda scores (negative β), indicating that the minor alleles were associated with worse clinical response to AED‐MS therapy.
Figure 1.
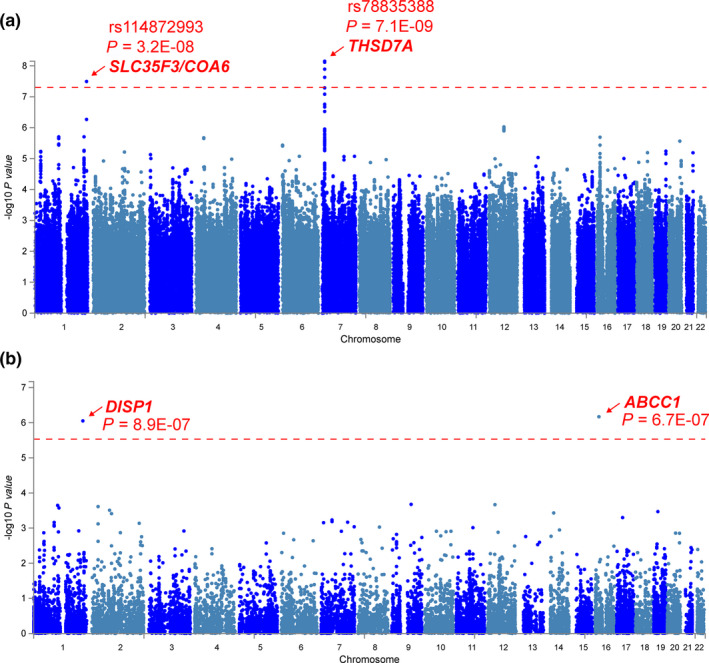
Manhattan plots for GWAS of AED‐MS treatment response in bipolar disorder. (a) At a SNP level, rs78835388 and rs114872993 are associated with AED‐MS response at a genome‐wide significant level (red line: P = 5E‐08). The SNP rs78835388 maps to an intron of THSD7A, while rs114872993 maps to an intergenic region between SLC35F3 and COA6. (b) At a gene level, ABCC1 and DISP1 show genome‐wide significant association with AED‐MS response (red line: P = 2.9E‐06; corrected for 17,049 protein‐coding genes). AED‐MS, antiepileptic drug–mood stabilizer; GWAS, genome‐wide association study; SNP, single‐nucleotide polymorphism.
Table 2.
Top five lead SNPs associated with total Alda scores of AED‐MS response in bipolar disorder (*P < 5E‐08)
| SNP | Chr | BP | Major allele | Minor allele | Gene | Variant location | Minor allele frequency | DosR 2 | β | P |
|---|---|---|---|---|---|---|---|---|---|---|
| rs78835388 a | 7 | 11814045 | A | G | THSD7A | intron | 0.021 | 0.97 | −6.6 | 7.1E‐09* |
| rs114872993 | 1 | 234482652 | A | G | SLC35F3 | 3'‐downstream | 0.029 | 0.88 | −4.5 | 3.2E‐08* |
| rs34701716 b | 1 | 223113809 | G | C | DISP1 | intron | 0.140 | 1 | 1.8 | 2.0E‐06 |
| rs13353016 | 1 | 108399495 | C | T | VAV3 | intron | 0.028 | 0.98 | −4.7 | 2.0E‐06 |
| rs875740 b | 16 | 16123048 | A | C | ABCC1 | intron | 0.340 | 1 | −1.3 | 2.0E‐06 |
AED‐MS, antiepileptic drug–mood stabilizer; BP, base pair; Chr, chromosome; SNP, single‐nucleotide polymorphism.
Other genome‐wide significant SNPs at the same locus: rs76802442, rs117511569, rs12056088; Dos R 2 = imputation (dosage) R 2, which provides a measure of genotype imputation quality.
Methylation quantitative trait loci (mQTL); see Table S1 for details.
Gene‐level and pathway‐level associations
Gene‐level association analysis considers multiple genetic markers in or near a gene simultaneously, taking into account linkage disequilibrium between markers, to determine evidence for their joint effect on a phenotype. 17 The Manhattan plot for gene‐level associations is shown in Figure 1b and the Q‐Q plot is displayed in Figure S2 b. The top five genes are listed in Table 3 . Associations with two genes, ABCC1 (P = 6.7E‐07) and DISP1 (P = 8.9E‐07), were statistically significant at the genome‐wide significance level for gene‐level tests (Figure 1b ). The minor alleles of ABCC1 top SNPs (MAF = 0.22–0.41) were associated with worse AED‐MS response, whereas the minor alleles of DISP1 top SNPs (MAF ≈ 0.14) were associated with better AED‐MS response. None of the investigated pathways were significantly enriched for association after Bonferroni correction.
Table 3.
Top five genes associated with AED‐MS Alda score in bipolar disorder patients at gene level (*P < 3E‐06)
| Gene | Chr | Start | Stop | # SNPs | P |
|---|---|---|---|---|---|
| ABCC1* | 16 | 16033434 | 16246931 | 695 | 6.7E‐07* |
| DISP1* | 1 | 222978406 | 223189337 | 579 | 8.9E‐07* |
| NTRK2 | 9 | 87273466 | 87648505 | 908 | 2.1E‐04 |
| PKP2 | 12 | 32933679 | 33059774 | 342 | 2.2E‐04 |
| NBPF4 | 1 | 108755963 | 108796689 | 24 | 2.3E‐04 |
AED‐MS, antiepileptic drug–mood stabilizer; Chr, chromosome; SNPs, single‐nucleotide polymorphisms.
For the genes with statistically significant gene‐level associations, we also tested the association of AED‐MS treatment response with gene expression estimated from SNP data using PrediXcan. 19 In these analyses, higher predicted gene expression levels in whole blood for ABCC1 and DISP1 were associated with lower total Alda scores indicating worse treatment response (ABCC1: odds ratio = −2.15, SE = 0.82, P = 0.0089; DISP1: odds ratio = −2.52, SE = 0.74, P = 0.00077; Figure 2 ).
Figure 2.
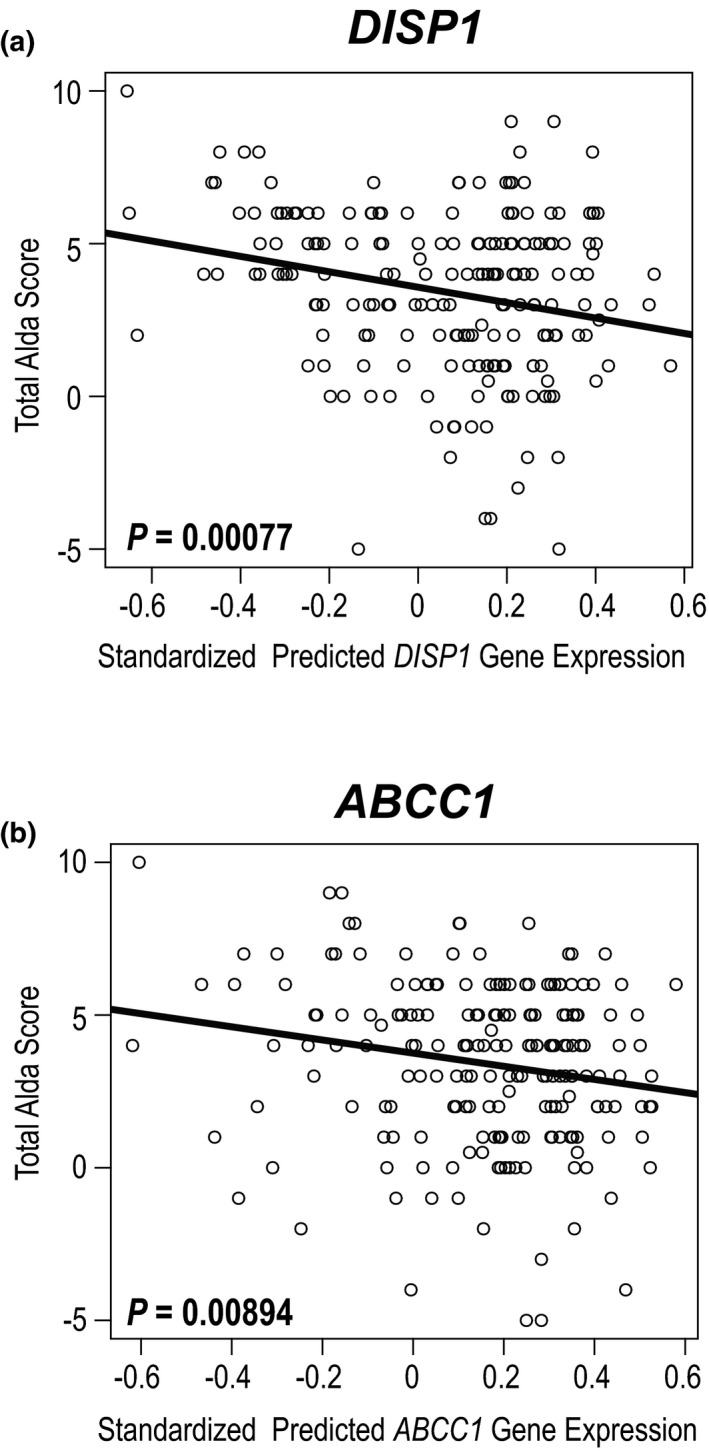
Correlations between total Alda score and the gene expression levels of (a) DISP1 and (b) ABCC1 in whole blood predicted by SNP data using PrediXcan. SNP, single‐nucleotide polymorphism.
Drug‐stratified analyses
We performed drug‐stratified association analyses of AED‐MS treatment response at the four genome‐wide significant loci (THSD7A, SLC35F3, ABCC1, and DISP1) to identify which AED‐MS (valproate/divalproex or lamotrigine) might be driving the observed genome‐wide significant associations (Figure 3 ). Although the statistical power was reduced in these analyses due to smaller sample sizes, the purpose of these analyses was to compare the associations (P values) between the two main treatment groups. SNPs in THSD7A appeared to be associated with the Alda scores for both valproate/divalproex and lamotrigine at similar levels (Figure 3a ). In contrast, SNPs in the SLC35F3 and DISP1 loci were mainly associated with Alda scores for valproate/divalproex rather than lamotrigine (Figure 3b,c ). The signal at ABCC1 was mainly due to SNPs associated with Alda scores for valproate/divalproex, but it is notable that Alda scores for lamotrigine appear to be associated with SNPs in the neighboring ABCC6 gene instead (red peak in Figure 3d ).
Figure 3.
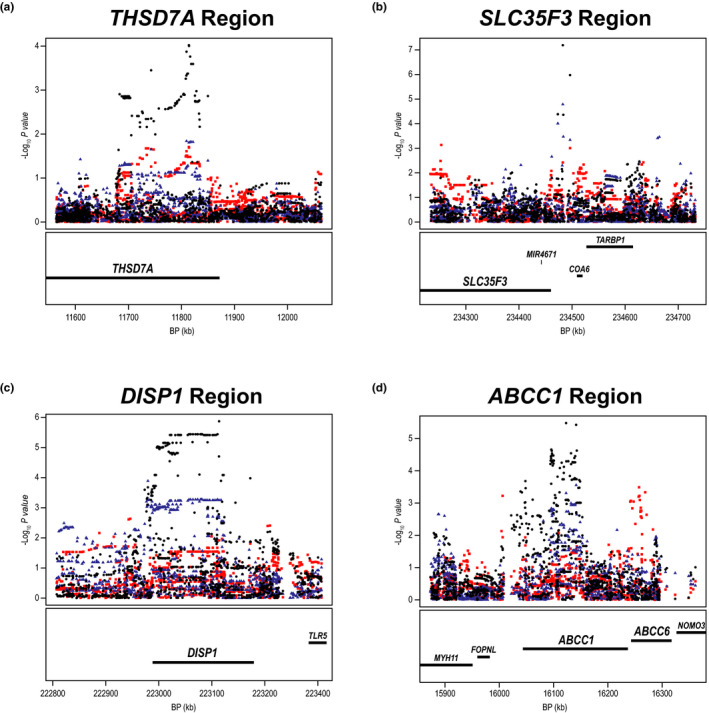
Drug‐stratified analyses of the top SNPs/genes (± 10 kb) associated with AED‐MS response in/near (a) THSD7A, (b) SLC35F3, (c) DISP1, and (d) ABCC1 showing allelic association with total Alda scores for valproate/divalproex (blue; n = 87), lamotrigine (red; n = 76), and combined AED‐MSs (black; n = 199). AED‐MS, antiepileptic drug–mood stabilizer; BP, base pair; kb, kilobase; SNP, single‐nucleotide polymorphism.
Functional Annotation of Genome‐wide Significant SNPs: rs78835388 and rs114872993
The two genome‐wide significant SNPs, rs78835388 and rs114872993, were relatively rare with MAF < 0.03 (Table 2 ); thus a large sample size is required for accurate eQTL analyses at these loci. Here, we used the GTEx project database, which currently includes eQTL analysis on various tissue types from 838 individuals. According to the GTEx database, THSD7A messenger RNA (mRNA) is predominantly expressed in the tibial nerve (Figure S3 a). The SNP rs78835388, which maps to intron 1 of THSD7A, is nominally associated with THSD7A mRNA levels in “Brain—Anterior Cingulate Cortex (BA24)” (P = 0.082, normalized effect size (NES) = −0.45, n = 147) and the association is statistically significant in tissues with a larger sample size such as “Nerve—Tibial” (P = 0.024, NES = −0.21, n = 532) and “Adipose—Subcutaneous” (P = 0.048, NES = −0.16, n = 581; Figure S3 b). SLC35F3 mRNA is predominantly expressed in brain tissues, while COA6 mRNA is expressed across different human tissues (Figure S4 a). The SNP rs114872993, which maps to an intergenic region between SLC35F3 and COA6 and is closer to the 3’‐downstream of SLC35F3, is associated with SLC35F3 mRNA levels in “Brain—Hippocampus” (P = 0.046, NES = −0.24, n = 165) and “Colon—Transverse” (P = 0.049, NES = 0.25, n = 368; Figure S4 b). This SNP was not significantly associated with COA6 mRNA expression in any human tissues.
Single‐cell RNA sequencing data from human brain samples 25 demonstrated that both THSD7A and SLC35F3 are highly expressed in neurons (Figure 4 ). Specifically, THSD7A is highly expressed in most types of inhibitory neurons and some types of excitatory neurons, while SLC35F3 is expressed in both inhibitory and excitatory neurons (Figure 4 ).
Figure 4.
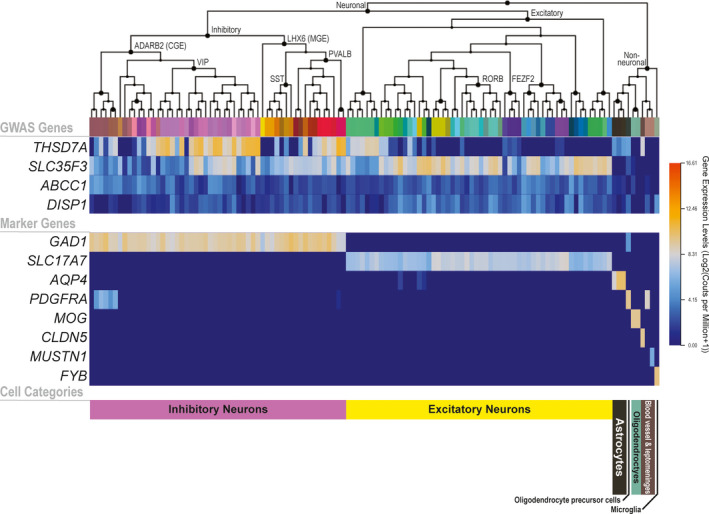
Heatmap for gene expression patterns of the four genome‐wide significant loci (THSD7A, SLC35F3, ABCC1, and DISP1) in various human brain cell types. RNA sequencing and transcriptomic clustering of sorted single‐cell nuclei from six human cortical regions were conducted by the Allen Institute of Brain Science. 25 The middle panel shows the gene expression patterns of cell type–specific marker genes for reference: GAD1 (glutamate decarboxylase 1) for inhibitory neurons; SLC17A7 (vesicular glutamate transporter 1) for excitatory neurons; AQP4 (aquaporin 4) for astrocytes; PDGFRA (platelet‐derived growth factor receptor A) for oligodendrocyte precursor cells and vascular and leptomeningeal cell; MOG (myelin oligodendrocyte glycoprotein) for oligodendrocytes; CLDN5 (claudin 5) for microvascular endothelial cells; MUSTN1 (musculoskeletal embryonic nuclear protein 1) for pericytes; FYB (FYN‐binding protein) for microglia. The lower panel shows general cell type categories. Increasing levels of gene expression are represented by a color gradient from dark blue to dark orange. The figure demonstrates that THSD7A is predominantly expressed in inhibitory neurons, while SLC35F3 is mostly expressed in excitatory neurons as well as in some inhibitory neurons; ABCC1 and DISP1 are expressed at much lower levels across the same cell types in comparison. The graphic was prepared using the Allen Brain Map Transcriptomics Explorer. 23
Functional annotation of genome‐wide significant genes: ABCC1 and DISP1
ABCC1 and DISP1 were significantly associated with AED‐MS treatment response at the gene level. According to GTEx, our most significantly associated SNP in ABCC1 (rs875740), which maps to intron 5 of ABCC1, is a strong sQTL for ABCC1 intron 6 (chr16:16,033,170‐16,036,472) in multiple gastrointestinal tissues (Figure S5 a), which might alter ABCC1‐mediated drug absorption. Similar to the ABCC1 SNP, the top DISP1 SNP (rs34701716) is a strong sQTL for DISP1 intron 1 (chr1:222,823,945‐222,835,119) in multiple human gut tissues (Figure S5 b). QTLbase search suggests that these top SNPs of DISP1 and ABCC1 are also methylation quantitative trait loci for intronic positions of DISP1 (rs34701716) and ABCC1 and ABCC6 (rs875740) in blood (Table S1 ), suggesting possible roles in regulating splice variants and gene expression through DNA methylation.
DISCUSSION
In this study, we performed the first GWAS for AED‐MS treatment response in BD patients and identified genome‐wide significant SNPs in the THSD7A gene and a locus near SLC35F3 (Figure 1a ). We also observed significant gene‐level associations of AED‐MS treatment response with ABCC1 and DISP1 (Figure 1b ). These results point to potential genetic markers for predicting AED‐MS treatment response in BD and provide novel candidate genes for further functional exploration of mechanisms underlying AED‐MS efficacy.
Neither THSD7A nor SLC35F3 have previously been reported to be involved in AED‐MS treatment response in BD, nor were they known to interact with AED‐MS compounds. THSD7A encodes N‐glycoprotein thrombospondin type 1 domain containing 7A, which mediates endothelial cell migration and tube formation in angiogenesis and is a potent neuroangiogenic factor. 27 Genetic polymorphisms in THSD7A were recently reported to be associated with BD in a large GWAS (rs113779084, P = 2.5E‐09), 28 and have also been implicated in schizophrenia 29 and BD. 28 , 29 , 30 Furthermore, SNPs in THSD7A have also been reported to be associated with plasma concentrations of kynurenine 31 and kynurenic acid, 32 two metabolites known to influence glutamatergic neurotransmission. 33 Finally, THSD7A protein expression has been shown to be altered in responders to antipsychotic drug therapy. 34 SLC35F3 encodes solute carrier family 35 member F3, which may transport thiamine, a vitamin that is crucial for normal brain functioning and has been implicated in brain disorders. 35 None of the AED‐MSs that were used to treat BD patients in this study are known to be substrates for SLC35F3, and no SLC35F3 substrates are currently known. The top SNPs in THSD7A and near SLC35F3 were associated with the respective genes’ mRNA expression in human brain and other tissues. Both THSD7A and SLC35F3 are expressed in glutamatergic and/or γ‐aminobutyric acidergic (GABAergic) neurons, which are thought to be targets for AED‐MS medications. 36 Our drug‐stratified association analysis at the THSD7A locus suggested association of THSD7A SNPs with response to both valproate/divalproex and lamotrigine (Figure 3a ), supporting the possibility that THSD7A might be related to pharmacodynamics for AED‐MSs.
Our gene‐level analyses demonstrated an association between AED‐MS response in BD and ABCC1 which encodes multidrug resistance‐associated protein 1 (MRP or MRP1), an efflux transporter that mediates ATP‐dependent transport of glutathione and glucuronide conjugates, methotrexate, and other xenobiotics. 37 The level of expression of this transporter might influence drug concentrations in target tissues/cells, resulting in variation in therapeutic efficacy. Previous pharmacogenetic studies have demonstrated the potential contribution of ABCC1 to variation in response to antidepressants and antipsychotics. 38 , 39 ABCC1 is thought to cause pharmacoresistance in the treatment of epilepsy, 40 but evidence that AEDs are ABCC1 substrates is inconsistent. 41 , 42 , 43 , 44 However, metabolites of those drugs could be substrates for ABCC1; for example, lamotrigine is not a substrate for ABCC1 but lamotrigine glutathione is. 45 Our findings that the ABCC1 rs875740 SNP variant allele (C), which is associated with altered splicing efficiency and DNA methylation of ABCC1 introns, was associated with better AED‐MS treatment response and that decreased ABCC1 gene expression was correlated with better AED‐MS treatment response support the notion that ABCC1 may influence the effect of AED‐MSs in the treatment of BD through pharmacokinetic mechanisms.
DISP1 also showed gene‐level significant association. It encodes the dispatched RND transporter family member 1, a membrane protein that is essential for normal Hedgehog signaling. 46 Microdeletion of a chromosomal region (1q41q42) that includes DISP1 was associated with developmental delay, seizures, and characteristic dysmorphic features. 46 The SNP rs17162912, which is proximal to the promoter region of DISP1, was reported to be associated with response to serotonin reuptake inhibitors in patients with obsessive‐compulsive disorder. 47 Future functional studies will be required to understand a possible role for DISP1 in AED‐MS response.
The main limitation of this study is the small sample size, which, together with the low MAFs of some of the top SNPs, means that some of the results might have been driven by a small number of patients carrying the effect alleles. Additionally, our analyses relied on retrospectively defined treatment outcomes, rather than a prospective assessment of treatment response. Nevertheless, the statistically significant associations with relevant genes support the possibility of important pharmacogenomic contributions to AED‐MS response, providing strong motivation for a larger GWAS in this area. The inclusion of multiple medications prevents the conclusion that the effects observed are necessarily medication specific. It remains possible that the findings may partially reflect general genetic effects on episode duration or other factors that contribute to treatment outcomes. However, the drug‐stratified analyses suggest that some of the observed associations are driven by genetic influences on response to specific medications. We also did not examine genetic effects on medication tolerability or side effects, which can contribute to treatment discontinuation and should be studied in the future. We did not exclude subjects with high Alda B scores whose A scores are considered less credible, but fewer than 6% of subjects in our sample had B scores > 4. Finally, because this study included only patients of European ancestry, the findings may not be generalizable to other populations.
In conclusion, we performed a pharmacogenomic GWAS of AED‐MS response in BD which revealed the association of AED‐MS treatment response with several loci, including THSD7A and ABCC1. These findings not only provide candidate genetic markers for predicting AED‐MS response in BD, but they also imply that individual genetic differences in the pharmacodynamics and pharmacokinetics of these medications may contribute to individual differences in their response. While our findings need to be replicated with larger samples, they offer key insights into AED‐MS pharmacogenomics and may contribute to the development of precision medicine for BD drug therapy.
Funding
Recruitment and genotyping for the Mayo Clinic Bipolar Disorder Biobank was supported by the Marriott Family Foundation and the Mayo Clinic Center for Individualized Medicine. This pharmacogenomics study was supported by a gift from Kent and Liz Dauten. R.M.W.’s effort was supported by NIH R01 AA27486.
Conflict of Interest
S.L.M. has been a consultant to or member of the scientific advisory board of Avanir, Bracket, F. Hoffmann‐La Roche Ltd., Idorsia, Mitsubishi Tanabe Pharma Corporation, Medibio, Myriad, Naurex, Neurocrine, Novo Nordisk, Opiant, Otsuka, Shire, Sunovion, and Takeda. She has also been a principal or coinvestigator on studies sponsored by Alkermes, Allergan, Avanir, Azevan Pharmaceuticals, Forest, Marriott Foundation, Myriad, National Institute of Mental Health, Naurex, Novo Nordisk, Otsuka, Shire, Sunovion, and Takeda Pharmaceutical Company Limited. She is also an inventor on United States Patent No. 6,323,236 B2, Use of Sulfamate Derivatives for Treating Impulse Control Disorders, and along with the patent’s assignee, University of Cincinnati, Cincinnati, Ohio, has received payments from Johnson and Johnson, which has exclusive rights under the patent. M.A.F. has received grant support from Assurex Health, Myriad, Pfizer, Marriott Foundation and Mayo Foundation; has been a consultant to Janssen Global Services, LLC, Mitsubishi Tanabe Pharma Corporation, Myriad, Sunovion, and Teva Pharmaceuticals; has received CME/Travel Support/presentation from CME Outfitters Inc. and Sunovion. B.S. had received research time support from Medibio. It is unrelated to the current study. R.M.W. is a cofounder of and a stockholder in OneOme LLC, a pharmacogenomics decision support company. All other authors declared no competing interests for this work.
Author Contributions
A.M.‐C.H., M.N., M.A.F., and J.M.B. designed the research. J.M.B., M.A.F., S.L.M., A.M.‐C.H., and B.S. performed the research. B.J.C., C.L.C., B.R.L., A.M.‐C.H., T.T.L.N., and D.L. analyzed the data. A.M.‐C.H., B.J.C., J.M.B., T.T.L.N., D.L., and R.M.W. wrote the manuscript.
Supporting information
Acknowledgments
We thank the participants in the Mayo Clinic Bipolar Disorder Biobank for their contribution to this research program. The Genotype‐Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by the National Cancer Institute (NCI), National Human Genome Research Institute (NHGRI), National Heart, Lung, and Blood Institute (NHLBI), National Institute on Drug Abuse (NIDA), National Institute of Mental Health (NIMH), and National Institute of Neurological Disorders and Stroke (NINDS). The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 03/25/20.
References
- 1. Gitlin, M. Lithium side effects and toxicity: prevalence and management strategies. Int. J. Bipolar Disord. 4, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frye, M.A. Clinical practice. Bipolar disorder—a focus on depression. N. Engl. J. Med. 364, 51–59 (2011). [DOI] [PubMed] [Google Scholar]
- 3. Rogawski, M.A. & Löscher, W. The neurobiology of antiepileptic drugs. Nat. Rev. Neurosci. 5, 553–564 (2004). [DOI] [PubMed] [Google Scholar]
- 4. Rogawski, M.A. & Löscher, W. The neurobiology of antiepileptic drugs for the treatment of nonepileptic conditions. Nat. Med. 10, 685–692 (2004). [DOI] [PubMed] [Google Scholar]
- 5. Stoll, A.L. & Severus, W.E. Mood stabilizers: shared mechanisms of action at postsynaptic signal‐transduction and kindling processes. Harv. Rev. Psychiatry 4, 77–89 (1996). [DOI] [PubMed] [Google Scholar]
- 6. Hou, L. et al Genetic variants associated with response to lithium treatment in bipolar disorder: a genome‐wide association study. Lancet 387, 1085–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee, H.‐Y. & Kim, Y.‐K. Catechol‐O‐methyltransferase Val158Met polymorphism affects therapeutic response to mood stabilizer in symptomatic manic patients. Psychiatry Res. 175, 63–66 (2010). [DOI] [PubMed] [Google Scholar]
- 8. Perlis, R.H. et al Genetic association study of treatment response with olanzapine/fluoxetine combination or lamotrigine in bipolar I depression. J. Clin. Psychiatry 71, 599–605 (2010). [DOI] [PubMed] [Google Scholar]
- 9. Manchia, M. et al Assessment of response to lithium maintenance treatment in bipolar disorder: A Consortium on Lithium Genetics (ConLiGen) report. PLoS One 8, e65636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frye, M.A. et al Development of a bipolar disorder biobank: differential phenotyping for subsequent biomarker analyses. Int. J. Bipolar Disord. 3, 30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. First, M.B. , Spitzer, R.L. , Gibbon, M. & Williams, J.B.W. Structured Clinical Interview for DSM‐IV‐TR Axis I Disorders, Research Version, Patient Edition with Psychotic Screen (SCID‐I/P W/ PSY SCREEN). In: Biometric Research (New York State Psychiatric Institute, New York, 2002).
- 12. Tighe, S.K. et al Test‐retest reliability of a new questionnaire for the retrospective assessment of long‐term lithium use in bipolar disorder. J. Affect. Disord. 174, 589–593 (2015). [DOI] [PubMed] [Google Scholar]
- 13. McElroy, S.L. et al Bipolar disorder with binge eating behavior: a genome‐wide association study implicates PRR5‐ARHGAP8. Transl. Psychiatry 8, 40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Purcell, S. et al PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. R Core Team . R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, 2013) <http://www.R‐project.org/> [Google Scholar]
- 16. Watanabe, K. , Taskesen, E. , van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Leeuw, C.A. , Mooij, J.M. , Heskes, T. & Posthuma, D. MAGMA: generalized gene‐set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fridley, B.L. & Biernacka, J.M. Gene set analysis of SNP data: benefits, challenges, and future directions. Eur. J. Hum. Genet. 19, 837–843 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gamazon, E.R. et al A gene‐based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Battle A. et al Characterizing the genetic basis of transcriptome diversity through RNA‐sequencing of 922 individuals. Genome Res. 24, 14–24 (2014). 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. GTex Consortium. The genotype‐tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. GTEx Consortium . GTEx Portal Release V8 <https://www.gtexportal.org/> (2020). Accessed March 16, 2020.
- 23. Allen Brain Map . Cell Types Database: RNA‐seq — Transcriptomics Explorer <http://celltypes.brain‐map.org/rnaseq/human_ctx_smart‐seq> (2020). Accessed March 25, 2020.
- 24. Hodge, R.D. et al Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tasic, B. et al Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng, Z. et al QTLbase: an integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res. 48, D983–D991 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuo, M.‐W. , Wang, C.‐H. , Wu, H.‐C. , Chang, S.‐J. & Chuang, Y.‐J. Soluble THSD7A is an N‐glycoprotein that promotes endothelial cell migration and tube formation in angiogenesis. PLoS One 6, e29000 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stahl, E.A. et al Genome‐wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bigdeli, T.B. et al Genome‐wide association study reveals greater polygenic loading for schizophrenia in cases with a family history of illness. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 276–289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Howard, D.M. et al Genome‐wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun. 9, 1470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu, D. et al Beta‐defensin 1, aryl hydrocarbon receptor and plasma kynurenine in major depressive disorder: metabolomics‐informed genomics. Transl. Psychiatry 8, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rhee, E.P. et al A genome‐wide association study of the human metabolome in a community‐based cohort. Cell Metab. 18, 130–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwarcz, R. , Bruno, J.P. , Muchowski, P.J. & Wu, H.‐Q. Kynurenines in the mammalian brain: when physiology meets pathology. Nat. Rev. Neurosci. 13, 465–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martins‐de‐Souza, D. , Solari, F.A. , Guest, P.C. , Zahedi, R.P. & Steiner, J. Biological pathways modulated by antipsychotics in the blood plasma of schizophrenia patients and their association to a clinical response. NPJ Schizophr. 1, 15050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Butterworth, R.F. Thiamin deficiency and brain disorders. Nutr. Res. Rev. 16, 277–284 (2003). [DOI] [PubMed] [Google Scholar]
- 36. Lee, Y. , Zhang, Y. , Kim, S. & Han, K. Excitatory and inhibitory synaptic dysfunction in mania: an emerging hypothesis from animal model studies. Exp. Mol. Med. 50, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cole, S.P.C. Multidrug resistance protein 1 (MRP1, ABCC1), a "multitasking" ATP‐binding cassette (ABC) transporter. J. Biol. Chem. 289, 30880–30888 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee, S.H. et al MRP1 polymorphisms associated with citalopram response in patients with major depression. J. Clin. Psychopharmacol. 30, 116–125 (2010). [DOI] [PubMed] [Google Scholar]
- 39. Piatkov, I. et al ABCB1 and ABCC1 single‐nucleotide polymorphisms in patients treated with clozapine. Pharmgenomics Pers. Med. 10, 235–242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Löscher, W. & Potschka, H. Drug resistance in brain diseases and the role of drug efflux transporters. Nat. Rev. Neurosci. 6, 591–602 (2005). [DOI] [PubMed] [Google Scholar]
- 41. Baltes, S. , Fedrowitz, M. , Tortós, C.L. , Potschka, H. & Loscher, W. Valproic acid is not a substrate for P‐glycoprotein or multidrug resistance proteins 1 and 2 in a number of in vitro and in vivo transport assays. J. Pharmacol. Exp. Ther. 320, 331–343 (2007). [DOI] [PubMed] [Google Scholar]
- 42. Luna‐Tortós, C. , Fedrowitz, M. & Löscher, W. Evaluation of transport of common antiepileptic drugs by human multidrug resistance‐associated proteins (MRP1, 2 and 5) that are overexpressed in pharmacoresistant epilepsy. Neuropharmacology 58, 1019–1032 (2010). [DOI] [PubMed] [Google Scholar]
- 43. Gibbs, J.P. , Adeyeye, M.C. , Yang, Z. & Shen, D.D. Valproic acid uptake by bovine brain microvessel endothelial cells: role of active efflux transport. Epilepsy Res. 58, 53–66 (2004). [DOI] [PubMed] [Google Scholar]
- 44. Ogawa, K. , Yumoto, R. , Hamada, N. , Nagai, J. & Takano, M. Interaction of valproic acid and carbapenem antibiotics with multidrug resistance‐associated proteins in rat erythrocyte membranes. Epilepsy Res. 71, 76–87 (2006). [DOI] [PubMed] [Google Scholar]
- 45. Chen, H. , Grover, S. , Yu, L. , Walker, G. & Mutlib, A. Bioactivation of lamotrigine in vivo in rat and in vitro in human liver microsomes, hepatocytes, and epidermal keratinocytes: characterization of thioether conjugates by liquid chromatography/mass spectrometry and high field nuclear magnetic resonance spectroscopy. Chem. Res. Toxicol. 23, 159–170 (2010). [DOI] [PubMed] [Google Scholar]
- 46. Roessler, E. et al Truncating loss‐of‐function mutations of DISP1 contribute to holoprosencephaly‐like microform features in humans. Hum. Genet. 125, 393–400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qin, H. et al Whole‐genome association analysis of treatment response in obsessive‐compulsive disorder. Mol. Psychiatry 21, 270–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


