Abstract
Chronic low-grade white adipose tissue (WAT) inflammation is a hallmark of metabolic syndrome in obesity. Here, we demonstrate that a subpopulation of murine WAT perivascular (PDGFRβ+) cells, termed “fibro-inflammatory progenitors” (FIPs), activate pro-inflammatory signaling cascades shortly after the onset of high-fat diet feeding and regulate pro-inflammatory macrophage accumulation in WAT in a TLR4-dependent manner. FIPs activation in obesity is mediated by the downregulation of ZFP423, identified here as a transcriptional co-repressor of NFκB. ZFP423 suppresses the DNA-binding capacity of the p65 subunit of NFκB by inducing a p300 to NuRD co-regulator switch. Doxycycline-inducible expression of Zfp423 in PDGFRβ+ cells suppresses inflammatory signaling in FIPs and attenuates metabolic inflammation of visceral WAT in obesity. Inducible inactivation of Zfp423 in PDGFRβ+ cells increases FIP activity, exacerbates adipose macrophage accrual, and promotes WAT dysfunction. These studies implicate perivascular mesenchymal cells as important regulators of chronic adipose tissue inflammation in obesity and identify ZFP423 as a transcriptional break on NFκB signaling.
Introduction
In the setting of chronic caloric excess, the expansion of visceral white adipose tissue (WAT) mass is often accompanied by the accumulation of pro-inflammatory macrophages. Adipose tissue macrophage (ATM) accrual is mediated by mechanisms promoting both macrophage recruitment 1,2 as well as macrophage proliferation 3. This unresolved low-grade meta-inflammatory state of visceral WAT is a defining feature of metabolic syndrome in obesity, and a contributor to the development of insulin resistance and vascular complications observed in type 2 diabetes 4.
Current efforts focus primarily on the role of adipocytes and classical immune cells in the development of metabolic WAT inflammation; however, the contribution of resident mesenchymal stromal cells in this regard have remained poorly defined. Our single-cell RNA sequencing efforts to define resident adipocyte precursor cells (APCs) revealed the existence of functionally distinct subpopulations of PDGFRβ+ perivascular cells within visceral WAT of adult mice 5. PDGFRβ+ marks the expression of mural cells (pericytes and vascular smooth muscle) as well as perivascular adventitial cells. PDGFRβ+ LY6C- CD9- cells are highly committed perivascular APCs and represent <20% of all PDGFRβ+ cells within murine visceral WAT. PDGFRβ+ LY6C+ cells are a distinct population of PDGFRβ+ cells that we refer to as “fibro-inflammatory progenitors” (herein, “FIPs”). FIPs display a fibrogenic, collagen-producing phenotype, lack adipogenic capacity, and can exert an anti-adipogenic effect on APCs through the production of secreted factors. Approximately 60–80% of all PDGFRβ+ cells within intra-abdominal WAT depots represent FIPs. Global gene expression profiling and unbiased pathway analysis of FIPs and APCs revealed that FIPs are enriched in the expression of genes related to classical pro-inflammatory signaling cascades, including genes encoding notable regulators of macrophage activation and accumulation in WAT. These data raise the hypothesis that FIPs can modulate macrophage homeostasis in WAT in vivo.
Here, we demonstrate that WAT FIPs are acutely activated under physiological conditions associated with metabolic inflammation. We present multiple inducible gain- and loss of function mouse models that indicate that the activation of inflammatory signaling pathways in FIPs is sufficient and necessary to drive sustained visceral adipose tissue macrophage accrual in obesity. Mechanistically, the activation of FIPs is mediated, at least in part, by the downregulation of the C2H2 zinc-finger transcriptional regulator, ZFP423, which we identify as a repressor of p65/NFκB activity in perivascular cells. These studies highlight an important role for perivascular mesenchymal cells in the regulation of chronic adipose tissue inflammation in obesity, and suggest pro-inflammatory stromal cells as possible targets for therapeutic interventions designed to promote healthy energy storage in adipose tissue.
Results
FIPs are physiologically activated in association with metabolic inflammation
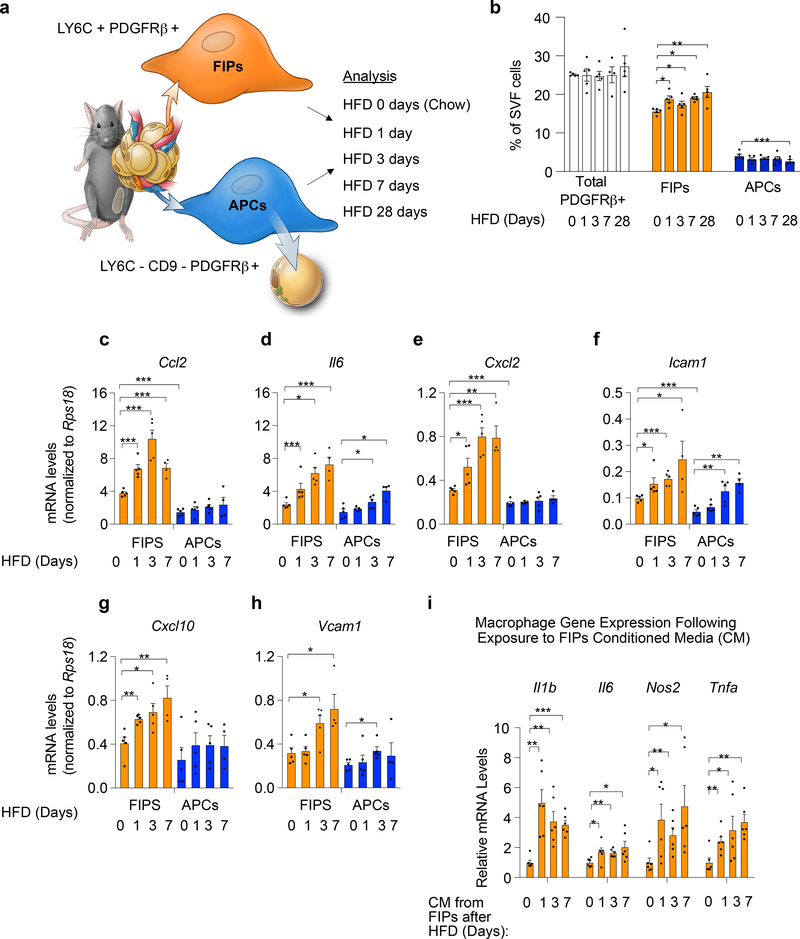
Given their abundance within the WAT stromal vascular fraction (SVF), we hypothesized that FIPs can sense pro-inflammatory stimuli and subsequently modulate WAT macrophage homeostasis in vivo. To begin to test this hypothesis, we examined the relative frequencies and gene expression profiles of FIPs and APCs in settings associated with metabolic inflammation of WAT. First, we quantified and isolated FIPs and APCs by FACS from epididymal WAT depots of 10 weeks-old mice maintained on either a chow diet or a high-fat diet (HFD) for 1, 3, or 7 days (Fig. 1a; Supplementary Fig. 1). Short-term HFD feeding led only to slight fluctuations in the relative proportions of FIPs and APCs; FIPs remained much more abundant than APCs during this time (Fig. 1b). 28 days of HFD feeding led to a statistically significant increase in the ratio of FIPs to APCs, with greater than 80% of PDGFRβ+ cells expressing markers of FIPs (Fig. 1b). Notably, short-term exposure to HFD led to a rapid and robust induction in mRNA levels of well-described pro-inflammatory chemokines (Ccl2, Cxcl2, Cxcl10), cytokines (Il6), and adhesion molecules (Icam1, Vcam1) within FIPs (Fig. 1c–h). On the contrary, mRNA levels of only some of the examined genes increased in the APCs during this time, and this occurred to a much lesser degree than observed in FIPs. We then asked whether the induction of these genes in FIPs following HFD feeding reflected a heightened functional pro-inflammatory phenotype. To test this, we exposed primary cultures of bone marrow-derived macrophages (BMDMs) to conditioned media collected from cultured FIPs obtained following HFD feeding. The expression levels of Il1b, Il6, Nos2, and Tnfa (indicators of activated macrophages), were elevated in the macrophages exposed to the conditioned media from FIPs harvested from HFD-fed mice (Fig. 1i). The activation of BMDMs is less robust when exposed to conditioned media from FIPs obtained from chow-fed mice. Collectively, these data provide evidence that FIPs exert a pro-inflammatory phenotype that is activated acutely upon HFD feeding.
Figure 1. FIPs are activated in association with adipose tissue inflammation.
a) Experimental design: APCs (LY6C- CD9- PDGFRβ+) and FIPs (LY6C+ PDGFRβ+) were isolated by FACS from gonadal WAT obtained from 10 weeks-old male C57BL/6 mice fed a standard chow diet (0 days HFD) or high-fat diet (HFD) for 1 day (HFD 1 day), 3 days (HFD 3 days) or 7 days (HFD 7 days) at 22 °C. Illustration is reproduced from Hepler et al. 5
b) Frequency of total PDGFRβ+ cells and PDGFRβ+ subpopulations within gonadal WAT at the indicated time points (days) following the onset of HFD-feeding. Data is presented as a percentage of all live SVF cells obtained from gonadal WAT.
c-h) Gene expression analysis of FIPs and APCs: mRNA levels of Ccl2 (c), Il6 (d), Cxcl2 (e), Icam1 (f), Cxcl10 (g) and Vcam1 (h) in FIPs and APCs isolated at the indicated time points. Representative data from a single experiment is shown.
Data in panels b-h were reproduced in 3 independent experiments. For representative experiment shown, n = 5 samples for HFD 0 days (chow). n = 5 for HFD 1 day. n = 5 for HFD 3 days. n = 4 for HFD 7 days. n=5 for HFD 28 days. Each sample (n) represents ~3×104 cells sorted from a pool of two fat depots from one mouse.
* denotes p< 0.05, ** denotes p<0.01, and *** denotes p<0.001 by one-way ANOVA.
i) Macrophage activation following exposure to FIPs conditioned media (CM): mRNA levels of genes associated with macrophage activation in cultured bone marrow-derived macrophages (BMDMs) following exposure to indicated FIPs conditioned media for 1.5 hours. n = 6 independent wells of macrophages examined per experiment.
Experiments in panel i were independently repeated 2 times. Representative data form two experiments is shown. * denotes p< 0.05, ** denotes p<0.01, and *** denotes p<0.001 by one-way ANOVA. In all panels, bars represent mean + s.e.m. Each dot represents value of individual measurement (n). Exact p values for each dataset can be found in Source Data Figure 1.
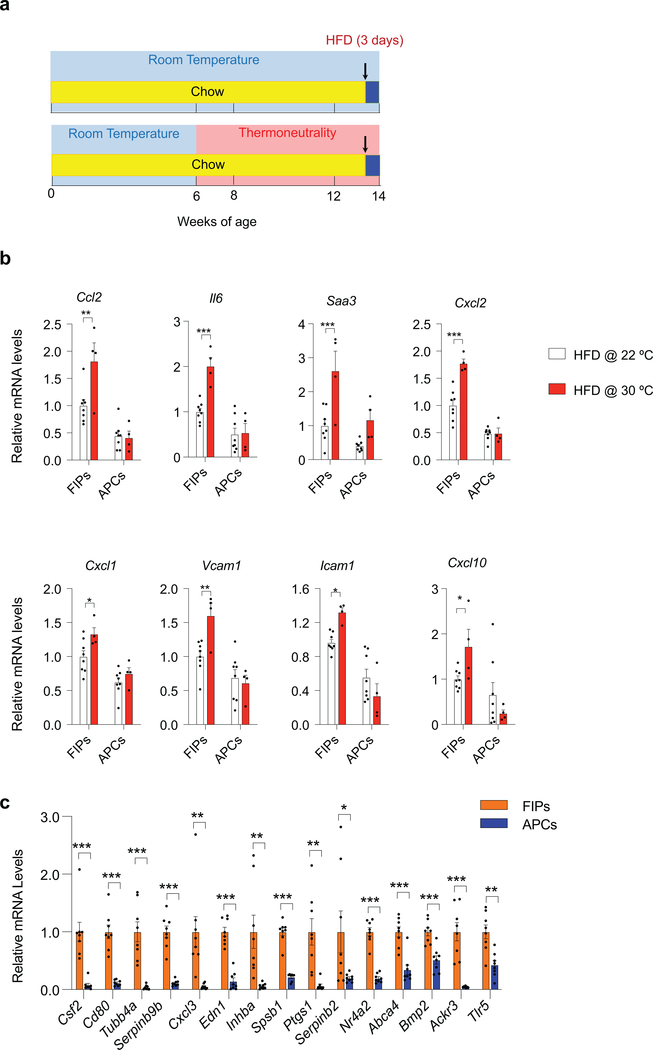
Thermoneutral (TN, 30°C) housing conditions accelerate the development of metabolic WAT inflammation in obesity 6,7. As such, we explored the effect of thermoneutrality on the inflammatory responses of FIPs (Extended Data Fig. 1a). mRNA levels of all of the inflammatory cytokines examined were higher in FIPs than in APCs under both housing conditions; however, FIPs isolated from animals maintained at TN expressed higher levels of inflammatory genes when compared to FIPs isolated from animals maintained at RT (Extended Data Fig. 1b). These data indicate that thermoneutral housing amplifies the activation of the pro-inflammatory phenotype of FIPs by HFD feeding, and provide further evidence that the pro-inflammatory phenotype of these cells is regulated physiologically.
We also performed unbiased bulk-RNA sequencing and Gene Set Enrichment Analysis of FIPs and APCs isolated from mice maintained on HFD at 30°C for 28 days. FIPs were enriched in a number of gene signatures associated with heightened pro-inflammatory signaling (Supplementary Table 1, 2). These data, along with our prior single-cell gene expression analysis, suggested that FIPs activate a number of classical pro-inflammatory signaling cascades that are implicated in the development of metabolic inflammation, including TNFα signaling, IL-6 signaling, and TLR (toll-like receptor) signaling 5 (Supplementary Table 1). Quantitative PCR analysis of FIPs and APCs isolated from HFD-diet fed mice validated the observed expression differences in the 15 most differentially expressed genes amongst the identified “HALLMARK_TNFA_SIGNALING_VIA_NFKB” signature.” All 15 signature genes reflective of heightened TNFα signaling were significantly and robustly enriched in FIPs when compared to APCs (Extended Data Fig. 1c).
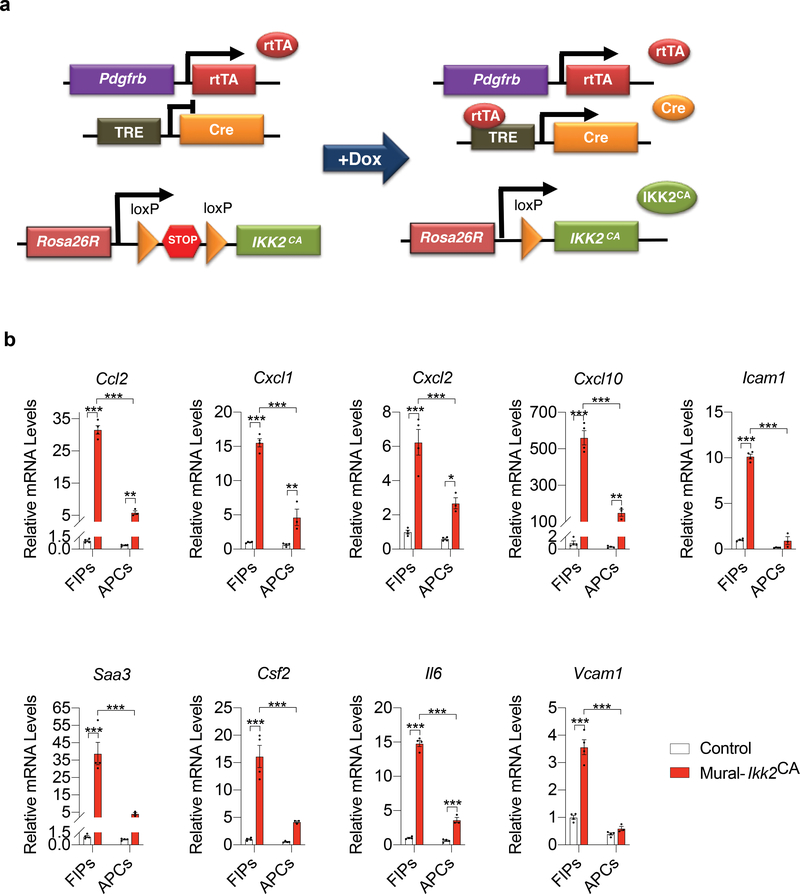
The NFκB signaling cascade is a classical signaling pathway that integrates many of the aforementioned pathways and is robustly activated within WAT upon HFD feeding 8,9. We asked whether the differential gene expression response of FIPs and APCs in pro-inflammatory settings might reflect an intrinsic difference in their potential to respond to NFκB activation. We derived a genetic “Tet-On” system in which the addition of doxycycline (“Dox”) results in expression of a constitutively active form of IKK2 (IKK2CA) in PDGFRβ+ cells (herein “Mural-Ikk2CA mice”) (Fig. 2a). IKK2CA expression leads to activation of NFκB signaling and is sufficient to drive the activation of several known NFκB transcriptional targets examined; however, genetic NFκB activation differentially impacts FIPs and APCs (Fig. 2b). mRNA levels of all of the genes encoding pro-inflammatory cytokines or adhesion molecules are induced much more robustly in FIPs than in APCs. These data provide additional evidence that FIPs are inherently more pro-inflammatory than APCs.
Figure 2. FIPs are more responsive than APCs to the activation of NFκB signaling.
a) Mural-Ikk2CA mice were derived by breeding PdgfrbrtTA transgenic mice to animals expressing Cre recombinase under the control of the tetracycline-response element (TRE-Cre) and carrying the Rosa26RIkk2CA allele. Littermates carrying only PdgfrbrtTA and Rosa26RIkk2CA alleles (i.e. Cre−) were used as the control animals (Control). The addition of doxycycline (Dox) leads to activation of the Rosa26RIkk2CA allele and constitutively active (CA) IKK2 in Pdgfrb-expressing cells. Constitutively active IKK2 expression leads to activation of NFκB signaling.
b) mRNA levels of indicated pro-inflammatory genes in cultured FIPs and APCs 1 day following the addition of doxycycline (1μg/ml) to culture medium. Data presented were reproduced in two independent experiments. Bars represent mean ± s.e.m., * denotes p< 0.05, ** denotes p<0.01, and *** denotes p<0.001 by two-way ANOVA. Exact p values can be found in Source Data Figure 2.
TLR4 signaling in PDGFRβ+ cells regulates WAT macrophage accrual in obesity
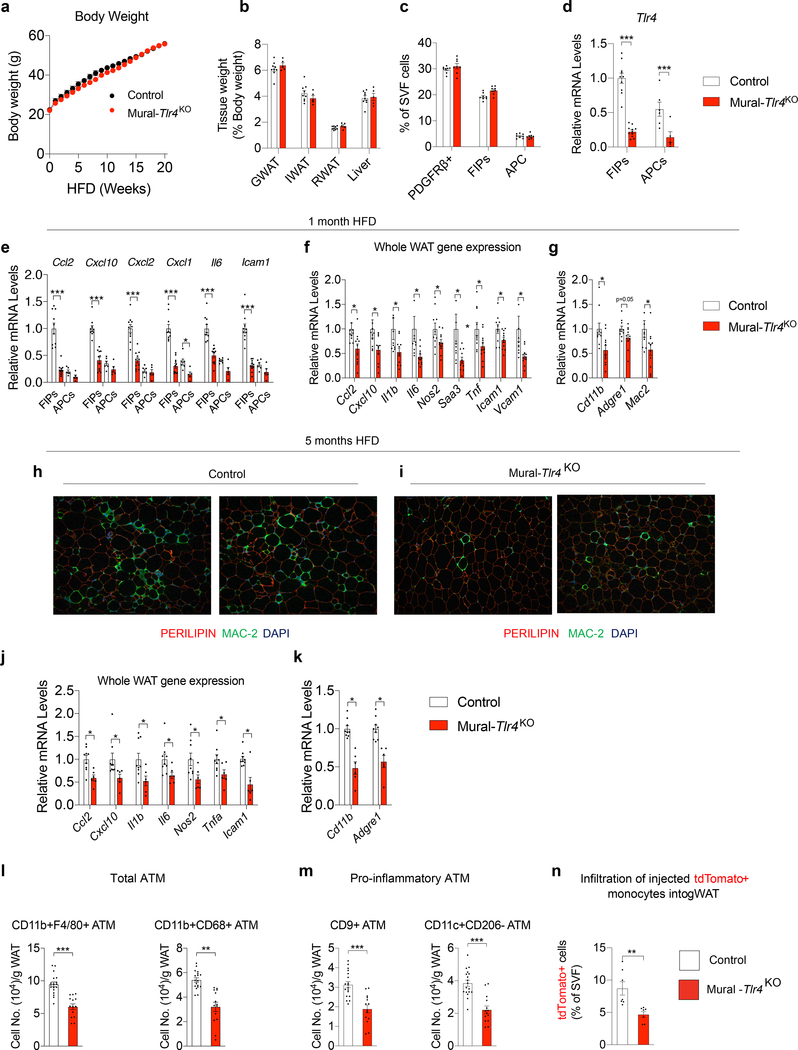
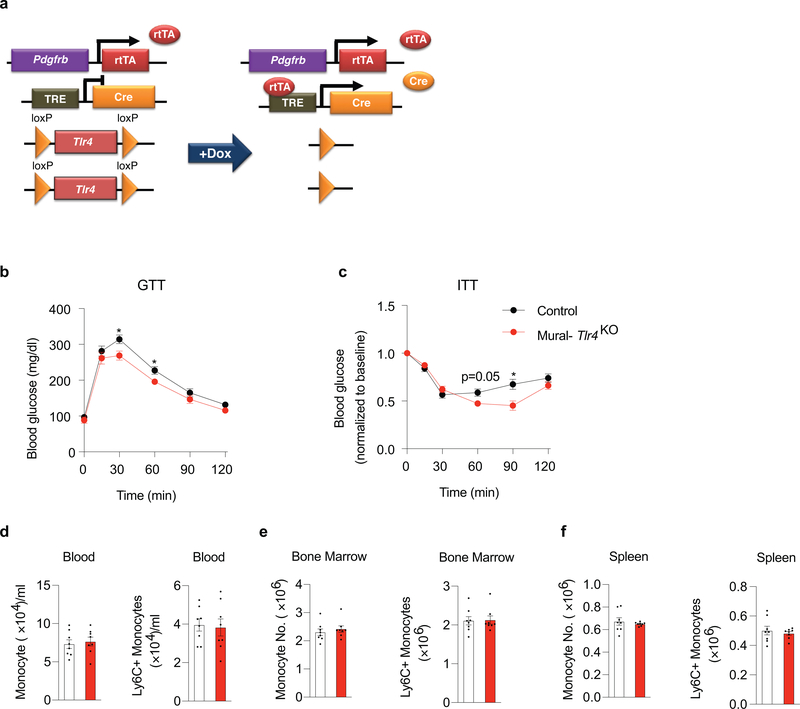
Whether the activation of pro-inflammatory signaling pathways in PDGFRβ+ cells impacts the development of metabolic WAT inflammation associated with obesity has remained unclear. To address this, we derived a loss-of-function model in which Tlr4 can be inactivated in PDGFRβ+ cells in a doxycycline-inducible manner (PdgfrbrtTA; TRE-Cre; Tlr4loxP/loxP; herein, “Mural-Tlr4KO“ mice) (Extended Data Fig. 2a). 8 weeks-old male Control [PdgfrbrtTA; Tlr4loxP/loxP mice (i.e. Cre -)] and Mural-Tlr4KO mice were switched to a doxycycline-containing HFD (Dox-HFD) diet for up to 5 months and maintained at room temperature. Thus, Cre expression is initiated at the onset of HFD feeding and maintained for the duration of the experiment. Over the 5-month period, body weights and WAT mass remained indistinguishable between the two groups (Fig. 3a,b). One month after the onset of Dox-HFD feeding, the frequencies of FIPs, APCs, and total PDGFRβ+ cells, were not impacted by the inactivation of Tlr4 (Fig. 3c). Gene expression analysis of freshly isolated FIPs and APCs confirmed the suppression of Tlr4 mRNA levels in both PDGFRβ+ subpopulations (Fig. 3d). The loss of Tlr4 specifically in PDGFRβ+ cells was associated with a statistically significant reduction in mRNA levels of many of the pro-inflammatory genes assayed within isolated FIPs, but not in APCs (Fig. 3e). Notably, levels of the pro-inflammatory transcripts analyzed with Tlr4-deficient FIPs were reduced to levels normally found in APCs. Moreover, loss of pro-inflammatory gene expression in FIPs is associated with reduced levels of notable pro-inflammatory cytokines and macrophage markers measured across adipose tissue as a whole (Fig. 3f,g).
Figure 3. Loss of Tlr4 in PDGFRβ+ cells attenuates WAT macrophage accumulation in obesity.
a) Control (n=7; black) and Mural-Tlr4KO (n=9; red) body weights following the onset of HFD feeding at room temperature.
b) WAT and liver mass of Control (n=9) and Mural-Tlr4KO (n=5) mice after 5 months of HFD feeding.
c) Frequency of total PDGFRβ+ cells, FIPs, and APCs, within gonadal WAT (gWAT) of Control (n=8) and Mural-Tlr4KO (n=8) mice after 1 month of HFD feeding.
d) Tlr4 mRNA levels within FIPs and APCs of Control (n=10) and Mural-Tlr4KO (n=11) mice after 1 month of HFD feeding. For Controls: FIPs n=10; APCs n=6. For Mural-Tlr4KO: FIPs n=11; APCs n=5.
e) Pro-inflammatory gene expression within gWAT FIPs and APCs of Control (n=10) and Mural-Tlr4KO (n=11) mice after 1 month of HFD feeding. For Controls: FIPs n=10; APCs n=6. For Mural-Tlr4KO: FIPs n=11; APCs n=5.
f) Pro-inflammatory gene expression in gWAT of Control (n=10) and Mural-Tlr4KO (n=11) mice after 1-month HFD feeding.
g) mRNA levels of macrophage-selective genes in gWAT of Control (n=10) and Mural-Tlr4KO (n=11) mice after 1-month HFD feeding.
h,i) 10x magnification images of PERILIPIN (red) and MAC-2 (green) expression in gWAT section from Control mice (h) and Mural-Tlr4KO mice (i) maintained on HFD for 5 months.
j) Pro-inflammatory genes expression in gWAT of Control (n=9) and Mural-Tlr4KO (n=6) mice after 5-months HFD feeding.
k) mRNA levels of macrophage-selective genes in gWAT of Control (n=9) and Mural-Tlr4KO (n=6) mice after 5-months HFD feeding.
l) Frequency of total adipose tissue macrophages (ATMs) within gWAT of Control (n=18) and Mural-Tlr4KO (n=14) mice after 5 months of HFD-feeding.
m) Frequency of pro-inflammatory ATMs within gWAT of Control (n=18) and Mural-Tlr4KO (n=14) mice after 5 months of HFD-feeding.
n) Frequency of tdTomato+ macrophages within gWAT SVF of obese mice following injection of tdTomato+ monocytes. n=7 for Control and n=7 for Mural-Tlr4KO mice.
Bars represent mean ± s.e.m., *p< 0.05, **p<0.01 or ***p<0.001 by unpaired two-tailed Student’s t-test (f,g, j-n) or one-way ANOVA (d, e). Exact p values and numbers of repetitions can be found in Source Data Figure 3.
Differences in the degree of systemic glucose homeostasis and gonadal adipose inflammation between the two genotypes were even more readily apparent after long-term HFD feeding. Mural-Tlr4KO mice were relatively more glucose tolerant and insulin sensitive than Control animals (Extended Data Fig. 2b,c). We observed fewer MAC-2+ immune cells and crown-like structures within gonadal WAT of Mural-Tlr4KO mice when compared to gonadal WAT of Control animals (Fig. 3h,i). In addition, mRNA levels of all pro-inflammatory cytokine/chemokines and macrophage selective genes examined were markedly lower in WAT of Mural-Tlr4KO mice (Fig. 3j,k). We analyzed the frequency of ATMs directly by flow cytometry, using multiple cell labeling strategies for ATM subtypes (Supplementary Fig. 2) 10,11. The total number of ATMs in Tlr4-deficient gonadal WAT was significantly lower than observed in gonadal WAT of Control mice (Fig. 3l). In particular, the numbers of pro-inflammatory ATMs within transgenic gonadal WAT were ~50% lower than observed in Controls (Fig. 3m). Notably, frequencies of pro-inflammatory CD9+ ATMs, which typically reside within crown-like structures of metabolically inflamed WAT 11, were strongly reduced within the gonadal WAT of transgenic mice. Importantly, levels of total and pro-inflammatory monocytes in the blood, bone marrow, and spleen were not impacted by the loss of Tlr4 in PDGFRβ+ cells (Extended Data Fig. 2d–f). This strongly suggests that the reduction in WAT macrophage accumulation in Mural-Tlr4KO mice is not secondary consequence to an altered monocyte/macrophage pool in another tissue. To confirm this, we assayed immune cell migration into WAT depots of obese mice following injection of labeled monocytes. Injected tdTomato+ monocytes migrate into WAT depots of HFD-fed Control animals and adopt an adipose tissue macrophage phenotype 2. In the Mural-Tlr4KO mice, the amount of monocyte migration into WAT is significantly lower than observed in Controls (Fig. 3n). These data highlight an important role for TLR4 signaling in mediating the pro-inflammatory response of FIPs, and provide evidence that pro-inflammatory signaling in PDGFRβ+ cells is necessary to drive sustained ATM accrual in obesity.
ZFP423 suppresses the pro-inflammatory phenotype of FIPs
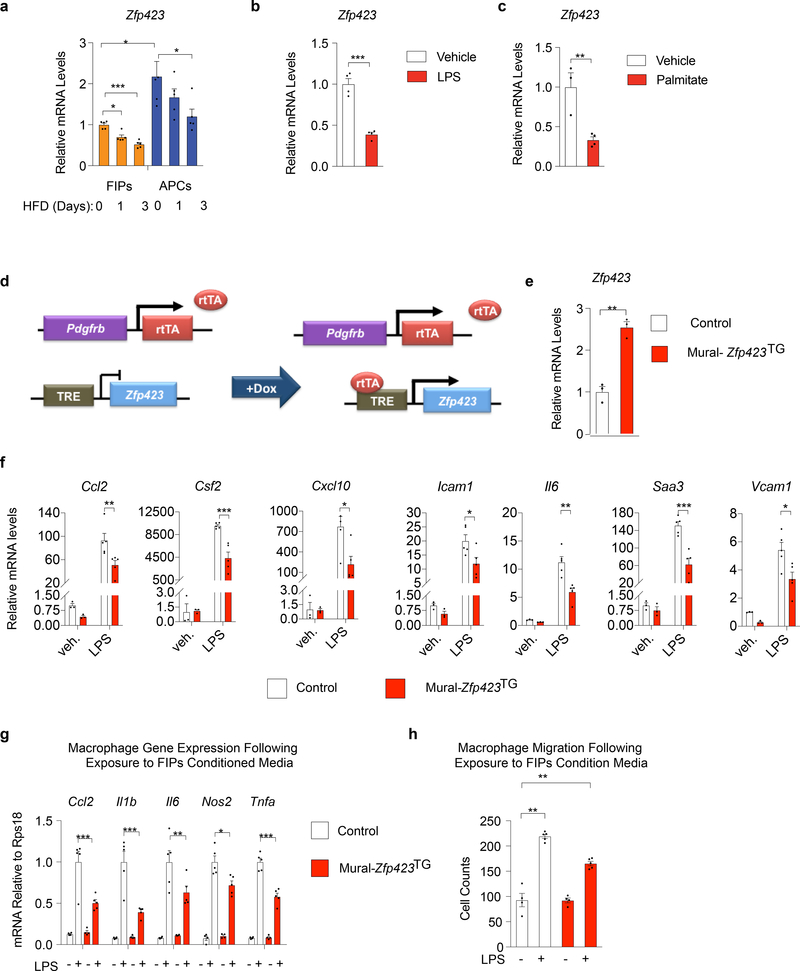
Zfp423 expression is one defining feature of committed preadipocytes in the perigonadal WAT of mice 5,12,13. Zfp423 encodes a multi-C2H2 zinc-finger transcriptional regulator that plays multiple roles in the establishment and maintenance of the adipocyte lineage 12,14–17. Using transgenic Zfp423GFP reporter mice, we previously determined that PDGFRβ+ cells expressing high mRNA levels of Zfp423 were enriched in markers of APCs, whereas cells expressing relatively lower levels of Zfp423 were enriched in markers of FIPs and gene signatures associated with inflammatory signaling 13. Our RNA-seq analysis described above was unable to confirm the enrichment of Zfp423 in APCs; however, quantitative PCR analysis with normalization to several housekeeping genes established that Zfp423 expression is enriched in APCs of mice maintained on a HFD at TN for 28 days (Supplementary Figure 3). FIPs express detectable levels of Zfp423; however, mRNA levels in these cells are quantitatively lower than observed in APCs. Interestingly, mRNA levels of Zfp423 were further reduced in association with the activation of the pro-inflammatory signaling pathways in FIPs. Within 1–3 days of HFD feeding, expression of Zfp423 declines in FIPs (Fig. 4a), in association with the aforementioned induction in mRNA levels of pro-inflammatory cytokines (see Fig. 1c–h). FIPs treated directly with LPS or palmitate expressed lower levels of Zfp423 when compared to vehicle treated cells (Fig. 4b,c). These data raise the hypothesis that ZFP423 exerts an anti-inflammatory effect in perivascular cells, with the downregulation of Zfp423 expression serving as a mechanism to activate the pro-inflammatory phenotype of these cells.
Figure 4. Zfp423 suppresses the pro-inflammatory phenotype of FIPs.
a) mRNA levels of Zfp423 in FIPs and APCs from gWAT of mice after HFD feeding for 0 days (chow) (n=5), 1 day (n=5), and 3 days (n=5).
b) mRNA levels of Zfp423 in primary FIPs cultures treated with vehicle (PBS) (n=4) or 100 ng/ml LPS (n=4) for 6 hours.
c) mRNA levels of Zfp423 in primary FIPs cultures treated with vehicle (BSA) (n=3) or 500 nmol/l palmitate (n=5) for 3 hours.
d) Mural-Zfp423TG (PdgfrbrtTA; TRE-Zfp423) mice were generated by breeding the PdgfrbrtTA transgenics to animals expressing Zfp423 under the control of the tetracycline-response element (TRE-Zfp423). Littermates carrying only PdgfrbrtTA were used as the controls (Control). In the presence of doxycycline, rtTA activates Zfp423 expression in Pdgfrb-expressing cells.
e) mRNA levels of Zfp423 within primary gWAT FIPs cultures from lean Control and Mural-Zfp423TG mice 24 hours after the addition of doxycycline (1μg/ml) to culture medium. FIPs were isolated from pooled depots from 5 mice and then plated immediately into triplicate wells for analysis (n=3 per group).
f) mRNA levels of indicated genes in primary FIPs cultures treated with vehicle (veh.) (n=3) or 100 ng/ml LPS (n=5) for 2 hours.
g) mRNA levels of genes associated with macrophage activation in cultured BMDMs following exposure to indicated FIPs conditioned media (CM) for 1.5 hours.
h) Macrophage migration following exposure to FIPs conditioned media (CM): cell counts of migrated macrophages following exposure to indicated CM for 3 hours.
For panels g,h FIPs (isolated from pooled depots of 6–8 mice per genotype) were treated with vehicle or LPS (100 ng/ml) for 2 hours and then incubated in serum-free medium for an additional 24 hours to produce conditioned media. n=4 (for groups with vehicle treatment) or n=5 (for groups with LPS treatment) independent wells of macrophages examined per experiment.
Experiments were independently repeated three times (a-c,e,f) or twice (g,h). Data in this figure are shown as the mean ± s.e.m., *p< 0.05, **p<0.01 or ***p<0.001 by two-tailed unpaired Student’s t-test (b,c), one-way ANOVA (a) or two-way ANOVA (f-h). Exact p values can be found in Source Data Figure 4.
We derived a genetic gain-of-function model in which Zfp423 expression can be induced in PDGFRβ+ cells in a doxycycline-inducible manner (PdgfrbrtTA; TRE-Zfp423; herein “Mural-Zfp423TG” mice) (Fig. 4d). We first explored the functional consequences of Zfp423 overexpression in vitro, using cultured FIPs isolated from gonadal WAT of chow-fed Control mice (mice carrying PdgfrbrtTA allele) and Mural-Zfp423TG mice. The addition of doxycycline to cultured FIPs led to a 2–3 fold induction in overall Zfp423 mRNA levels (Fig. 4e). This modest induction was sufficient to attenuate LPS-driven activation of pro-inflammatory gene expression (Fig. 4f). Zfp423 overexpression also diminished the capacity for FIPs to promote macrophage activation and migration. BMDMs exposed to conditioned media from LPS-treated Mural-Zfp423TG FIPs expressed relatively lower levels of Ccl2, Il1b, Il6, Nos2, and Tnfa than BMDMs exposed to conditioned media from LPS-treated Control FIPs (Fig. 4g). Moreover, the migratory response of BMDMs to conditioned media from LPS-treated Mural-Zfp423TG FIPs was attenuated in comparison to the response elicited by conditioned media from LPS-treated Control FIPs (Fig. 4h). Thus, ZFP423 overexpression leads to a suppression of the FIPs pro-inflammatory phenotype.
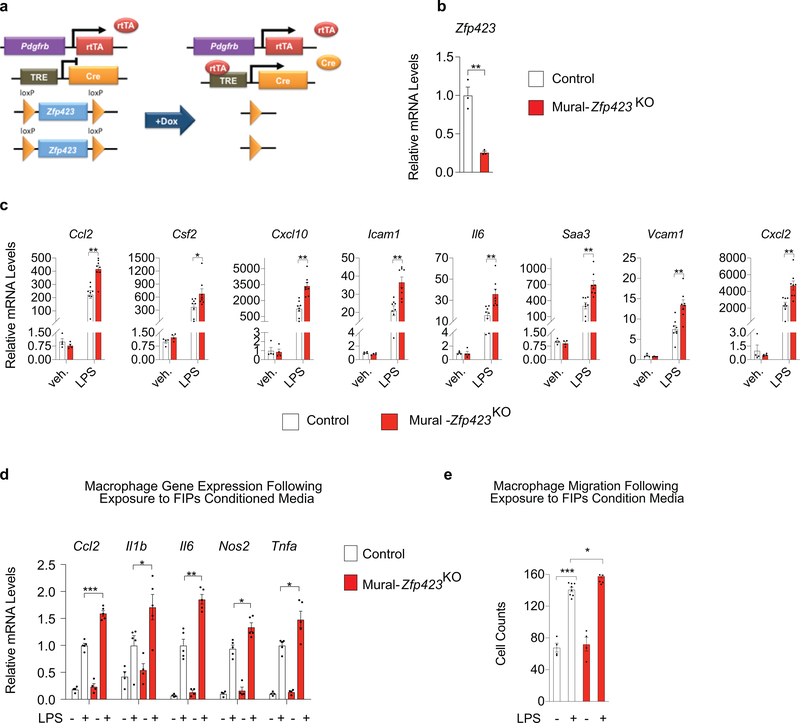
We also examined whether inactivation of Zfp423 can amplify the pro-inflammatory phenotype of FIPs. We utilized our previously reported model of doxycycline-inducible Zfp423 ablation in Pdgfrb-expressing cells (PdgfrbrtTA; TRE-Cre; Zfp423loxP/loxP; herein, “Mural-Zfp423KO” mice) (Extended Data Fig. 3a) 15. After 7 days of doxycycline-containing chow diet feeding, mRNA levels of Zfp423 are reduced by ~75% in isolated FIPs from Mural-Zfp423KO mice (Extended Data Fig. 3b). Zfp423 inactivation in FIPs led to a heightened pro-inflammatory phenotype in vitro. In response to LPS, Zfp423-deficient FIPs expressed higher mRNA levels of notable pro-inflammatory genes than LPS-treated Control FIPs (Extended Data Fig. 3c). BMDMs exposed to conditioned media from LPS-treated Mural-Zfp423KO FIPs exhibited a more “activated” gene expression profile in comparison to BMDMs exposed to conditioned media from LPS-treated Control FIPs (Extended Data Fig. 3d). The migratory response of BMDMs to conditioned media from LPS-treated Control FIPs is already fairly robust; however, the response to conditioned media from LPS-treated Mural-Zfp423KO FIPs was slightly, but reproducibly, elevated (Extended Data Fig. 3e). Taken together, these Zfp423 gain- and loss of function studies indicate that Zfp423 suppresses the pro-inflammatory phenotype of FIPs in a cell-autonomous manner.
Zfp423 overexpression in PDGFRβ+ cells attenuates LPS-driven WAT inflammation
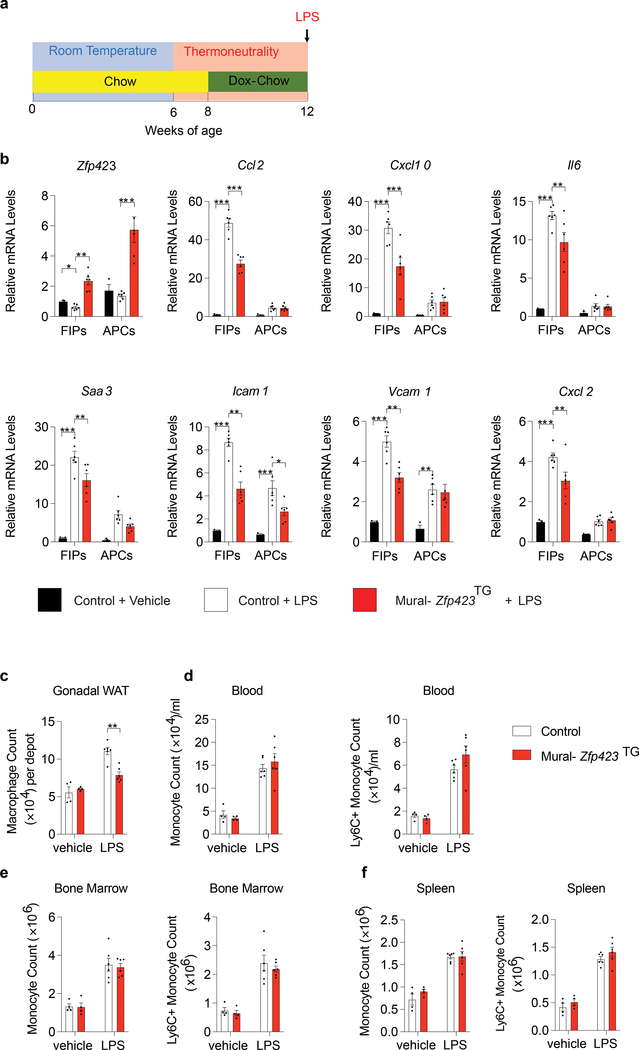
Next, we sought to determine whether ZFP423 regulates the pro-inflammatory phenotype of native FIPs in vivo, and whether modulating Zfp423 expression can impact the development of WAT inflammation. To this end, we utilized mice that were maintained at thermoneutrality due to the heightened inflammatory response FIPs exhibit under these housing conditions. Control and Mural-Zfp423TG male mice were switched to a doxycycline-containing chow diet for 4 weeks before being administered a single dose of LPS (Fig. 5a). LPS induced pro-inflammatory gene expression within gonadal WAT FIPs of Control mice. The overall response was more robust in FIPs than in APCs, providing additional evidence that FIPs exert a stronger transcriptional response to pro-inflammatory signals than APCs (Fig. 5b). Four weeks of doxycycline treatment led to an increase in overall Zfp423 mRNA levels within both FIPs and APCs of gonadal WAT of Mural-Zfp423TG mice (Fig. 5b). Importantly, transgene activation increased Zfp423 mRNA levels in FIPs to levels typically observed in APCs of Control mice. This modest level of Zfp423 induction was sufficient to significantly blunt the transcriptional response to LPS within FIPs, whole gonadal WAT, and whole retroperitoneal WAT, from Mural-Zfp423TG mice (Fig. 5b; Supplementary Fig. 4a,c). On the contrary, the expression levels of assayed pro-inflammatory genes in the liver were not impacted by transgenic expression of Zfp423 in PDGFRβ+ cells (Supplementary Fig. 4e). 24 hours post-LPS injection, the expression of macrophage-selective genes is elevated within WAT depots and livers of Control animals (Supplementary Fig. 4b,d,f); however, the induction of these same genes is less robust in the WAT depots, but not in the livers, of Mural-Zfp423TG mice (Supplementary Fig. 4b,d,f). Flow cytometry analysis revealed that the numbers of CD45+ CD11b+ F4/80+ ATMs were lower within the gonadal WAT depot of Mural-Zfp423TG mice than observed in the Control tissues (Fig. 5c). These changes occurred in the absence of changes in circulating blood monocytes, resident bone marrow monocytes, or splenic monocytes (Fig. 5d–f). These results confirm the anti-inflammatory function of Zfp423 in native FIPs in vivo, and provide evidence that inflammatory signaling in PDGFRβ+ cells contribute to the overall response of WAT to pro-inflammatory stimuli.
Figure 5. Zfp423 overexpression in PDGFRβ+ cells limits LPS-induced WAT inflammation.
a) Experimental design: Mural-Zfp423TG or Control mice were maintained at room temperature (RT) and then switched to thermoneutral housing conditions (TN) at the age of 6 weeks. 8-week old mice were administrated with Doxycycline-containing chow diet (Dox-Chow) for another 4 weeks before being i.p. injected with LPS (0.5 mg/Kg body weight) or vehicle (PBS). Gonadal WAT was harvested either 2 hours (b) or 24 hours (c-f) post-injection.
b) mRNA levels of Zfp423 and indicated pro-inflammatory genes in freshly isolated FIPs and APCs. n=3 individual mice for Control + Vehicle group; n=6 for Control + LPS group; and n=6 for Mural-Zfp423TG+LPS group.
c) Flow cytometry analyses of CD45+ CD11b+ F4/80+ macrophages in gonadal WAT.
d) Frequency of total monocytes (CD45+ CD11b+ CD115+) and pro-inflammatory monocytes (CD45+ CD11b+ CD115+ Ly6C+) in blood.
e) Frequency of total monocytes and pro-inflammatory monocytes in bone marrow.
f) Frequency of total monocytes and pro-inflammatory monocytes in spleen.
For c-f, n=4 individual mice for Control + Vehicle group; n=4 individual mice for Mural-Zfp423TG + Vehicle group; n=6 for Control + LPS group; and n=6 for Mural-Zfp423TG + LPS group.
All data are shown as the mean ± s.e.m. For b,c *p< 0.05, **p<0.01 or ***p<0.001 by two-way ANOVA. Exact p values can be found in Source Data Figure 5. All data presented were reproduced in two independent animal cohorts.
Zfp423 overexpression in PDGFRβ+ cells attenuates metabolic WAT inflammation
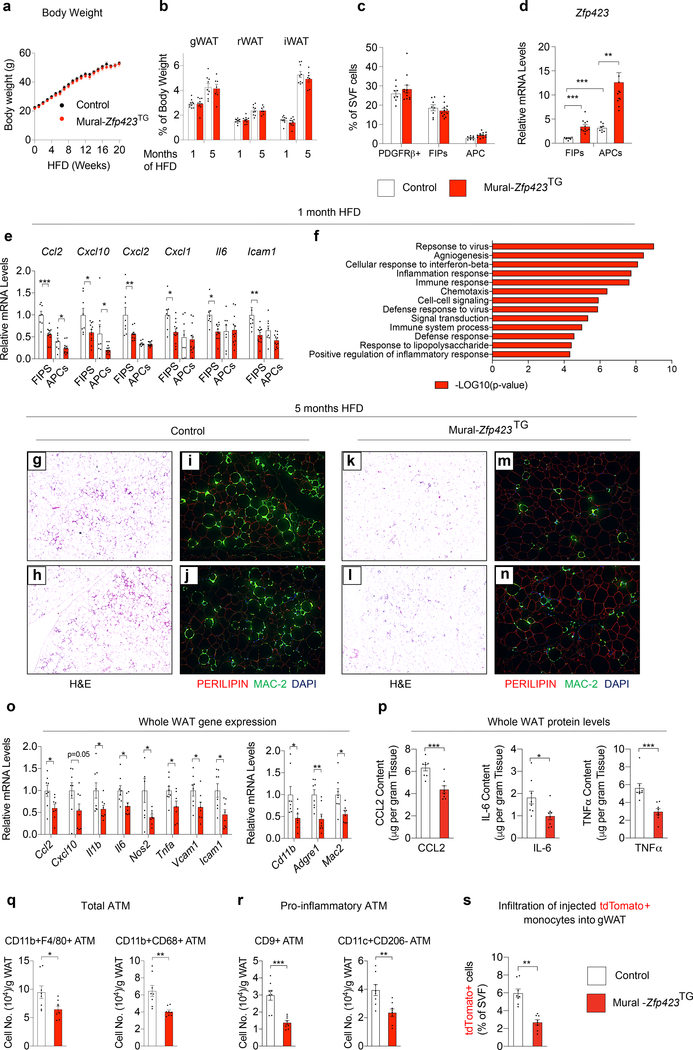
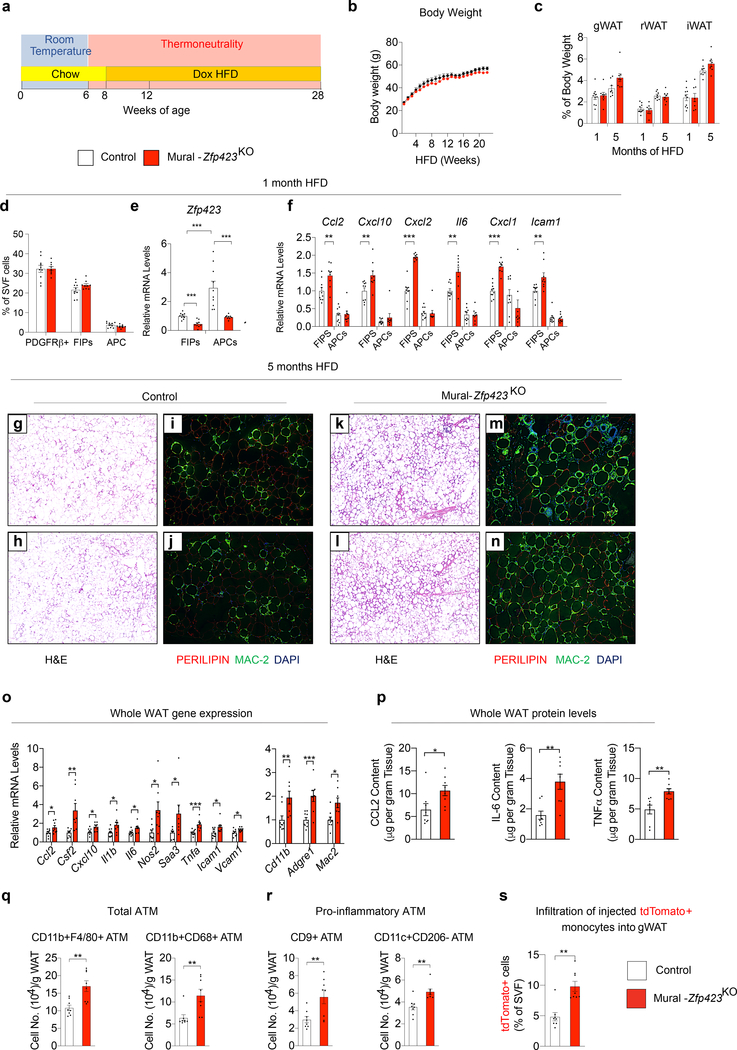
We also examined whether overexpression of Zfp423 in PDGFRβ+ cells can suppress the pro-inflammatory response of FIPs associated with HFD feeding, and elicit a phenotype similar to that of obese Mural-Tlr4KO mice. Control and Mural-Zfp423TG male mice were switched to a Dox-HFD diet for up to 5 months. Over this period, body weights remained indistinguishable between the two groups (Fig. 6a). One month after the onset of HFD feeding, adipose tissue mass and the the frequencies of the PDGFRβ+ subpopulations did not significantly differ between Control and Mural-Zfp423TG mice (Fig. 6b,c). Total levels of Zfp423 mRNA were increased ~3–4 fold in both FIPs and APCs, with Zfp423 in FIPs reaching levels observed normally in APCs (Fig. 6d). This modest, yet physiological, level of Zfp423 overexpression for 4 weeks of HFD feeding led to a significant reduction in expression of many of the pro-inflammatory genes assayed within FIPs. The mRNA levels of these transcripts were now reduced to levels observed naturally in APCs. The effects on gene expression in transgenic APCs were much more modest (Fig. 6e). Unbiased bulk RNA-seq analysis of Control and Zfp423-transgenic FIPs confirmed and extended these findings. Amongst the most differentially expressed gene signatures in Zfp423-overexpressing FIPs after 1 month of HFD feeding were several of those related to pro-inflammatory signaling (Fig. 6f; Supplementary Table 3,4).
Figure 6. Zfp423 overexpression in PDGFRβ+ cells attenuates WAT inflammation in obesity.
a) Control (n=9; black) and Mural-Zfp423TG (n=7; red) body weights following the onset of HFD feeding at thermoneutrality.
b) WAT mass of Control and Mural-Zfp423TG mice after 1 month (n=10 for Control; n=9 for Mural-Zfp423TG) and 5 months (n=9 for Control; n=7 for Mural-Zfp423TG) of HFD feeding.
c) Frequency of total PDGFRβ+ cells, FIPs, and APCs, within gonadal WAT (gWAT) of Control (n=8) and Mural-Zfp423TG (n=12) mice after 1 month of HFD feeding.
d) Zfp423 mRNA levels within gWAT FIPs and APCs from Control (n=8) and Mural-Zfp423TG (n=12) mice after 1 month of HFD feeding.
e) Pro-inflammatory gene expression in gWAT FIPs and APCs from Control (n=8) and Mural-Zfp423TG (n=12) mice after 1 month of HFD feeding.
f) Pathway analysis of genes with differential expression (FDR q-value<0.05 and Log2 fold change>0.5) between Control and Mural-Zfp423TG FIPs after 1 month of HFD feeding.
g,h) 4x magnification images of H&E stained gWAT from Control mice maintained on HFD for 5 months.
i,j) 10x magnification images of PERILIPIN (red) and MAC-2 (green) expression in gWAT of Control mice maintained on HFD for 5 months.
k,l) 4x magnification images of H&E stained gWAT from Mural-Zfp423TG mice maintained on HFD for 5 months.
m,n) 10x magnification images of PERILIPIN (red) and MAC-2 (green) expression in gWAT of Mural-Zfp423TG mice maintained on HFD for 5 months.
o) Pro-inflammatory- and macrophage-selective gene expression in gWAT of Control (n=8) and Mural-Zfp423TG (n=8) mice after 5-months HFD feeding.
p) Cytokine protein levels in gWAT of Control (n=8) and Mural-Zfp423TG (n=8) mice after 5-months HFD feeding.
q) Frequency of total adipose tissue macrophages (ATMs) within gWAT of Control (n=8) and Mural-Zfp423TG (n=8) mice after 5 months of HFD-feeding.
r) Frequency of pro-inflammatory ATMs within gWAT of Control (n=8) and Mural-Zfp423TG (n=8) mice after 5 months of HFD-feeding.
s) Frequency of tdTomato+ macrophages within gWAT SVF of obese mice following injection of tdTomato+ monocytes. n=8 for Control and n=8 for Mural-Zfp423TG.
Bars represent mean ± s.e.m., *p< 0.05, **p<0.01 or ***p<0.001 by unpaired two-tailed Student’s t-test (o-s) or one-way ANOVA (d, e). Exact p values and numbers of repetitions can be found in Source Data Figure 6.
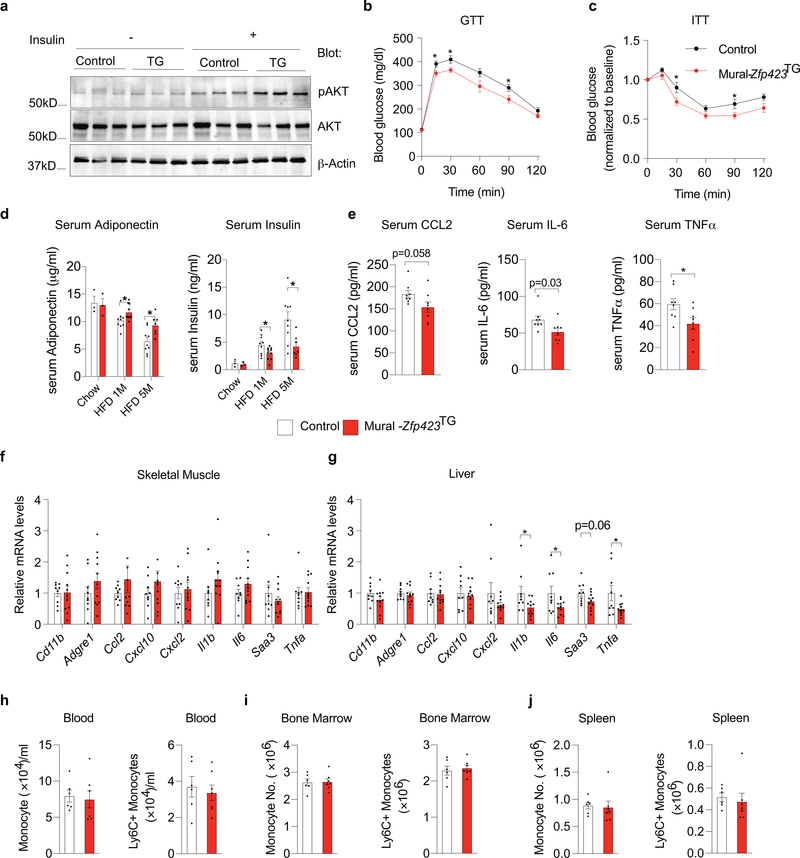
Mural-Zfp423TG mice maintained long-term on HFD indeed exhibited a phenotype very similar to that of Mural-Tlr4KO mice. After 5 months of HFD feeding, adipose tissue mass remained comparable between obese Control and Mural-Zfp423TG mice (Fig. 6b); however, differences in the degree of adipose inflammation were readily apparent. The gonadal WAT depot of Mural-Zfp423TG mice exhibited less metabolic inflammation than observed in Control mice, evidenced by fewer MAC-2+ immune cells and crown-like structures (Fig. 6g–n) and lower mRNA and protein levels of key pro-inflammatory cytokine/chemokines (Fig. 6o,p). The numbers of total and pro-inflammatory ATMs within transgenic gonadal WAT were ~50% lower than observed in Controls (Fig. 6q,r). Importantly, in vivo cell migrations assays indicated that ~50% fewer injected tdTomato+ monocytes infiltrate the gonadal WAT depot of Mural-Zfp423TG mice in comparison to Controls (Fig. 6s). These data indicate that transgenic expression of Zfp423 in PDGFRβ+ cells, much like the inactivation of TLR4 signaling in PDGFRβ+ cells, suppress adipose tissue macrophage accumulation in obesity.
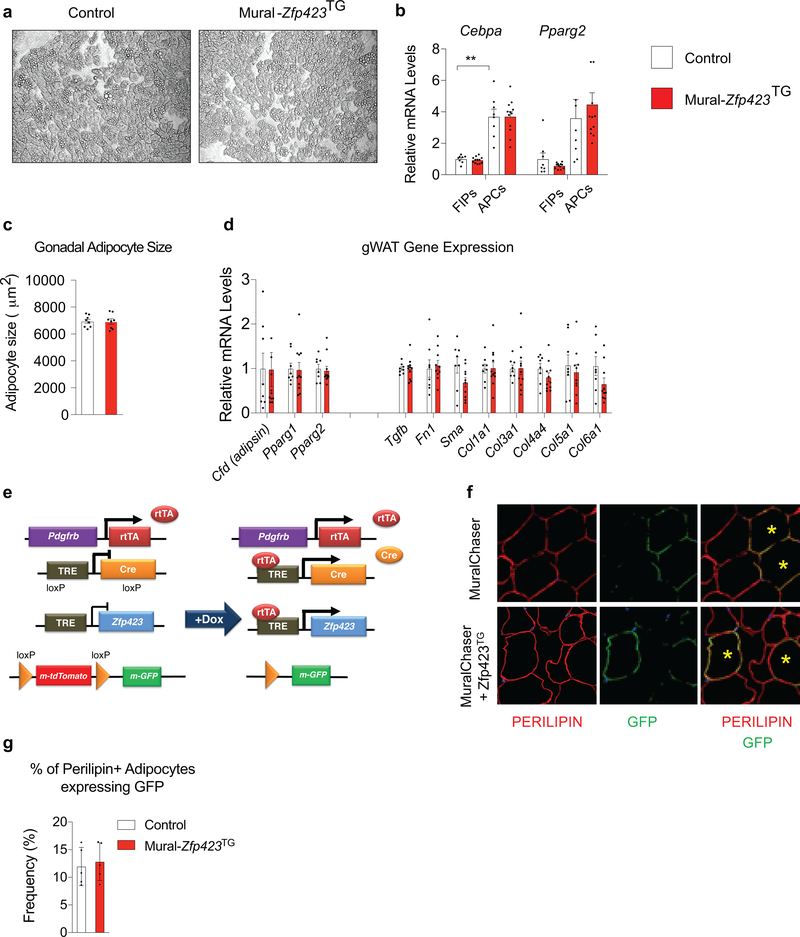
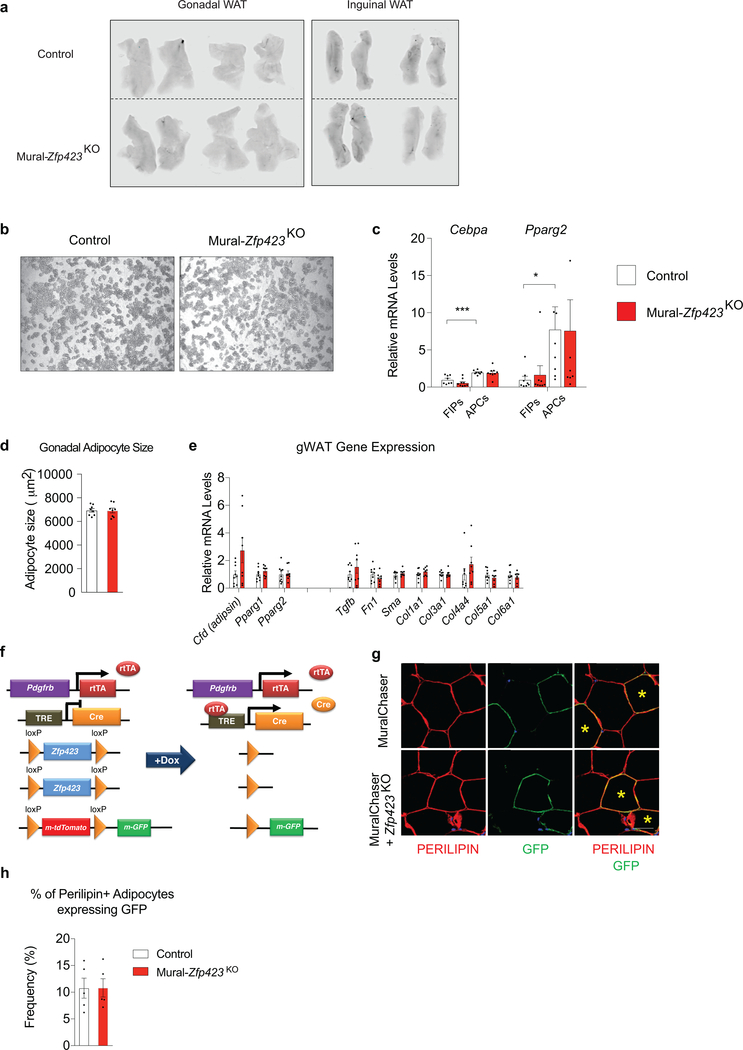
Our prior studies of inducible Pparg expression in progenitors demonstrated that increased de novo adipogenesis can drive healthy WAT remodeling in the setting of diet-induced obesity 18. In vitro, Zfp423 overexpression can drive Pparg expression and adipogenesis 14; therefore, we evaluated whether the 4-fold increase in Zfp423 expression in APCs is similarly sufficient to drive adipocyte differentiation and improve other aspects of adipose tissue health in Mural-Zfp423TG mice. In vitro, We did not observe any substantial increase in the extent to which APCs undergo adipogenesis when Zfp423 is expressed (Extended Data Fig. 4a). In vivo, mRNA levels of pro-adipogenic transcription factors within FIPs and APCs were comparable between obese Control and Mural-Zfp423TG mice after 1 month of HFD feeding (Extended Data Fig. 4b). After 5 months of HFD feeding, mean adipocyte size, whole adipose tissue mRNA levels of adipocyte-selective genes, and genes associated with WAT fibrosis, were also comparable between the two groups (Extended Data Fig. 4c,d). We assessed de novo adipogenesis directly using our previously described pulse-chase lineage tracing system, termed the “MuralChaser” model (PdgfrbrtTA; TRE-Cre; Rosa26RmT/mG) 13. The addition of doxycycline to these transgenic animals triggers permanent activation of membrane-bound GFP (mGFP) expression in Pdgfrb-expressing cells. This system can be used to quantitatively track the formation of new gonadal adipocytes (i.e. mGFP+ adipocytes) that emerge from PDGFRβ+ cells in association with HFD feeding. We bred the TRE-Zfp423 allele into the MuralChaser background to derive animals in which the addition of doxycycline drives Zfp423 overexpression and indelible mGFP labeling of PDGFRβ+ cells (herein “MuralChaser-Zfp423TG” mice) (Extended Data Fig. 4e). Following Dox-HFD feeding, 4e did not observe any significant difference in the numbers of gonadal WAT mGFP+ adipocytes between Control and MuralChaser-Zfp423TG mice (Extended Data Fig. 4f,g). Therefore, Zfp423 overexpression, at least to the modest levels induced in this model, is not driving appreciable levels of de novo adipogenesis under these conditions. As such, the reduction in WAT inflammation occurring in this setting is mostly likely driven by the anti-inflammatory function of ZFP423 in PDGFRβ+ cells, rather than any direct pro-adipogenic function or influence on Pparg expression.
Chronic WAT inflammation, defined by the presence of pro-inflammatory macrophages, is tightly linked to insulin resistance in mice. We assessed the response of gonadal WAT in Control and Mural-Zfp423TG mice to the actions of insulin, as reflected by the phosphorylation of its downstream signal transducer, Akt (pAKT). Indeed, levels of pAKT were significantly higher in the gonadal WAT of obese Mural-Zfp423TG mice following insulin stimulation when compared to pAKT levels in gonadal WAT from obese control mice (Extended Data Fig. 5a). Obese Mural-Zfp423TG mice exhibited relatively better glucose tolerance and systemic insulin sensitivity in comparison to obese Control animals (Extended Data Fig. 5b,c). Serum protein analysis supported these observations. Levels of the adipokine, ADIPONECTIN, are often indicative of adipose tissue health and insulin sensitivity in obesity 19. ADIPONECTIN levels progressively decline as Control animals became obese over the 5-month period of HFD feeding. This decline occurred to a lower extent in Mural-Zfp423TG mice (Extended Data Fig. 5d). Moreover, after 5 months of HFD feeding, serum insulin levels in the transgenic mice are nearly 50% of the levels observed in Control mice (Extended Data Fig. 5d). Correlating with these phenotypes was a slight reduction in circulating protein levels of CCL2, IL-6, and TNFα (Extended Data Fig. 5e). Importantly, most of the anti-inflammatory effects observed were limited to the adipose tissue, despite the overexpression of Zfp423 in PDGFRβ+ cells of other tissues. Levels of the expression of macrophage-selective genes (Cd11b, Adgre1) and most pro-inflammatory genes assayed within skeletal muscle and liver were not substantially impacted by Zfp423 overexpression in PDGFRβ+ cells, suggesting that the anti-inflammatory effects in this model were largely restricted to adipose tissue (Extended Data Fig. 5f,g). In fact, levels of total and pro-inflammatory monocytes in the blood, bone marrow, and spleen, were comparable between Control and transgenic animals (Extended Data Fig. 5h–j). Collectively, these data are highly consistent with the notion that adipose tissue inflammation is linked to systemic insulin sensitivity.
Zfp423 inactivation in PDGFRβ+ cells exacerbates metabolic WAT inflammation
Based on the data above, it would be predicted that loss of Zfp423 in PDGFRβ+ cells would lead to elevated pro-inflammatory signaling and exacerbated adipose tissue inflammation upon HFD feeding. We previously examined adipose depots of Mural-Zfp423KO mice undergoing HFD feeding, with a strict focus on the impact of Zfp423-deficiency on adipocyte differentiation 15. We found that visceral WAT PDGFRβ+ cells lacking Zfp423 are able to undergo adipogenesis in response to HFD feeding, indicating that Zfp423 is dispensable for visceral adipocyte differentiation in adulthood. We did not examine adipose tissue inflammation directly in this prior study; however, obese Mural-Zfp423KO mice did not appear to exhibit any impairment in glucose tolerance when compared to obese control animals.
Our prior study relied on Mural-Zfp423KO mice housed at 22 °C and maintained on HFD for only 8 weeks 15. As described above, the pro-inflammatory phenotype of FIPs is significantly heightened under thermoneutral housing conditions. Therefore, we reassessed the impact of mural Zfp423 inactivation on adipose tissue health in animals maintained at 30 °C for the duration of HFD feeding. For these experiments, we followed the same experimental paradigm utilized for the study of Zfp423 transgenic animals (Extended Data Fig. 6a). Inactivation of Zfp423 in these cells did not appear to significantly impact body or adiposity over the course of HFD feeding (Extended Data Fig. 6b,c); however, the local adipose tissue phenotype and systemic consequences were almost perfectly symmetrical to those observed in the corresponding Zfp423 gain of function model. After 1 month of HFD feeding, the frequencies of gonadal WAT PDGFRβ+ subpopulations were comparable between Control and Mural-Zfp423KO mice (Extended Data Fig. 6d). Gene expression analysis of isolated FIPs and APCs indicated that Zfp423 expression was significantly diminished in both PDGFRβ+ subpopulations within the gonadal WAT from Mural-Zfp423KO mice (Extended Data Fig. 6e). The reduction in Zfp423 levels in FIPs led to increased pro-inflammatory gene expression in these cells, consistent with the in vitro results (Extended Data Fig. 6f). Importantly, inactivation of Zfp423 in APCs did not lead to increased pro-inflammatory gene expression. This may suggest that APCs utilize additional factors to suppress pro-inflammatory signaling in these cells, and/or require the presence of additional molecules to drive the activation of these pathways.
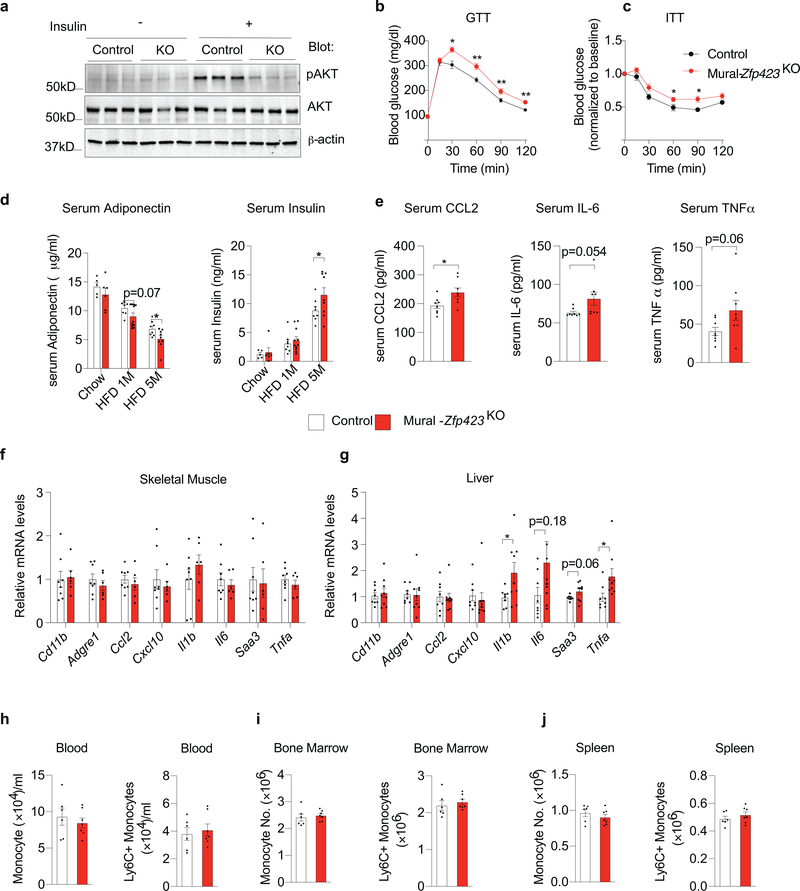
Histological analysis of WAT depots after 5 months of HFD feeding revealed a substantial increase in the degree of inflammation in Mural-Zfp423KO mice. In comparison to gonadal WAT of Control mice, gonadal WAT depots of Mural-Zfp423KO mice exhibited more MAC-2+ cells and crown-like structures (Extended Data Fig. 6g–n), higher tissue mRNA levels of several pro-inflammatory cytokines and adhesion molecules, and higher tissue protein levels of CCL2, IL-6, and TNFα (Extended Data Fig. 6o,p). Moreover, numbers of total and pro-inflammatory ATMs were approximately 2-fold higher in gonadal WAT of Mural-Zfp423KO mice in comparison to Controls (Extended Data Fig. 6q,r). Importantly, injected tdTomato+ monocytes migrate and adopt a macrophage phenotype more readily in obese Mural-Zfp423KO mice in comparison to obese Controls (Extended Data Fig. 6s).
A trivial explanation for the pathologic WAT expansion observed in Mural-Zfp423KO mice could be the development of vascular instability in face of mural cell Zfp423 inactivation. To test this possibility, we performed Evans Blue staining of gonadal and inguinal WAT depots from Control and Mural-Zfp423KO mice that were maintained for 2 months on HFD diet under thermoneutral housing conditions. We chose this 2-months timepoint in order to determine if any changes in vascular permeability precede the overt inflammation that is observed at the 5-months timepoint. Under these conditions, we did not observe any differences in Evans Blue incorporation between the two genotypes (Extended Data Fig. 7a); this suggests that there is no gross difference in the degree of vascular permeability. We also reassessed the requirement of Zfp423 in adipocyte differentiation from mural cells in these thermoneutral-housed animals. Consistent with our previous studies, we did not observe any substantial change in the adipogenic potential or levels of Cebpa and Pparg within APCs when Zfp423 is inactivated (Extended Data Fig. 7b,c). After 5 months of HFD feeding, mean adipocyte size and whole adipose tissue mRNA levels of adipocyte-selective genes and genes associated with WAT fibrosis were also comparable between obese Control and Mural-Zfp423KO mice (Extended Data Fig. 7d,e). Using lineage-tracing, we confirmed that Zfp423 inactivation is not influencing de novo adipogenesis within these depots of obese Mural-Zfp423KO mice (Extended Data Fig. 7f–h). All together, these data support the notion that the increased adipose tissue inflammation within Mural-Zfp423KO mice is likely mediated by the loss of the anti-inflammatory function of ZFP423 in PDGFRβ+ cells, rather than by detrimental effects on vascular integrity or the perivascular progenitor cell compartment.
The local and systemic effects of inactivating Zfp423 in PDGFRβ+ cells at the onset of HFD feeding were also perfectly symmetrical to the effects observed in the Zfp423-transgenic model. Increased pro-inflammatory macrophage accrual in WAT of Mural-Zfp423KO mice was associated with impairments in insulin-stimulated AKT phosphorylation in WAT (Extended Data Fig. 8a). Moreover, in comparison to Control animals, Mural-Zfp423KO mice were more glucose intolerant and insulin resistant (Extended Data Fig. 8b,c). Consistent with these observations, levels of ADIPONECTIN were lower, and serum insulin and CCL2 levels were higher, in Mural-Zfp423KO mice than in Controls (Extended Data Fig. 8d,e). Similar to what was observed in the Zfp423-transgenic mice, these phenotypes occurred without any substantial impact on the expression of macrophage-selective genes within the skeletal muscle and liver (Extended Data Fig. 8f,g), or changes in the amount of monocytes in the blood, bone marrow, or spleen (Extended Data Fig. 8h–j These loss of function data provide additional evidence that Zfp423 exerts an anti-inflammatory function in adipose PDGFRβ+ cells, and further suggest that the activation of pro-inflammatory signaling cascades in mural cells can heavily influence adipose tissue health in obesity.
ZFP423 represses the transcriptional activity of NFκB
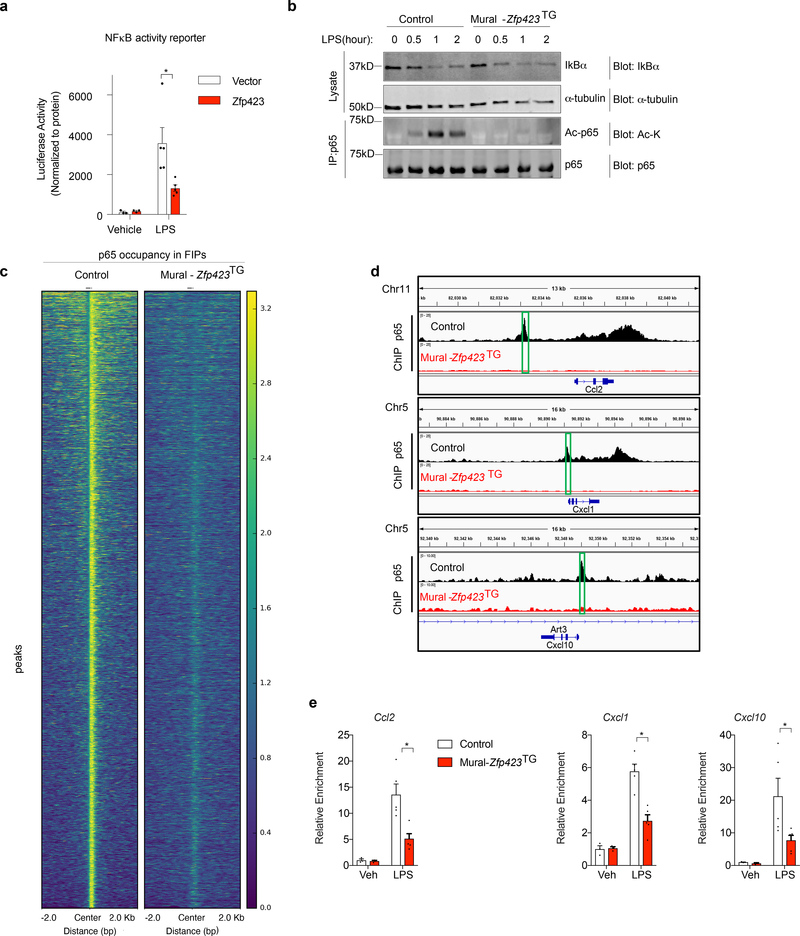
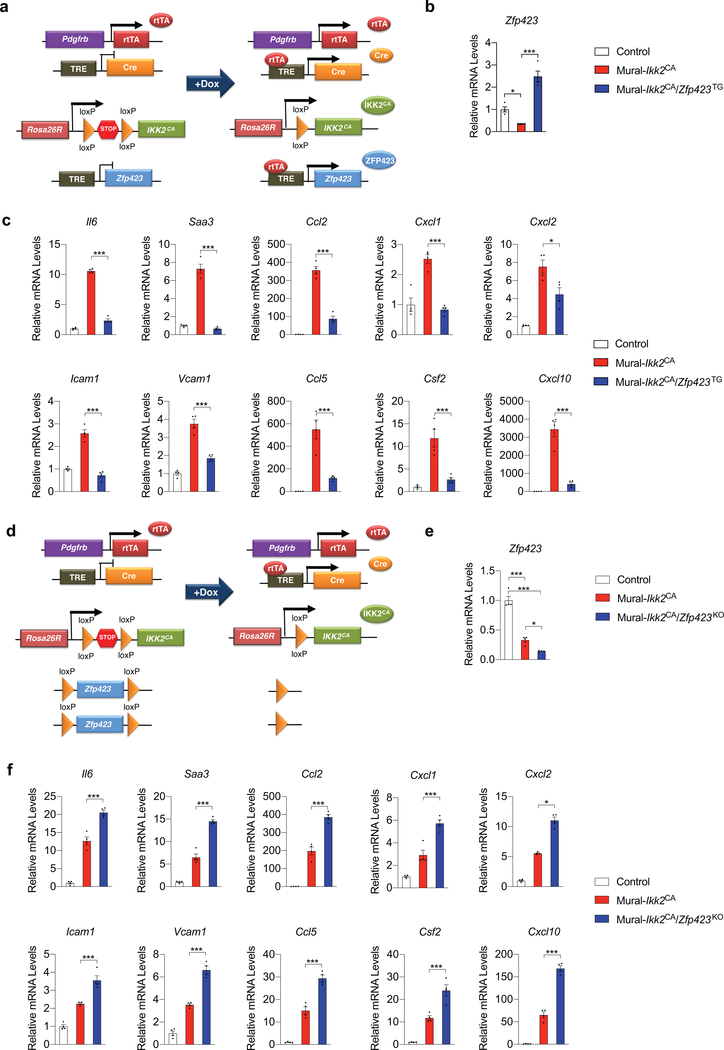
As described above, cultured FIPs and APCs differ in their ability to respond to the genetic activation of NFκB signaling. This prompted us to ask whether the molecular mechanism by which ZFP423 exerted its anti-inflammatory effect involved modulation of this pathway. We reconstituted the TRE-Zfp423 transgene into the Mural-Ikk2CA background to derive animals in which the addition of doxycycline leads to the expression of both IKK2CA and ZFP423 (Mural-Ikk2CA/Zfp423TG mice) (Extended Data Fig. 9a). Remarkably, a modest 2–3 fold increase in the mRNA levels of Zfp423 in isolated FIPs was sufficient to almost fully suppress all of IKK2CA-induced NFκB transcriptional targets examined (Extended Data Fig. 9b,c). We also derived an additional model where the floxed Zfp423 alleles are incorporated into the Mural-Ikk2CA background. The addition of doxycycline thus leads to the simultaneous expression of IKK2CA and inactivation of Zfp423 alleles (Mural-Ikk2CA/Zfp423KO mice) (Extended Data Fig. 9d,e Isolated Zfp423-deficient FIPs from this model show an exaggerated transcriptional response to the activation of NFkB signaling by IKK2CA expression (Extended Data Fig. 9f). These data suggest that ZFP423 suppresses the transcriptional activity of NFκB. We performed NFκB-dependent gene reporter assays in commercially available HEK 293 cells engineered to stably express TLR signaling components, which mediate LPS-induced pro-inflammatory responses (herein “293-TLR4 cells”). 293-TLR4 cells responded to LPS with a robust activation of the NFκB-dependent reporter gene (Fig. 7a). Overexpression of Zfp423 in these cells diminished the activation of the NFκB-dependent gene reporter by > 2-fold, providing evidence that ZFP423 indeed regulates the activity of NFκB (Fig. 7a).
Figure 7. ZFP423 suppresses the DNA-binding capacity and activity of NFκB in FIPs.
a) NFκB-dependent luciferase gene reporter activity in 293-TLR4 cells co-transfected with Zfp423-expressing vector (Zfp423) or empty vector (Vector). Cells were treated with LPS (100ng/ml) (n=5 per group) or vehicle (PBS) (n=3 per group) for 8 hours prior to harvest.
b) Western blot analysis of IκBα, acetylated lysine (Ac-K), and total p65 levels in whole cell lysates or p65 immunoprecipitates from Control and Mural-Zfp423TG FIPs treated with LPS (100 ng/ml) for indicated times. α-tubulin was used as an internal control for protein loading.
c) Heat map illustrating p65-occupied regions in Control and Mural-Zfp423TG FIPs following LPS treatment.
d) p65 enrichment at Ccl2, Cxcl1 and Cxcl10 loci in Control and Mural-Zfp423TG FIPs treated with 100 ng/ml LPS for 2 hours. Green boxes indicate the putative p65 binding motif (GGGRNYYYCC) containing regions targeted by ChIP-qPCR primers.
e) Confirmation of ChIP-seq data by ChIP-PCR. qPCR analysis of p65 occupancy at indicated loci in Control and Mural-Zfp423TG FIPs treated with 100ng/ml LPS for 2 hours. PCR primers spanning the green boxed regions indicated in panel d were used. n= 3 for vehicle group; n=5 for LPS group. Each sample (n) represents individual p65 chromatin immunoprecipitation from ~1,000,000 cells of 8 mice.
For a,e, data are shown as the mean ± s.e.m., * denotes p<0.05 by by two-way ANOVA. Exact p values, numbers of repetitions, and uncropped western blots, can be found in Source Data Figure 7.
NFκB, consisting of the p50 and p65 protein subunits, is highly regulated at the post-translational level 20. Under basal conditions, IkBα is bound to p50/p65 to mediate cytoplasmic sequestration of the protein complex. Pro-inflammatory signals trigger IkBα degradation, allowing p50/p65 to enter the nucleus to activate transcription. This pathway is operative in FIPs from Control mice. LPS treatment leads to rapid degradation of IkBα and nuclear localization of p65. Analysis of cultured FIPs from Mural-Zfp423TG mice indicates that neither IkBα degradation nor nuclear localization of p65 are overtly impacted by Zfp423 overexpression (Fig. 7b; Supplementary Fig. 5). The p65 subunit of NFκB is also highly regulated by post-translational modifications, including protein acetylation 21,22. In response to LPS treatment, p65 acetylation is pronounced in FIPs from Control mice; however, under the same conditions, levels of acetylated-p65 are strongly reduced in Zfp423-overexpressing FIPs (Fig. 7b). These data indicate that the presence of ZFP423 impacts NFκB signaling at the level of p65 acetylation.
Acetylation of p65 is mediated, at least in part, through interaction with CBP/p300. Acetylation of p65 can impact DNA-binding and/or transcriptional activity and enhancer selectivity 20. As such, we tested whether p65 binding to its target loci would be impacted in cells overexpressing Zfp423. We utilized ChIP-seq to assess p65 occupancy genome-wide in LPS-treated FIPs from Control and Mural-Zfp423TG mice (Supplementary Fig. 6a). Our analysis identified 26,377 regions occupied by p65 within LPS-treated FIPs from Control mice (Fig. 7c; Supplementary Table 5). Computational analysis confirmed the enrichment of NFκB/p65 binding motifs within these regions (Supplementary Fig. 6b). Remarkably, the p65 occupancy at target loci was reduced globally within LPS-treated Zfp423-expressing FIPs (Fig. 7c). We validated these data in independent p65 ChIP experiments, utilizing PCR primers spanning predicted p65 response elements proximal to key inflammatory cytokine genes (Fig. 7d). ChIP, followed by PCR analysis, confirmed the presence of p65 at these individual loci within Control FIPs, with a significant reduction in occupancy within FIPs of Mural-Zfp423TG mice (Fig. 7e). Thus, ZFP423 expression leads to a disruption of NFκB binding to target loci in FIPs.
ZFP423 induces a NFκB co-regulator switch in FIPs
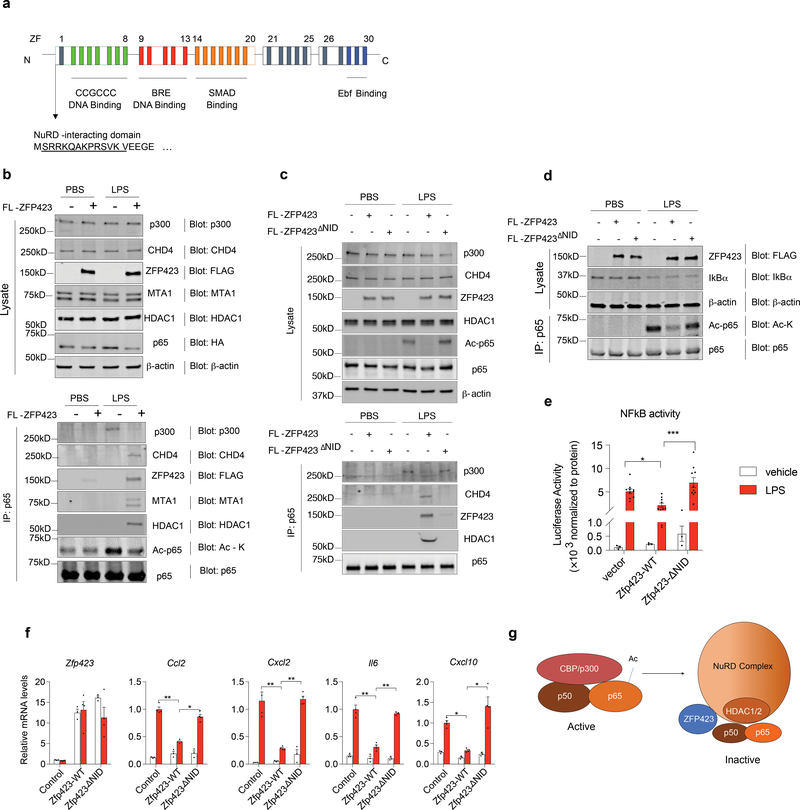
ZFP423 contains 30 C2H2 zinc-finger motifs that mediate interactions with many notable transcription factors. In an effort to understand how ZFP423 may impact p65 protein acetylation, we sought to identify ZFP423-interacting partners in an unbiased manner by employing affinity purification of ZFP423-complexes and mass-spectrometry analysis (Supplementary Fig. 7a). For the initial experiment, we transiently transfected HEK293 cells with plasmid encoding FLAG-tagged ZFP423 protein. LC-MS/MS analysis revealed the presence of all core components of the NuRD (Nucleosome Remodeling Deacetylase) co-repressor complex in association with ZFP423 (Supplementary Fig. 7b,c; Supplementary Table 6). This complex includes several chromatin remodeling factors, (CHD4, MTA1/2, RBAP46/48, and MBD3) along with histone deacetylases 1 and 2 (HDAC1 and HDAC2) 23. Prior studies of NuRD interacting proteins identified a consensus “NuRD interaction domain (NID)” which consists of 12 amino acids (aa) that directly interact with the RBAP48 subunit of the NuRD complex 24. The 12 aa motif is present in a number of C2H2 finger transcription factors, including the N-terminus of ZFP423 (Fig. 8a). All core components of the NuRD complex are readily detected by western blot analysis of primary FIPs (Supplementary Fig. 7d, 8). Endogenous NuRD components associate with virally expressed FLAG-ZFP423 protein in FIPs, but not with a FLAG-tagged variant of ZFP423 that is expressed but lacks the 12 aa NID (FLAG-ZFP423ΔNID) (Supplementary Fig. 7d). These data validate the mass spectrometry dataset and confirm that the ZFP423-NuRD interaction can occur within FIPs. Interestingly, we found that endogenous p65 protein is physically associated with the NuRD complex in a ZFP423- and LPS-dependent manner (Fig. 8b). In cells overexpressing ZFP423, LPS no longer triggers a p65-p300 interaction. Instead, p65 is associated with ZFP423 and the NuRD complex (Fig. 8b). Thus, ZFP423 expression appears to mediate a p65 co-regulator switch in FIPs. The NID is essential for the ability of ZFP423 to exert these inhibitory effects on NFκB signaling. FLAG-ZFP423ΔNID does not interact with p65 in FIPs (Fig. 8c), induce a p300-NuRD co-regulator switch (Fig. 8c), or block p65 acetylation (Fig. 8d). Moreover, ZFP423ΔNID is unable to inhibit NFκB reporter activity (Fig. 8e) or suppress pro-inflammatory gene expression in these cells (Fig. 8f). ChIP-seq analysis of CHD4, an obligate component of the NuRD complex, did not reveal an increased level of occupancy at p65 target loci in ZFP423-expressing cells (Supplementary Fig. 6c,d). Thus, it does not appear that the ZFP423-NuRD complex is necessarily replacing p65/p300 at target loci, but instead it may be tethering the active p65 complex away from DNA. Collectively, these data suggest a model in which ZFP423 suppresses NFκB signaling by triggering a p65 co-regulator switch: ZFP423 expression leads to the recruitment of the NuRD co-repressor complex to p65, with a concomitant loss of co-activator (p300) recruitment, p65 acetylation, and p65 DNA-binding capacity (Fig. 8g).
Figure 8. ZFP423 induces a NFκB/p65 co-regulator switch.
a) Schematic of ZFP423: ZFP423 contains 30 C2H2 zinc-fingers (ZF) clustered into five domains. Amino acids 2–13 represent a defined NuRD-interaction domain that resides within numerous zinc-finger proteins that interact directly with the NuRD co-repressor complex.
b) Western blot of indicated protein expression in FIPs whole cell lysates (left) and p65 immunoprecipitates (right) from primary FIPs transduced with control retrovirus or retrovirus expressing FLAG-tagged ZFP423 (FLAG-ZFP423). Cells were treated with LPS (100ng/ml) or vehicle for 2 hours prior to harvest.
c) Western blot of indicated protein expression in FIPs whole cell lysates (top) and p65 immunoprecipitates (bottom) from primary FIPs transduced with control retrovirus or retrovirus expressing FLAG-tagged ZFP423 (FL-ZFP423) or FLAG-tagged ZFP423 lacking the NuRD interaction domain (NID) (FL-ZFP423ΔNID). Cells were treated with LPS (100ng/ml) or vehicle for 2 hours prior to harvest.
d) Western blot of indicated protein expression within whole cell lysates of FIPs transduced with control retrovirus, wild-type FLAG-tagged ZFP423 (FLAG-ZFP423WT), or retrovirus expressing FLAG-ZFP423 lacking the 12 amino acid NuRD-interaction domain (NID) (FLAG-ZFP423ΔNID) and treated with LPS (100ng/ml) or vehicle. β-actin expression was used as loading control.
e) NFκB-dependent gene reporter activity in 293-TLR4 cells co-transfected with control, FLAG-ZFP423WT, or FLAG-ZFP423ΔNID plasmids. Cells were treated with LPS (100ng/ml) (n=10 per group) or vehicle (PBS) (n=3 per group) for 8 hours prior to harvest. Bars represent mean ± s.e.m., ** denotes p<0.01 by two-way ANOVA.
f) mRNA levels of indicated genes in FIPs transduced with control, FLAG-ZFP423WT, or FLAG-ZFP423ΔNID, retroviruses and treated with vehicle (PBS) (n=3 per group) or 100 ng/ml LPS (n=4 per group) for 2 hours. * denotes p<0.05, ** denotes p<0.01 by two-way ANOVA.
g) Proposed model: The nuclear p65 subunit of NFκB is acetylated (Ac) and activated by its interaction with CBP/p300. The presence of ZFP423 triggers a transcriptional co-regulator switch; ZFP423 facilitates the recruitment of the NuRD co-repressor complex to p65, thereby excluding CBP/p300 association and p65 acetylation.
Exact p values, numbers of repetitions, and uncropped western blots can be found in Source Data Figure 8.
Discussion
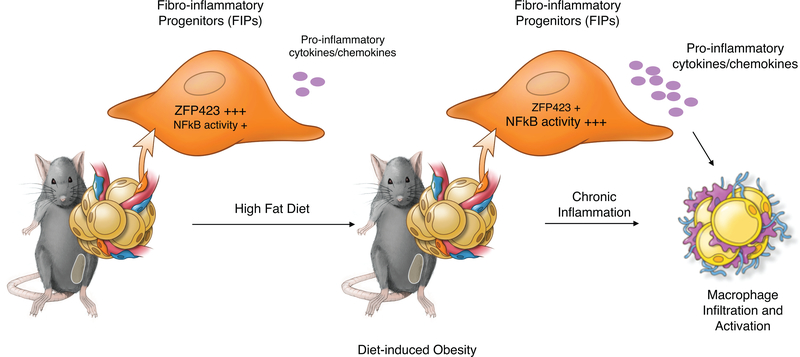
The data here highlight an important role for perivascular mesenchymal cells in the regulation of adipose tissue inflammation and health. Our studies identify FIPs as a specialized subpopulation of perivascular cells that are functionally distinct from APCs and differentially regulated in association with physiological settings of metabolic inflammation. We propose a model in which FIPs serve, in part, as critical regulators of macrophage accrual in obesity (Extended Data Figure 10). In lean mice, FIPs appear to maintain some basal inflammatory signaling. The presence of ZFP423 may serve to moderate NFκB activity in order to maintain inflammatory balance. In response to HFD feeding, the downregulation of Zfp423 expression facilitates further activation of NFκB signaling and the induction of pro-inflammatory gene expression. The congruence of the phenotypes of the various mouse models presented here support the underlying hypothesis that pro-inflammatory signaling events within perivascular mesenchymal cells significantly impact chronic WAT inflammation in obesity.
FIPs exert a pro-inflammatory response and phenotype within 1–3 days of HFD, likely placing them amongst the earliest of responders in WAT. Going forward, a tremendous challenge will be in identifying the factors associated with HFD feeding that trigger the activation of FIPs. A number of possible triggers may activate a pro-inflammatory response in WAT 25. This includes gut-derived antigens (e.g. LPS), dietary lipids, hypoxia, and even mechanical stress. Furthermore, it will be of interest to understand whether and how other cells types, particularly adipocytes, coordinate this response with FIPs.
The pro-inflammatory phenotype of FIPs is dependent on TLR4 signaling, a key mediator of pro-inflammatory responses to LPS and other stimuli. Mice lacking Tlr4 are protected from diet-induced insulin resistance and inflammation; however, which Tlr4-expressing cells mediate this effect have been unknown. Tissue-specific knockout models highlight hepatocyte TLR4 signaling as a key mediator of metabolic inflammation and insulin resistance 26. On the other hand, adipocyte TLR4 signaling is more complex and may even be beneficial for appropriate WAT expansion in obesity 27. Our studies here highlight TLR4 signaling in perivascular stromal cells as important mediators of pro-inflammatory macrophage accumulation in WAT.
ZFP423 appears to be amongst a select group of transcriptional components that safeguard against hyperinflammatory responses 28–30. The downregulation of Zfp423 at the onset of HFD may thus represent a mechanism by which FIPs can activate their pro-inflammatory phenotype. It is notable that the activation of NFκB signaling in FIPs via IKK2CA expression led to a downregulation of Zfp423 expression (see Extended Data Fig. 9b,e). This suggests that ZFP423 and NFkB may be part of a regulatory loop that maintains inflammatory balance in these cells. Zfp423 is also expressed in various regions of the brain, proliferating muscle satellite cells, and in endothelial cells 12,31–34. Whether ZFP423 exerts an anti-inflammatory function in other tissues is unclear. The presence of such tissue-selective modifiers of NFκB signaling may allow for spatial control of inflammatory responses in the body.
The p65 subunit of NFκB is heavily regulated by phosphorylation and acetylation. The latter plays an important role in enhancing the DNA binding capacity and/or transcriptional activity of p65. CBP/p300 is a well-characterized acetyltransferase for p65, whereas HDAC3 and Sirt1 are amongst the known deacetylases of the protein 20,29,35,36. Our biochemical data suggest a model in which ZFP423 expression leads to the recruitment of the NuRD co-repressor complex to p65. The NuRD repressor complex contains HDAC1 and HDAC2; however, prior studies suggest that these proteins do not directly mediate p65 deacetylation. As such, it is likely that ZFP423 is mediating a co-regulator switch, in which the presence of the NuRD complex displaces CBP/p300 from p65 to limit its activation. Consistent with this model, an engineered variant of mouse ZFP423 lacking the NuRD interaction domain is unable to suppress NFκB activity and the pro-inflammatory phenotype of FIPs.
A limitation to our study is the inability to specifically target subpopulations of PDGFRβ+ cells in mice. The PdgfrbrtTA allele targets both FIPs and APCs; however, it should be noted that FIPs represent a vast majority of all PDGFRβ+ cells within WAT. FIPs are naturally more pro-inflammatory than APCs, and the most robust changes in pro-inflammatory genes expression occur in FIPs rather than APCs following Tlr4 or Zfp423 manipulation. Furthermore, visceral WAT APCs retain their normal differentiation capacity following loss- or gain of Zfp423 function in these cells. Importantly, we did not observe substantial changes in pro-inflammatory responses in non-adipose tissues of our models following LPS-treatment or HFD-feeding. These data suggest that the manipulation of Tlr4 or Zfp423 levels within PDGFRβ+ cells did not elicit broad inflammatory effects, but rather local impacts on WAT inflammation. Our work highlights a maladaptive mechanism by which FIPs are activated in adipose tissue to promote chronic metabolic inflammation; however, these data cannot exclude a role for these cells in lean animals, perhaps serving in the homeostatic control of adipose tissue mass and function. FIPs also express IL-33, a key cytokine important in maintaining an anti-inflammatory state in WAT 37,38. A deeper understanding of the heterogeneity of adipose stromal cells may identify additional mechanisms controlling both physiological WAT homeostasis and pathophysiological expansion of fat tissue in the face of caloric excess. Deciphering such mechanisms may inform novel strategies to improve WAT health and uncouple the development of metabolic disease from obesity.
Materials and Methods
Animals
All animal experiments were performed according to procedures approved by the UTSW Institutional Animal Care and Use Committee. PdgfrbrtTA transgenic mice (C57BL/6-Tg(Pdgfrb-rtTA)58Gpta/J; JAX028570; RRID: IMSR_JAX:028570), TRE-Cre (B6.Cg-Tg(tetO-cre)1Jaw/J; JAX 006234; RRID:IMSR_JAX:006234), and Rosa26RIKK2CA (B6(Cg)-Gt(ROSA)26Sortm4(Ikbkb)Rsky/J; JAX008242) mice were obtained from Jackson Laboratories. Zfp423loxp/loxp mice were previously described 15 and generously provided by S. Warming (Genentech). Tlr4loxp/loxp mice were generated at UTSW and previously described 26. TRE-Zfp423 transgenic mice were derived by the UTSW Transgenic Core Facility and previously described 17. The animals used in this study were all male mice on a pure C57BL/6 background. The mice maintained on a 12hr light/dark cycle in a temperature-controlled environment (room temperature, 22°C; thermoneutrality, 30°C) and given free access to water and food.
Rodent diets and drug treatments
Mice were maintained on a standard rodent chow diet or chow diet containing 600 mg/kg doxycycline (DOX) (Bio-Serv, #S4107). For thermoneutrality experiments, mice were housed in thermoneutral chamber (30°C) beginning at the age of 6 weeks and maintained on a standard chow diet 2 weeks before being switched to indicated high-fat diets. For high-fat diet studies, mice (8 weeks old) were fed a standard HFD (HFD; 60 kcal% fat; Research Diets, #D12492i) or DOX-containing HFD (600 mg/kg DOX, 60% kcal% fat, Bio-Serv, #S5857) as described in the main text. For lipopolysaccharide administration, mice were i.p. injected with vehicle (PBS) or LPS (0.3 mg per kg body weight; Sigma, #L3024) and sacrificed after 2 hours or 24 hours as indicated in the main text.
Serum measurements
Serum levels of adiponectin and insulin were measured using the mouse adiponectin ELISA (Millipore, #EZMADP-60K) and mouse insulin ELISA kit (Crystal Chem, #90080), respectively. Assays were performed according to manufacturer instructions.
Histological analysis
Dissected tissues were fixed in freshly prepared 4% paraformaldehyde for 48 hours and then maintained in 50% ethanol solution. Paraffin embedding, sectioning, and hematoxylin and eosin (H&E) staining, were conducted by the Molecular Pathology Core Facility at UT Southwestern. Bright-field and fluorescent images were acquired using a Keyence BZ-X710 microscope. Adipocyte size was measured as previously described 18. In brief, bright-field images of H&E-stained paraffin sections were analyzed using of Keyence BZ-X Analyzer software. More than 200 adipocytes were quantified in each individual animal.
Indirect immunofluorescence
Indirect immunofluorescence was performed as previously described13. Antigen retrieval of the rehydrated sections was performed using Antigen Retriever 2100 (Electron Microscopy Sciences) and R-Buffer A pH 6.0 solution (Electron Microscopy Sciences, #62706). For GFP immunostaining, animals were perfused with 4% PFA before dissection. For quantification of adipocyte hyperplasia, indirect immunofluorescence assay of PERILIPIN and GFP expression was performed as previously described15. Cell numbers of PERILIPIN+ GFP+ and PERILIPIN+ GFP- adipocytes were counted on 8 randomly selected 10× images of stained WAT depots. A total of 3000–4000 PERILIPIN + adipocytes were counted for each mouse and each data point represents the percentage of PERILIPIN+ adipocytes expressing GFP adipocytes. Antibodies and concentrations used for immunofluorescence include: guinea pig anti-PERILIPIN 1:1000 (Fitzgerald #20R-PP004); rabbit anti-MAC-2 1:500 (Cedarlane, Clone M3/38, #CL8942AP); chicken anti-GFP 1:500 (Abcam, #ab13970); goat anti-guinea pig Alexa flour 647 1:200 (Invitrogen, #A21450); goat anti-rat Alexa flour 488 1:200 (Invitrogen, #A11006); goat anti-chicken Alexa flour 488 1:200 (Invitrogen, #A11039).
Evans Blue Staining
Vascular permeability of white adipose tissue was determined by Evans Blue injection as previously described 39. In brief, the mice were injected with 160mg per kg body weight Evans Blue dye (Sigma, #E2129) via tail vein. Thirty minutes after injection, the mice were anesthetized and then perfused with PBS. Harvested whole gonadal and inguinal fat depots were fixed in 4% PFA overnight. Rinsed fat pads (PBS) were imaged for Evans Blue auto fluorescence at 700 nm using a LI-COR Odyssey infrared imaging system.
Gene expression analysis by quantitative PCR
Total RNA from tissue and cultured cells was extracted and purified using the TRIzol reagent (Invitrogen, #15596026). Total RNA from freshly FACS-sorted cells was extracted and purified using the RNAqueous Micro total RNA isolation kit (ThermoFisher Scientific, #AM1931). cDNA was synthesized using random hexamer primers (ThermoFisher Scientific, #N8080127) and M-MLV reverse transcriptase (ThermoFisher Scientific, #28025013). Relative levels of mRNAs were determined by quantitative real-time PCR using SYBR Green PCR system (Applied Biosystems) and Rps18 was used as an internal control for calculation using the ΔΔ-Ct method. All primers sequences used in this study are listed in Supplementary Table 7.
Isolation of adipose stromal vascular fraction and flow cytometric assays
The stromal vascular fraction of white adipose tissue was isolated as previously described18. Briefly, minced gonadal white fat depots were incubated for 1 hr in digestion buffer (1×Hank’s Balanced Salted Solution, 1.5% bovine serum albumin and 1 mg/ml Collagenase D [Roche, #11088882001]) at 37°C within a shaking water bath. The digested mixture was sequentially filtered through a 100 μm cell strainer and then a 40 μm cell strainer. The red blood cells in SVFs were lysed by short incubation in 1 ml 1×RBC lysis buffer (eBioscience, #00–4300-54) and the SVF cells were then resuspended in blocking buffer (2% FBS/PBS containing anti-mouse CD16/CD32 Fc Block 1:200). For FACS isolation of FIPs and APCs, primary antibodies were added to the cells in blocking buffer for 15 min while incubating at 4°C. The cells were then washed once and resuspended in 2% FBS/PBS before sorting. FACS was performed using a BD Biosciences FACSAria cytometer at the Flow Cytometry Core Facility at UT Southwestern. The primary antibodies and the working concentrations are as following: CD45-PerCP/Cyanine5.5 1:400 (Biolegend, clone 30-F11, #103132), CD31-PerCP/Cyanine 5.5 1:400 (Biolegend, clone 390, #102420), PDGFRβ-PE 1:75 (Biolegend, clone APB5, #136006), LY6C-APC 1:400 (Biolegend, clone HK1.4, #128016), CD9-FITC 1:400 (Biolegend, clone MZ3, #124808).
For the analysis of adipose tissue macrophages, isolated SVF cells were incubated with flow antibodies for 1hr at 4°C. The primary antibodies and the working concentrations are as following: CD45-PerCP/Cyanine5.5 1:400 (Biolegend, clone 30-F11, #103132), CD11b-Pacific Blue 1:200 (Biolegend, clone M1/70, #101224), F4/80-PE 1:200 (Biolegend, clone BM8, #123110), CD11c-APC 1:200 (Biolegend, clone N418, #117310), CD206-FITC 1:200 (Biolegend, clone C068C2, #124808), CD68-FITC 1:200 (Biolegend, clone FA-11, #137005), CD68-APC 1:200 (Biolegend, clone FA-11, #137007), CD9-FITC 1:400 (Biolegend, clone MZ3, #124808). For the analyses of monocytes in blood, peripheral blood mononuclear cells were prepared using Histopaque-1077 (Sigma, #10771) according to the manufacturer’s instructions. Meanwhile, cells were prepared from bone marrow and spleen for monocytes analysis. The cells were incubated with flow antibodies for 1hr at 4°C. The primary antibodies and the working concentrations are as following: CD45-PerCP/Cyanine5.5 1:400 (Biolegend, clone 30-F11, #103132), CD11b-Pacific Blue 1:200 (Biolegend, clone M1/70, #101224), CD115-APC/Cyanine7 1:200 (Biolegend, clone AFS98, #135531), LY6C-APC 1:400 (Biolegend, clone HK1.4, #128016). After twice wash with 2% FBS/PBS, cells were fixed by incubation with BD Cytofix (BD Biosciences, #554655) for ≥ 30 min. Flow cytometry analysis was performed using a BD Biosciences LSR Fortessa. All the flow cytometry datasets were analyzed and graphed using FlowJo Version 10.6.1. Detailed information of commercial manufacturers and validation of the antibodies can be found in the Reporting Summary.
Cellular assays
Culture of PDGFRβ+ subpopulations
Freshly isolated FIPs and APCs were cultured in previously defined “ITS media,” consisting of 60% low glucose DMEM, 40% MCDB-201 media, 2% FBS, 1% ITS premix (Insulin-Transferrin-Selenium) (BD Bioscience, #354352), 0.1 mM L-ascorbic acid-2–2phosphate (Sigma, #A8960–5G), 10 ng/mL FGF basic (R&D systems, #3139-FB-025/CF), Pen/Strep, and gentamicin. For gain of function cellular studies, cultures from Control and Mural-Zfp423TG mice were incubated with doxycycline (1 μg/ml) in ITS media for more than 24 hours before beginning treatment. For loss of function studies in vitro, PDGFRβ+ cells were isolated from Control and Mural-Zfp423KO mice maintained on doxycycline-containing chow diet for 10 days and then placed in culture.
Adipocyte differentiation
For adipogenesis assays, sorted primary cells were plated in 48-well plates at a density of 4×104 cells/well in ITS-media containing 2% FBS (Millipore, #TMS-013-B) and incubated at 37°C in 10% CO2 for 7 days. Media was replaced every other day. APCs differentiate spontaneously into adipocytes upon reaching confluence.
LPS and palmitate treatment of cells
FIPs were plated in 24-well plates at 5×105 cells/well in ITS media containing 2% endotoxin-free FBS (GIBICO, #16000–044) and cultured until the cells reached confluence. After overnight incubation with serum-free ITS media, the cells were treated with 100 ng/ml LPS (PBS was used as vehicle control) or 0.5 mM Palmitate (Sigma, #P9676; BSA was used as vehicle control) for 2 hours before harvest for RNA isolation.
Preparation of FIPs conditioned medium
FIPs were isolated from gonadal WAT of wildtype C57BL/6 male mice maintained on HFD for 0 days (chow diet), 1 day, 3 days, or 7 days and then plated in 24-well plates (1×105 cells/well) containing serum-free ITS media. 24 hours after plating, conditioned media was harvested. For conditioned medium derived from LPS-treated cells, FIPs were first maintained in serum-free ITS media overnight and then treated with 100 ng/ml LPS for 3 hours. After three washes with PBS (to remove LPS), cells were replaced with fresh serum-free ITS media and incubated for 24 hours. After 24 hours, medium was collected and briefly centrifuged (3000g for 15min) to remove debris. For all experiments conditioned medium was mixed 1:1 with fresh serum-free ITS media before being placed on target cells.
Macrophage differentiation and assays
Bone marrow derived macrophages (BMDMs) used in this study were derived from bone marrow stem cells (BMSCs) isolated from the femurs and tibias of male mice as previously described40. To induce the differentiation of mature macrophages, BMSCs were maintained in L929 cell-derived differentiation medium for 7 days. Mature BMDMs were maintained in 2%FBS-containing ITS media for at least 24 hours, and then switched to serum-free ITS media for more than 6 hours before further experiments. For conditioned medium experiments, BMDMs were incubated in conditioned medium for 2 hours and then harvested for RNA analysis. In vitro chemotaxis assay was performed using Transwell plate (Corning, #3428) as previously described 41. Briefly, BMDMs (1×105 cells/100μl/well) were added into the upper chamber of an 8 μm polycarbonate filter (24-tranwell format), with 600 μl conditioned media placed in the lower chamber. After 3 hours of incubation, migrated cells were fixed with methanol and stained with DAPI. DAPI+ cells were counted on 4 randomly selected 20× images of stained membrane.
In vivo monocytes infiltration assays
Leukocyte pools from the blood of 10-week-old Rosa26RmTmG mice were subjected to erythrocyte lysis prior to staining with flow antibodies. Then, CD45+ CD11b+ CD115+ circulating monocytes were isolated by FACS. The monocytes were counted, and ~1×106 viable cells were resuspended in 200ul PBS and injected into each obese mouse (i.e. after 5 months of HFD feeding) via tail vein. Two days later, the tdTomato+ cells in the SVF of gonadal WAT were analyzed using flow cytometry.
Retroviral Production and Transduction
Retrovirus was packaged in Phoenix cells as previously described 17. Briefly, Phoenix packaging cells (ATCC CRL-3213) were co-transfected with 10 μg of the pMSCV overexpression plasmids (pMSCV-Zfp423-WT, pMSCV-Zfp423-ΔNID or empty vector) and 5 μg gag-pol and 5 μg VSV-g plasmids using Lipofectamine LTX (ThermoFisher Scientific, #15338100). Subsequently, FIPs were transduced with diluted virus (1:10 ratio) in ITS media containing 8 μg/ml polybrene (Sigma, #TR-1003) for 24 hours. Following transduction, cells were maintained in ITS media for ≥ 48 hours before beginning treatments.
Co-immunoprecipitation assay
Co-immunoprecipitation was performed as previously described17. In brief, cells were lysed in Pierce IP lysis buffer (ThermoFisher Scientific, #87787) supplemented with 1% Protease Inhibitor Cocktail (Sigma, #P8340). Following the overnight incubation with the indicated antibodies at 4°C, the cell lysates were mixed with Protein G Sepharose 4 Fast Flow (GE Healthcare Bio-sciences, #17–0618-01) for 1 hour at room temperature to capture immune complexes. After three sequential washes with Pierce IP lysis buffer, samples were eluted by boiling in 2×SDS loading buffer and resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
Luciferase Gene Reporter assay
293/hTLR4A-MD2-CD14 cells (Invivogen, #293-htlr4md2cd14) were maintained in growth media consisting of high glucose DMEM containing 10% FBS, Normocin (100μg/ml), Blasticidin (10μg/ml), and Hygromycin B Gold (50μg/ml). Cells were transfected in 24-well plates using Lipofectamine LTX with 200 ng NFκB activity luciferase reporter 42 and 300 ng of expression plasmid (pcDNA3-FLAG-ZFP423-WT, pcDNA3-FLAG-ZFP423-ΔNID, or empty vector (pcDNA3.1). 48 hours after transfection, cells were replaced with serum-free growth media and incubated overnight. Following 100ng/ml LPS treatment for 8 hours, cells were harvested into 50μl of passive lysis buffer (Promega, #E1941). Luciferase activity of cell extract was evaluated using Bright-Glo Luciferase Assay System (Promega, #E2610) with the GluMax Discover Microplate Reader (Promega). Protein concentration of cell lysis was determined by Bradford Reagent (Sigma, #B6916) and used for internal normalization.
Affinity purification and proteomics analysis
293 cells transfected with pMSCV-GFP or pMSCV-FLAG-ZFP423 plasmids were lysed in Pierce IP lysis buffer (ThermoFisher Scientific, #87787) supplemented with Protease Inhibitor Cocktail (Sigma, #P8340). Cell lysate was incubated with anti-FLAG (Sigma, #F1804, 1:100) or anti-ZFP423 (Santa Cruz, #sc-48785, 1:100) overnight at 4°C. Lysates were then incubated with Protein G Sepharose 4 Fast Flow (GE Healthcare Bio-sciences, #17–0618-01) for 2 hours at 4°C to capture immune complexes. Purified immune complexes were washed sequentially three times with washing buffer (50 mM HEPES [pH 7.4], 10% glycerol, 0.05% NP40, 1mM DTT, 0.25 mM PMSF, 150 mM KCl) supplemented with Protease Inhibitor Cocktail (Sigma, #P8340). Proteins were eluted with Pierce IgG Elution Buffer (ThermoFisher Scientific, #21004) and briefly resolved by SDS-PAGE prior to the submission to the UT Southwestern Proteomics Core. For proteomics analysis, gel fragments were reduced and alkylated with DTT (20 mM) and iodoacetamide (27.5 mM). Gel fragments were then incubated on ice with a 0.1 μg/μL solution of trypsin in 50 mM triethylammonium bicarbonate (TEAB). 50 μL of 50 mM TEAB was then added to the gel pieces for overnight digestion (Pierce). Following solid-phase extraction with an Oasis MCX μelution plate (Waters), the resulting peptides were reconstituted in 10 μL of 2% (v/v) acetonitrile (ACN) and 0.1% trifluoroacetic acid in water. 2 μL of this was injected onto an Orbitrap Fusion Lumos mass spectrometer (Thermo Electron) coupled to an Ultimate 3000 RSLC-Nano liquid chromatography systems (Dionex). Samples were injected onto a 75 μm i.d., 75-cm long EasySpray column (Thermo), and eluted with a gradient from 1–28% buffer B over 90 min. Buffer A contained 2% (v/v) ACN and 0.1% formic acid in water, and buffer B contained 80% (v/v) ACN, 10% (v/v) trifluoroethanol, and 0.1% formic acid in water. The mass spectrometer operated in positive ion mode with a source voltage of 2.4 kV and an ion transfer tube temperature of 275 °C. MS scans were acquired at 120,000 resolution in the Orbitrap and up to 10 MS/MS spectra were obtained in the ion trap for each full spectrum acquired using higher-energy collisional dissociation (HCD) for ions with charges 2–7. Dynamic exclusion was set for 25 s after an ion was selected for fragmentation.
Raw MS data files were analyzed using Proteome Discoverer v2.2 (Thermo), with peptide identification performed using Sequest HT searching against the human protein database from UniProt. Fragment and precursor tolerances of 10 ppm and 0.6 Da were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification and oxidation of Met was set as a variable modification. The false-discovery rate (FDR) cutoff was 1% for all peptides.
Immunoblotting and antibodies
Cells and tissues were lysed by homogenization in RIPA lysis buffer (ThermoFisher Scientific, #89900). Protein extracts were separated by SDS-PAGE and then transferred onto PVDF membrane (Millipore, #IPVH00010). Following overnight incubation with the indicated primary antibodies at 4°C, membranes were incubated with IR dye-coupled secondary antibodies (LI-COR) and then visualized by using the LI-COR Odyssey infrared imaging system (LI-COR). The primary antibodies and diluted ratio include: anti-phospho-AKT 1:1000 (Ser473; Cell Signaling Technology, #9271); anti-AKT 1:1000 (Cell Signaling Technology, #2920); anti-β-ACTIN 1:5000 (Sigma, #A1978); anti-IκBα 1:1000 (Cell signaling Technology, #4814); anti-α-Tubulin 1:1000 (Cell Signaling Technology, #3873); anti-Acetylated Lysine 1:1000 (Cell Signaling Technology, #9441); anti-NFκB p65 1:1000 (Cell Signaling Technology, #8242); anti-p300 1:1000 (Cell Signaling Technology, #86377); anti-FLAG (Sigma, #F1804); anti-Acetyl NFκB p65 (Lys310) 1:1000 (Cell Signaling Technology, #12629). NuRD complex components (CHD4, MTA1, HDAC1, RbAP46) antibodies were all from NuRD Complex Antibody Sampler Kit (Cell Signaling Technology, #8349) and diluted 1:1000 for primary antibody incubation. Detailed information of commercial manufacturers and validation of the antibodies can be found in the Reporting Summary.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) was performed as described 39. FIPs from control or Mural-Zfp423TG mice were cross-linked with 1% formaldehyde in PBS for 10 minutes at 37 °C and quenched in 125 mM glycine in PBS for 5 minutes at 4 °C. FIPs were then lysed in Farnham lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40, 1 mM DTT, and Protease inhibitor cocktail [Sigma, #P8340]) to obtain nuclear material. Crude nuclear pellets were collected by centrifugation and then lysed by incubation in lysis buffer containing 5 mM Tris-HCl pH 7.9, 1% SDS, 10 mM EDTA, 1mM DTT, and Protease inhibitor cocktail [Sigma, #P8340]. Chromatin fragmentation (200–500 bp length) was performed at 4 °C by Bioruptor 300 using the setting of 10 cycles of 30 seconds on and 60 seconds off. Soluble chromatin was diluted 1:10 with dilution buffer (20 mM Tris-HCl pH 7.9, 0.5% Triton X-100, 2 mM EDTA, 150 mM NaCl, 1 mM DTT, and Protease inhibitor cocktail [Sigma, #P8340]) and pre-cleared using Protein G Sepharose 4 Fast Flow (GE Healthcare Bio-sciences, #17–0618-01) for 1 hour at 4 °C. Pre-cleared samples were incubated with anti-p65 antibody (1:100 in dilution, Cell Signaling technology, #8242) or anti-CHD4 antibody (1:100 in dilution, Cell Signaling Technology, #12011) overnight at 4 °C. Antibody-protein-DNA complexes were captured by incubation with Protein G Sepharose 4 Fast Flow (GE Healthcare Bio-sciences, #17–0618-01) at 4 °C for 2 hours. Immunoprecipitated material was consecutively washed with low salt wash buffer (20 mM Tris-HCl pH 7.9, 2 mM EDTA, 125 mM NaCl, 0.05% SDS, 1% Triton X-100, and Protease inhibitor cocktail [Sigma, #P8340]), high salt wash buffer (20 mM Tris-HCl pH 7.9, 2mM EDTA, 500 mM NaCl, 0.05% SDS, 1% Triton X-100, and Protease inhibitor cocktail [Sigma, #P8340]), LiCl wash buffer (10 mM Tris-HCl pH 7.9, 1 mM EDTA, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, and Protease inhibitor cocktail [Sigma, #P8340]), and 1x Tris-EDTA (TE). After elution (100 mM NaHCO3, 1% SDS), the immunoprecipitated material was digested with RNase (Roche, #11119915001) and proteinase K (ThermoFisher Scientific, #EO0491) prior to the purification and concentration of the immunoprecipitated genomic DNA by ChIP DNA Clean & Concentrator kit (Zymo Research, #D5201). ChIP-isolated DNA was subjected to qPCR (ChIP-qPCR) or library production (ChIP-seq) using Nebnext NGS DNA Library Preparation for Illumina kit (New England BioLabs, #E7645). Sequencing was performed with Illumina NextSeq 500 Mid Output (130M) by the UT Southwestern McDermott Center Next Generation Sequencing Core.
RNA-seq analysis
Reads with phred quality scores less than 20 and less than 35 bp after trimming were removed from further analysis using trimgalore version 0.4.1. Quality-filtered reads were then aligned to the mouse reference genome GRCm38 (mm10) using the HISAT (v 2.0.1) 43 aligner using default settings and marked duplicates using Sambamba version 0.6.6 44. Aligned reads were quantified using ‘featurecount’ (v1.4.6) 45 per gene ID against mouse Gencode version 20 46. Differential gene expression analysis was done using the R package DEseq2 (v 1.6.3) 47. Cutoff values of absolute fold change greater than 1.0 and FDR <= 0.05 were then used to select for differentially expressed genes between sample group comparisons. Unadjusted and adjusted p values of all comparisons are provided in Supplementary tables 1–4. All RNA-seq data have been deposited to GEO.
ChIP-seq analysis
Trimgalore version 0.4.1 (Martin, 2011) was used to remove adapter sequences and to remove reads shorter than 35bp or with phred quality scores less than 20. Trimmed reads were then aligned to the mouse reference genome (GRCm38/mm10) using default parameters in BWA version 0.7.12 48. The aligned reads were subsequently filtered for quality and uniquely mappable reads were retained for further analysis using Samtools version 1.3 49 and Sambamba version 0.6.6 44. Library complexity was measured using BEDTools version 2.26.0 50 and meets ENCODE data quality standards 51. Relaxed peaks were called using MACS version 2.1.0 52 with a p-value of 1×10−2. Gene annotation of the regions bound by p65 were performed by GREAT version 3.0 53. For heatmaps and profiles of ChIP-seq intensities, we used deepTools version 2.5.0 54 to generate read abundance from all ChIP-seq datasets around peak center (± 2.0 kb), using ‘computeMatrix’. These matrices were then used to create heatmaps and profiles, using deepTools commands ‘plotHeatmap’ and ‘plotProfile’ respectively. To identify the primary motif present in p65-occupied peaks, we performed de novo motif discovery using Homer version 4.9 with the ‘findMotifsGenome.pl’ script using default parameters 55,56. Additional detail regarding software and analysis can be found in the Reporting Summary.
In vivo insulin stimulation
After overnight fasting, anesthetized animals were injected with insulin (2 U per kg body weight; Eli Lily, #0002751001) through the portal vein. Gonadal WAT was immediately frozen 5 min after insulin injection. Tissues collected before insulin treatment were used as untreated controls for each mouse.
Metabolic phenotyping
For glucose tolerance tests (GTT), mice were subjected to overnight fast and then i.p. injected with glucose (Sigma, #G7021) at the dosage of 1g per kg body weight. For insulin tolerance tests (ITT), mice were i.p. administered with insulin (0.75 U per kg body weight; Eli Lilly, #0002751001) after a 6 hour fast. Glucose concentrations were determined using Bayer Contour glucometers.
Statistics and Reproducibility
No statistical method was used to predict sample sizes. We determined samples sizes on the basis of experience and reported experimental designs. No randomization or blinding was performed to allocate the samples. No criteria of inclusion or exclusion of data were used. Statistical analysis was carried out as indicated in figure legends. All data are presented as the mean + s.e.m. unless otherwise indicated in figure legends. Data variance was examined by F-test or Bartlett’s test. The data meet the assumptions of the indicated statistical analysis. All tests were performed as two sided. P value less than 0.05 was considered statistically significant. All statistical analyses were performed using Microsoft Excel or GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, CA, USA). All statistical information, including p values, samples sizes, and repetitions, are provided in Source Data associated with each figure.
Data Availability
All animal models, reagents, and datasets are freely available to investigators upon reasonable request. ChIP-seq data have been deposited to Gene Expression Omnibus (GEO Accession GSE134868). (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134868) RNA-seq data have been deposited to Gene Expression Omnibus (GEO Accession GSE151092). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151092 Mass spectrometry data has been deposited to MAssIVE (http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=af5cbb331b004854ac6c182936148168)
Extended Data
Extended Data Fig. 1: Thermoneutral housing amplifies the pro-inflammatory phenotype of FIPs.
a) Experimental design: 6 weeks-old male C57BL/6 mice were transferred to thermoneutral housing conditions (TN, 30 °C) or maintained at room temperature (RT, 22 °C) for two months. Then animals were fed HFD for 3 days prior to gene expression analysis of FIPs and APCs.
b) mRNA levels of indicated genes in freshly isolated FIPs and APCs obtained following 3 days of HFD. For each experiment, n = 8 for FIPs and APCs at 22 °C. n = 4 for FIPs and APCs at 30 °C. Data presented were reproduced in two independent experiments. Bars represent mean ± s.e.m., * denotes p< 0.05, ** denotes p<0.01, and *** denotes p<0.001 by two-way ANOVA.
c) mRNA levels of TNF-signaling signature genes (see Supplementary Table 1) in freshly isolated FIPs (n=8) and APCs (n=8) obtained following 1 month of HFD at 30 °C. Bars represent mean ± s.e.m., * denotes p< 0.05, ** denotes p<0.01, and *** denotes p<0.001 by unpaired two-tailed Student’s t-test.
Exact p values and numbers of repetitions can be found in Source Data Extended Data Figure 1.
Extended Data Fig. 2: Derivation of Mural-Tlr4KO mice and systemic phenotype following high fat diet feeding.
a) Mural-Tlr4KO (PdgfrbrtTA; TRE-Cre; Tlr4loxP/loxP) mice were generated by breeding the PdgfrbrtTA transgenic mice to animals expressing Cre recombinase under the control of the tetracycline-response element (TRE-Cre) and carrying floxed Tlr4 alleles (Tlr4loxP/loxP). Littermates carrying only PdgfrbrtTA and Tlr4loxP/loxP alleles (i.e. Cre-) were used as the control animals (Control). The addition of doxycycline (Dox) leads to inactivation of Tlr4 in Pdgfrb-expressing cells.
b) Intraperitoneal glucose tolerance tests (GTT) of RT-housed Control (n=9) and Mural-Tlr4KO (n=6) mice after HFD feeding. * denotes p< 0.05 by unpaired two-tailed Student’s t-test.
c) Intraperitoneal insulin tolerance tests (ITT) of RT-housed Control (n=9) and Mural-Tlr4KO (n=6) mice after HFD feeding. * denotes p< 0.05 by unpaired two-tailed Student’s t-test.
d-f) Levels of total monocytes and pro-inflammatory monocytes (LY6C+) in blood (d), bone marrow (e), and spleen (f) of Control (n=8) and Mural-Tlr4KO (n=8) mice after 5 months of HFD feeding. Bars represent mean ± s.e.m.
Exact p values can be found in Source Data Extended Data Figure 2. Data were reproduced two times in independent experiments.
Extended Data Fig. 3: Inactivation of Zfp423 exacerbates FIPs inflammatory responses.
a) Mural-Zfp423KO (PdgfrbrtTA; TRE-Cre; Zfp423loxP/loxP) mice were generated by breeding the PdgfrbrtTA transgenic mice to animals expressing Cre recombinase under the control of the tetracycline-response element (TRE-Cre) and carrying floxed Zfp423 alleles (Zfp423loxP/loxP). Littermates carrying only PdgfrbrtTA and Zfp423loxP/loxP alleles (i.e. Cre−) were used as the control animals (Control). The addition of doxycycline (Dox) leads to inactivation of Zfp423 in Pdgfrb-expressing cells.
b) mRNA levels of Zfp423 in FIPs of Control (n=3) and Mural-Zfp423KO (n=3) mice fed Dox-containing chow diet for 10 days.
c) mRNA levels of indicated pro-inflammatory genes in indicated FIPs treated with vehicle (veh.) (n=4) or 100 ng/ml LPS (n=8) for 2 hours.
d) mRNA levels of genes associated with macrophage activation in cultured BMDMs following exposure to indicated FIPs conditioned media (CM) for 1.5 hours. n=4 (for groups with vehicle treatment) or n=5 (for groups with LPS treatment) independent wells of macrophages examined per experiment.
e) Macrophage migration following exposure to FIPs conditioned media (CM): cell counts of migrated macrophages following exposure to indicated CM for 3 hours. n=4 (for groups with vehicle treatment) or n=9 (for groups with LPS treatment) independent wells of macrophages examined per experiment.
For panels d,e FIPs (isolated from pooled depots of 6–8 mice per genotype) were treated with vehicle or LPS (100 ng/ml) for 2 hours and then incubated in serum-free medium for an additional 24 hours to produce conditioned media.
Experiments in this figure were independently repeated three times. Data in this figure are shown as the mean ± s.e.m., *p< 0.05, **p<0.01 or ***p<0.001 by two-tailed unpaired Student’s t-test (b) or two-way ANOVA (c-e). Exact p values can be found in Source Data Extended Data Figure 3.
Extended Data Fig. 4: Adipogenesis from PDGFRβ+ progenitors is not impacted in Mural-Zfp423TG mice.
a) Representative 20x magnification images of APCs from gonadal WAT of Control (pooled from 4 mice) and Mural-Zfp423TG (pooled from 4 mice) animals maintained for 8 days in growth media. No overt difference in the degree of spontaneous adipogenesis is observed.
b) mRNA levels of key regulators of adipogenesis (Cebpa and Pparg2) in freshly isolated FIPs and APCs from gonadal WAT of Control (n=8) and Mural-Zfp423TG (n=12) mice after 1-month HFD feeding.
c) Average adipocyte size within gonadal WAT sections of control (n=8) and Mural-Zfp423TG (n=8) mice after 5-month Dox-HFD feeding.
d) mRNA levels of adipocyte-selective genes and fibrogenic genes in whole gonadal WAT of control (n=8) and Mural-Zfp423TG (n=11) mice after 5-month Dox-HFD feeding.
In b-d, bars represent mean ± s.e.m. ** denotes p <0.01 by one-way ANOVA (b).
e) MuralChaser-Zfp423TG (PdgfrbrtTA; TRE-Cre; TRE-Zfp423; Rosa26RmT/mG) mice were generated by reconstituting the Rosa26RmT/mG allele into the Mural-Zfp423TG background. The addition of doxycycline results in overexpression of Zfp423 and indelible labeling of Pdgfrb-expressing cells with mGFP expression.
f) Representative 63x magnification confocal immunofluorescence images of gonadal WAT sections from MuralChaser (PdgfrbrtTA; TRE-Cre; Rosa26RmT/mG) and MuralChaser-Zfp423TG mice after HFD feeding. 8 weeks-old animals were maintained on doxycycline-containing HFD feed for 8 weeks. Sections were stained with anti-PERILIPIN (PERILIPIN; red) and anti-GFP (green) antibodies and counterstained with DAPI (blue; nuclei). Composite images were generated by digital overlay.
g) Quantification of de novo adipogenesis: the frequency of GFP+ PERILIPIN+ cells observed by immunostaining was quantified by assaying ~3000–4000 adipocytes total from 5 mice per genotype. Bars represent mean ± s.e.m.
Exact p values can be found in Source Data Extended Data Figure 4. Data were reproduced two times in independent experiments.
Extended Data Fig. 5: Zfp423 overexpression in PDGFRβ+ cells confers metabolic benefits in obesity.
a) Western blot of phosphorylated AKT (pAKT), total AKT, and β-Actin protein expression in tissue extracts of gonadal WAT from Control (n=3) and Mural-Zfp423TG (n=3) mice after 5-month HFD feeding. Tissues were collected before (- insulin) and after insulin stimulation (+ insulin).
b) Intraperitoneal glucose tolerance tests (GTT) of thermoneutral-housed Control (n=9) and Mural-Zfp423TG (n=11) mice after HFD feeding. * denotes p< 0.05 by unpaired two-tailed Student’s t-test.
c) Intraperitoneal insulin tolerance tests (ITT) of thermoneutral-housed Control (n=9) and Mural-Zfp423TG (n=11) mice after HFD feeding. * denotes p< 0.05 by unpaired two-tailed Student’s t-test.
d) Levels of serum ADIPONECTIN and serum INSULIN (six-hour fasted) in Control and Mural-Zfp423TG mice maintained on doxycycline-containing chow diet or doxycycline-containing HFD for 1 month or 5 months. For control mice, n=3 of chow, n=9 of 1-month HFD, n=9 of 5-month HFD; for Mural-Zfp423TG mice, n=3 of chow, n=10 of 1-month HFD, n=7 of 5-month HFD.
e) Serum levels of indicated pro-inflammatory cytokines (CCL2, IL-6, TNFα) in Control (n=8) and Mural-Zfp423TG (n=8) mice maintained on doxycycline-containing HFD for 5 months.
f) mRNA levels of pro-inflammatory genes in skeletal muscle of Control (n=8) and Mural-Zfp423TG (n=8) mice after 5-months HFD feeding.
g) mRNA levels of pro-inflammatory genes in livers of Control (n=8) and Mural-Zfp423TG (n=8) mice after 5-months HFD feeding.
h-j) Levels of total monocytes and pro-inflammatory monocytes (LY6C+) in blood (h), bone marrow (i), and spleen (j) of Control (n=6) and Mural-Zfp423TG (n=7) mice after 5 months of HFD feeding.
For panels d-j, bars represent mean ± s.e.m., and * denotes p< 0.05 by unpaired two-tailed Student’s t-test. Exact p values, numbers of repetitions, and uncropped western blots can be found in Source Data Extended Data Figure 5.
Extended Data Fig. 6: Zfp423 inactivation in PDGFRβ+ cells exacerbates chronic WAT inflammation.
a) Approach: 6 weeks-old mice were preconditioned at thermoneutrality for two weeks then switched to a doxycycline-containing HFD for indicated period.
b) Control (n=8; black) and Mural-Zfp423KO (n=9; red) body weights following the onset of HFD feeding.
c) WAT mass mice after 1 month (Control, n=10; Mural-Zfp423KO, n=8) and 5 months (Control, n=8; Mural-Zfp423KO, n=8) of HFD feeding.
d) Frequency of total PDGFRβ+ cells, FIPs, and APCs within gonadal WAT (gWAT) of Control (n=10) and Mural-Zfp423KO (n=9) mice after 1 month of HFD feeding.
e) Zfp423 mRNA levels within freshly isolated FIPs and APCs from gWAT of Control (n=10) and Mural-Zfp423KO (n=9) mice after 1 month of HFD feeding.
f) Pro-inflammatory gene expression within freshly isolated FIPs and APCs from gWAT of Control (n=10) and Mural-Zfp423KO (n=9) mice after 1 month of HFD feeding.
g,h) 4x magnification images of H&E stained gWAT sections from Control mice maintained on HFD for 5 months.
i,j) 10x magnification images of PERILIPIN (red) and MAC-2 (green) expression within gWAT sections from Control mice maintained on HFD for 5 months.
k,l) 4x magnification images of H&E stained gWAT sections from from Mural-Zfp423KO mice maintained on HFD for 5 months.
m,n) 10x magnification images of PERILIPIN (red) and MAC-2 (green) expression within gWAT sections from Mural-Zfp423KO mice maintained on HFD for 5 months.
o) Pro-inflammatory- and macrophage-selective gene expression in gWAT of Control (n=9) and Mural-Zfp423KO (n=8) mice after 5-months HFD feeding.
p) Protein levels of indicated cytokines in gWAT od Control (n=8) and Mural-Zfp423KO (n=8) mice after 5-months HFD feeding.
q) Frequency of total adipose tissue macrophages (ATMs) within gWAT of Control (n=8) and Mural-Zfp423KO (n=8) mice after 5 months of HFD-feeding.
r) Frequency of pro-inflammatory ATMs within gWAT of Control (n=8) and Mural-Zfp423KO (n=8) mice after 5 months of HFD-feeding.
s) Frequency of tdTomato+ macrophages within gWAT SVF of obese mice following injection of tdTomato+ monocytes. n=8 for Control; n=8 for Mural-Zfp423TG.
Bars represent mean ± s.e.m., *p< 0.05, **p<0.01 or ***p<0.001 by unpaired two-tailed Student’s t-test (o-s) or one-way ANOVA (e, f). Exact p values and numbers of repetitions can be found in Source Extended Data Figure 6.
Extended Data Fig. 7: Vascular integrity and adipocyte differentiation from PDGFRβ+ perivascular progenitors are not impacted in Mural-Zfp423KO mice.
a) Representative images of gonadal and inguinal WAT depots of Control and Mural-Zfp423KO mice after Evans blue injection. Fixed tissues were imaged for Evans Blue auto fluorescence at 700 nm using a LI-COR Odyssey infrared imaging system.
b) Representative 10x magnification images of APCs from gonadal WAT of Control (pooled from 4 mice) and Mural-Zfp423KO (pooled from 4 mice) animals maintained for 8 days in growth media. No overt difference in the degree of spontaneous adipogenesis is observed.
c) mRNA levels of key regulators of adipogenesis (Cebpa and Pparg2) in freshly isolated FIPs and APCs from gonadal WAT of control (n=10) and Mural-Zfp423KO (n=9) mice after 1-month HFD feeding. Bars represent mean ± s.e.m., * denotes p<0.05, *** denotes p<0.001 by one-way ANOVA.
d) Average adipocyte size within gonadal WAT sections of control (n=11) and Mural-Zfp423KO (n=8) mice after 5-month Dox-HFD feeding.
e) mRNA levels of adipocyte-selective genes and fibrogenic genes in whole gonadal WAT of control (n=9) and Mural-Zfp423KO (n=8) mice after 5-month Dox-HFD feeding.
f) MuralChaser-Zfp423KO (PdgfrbrtTA; TRE-Cre; Zfp423loxP/loxP; Rosa26RmT/mG) mice were generated by reconstituting the Rosa26RmT/mG allele into the Mural-Zfp423KO background. The addition of doxycycline results in inactivation of Zfp423 and indelible labeling of Pdgfrb-expressing cells with mGFP expression.
g) Representative 63x magnification confocal immunofluorescence images of gonadal WAT sections from MuralChaser (PdgfrbrtTA; TRE-Cre; Rosa26RmT/mG) and MuralChaser-Zfp423KO mice after HFD feeding. 8 weeks-old animals were administered doxycycline-containing chow diet for 7 days and then switched to HFD feed (without doxycycline) for another 8 weeks. Sections were stained with anti-PERILIPIN (PERILIPIN; red) and anti-GFP (green) antibodies and counterstained with DAPI (blue; nuclei). Composite images were generated by digital overlay.
h) Quantification of de novo adipogenesis: the frequency of GFP+ PERILIPIN+ cells observed by immunostaining was quantified by assaying ~3000–4000 adipocytes total from 5 mice per genotype. Bars represent mean ± s.e.m.
Exact p values and numbers of repetitions can be found in Source Data Extended Data Figure 7.
Extended Data Fig. 8: Systemic consequences of inactivating mural cell Zfp423 in obese mice housed at thermoneutrality.
a) Western blot of phosphorylated AKT (pAKT), total AKT, and β-actin protein expression in tissue extracts of gonadal WAT from Control (n=3) and Mural-Zfp423KO (n=3) mice after 5-month HFD feeding. Tissues were collected before (- insulin) and after insulin stimulation (+ insulin) of anesthetized animals.
b) Intraperitoneal glucose tolerance tests (GTT) of thermoneutral-housed Control (n=12) and Mural-Zfp423KO (n=16) mice after HFD feeding. * denotes p< 0.05, ** denotes p< 0.01 by unpaired two-tailed Student’s t-test.
c) Intraperitoneal insulin tolerance tests (ITT) of thermoneutral-housed Control (n=8) and Mural-Zfp423KO (n=14) mice after HFD feeding. * denotes p< 0.05 by unpaired two-tailed Student’s t-test.
d) Serum levels of ADIPONECTIN (left) and INSULIN (six-hour fasting) (right) in Control and Mural-Zfp423KO mice maintained on doxycycline-containing chow diet or doxycycline-containing HFD for 1 month or 5 months. For control, n=5 of chow, n=8 of 1-month HFD, n=8 of 5-month HFD; for Mural-Zfp423KO, n=6 of chow, n=11 of 1-month HFD, n=9 of 5-month HFD.
e) Serum levels of indicated pro-inflammatory cytokines in Control (n=8) and Mural-Zfp423KO (n=8) mice maintained on doxycycline-containing HFD for 5 months
f) mRNA levels of pro-inflammatory genes in skeletal muscle of Control (n=8) and Mural-Zfp423KO (n=6) mice after 5-months HFD feeding.
g) mRNA levels of pro-inflammatory genes in livers of Control (n=8) and Mural-Zfp423KO (n=9) mice after 5-months HFD feeding.
h-j) Levels of total monocytes and pro-inflammatory monocytes (LY6C+) in blood (h), bone marrow (i), and spleen (j) of Control (n=6) and Mural-Zfp423KO (n=7) mice after 5 months of HFD feeding.
Bars represent mean ± s.e.m.. * denotes p< 0.05 by unpaired two-tailed Student’s t-test (d,e,g). Exact p values, numbers of repetitions, and uncropped western blots can be found in Source Data Extended Data Figure 8.
Extended Data Fig. 9: Modulating Zfp423 expression impacts the activation of NFκB signaling in FIPs.
a) Mural-Ikk2CA/Zfp423TG (PdgfrbrtTA; TRE-Cre; TRE-Zfp423; Rosa26RIkk2CA) mice were generated by breeding the Rosa26RIkk2CA allele into the Mural-Zfp423TG background. The addition of doxycycline (Dox) leads to overexpression of Zfp423 and simultaneous activation of constitutively active (CA) IKK2 in Pdgfrb-expressing cells.
b) mRNA levels of Zfp423 in cultured FIPs from Control (n=4), Mural-Ikk2CA (n=4), and Mural-Ikk2CA/Zfp423TG (n=4) mice following exposure to doxycycline (1μg/ml) in vitro for 1 day.
c) mRNA levels of indicated pro-inflammatory genes in cultured FIPs from Control (n=4), Mural-Ikk2CA (n=4), and Mural-Ikk2CA/Zfp423TG (n=4) mice following exposure to doxycycline (1μg/ml) in vitro for 1 day.
d) Mural-Ikk2CA/Zfp423KO (PdgfrbrtTA; TRE-Cre; Zfp423loxP/loxP; Rosa26RIkk2CA) mice were generated by breeding the Rosa26RIkk2CA allele into the Mural-Zfp423KO background. The addition of doxycycline (Dox) leads to inactivation of Zfp423 and simultaneous activation of constitutively-active (CA) IKK2 in Pdgfrb-expressing cells.
e) mRNA levels of Zfp423 in cultured FIPs from Control (n=4), Mural-Ikk2CA (n=4), and Mural-Ikk2CA/Zfp423KO (n=4) mice following exposure to doxycycline (1μg/ml) in vitro for 1 day.
f) mRNA levels of indicated pro-inflammatory genes in cultured FIPs from Control (n=4), Mural-Ikk2CA (n=4), and Mural-Ikk2CA/Zfp423KO (n=4) mice following exposure to doxycycline (1μg/ml) in vitro for 1 day.
Experiments in this figure were independently repeated three times. Data in this figure are shown as the mean ± s.e.m., *p< 0.05 or ***p<0.001 by one-way ANOVA. Exact p values can be found in Source Data Extended Data Figure 9.
Extended Data Fig. 10: Proposed Model: PDGFRβ+ perivascular cells regulate WAT macrophage accumulation in obesity.
FIPs are a subpopulation of PDGFRβ+ perivascular cells that are capable of exerting a pro-inflammatory phenotype. In lean (chow-fed) mice, the expression of ZFP423 in FIPs serves to moderate the activation of NFκB signaling and adipose tissue inflammation, through a molecular mechanism depicted in Fig. 8g. In response to high fat diet feeding, Zfp423 expression is reduced, facilitating the activation of NFκB signaling in FIPs and the production of pro-inflammatory cytokines/chemokines required to drive the chronic macrophage accumulation and activation that occurs during prolonged high fat diet feeding.
Supplementary Material
Acknowledgements
The authors are grateful to P. Scherer and C. Kusminski for critical reading of the manuscript and members of the UTSW Touchstone Diabetes Center for useful discussions. The authors thank Charlotte Lee, the UTSW Animal Resource Center, Metabolic Phenotyping Core, Pathology Core, Live Cell Imaging Core, Flow Cytometry Core, McDermott Sequencing Center, and Proteomics Core, for excellent guidance and assistance with experiments performed here. This study and/or personnel were supported in part by the NIH NIDDK F31DK113696 to C.H., NIDDK R01 DK104789, R56 DK119163, and R01 DK119163 to R.K.G., the American Heart Association postdoctoral fellowship 16POST26420136 to M.S, NIDDK R01 DK115477 to D.W.S, NIDDK R01 DK108773 and AHA 14SDG19880020 to D.O., American Diabetes Association 1–18-PMF-030 to V.A.P., NIH R03 DK101865 to O.T.G, the Cancer Prevention and Research Institute of Texas (RP150596) to S.D.B and V.S.M, NIH NIAAA K01AA024809 to L.J., and NIDDK PO1 DK088761to J.K.E.
Footnotes
Competing interests statement. The authors declare that they have no competing financial interests.
References
- 1.Weisberg SP et al. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J Clin Invest 116, 115–124, doi: 10.1172/JCI24335 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh DY, Morinaga H, Talukdar S, Bae EJ & Olefsky JM Increased macrophage migration into adipose tissue in obese mice. Diabetes 61, 346–354, doi: 10.2337/db11-0860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano SU et al. Local proliferation of macrophages contributes to obesity-associated adipose tissue inflammation. Cell Metab 19, 162–171, doi: 10.1016/j.cmet.2013.11.017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saltiel AR & Olefsky JM Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest 127, 1–4, doi: 10.1172/JCI92035 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hepler C et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife 7, doi: 10.7554/eLife.39636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giles DA et al. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat Med 23, 829–838, doi: 10.1038/nm.4346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian XY et al. Thermoneutral Housing Accelerates Metabolic Inflammation to Potentiate Atherosclerosis but Not Insulin Resistance. Cell Metab 23, 165–178, doi: 10.1016/j.cmet.2015.10.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill AA et al. Activation of NF-kappaB drives the enhanced survival of adipose tissue macrophages in an obesogenic environment. Mol Metab 4, 665–677, doi: 10.1016/j.molmet.2015.07.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang SH et al. The protein kinase IKKepsilon regulates energy balance in obese mice. Cell 138, 961–975, doi: 10.1016/j.cell.2009.06.046 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho KW, Morris DL & Lumeng CN Flow cytometry analyses of adipose tissue macrophages. Methods Enzymol 537, 297–314, doi: 10.1016/B978-0-12-411619-1.00016-1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill DA et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc Natl Acad Sci U S A 115, E5096–E5105, doi: 10.1073/pnas.1802611115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RK et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab 15, 230–239, doi: 10.1016/j.cmet.2012.01.010 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vishvanath L et al. Pdgfrbeta+ Mural Preadipocytes Contribute to Adipocyte Hyperplasia Induced by High-Fat-Diet Feeding and Prolonged Cold Exposure in Adult Mice. Cell Metab 23, 350–359, doi: 10.1016/j.cmet.2015.10.018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RK et al. Transcriptional control of preadipocyte determination by Zfp423. Nature 464, 619–623, doi: 10.1038/nature08816 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hepler C et al. Directing visceral white adipocyte precursors to a thermogenic adipocyte fate improves insulin sensitivity in obese mice. Elife 6, doi: 10.7554/eLife.27669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao M et al. Fetal development of subcutaneous white adipose tissue is dependent on Zfp423. Mol Metab 6, 111–124, doi: 10.1016/j.molmet.2016.11.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao M et al. Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell Metab 23, 1167–1184, doi: 10.1016/j.cmet.2016.04.023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao M et al. De novo adipocyte differentiation from Pdgfrbeta(+) preadipocytes protects against pathologic visceral adipose expansion in obesity. Nat Commun 9, 890, doi: 10.1038/s41467-018-03196-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloting N et al. Insulin-sensitive obesity. Am J Physiol Endocrinol Metab 299, E506–515, doi:ajpendo.00586.2009 [pii] 10.1152/ajpendo.00586.2009 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Huang B, Yang XD, Lamb A & Chen LF Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal 22, 1282–1290, doi: 10.1016/j.cellsig.2010.03.017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LF, Mu Y & Greene WC Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J 21, 6539–6548, doi: 10.1093/emboj/cdf660 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Fischle W, Verdin E & Greene WC Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 293, 1653–1657, doi: 10.1126/science.1062374 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Torchy MP, Hamiche A & Klaholz BP Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci 72, 2491–2507, doi: 10.1007/s00018-015-1880-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauberth SM & Rauchman M A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. J Biol Chem 281, 23922–23931, doi: 10.1074/jbc.M513461200 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Reilly SM & Saltiel AR Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol 13, 633–643, doi: 10.1038/nrendo.2017.90 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Jia L et al. Hepatocyte Toll-like receptor 4 regulates obesity-induced inflammation and insulin resistance. Nat Commun 5, 3878, doi: 10.1038/ncomms4878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao C et al. Short-Term Versus Long-Term Effects of Adipocyte Toll-Like Receptor 4 Activation on Insulin Resistance in Male Mice. Endocrinology 158, 1260–1270, doi: 10.1210/en.2017-00024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardamone MD et al. A protective strategy against hyperinflammatory responses requiring the nontranscriptional actions of GPS2. Mol Cell 46, 91–104, doi: 10.1016/j.molcel.2012.01.025 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillum MP et al. SirT1 regulates adipose tissue inflammation. Diabetes 60, 3235–3245, doi: 10.2337/db11-0616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saijo K et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 137, 47–59, doi: 10.1016/j.cell.2009.01.038 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Addison WN et al. Zfp423 Regulates Skeletal Muscle Regeneration and Proliferation. Mol Cell Biol 39, doi: 10.1128/MCB.00447-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng LE, Zhang J & Reed RR The transcription factor Zfp423/OAZ is required for cerebellar development and CNS midline patterning. Dev Biol 307, 43–52, doi: 10.1016/j.ydbio.2007.04.005 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng LE & Reed RR Zfp423/OAZ participates in a developmental switch during olfactory neurogenesis. Neuron 54, 547–557, doi: 10.1016/j.neuron.2007.04.029 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warming S, Rachel RA, Jenkins NA & Copeland NG Zfp423 is required for normal cerebellar development. Mol Cell Biol 26, 6913–6922, doi: 10.1128/MCB.02255-05 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiernan R et al. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem 278, 2758–2766, doi: 10.1074/jbc.M209572200 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Yeung F et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J 23, 2369–2380, doi: 10.1038/sj.emboj.7600244 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spallanzani RG et al. Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci Immunol 4, doi: 10.1126/sciimmunol.aaw3658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahlakoiv T et al. Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci Immunol 4, doi: 10.1126/sciimmunol.aax0416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo X et al. PARP-1 Controls the Adipogenic Transcriptional Program by PARylating C/EBPbeta and Modulating Its Transcriptional Activity. Mol Cell 65, 260–271, doi: 10.1016/j.molcel.2016.11.015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan B et al. The metabolic ER stress sensor IRE1alpha suppresses alternative activation of macrophages and impairs energy expenditure in obesity. Nat Immunol 18, 519–529, doi: 10.1038/ni.3709 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Oh DY et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142, 687–698, doi: 10.1016/j.cell.2010.07.041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh DY et al. A Gpr120-selective agonist improves insulin resistance and chronic inflammation in obese mice. Nat Med 20, 942–947, doi: 10.1038/nm.3614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360, doi: 10.1038/nmeth.3317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarasov A, Vilella AJ, Cuppen E, Nijman IJ & Prins P Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034, doi: 10.1093/bioinformatics/btv098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, doi: 10.1093/bioinformatics/btt656 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Frankish A et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47, D766–D773, doi: 10.1093/nar/gky955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842, doi: 10.1093/bioinformatics/btq033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landt SG et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22, 1813–1831, doi: 10.1101/gr.136184.111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng J, Liu T, Qin B, Zhang Y & Liu XS Identifying ChIP-seq enrichment using MACS. Nat Protoc 7, 1728–1740, doi: 10.1038/nprot.2012.101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLean CY et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501, doi: 10.1038/nbt.1630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramirez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165, doi: 10.1093/nar/gkw257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinz S, Romanoski CE, Benner C & Glass CK The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16, 144–154, doi: 10.1038/nrm3949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All animal models, reagents, and datasets are freely available to investigators upon reasonable request. ChIP-seq data have been deposited to Gene Expression Omnibus (GEO Accession GSE134868). (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134868) RNA-seq data have been deposited to Gene Expression Omnibus (GEO Accession GSE151092). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151092 Mass spectrometry data has been deposited to MAssIVE (http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=af5cbb331b004854ac6c182936148168)