Abstract
The DNA damage response (DDR) is necessary to maintain genome integrity and prevent the accumulation of oncogenic mutations. Consequently, proteins involved in the DDR often serve as tumor suppressors, carrying out the crucial task of keeping DNA fidelity intact. Mediator of DNA damage checkpoint 1 (MDC1) is a scaffold protein involved in the early steps of the DDR. MDC1 interacts directly with γ-H2AX, the phosphorylated form of H2AX, a commonly used marker for DNA damage. It then propagates the phosphorylation of H2AX by recruiting ATM kinase. While the function of MDC1 in the DDR has been reviewed previously, its role in cancer has not been reviewed, and numerous studies have recently identified a link between MDC1 and carcinogenesis. This includes MDC1 functioning as a tumor suppressor, with its loss serving as a biomarker for cancer and contributor to drug sensitivity. Studies also indicate that MDC1 operates outside of its traditional role in DDR, and functions as a co-regulator of nuclear receptor transcriptional activity, and that mutations in MDC1 are present in tumors and can also cause germline predisposition to cancer. This review will discuss reports that link MDC1 to cancer and identify MDC1 as an important player in tumor formation, progression, and treatment. We also discuss mechanisms by which MDC1 levels are regulated and how this contributes to tumor formation.
Keywords: MDC1, cancer, DNA damage response, γ-H2AX, DNA damaging agents, transcriptional co-regulator
Introduction
The DNA damage response (DDR) is crucial for the maintenance of genome integrity and cell survival. A number of proteins involved in the DDR have been associated with carcinogenesis, due to the potential for unresolved DNA damage to contribute to genomic aberrations and malignant transformation. Mediator of DNA damage checkpoint 1 (MDC1), also known as nuclear factor with BRCT domains 1 (NFBD1), is a scaffold protein involved in the early steps of the DDR, accumulating at sites of DNA double-stranded breaks within one minute of genomic insult [1]. There are several known sensors of DNA double-stranded breaks, including poly ADP-ribose polymerase-1 (PARP-1), the MRN complex (made up of MRE11, RAD50, and NBS1), the Ku70/80 complex, and most recently identified, SIRT6 [2–5]. Although the MRN complex is lower in abundance and may arrive more slowly than PARP-1 and Ku70/80, it is required for activation of the ATM kinase, a critical step in double stranded break repair [6]. When a DNA double-stranded break is sensed by the MRN complex (Fig. 1A), the free DNA ends directly interact with RAD50 [7, 8]. The MRN complex then recruits the ATM kinase [9, 10], which is responsible for the phosphorylation of histone H2AX at serine 139 to form γ-H2AX [11, 12] (Fig. 1B). Histone H2AX is an H2A variant which makes up 2–25% of the H2A in mammalian cells, and its phosphorylation plays a crucial role in the DDR [12, 13]. While the MRN complex is sufficient for the initial recruitment of ATM and phosphorylation of the closest H2AX molecules, this phosphorylation is propagated to neighboring molecules of H2AX via MDC1.
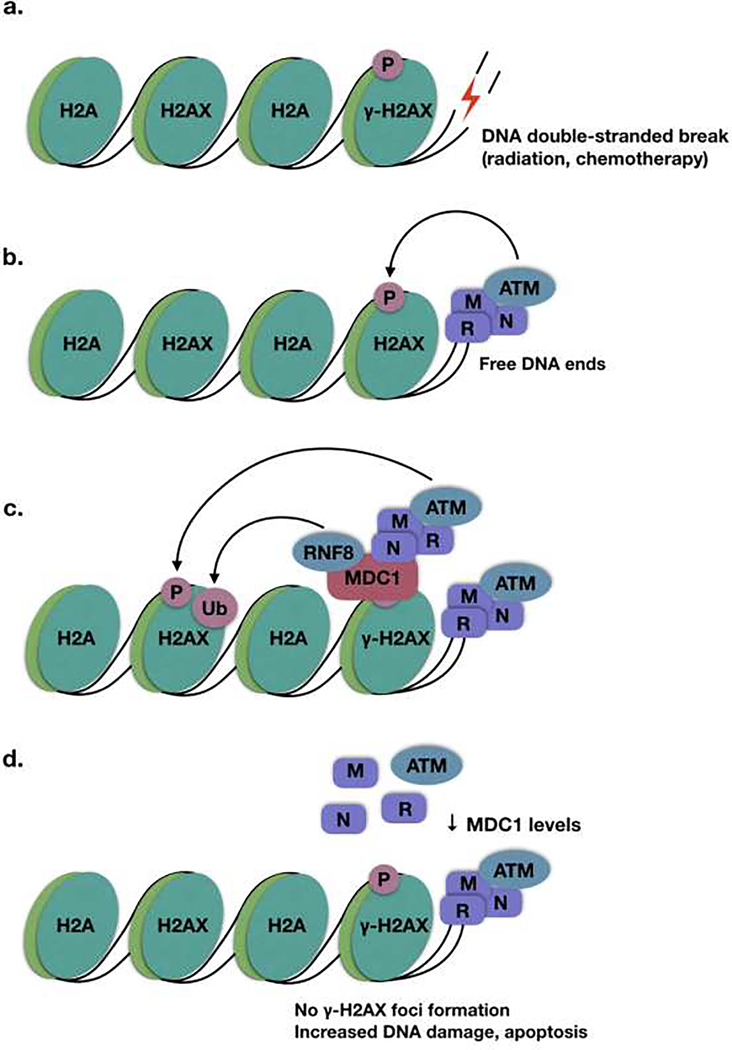
Figure 1. MDC1 is a scaffold protein which propagates the phosphorylation of H2AX to form γ-H2AX.
A. DNA double-stranded breaks form free DNA ends. B. Free DNA ends are detected by the MRN complex, which recruits ATM kinase to phosphorylate H2AX to form γ-H2AX. C. MDC1 interacts directly with γ-H2AX to recruit additional MRN complex and ATM, amplifying the γ-H2AX signal. It also recruits RNF8, resulting in the ubiquitination of γ-H2AX. D. In the absence of MDC1, free ends are still detected by the MRN complex, but the γ-H2AX signal is not propagated onto nearby chromatin to form visible γ-H2AX foci.
MDC1 functions to amplify the phosphorylation of H2AX to form γ-H2AX. γ-H2AX can extend as much as 2 Mb beyond the location of the DNA double-stranded break [14]. It has been shown that MDC1 is necessary for the high density accumulation of γ-H2AX in the 200 kb surrounding the DNA double-stranded break. Although free, activated ATM is able to phosphorylate additional H2AX between 200 and 600 kb from the double-stranded break, MDC1 is necessary for the high density accumulation of γ-H2AX directly surrounding the DNA lesion [15]. To achieve this, it interacts directly with γ-H2AX via its BRCT domain and recruits additional MRN complex [16, 17]. In turn, additional ATM is recruited by the MRN complex to propagate the phosphorylation of H2AX, resulting in an amplification of the γ-H2AX signal (Fig. 1c). The amplification of this signal and accumulation of additional DDR proteins results in DDR foci, which are visible by fluorescence microscopy. In addition to mediating the phosphorylation of H2AX, MDC1 itself is phosphorylated by ATM, and this phosphorylation is required for the ubiquitylation of γ-H2AX by RNF8, which is a necessary step in the response to genotoxic stress that is required for homologous recombination (Fig. 1b) [1, 18, 19]. This ubiquitylation has been shown to initiate a ubiquitylation cascade, with the E3 ligase RNF168 directly binding to RNF8-ubiquitylated histones [20]. This is required for the recruitment of downstream factors and the resolution of double-stranded breaks.
As an early effector in the DDR, MDC1 is required for homologous recombination repair of DNA double-stranded breaks [21]. In many respects, mice lacking MDC1 phenocopy H2AX knockout mice, displaying radiation sensitivity, chromosomal instability, growth defects, male infertility, and immune defects [16, 22, 23]. Although tumorigenesis has not been studied in MDC1 knockout mice, H2AX homozygous null and haploinsufficient mice show increased tumor burden (mainly lymphomas), particularly in the absence of p53 [24]. Similarly, RNF8 knockout mice show radiation sensitivity, growth defects, and tumor susceptibility (mainly lymphoma and thymoma), as well as immune deficiency due to a defect in IgH class switch recombination [25, 26].
While the role of MDC1 in the DNA damage response has been reviewed previously [27–31], these reviews focused on the molecular action of MDC1, rather than its potential role in carcinogenesis. This is likely because many of the associations between MDC1 and cancer have only been identified in the last 5–10 years. In this review, we will focus on new discoveries which relate MDC1 to cancer development, progression, and treatment.
MDC1 expression in cancer
Due to the role of MDC1 in the DDR, and the known associations between the DDR and oncogenesis, a number of studies have evaluated the expression of MDC1 in tumors relative to benign tissue. It has been hypothesized that the expression of MDC1, like other DDR proteins, may have either pro- or anti-oncogenic effects. Because MDC1 is crucial for the DNA damage response, the loss of MDC1 may result in mutagenesis and malignant transformation. However, MDC1 also interacts directly with p53 to inhibit its activity and prevent apoptosis [32, 33]. Therefore, the presence of MDC1 could oppose apoptosis, keeping early tumor cells viable.
Initial studies of breast, lung, and testicular germ cell tumors evaluated the expression of MDC1 and its downstream effector, 53BP1 [34], by immunohistochemistry (IHC) [35]. Bartkova et al. found ubiquitous expression of both proteins in benign tissue, but reduction or loss of MDC1 in 30% of breast cancer and 26% of lung cancer samples tested. Similarly, 53BP1 was reduced in 26% and 24% of breast and lung cancer samples tested, respectively. This loss was, in most cases, mutually exclusive, meaning that over half of each cancer type had reduced expression of either MDC1 or 53BP1, consistent with studies showing that they function in the same pathway [34]. In contrast, there was no evidence of MDC1 or 53BP1 loss in testicular germ cell tumors. A lower, but substantial percentage of endometrial cancer samples (~9%)show MDC1 loss, despite observed ubiquitous expression in benign endometrial tissue [36], and a small percentage of ovarian carcinomas have been found to lack MDC1 [37]. While low grade prostate tumors show a slight increase in MDC1 levels, higher grade tumors show MDC1 protein loss [38]. MDC1 protein expression has not been evaluated in gliomas or bladder cancer, but they both show a reduction in MDC1 RNA levels relative to local tissue [39, 40]. Overall, studies across a variety of tumors show MDC1 loss relative to benign tissue at the RNA and protein levels, indicating a potential tumor-suppressive role for MDC1.
Additional studies have found a correlation between MDC1 loss and tumor aggression. For example, MDC1 loss can predict recurrent nodal metastases in breast cancer patients [41]. This was consistent with a later study, which found that MDC1 expression decreased with increasing stage in breast cancer [42].Similarly, the Sox9-mediated up-regulation of MDC1 expression was associated with reduced perineural and lymph node invasion in pancreatic cancer [43]. Altogether, the consensus of a number of studies is that low MDC1 expression correlates with more invasive, later stage tumors, indicating that MDC1 expression may serve to limit the progression of disease.
However, there are also cases in which MDC1 overexpression appears to play a role in cancer initiation or progression. The first of these studies, by Yuan et al. in 2011, found that MDC1 is upregulated in about 60% of cervical cancer samples, using small datasets of 40–60 tumors with case matched normal tissues [44]. Similarly, in laryngeal squamous cell carcinoma (SCC), MDC1 protein levels are increased on immunohistochemistry by 56% of tumor samples tested compared with 27% of benign tissues [45]. MDC1 expression is also found in only 20% of benign nasopharyngeal tissue, but in 72% of nasopharyngeal carcinoma [46]. Although a correlation has been found between high MDC1 expression in ovarian carcinoma and lower overall survival, the levels of MDC1 in benign tissue were not evaluated in this study [47]. These findings suggest that MDC1 expression is actually higher in certain cancers. Notably, the majority of cervical cancer and some laryngeal SCC are usually driven by human papillomavirus infection (HPV), and nasopharyngeal carcinoma by Epstein-Barr virus (EBV) [48–50]. Viral carcinogenesis activates the DNA damage response via several mechanisms, both via the viral infection itself and through expression of viral proteins [51, 52]. Therefore, it is possible that the higher levels of MDC1 in these cancers is a passenger event rather than a driver in tumorigenesis, due to the viral integration and subsequent activation of the DNA damage response.
Taken together, the work characterizing MDC1 expression in cancer indicates that MDC1 often functions as a tumor suppressor, with MDC1 loss being associated with tumor development. However, a subset of cancers, and primarily virally-driven tumors, have up-regulation of MDC1. These findings are summarized in Table 1. This association between MDC1 expression and potentially both oncogenic and tumor suppressive roles indicates an opportunity for further research to understand its complex role in cancer.
Table 1.
Summary of MDC1 expression and associated treatment sensitivities in cancer. This table summarizes the MDC1 levels in different types of benign tissue and tumors, as well as demonstrated therapeutic sensitivities with MDC1 loss in these tumors.
| Cancer type | MDC1 levels in benign tissue | MDC1 levels in tumor | Therapeutic sensitivities with MDC1 loss/knockdown |
|---|---|---|---|
| Pancreatic | High (IHC) [43] | Reduced in invasive tumors (IHC) [43] | Unknown |
| Breast | High (IHC) [35] | Reduced, particularly in high stage and nodal metastases (IHC) [35, 41, 42] | Doxorubicin, cisplatin [57] |
| Lung | High (IHC) [35] | Reduced (IHC) [35] | Unknown |
| Glioma | Both MDC1 and MDC1-AS are high (qPCR, Western blot) [39] | Both MDC1 and MDC1-AS are reduced (qPCR, Western blot) [39] | Unknown |
| Bladder | Both MDC1 and MDC1-AS are high (RNA) [40] | Both MDC1 and MDC1-AS are reduced (RNA) [40] | Unknown |
| Gastric | MDC1-AS is high (RNA) [87] | MDC1-AS is reduced (RNA) [87] | Unknown |
| Prostate | Moderate [38] | Increased in well-differentiated tumors (IHC) Reduced in poorly differentiated tumors (IHC) [38] |
Unknown |
| Cervical | Low (IHC) [44] | Increased (RNA and IHC) [44] | Doxorubicin [44], cisplatin [58] |
| Squamous cell | Low (laryngeal, IHC) [45] | Increased (laryngeal, IHC) [45] Increased in lymph node mets (oral, IHC) [107] |
Radiation (laryngeal) [61] |
| Nasopharyngeal | Low (IHC) [46] | Increased (IHC) [58] | Doxorubicin [46], cisplatin [46, 59], 5-FU [46, 59], radiation [62], olaparib [64] |
| Endometrial | Unknown | Lost in a subset of patients (IHC) [36] | Unknown |
| Hepatocellular | Unknown | Increased expression of repressive miRNA [90] | Unknown |
| Ovarian | Unknown | Increased expression correlates with low survival (IHC) [47] Low expression in small subset of patients (IHC) [37] |
Unknown |
| Esophageal | Unknown | Unknown | Doxorubicin, cisplatin [56] |
MDC1 loss and sensitivity to DNA damaging agents
As previously discussed, several studies have found associations between MDC1 loss and cancer incidence as well as higher grade, more aggressive tumors [35–37, 41–43]. However, even prior to its association with cancer, early studies on the function of MDC1 in the DDR noted that MDC1 knockdown resulted in increased radiosensitivity and sensitivity to DNA-damaging chemotherapeutics [32, 53]. After its identification as a tumor suppressor, which is lost in a subset of cancers, this observation was used to hypothesize that tumors lacking MDC1 may be highly sensitive to these treatments.
In particular, the effect of MDC1 loss on Adriamycin (doxorubicin), CDDP (cisplatin), and radiation sensitivity has been extensively studied. Doxorubicin is a topoisomerase II inhibitor which results in the induction of single- and double-stranded DNA breaks [54]. In addition, it is responsible for generating free radicals which contribute to additional DNA damage, as well as increased cell stress and induction of apoptosis [55]. A number of studies have observed that MDC1 loss increases sensitivity to doxorubicin. This has been demonstrated in esophageal [56], nasopharyngeal [46], breast [57], and cervical carcinoma [44]. Several of these studies, as well as others, also indicate enhanced sensitivity to cisplatin with MDC1 knockdown [56–59]. Like doxorubicin, the primary mechanism of action for cisplatin therapy is the induction of DNA damage. Cisplatin crosslinks to purine bases of DNA, halting cell cycle progression, and ultimately resulting in apoptosis if the damage is not repaired. However, cisplatin also causes DNA damage through the formation of free radicals, and promotes apoptosis via other pathways, including modulation of calcium signaling and activation of ERK [60]. It is likely that with loss of MDC1, the defect in the DDR results in an inability to repair the DNA damage caused by doxorubicin and cisplatin, resulting in increased apoptosis in the presence of these chemotherapeutic agents. This is true as well for radiation, where MDC1 loss has been shown to increase radiation-induced apoptosis in a variety of cancer types, including laryngeal squamous cell carcinoma [61] and nasopharyngeal carcinoma [62]. Finally, two studies in nasopharyngeal carcinoma find increased sensitivity to the pyrimidine analog 5-fluorouracil (5-FU) upon MDC1 knockdown [46, 59].
Radiation, doxorubicin, and cisplatin work by inducing DNA damage in all cells, and preferentially killing faster growing cells or cells with an impaired DDR. However, ultimately, all cells including normal cells will be affected, resulting in a low therapeutic index and high toxicity for these drugs. An alternative therapy emerging for tumors with DDR defects is Poly(ADP-ribose) polymerase (PARP) inhibition. PARP inhibitors block the repair of endogenous single-stranded DNA breaks by PARP enzymes, resulting in cytotoxic double-stranded DNA breaks [63]. Therapeutically, they are synthetically lethal with Breast Cancer gene (BRCA) 1 or 2 deficiency, which causes a defect in homologous recombination. PARP inhibition is therefore significantly less toxic than conventional chemotherapeutics to normal cells. Wang et al. reveals that MDC1 loss may result in increased sensitivity to PARP inhibitors in nasopharyngeal carcinoma [64]. Although more work is required to determine whether this is a therapeutically viable option, it is promising evidence that the use of PARP inhibitors may be expanded beyond BRCA mutated tumors.
An interesting observation regarding MDC1 loss and sensitivity to DNA-damaging chemotherapeutics is the signature combination of increased DNA damage with a failure to accumulate γ-H2AX foci. Certain chemotherapeutics, including doxorubicin and cisplatin, and radiation induce DNA double-stranded breaks. γ-H2AX, represents an early step in DNA damage recognition. Recall that MDC1 directly interacts with γ-H2AX to recruit ATM, the kinase which propagates the phosphorylation of H2AX to form γ-H2AX [16]. Many studies use γ-H2AX as a surrogate marker for DNA damage, due to its early role in the DDR. However, in the case of MDC1 loss, the DDR is interrupted upstream of γ-H2AX foci formation, resulting in a paradoxical increase in DNA damage with a reduction in the formation of γ-H2AX (Fig. 1d). This observation is consistently seen across several of the studies reviewed above [46, 57–59, 62, 64]. This phenomenon is important to note: when evaluating systems for an increase or decrease in DNA damage, it may be misleading to rely on γ-H2AX as a marker for DNA damage, particularly in the case where upstream mediators, such as MDC1, may be perturbed. Instead, it is important to combine γ-H2AX levels with an assay for directly detecting DNA double stranded breaks, such as a neutral comet assay. In this assay, cells are run on an electrophoretic field. Intact DNA stays inside the nucleus, while DNA with double-stranded breaks migrates and forms a comet-like tail [65, 66]. This is a direct measure of DNA double-stranded breaks. The combination of increased DNA damage via comet assay with a reduction in γ-H2AX foci formation may be a useful fingerprint of an upstream DDR defect, such as loss of MDC1.
MDC1 as a transcriptional co-regulator in cancer
It has been clearly demonstrated that the role of MDC1 in the DDR has potential to impact carcinogenesis and cancer treatment. However, MDC1 has also been reported to function as a transcriptional co-regulator for the nuclear hormone receptors, Estrogen Receptor α (ERα) and Androgen Receptor (AR) in breast and prostate cancer, respectively [38, 42].
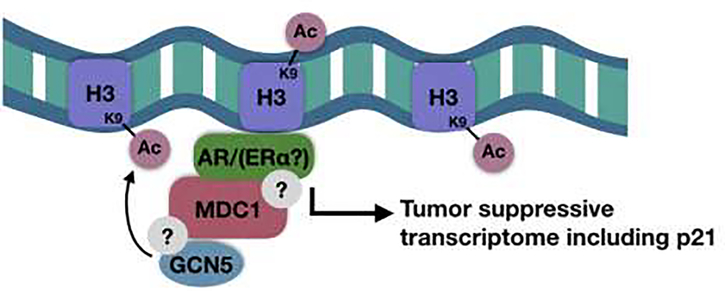
Zou et al. found that in breast cancer, MDC1 directly interacts with ERα, resulting in its co-activation and up-regulation of ERα target genes, including p21, a negative cell cycle regulator. The authors propose that this MDC1 co-activation and resulting increase in p21 expression serves as a barrier to prevent malignant transformation in breast tissue. Consistent with this model, they observed loss of MDC1 in later stage breast cancer [42]. This is also consistent with Patel et al., which showed MDC1 loss in metastatic breast cancer [41]. A similar model has been proposed by Zhao et al. in a study of MDC1 in prostate cancer, where MDC1 was identified as an AR co-activator in a drosophila-based screen [38]. The screen used the position effect variegation (PEV) system in drosophila, which uses GFP and white expression to identify euchromatic and heterochromatic regions [67]. Using this method, transcriptional co-regulators can be identified via monitoring the chromatin remodeling as expression levels of GFP and white fluctuate [68]. Upon the identification of the MDC1 homolog mu2 as an AR co-activator, MDC1 co-activation of AR was validated in human prostate cancer cells. MDC1 was shown to associate with AR via immunoprecipitation, and MDC1 knockdown resulted in decreased expression of a subset of AR target genes, including p21. To determine how MDC1 loss may be altering AR transcription, it was shown that MDC1 knockdown reduces H3K9 acetylation at androgen response elements (AREs). H3K9 acetylation mediates the transition between transcriptional initiation and elongation by facilitating the release of RNA Pol II [69]. MDC1 knockdown results in impaired recruitment of GCN5, the histone acetyltransferase that has been shown to acetylate H3K9 on AREs, resulting in reduced H3K9 acetylation and reduced AR-mediated transcription in the absence of MDC1 (Fig. 2). However, this study did not determine whether the interaction between MDC1 and either the AR or GCN5 is a direct interaction, or if they exist in complex with other potential binding partners. In fact, it has been shown that GCN5 is recruited and functions to acetylate histones during DNA double-stranded break repair [70]. Therefore, it is possible that AR co-activation by MDC1 is actually occurring in the context of DNA damage. However, it is proposed that, similarly to its function in ER-positive breast cancer, MDC1 functions to maintain a tumor-suppressive AR gene transcriptome and, when lost, may contribute to tumor development [38, 42]. Still, it is currently unknown whether the co-activation of ERα also occurs via recruitment of GCN5, or if this is through another mechanism entirely.
Figure 2. Potential model for MDC1-mediated co-activation of AR and ERα.
MDC1 has been shown to co-immunoprecipitate with both AR and ERα, and there is evidence that it plays a role in recruiting GCN5 to acetylate H3K9, resulting in increased transcription of p21 [38, 42].
A multitude of studies have described crosstalk between DDR proteins and nuclear hormone receptor transcriptional activation [71]. The nuclear hormone receptors AR and ER are known to influence the transcription of proteins involved in a variety of DDR pathways. For instance, it has been shown that AR signaling promotes DNA repair in prostate cancer via direct induction of DDR gene expression, including upregulation of DNA PK-cs, ATR, and PARP1. [72, 73]. Additionally, both AR and ER have been shown to utilize topoisomerase II β in order to induce double-stranded breaks, relieving torsional stress for transcriptional activation [74–76]. These transcription-associated double-stranded breaks result in the recruitment of DDR proteins, and it is possible that these factors are then locally available to act as transcriptional co-regulators. Alternatively, the repair of the damaged ER or AR target genes may indirectly result in their up-regulation, simply by allowing for transcription of genes that were previously interrupted by double-stranded breaks.
This interplay between the DDR and nuclear hormone receptor signaling is of great interest for therapeutic purposes. For instance, hormonal therapy in combination with radiotherapy is a common treatment in prostate cancer and serves to improve survival [77, 78]. Additionally, there is emerging evidence that treatment with androgen deprivation therapy in prostate cancer confers a functional inhibition of homologous recombination, impairing the DNA damage response and rendering cells sensitive to PARP inhibition [79, 80]. This has led to a variety of clinical trials attempting to combine PARP inhibition with AR-targeting therapies [81]. Fully understanding how nuclear receptors influence DDR proteins and vice versa may be critical to utilization of these therapeutic strategies.
Regulation of MDC1 levels in cancer
The abundance of studies describing the impact of MDC1 loss or overexpression in various types of cancer raises the question of what regulates MDC1 transcript and protein levels. Wang et al. examined MDC1 in breast cancer, and found that Liver receptor homolog-1 (LRH1) is a transcription factor responsible for promoting MDC1 expression and subsequent chemoresistance in breast cancer cells [57]. Additionally, the MDC1 promoter contains transcription factor binding sites for STAT-1, Sp1, and NF-Y, although only Sp1 and NF-Y have been shown to bind to the promoter, and Sp1 binding was shown to be most important for MDC1 transcription [82]. In addition to identifying these transcription factors, several studies have attempted to address the question of what regulates MDC1 expression by examining non-coding RNAs. Several non-coding RNAs have been shown to alter MDC1 transcript levels, serving to either up- or down-regulate MDC1 mRNA.
Over 90% of the human genome is actively transcribed into noncoding RNA, compared with the 2% that represent protein-coding genes [83, 84]. For this reason, the role of these noncoding RNAs in carcinogenesis is of great interest, with a variety of long noncoding RNAs (lncRNAs) being associated with both tumor suppression and oncogenesis [85]. Noncoding RNA transcripts which are transcribed from the opposite DNA strand of a protein-coding region are referred to as antisense transcripts. Over 70% of transcribed RNAoverlaps with opposite strand coding regions [86]. Katayama et al. found that these antisense transcripts are often regulated similarly to their opposite strand coding transcripts, and that antisense RNAs can alter the transcription of their sense strand counterparts. This suggests a role for antisense RNA in coding gene regulation.
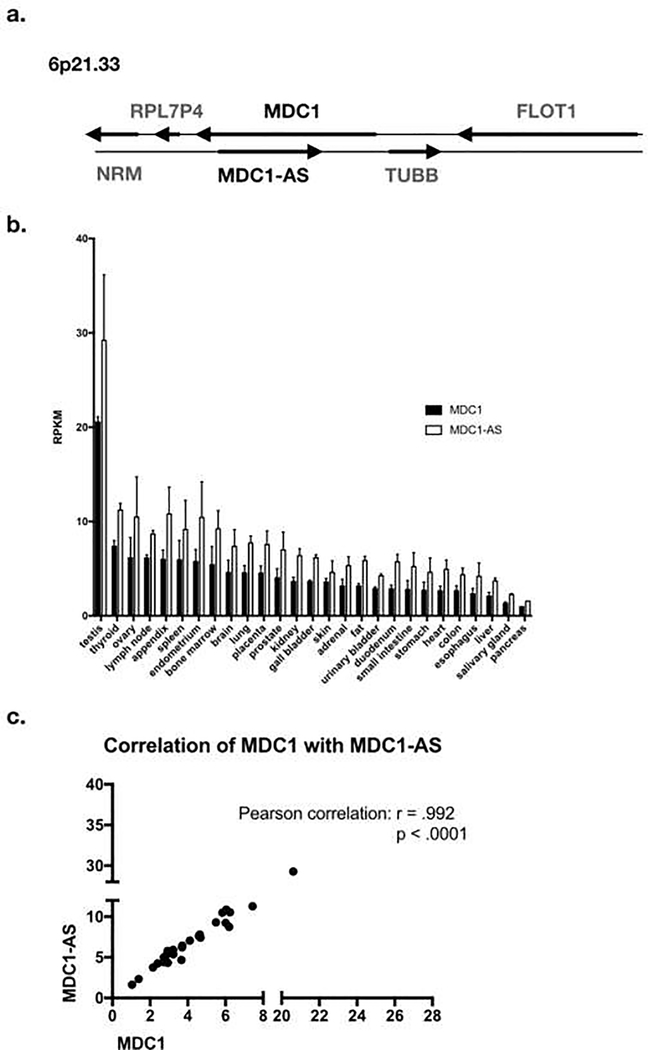
MDC1-AS (antisense) is a lncRNA that has been shown to be down-regulated in gastric cancer, bladder cancer, and glioma [39, 40, 87]. It is transcribed on the antisense strand of the MDC1 transcript (Fig. 3A). In accordance with the above findings, MDC1-AS appears to play a part in the regulation of MDC1 levels. A microarray screen for differentially expressed lncRNAs in bladder cancer identified MDC1-AS as a downregulated lncRNA [40]. They found that this MDC1-AS tumor-suppressive activity was dependent on the presence of MDC1, and that MDC1-AS expression promoted up-regulation of MDC1 mRNA itself. The mechanism by which this occurs is unclear, although one study has found that antisense RNA may increase the stability of the sense mRNA counterpart, in the case of β-secretase expression in Alzheimer’s [88]. These findings have been recapitulated in glioma and gastric cancer, finding lower levels of MDC1-AS and MDC1 itself in tumors compared with benign tissue [39, 87]. Consistent with this, analysis of RNA-seq data of normal tissues from the Human Protein Atlas (HPA) indicates that expression of MDC1 mRNA strongly correlates with expression of MDC1-AS in normal tissues, with r=.992 and p<.0001 (Fig. 3B, 3C) [89]. Therefore, it is reasonable to suggest that tumors with low levels of MDC1-AS will also have low levels of MDC1, and the idea that MDC1-AS regulates MDC1 transcript is validated in these studies. However, it begs the question of what regulates MDC1-AS levels, and whether other factors govern MDC1 expression in different tissues and cancer types.
Figure 3. MDC1-AS is the antisense transcript of MDC1, and its transcription correlates with MDC1 transcription.
A. Map of MDC1-AS gene relative to MDC1. B. Summary of MDC1 and MDC1-AS RNA expression in benign tissue types. RPKM is reads per kilobase per million mapped reads, a normalized unit of transcription expression. Data from the Human Protein Atlas [89]. C. MDC1 and MDC1-AS levels strongly correlate (r=.992, p<.0001, Pearson correlation).
In addition to lncRNAs, Zhang et al. identified the microRNA, mir-300, as a regulatory RNA which suppresses MDC1 expression in hepatocellular carcinoma [90]. miRNAs are shown to repress their target mRNAs through a variety of mechanisms, inducing both translational repression and mRNA degradation [91, 92]. In addition, they have been shown to promote the expression of target genes in certain instances [93]. Mir-300 has been shown to regulate a variety of genes in cancer, including p53 and focal adhesion kinase (FAK), a metastasis suppressor [94, 95]. In the case of MDC1, it was shown that mir-300 plays a repressive role, with its levels inversely correlated with levels of MDC1 in hepatocellular carcinoma. It was also shown to directly target the MDC1 3’ UTR in a luciferase assay. The repression of MDC1 by mir-300 was associated with reduced cell growth and viability, supporting a role for MDC1 as a tumor suppressor. However, mir-300 has also been shown to target and reduce the expression of Sp1, a transcription factor that promotes expression of MDC1 [82, 96]. Therefore, mir-300 may both directly and indirectly regulate MDC1 transcript levels. Taken together, the upregulation of MDC1 by MDC1-AS lncRNA serves as a tumor suppressor, while its repression by mir-300 is oncogenic. This indicates that the regulation of MDC1 by non-coding RNAs may be an important mechanism by which MDC1 loss contributes to tumorigenesis.
In addition to these mechanisms of MDC1 transcriptional regulation, it is notable that despite certain studies finding relatively low or high levels of MDC1 protein on IHC, these do not always correspond with mRNA levels (Table 1, Fig. 3B). For instance, although Zhao et al. found high MDC1 expression in benign pancreatic tissue, mRNA levels of MDC1 are actually relatively low in the pancreas when compared with other tissues [43, 89]. This discordance between MDC1 mRNA levels and MDC1 protein abundance suggests regulation of MDC1 at the posttranscriptional and/or post-translational level. MDC1 has been shown to be ubiquitylated and degraded via the ubiquitin-proteasome system as a normal part of DNA damage foci disassembly, but this may or may not contribute to overall levels of MDC1 in tissues [97]. Additionally, the ligase that ubiquitylates MDC1 has not been identified, so further work must be done in order to analyze whether this ubiquitylation impacts overall MDC1 protein levels, and if the E3 ligase has a role in cancer. Additional studies are needed to fully understand both the transcriptional and posttranscriptional mechanisms by which MDC1 levels are regulated in cancer.
MDC1 Mutations in Cancer
The majority of focused studies on MDC1 in cancer have noted alterations of MDC1 protein and RNA levels, with the loss of MDC1 in most cases associated with tumor formation. However, in addition to alterations in MDC1 expression, there are also a number of MDC1 mutations found in patient samples.
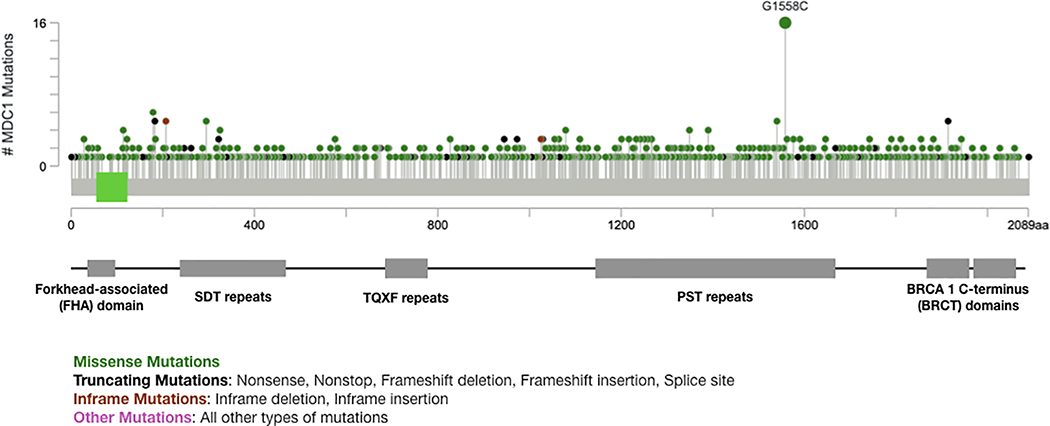
Mutations are found throughout the length of the MDC1 protein sequence (Fig. 4). The most frequent mutation identified in MDC1 is G1558C, which has been found in melanoma, bladder cancer, prostate cancer, and non-small cell lung cancer patient samples [98–103]. Although there have been no targeted functional studies of G1558 mutations, G1558 lies in the proline-serine-threonine (PST)-rich repeat region of MDC1, which contains 13 imperfect repeats of about 41 amino acids from AA 1141–1662 [29]. This region is one of the less characterized domains of MDC1, and it is not required for MDC1 to accumulate at DNA double-stranded breaks or for its recruitment of the MRN complex [104, 105]. However, despite this domain not playing a role in MDC1’s recruitment or interactions, constructs lacking this domain are defective in homologous recombination. The precise explanation for the importance of this domain is unclear. With it being a frequently mutated site in tumors, further investigation of the function of the PST-repeats and G1558 specifically would be worthwhile. The frequency of these somatic mutations in MDC1 in cancers likely represents defects in the tumor-suppressive functions of MDC1.
Figure 4. Map of MDC1 mutations in human cancer.
MDC1 somatic mutations occur across the entire length of the MDC1 protein, but G1558C, within the PST repeats, is the most common MDC1 mutation observed (Fig. generated in cBioPortal [98, 99]).
In addition to the somatic mutations identified in MDC1 in cancer tissue, Wang et al. evaluated single-nucleotide polymorphisms (SNPs) in the MDC1 gene and risk of lung cancer in a Chinese population [106]. This study showed that a SNP in the 5’ UTR of MDC1, rs4713354A>C, results in about a 1.3-fold increased risk of lung cancer. Additionally, they found that this SNP was associated with overall reduced MDC1 mRNA expression. This is consistent with other studies of MDC1 in lung cancer: tumor tissues have reduced levels of MDC1 relative to benign tissues, and the 5’ UTR SNP in MDC1 may result in a low-MDC1 phenotype, resulting in increased cancer risk [35].
Conclusions
The DNA damage response has been widely studied in relation to carcinogenesis, with many DDR proteins playing roles in tumor suppression. MDC1, a scaffold protein involved in responding to DNA double-stranded breaks, has been shown to be crucial to the formation of DDR foci, specifically the propagation of γ-H2AX [16, 17, 21]. As detailed above, numerous studies have linked MDC1 to carcinogenesis. MDC1 has primarily been characterized as a tumor suppressor, with studies showing MDC1 loss in a variety of cancers across organ systems [35, 36, 38–43, 87]. The model proposed is that the increased genomic instability resulting from MDC1 loss allows the potential for malignant transformation. However, an exception may be a subset of tumors that show increased MDC1 expression, possibly due to the up-regulation of the DNA damage response in virally driven tumors [44–46].
Although MDC1 clearly has a tumor suppressive function, one remaining question is what regulates the levels of MDC1. Several transcription factors and non-coding RNAs have been shown to regulate MDC1 transcript levels, but it is also possible that MDC1 levels are controlled at the post-transcriptional/post-translational level. With so many factors potentially contributing to MDC1 regulation, studies need to be conducted to demonstrate what causes loss of MDC1 in individual tumor types.
Additionally, there are opportunities for potential therapeutic interventions in MDC1-low or MDC1-mutated tumors. These genomically unstable cells have been shown to be more susceptible to DNA damaging chemotherapeutics, radiation, and PARP inhibitors, due to the lack of adequate DNA double-stranded break repair [44, 46, 56–59, 62]. This may provide an opportunity to use MDC1 levels as a biomarker for patients who may benefit from these therapies.
Altogether, MDC1 has been associated with so many different types of cancer that further research into its mechanism of action in tumorigenesis may be particularly useful, since the implications could be broadly applicable. Future studies of MDC1 in cancer should focus on its regulation and how to target MDC1 low or MDC1 high tumors.
Highlights.
MDC1 is a scaffold protein that functions in DDR to maintain genome integrity
MDC1 loss sensitizes cancers to chemotherapies
MDC1 also shows potential roles as a nuclear receptor transcriptional co-regulator
Mutations to MDC1 and changes in MDC1expression occurs in cancer
MDC1 plays important roles in tumor formation, progression, and treatment
Acknowledgments
This work was supported by NIH T32 CA009161 and T32 GM136573 to SR. We would like to thank Susan Ha for critical reading and editing of this article. The authors declare no conflicts or competing interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J, RNF8 Ubiquitylates Histones at DNA Double-Strand Breaks and Promotes Assembly of Repair Proteins, Cell, 131 (2007) 887–900. [DOI] [PubMed] [Google Scholar]
- [2].Yang G, Liu C, Chen S-H, Kassab MA, Hoff JD, Walter NG, Yu X, Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors, Nucleic Acids Res, 46 (2018) 3446–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sung S, Li F, Park YB, Kim JS, Kim AK, Song OK, Kim J, Che J, Lee SE, Cho Y, DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA, The EMBO journal, 33 (2014) 2422–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Woods DS, Sears CR, Turchi JJ, Recognition of DNA Termini by the C-Terminal Region of the Ku80 and the DNA-Dependent Protein Kinase Catalytic Subunit, PloS one, 10 (2015) e0127321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Onn L, Portillo M, Ilic S, Cleitman G, Stein D, Kaluski S, Shirat I, Slobodnik Z, Einav M, Erdel F, Akabayov B, Toiber D, SIRT6 is a DNA double-strand break sensor, Elife, 9 (2020) e51636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lee JH, Paull TT, ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex, Science (New York, N.Y.), 308 (2005) 551–554. [DOI] [PubMed] [Google Scholar]
- [7].de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C, Human Rad50/Mre11 is a flexible complex that can tether DNA ends, Molecular cell, 8 (2001) 1129–1135. [DOI] [PubMed] [Google Scholar]
- [8].van den Bosch M, Bree RT, Lowndes NF, The MRN complex: coordinating and mediating the response to broken chromosomes, EMBO Rep, 4 (2003) 844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y, Requirement of the MRN complex for ATM activation by DNA damage, The EMBO journal, 22 (2003) 5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Falck J, Coates J, Jackson SP, Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage, Nature, 434 (2005) 605–611. [DOI] [PubMed] [Google Scholar]
- [11].Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ, ATM phosphorylates histone H2AX in response to DNA double-strand breaks, The Journal of biological chemistry, 276 (2001) 42462–42467. [DOI] [PubMed] [Google Scholar]
- [12].Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA, ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation, Cancer research, 64 (2004) 2390–2396. [DOI] [PubMed] [Google Scholar]
- [13].Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM, DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139, The Journal of biological chemistry, 273 (1998) 5858–5868. [DOI] [PubMed] [Google Scholar]
- [14].Rogakou EP, Boon C, Redon C, Bonner WM, Megabase Chromatin Domains Involved in DNA Double-Strand Breaks in Vivo, Journal of Cell Biology, 146 (1999) 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH, Formation of Dynamic γ-H2AX Domains along Broken DNA Strands Is Distinctly Regulated by ATM and MDC1 and Dependent upon H2AX Densities in Chromatin, Molecular cell, 34 (2009) 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J, MDC1 Maintains Genomic Stability by Participating in the Amplification of ATM-Dependent DNA Damage Signals, Molecular cell, 21 (2006) 187–200. [DOI] [PubMed] [Google Scholar]
- [17].Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP, MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks, Cell, 123 (2005) 1213–1226. [DOI] [PubMed] [Google Scholar]
- [18].Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J, RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly, Cell, 131 (2007) 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang F, Bick G, Park J-Y, Andreassen PR, MDC1 and RNF8 function in a pathway that directs BRCA1-dependent localization of PALB2 required for homologous recombination, Journal of Cell Science, 125 (2012) 6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C, RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins, Cell, 136 (2009) 435–446. [DOI] [PubMed] [Google Scholar]
- [21].Zhang J, Ma Z, Treszezamsky A, Powell SN, MDC1 interacts with Rad51 and facilitates homologous recombination, Nature Structural & Molecular Biology, 12 (2005) 902–909. [DOI] [PubMed] [Google Scholar]
- [22].Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW, Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX, Proceedings of the National Academy of Sciences of the United States of America, 99 (2002) 8173–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A, Genomic instability in mice lacking histone H2AX, Science (New York, N.Y.), 296 (2002) 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A, H2AX Haploin-sufficiency Modifies Genomic Stability and Tumor Susceptibility, Cell, 114 (2003) 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li L, Halaby M-J, Hakem A, Cardoso R, El Ghamrasni S, Harding S, Chan N, Bristow R, Sanchez O, Durocher D, Hakem R, Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer, Journal of Experimental Medicine, 207 (2010) 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Santos MA, Huen MSY, Jankovic M, Chen H-T, López-Contreras AJ, Klein IA, Wong N, Barbancho JLR, Fernandez-Capetillo O, Nussenzweig MC, Chen J, Nussenzweig A, Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8, Journal of Experimental Medicine, 207 (2010) 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stucki M, Jackson SP, γH2AX and MDC1: Anchoring the DNA-damage-response machinery to broken chromosomes, DNA Repair, 5 (2006) 534–543. [DOI] [PubMed] [Google Scholar]
- [28].Stucki M, Jackson SP, MDC1/NFBD1: a key regulator of the DNA damage response in higher eukaryotes, DNA Repair, 3 (2004) 953–957. [DOI] [PubMed] [Google Scholar]
- [29].Jungmichel S, Stucki M, MDC1: The art of keeping things in focus, Chromosoma, 119 (2010) 337–349. [DOI] [PubMed] [Google Scholar]
- [30].Coster G, Goldberg M, The cellular response to DNA damage: A focus on MDC1 and its interacting proteins, Nucleus, 1 (2010) 166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ozaki T, NFBD1/MDC1: DNA damage response, cell cycle regulation and carcinogenesis, Cancer Research Frontiers, 1 (2015) 49–59. [Google Scholar]
- [32].Nakanishi M, Ozaki T, Yamamoto H, Hanamoto T, Kikuchi H, Furuya K, Asaka M, Delia D, Nakagawara A, NFBD1/MDC1 Associates with p53 and Regulates Its Function at the Crossroad between Cell Survival and Death in Response to DNA Damage, Journal of Biological Chemistry, 282 (2007) 22993–23004. [DOI] [PubMed] [Google Scholar]
- [33].Shahar OD, Gabizon R, Feine O, Alhadeff R, Ganoth A, Argaman L, Shimshoni E, Friedler A, Goldberg M, Acetylation of lysine 382 and phosphorylation of serine 392 in p53 modulate the interaction between p53 and MDC1 in vitro, PloS one, 8 (2013) e78472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Eliezer Y, Argaman L, Rhie A, Doherty AJ, Goldberg M, The Direct Interaction between 53BP1 and MDC1 Is Required for the Recruitment of 53BP1 to Sites of Damage, Journal of Biological Chemistry, 284 (2009) 426–435. [DOI] [PubMed] [Google Scholar]
- [35].Bartkova J, Hořejš í Z, Sehested M, Nesland JM, Rajpert-De Meyts E, Skakkebæk NE, Stucki M, Jackson S, Lukas J, Bartek J, DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours, Oncogene, 26 (2007) 7414–7422. [DOI] [PubMed] [Google Scholar]
- [36].MERENTITIS D, NGUYEN BD, SAMARTZIS EP, NOSKE A, BRANDT S, DEDES KJ, Loss of MDC1 in Endometrial Carcinoma Is Associated With Loss of MRN Complex and MMR Deficiency, Anticancer Research, 39 (2019) 6547–6553. [DOI] [PubMed] [Google Scholar]
- [37].Ye Q, Chen L, Yin X, Liu YJC, Ji Q, Zhao E, Development of Serous Ovarian Cancer is Associated with the Expression of Homologous Recombination Pathway Proteins, Pathology & Oncology Research, 20 (2014) 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang C, Sun H, Zou R, Zhou T, Wang S, Sun S, Tong C, Luo H, Li Y, Li Z, Wang E, Chen Y, Cao L, Li F, Zhao Y, MDC1 functionally identified as an androgen receptor co-activator participates in suppression of prostate cancer, Nucleic Acids Res, 43 (2015) 4893–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yue H, Zhu J, Xie S, Li F, Xu Q, MDC1-AS, an antisense long noncoding RNA, regulates cell proliferation of glioma, Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 81 (2016) 203–209. [DOI] [PubMed] [Google Scholar]
- [40].Xue Y, Ma G, Zhang Z, Hua Q, Chu H, Tong N, Yuan L, Qin C, Yin C, Zhang Z, Wang M, A novel antisense long noncoding RNA regulates the expression of MDC1 in bladder cancer, Oncotarget, 6 (2015) 484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Patel AN, Goyal S, Wu H, Schiff D, Moran MS, Haffty BG, Mediator of DNA damage checkpoint protein 1 (MDC1) expression as a prognostic marker for nodal recurrence in early-stage breast cancer patients treated with breast-conserving surgery and radiation therapy, Breast Cancer Research and Treatment, 126 (2011) 601–607. [DOI] [PubMed] [Google Scholar]
- [42].Zou R, Zhong X, Wang C, Sun H, Wang S, Lin L, Sun S, Tong C, Luo H, Gao P, Li Y, Zhou T, Li D, Cao L, Zhao Y, MDC1 Enhances Estrogen Receptor-mediated Transactivation and Contributes to Breast Cancer Suppression, International journal of biological sciences, 11 (2015) 992–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhou H, Qin Y, Ji S, Ling J, Fu J, Zhuang Z, Fan X, Song L, Yu X, Chiao PJ, SOX9 activity is induced by oncogenic Kras to affect MDC1 and MCMs expression in pancreatic cancer, Oncogene, 37 (2018) 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yuan C, Bu Y, Wang C, Yi F, Yang Z, Huang X, Cheng L, Liu G, Wang Y, Song F, NFBD1/MDC1 is a protein of oncogenic potential in human cervical cancer, Molecular and Cellular Biochemistry, 359 (2012) 333–346. [DOI] [PubMed] [Google Scholar]
- [45].Liu X, Qiu Z, Wang Z, Zuo W, Gong Z, Liu C, Zeng Q, Qian Y, Jiang L, Li Y, Bu Y, Hu G, NFBD1/MDC1 participates in the regulation of proliferation and apoptosis in human laryngeal squamous cell carcinoma, Clinical and Translational Oncology, 20 (2018) 534–541. [DOI] [PubMed] [Google Scholar]
- [46].Wang Z, Liao K, Zuo W, Liu X, Qiu Z, Gong Z, Liu C, Zeng Q, Qian Y, Jiang L, Bu Y, Hong S, Hu G, Depletion of NFBD1/MDC1 Induces Apoptosis in Nasopharyngeal Carcinoma Cells Through the p53-ROS-Mitochondrial Pathway, Oncology research, 25 (2017) 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu X, Dong R, Jiang Z, Wei Y, Li Y, Wei L, Sun H, Li Y, Yang N, Yang Q, Liu Z, Kong B, MDC1 promotes ovarian cancer metastasis by inducing epithelial-mesenchymal transition, Tumor Biology, 36 (2015) 4261–4269. [DOI] [PubMed] [Google Scholar]
- [48].Durst M, Gissmann L, Ikenberg H, zur Hausen H, A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions, Proceedings of the National Academy of Sciences of the United States of America, 80 (1983) 3812–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Torrente MC, Rodrigo JP, Haigentz M Jr., Dikkers FG, Rinaldo A, Takes RP, Olofsson J, Ferlito A, Human papillomavirus infections in laryngeal cancer, Head & neck, 33 (2011) 581–586. [DOI] [PubMed] [Google Scholar]
- [50].Tsao SW, Tsang CM, Lo KW, Epstein-Barr virus infection and nasopharyngeal carcinoma, Philos Trans R Soc Lond B Biol Sci, 372 (2017) 20160270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen Y, Williams V, Filippova M, Filippov V, Duerksen-Hughes P, Viral carcinogenesis: factors inducing DNA damage and virus integration, Cancers (Basel), 6 (2014) 2155–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Weitzman MD, Lilley CE, Chaurushiya MS, Genomes in Conflict: Maintaining Genome Integrity During Virus Infection, Annual Review of Microbiology, 64 (2010) 61–81. [DOI] [PubMed] [Google Scholar]
- [53].Peng A, Chen P-L, NFBD1, Like 53BP1, Is an Early and Redundant Transducer Mediating Chk2 Phosphorylation in Response to DNA Damage, Journal of Biological Chemistry, 278 (2003) 8873–8876. [DOI] [PubMed] [Google Scholar]
- [54].Tewey K, Rowe T, Yang L, Halligan B, Liu L, Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II, Science (New York, N.Y.), 226 (1984) 466–468. [DOI] [PubMed] [Google Scholar]
- [55].Doroshow JH, Role of hydrogen peroxide and hydroxyl radical formation in the killing of Ehrlich tumor cells by anticancer quinones, Proceedings of the National Academy of Sciences of the United States of America, 83 (1986) 4514–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yang Z, Bu Y, Wang C, Liu G, Song F, Growth inhibition, morphology change, and cell cycle alterations in NFBD1-depleted human esophageal cancer cells, Molecular and Cellular Biochemistry, 342 (2010) 1–6. [DOI] [PubMed] [Google Scholar]
- [57].Wang S, Zou Z, Luo X, Mi Y, Chang H, Xing D, LRH1 enhances cell resistance to chemotherapy by transcriptionally activating MDC1 expression and attenuating DNA damage in human breast cancer, Oncogene, 37 (2018) 3243–3259. [DOI] [PubMed] [Google Scholar]
- [58].Singh N, Bhakuni R, Chhabria D, Kirubakaran S, MDC1 depletion promotes cisplatin induced cell death in cervical cancer cells, BMC Research Notes, 13 (2020) 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zeng Q, Wang Z, Liu C, Gong Z, Yang L, Jiang L, Ma Z, Qian Y, Yang Y, Kang H, Hong S, Bu Y, Hu G, Knockdown of NFBD1/MDC1 enhances chemosensitivity to cisplatin or 5-fluorouracil in nasopharyngeal carcinoma CNE1 cells, Molecular and Cellular Biochemistry, 418 (2016) 137–146. [DOI] [PubMed] [Google Scholar]
- [60].Dasari S, Bernard Tchounwou P, Cisplatin in cancer therapy: Molecular mechanisms of action, European Journal of Pharmacology, 740 (2014) 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gou Q, Xie Y, Liu L, Xie K, Wu Y, Wang Q, Wang Z, Li P, Downregulation of MDC1 and 53BP1 by short hairpin RNA enhances radiosensitivity in laryngeal carcinoma cells, Oncology reports, 34 (2015) 251–257. [DOI] [PubMed] [Google Scholar]
- [62].Wang Z, Zeng Q, Chen T, Liao K, Bu Y, Hong S, Hu G, Silencing NFBD1/MDC1 enhances the radiosensitivity of human nasopharyngeal cancer CNE1 cells and results in tumor growth inhibition, Cell Death & Disease, 6 (2015) e1849–e1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dziadkowiec KN, Gąsiorowska E, Nowak-Markwitz E, Jankowska A, PARP inhibitors: review of mechanisms of action and BRCA1/2 mutation targeting, Prz Menopauzalny, 15 (2016) 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wang Z, Zuo W, Zeng Q, Qian Y, Li Y, Liu C, Wang J, Zhong S, Bu Y, Hu G, Loss of NFBD1/MDC1 disrupts homologous recombination repair and sensitizes nasopharyngeal carcinoma cells to PARP inhibitors, Journal of Biomedical Science, 26 (2019) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Collins AR, The comet assay for DNA damage and repair, Molecular Biotechnology, 26 (2004) 249. [DOI] [PubMed] [Google Scholar]
- [66].Ostling O, Johanson KJ, Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells, Biochemical and biophysical research communications, 123 (1984) 291–298. [DOI] [PubMed] [Google Scholar]
- [67].Zhao Y, Takeyama K.-i., Sawatsubashi S, Ito S, Suzuki E, Yamagata K, Tanabe M, Kimura S, Fujiyama S, Ueda T, Murata T, Matsukawa H, Shirode Y, Kouzmenko AP, Li F, Tabata T, Kato S, Corepressive action of CBP on androgen receptor transactivation in pericentric heterochromatin in a Drosophila experimental model system, Mol Cell Biol, 29 (2009) 1017–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Elgin SCR, Reuter G, Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila, Cold Spring Harb Perspect Biol, 5 (2013) a017780–a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, Sagum CA, Jung SY, Qin J, Tsai M-J, Tsai SY, Li W, Foulds CE, O’Malley BW, Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation, The Journal of biological chemistry, 292 (2017) 14456–14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tamburini BA, Tyler JK, Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair, Mol Cell Biol, 25 (2005) 4903–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Schiewer MJ, Knudsen KE, Linking DNA Damage and Hormone Signaling Pathways in Cancer, Trends Endocrinol Metab, 27 (2016) 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, Arora VK, Yen W-F, Cai L, Zheng D, Carver BS, Chen Y, Watson PA, Shah NP, Fujisawa S, Goglia AG, Gopalan A, Hieronymus H, Wongvipat J, Scardino PT, Zelefsky MJ, Jasin M, Chaudhuri J, Powell SN, Sawyers CL, Androgen receptor signaling regulates DNA repair in prostate cancers, Cancer Discov, 3 (2013) 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, Ma T, Den RB, Dicker AP, Feng FY, Knudsen KE, A hormone-DNA repair circuit governs the response to genotoxic insult, Cancer Discov, 3 (2013) 1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription, Science (New York, N.Y.), 312 (2006) 1798–1802. [DOI] [PubMed] [Google Scholar]
- [75].Haffner MC, De Marzo AM, Meeker AK, Nelson WG, Yegnasubramanian S, Transcription-induced DNA double strand breaks: both oncogenic force and potential therapeutic target?, Clin Cancer Res, 17 (2011) 3858–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, Meeker AK, Netto G, De Marzo AM, Nelson WG, Yegnasubramanian S, Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements, Nat Genet, 42 (2010) 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bolla M, Gonzalez D, Warde P, Dubois JB, Mirimanoff RO, Storme G, Bernier J, Kuten A, Sternberg C, Gil T, Collette L, Pierart M, Improved survival in patients with locally advanced prostate cancer treated with radiotherapy and goserelin, The New England journal of medicine, 337 (1997) 295–300. [DOI] [PubMed] [Google Scholar]
- [78].Warde P, Mason M, Ding K, Kirkbride P, Brundage M, Cowan R, Gospodarowicz M, Sanders K, Kostashuk E, Swanson G, Barber J, Hiltz A, Parmar MK, Sathya J, Anderson J, Hayter C, Hetherington J, Sydes MR, Parulekar W, Combined androgen deprivation therapy and radiation therapy for locally advanced prostate cancer: a randomised, phase 3 trial, Lancet (London, England), 378 (2011) 2104–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Asim M, Tarish F, Zecchini HI, Sanjiv K, Gelali E, Massie CE, Baridi A, Warren AY, Zhao W, Ogris C, McDuffus L-A, Mascalchi P, Shaw G, Dev H, Wadhwa K, Wijnhoven P, Forment JV, Lyons SR, Lynch AG, O’Neill C, Zecchini VR, Rennie PS, Baniahmad A, Tavaré S, Mills IG, Galanty Y, Crosetto N, Schultz N, Neal D, Helleday T, Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer, Nature Communications, 8 (2017) 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Li L, Karanika S, Yang G, Wang J, Park S, Broom BM, Manyam GC, Wu W, Luo Y, Basourakos S, Song JH, Gallick GE, Karantanos T, Korentzelos D, Azad AK, Kim J, Corn PG, Aparicio AM, Logothetis CJ, Troncoso P, Heffernan T, Toniatti C, Lee H-S, Lee J-S, Zuo X, Chang W, Yin J, Thompson TC, Androgen receptor inhibitor–induced “BRCAness” and PARP inhibition are synthetically lethal for castration-resistant prostate cancer, Science Signaling, 10 (2017) eaam7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Pezaro C, PARP inhibitor combinations in prostate cancer, Ther Adv Med Oncol, 12 (2020) 1758835919897537–1758835919897537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bu Y, Suenaga Y, Ono S, Koda T, Song F, Nakagawara A, Ozaki T, Sp1-mediated transcriptional regulation of NFBD1/MDC1 plays a critical role in DNA damage response pathway, Genes to Cells, 13 (2008) 53–66. [DOI] [PubMed] [Google Scholar]
- [83].Qi P, Du X, The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine, Modern Pathology, 26 (2013) 155–165. [DOI] [PubMed] [Google Scholar]
- [84].Ponting CP, Belgard TG, Transcribed dark matter: meaning or myth?, Human Molecular Genetics, 19 (2010) R162–R168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Huarte M, Rinn JL, Large non-coding RNAs: missing links in cancer?, Hum Mol Genet, 19 (2010) R152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, Antisense Transcription in the Mammalian Transcriptome, Science (New York, N.Y.), 309 (2005) 1564. [DOI] [PubMed] [Google Scholar]
- [87].Qin Y, Zhuang S, Wen J, Zheng K, Long non-coding RNA MDC1-AS inhibits human gastric cancer cell proliferation and metastasis through an MDC1-dependent mechanism, Exp Ther Med, 15 (2018) 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C, Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase, Nat Med, 14 (2008) 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlen M, Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics, Molecular & cellular proteomics : MCP, 13 (2014) 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhang J, Luo H, Du J, Liu Y, MicroRNA-300 plays as oncogene by promoting proliferation and reducing apoptosis of liver cancer cells by targeting MDC1, Int J Clin Exp Pathol, 9 (2016) 1231–1239. [Google Scholar]
- [91].O’Brien J, Hayder H, Zayed Y, Peng C, Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation, Frontiers in Endocrinology, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Huntzinger E, Izaurralde E, Gene silencing by microRNAs: contributions of translational repression and mRNA decay, Nature Reviews Genetics, 12 (2011) 99–110. [DOI] [PubMed] [Google Scholar]
- [93].Vasudevan S, Posttranscriptional upregulation by microRNAs, Wiley interdisciplinary reviews. RNA, 3 (2012) 311–330. [DOI] [PubMed] [Google Scholar]
- [94].Xu X-H, Li D-W, Feng H, Chen H-M, Song Y-Q, MiR-300 regulate the malignancy of breast cancer by targeting p53, Int J Clin Exp Med, 8 (2015) 6957–6966. [PMC free article] [PubMed] [Google Scholar]
- [95].Wang R, Yu Z, Chen F, Xu H, Shen S, Chen W, Chen L, Su Q, Zhang L, Bi J, Zeng W, Li W, Huang X, Wang Q, miR-300 regulates the epithelial-mesenchymal transition and invasion of hepatocellular carcinoma by targeting the FAK/PI3K/AKT signaling pathway, Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie, 103 (2018) 1632–1642. [DOI] [PubMed] [Google Scholar]
- [96].Yan H, Li J, Ying Y, Xie H, Chen H, Xu X, Zheng X, MIR-300 in the imprinted DLK1-DIO3 domain suppresses the migration of bladder cancer by regulating the SP1/MMP9 pathway, Cell cycle (Georgetown, Tex.), 17 (2018) 2790–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Shi W, Ma Z, Willers H, Akhtar K, Scott SP, Zhang J, Powell S, Zhang J, Disassembly of MDC1 Foci Is Controlled by Ubiquitin-Proteasome-dependent Degradation, Journal of Biological Chemistry, 283 (2008) 31608–31616. [DOI] [PubMed] [Google Scholar]
- [98].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Cancer Discov, 2 (2012) 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal, Sci Signal, 6 (2013) pl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV, Hodis E, Rosenberg M, McKenna A, Cibulskis K, Farlow D, Zimmer L, Hillen U, Gutzmer R, Goldinger SM, Ugurel S, Gogas HJ, Egberts F, Berking C, Trefzer U, Loquai C, Weide B, Hassel JC, Gabriel SB, Carter SL, Getz G, Garraway LA, Schadendorf D, The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma, Cancer Discov, 4 (2014) 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Faltas BM, Prandi D, Tagawa ST, Molina AM, Nanus DM, Sternberg C, Rosenberg J, Mosquera JM, Robinson B, Elemento O, Sboner A, Beltran H, Demichelis F, Rubin MA, Clonal evolution of chemotherapy-resistant urothelial carcinoma, Nat Genet, 48 (2016) 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, Tomlins SA, Nanus DM, Tagawa ST, Van Allen EM, Elemento O, Sboner A, Garraway LA, Rubin MA, Demichelis F, Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer, Nat Med, 22 (2016) 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB, Abu-Akeel M, Liu C, Sauter JL, Rekhtman N, Chang E, Callahan MK, Chaft JE, Voss MH, Tenet M, Li XM, Covello K, Renninger A, Vitazka P, Geese WJ, Borghaei H, Rudin CM, Antonia SJ, Swanton C, Hammerbacher J, Merghoub T, McGranahan N, Snyder A, Wolchok JD, Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer, Cancer cell, 33 (2018) 843–852.e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Shang YL, Bodero AJ, Chen P-L, NFBD1, a Novel Nuclear Protein with Signature Motifs of FHA and BRCT, and an Internal 41-Amino Acid Repeat Sequence, Is an Early Participant in DNA Damage Response, Journal of Biological Chemistry, 278 (2003) 6323–6329. [DOI] [PubMed] [Google Scholar]
- [105].Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, Jackson SP, Scully R, Distinct Roles of Chromatin-Associated Proteins MDC1 and 53BP1 in Mammalian Double-Strand Break Repair, Molecular cell, 28 (2007) 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Wang B, Zhang L, Qiu F, Fang W, Deng J, Zhou Y, Lu J, Yang L, A Newfound association between MDC1 functional polymorphism and lung cancer risk in Chinese, PloS one, 9 (2014) e106794–e106794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dave JH, Vora HH, Ghosh NR, Trivedi TI, Mediator of DNA damage checkpoint protein 1 (MDC1) as a prognostic marker for patients with oral squamous cell carcinoma, Journal of Oral Pathology & Medicine, 46 (2017) 253–258. [DOI] [PubMed] [Google Scholar]