Abstract
Targeted covalent inhibitors are currently showing great promise for systems that are normally difficult to target with small molecule therapies. This renewed interest has spurred the refinement of existing computational methods as well as the design of new ones, expanding the toolbox for discovery and optimization of selective and effective covalent inhibitors. Commonly applied approaches are covalent docking methods that predict the conformation of the covalent complex with known residues. More recently, a new predictive method, reactive docking, was developed building on the growing corpus of data generated by large proteomics experiments. This method was successfully used in several “Inverse Drug Discovery” programs that use high-throughput techniques to isolate effective compounds based on screening of entire compound libraries based on desired phenotypes.
Keywords: covalent docking, structure-based drug design, covalent virtual screening, irreversible binding, TCI
A New Paradigm in Drug Discovery
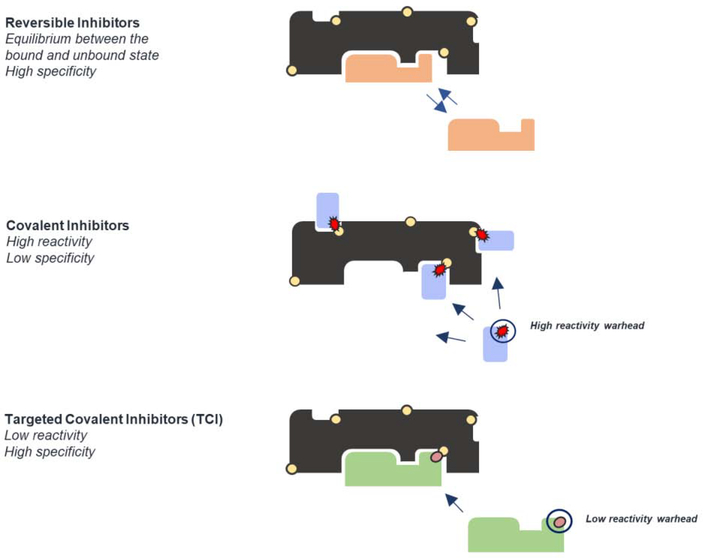
The discovery of Targeted Covalent Inhibitors (TCI) has reignited interest in covalent compounds as drug candidates [1–4] (Figure 1). While the strict definition provided by Singh et al [1] is limited to reactive binders targeting non-catalytic residues, we believe a more broader definition of TCI should include all covalent inhibitors that show selectivity against the target for which they have been designed. Of particular interest, these compounds are being used as exploratory tools in the early phases of drug discovery [5,6] and in “Inverse Drug Discovery” approaches that identify targets based on interventions to phenotype. These compounds can simplify the identification of the molecular target responsible for phenotypic changes in cells when screening compound libraries. The hit-to-lead refinement process can then combine high specificity with covalent modification, leading to optimal inhibitors. They are typically developed by screening focused libraries of electrophilic fragments, but they can also be performed by attaching warheads to existing non-covalent-binding ligands, or functionalization of DNA-encoded libraries [7].
Figure 1. Conceptual approaches for inhibitor design.
Different binding mechanism of action for reversible inhibitors, covalent inhibitors, and Targeted Covalent Inhibitors (TCI)
Covalent site- or residue-specific molecules can be used as molecular probes to characterize biological role or location of their targets [8,9] or modulate their activity and function for therapeutic purposes. In these cases, residues targeted can play a direct role in catalytic sites, like in caspases [10], glutathione transferase [11], and cathepsin K [12], or aberrant mutations that can be opportunistically exploited, such as for the KRAS G12C mutant [13]. Many natural compounds also exhibit an irreversible binding mechanism [14], with a variety of warheads and chemically diverse scaffolds. The TCI approach has already shown significant successes: kinase TCI have been approved by the FDA for the treatment of cancer, including ibrutinib and acalabrutinib targeting BTK and afatinib and osimertinib against EGFR [2]. Drug candidates against KRAS G12C mutation are undergoing clinical trials for cancer treatment (Clinical Trial Numberi: NCT03785249, NCT03600883, NCT04006301). Ibrutinib is also being evaluated for the treatment of Chronic Graft Versus Host Disease (cGVHD) occurring in bone marrow or stem cell transplants (Clinical Trial Number: NCT04294641).
Lately, there has been interest in reversible covalent inhibitors as well [15], which associate some of the advantages of irreversible binders with reduced chances of side effects [16]. Computational approaches are being developed hand-in-hand with high-throughput screening approaches, to reconcile experimental results and to drive innovation into new chemical spaces.
Unique Features of Targeted Covalent Inhibitors
Design of a TCI is always a careful balancing act, to find the right mix of “treatment and toxicity” [17], optimizing the potency of the compound while minimizing off-target reactivity. Over the years, there has been an on-going debate on the liability of idiosyncratic toxicity [1] with covalent drugs. It has been argued that unlike conventional binding drugs, in which toxicity is often the result of an unbalanced pharmacological response, covalent drugs are more likely to induce idiosyncratic toxicity [18,19] resulting from binding to off-target proteins which often results in adverse immune response. Although, it has been shown that ideal TCI can have relatively high selectivity and low off-target interactions. Looking to Nature, penicillin achieves this balance with a typical drug-sized molecule that closely mimics bacterial peptidoglycans, and the intrinsic reactivity of the β-lactam ring [20] is boosted significantly in the proper context, becoming a highly active warhead [21]. There are is number of components in both the irreversible binder molecule and its target that dictate the final outcome of the interaction, and TCI combine and exploit several of these factors.
Target components.
Structural and physico-chemical characteristics affect and modulate the intrinsic reactivity of amino acids, which will deviate from what is measured in isolated conditions. For example, the structural context in which residues are found in folded proteins can alter the pKa of side chain moieties, making them more prone to participate in reactions. A clear example of residue selectivity is represented by the sulfur(VI)-fluoride exchange (SuFEx) warheads. Tyrosine phenols that can be targeted very selectively with fluorosulfate SuFEx warheads [22] when flanked by positively charged amino acids such as lysine, arginine, and histidine [23,24]. Similar residue propensity can be found for sulfonyl fluorides targeting pKa -perturbed lysines [22]. The same residues have been proven not to react when the protein is not found in its naturally folded state because flanking interactions are lost [25]. Several laboratories have characterized the intrinsic reactivity of different warheads, using theoretical methods[26], MS [27,28], and NMR [29], which can be used to predict their interaction with amino acids and quantify possible reactivity-driven off-target interaction. Similarly, a number of attempts have been made to use predictive computational models to estimate the reactivity of given classes of functional groups [30–34].
Beside cysteines [35–37], a number of studies addressed the proteome-wide reactivity of specific residues, such as serines [38], lysines [39], and others [37]. Due to the irreversible nature of the interaction, low initial affinity is sufficient to stabilize the non-covalent adduct and facilitate the formation of the new covalent bond. This represents a clear advantage over conventional binders because it allows targeting of less druggable sites [40]. That said, while not as indispensable as for reversible binders, the proximity of cavities and pockets on the macromolecular surface near the targeted residues further increases the likelihood of the reaction occurring [41]. The shape of such cavities can be exploited for increasing specificity and affinity of the binders, hence allowing use of less reactive warheads. The application of any structure-based methods relies on the availability of accurate models of the targets. Therefore very flexible proteins represent a major challenge for reversible and irreversible binders alike, and the accuracy of predictions is reduced dramatically when using a single conformation, apo structures with no well-defined binding site pockets, or allosteric/cryptic pockets. Although, since modeling of irreversible binding events relies on the accurate description of the reaction geometries, the issue of target flexibility could arguably have a higher impact on the modeling of covalent binders.
Ligand components.
On the binder side, for it to be sufficiently specific it is essential that the warhead has the proper balance of low reactivity and orientational bias, so that it will react only when in close proximity and in optimal relative orientation with the desired target. Orientational optimization depends on several factors, including tuning of the solution conformation to match the bound conformation, and orienting the warhead to promote the desired chemical reaction mechanism. Most effort has been focused on targeting cysteines with ⍺-β-unsaturated carbonyl compounds, although many additional approaches, with different chemical warheads targeting cysteine, lysine, tyrosine, and many other amino acids are being explored [3,42,43]. Remarkably, with an opportune warhead even the protein N-terminus can be targeted [44] as shown in a recently approved drug for sickle cell disease [45]. SuFEx chemistry has gathered a significant amount of interest for the design of TCI, lately, due to their relatively low reactivity and their precise geometric and structural constraints. These constraints are ideal characteristics for design of TCI, greatly narrowing the odds of off-target sites with similar target and pocket geometry. Overviews of different warheads and their selectivity are discussed in detail in several recent reviews [3,46]. The intrinsic reactivity of warheads can be assessed experimentally or computationally using a non-specific covalent modifier such as glutathione. For a comprehensive review on assessing electrophilic reactivity, refer to Schwobel [28]. Lonsdale [31] presents a novel approach using pKa assessment, which is compared with previously reported approaches such as NMR chemical shifts and quantum mechanical (QM) model calculations, to estimate reactivity of a series of acrylamides using glutathione.
Given the stringent characteristics of TCI, the identification of drug design leads depends on the opportune design of the library to be screened. On one hand, the choice of the opportune warheads controls the selectivity and provides the basic intrinsic reactivity of the molecule. On the other hand, ultimate reactivity is also modulated by the chemical features of the non-reactive components, which can be discriminated by the target prior to the reaction conditions to be satisfied. Parker and Cravatt showed that stereoisomers can be discriminated even by non-enzymatic binding pockets [47], hence can be used to provide further reactivity fine tuning. Irreversible warheads can be the starting point for the design of reversible binders: by introducing opportune chemical modifications, the reactivity of irreversible warheads can be modulated to turn them into reversible ones [48,49]. Zhang [50] applied Free Energy Perturbation (FEP) protocols based on molecular dynamics (MD) to predict the binding free energies of reversible covalent drugs and rank their reactivity. Schirmeister [49] applied QM methods and hybrid QM/MM methods to present a proof of principle of rational design of reversible covalent vinylsulfone inhibitors.
Kinetics.
The kinetics of TCI are unique, a fact that must be reflected in any rational design effort [51,52]. They do not follow typical binding kinetics, so measures such as KD and IC50 alone are not sufficient to describe their kinetics. Rather, the kinetics are dependent on a two-step mechanism. First, the reversible binding of the molecule to the site is characterized by the binding constant KI. Then, the reaction of the warhead with the target is based on kinact, the rate of reaction. The overall course of the reaction is then related to the second-order rate constant kinact/KI, and occupancy steadily increases with time. The dual contributions of these kinetics are reflected in observed properties of TCI. A recent high-throughput screen of fragments decorated with several mildly electrophilic warheads gave encouraging results, increasing normally-weak binding strength of these small molecules through covalent attachment to cysteine, while also revealing that promiscuous inhibitors were surprisingly rare [53]. Studies with EGFR TCI have revealed that the efficacy of these compounds is closely tied to the reversible binding strength [54], underscoring the desirable ability to tune specificity by tailoring the traditional binding characteristics of the molecule to the active site.
Kinetics of reversible covalent inhibitors is further complicated due to the fact that the formation of the new covalent bond is under thermodynamic control [50,55].
Their residence times can vary significantly from a few minutes to multiple days [48,56].
Computational Methods
Design of TCI is particularly amenable to structure-based approaches, because their specificity is largely a function of (1) the binding strength of the groups carrying the warhead, and (2) the molecular context of the target that results in the positioning of susceptible groups in proximity to the warhead. If the reactivity of the species involved is properly characterized, it is possible to apply all of the traditional approaches for structure-based drug discovery to this context, including fragment screening, modification of an existing lead compound with a warhead, and brute-force virtual screening of entire libraries of synthetically accessible compounds. A variety of computational methods have been developed to allow a structure-based approach to discovery of covalent inhibitors. These methods face several challenges --prediction of binding poses and prediction of reaction rates--and progress has been made addressing both of them. Highly computationally-intensive methods, such as hybrid MM/QM methods presented by Schmidt [57] and Yu [58] seek to characterize the entire process of binding and covalent attachment [52]. We focus here on simpler, faster docking methods that are designed for virtual screens and compound discovery.
Site Prediction.
The structural context of the target amino acid is a key component determining reactivity. Typically, deprotonated states of the sidechain provide the nucleophile for the reaction, so nearby cationic residues can promote formation of the reactive form. AwoonorWilliams [52] reviews several methods for predicting the pKa of target amino acids. Since kinases have emerged as a particularly amenable target for development of TCI, several studies have focused on analyzing the entire body of available structures to identify possible targets within the kinase “Cysteinome” [59,60]. Zhao [61] analyzed 1599 structures of 169 unique kinases that had one or more cysteines in the active site, characterizing five potential structural sites for covalent attachment by TCI. The characteristics of these sites were analyzed by function-site interaction fingerprinting (Fs-IFP), which includes information on the directionality of target thiols and if they are consistent with potential covalent reactions. Density functional theory (DFT) calculations were further used to characterize the reactivity.
A retrospective analysis based on a structure-based pocket prediction tool [62] was performed on 68 diverse protein structures, for which isoTOP-ABPP and TMT-ABPP experiments identified the labeling of 74 cysteines. The results showed that 68% of the cysteines are located in close proximity to a druggable pocket [41]. Similarly, Zhang [50] analyzed 1462 sites of covalent attachment to cysteine by both small, highly active covalent ligands and TCI, finding that these cysteines have lower pKa and higher exposure. The study also showed that druggable pockets are found close to cysteines in 88.1% of the TCI complexes, whereas local pockets were found in only 64.3% of complexes with highly active covalent inhibitors and 60.8% of non-covalent inhibitors. Based on these observations, they were able to formulate a predictive algorithm for finding potential covalent binding sites. These results confirm the hypothesis that the noncovalent binding affinity and the geometry of pockets are essential to drive the covalent bond formation in TCI.
Covalent Docking
Most current docking methods focus on modeling the conformation of covalent inhibitors in the bound covalent complex. Several detailed reviews of methods are available, so individual methods are not compared here [52,63,64]. Three conceptual methods are typically employed: biased (or constrained), tethered and hybrid.
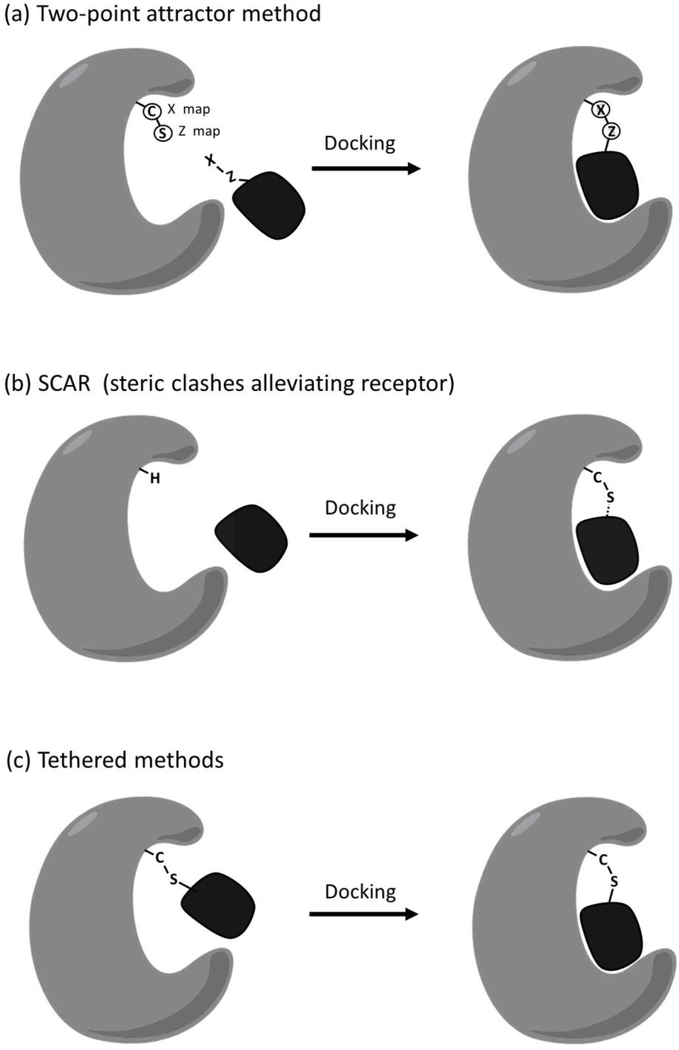
Biased methods modify the site of attachment in the target and the ligand and add new potentials to the force field to favor or constrain conformations consistent with the covalent bond. Examples include CovalentDock [65] and the two-point attractor method of AutoDock [66]. In a simpler variation, termed SCAR (steric clashes alleviating receptor), the cysteine may be removed entirely, and the bond inferred after traditional docking [67] (Figure 2).
Figure 2. Schematic of covalent docking methods.
Overview of docking methods to model covalent binding. (a) Two-point attractor method uses two dummy atom types (X and Z) to superimpose and constrain atoms involved in the bond. (b) SCAR method removes the atoms that form the bond during docking, then adds them to the predicted complex. (c) Tethered methods start with a trial conformation for the covalent complex, then optimize the pose by exploring the available conformational space.
Tethered approaches, employed in popular tools such as GOLD [68] and many others [66] pre-form the covalent complex based on the known chemistry and geometry of the bond, and use the docking engine to optimize the pose and evaluate non-covalent interactions for the remaining portion of the ligand. This has the great advantage of speeding up the entire docking simulation, since translational degrees of freedom can be ignored (Figure 2). Both approaches ignore the energetics of covalent bond formation, enforcing an idealized covalent bond and scoring docked complexes primarily on the non-covalent interactions.
Hybrid methods combine conventional methods with biased or tethered methods and are relatively more articulated and complex. Multi-step protocols are performed to evaluate first the strength and specificity of the non-covalent interaction, and then probability of reaction is estimated based on the proximity and relative orientation of warhead and target, often after a refinement process (e.g. MD). Examples include CovDock, DUckCov, and EnzyDock.
CovDock [69] ranks TCI by predicting poses of non-covalent and covalently-bound states and defining an apparent binding affinity score that is the average of the two. This assumes that the energetics of bond formation are the same across a given set of trial TCI, allowing ranking of compounds with identical or similar warheads. A multistep process is used, positioning the ligand in an alanine-mutated site with Glide [70] with a constraint to keep the warhead within 8 Angstroms of the desired position, restoring the amino acid and choosing poses with acceptable covalent geometry, and refining and scoring the covalent complex. The method shows a 29/38 success rate in pose prediction in a retrospective study of two classes of inhibitors, and affinity predictions with R-squared correlations of 0.62 for 11 acrylamides with cSrc kinase and 0.32 for 27 peptidyl ketoamides with HIV serine protease. The method is compute-intensive (1–3 hours/ligand), so a faster method, CovDock-VS [71], has also been developed that speeds up the process by incorporating approximations at several steps.
DuckCov [72] is a rapid pipeline for virtual screening of TCI. First, the non-covalent complex is predicted using rDock [73] to place ligands based on pharmacophores and constraining the warhead to be in proximity to the target amino acid. Then, hydrogen bonding is evaluated using DUck, a dynamic undocking approach [74]. Finally, the best candidates are subjected to compute-intensive evaluation using CovDock. The method was validated by virtual screening of ~55,000 acrylamides against specific sites in two kinases, with 3/5 successful hits in JAK3 confirmed by an enzyme-based activity assay and 4/9 hits for KRas confirmed by NMR. EnzyDock [75] uses a CHARMM-based molecular mechanics approach to explore chemical states along a reaction pathway. One state is first docked to the desired active site, then other states are docked and subjected to molecular dynamics in similar conformations. The method was applied to model Michael addition of covalent inhibitors on cysteines in four proteins, incorporating the non-bonded state and the covalent state into the process. Validation was performed by comparing experimental and predicted conformations of the covalent state. Retrospective comparisons of docking methods show that they are effective in reproducing observed covalent complexes about half the time. Wen [76] compared four popular methods, docking 330 known complexes with serine or cysteine, and results were assessed using a typical RMS threshold, giving success rates of 44–57%. Scarpino [77] compared six methods using 207 complexes with cysteine in 54 targets, and found success rates of 37–62%. RMSD<2.0A for best scored pose was used as the criterion for success in both studies. As with traditional docking methods, these studies found that the success was dependent on the size and flexibility of the ligands, as well as the accessibility and flexibility of the target amino acid side chain, with significant differences between different types of warheads.
Wen [76] also makes the compelling statement: “The performance of an ideal covalent docking tool should be independent of receptor class, warhead type and reaction mechanisms.” This is often the goal for development of traditional computational docking tools: to create a single tool that is parameterized and calibrated to be effective for all possible applications. In practice, however, this is rarely the case, and most existing tools are effective for drug-like molecules with characteristics roughly corresponding to Lipinski’s Rule-of-Five, binding to relatively rigid, most often protein, targets. When we add the rapidly-growing diversity of covalent warheads, this goal becomes even harder to achieve. The current solution to this conundrum is two-fold: developers of methods do their best to provide general tools, but at the same time, engineer in enough flexibility in methods to allow customization for challenging systems. Given the moderate performance of current force fields in predicting binding affinities, many workers currently tune force fields to optimize performance for their desired application. In addition, search algorithms often require significant customization if the application strays from well-behaved ligands and largely-rigid receptors. The recent report of lysine-targeted inhibitors to a challenging target, eIF4E, is a case in point [78]: the authors modified DOCKovalent [79] (originally designed to model cysteine or serine modifications) to accommodate covalent attachment to lysine,and added a minimization and rescoring step to rank the best hits, leading to two successful leads for further development.
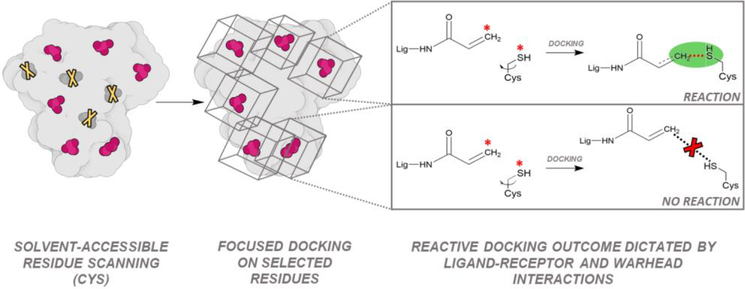
The availability of effective TCI has opened an exciting new approach to high-throughput discovery, termed “phenotypic drug discovery” [80,81] or “inverse drug discovery” [82]. In approaches such as variations of “activity based protein profiling” (ABPP) [35], whole cells are subjected to a cocktail of compounds, and new phenotypes are discovered and correlated directly with the target and the site of binding/inhibition by using tandem mass spectroscopy [34,35,51]. These proteomics experiments are very powerful and can generate huge amounts of data useful for the design and training of new computational methods. To exploit this data, and provide a predictive tool to expand these studies, we have developed a new approach, Reactive Docking (Figure 3) that does not use any of the methods in the previously defined categories [24,36,82]. Reactive Docking does not use constraints and the ligand is modeled as completely free and untethered while interacting with the receptor, including the potentially modifiable residue that is modeled as flexible (Figure 3). Residues more likely to be modified are also predicted by the protocol. Then, the ultimate reaction outcome is predicted by using conventional docking approach opportunely modified to include an additional soft potential [70] that explores energetics and geometric neighbors of the near-attack conformation (NAC) [83,84]. When the NAC-like conformation is not the best energy result, the reaction is predicted not to happen. While it is not necessary to specify the residues that have high likelihood of being modified, we developed a pre-processing protocol to facilitate their identification, pruning residues unlikely to be modified, such as those embedded below the protein surface (i.e. solvent inaccessible). Reactive docking was initially developed to model cysteine modifications by acrylamide and chloroacetamide warheads [36], but virtually any chemical reaction can be modeled. Since its initial application, more data has been used to adapt it to model other reactions, allowing prospective prediction of covalent binding sites on a protein or proteins of interest.
Figure 3. Reactive Docking Pipeline.
(Left) Scanning of all the solvent accessible residues and independent docking on each residue. (Right) Simulation of the reaction event: a custom soft potential is defined between the reactive atoms. If reactive atoms in docking results are found at appropriate bonding distance, the reaction is considered likely to occur
In order to be applicable in this context, any computational method needs to be fast, nimble, and easily deployable in a very high throughput fashion. Reactive Docking has been designed to address the challenges posed by such large experimental settings, in which thousands of potentially modifiable residues in many diverse protein targets are evaluated concurrently. Since it requires only the initial unmodified structures of the ligands and target, and the information on which atoms are involved in the initial reaction mechanism, the method can be used in a fully predictive fashion. Conversely, it can be also used to complement the experimental data, helping address methodological limitations. For example, when multiple potentially modifiable residues (i.e. cysteines) are present in the same proteolytic peptide, mass spectrometry analysis can detect the modification events, but cannot be used to resolve the location of such modifications at the single residue level. Reactive Docking has been used successfully to resolve this ambiguity in ABPP experiments on human T cells [41], identifying the residue most likely to be modified based on structural data. The method has been extended and applied to other reactions, such as those involving SuFEx warheads [25,82]. The residue scanning tool has been modified to recognize residues targetable by SuFEx that satisfy the geometric constraints (i.e., flanking residues). The approach has been then used prospectively in an “inverse drug discovery” campaign [82], in which tyrosine and lysine in the human proteome were targeted with three chemical probes functionalized with fluorosulfate groups. Reactive docking was used to determine the structural components determining protein target preference, as well as to explain the single residue discrimination power of the probes with competing tyrosines or lysine interactions [82]. As target diversity and selectivity of the SuFEx warheads has continued to expand, reactive docking has also been used to model serine [25]. The versatility in modeling different types of reactions, its predictive power, and the possibility of running in a high-throughput fashion make this method applicable in large prospective drug discovery campaigns. Recently, Reactive Docking has been deployed in the OpenPandemics - COVID-19ii, a large screening effort on the IBM World Community Gridiii targeting the SARS-CoV2 virus. The protocol is being used to target cysteine, tyrosine, and lysine residues in the whole viral proteome, screening libraries containing millions of compounds functionalized with diverse warheads. Building on the previous work with large proteomics experiments, the goal is to exploit its predictive power to perform virtual screenings of covalent targets across the viral proteome, expanding the range of targets from the well-characterized catalytic sites.
Concluding Remarks
The clear benefit of covalent inhibitors is their irreversible mechanism of action, making it possible to target traditionally undruggable sites and allowing innovative new phenotypic screening approaches. TCI remove the major impediment to use of covalent inhibition, by allowing fine-tuning of the specificity of these inhibitors and greatly reducing unwanted off-target effects. Computational approaches are working hand-in-hand with high-throughput screening to help answer these challenges. Modeling is instrumental in the discovery and deployment of these new inhibitors, helping to increase specificity and affinity of the binders, hence allowing use of less reactive, safer warheads. The availability of a large amount of experimental data laid the foundations for the development of more advanced and fully predictive computational methods for high-throughput screening of irreversible inhibitors. While there are experimental high-throughput approaches, such as DNA-encoded libraries, and covalent fragment-based screenings, they require significant amount of resources to be performed (equipment, reagents, facilities, trained personnel, etc.). Conversely, continuing the trend [85] set for conventional virtual screening methods, computational methods are the optimal tool to efficiently explore broad regions of the chemical space [86], while requiring very limited resources as many computational approaches can be run on commodity hardware. Computational methods are well-suited for assessing the potential druggability of targets in very high-throughput fashion [85], and provide an effective way to reduce the number of compounds to test. Ultimately, they represent a valuable complement to experimental efforts, reducing considerably the resources required to identify new molecular modulators of biological targets, enriching the number of hits [85], and effectively increasing the efficiency of experimental resources.
The field faces several outstanding questions that need to be addressed in the near future (see Outstanding Questions). New selective warheads are being designed [87] and we anticipate many more to be developed, possibly exploiting novel mechanisms. Computational methods will need to anticipate and promote discovery of such new warheads and mechanisms, providing new effective tools that evolve at the same pace as this very energetic field. This will involve continued enhancement of the basic methods of reactive docking to incorporate these new reaction models. In addition, effective approaches to large proteome-wide virtual screens are still under intense development, looking for a sweet spot that balances detail and accuracy of the method with computational feasibility. Methods will include new developments in the underlying physics of the interaction, incorporating mechanistic details with more accuracy, as well as moving to platforms that allow deployment of very large screening efforts, such as distributed computing and HPC platforms.
Outstanding questions.
How can computation spur the discovery of new warheads with new mechanisms?
How can computational methods be improved and optimized to overcome the current limitations in docking accuracy and ranking ability?
How can high-throughput virtual screening methods can be applied to irreversible inhibitors?
How can computational methods be used to predict warhead reactivity and binding kinetics?
How can computation help to fine tune the balance of warhead strength with specificity of non-covalent moieties in TCI?
How can computation address the extra complexity coming from the target flexibility and the identification of cryptic sites?
The final challenge is to improve TCI, and methods to discover them, to the level where they are common solutions to therapeutic challenges. Looking to previous structure-based efforts with conventional binders, this will involve a strong synergistic dialog between computational and experimental efforts. Drug design efforts are typically cyclic, with computational efforts alternating with experimental efforts, each improving at each turn of the cycle. Enhancement to the methodology is often concurrent with this process, as the effectiveness of both the method of discovery and the efficacy of the compounds is evaluated at each step. The field of TCI design is in its infancy, and will greatly benefit from this same course of cyclic development. The growing flow of experimental data generated will be essential to improve the quality of the predictive models, which in turn will provide reliable and more accurate high-throughput approaches. Computation will certainly play a central role, contributing to driving the innovation in experimental approaches, sharpening specificity, and providing new structural insight to target most intractable sites.
Highlights.
Molecular modeling is widely applied in drug discovery, with a number of successful approaches. Only recently there have been applications to the characterization of covalent inhibitors.
Several methods have been proposed, but a number of challenges arise from the modeling of the warhead reactivity, predict residue propensity to react, and ultimately the prediction of reaction outcome.
The recent resurgence of interest in covalent inhibitors is generating large amounts of experimental data that can be used to train computational models and improve the accuracy of predictive methods.
In order to be useful in drug discovery campaigns, computational methods need to achieve an ideal balance between accuracy and speed, in order to reduce experimental resources required to identify new hits while being applicable in high-throughput fashion.
Acknowledgments
This work was supported by Grants R01-GM069832 and U54AI150472 from the National Institutes of Health. This is manuscript 29986 from the Scripps Research Institute.
Footnotes
Disclaimer Statement
None
Resources
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singh J et al. (2011) The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 [DOI] [PubMed] [Google Scholar]
- 2.Lonsdale R and Ward RA (2018) Structure-based design of targeted covalent inhibitors. Chem. Soc. Rev. 47, 3816–3830 [DOI] [PubMed] [Google Scholar]
- 3.Gehringer M and Laufer SA (2019) Emerging and Re-Emerging Warheads for Targeted Covalent Inhibitors: Applications in Medicinal Chemistry and Chemical Biology. J. Med. Chem. 62, 5673–5724 [DOI] [PubMed] [Google Scholar]
- 4.Dalton SE and Campos S (2020) Covalent Small Molecules as Enabling Platforms for Drug Discovery. Chembiochem Eur. J. Chem. Biol. 21, 1080–1100 [DOI] [PubMed] [Google Scholar]
- 5.Barglow KT and Cravatt BF (2007) Activity-based protein profiling for the functional annotation of enzymes. Nat. Methods 4, 822–827 [DOI] [PubMed] [Google Scholar]
- 6.Speers AE and Cravatt BF (2009) Activity-Based Protein Profiling (ABPP) and Click Chemistry (CC)–ABPP by MudPIT Mass Spectrometry. Curr. Protoc. Chem. Biol. 1, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zambaldo C et al. (2016) Screening for covalent inhibitors using DNA-display of small molecule libraries functionalized with cysteine reactive moieties. MedChemComm 7, 1340–1351 [Google Scholar]
- 8.Mendoza VL and Vachet RW (2009) Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 28, 785–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojima H et al. (2020) Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins. Cell Chem. Biol. 27, 334–349.e11 [DOI] [PubMed] [Google Scholar]
- 10.Thornberry NA (1997) The caspase family of cysteine proteases. Br. Med. Bull. 53, 478–490 [DOI] [PubMed] [Google Scholar]
- 11.Board PG et al. (2000) Identification, Characterization, and Crystal Structure of the Omega Class Glutathione Transferases. J. Biol. Chem. 275, 24798–24806 [DOI] [PubMed] [Google Scholar]
- 12.Thompson SK et al. (1997) Design of potent and selective human cathepsin K inhibitors that span the active site. Proc. Natl. Acad. Sci. 94, 14249–14254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janes MR et al. (2018) Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 172, 578–589.e17 [DOI] [PubMed] [Google Scholar]
- 14.Drahl C et al. (2005) Protein-reactive natural products. Angew. Chem. Int. Ed Engl. 44, 5788–5809 [DOI] [PubMed] [Google Scholar]
- 15.Serafimova IM et al. (2012) Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles. Nat. Chem. Biol. 8, 471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senkane K et al. (2019) The Proteome-Wide Potential for Reversible Covalency at Cysteine. Angew. Chem. 131, 11507–11511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barf T and Kaptein A (2012) Irreversible Protein Kinase Inhibitors: Balancing the Benefits and Risks. J. Med. Chem. 55, 6243–6262 [DOI] [PubMed] [Google Scholar]
- 18.Uetrecht J and Naisbitt DJ (2013) Idiosyncratic adverse drug reactions: current concepts. Pharmacol. Rev. 65, 779–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer RA (2015) Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies. Drug Discov. Today 20, 1061–1073 [DOI] [PubMed] [Google Scholar]
- 20.Meng X et al. (2017) Definition of the Nature and Hapten Threshold of the β-Lactam Antigen Required for T Cell Activation In Vitro and in Patients. J. Immunol. 198, 4217–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mucsi Z et al. (2013) Penicillin’s catalytic mechanism revealed by inelastic neutrons and quantum chemical theory. Phys. Chem. Chem. Phys. 15, 20447–20455 [DOI] [PubMed] [Google Scholar]
- 22.Jones LH and Kelly JW (2020) Structure-based design and analysis of SuFEx chemical probes. RSC Med. Chem. 11, 10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W et al. (2016) Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J. Am. Chem. Soc. 138, 7353–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narayanan A and Jones LH (2015) Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci. 6, 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Q et al. (2019) SuFEx-enabled, agnostic discovery of covalent inhibitors of human neutrophil elastase. Proc. Natl. Acad. Sci. U. S. A. 116, 18808–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Northrop BH and Coffey RN (2012) Thiol–Ene Click Chemistry: Computational and Kinetic Analysis of the Influence of Alkene Functionality. J. Am. Chem. Soc. 134, 13804–13817 [DOI] [PubMed] [Google Scholar]
- 27.Bian Y et al. (2020) Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS. Nat. Commun. 11, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwöbel JAH et al. (2011) Measurement and Estimation of Electrophilic Reactivity for Predictive Toxicology. Chem. Rev. 111, 2562–2596 [DOI] [PubMed] [Google Scholar]
- 29.Martin JS et al. (2019) Characterising covalent warhead reactivity. Bioorg. Med. Chem. 27, 2066–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palazzesi F et al. (2020) BIreactive: A Machine-Learning Model to Estimate Covalent Warhead Reactivity. J. Chem. Inf. Model. DOI: 10.1021/acs.jcim.9b01058 [DOI] [PubMed] [Google Scholar]
- 31.Lonsdale R et al. (2017) Expanding the Armory: Predicting and Tuning Covalent Warhead Reactivity. J. Chem. Inf. Model. 57, 3124–3137 [DOI] [PubMed] [Google Scholar]
- 32.Smith JM and Rowley CN (2015) Automated computational screening of the thiol reactivity of substituted alkenes. J. Comput. Aided Mol. Des. 29, 725–735 [DOI] [PubMed] [Google Scholar]
- 33.Soylu İ and Marino SM (2016) Cy-preds: An algorithm and a web service for the analysis and prediction of cysteine reactivity. Proteins 84, 278–291 [DOI] [PubMed] [Google Scholar]
- 34.Wang H et al. (2018) Sequence-Based Prediction of Cysteine Reactivity Using Machine Learning. Biochemistry 57, 451–460 [DOI] [PubMed] [Google Scholar]
- 35.Weerapana E et al. (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Backus KM et al. (2016) Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weerapana E et al. (2008) Disparate proteome reactivity profiles of carbon electrophiles. Nat. Chem. Biol. 4, 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidd D et al. (2001) Profiling Serine Hydrolase Activities in Complex Proteomes. Biochemistry 40, 4005–4015 [DOI] [PubMed] [Google Scholar]
- 39.Hacker SM et al. (2017) Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 9, 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang CV et al. (2017) Drugging the “undruggable” cancer targets. Nat. Rev. Cancer 17, 502–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vinogradova EV et al. (2020) An Activity-Guided Map of Electrophile-Cysteine Interactions in Primary Human T Cells. Cell 182, 1009–1026.e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettinger J et al. (2017) Lysine-Targeting Covalent Inhibitors. Angew. Chem. Int. Ed Engl. 56, 15200–15209 [DOI] [PubMed] [Google Scholar]
- 43.Mukherjee H and Grimster NP (2018) Beyond cysteine: recent developments in the area of targeted covalent inhibition. Curr. Opin. Chem. Biol. 44, 30–38 [DOI] [PubMed] [Google Scholar]
- 44.Cuesta A and Taunton J (2019) Lysine-Targeted Inhibitors and Chemoproteomic Probes. Annu. Rev. Biochem. 88, 365–381 [DOI] [PubMed] [Google Scholar]
- 45.Metcalf B et al. (2017) Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin. ACS Med. Chem. Lett. 8, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon DA and Weerapana E (2015) Covalent protein modification: the current landscape of residue-specific electrophiles. Curr. Opin. Chem. Biol. 24, 18–26 [DOI] [PubMed] [Google Scholar]
- 47.Wang Y et al. (2019) Expedited mapping of the ligandable proteome using fully functionalized enantiomeric probe pairs. Nat. Chem. 11, 1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradshaw JM et al. (2015) Prolonged and tunable residence time using reversible covalent kinase inhibitors. Nat. Chem. Biol. 11, 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schirmeister T et al. (2016) Quantum Chemical-Based Protocol for the Rational Design of Covalent Inhibitors. J. Am. Chem. Soc. 138, 8332–8335 [DOI] [PubMed] [Google Scholar]
- 50.Zhang H et al. (2019) Ranking Reversible Covalent Drugs: From Free Energy Perturbation to Fragment Docking. J. Chem. Inf. Model. 59, 2093–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strelow JM (2017) A Perspective on the Kinetics of Covalent and Irreversible Inhibition. SLAS Discov. Adv. Sci. Drug Discov. 22, 3–20 [DOI] [PubMed] [Google Scholar]
- 52.Awoonor-Williams E et al. (2017) Modeling covalent-modifier drugs. Biochim. Biophys. Acta Proteins Proteomics 1865, 1664–1675 [DOI] [PubMed] [Google Scholar]
- 53.Resnick E et al. (2019) Rapid Covalent-Probe Discovery by Electrophile-Fragment Screening. J. Am. Chem. Soc. 141, 8951–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz PA et al. (2014) Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. Proc. Natl. Acad. Sci. 111, 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krenske EH et al. (2016) Kinetics and Thermodynamics of Reversible Thiol Additions to Mono- and Diactivated Michael Acceptors: Implications for the Design of Drugs That Bind Covalently to Cysteines. J. Org. Chem. 81, 11726–11733 [DOI] [PubMed] [Google Scholar]
- 56.Mah R et al. (2014) Drug discovery considerations in the development of covalent inhibitors. Bioorg. Med. Chem. Lett. 24, 33–39 [DOI] [PubMed] [Google Scholar]
- 57.Schmidt TC et al. (2014) Protocol for Rational Design of Covalently Interacting Inhibitors. ChemPhysChem 15, 3226–3235 [DOI] [PubMed] [Google Scholar]
- 58.Yu HS et al. (2019) Toward Atomistic Modeling of Irreversible Covalent Inhibitor Binding Kinetics. J. Chem. Inf. Model. 59, 3955–3967 [DOI] [PubMed] [Google Scholar]
- 59.Liu Q et al. (2013) Developing Irreversible Inhibitors of the Protein Kinase Cysteinome. Chem. Biol. 20, 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaikuad A et al. (2018) Inside Back Cover: The Cysteinome of Protein Kinases as a Target in Drug Development (Angew. Chem. Int. Ed. 16/2018). Angew. Chem. Int. Ed. 57, 4429–4429 [DOI] [PubMed] [Google Scholar]
- 61.Zhao Z et al. (2017) Determining Cysteines Available for Covalent Inhibition Across the Human Kinome. J. Med. Chem. 60, 2879–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ravindranath PA and Sanner MF (2016) AutoSite: an automated approach for pseudo-ligands prediction-from ligand-binding sites identification to predicting key ligand atoms. Bioinforma. Oxf. Engl. 32, 3142–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sotriffer C (2018) Docking of Covalent Ligands: Challenges and Approaches. Mol. Inform. 37, 1800062. [DOI] [PubMed] [Google Scholar]
- 64.Kumalo HM et al. (2015) Theory and Applications of Covalent Docking in Drug Discovery: Merits and Pitfalls. Molecules 20, 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ouyang X et al. (2013) CovalentDock: Automated covalent docking with parameterized covalent linkage energy estimation and molecular geometry constraints. J. Comput. Chem. 34, 326–336 [DOI] [PubMed] [Google Scholar]
- 66.Bianco G et al. (2016) Covalent docking using autodock: Two-point attractor and flexible side chain methods. Protein Sci. Publ. Protein Soc. 25, 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ai Y et al. (2016) Discovery of Covalent Ligands via Noncovalent Docking by Dissecting Covalent Docking Based on a “Steric-Clashes Alleviating Receptor (SCAR)” Strategy. J. Chem. Inf. Model. 56, 1563–1575 [DOI] [PubMed] [Google Scholar]
- 68.Schröder J et al. (2013) Docking-Based Virtual Screening of Covalently Binding Ligands: An Orthogonal Lead Discovery Approach. J. Med. Chem. 56, 1478–1490 [DOI] [PubMed] [Google Scholar]
- 69.Zhu K et al. (2014) Docking covalent inhibitors: a parameter free approach to pose prediction and scoring. J. Chem. Inf. Model. 54, 1932–1940 [DOI] [PubMed] [Google Scholar]
- 70.Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes | Journal of Medicinal Chemistry.. [Online]. Available: https://pubs.acs.org/doi/10.1021/jm051256o. [Accessed: 07-Oct-2020] [DOI] [PubMed] [Google Scholar]
- 71.Toledo Warshaviak D et al. (2014) Structure-based virtual screening approach for discovery of covalently bound ligands. J. Chem. Inf. Model. 54, 1941–1950 [DOI] [PubMed] [Google Scholar]
- 72.Rachman M et al. (2019) DUckCov: a Dynamic Undocking Based Virtual Screening Protocol for Covalent Binders. Chemmedchem 14, 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruiz-Carmona S et al. (2014) rDock: A Fast, Versatile and Open Source Program for Docking Ligands to Proteins and Nucleic Acids. PLOS Comput. Biol. 10, e1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ruiz-Carmona S et al. (2017) Dynamic undocking and the quasi-bound state as tools for drug discovery. Nat. Chem. 9, 201–206 [DOI] [PubMed] [Google Scholar]
- 75.Das S et al. (2019) EnzyDock: Protein–Ligand Docking of Multiple Reactive States along a Reaction Coordinate in Enzymes. J. Chem. Theory Comput. 15, 5116–5134 [DOI] [PubMed] [Google Scholar]
- 76.Wen C et al. (2019) Systematic Studies on the Protocol and Criteria for Selecting a Covalent Docking Tool. Molecules 24, 2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scarpino A et al. (2018) Comparative Evaluation of Covalent Docking Tools. J. Chem. Inf. Model. 58, 1441–1458 [DOI] [PubMed] [Google Scholar]
- 78.Wan X et al. (2020) Discovery of Lysine-Targeted eIF4E Inhibitors through Covalent Docking. J. Am. Chem. Soc. 142, 4960–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.London N et al. (2014) Covalent docking of large libraries for the discovery of chemical probes. Nat. Chem. Biol. 10, 1066–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dominguez E et al. (2014) Integrated phenotypic and activity-based profiling links Ces3 to obesity and diabetes. Nat. Chem. Biol. 10, 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moffat JG et al. (2017) Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 16, 531–543 [DOI] [PubMed] [Google Scholar]
- 82.Mortenson DE et al. (2018) “Inverse Drug Discovery” Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates. J. Am. Chem. Soc. 140, 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bruice TC (2002) A view at the millennium: the efficiency of enzymatic catalysis. Acc. Chem. Res. 35, 139–148 [DOI] [PubMed] [Google Scholar]
- 84.Benkovic SJ and Hammes-Schiffer S (2003) A perspective on enzyme catalysis. Science 301, 1196–1202 [DOI] [PubMed] [Google Scholar]
- 85.Irwin JJ and Shoichet BK (2016) Docking Screens for Novel Ligands Conferring New Biology. J. Med. Chem. 59, 4103–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lyu J et al. (2019) Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vantourout JC et al. (2020) Serine-Selective Bioconjugation. J. Am. Chem. Soc.(ASAP article) [DOI] [PMC free article] [PubMed] [Google Scholar]