Abstract
KRAS mutations are among the most common genetic alterations in lung, colorectal and pancreatic cancers. Direct inhibition of KRAS oncoproteins has been a longstanding pursuit in precision oncology, one established shortly after the discovery of RAS mutations in human cancer cells nearly 40 years ago. Recent advances in medicinal chemistry have established inhibitors targeting KRAS(G12C), a mutation found in ~13% of lung adenocarcinomas and, at a lower frequency, in other cancers. Preclinical studies describing their discovery and mechanism of action, coupled with early clinical data from patients treated with these drugs, have sparked a renewed enthusiasm in the study of KRAS and its therapeutic potential. Here we discuss how these advances are reshaping the fundamental aspects of KRAS oncoprotein biology and the strides being made towards improving patient outcomes in the clinic.
KRAS and the highly-related NRAS and HRAS GTPases hydrolyze guanosine triphosphate (GTP) to guanosine diphosphate (GDP). They control diverse cellar functions by cycling between an active, GTP-bound, and an inactive, GDP-bound, conformation (reviewed in Hobbs et al., 2016; Pylayeva-Gupta et al., 2011). The GTPase activity of KRAS is enhanced by GTPase-activating proteins (GAP), such as NF1, and the exchange of GDP for GTP is enhanced by guanine-nucleotide exchange factors (GEF), such as SOS1/2 (Bos et al., 2007; Margarit et al., 2003; Trahey and McCormick, 1987). The balance between hydrolysis and exchange determines the levels of active KRAS in cells.
KRAS activates several effector proteins to control diverse cellular functions (reviewed in Downward, 2003; Malumbres and Barbacid, 2003). The best studied effectors include RAF kinases and the catalytic subunit of PI3K. KRAS-GTP binding to RAF kinases stimulates their dimerization and activation, which triggers the step-wise activation of MEK and ERK, a pathway that drives cell-cycle progression and proliferation. By binding to PI3K, KRAS helps activate AKT and mTOR, which regulate apoptosis, metabolism and translation.
Effective targeting of KRAS signaling has been difficult to achieve in patients (reviewed in Ostrem and Shokat, 2016; Papke and Der, 2017). Competitive inhibition of nucleotide binding, as that afforded for kinases, has not been feasible for RAS GTPases because of their high affinity for the nucleotide. Blocking the localization of KRAS at the plasma membrane, a key component for its activation, has been ineffective because multiple compensatory pathways regulate this process. Similarly, targeting effector signaling downstream of KRAS has not yielded significant clinical benefit (reviewed in Karoulia et al., 2017; Lito et al., 2013), either because of paradoxical signaling activation triggered by the inhibitor (e.g. RAF inhibitors) or because on-target toxicity limiting the maximum tolerated dose in patients (e.g. MEK or AKT inhibitors). Considering this historical context and the prevalence of KRAS activating mutations in cancer patients, the discovery and clinical development of KRAS(G12C) selective inhibitors is heralding a new era in precision oncology. Here we review the evolution of these exciting new therapies, their mechanism of action and cellular effects in preclinical studies, as well as the initial observations from patients treated with KRAS(G12C) inhibitors in clinical trials.
KRAS activation in cancer
KRAS mutations are found predominantly in lung (approximately 25% of cases and estimated to affect 57,000 patients per year in the US), pancreatic (~95% of cases and ~54,000 patients) and colorectal (~35% of cases and ~36,000 patients) cancers. Mutations in NRAS and HRAS are also found in cancer patients, but at a lower frequency than KRAS. KRAS mutations result in single amino acid substitutions that activate the oncoprotein by hindering its ability to hydrolyze GTP (reviewed in Simanshu et al., 2017). Substitutions at the G12 (e.g. D/C/V/R/A/S) or G13 (e.g. D/C/V) residues prevent the stabilization of the hydrolysis transition state by a critical arginine residue in GAPs (Bollag and McCormick, 1991; Scheffzek et al., 1997). Less common variants, such as Q61H/L/R/K, interfere with the coordination of the water molecule involved in hydrolysis (Scheidig et al., 1999), whereas A146T enhances the propensity for nucleotide-exchange (Poulin et al., 2019). The distribution of KRAS mutant alleles differs across tumors, with G12C comprising ~50% of KRAS mutations in lung cancer and G12D being the most common allele in pancreatic and colorectal cancers (Li et al., 2018a; Riely et al., 2008). In addition to being found in ~13% of patients with lung adenocarcinoma, the KRAS(G12C) allele is found at a lower frequency in those with colorectal (~3%), uterine (~2%), mesothelioma (~1%), pancreatic (<1%), cervical (<1%), bladder (<1%) or gastric (<1%) cancers (the frequency estimates are from the cancer genome atlas (TCGA) and www.cbioportal.org).
While insensitive to GAP-assisted hydrolysis, RAS oncoproteins have measurable intrinsic GTP hydrolysis rates in biochemical assays (Gibbs et al., 1988; Hunter et al., 2015; Smith et al., 2013; Wey et al., 2013). This was believed to be inconsequential for cellular function and KRAS oncoproteins were traditionally viewed as being ‘constitutively’ active. Recent studies by us and others are reshaping this notion, by showing that some KRAS oncoproteins cycle between their active and inactive states in cancer cells and are dependent on nucleotide-exchange for activation (Lito et al., 2016; Nichols et al., 2018; Patricelli et al., 2016; Poulin et al., 2019; Rabara et al., 2019). As discussed below, this has important implications for their susceptibility to therapy.
Allele-specific inhibitors targeting a mutated cysteine residue
Direct KRAS oncoprotein inhibition has been a long-standing objective in precision medicine. The earliest efforts track back to the 1980’s, when RAS activating mutations were found in human cancer cells and not long after the discovery of RAS oncogenes in transforming viruses. The discovery of inhibitors that selectively target KRAS(G12C) (hereafter referred to as G12Ci), while sparing wild-type or other mutant KRAS (Ostrem et al., 2013), was a groundbreaking advance in this quest. The inhibitors bind covalently to the mutant cysteine residue, and occupy a pocket in the switch-II region (SIIP), when KRAS(G12C) is in its inactive, GDP-bound, state (Figure 1A). The drugs are expected to also bind to NRASG12C and HRASG12C; although these variants are very rare: NRASG12C is estimated to be present in ~0.5% of acute myeloid leukemias and colorectal cancers, and HRASG12C in ~0.4% of head and neck cancers. As such, the effect of G12Ci on these oncoproteins has not been scrutinized experimentally.
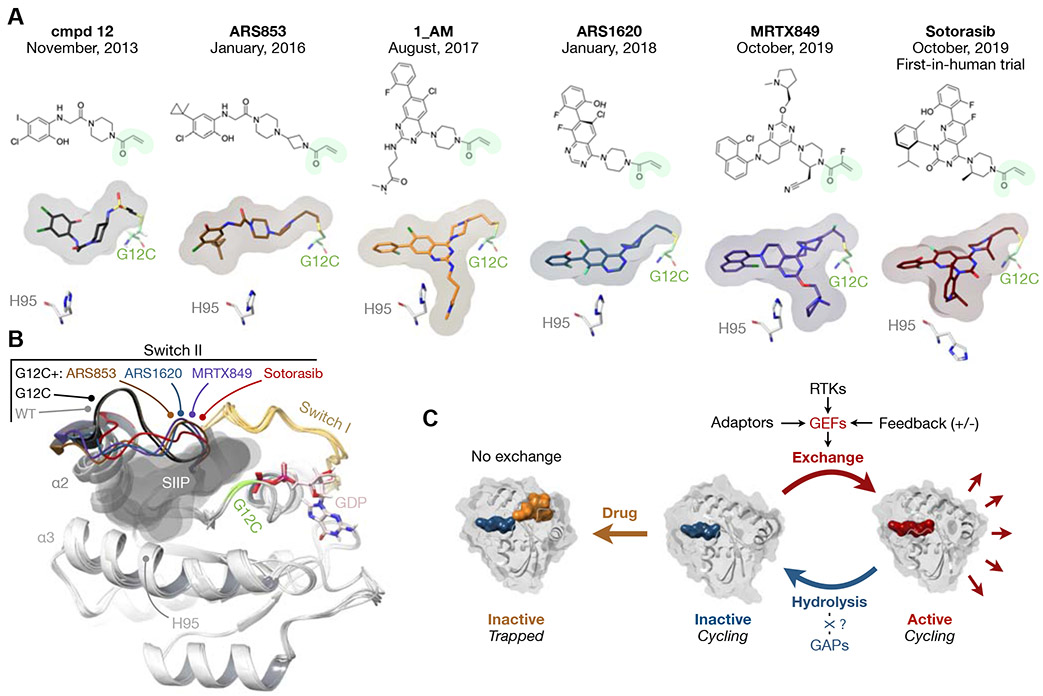
Figure 1. G12C inhibitors and their mechanism of action.
A) Chemical structures of G12C inhibitors with their initial publication date. The surface plot denotes the orientation of each inhibitor in the SIIP of mutant KRAS. The acrylamide warhead that engages the G12C residue is shown in green. The orientation of the histidine (H) 95 residue on KRAS is also shown. An interaction with this residue is thought to enhance the potency of inhibition.
B) Superimposed structures of KRAS(G12C) bound to GDP alone, or in the presence of the indicated inhibitors. KRAS WT is included as a comparison. The surfaces in gray denote the area occupied by the inhibitors. Note the displacement of switch II in drug-bound structures. From 4LYF, 5F2E, 5V9L, 5V9U, 60IM, 6UT0, 4OBE and 4LDJ.
C) Inactive state-selective KRAS(G12C) inhibition requires intact GTPase activity and occurs by preventing nucleotide-exchange to the active state. KRAS oncoproteins are insensitive to GAPs (dotted line, see text) and a relatively high intrinsic hydrolysis rate by KRAS(G12C) is thought to be sufficient for inhibition. The alternative possibility is that GTP hydrolysis by KRAS oncoproteins is aided by unidentified cellular factors (question mark). Space-filling models of GTP, GDP and G12Ci are respectively colored in red, blue and orange.
Structure-based optimization led to inhibitors with progressively higher affinities for KRAS(G12C), including ARS853 (Lito et al., 2016; Patricelli et al., 2016), 1_AM (Zeng et al., 2017), ARS1620 (Janes et al., 2018). These suppress KRAS(G12C) activation, attenuate proliferation and induce cell-death to varying degrees across tumor models. Importantly, ARS1620 inhibits tumor growth in xenograft models (Janes et al., 2018), consolidating inactive state-selective inhibition as an effective approach for suppressing KRAS(G12C) signaling.
Being made publicly available from early on, the structure-activity relationships of the ARS compounds furthered the development of even more potent G12Ci. While similar in that they share an acrylamide warhead that engages G12C, these inhibitors have distinct chemical structures (Figure 1A). Two of these, sotorasib (also known as AMG510, Canon et al., 2019) and MRTX849 (Hallin et al., 2020), have antiproliferative IC50s in the low nanomolar range and potently suppress xenograft tumor growth in mice. Their enhanced potency reportedly owes to an enhanced interaction with the H95 residue in the α3 helix of KRAS(G12C) (Fell et al., 2020; Lanman et al., 2020), an interaction that is also reported to be responsible for the improved potency of ARS1620 and 1_AM over ARS853 (Janes et al., 2018; Zeng et al., 2017). Excitingly, sotorasib and MRTX849 now show clinical activity in patients with KRAS(G12C)-mutant tumors.
Other evolving approaches to target KRAS(G12C) include GDP-analogues (Hunter et al., 2014), compounds that displace effector-binding (Kessler et al., 2019) and ‘tri-complex’, KRAS(G12C)-GTP:cyclophylin A (CYPA) inhibitors (Nichols et al., 2020). The cellular effects of these emerging compounds are under evaluation and it remains to be seen if they achieve the same or better KRAS inhibition than SIIP inhibitors.
Mechanism of inactive state-selective KRAS(G12C) inhibition
How do inactive state-selective drugs inhibit KRAS(G12C), an oncoprotein conventionally thought to be constitutively active? Key to answering this question is the realization that KRAS(G12C) undergoes nucleotide cycling in cancer cells and fluctuates between its active and inactive states (Lito et al., 2016). When introduced alongside KRAS(G12C), secondary mutations that completely block hydrolysis, such as A59G or Q61L, increase baseline KRAS(G12C) activation and attenuate the response to G12Ci-treatment. Experiments using mass spectrometry to quantify drug-bound KRAS(G12C) following perturbations of its nucleotide cycle in cells and studies utilizing more potent inhibitors, reached a similar conclusion (Hallin et al., 2020; Patricelli et al., 2016). Thus, an intact GTPase activity is required for KRAS(G12C) inhibition.
Drug-binding to KRAS(G12C) prevents its reactivation by nucleotide-exchange and traps the oncoprotein in the inactive state (Figure 1C). This is supported by experiments in which drug-bound KRAS(G12C) cannot undergo SOS1- or EDTA- mediated nucleotide-exchange (Canon et al., 2019; Janes et al., 2018; Lito et al., 2016; Patricelli et al., 2016). Initially, drug-binding was thought to lower the affinity of KRAS(G12C) for GTP, however, the drug also attenuates the exchange of GDP for GDP (Janes et al., 2018; Patricelli et al., 2016), suggesting a primary effect on nucleotide-exchange rather than affinity. Indeed, the potency of G12Ci-treatment is attenuated when secondary mutations enhancing nucleotide-exchange are engineered alongside KRAS(G12C) (Lito et al., 2016). In a similar manner, inhibition of SOS1 enhances KRAS(G12C) inhibition (Hillig et al., 2019).
A question yet to be answered is how does KRAS(G12C) undergo sufficient hydrolysis to enable inhibition by inactive state-selective drugs? It has been proposed that KRAS(G12C) is unique among commonly occurring KRAS mutants, in that it has a high intrinsic hydrolysis rate (Hunter et al., 2015; Simanshu et al., 2017). Whether this is sufficient to satisfy the kinetics of drug trapping in cells, however, remains unknown.
Initial clinical effects
Indicative of the tremendous progress made in the seven years since their discovery, several inactive state-selective KRAS(G12C) inhibitors are now in clinical trials (Table I). These include sotorasib, MRTX849, GDC6036, LY3499446 and JNJ74699157 (ARS3248). Others are anticipated to enter the clinic in the coming year.
Table I.
KRAS(G12C) inhibitors in clinical trials
| Drug | Status | Phase | Design | Trial |
|---|---|---|---|---|
| Sotorasib (AMG510) | Recruiting | I/II/III | Monotherapy or combination with: AMG404 (anti-PDl) Panitumumab RMC4630 Trametinib Afatinib |
NCT04380753 NCT03600883 NCT04303780 NCT04185883 |
| MRTX849 | Recruiting | I/II | Monotherapy or combination with: Pembrolizumab Cetuximab Afatinib TNO155 |
NCT03785249 NCT04330664 |
| GDC6036 | Recruiting | I | Monotherapy | NCT04449874 |
| LY3499446 | Not recruiting * | I | Monotherapy | NCT04165031 |
| JNJ74699157 | Not recruiting * | I | Monotherapy | NCT04006301 |
Terminated due to toxicity.
The clinical data so far suggest a wide-therapeutic index for some G12Ci, namely sotorasib and MRTX849. Treatment with these drugs appears well-tolerated by most patients, although side effects including diarrhea, anemia or liver enzyme abnormalities have been reported (Christensen et al., 2019; Govindan et al., 2019). It is important to note that unanticipated toxicity in animal or early-phase clinical studies has halted the clinical development of other compounds. Almost all available G12Ci have a selectivity threshold in preclinical studies, i.e., a concentration above which they also inhibit the growth of KRAS wild-type cells. The covalent nature of drug binding may lead to off-target effects caused by non-selective interaction with cysteine residues in other proteins. Global proteomic analyses aiming to identify all cysteine residues that are covalently-modified by the drug (first described by Patricelli et al., 2016) show a clear selectivity for KRAS(G12C) in cells harboring this mutation. Were such studies to also be performed in wild-type KRAS cells, a setting where KRAS(G12C) cannot outcompete weaker targets, they would perhaps more accurately predict off-target effects in normal tissue and help understand the sources of G12Ci-toxicity. Factors beyond covalent-binding may also contribute to toxicity, especially considering that the severity of toxicity varies between drugs.
Efficacy data are beginning to emerge from the clinical trial investigating sotorasib, the first G12Ci to advance past dose-escalation phase. Of the lung cancer patients treated with sotorasib, nearly half had at least a partial response at initial evaluation (Govindan et al., 2019). Only a subset of these responses (~30%) were confirmed on subsequent evaluation, suggesting that early adaptation limits the effect of therapy. The vast majority of colorectal cancer patients did not have radiologic response to treatment, although considerable symptomatic improvement has been reported. MRTX849 appears to have similar effects in lung and colorectal cancer patients, yet more mature data are needed to validate early results (Christensen et al., 2019). While similar in inhibitory mechanism, sotorasib and MRTX849 differ in their elimination half-lives (sotorasib: 6h and MRTX849: 25h) and administration schemes (with sotorasib dosed daily and MRTX849 twice daily).
The lack of a G12Ci-treatment response in most patients with colorectal cancer (and in some patients with lung cancer) invokes studies suggesting that not all KRAS-mutant cancers are dependent on this oncoprotein for growth (Singh et al., 2012). Alternatively, it may be that the ability to target the oncoprotein in some settings is compromised. For example, BRAF V600-mutant colorectal cancers are insensitive to RAFi-treatment (Corcoran et al., 2012; Prahallad et al., 2012), owing to a higher activation of EGFR and the formation of RAFi-insensitive RAF dimers (Poulikakos et al., 2010). These tumors retain dependency on BRAF and targeting multiple nodes of the pathway results in better antitumor effects (Kopetz et al., 2020). A conclusion that cancers are independent of KRAS(G12C) thus requires evidence of potent and sustained target inhibition without an effect on phenotypes associated with a selective growth-advantage.
Adaptation to treatment
G12Ci-treatment only briefly suppresses KRAS signaling in cells (Janes et al., 2018; Lito et al., 2016; Misale et al., 2019; Ryan et al., 2019). Within 24-72h of treatment, an initial oncoprotein signaling inhibition is accompanied by a re-accumulation of active KRAS and a reactivation of ERK signaling (Xue et al., 2020). This pattern is consistent with adaptation, a process that is well-described in response to RAF or MEK inhibitors and one that limits their therapeutic potential (Lamba et al., 2014; Lito et al., 2012; Lito et al., 2014; Manchado et al., 2016; Montero-Conde et al., 2013; Sun et al., 2014). The accumulation of active KRAS suggests that a compensatory activation of receptor tyrosine kinases (RTK) and SOS1/2, which are both feedback suppressed by ERK, is in large part responsible for the adaptive changes noted during G12Ci-treatment. However, inhibition of ERK also leads to down-regulated expression of MAPK phosphatases (e.g., DUSP4, DUSP6 etc.), which in turn results in increased ERK activation (Eblaghie et al., 2003). Although, in principle, this effect may also contribute to G12Ci adaptation, it has not yet been experimentally validated.
RTKs may modulate adaptation to G12Ci-treatment in two ways (Figure 2). By stimulating SOS1/2-mediated nucleotide exchange, RTK activation shifts KRAS(G12C) to its GTP-bound conformation, which is insensitive to the drug. RTKs can also bypass inhibition in a G12C-independent manner, i.e., through the activation of wild-type RAS, PI3K/AKT/mTOR or other pathways (Janes et al., 2018; Misale et al., 2019; Ryan et al., 2019). The role of PI3K/mTOR is supported by the presence of antiproliferative synergy when the G12Ci is combined with PI3K or mTOR inhibitors (Hallin et al., 2020; Misale et al., 2019). While it is evident that wild-type RAS proteins are feedback activated following G12Ci treatment, it is not clear whether this represents a compensatory response or one that drives adaptation. To this end, further testing is needed to show that wild-type RAS down-regulation enhances G12C inhibition and to determine the relative contribution of NRAS, HRAS and/or wild type KRAS (in cells harboring a heterozygous KRAS(G12C) mutation).
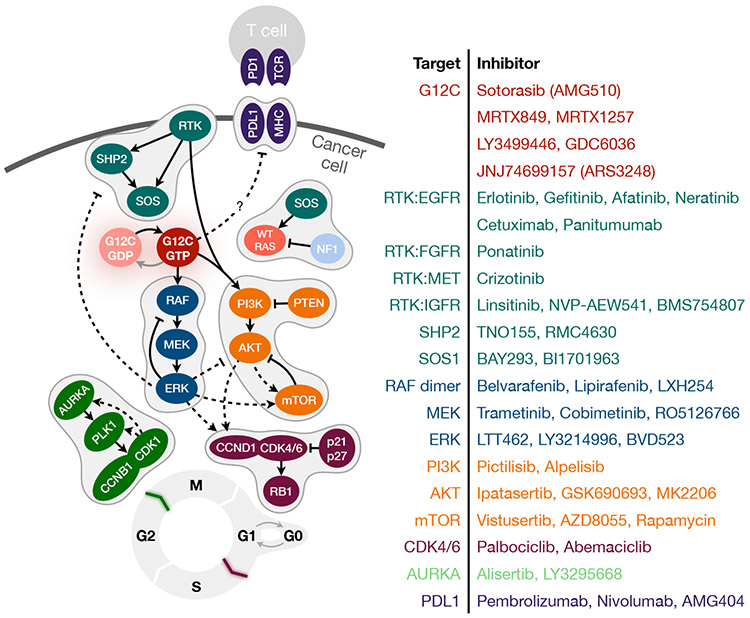
Figure 2. Opportunities for combination therapy.
A simplified schematic of KRAS signaling along with inhibitors targeting various intermediates reported to modulate KRAS(G12C) inhibition. Solid lines indicate a direct effect whereas dotted lines indicate indirect effects. Colors denote signaling intermediates along the same pathway.
The adaptive signaling changes noted above have been integrated with single-cell modeling to show that adaptation to G12Ci-treatment occurs rapidly and in a non-uniform manner across cancer cell populations (Xue et al., 2020). Shortly after treatment, some cells are sequestered in a quiescent (G0) state with low KRAS activity, whereas others bypass this effect to resume proliferation. The divergent response is modulated by the production of new KRAS(G12C) and its distribution between the active/drug-insensitive and inactive/drug-sensitive states. New KRAS(G12C) is synthesized in response to suppressed ERK signaling and it is more susceptible to activation by nucleotide-exchange than baseline KRAS(G12C), which signals in the presence of feedback-suppressed SOS1/2. This explains why G12Ci-rechallenge is less effective than initial treatment and how active KRAS can re-accumulate during G12Ci-treatment, even though the drug binds in covalent/irreversible manner. Taken together these findings suggest that sustained inactive state-selective KRAS(G12C) inhibition can be achieved by blocking the production of new KRAS, enhancing its degradation or preventing its conversion to the active/drug-insensitive state.
Acquired resistance
Acquired resistance limits the clinical benefit of almost all targeted therapies and it is highly likely to also limit the effect of G12Ci. Although models with acquired resistance have not yet been reported, the mechanisms of inhibition and adaptation make several predictions on how acquired resistance to G12Ci-treatment might emerge.
Events predicted to attenuate target inhibition include: a) Secondary mutations on KRAS(G12C) that impede drug binding or enhance the propensity for the active/drug-insensitive conformation. b) Genetic events that consolidate the up-regulation of KRAS, such as KRAS amplification (Sale et al., 2019; Wong et al., 2018) or alterations leading to its diminished degradation (Bigenzahn et al., 2018). c) Activation of nucleotide-exchange through alterations in GEFs, like SOS1/2, or other upstream intermediates, such as RTKs or SHP2.
Events predicted to mediate resistance while the target remains inhibited, include: a) Activation of downstream signaling through gain-of-function events in RAF, MEK or ERK as well as loss-of-function events in RB1 or cyclin-dependent kinase inhibitors (e.g., p21, p27). SHP2. b) Activation of parallel signaling pathways through loss of NF1 or PTEN or activating mutations in other RAS family GTPases.
Studies investigating the effects of genetic KRAS down-regulation and those describing resistance to MEKi treatment provide additional insights into events that may lead to G12Ci-resistance. These include epithelial-to-mesenchymal transformation, through ZEB1 or YAP1 (Peng et al., 2019; Shao et al., 2014) as well as alterations in metabolic dependencies, including glycolysis (Amendola et al., 2019; Kerr et al., 2016), glutaminolysis (Romero et al., 2017, macropinocytosis (Commisso et al., 2013) and autophagy (Bryant et al., 2019; Kinsey et al., 2019).
Maximizing therapeutic impact
Clinically-active KRAS(G12C) inhibitors have a tremendous potential for impacting patient care, especially in patients with lung cancer, a disease affecting approximately 230,000 people each year in the US. Below we present several approaches aimed at maximizing their clinical benefit.
Optimizing monotherapy.
Drug-exposure directly correlates with the magnitude of ERK inhibition and response in patients with BRAF(V600) mutant tumors treated with a RAFi (Bollag et al., 2010). In this setting, a greater than 80% inhibition of ERK phosphorylation is needed to achieve clinical responses. Thus, efforts to establish inhibitors with an even higher affinity for the inactive-state of KRAS(G12C) will ensure a more potent and durable inhibition by delaying adaptation. Compounds that directly target active KRAS(G12C), such as the ‘tricomplex’ active state-selective inhibitors described above, also hold promise in this regard (provided that they retain a therapeutic index and can be translated to clinical trials).
Where feasible, alternative formulations, or administration schemes, enabling high drug exposures in patients may enhance the response rate and/or its magnitude. For example, very few adverse-effects leading to drug-discontinuation were observed during dose-escalation for sotorasib. It will be interesting to determine if sotorasib produces stronger antitumor effects when administered to patients on a twice-daily schedule.
Intermittent or pulsatile therapy has been suggested to prolong the response to inhibition of BCR-ABL, EGFR or BRAF (Das Thakur et al., 2013; Shah et al., 2008), to enable multimodal pathway inhibition (Xue et al., 2017) and to improve antitumor immunity (Choi et al., 2019). Intermittent KRAS(G12C) inhibition may have merit in principle, but the expected benefits need to be leveraged against the irreversible nature of drug-binding. More experimentation is required to determine the duration of target-engagement in wash-out studies and if intermittent dosing mitigates the adaptive changes noted above.
Pharmacodynamic endpoints must be implemented in the clinical assessment of the G12Ci-treatment response. Irreversible drug-binding affords the direct quantification of drug-bound KRAS(G12C) in tumor biopsies. Coupled with standard approaches to measure the phosphorylation of ERK or other KRAS signaling intermediates, this can provide invaluable clinical insight. Non-invasive cell-free DNA assays offer an alternative approach to estimate the effect of treatment on KRAS(G12C) allele burden over time. Such interventions are particularly important if we are to distinguish tumors that grow independently of KRAS(G12C) from those where KRAS(G12C) is not susceptible to inactive state-selective inhibition. They can also help understand the determinants of the transient response in some lung cancer patients.
Rational drug combinations.
Owing to a wide therapeutic-index, G12Ci are well-suited for combination therapy. Emerging paradigms include co-targeting upstream, downstream or parallel signaling pathways, as well as cell-cycle or immune checkpoints (Figure 2).
Co-targeting upstream signaling.
Suppressing nucleotide-exchange increases the residency of KRAS(G12C) in its inactive/drug-sensitive state and enhances the therapeutic effect of G12Ci (Figure 1C). Since RTK activation is a key stimulus for exchange, the first invocation of this idea was in experiments demonstrating enhanced antiproliferative effects by co-targeting RTKs (Lito et al., 2016; Patricelli et al., 2016). RTKi directly increase the ability of the G12Ci to engage its target (Patricelli et al., 2016). However, the ‘dominant’ RTK that drives GTP-loading of KRAS varies across models (Lito et al., 2016) and to be effective, this approach requires an a priori knowledge of which RTK to target. Not surprisingly, EGFR has emerged as an early lead and G12Ci combinations with cetuximab or erlotinib are entering clinical testing in patients with colorectal (Amodio et al., 2020) or lung cancer, respectively.
Direct suppression of nucleotide-exchange downstream of multiple RTKs is also effective, as evidenced by using SOS1-specific siRNAs (Lito et al., 2016) or emerging SOS1-selective inhibitors, such as BAY293 or BI1701963 (Hillig et al., 2019; Hofmann et al., 2019). The G12Ci/SOS1 combination is an attractive candidate for clinical testing; yet it remains to be seen if compensatory activity by SOS2, or other RAS-specific GEFs, hinders selective SOS1 inhibition.
An alternative approach is to co-target SHP2, an adaptor phosphatase which, among other functions, has also been implicated in the RTK-dependent activation of SOS1/2 (Fedele et al., 2018). Several studies demonstrate that SHP2i (such as RMC4630 and TNO155) enhance the antiproliferative and antitumor effects of G12Ci (Hallin et al., 2020; Lou et al., 2019; Ryan et al., 2019; Xue et al., 2020). On the basis of these findings clinical trials evaluating the benefit of MRTX849/TNO155 or sotorasib/RMC4630 are imminent.
Co-targeting parallel signaling.
While G12Ci-treatment potently inhibits RAF/MEK/ERK, the effect on PI3K/AKT/mTOR signaling is more subtle. Since the activation of PI3K requires input from RAS and RTKs, both signals may need to be inhibited for complete AKT blockade in KRAS(G12C) mutant cells. Furthermore, models wherein pERK inhibition is coupled with suppressed pS6K/pS6 exhibit a more pronounced antiproliferative effect, as compared to those with pERK inhibition alone (Janes et al., 2018; Misale et al., 2019). S6K is a known substrate of mTOR, which is activated downstream of AKT, but it can also be activated by RSK, downstream of ERK. Despite the unclear mechanism of PI3K, AKT and mTOR activation in KRAS(G12C) mutant cells, the combined inhibition of PI3K or mTOR alongside KRAS(G12C) has more pronounced antitumor effects than either drug alone (Hallin et al., 2020; Misale et al., 2019). Similarly, a three-drug combination targeting KRAS(G12C), IGF1R and mTOR led to potent antitumor effect (Molina-Arcas et al., 2019), supporting RTK/PI3K/AKT/mTOR as a target pathway for combination therapy.
Co-targeting downstream signaling.
An intuitive approach to enhance KRAS(G12C) inhibition is by targeting RAF dimers, MEK or ERK. However, inhibition of ERK output releases the feedback suppression of SOS1/2, which may impede target engagement by the drug. The MEKi trametinib does not significantly enhance the effect of ARS1620 or MRTX849 but its reported to enhance the antitumor effect of sotorasib (Canon et al., 2019). The sotorasib/trametinib combination is now in clinical testing and data from this trial will inform the potential benefit of co-targeting RAF/MEK/ERK alongside KRAS(G12C).
Co-targeting cell-cycle checkpoints.
A key output of KRAS signaling is the activation of the CyclinD:CDK4/6 leading to RB1 hyper-phosphorylation and progression through the G1/S cell-cycle checkpoint. G12Ci-treatment sequesters cells in a quiescent (G0) state, with adapting cells progressing through the G1/S and G2/M checkpoints. Two approaches that enhance the effect of G12Ci by maximizing cell-cycle arrest, include targeting CDK4/6 with palbociclib (Hallin et al., 2020; Lou et al., 2019) or AURKA with alisertib (Xue et al., 2020).
Co-targeting immune checkpoints.
KRAS signaling has an immunosuppressive effect on the tumor microenvironment (Coelho et al., 2017; Pylayeva-Gupta et al., 2012; Ruscetti et al., 2020). KRAS(G12C) inhibition might therefore enhance immune checkpoint inhibition, a hypothesis tested in an immune-competent colorectal cancer model (Canon et al., 2019). Indeed, sotorasib enhanced the expression of inflammatory chemokines leading to enhanced T cells infiltration. When combined with a programmed cell-death protein 1 (PD1) antibody, sotorasib led to a complete and durable antitumor effect. More work is needed to understand how KRAS(G12C) leads to immunosuppression, an effort that will be aided by the generation of immune-competent KRAS(G12C) mutant mice (Li et al., 2018b; Zafra et al., 2019). Perhaps even more informative will be the results from an ongoing clinical trial testing the benefit of sotorasib in combination with PD1 blockade in patients.
Summary and future directions
The last seven years have seen tremendous progress leading to clinically-active inhibitors and a better understanding of how KRAS oncoproteins signal in cancer. We now recognize that KRAS(G12C), one of the most common driver oncoproteins in lung cancer, exists in an excitable state and undergoes nucleotide-cycling between its active and inactive conformations in cancer cells. As a consequence, it is dependent on nucleotide-exchange for activation and susceptible to drugs that block this process. Inactive state-selective inhibitors disrupt this cycle and trap KRAS(G12C) in its GDP-bound state to suppress tumor growth in cancer patients.
How does KRAS(G12C) hydrolyze sufficient GTP to enable cycling in cancer cells? The consensus view from recent reviews suggests that KRAS(G12C) is unique among KRAS oncoproteins, in that it has a high intrinsic GTP hydrolysis rate. An alternative possibility is that GTP-hydrolysis by KRAS mutants is subject to regulation by yet-to-be-determined cellular factors. If true, the latter would suggest that KRAS oncoproteins are broadly susceptible to inactive state-selective inhibition (provided the identification of chemical scaffolds that engage KRAS oncoproteins in a non-cysteine dependent manner).
The duration of KRAS(G12C) inhibition is limited by adaptation, a process that may be responsible for the less than maximal therapeutic effects observed in clinical trials. Other clinically relevant modulators of therapeutic response, including mechanisms of acquired resistance, tissue specific dependencies or the effect of alterations that co-occur alongside KRAS(G12C), are still under investigation. Nevertheless, emerging combination therapies that ensure maximal and durable inhibition of KRAS(G12C) signaling, along with those that enhance the effect of the immune system, are currently being translated into the clinic and are highly likely to enhance the therapeutic benefit of KRAS(G12C) inhibition.
The success of inactive state-selective KRAS(G12C) inhibitors is paving the way for inhibitors with broader therapeutic utility. These include inactive state-selective inhibitors targeting non-G12C mutants, active-state-selective G12C inhibitors as well as compounds that prevent the membrane localization or induce the degradation of KRAS oncoproteins. In theory, inactive state-selective non-G12C inhibitors will likely be susceptible to adaptive mechanisms similar to those described for sotorasib, MRTX849 and ARS1620. Active-state-selective drugs, exemplified by the ‘tri-complex’ KRAS(G12C)-GTP:CYPA inhibitors, have potential advantages. By directly binding to and preventing effector interaction with active KRAS they are predicted to withstand some of the adaptive pressures that limit the effect of inactive state-selective compounds. However, the dependency on CYPA for inhibition may pose its own limitations that need to be addressed experimentally. Lastly, the cellular effects of degraders will depend on which KRAS conformation is targeted by the compound. Early KRAS degraders couple chemical moieties that stimulate degradation with warheads that engage G12C in a covalent manner and bind only to its inactive state.
Whatever their strengths or limitations, emerging KRAS-directed therapies will undoubtedly enrich our understanding of KRAS oncoprotein biology. At the same time, they hold promise in providing much needed therapeutic options for patients with KRAS mutant cancers. In order to maximize the therapeutic potential of these exciting new therapies, preclinical evidence must be coupled with appropriate clinical trial designs, incorporating pharmacodynamic endpoints and evidence-based administration schemes. With optimal integration of basic, translational and clinical research activities, our collective efforts have the potential to bring forth unprecedented success in precision oncology.
Acknowledgments
The authors thank Megan Mroczkowski for discussing the manuscript. P.L. is supported in part by the NIH/NCI (1R01CA23074501, 1R01CA23026701A1, K08CA191082-01A1), the Pew Charitable Trusts, the Damon Runyon Cancer Research Foundation and the American Lung Association. J.Y.X. is supported in part by the NIH/NCI (1F30CA232549-01) and a Medical Scientist Training Program grant to the Weill Cornell-Rockefeller-Sloan Kettering Tri-Institutional MD-PhD Program (T32GM007739).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interests
MSKCC has received research funds from Amgen, Mirati and Revolution Medicines and has confidentiality agreements with these companies. A part of these funds is allocated for research to be conducted under the supervision of P.L. P.L. has not received honoraria, consultation fees, stock options or travel reimbursement from any company. P.L. is listed as an inventor on a patent application filed by MSKCC that describes an approach to treat KRAS mutant cancers.
References
- Amendola CR, Mahaffey JP, Parker SJ, Ahearn IM, Chen WC, Zhou M, Court H, Shi J, Mendoza SL, Morten MJ et al. (2019). KRAS4A directly regulates hexokinase 1. Nature 576, 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, Arena S, Montone M, Mussolin B, Bian Y, et al. (2020). EGFR Blockade Reverts Resistance to KRAS(G12C) Inhibition in Colorectal Cancer. Cancer Discov 10, 1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigenzahn JW, Collu GM, Kartnig F, Pieraks M, Vladimer GI, Heinz LX, Sedlyarov V, Schischlik F, Fauster A, Rebsamen M, et al. (2018). LZTR1 is a regulator of RAS ubiquitination and signaling. Science 362, 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. (2010). Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, and McCormick F (1991). Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature 351, 576–579. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, and Wittinghofer A (2007). GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877. [DOI] [PubMed] [Google Scholar]
- Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM, George SD et al. (2019). Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 25, 628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, Gaida K, Holt T, Knutson CG, Koppada N, et al. (2019). The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223. [DOI] [PubMed] [Google Scholar]
- Choi H, Deng J, Li S, Silk T, Dong L, Brea EJ, Houghton S, Redmond D, Zhong H, Boiarsky J, et al. (2019). Pulsatile MEK Inhibition Improves Anti-tumor Immunity and T Cell Function in Murine Kras Mutant Lung Cancer. Cell Rep 27, 806–819 e805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JG, Fell JB, Hallin J, Baer B, engstrom L, Blake J, Briere D, Ballard J, Burkhard M, Fischer J, et al. (2019). Abstract C069: The identification of MRTX849, a novel KRASG12C inhibitor under clinical investigation, provides insight toward therapeutic susceptibility of KRAS mutant cancers. Molecular Cancer Therapeutics 18, C069–C069. [Google Scholar]
- Coelho MA, de Carne Trecesson S, Rana S Zecchin D, Moore C Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K, et al. (2017). Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity 47, 1083–1099 e1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. (2013). Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, Brown RD, Della Pelle P, Dias-Santagata D, Hung KE, et al. (2012). EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2, 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M, and Stuart DD (2013). Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J (2003). Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3, 11–22. [DOI] [PubMed] [Google Scholar]
- Eblaghie MC, Lunn JS, Dickinson RJ, Munsterberg AE, Sanz-Ezquerro JJ, Farrell ER, Mathers J, Keyse SM, Storey K, and Tickle C (2003). Negative feedback regulation of FGF signaling levels by Pyst1/MKP3 in chick embryos. Curr Biol 13, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Fedele C, Ran H, Diskin B, Wei W, Jen J, Geer MJ, Araki K, Ozerdem U, Simeone DM, Miller G et al. (2018). SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models. Cancer Discov 8, 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell JB, Fischer JP, Baer BR, Blake JF, Bouhana K, Briere DM, Brown KD, Burgess LE, Burns AC, Burkard MR et al. (2020). Identification of the Clinical Development Candidate MRTX849, a Covalent KRAS(G12C) Inhibitor for the Treatment of Cancer. J Med Chem. [DOI] [PubMed] [Google Scholar]
- Gibbs JB, Schaber MD, Allard WJ, Sigal IS, and Scolnick EM (1988). Purification of ras GTPase activating protein from bovine brain. Proc Natl Acad Sci U S A 85, 5026–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan R, Fakih M, Price T, Falchook G, Desai J, Kuo J, Strickler J, Krauss J, Li B, Denlinger C et al. (2019). OA02.02 Phase 1 Study of Safety, Tolerability, PK and Efficacy of AMG 510, a Novel KRASG12C Inhibitor, Evaluated in NSCLC. Journal of Thoracic Oncology 14, S208. [Google Scholar]
- Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, Sudhakar N, Bowcut V, Baer BR, Ballard JA et al. (2020). The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 10, 54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig RC, Sautier B, Schroeder J, Moosmayer D, Hilpmann A, Stegmann CM, Werbeck ND, Briem H, Boemer U, Weiske J, et al. (2019). Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc Natl Acad Sci U S A 116, 2551–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs GA, Der CJ, and Rossman KL (2016). RAS isoforms and mutations in cancer at a glance. J Cell Sci 129, 1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann MH, Gmachl M, Ramharter J, Savarese F, Gerlach D, Marszalek JR, Sanderson MP, Trapani F, Kessler D, Rumpel K, et al. (2019). Abstract PL06-01: Discovery of BI-3406: A potent and selective SOS1::KRAS inhibitor opens a new approach for treating KRAS-driven tumors. Molecular Cancer Therapeutics 18, PL06-01–PL06-01. [Google Scholar]
- Hunter JC, Gurbani D, Ficarro SB, Carrasco MA, Lim SM, Choi HG, Xie T, Marto JA, Chen Z, Gray NS et al. (2014). In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C. Proc Natl Acad Sci U S A 111, 8895–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, and Westover KD (2015). Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol Cancer Res 13, 1325–1335. [DOI] [PubMed] [Google Scholar]
- Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, et al. (2018). Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 172, 578–589 e517. [DOI] [PubMed] [Google Scholar]
- Karoulia Z, Gavathiotis E, and Poulikakos PI (2017). New perspectives for targeting RAF kinase in human cancer. Nat Rev Cancer 17, 676–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr EM, Gaude E, Turrell FK, Frezza C, and Martins CP (2016). Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 531, 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D, Gmachl M, Mantoulidis A, Martin LJ, Zoephel A, Mayer M, Gollner A, Covini D, Fischer S, Gerstberger T, et al. (2019). Drugging an undruggable pocket on KRAS. Proc Natl Acad Sci U S A 116, 15823–15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT et al. (2019). Protective autophagy elicited by RAF-->MEK-->ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med 25, 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopetz S, Grothey A, and Tabernero J (2020). Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. Reply. N Engl J Med 382, 877–878. [DOI] [PubMed] [Google Scholar]
- Lamba S, Russo M, Sun C, Lazzari L, Cancelliere C, Grernrum W, Lieftink C, Bernards R, Di Nicolantonio F, and Bardelli A (2014). RAF suppression synergizes with MEK inhibition in KRAS mutant cancer cells. Cell Rep 8, 1475–1483. [DOI] [PubMed] [Google Scholar]
- Lanman BA, Allen JR, Allen JG, Amegadzie AK, Ashton KS, Booker SK, Chen JJ, Chen N, Frohn MJ, Goodman G et al. (2020). Discovery of a Covalent Inhibitor of KRAS(G12C) (AMG 510) for the Treatment of Solid Tumors. J Med Chem 63, 52–65. [DOI] [PubMed] [Google Scholar]
- Li S, Balmain A, and Counter CM (2018a). A model for RAS mutation patterns in cancers: finding the sweet spot. Nat Rev Cancer 18, 767–777. [DOI] [PubMed] [Google Scholar]
- Li S, Liu S, Deng J, Akbay EA, Hai J, Ambrogio C, Zhang L, Zhou F, Jenkins RW, Adeegbe DO et al. (2018b). Assessing Therapeutic Efficacy of MEK Inhibition in a KRAS(G12C)-Driven Mouse Model of Lung Cancer. Clin Cancer Res 24, 4854–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, Huang A, Wong WL, Callahan MK, Merghoub T et al. (2012). Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22, 668–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Rosen N, and Solit DB (2013). Tumor adaptation and resistance to RAF inhibitors. Nat Med 19, 1401–1409. [DOI] [PubMed] [Google Scholar]
- Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, Saborowski M, Kastenhuber E, Fellmann C, Ohara K, et al. (2014). Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25, 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Solomon M, Li LS, Hansen R, and Rosen N (2016). Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou K, Steri V, Ge AY, Hwang YC, Yogodzinski CH, Shkedi AR, Choi ALM, Mitchell DC, Swaney DL, Hann B et al. (2019). KRAS(G12C) inhibition produces a driver-limited state revealing collateral dependencies. Sci Signal 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, and Barbacid M (2003). RAS oncogenes: the first 30 years. Nat Rev Cancer 3, 459–465. [DOI] [PubMed] [Google Scholar]
- Manchado E, Weissmueller S, Morris J.P.t., Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, et al. (2016). A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534, 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M, Bar-Sagi D, and Kuriyan J (2003). Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695. [DOI] [PubMed] [Google Scholar]
- Misale S, Fatherree JP, Cortez E, Li C, Bilton S, Timonina D, Myers DT, Lee D, Gomez-Caraballo M, Greenberg M et al. (2019). KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin Cancer Res 25, 796–807. [DOI] [PubMed] [Google Scholar]
- Molina-Arcas M, Moore C, Rana S, van Maldegem F, Mugarza E, Romero-Clavijo P, Herbert E, Horswell S, Li LS, Janes MR, et al. (2019). Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Conde C, Ruiz-Llorente S, Dominguez JM, Knauf JA, Viale A, Sherman EJ, Ryder M, Ghossein RA, Rosen N, and Fagin JA (2013). Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov 3, 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R, Schulze C, Bermingham A, Choy T, Cregg J, Kiss G, Marquez A, Reyes D, Saldajeno-Concar M, Weller C et al. (2020). A06 Tri-complex Inhibitors of the Oncogenic, GTP-Bound Form of KRASG12C Overcome RTK-Mediated Escape Mechanisms and Drive Tumor Regressions in Preclinical Models of NSCLC. Journal of Thoracic Oncology 15, S13–S14. [Google Scholar]
- Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, Tzitzilonis C, Mordec K, Marquez A, Romero J, et al. (2018). RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol 20, 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, and Shokat KM (2013). K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, and Shokat KM (2016). Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discov 15, 771–785. [DOI] [PubMed] [Google Scholar]
- Papke B, and Der CJ (2017). Drugging RAS: Know the enemy. Science 355, 1158–1163. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T et al. (2016). Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov 6, 316–329. [DOI] [PubMed] [Google Scholar]
- Peng DH, Kundu ST, Fradette JJ, Diao L, Tong P, Byers LA, Wang J, Canales JR, Villalobos PA, Mino B et al. (2019). ZEB1 suppression sensitizes KRAS mutant cancers to MEK inhibition by an IL17RD-dependent mechanism. Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, and Rosen N (2010). RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin EJ, Bera AK, Lu J, Lin YJ, Strasser SD, Paulo JA, Huang TQ, Morales C, Yan W, Cook J, et al. (2019). Tissue-Specific Oncogenic Activity of KRAS(A146T). Cancer Discov 9, 738–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, and Bernards R (2012). Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, and Bar-Sagi D (2011). RAS oncogenes: weaving a tumorigenic web. Nature reviews Cancer 11, 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, and Bar-Sagi D (2012). Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 21, 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabara D, Tran TH, Dharmaiah S, Stephens RM, McCormick F, Simanshu DK, and Holderfield M (2019). KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis. Proc Natl Acad Sci U S A 116, 22122–22131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, Nafa K, Riedel ER, Hsu M, Pao W et al. (2008). Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 14, 5731–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A, Sanchez-Rivera FJ, et al. (2017). Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 23, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti M, Morris J.P.t., Mezzadra R, Russell J, Leibold J, Romesser PB, Simon J, Kulick A, Ho YJ, Fennell M, et al. (2020). Senescence-Induced Vascular Remodeling Creates Therapeutic Vulnerabilities in Pancreas Cancer. Cell 181, 424–441 e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, Hong CB, and Corcoran RB (2019). Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin Cancer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale MJ, Balmanno K, Saxena J, Ozono E, Wojdyla K, McIntyre RE, Gilley R, Woroniuk A, Howarth KD, Hughes G et al. (2019). MEK1/2 inhibitor withdrawal reverses acquired resistance driven by BRAF(V600E) amplification whereas KRAS(G13D) amplification promotes EMT-chemoresistance. Nat Commun 10, 2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, and Wittinghofer A (1997). The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338. [DOI] [PubMed] [Google Scholar]
- Scheidig AJ, Burmester C, and Goody RS (1999). The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure 7, 1311–1324. [DOI] [PubMed] [Google Scholar]
- Shah NP, Kasap C, Weier C, Baibas M, Nicoll JM, Bleickardt E, Nicaise C, and Sawyers CL (2008). Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell 14, 485–493. [DOI] [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW et al. (2014). KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu DK, Nissley DV, and McCormick F (2017). RAS Proteins and Their Regulators in Human Disease. Cell 170, 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, Haber DA, and Settleman J (2012). TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell 148, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, Neel BG, and Ikura M (2013). NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc Natl Acad Sci U S A 110, 4574–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, et al. (2014). Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep 7, 86–93. [DOI] [PubMed] [Google Scholar]
- Trahey M, and McCormick F (1987). A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238, 542–545. [DOI] [PubMed] [Google Scholar]
- Wey M, Lee J, Jeong SS, Kim J, and Heo J (2013). Kinetic mechanisms of mutation-dependent Harvey Ras activation and their relevance for the development of Costello syndrome. Biochemistry 52, 8465–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GS, Zhou J, Liu JB, Wu Z, Xu X, Li T, Xu D, Schumacher SE, Puschhof J, McFarland J, et al. (2018). Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat Med 24, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue JY, Zhao Y, Aronowitz J, Mai TT, Vides A, Qeriqi B, Kim D, Li C, de Stanchina E, Mazutis L, et al. (2020). Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 577, 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Martelotto L, Baslan T, Vides A, Solomon M, Mai TT, Chaudhary N, Riely GJ, Li BT, Scott Kv et al. (2017). An approach to suppress the evolution of resistance in BRAF(V600E)-mutant cancer. Nat Med 23, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafra MP, Alonso-Curbelo D, Goswami S, Schatoff EM, Han T, Wilkinson JE, and Dow LE (2019). An in vivo KRAS allelic series reveals distinct phenotypes of common oncogenic variants. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Lu J, Li L, Feru F, Quan C, Gero TW, Ficarro SB, Xiong Y, Ambrogio C, Paranal RM et al. (2017). Potent and Selective Covalent Quinazoline Inhibitors of KRAS G12C. Cell Chem Biol 24, 1005–1016 e1003. [DOI] [PubMed] [Google Scholar]