Abstract
Although lower-grade gliomas are driven by mutations in the isocitrate dehydrogenase 1 (IDH1) gene and are less aggressive than primary glioblastoma, they nonetheless generally recur. IDH1 mutant patients are increasingly being treated with temozolomide (TMZ), but early detection of response remains a challenge and there is a need for complementary imaging methods to assess response to therapy prior to tumor shrinkage. The goal of this study was to determine the value of magnetic resonance spectroscopy (MRS)-based metabolic changes for detection of response to TMZ in both genetically engineered and patient-derived mutant IDH1 models. Using 1H MRS in combination with chemometrics identified several metabolic alterations in TMZ-treated cells, including a significant increase in steady-state glutamate levels. This was confirmed in vivo, where the observed 1H MRS increase in glutamate/glutamine occurred prior to tumor shrinkage. Cells labeled with [1-13C]glucose and [3-13C]glutamine, the principal sources of cellular glutamate, showed that flux to glutamate both from glucose via the tricarboxylic acid cycle and from glutamine were increased following TMZ treatment. In line with these results, hyperpolarized [5-13C]glutamate produced from [2-13C]pyruvate and hyperpolarized [1-13C]glutamate produced from [1-13C]α-ketoglutarate were significantly higher in TMZ-treated cells compared to controls. Collectively, our findings identify 1H MRS-detectable elevation of glutamate and hyperpolarized 13C MRS-detectable glutamate production from either pyruvate or α-ketoglutarate as potential translatable metabolic biomarkers of response to TMZ treatment in mutant IDH1 glioma.
Keywords: IDH1 mutation, glioma, temozolomide, metabolism, biomarker
INTRODUCTION
Gliomas are the most commonly occurring primary malignant tumors in the central nervous system with an annual incidence of 17,000 in the United States (1). Historically, glioblastoma (GBM), astrocytoma and oligodendroglioma were classified based on histology. The more recent World Health Organization (WHO) classification also includes molecular parameters, which characterize astrocytoma and oligodendroglioma primarily by mutations in the isocitrate dehydrogenase (IDH, mostly IDH1) gene (2). Primary GBM is IDH wild-type, grade IV and the most aggressive glioma subtype, with a median survival of 15 months (3). In contrast, grade II and III astrocytoma and oligodendroglioma (lower grade glioma or LGG) are IDH mutant and are characterized by a slower growth rate and a longer survival of ~10 years (4). While management of adult LGGs is evolving, the standard of care typically includes maximal surgical resection potentially followed by radiotherapy and chemotherapy with PCV (Procarbazine, CCNU, and Vincristine) or temozolomide (TMZ). The utility of PCV has been demonstrated in clinical trials, and although its benefits compared to TMZ remain under investigation, TMZ is currently more commonly used because of its improved adverse effect/toxicity profile [NCT00887146] (5–8).
Tumor detection, as well as assessment of treatment response, are performed in the clinic using magnetic resonance imaging (MRI) (9,10). Post-Gadolinium (Gd) T1-weighted imaging, T2-weighted imaging, fluid attenuation inversion recovery (FLAIR) imaging, perfusion MRI and diffusion-weighted MRI have all been used to assess tumor size, vascularity, cellularity, and more (10,11). However, imaging studies to detect response to TMZ prior to LGG tumor shrinkage have been limited. In GBM, one study used amide proton transfer imaging and detected response prior to tumor shrinkage in an animal model (12) and a preliminary clinical study reported on the utility of O-(2–18F-fluoroethyl)-L-tyrosine positron emission tomography for early detection of response to TMZ (13). In LGG, two clinical studies investigated the effects of TMZ using 1H magnetic resonance spectroscopy (MRS), a metabolic imaging method that reports on steady state metabolite levels. Both studies showed that choline levels dropped in tumors responsive to treatment, although the drop in choline mostly paralleled tumor shrinkage (14,15).
We have previously used 1H MRS to study mutant IDH1 glioma and showed that in addition to elevated levels of 2-hydroxyglutarate (2-HG), these cells have lower glutamate, lactate and phosphocholine when compared to wild-type IDH1 cells (16). We also investigated mutant IDH1 glioma using another translational metabolic imaging method, namely hyperpolarized 13C MRS, which provides complementary information by probing metabolic fluxes (17,18). Using this method, we showed that mutant IDH1 cells have a lower flux to glutamate from both pyruvate and α-ketoglutarate (α-KG) (19,20). The goal of this study was to determine whether MRS-based metabolic imaging biomarkers could be used to assess the response of mutant IDH1 glioma to TMZ treatment.
We investigated genetically engineered and patient-derived mutant IDH1 cell and animal models. We first investigated the effects of TMZ on the global metabolic profile of our cells using 1H MRS combined with chemometrics, and then probed specific metabolic pathways using thermal equilibrium 13C MRS and hyperpolarized 13C MRS. We found that across our models, response to TMZ treatment was associated with an increase in 1H MRS glutamate levels as well as an increase in glutamate production from glucose and glutamine, resulting in hyperpolarized 13C MRS-detectable elevated flux from both pyruvate and α-KG. Our findings might assist in early detection of tumor response to TMZ and thus help improve personalized treatment strategies for LGG patients.
METHODS
Cell culture and drug treatment
NHAIDHmut and U87IDHmut cells (immortalized normal human astrocytes and glioblastoma engineered to express the IDH1 R132H mutant gene) were maintained in culture as described (16). SF10602 and SF10417, patient-derived mutant IDH1 astrocytoma and oligodendroglioma respectively, courtesy of the Costello Lab (UCSF), were cultured as described (21). BT142 astrocytoma, courtesy of the Weiss lab (University of Calgary) were cultured as described (22,23). All mutant IDH1 lines used were MGMT promoter methylated (personal communications Drs. Pieper (NHAIDHmut, U87IDHmut (24)), Costello (SF10602/SF10417) and Luchman (BT142). Cells were treated with TMZ (Sigma-Aldrich) doses based on literature (25) that were confirmed to result in ~50% reduction in cell number: NHAIDHmut and U87IDHmut-100μM for 3 days; SF10417–400μM for 3 days; SF10602–400μM for 7 days. DMSO was used as solvent control (<0.2%). In parallel, to generate TMZ-resistant cells, NHAIDHmut cells were exposed to increasing concentrations of TMZ (10–400μM) for 3 months. The surviving colonies were selected and established as NHAIDHmut TMZ-resistant cells (NHAIDHmut-TR). For studies investigating the contribution of glucose and glutamine as carbon sources, cells were cultured in media in which glucose or glutamine were replaced in part with [1-13C]glucose (5.5mM/25mM total glucose; Sigma-Aldrich) or [3-13C]glutamine (3mM/6mM total glutamine; Sigma-Aldrich). Authentication of cell lines by short tandem repeat fingerprinting and routine testing for mycoplasma were performed within 6 months of study.
Western blot analysis
Cells were lysed using cell lysis buffer (ThermoFisher Scientific) with 1μl/ml protease inhibitor (Calbiochem) and 10μl/ml phosphatases inhibitor (Sigma-Aldrich). Extracted proteins were quantified using Bradford assay and electrophoresed on 4–20% SDS-PAGE gels and transferred to nitrocellulose membrane. After blocking with 5% fat-free milk, membranes were incubated with primary antibodies (Phospho-H2AX, H2AX and β-actin, Cell Signaling Technology) overnight at 4°C, washed and incubated for 60min with HRP-conjugated secondary antibodies (Cell Signaling Technology). Immunoblots were visualized using Pierce ECL-Western blotting substrate and quantification of bands performed with ImageJ software (Rasband WS, NIH).
Cell extraction, MRS data acquisition and analysis
Cell extracts:
Metabolites were extracted from cells using the dual-phase extraction (16). Lyophilized aqueous phase was resuspended in 400μl D2O and pH adjusted with phosphate buffer. Sodium 3-(trimethylsilyl)propionate-2,2,3,3-d4 (TSP) was used as an internal reference.
MRS data acquisition:
MRS spectra were acquired using a 500MHz Bruker spectrometer equipped with a triple resonance cryoprobe. 1H MRS spectra were recorded using a 90° flip angle, 3s TR, 256 acquisitions and ZGPR water presaturation. For 13C MRS a 30° flip angle, 3s TR and 2048 scans were used.
MRS data analysis:
1H spectra were Fourier transformed, phased and baseline corrected using MNova7.1.0 (Mestrelab). Residual water signal (4.5–5.1ppm) and TSP were excluded from analysis. MATLAB-based iCoshift tool was used to minimize chemical variations of the spectral regions 0.5–4.5ppm and 5.1–10ppm (26). After normalizing to cell number and TSP signal, spectra were scaled with unit variance scaling to give equal weighting to all variables and multivariate analysis was performed using SIMCA15.0 (Umetrics) (27). Principal component analysis (PCA) was applied to identify inherent clustering and distribution of samples between TMZ- and vehicle-treated samples. Partial least squares discriminant analysis (PLS-DA) and orthogonal PLS-DA (OPLS-DA) models were then generated for visualizing the improved class separation. R2 (goodness of fit) and Q2 (goodness of prediction) were used to validate the robustness of the models. Significant metabolites contributing to class discrimination were identified based on S-line correlation plots (|r|≥0.6).
For univariate analysis of 13C spectra and of 1H MRS peaks identified as contributing to class discrimination, spectra were analyzed using Mnova7.1.0. Metabolites of interest were integrated, corrected for saturation and NOE, normalized to TSP and cell number, and samples compared using a Student’s t-test.
Animal studies
Mutant IDH1 mouse models and treatment:
All studies were performed under UCSF Institutional Animal Care and Use Committee approval. Orthotopic tumors were implanted in mice as previously (28–30) by intracranially injecting 3×105 U87IDHmut cells into 6–8 weeks old female athymic nude nu/nu homozygous mice, or 1×105 BT142 and SF10417 cells into 6–8 weeks old female Fox Chase SCID mice (Charles River) or treatment, TMZ (Sigma-Aldrich) was freshly resuspended in ORA-plus (Perrigo) and mice treated once daily per os (p.o.) with either TMZ (5mg/kg, 4ml/kg) or Ora-plus (4ml/kg). Mice were randomly divided into control and TMZ-treated groups when tumor size was comparable and large enough to perform the MRS study: for U87IDHmut xenografts at 0.06±0.03cm3 (33±11 days post-implantation, n=16), for SF10417 xenografts at 0.07±0.04cm3 (257±26 days post-implantation, n=13) and for BT142 at 0.08±0.03cm3 (124±5 days post-implantation, n=19).
In vivo MR studies:
Studies were performed using a vertical wide-bore 14.1T scanner (1000mT/cm gradients, Agilent) equipped with a millipede 40mm 1H coil for T2-weighted anatomical imaging and 1H MRS experiments. Axial T2-weighted images were acquired using 2D spin-echo multi-slice sequence: TE=20ms, TR=1200ms, FOV=30×30mm2, matrix=256×256, slice thickness=1mm, averages=2. Tumor volume was determined using in-house software as the sum of manually contoured tumor areas in each slice and multiplied by slice thickness. In vivo 1H MRS spectra were acquired from a 2×2×2mm3 voxel placed in the center of the tumor avoiding necrotic areas and ascertaining that over 80% of the voxel contains tumor. Spectra were acquired using the Point-RESolved Spectroscopy (PRESS) sequence: TE=20ms, TR=4000ms, averages=512, datapoints=10000, spectral width=10000Hz.
Spectra (0.6–4.2ppm) were analyzed using LCModel (31) with a basis set that includes 2-HG, 4-aminobutyrate, alanine, N-acetylaspartate, N-acetylaspartylglutamate, glutamine, glutamate, glutathione, creatine, serine, citrate, phosphocreatine, choline, phosphocholine, glycerophosphocholine, phosphoethanolamine, aspartate, scyllo-inositol, lactate, taurine, glycine, myo-inositol, following normalization to total signal (29). The average concentration for each metabolite, , and standard deviation for each metabolite, , were calculated using the concentrations for each animal, , and its associated Cramer Rao Lower Bounds (CRLB) determined by LCModel as follows (29,31):
where and , and j corresponds to an individual animal.
Immunohistochemistry:
At the end of MR studies, animals were euthanized, and the brain fixed for 24h in formalin, followed by dehydration and embedding in paraplast plus wax (McCormick Scientific). Immunohistochemistry was performed using Phospho-H2AX and caspase-3 antibodies (Cell signaling).
Ex vivo MRS of tumor extracts:
For SF10417, 10–30mg of frozen tumor were extracted using the dual-phase extraction and 1H MRS spectra recorded as described except that metabolite levels were normalized to total signal.
Hyperpolarized 13C MRS studies
[2-13C]pyruvate and [1-13C]α-KG (Sigma-Aldrich) were polarized using a Hypersense DNP polarizer (Oxford) as previously (19,20). For hyperpolarized [2-13C]pyruvate, metabolism was investigated in live cells encapsulated in agarose beads using an MR-compatible perfusion systems into which 1.5ml of hyperpolarized [2-13C]pyruvate mixed in Tris-based isotonic buffer was injected to a final concentration of 5mM as previously (20). Data was acquired (depending on scanner availability) on either 500MHz Agilent or 600MHz Bruker spectrometer using 5° flip angle and 3s TR over 300s. For hyperpolarized [1-13C]α-KG, metabolism was investigated in live cell suspensions (~5–10×107 in 200μl saline) into which 300μl of hyperpolarized [1-13C]α-KG in isotonic buffer was injected to a final concentration of 15mM (19,32) and data acquired (due to scanner availability) using 60MHz (Oxford) benchtop NMR using a 20° flip angle and 3s TR over 300s. All data were analyzed using MNova and product signals normalized to substrate signal and to cell number.
Spectrophotometric enzyme activity assays
Pyruvate dehydrogenase (PDH) (Abcam), alanine aminotransferase (ALT), glutamate dehydrogenase (GDH) and aspartate transaminase (AST) activities were assessed using commercially available kits (BioVision) following manufacturer instructions with absorbance measured via Spectrophotometer (Tecan).
Statistical analysis
All studies were performed at least 3 times unless otherwise stated. Data is presented as mean±standard deviation and the statistical significance of differences determined using unpaired two-tailed Student’s t-tests (GraphPad Prism) with p-value<0.05 considered significant.
RESULTS
TMZ treatment leads to 1H MRS-detectable changes in metabolite levels
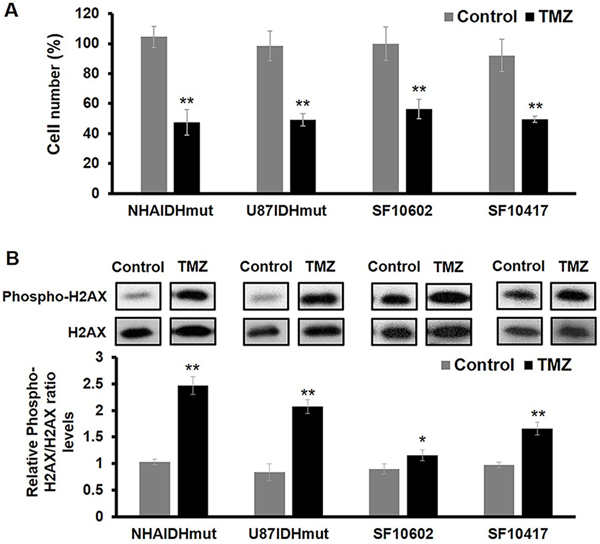
First, we confirmed that treatment of our cells with TMZ doses expected, based on literature (25), to lead to ~50% drop in cell number, had the anticipated effect. As illustrated in Figure 1A, treatment of NHAIDHmut, U87IDHmut, SF10602 and SF10417 cells led to a reduction in cell number by 48% (p-value<0.01), 49% (p-value<0.01), 56% (p-value<0.01) and 50% (p-value<0.01), respectively. In contrast, the resistant NHAIDHmut-TR cells showed no reduction in cell number following treatment with the dose regimen used for NHAIDHmut (97.15±4.22% in control cells and 97.44±3.42% in TMZ-treated cells, p>0.05). In addition, assessment of phospho-H2AX (Fig.1B and S1), an early indicator of TMZ-induced DNA double-strand breaks (DSBs), indicated that TMZ treatment was associated with a significant increase in phospho-H2AX (p-value<0.01 in all cell lines; Fig.1B).
Fig.1:
Cytotoxic effects of TMZ in mutant IDH1 cell lines. (A) Quantification of cell number after TMZ treatment showed ~50% inhibition of cell proliferation. (B) Western blots of Phospho-H2AX and H2AX (top panel) and quantification of Phospho-H2AX normalized to H2AX levels (bottom panel) indicate the DNA damage effect of TMZ. See Fig.S1 for complete blots of all the samples. *p-value<0.05; **p-value<0.01.
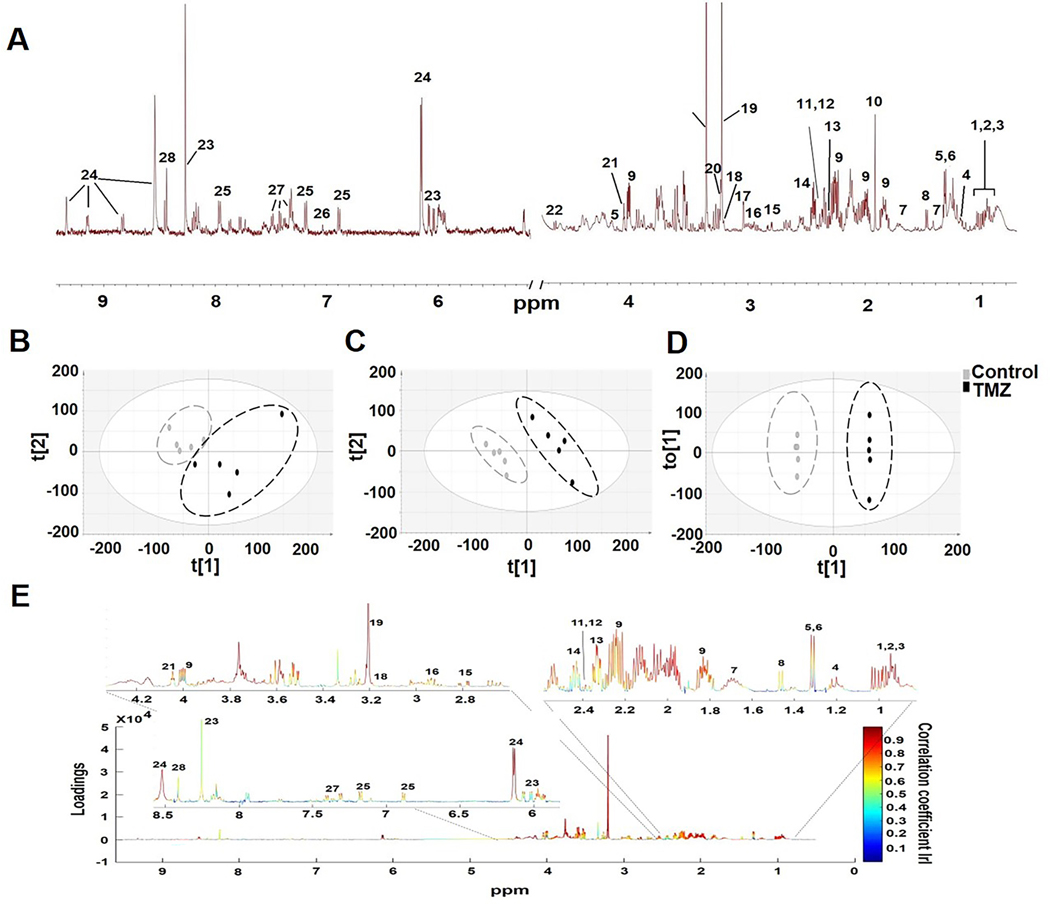
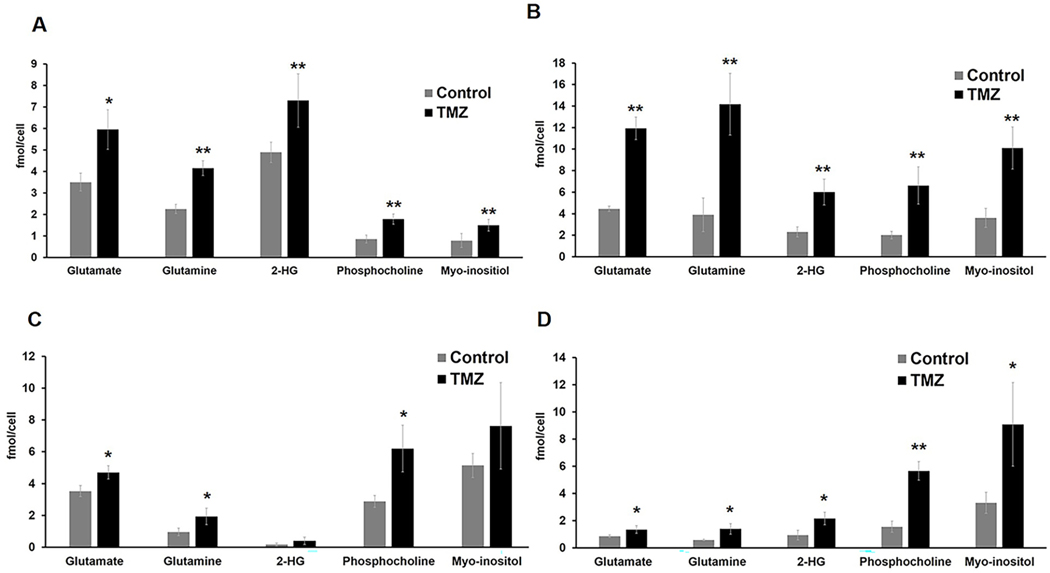
Next, we assessed the 1H MRS data in which twenty-eight metabolites could be identified based on the Human Metabolome Data Base and literature (Fig.2A for NHAIDHmut). Chemometric analysis was then applied to the 1H MRS data. Unsupervised PCA score plots visualized the inherent clustering of control and TMZ-treated groups for both NHAIDHmut (R2X=0.488, Q2=0.0363, n=5 for controls and TMZ; Fig.2B) and U87IDHmut cells (R2X=0.628, Q2=0.256, n=5 for controls and TMZ; Fig.S2A). Improved separation between the groups was obtained by supervised classification using PLS-DA (R2Y=0.998, Q2=0.917; Fig.2C for NHAIDHmut and R2Y=0.997, Q2=0.96; Fig.S2B for U87IDHmut) and OPLS-DA (Fig.2D for NHAIDHmut and Fig.S2C for U87IDHmut). Since supervised statistical models tend to overfit the data, cross-validation of the OPLS-DA model was used to validate the predictability and robustness of the model. High R2 and Q2 values (R2Y=1, Q2=0.764 for NHAIDHmut; R2Y=1, Q2=0.962 for U87IDHmut) confirmed the good predictive ability and accuracy of the model for discrimination between the control and TMZ-treated NHAIDHmut and U87IDHmut cells. We then used the OPLS-DA model to extract the S-line plot. Most significant metabolites contributing to class separation were identified using a correlation coefficient threshold >±0.60 (Fig.2E and Fig.S2D). Finally, univariate analysis was applied to the integral values of the metabolites identified in this manner as being altered with TMZ treatment (Table S1). Most notable were alterations in metabolites that can also be readily detected in vivo namely a significant increase in glutamate, glutamine, 2-HG, phosphocholine and myo-inositol (Fig.3A and 3B). Importantly, when we then investigated our two clinically relevant patient-derived mutant IDH1 cell lines, SF10602 and SF10417, we observed similar significant changes in glutamate, glutamine and phosphocholine following treatment, however SF10602 showed no change in 2-HG and myo-inositol (Fig.3C and 3D). Thus, metabolic alterations associated with TMZ and commonly observed in all of our genetically engineered and patient-derived cell lines were increased glutamate, glutamine and phosphocholine. Importantly, our TMZ-resistant cells, NHAIDHmut-TR, showed no change in glutamate, glutamine or 2-HG, although a significant increase in phosphocholine and myo-inositol was observed (Fig.S3).
Fig.2:
(A) A typical 1H MRS spectrum of NHAIDHmut cell line. Metabolites are identified as follows: 1.Isoleucine, 2.Leucine, 3.Valine, 4.3-hydroxybutyrate, 5.Lactate, 6.Threonine, 7.Lysine, 8.Alanine, 9.2-hydroxyglutarate, 10.Acetate, 11.Pyruvate, 12.Succinate, 13. Glutamate, 14.Glutamine, 15.Aspartate, 16.Glutathione, 17.Creatine, 18.Choline, 19.Phosphocholine, 20.Glycerophosphocholine, 21.Myo-inositol, 22.Glucose, 23.ATP/ADP/AMP, 24.NAD/NADP+, 25.Tyrosine, 26.Histidine, 27.Phenylalanine, 28.Formate. (B) Two-dimensional scatter plots of PCA, (C) PLS-DA and (D) OPLS-DA show discrimination between control and TMZ-treated groups. (E) Color map representing S-line plot identifies metabolites with correlation coefficient values >±0.6 responsible for discriminating TMZ-treated and control cells.
Fig.3:
Quantification of 1H MRS-detected glutamate, glutamine, 2-HG, phosphocholine and myo-inositol levels in (A) NHAIDHmut, (B) U87IDHmut, (C) SF10602 and (D) SF10417 cell lines. *p-value<0.05, **p-value<0.01.
In vivo studies confirm that TMZ treatment increases glutamate/glutamine levels prior to visible tumor shrinkage associated with improved survival
To assess the potential translational value of our findings, we next evaluated the impact of TMZ treatment on 1H MRS-detectable metabolites in vivo. To that end we investigated orthotopically implanted tumors of U87IDHmut and SF10417. In addition, because SF10602 cells do not form tumors in vivo, we investigated a different patient-derived astrocytoma model - BT142 (22,23,30).
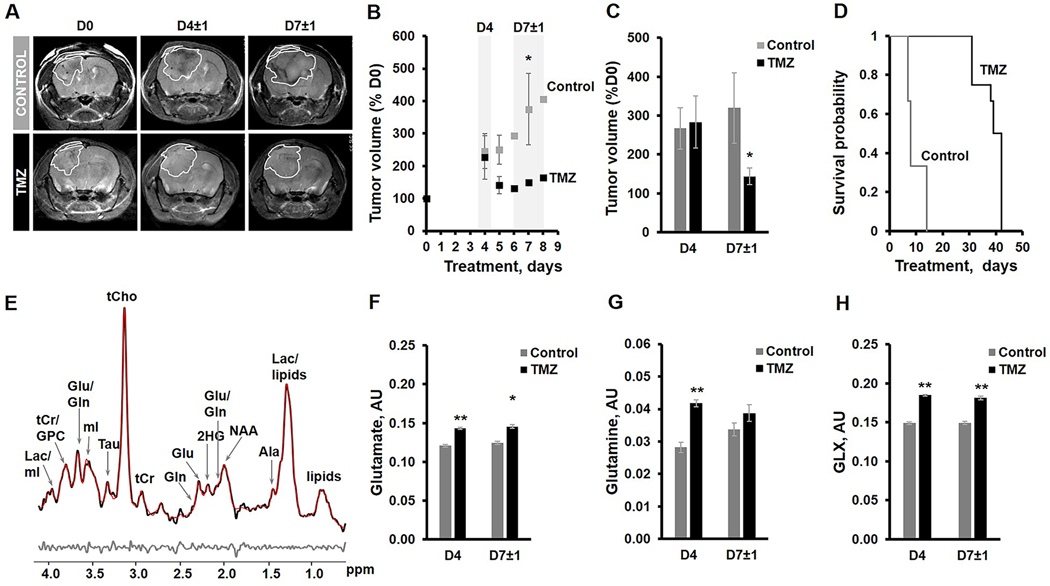
Figure 4A illustrates anatomical T2-weighted images of control and TMZ-treated U87IDHmut tumor-bearing mice at D0, D4±1 and D7±1 of treatment. Temporal evolution of average tumor volume during treatment (Fig.4B) and quantification of average U87IDHmut tumor volume as a percentage of D0 (Fig.4C) demonstrate that there is no difference in tumor volume at D4 (267% of D0, n=4 for control and 283% of D0, n=4 for TMZ-treated, p-value>0.05), but by D7±1 TMZ-treated U87IDHmut tumors were significantly smaller than controls (320%, n=4 for control and 144%, n=3 for TMZ-treated, p-value<0.05; Fig.4C). The Kaplan-Meyer survival plot illustrates that treatment with TMZ led to a significantly longer survival of 38±5 days in the TMZ-treated group compared to 10±4 days in the control (Chi-square value=42.2; p-value<0.01; Fig.4D).
Fig.4:
(A) Anatomical T2-weighted images of control and TMZ-treated U87IDHmut tumor-bearing mice. (B) Temporal evolution of tumor volume presented as a percentage of D0. (C) Quantification of average tumor volume in each treatment group as percentage of D0 value at D4 and D7±1 of treatment. (D) Kaplan-Meyer survival plot. (E) A Representative LCModel fitted 1H MRS spectrum from the 2×2×2mm3 voxel placed inside control U87IDHmut tumor. Raw data-black; LCmodel fit-red; Residual-grey. (F-H) Quantification of 1H MRS-detected metabolites of interest at D4, prior to visible tumor changes, and at D7±1, when tumor shrinkage was observed. Sum of glutamate and glutamine-GLX. *p-value<0.05, **p-value<0.01.
Figure 4E exhibits the in vivo 1H MRS spectrum from the U87IDHmut tumor voxel. Metabolites from spectra acquired at D4, prior to visible differences in tumor size, and at D7±1, when TMZ-treated tumors were significantly smaller, were quantified (Table S2). Consistent with the data commonly observed in our cell lines, significant increases were observed at D4 in glutamate (by 19%; p-value<0.01), glutamine (by 48%; p-value<0.01), and the sum of glutamate and glutamine (GLX by 24%;p-value<0.01) in response to TMZ treatment (Fig.4F-H) (controls: n=3, TMZ-treated: n=4). This effect was slightly reduced by D7±1 but still demonstrated a significant increase in the concentration of glutamate (by 17%; p-value<0.01) and GLX (by 22%; p-value<0.01) although the concentration of glutamine was comparable (p-value>0.05) (controls: n=4, TMZ-treated: n=4) (Fig.4F-H). The concentration of total choline-containing metabolites (tCho) was also significantly increased (by 62%; p-value<0.01) in TMZ-treated U87IDHmut tumors at D4 (controls: n=3, TMZ-treated: n=4), but this effect was reversed by D7±1 demonstrating a significant decrease of tCho (by 34%; p-value<0.01) in response to TMZ treatment (controls: n=3, TMZ-treated: n=4) (Table S2 and S3). Additionally, we also detected a significant decrease in myo-inositol in TMZ-treated tumors at D4 (by 21%; p-value<0.01) and D7±1 (by 46%; p-value<0.01) (Table S2 and S3) and other metabolites were modulated differentially (Table S2 and S3).
Data for the patient-derived BT142 xenografts is illustrated in Figure S4. As in the case of the U87IDHmut, quantification of average tumor volume at D6±1 showed no difference in tumor size following TMZ treatment, 132% of D0 for control and 120% for TMZ-treated (n=9 for both groups, p-value>0.05), but by D15±1 TMZ treatment resulted in a significant difference compared to controls growing to 176% of D0, and treated tumors shrinking to 88% of D0 (n=6 for both groups, p-value<0.01; Fig.S4A-C).
1H MRS spectra and quantification of the metabolite concentrations at D6±1, prior to visible tumor changes, and at D15±1, when tumor shrinkage was observed are shown in Figure S4D-G. Consistent with results from U87IDHmut tumors, at D6±1 BT142 tumors showed significant increases in glutamate by 4% (p-value<0.01), glutamine by 50% (p-value<0.01) and GLX by 17% (p-value<0.01) (controls: n=9, TMZ-treated: n=11) (Fig.S4D-G). By D15±1 a generally smaller but still significant increase in the concentration of glutamate (by 8%; p-value<0.01), glutamine (by 13%; p-value<0.01) and GLX (by 7%; p-value<0.01) was observed (controls: n=6, TMZ-treated: n=6). tCho was significantly reduced in TMZ-treated tumors by 13% (p-value<0.01) at D6±1 and by 27% (p-value<0.01) at D15±1 (Table S4 and S5). We also detected a significant decrease in myo-inositol at D6±1 (by 18%; p-value<0.01) and D15±1 (by 24%; p-value<0.01) and several other metabolites were modulated in different ways (Table S4 and S5).
Finally, we also confirmed our findings in a small scale study of tumor extracts from the patient-derived SF10417 model for which we only acquired data at D7±1 when average tumor volume was comparable in control and TMZ-treated tumors (controls: 132% of D0, n=5; TMZ-treated: 146%, n=4, p-value>0.05) (Fig.S5A-B). Similar to the other in vivo models, TMZ-treated SF10417 tumors showed a significant increase in the concentration of glutamate by 80% (p-value<0.01) and GLX by 58% (p-value<0.01), while the increase in glutamine concentration did not reach significance (p-value>0.05) and tCho levels dropped by 41% (p-value<0.05) (Table S6, Fig.S5C-F).
We confirmed DNA damage in response to TMZ treatment by performing immunohistochemical staining for phospho-H2AX and confirmed cell death by assessing caspase-3 levels. In line with the effects of TMZ on tumor size and animal survival we observed higher levels of phospho-H2AX and caspase-3 in TMZ-treated U87IDHmut (Fig.S6) and BT142 tumors (Fig.S7) when compared to their controls.
Flux from both glucose and glutamine to glutamate is increased in TMZ-treated cells
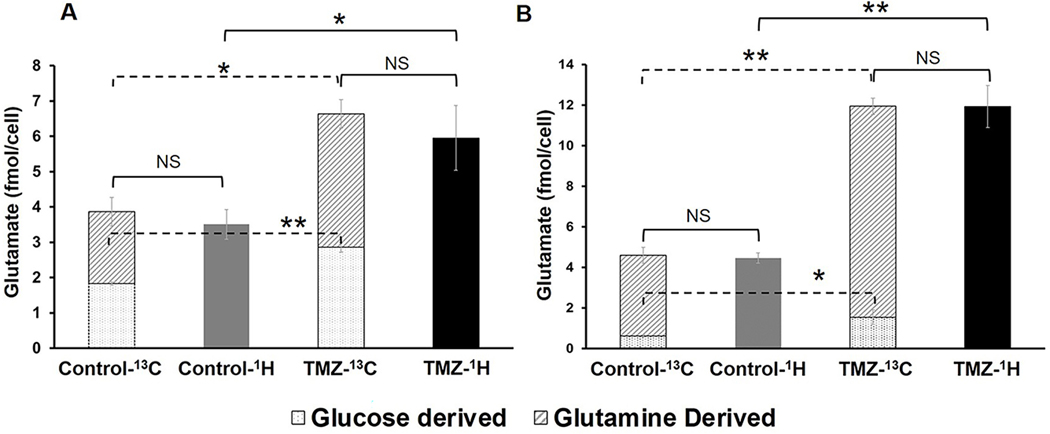
Because our 1H MRS studies identified an increase in glutamate/glutamine, which are in equilibrium with each other, as a consistent early metabolic alteration associated with response to TMZ-treatment, we next wanted to assess the underlying metabolic pathways that contribute to this increase. We used 13C MRS to probe the contributions of [1-13C]glucose and [3-13C]glutamine to glutamate production (20). As illustrated in Figure 1, we observed that the increase in total glutamate levels following TMZ treatment was due to a significant increase in flux from both glucose and glutamine in NHAIDHmut and U87IDHmut cells. In NHAIDHmut cells, we detected an increase in [1-13C]glucose derived-glutamate by 56% (p-value<0.01) while [3-13C]glutamine-derived glutamate increased by 85% in TMZ-treated cells (p-value<0.05; Fig.5A). Similarly, in U87IDHmut cells, glutamate produced from [1-13C]glucose increased by 147% (p-value<0.05) in TMZ-treated cells, while flux from [3-13C]glutamine to glutamate increased by 161% (p-value<0.01; Fig.5B) following TMZ treatment. Within experimental error, these changes fully accounted for the observed increase of the total glutamate pools observed by 1H MRS.
Fig.5:
Comparison of total glutamate pools (as detected by 1H MRS) with [1-13C]glucose and [3-13C]glutamine-derived glutamate (as detected by 13C MRS) in control and TMZ-treated NHAIDHmut (A) and U87IDHmut (B) cell lines. *p-value<0.05, **p-value<0.01, NS-not significant.
Elevated flux to [5-13C]glutamate can be detected in TMZ-treated cells using hyperpolarized [2-13C]pyruvate
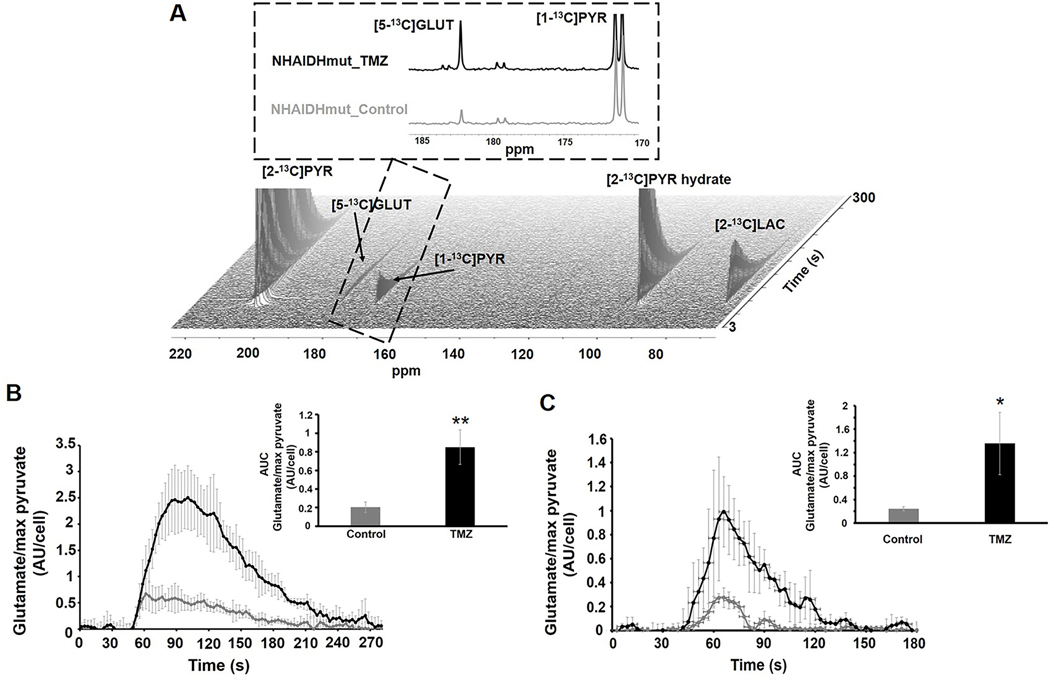
To determine whether hyperpolarized 13C MRS could be used as a complementary method by dynamically assessing glutamate synthesis, we next probed the metabolism of hyperpolarized [2-13C]pyruvate, an intermediate in the metabolic pathways converting glucose to glutamate. Following injection of [2-13C]pyruvate, dynamic production of [5-13C]glutamate was detected in both the NHAIDHmut (Fig.6A and 6B) and U87IDHmut (Fig.6C) cell lines and demonstrated that hyperpolarized [2-13C]pyruvate-derived [5-13C]glutamate was significantly higher in TMZ-treated cells compared to controls. [5-13C]glutamate levels were increased by 318% (p<0.01) in NHAIDHmut (control: n=5, TMZ: n=6; Fig.6B insert) and 453% (p<0.05) in U87IDHmut (control: n=3, TMZ: n=3; Fig.6C insert). In line with this observation, the activity of pyruvate dehydrogenase (PDH), the rate limiting enzyme for pyruvate metabolism to glutamate (20), was significantly increased by TMZ-treatment in both our cell lines (Fig.S8A and S8B). Production of lactate from pyruvate could also be observed, but was not consistent across our two cell lines. In NHAIDHmut cells it increased by 312% (from 0.85±0.22 AU/cell of lactate in control cells to 3.5±0.62 in TMZ-treated, p<0.01) but in U87IDHmut cells the difference in lactate production was not significant (1.13±0.61 AU/cell of lactate in controls and 2.09±1.38 in TMZ-treated, p=0.33).
Fig.6:
(A) Dynamic 13C MRS array showing metabolism of hyperpolarized [2-13C]pyruvate in NHAIDHmut cell line treated with TMZ or DMSO for 72h. Inserts show the sum of all spectra within the dotted region. [2-13C]PYR: [2-13C]pyruvate, [2-13C]PYR hydrate: [2-13C]pyruvate hydrate, [1-13C]PYR: [1-13C]pyruvate, [5-13C]GLUT: [5-13C]glutamate. (B) Temporal evolution and quantification (insert) of hyperpolarized [5-13C]glutamate production in NHAIDHmut cell line. (C) Temporal evolution and quantification (insert) of hyperpolarized [5-13C]glutamate production in U87IDHmut cell line. *p-value<0.05, **p-value<0.01.
Elevated flux to [1-13C]glutamate can be detected in TMZ-treated cells using hyperpolarized [1-13C]α-KG
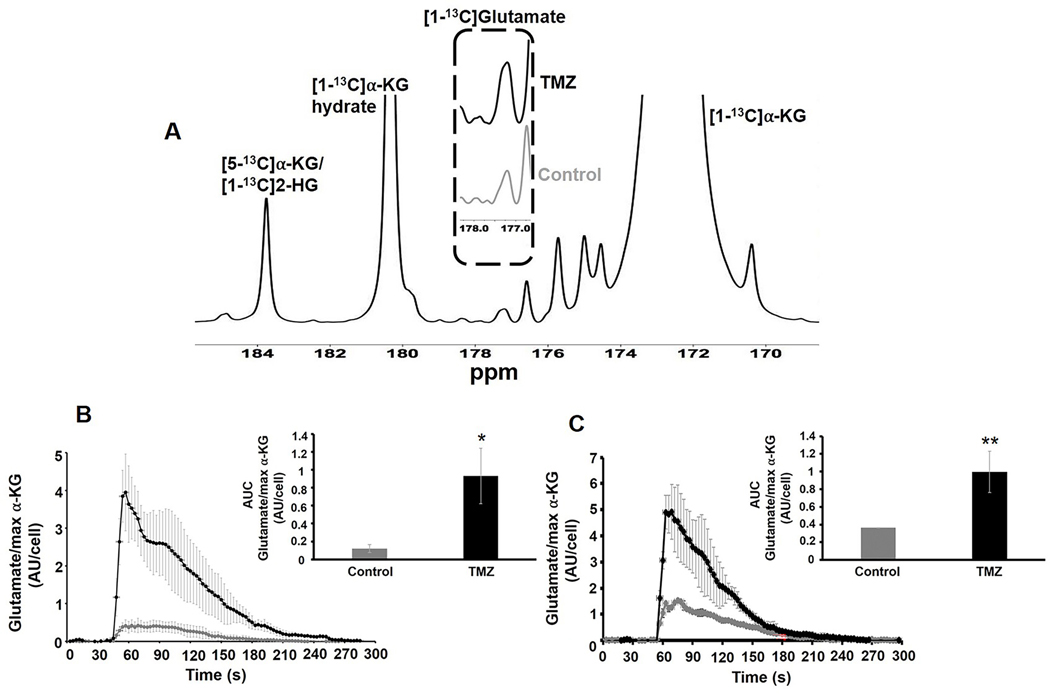
Since glutamate can also be produced from α-KG, downstream of pyruvate, we also probed the fate of hyperpolarized [1-13C]α-KG. As illustrated in Figure 7A build-up of hyperpolarized [1-13C]glutamate could be detected following injection of hyperpolarized [1-13C]α-KG and [1-13C]glutamate accumulation was significantly higher in both NHAIDHmut (p-value<0.05; Fig.7B;) and U87IDHmut cells (p-value<0.01; Fig.7C) following TMZ-treatment. Consistent with this observation the activities of enzymes known to catalyze the α-KG to glutamate conversion, namely aspartate transaminase (AST), glutamate dehydrogenase (GDH) and alanine aminotransferase (ALT) were significantly higher in cells treated with TMZ when compared with controls (Fig.S9A-F).
Fig.7:
(A) Representative sum spectrum of 13C MRS data from live cells showing the metabolism of hyperpolarized [1-13C]α-ketoglutarate. The dotted region compares glutamate production in NHAIDHmut cell line treated with TMZ or DMSO for 72h. [1-13C]αKG: [1-13C]α-ketoglutarate, [1-13C]αKG Hydrate: [1-13C]α-ketoglutarate hydrate. (B) Temporal evolution and quantification (insert) of production of hyperpolarized [1-13C]glutamate in NHAIDHmut cell line. (C) Temporal evolution and quantification (insert) of production of hyperpolarized [1-13C]glutamate in U87IDHmut cell line. *p-value<0.05, **p-value<0.01.
DISCUSSION
Chemotherapeutic treatment, including with TMZ, which had previously been reserved for the treatment of GBM, is increasingly being used in the treatment of mutant IDH1 LGG alone or in combination with other treatments (6,7). When assessing response to treatment, conventional MRI methods provide anatomic information but fail to predict response (33). Furthermore, concurrent radiotherapy and TMZ treatment increase radiographic pseudo-progression in LGG, creating a challenge to the interpretation of images (34). Metabolic imaging provides complementary information and, as such, has the potential to detect tumor response and distinguish pseudo-response or pseudo-progression from true progressive disease (35). Early detection of response to TMZ was shown in GBM models (12,36). In addition, studies have demonstrated the utility of MRS for monitoring response to other treatments including inhibitors of mutant IDH (37), glutaminase (38), HDAC (39) and PI3K/mTOR (29).
To model the behavior of mutant IDH1 gliomas, we initially investigated NHAIDHmut and U87IDHmut cells genetically engineered to express mutant IDH1 (16,20,29). NHAIDHmut cells provide a good model for mutant IDH1 but do not reproduce the full set of oncogenic events that occur in mutant IDH1 patients (40). U87IDHmut cells were engineered using a GBM cell line, and as such are likely to harbor oncogenic events that are not typically observed in LGG. There are further potential limitations to these cells, as illustrated by the fact that they still express lactate dehydrogenase A (LDHA), in contrast to most LGG (41). We therefore further confirmed our findings by expanding our investigations to patient-derived mutant IDH1 models derived from both oligodendroglioma and astrocytoma. Models derived from patients are also limited by the fact that patients have likely upgraded and/or undergone treatment. Indeed, one of our models, BT142 was generated from an IDH1-mutant astrocytoma which retained its endogenous mutant IDH1 and aggressive tumor-initiating capacity but lost its wild-type allele, and therefore no longer produces detectable 2-HG levels (22,23). Importantly, the loss of either wild-type or mutant IDH1 allele was reported in over 10% of mutant IDH1 glioma upon recurrence (42). Collectively therefore, our investigations in multiple different models lends support to the fact that our common MRS-detectable metabolic alterations are likely to be clinically useful for non-invasive assessment of early response to TMZ treatment in LGG. Of note all our models were MGMT promoter methylated consistent with data showing an association between MGMT status and TMZ sensitivity and survival in patients (43).
Using 1H MRS, we observed alterations in several metabolites. However, the metabolic changes observed in choline, 2-HG and myo-inositol were not consistent across our cell lines and in vivo models. We observed elevated levels of phosphocholine in TMZ treated cells similar to one study in TMZ-treated GBM cells (44). However, this finding was not reproduced in our patient-derived tumor studies: at the early time point tCho increased in U87IDHmut but decreased in BT142 and SF10417 LGG tumors. When tumor shrinkage was visible, all models showed a decrease in tCho. Interestingly, a drop in tCho was also reported in clinical studies of LGG following temozolomide therapy, but this drop occurred in parallel to a decrease in tumor volume, possibly limiting the value of tCho as a predictor of response (14,15). Similarly, the increase in 2-HG observed in our cells was not reproduced in the patient-derived tumors. Finally, although myo-inositol was reported to be elevated in LGG (45) and a predictor of overall survival in patients with recurrent glioblastoma treated with the anti-angiogenic Bevacizumab (46), the increase in myo-inositol observed in our cell studies was not observed in any of the tumor models.
In contrast, however, we found that in all of our cell lines as well as our in vivo orthotopic tumor models, TMZ treatment is associated with a significant increase in glutamate/glutamine and the composite GLX peak. Furthermore, the increase in glutamate/glutamine and GLX occurred prior to any MRI-detectable change in tumor size when comparing control and TMZ-treated tumors. The in vivo finding that TMZ treatment reduced LGG tumor size and enhanced survival is consistent with clinical observations (14,15) and in line with a previous BT142 animal study (30). Importantly, the increase in GLX is likely specifically associated with response to TMZ in mutant IDH1 tumors. This is supported by our findings in TMZ-resistant NHAIDHmut-TR cells that showed no alteration in glutamate or glutamine (Fig.S3). It is also supported by our previous work in which we used MRS to study the effect of TMZ in wild-type IDH GS-2 and U87 GBM tumors, and found that response was associated with a drop in 13C MRS-detectable changes pyruvate to lactate conversion but no changes in 1H MRS-detectable GLX ((30); values in GS-2 GBM tumors were 0.18±0.02 in TMZ-treated versus 0.15±0.07 in controls; p-value>0.1).
1H MRS is clinically translatable and its value for investigating brain tumors has been established (9,35). The relatively small changes observed in this study in glutamate/glutamine/GLX, and the fact that our studies used a relatively high field strength point to a potential limitation for future clinical implementation. However, previous investigations have demonstrated the feasibility of detecting relatively small changes in brain metabolites (45,47). Furthermore, optimized 1H MRS sequences that help resolve the potential overlap of 2-HG, glutamate, glutamine and GABA in patients with mutant IDH1 tumors have been developed (48), pointing to the potential utility of glutamate/glutamine/GLX to assess the early effects of TMZ in the clinic.
To understand the underlying mechanism for the elevated levels of glutamate, and to assess the potential utility of hyperpolarized 13C MRS as a complementary method for probing response to TMZ, we investigated flux from glucose and glutamine to glutamate using 13C MRS, as well as flux from pyruvate and α-KG to glutamate using hyperpolarized 13C MRS. We found that the increase in glutamate was mediated by a greater flux from both glucose and glutamine. We also confirmed that the elevated flux from glucose via the TCA cycle to glutamate could be detected using hyperpolarized pyruvate and hyperpolarized α-KG. These metabolic alterations are mediated, at least in part, by an increase in PDH activity, which controls pyruvate entry into the TCA cycle as well as an increase in the activities of ALT, GDH and AST, which control the flux of α-KG to glutamate. In GBM, TMZ was shown to affect the expression of pyruvate kinase (PKM2) (49), and in mutant IDH1 tumors, various mechanisms of TMZ-induced metabolic stress were described (25). However, further studies are needed to determine how TMZ affects the activities of PDH, ALT, GDH and AST to fully explain our MRS-detectable observations. In addition, since the increase in glutamate levels observed in our study reflects intracellular levels, further studies are also needed to assess the potential impact of TMZ on the glutamate/glutamine cycle and on glutamate receptors (50).
Previous studies in glioblastoma have shown that a drop in hyperpolarized [1-13C]pyruvate to [1-13C]lactate conversion (catalyzed by LDHA), can serve as a predictive biomarker of tumor response to TMZ (36,49). However, in the case of LGG, our group has found that hyperpolarized [1-13C]lactate production was not significantly altered in response to TMZ treatment in BT142 tumor-bearing mice (30), in line with the mutant IDH1-mediated promoter hypermethylation and downregulation of LDHA expression demonstrated in mutant IDH1 tumors (41). Furthermore, as shown here with [2-13C]pyruvate, even in our genetically engineered cells that do express LDHA, TMZ did not affect hyperpolarized lactate production consistently in our two cell lines, confirming the limited utility of hyperpolarized lactate production as a biomarker of response to TMZ in mutant IDH1 cells.
In contrast, we show here that TMZ treatment is associated with an increase in hyperpolarized 13C MRS metabolism of [2-13C]pyruvate and [1-13C]α-KG to glutamate. Our findings thus identify an alternate approach, and point to the potential utility of [2-13C]pyruvate and [1-13C]α-KG as probes for assessing response to TMZ in LGG. Importantly, the in vivo implementation of hyperpolarized [1-13C]α-KG has been shown in rats (32) and multiple studies have now demonstrated the translational potential of hyperpolarized 13C MRS for non-invasive monitoring of metabolic fluxes (17). In the case of human brain studies, the utility of hyperpolarized [1-13C]pyruvate was shown in GBM patients (36) and more recently the implementation of hyperpolarized [2-13C]pyruvate was demonstrated in normal volunteers (18).
In summary, our findings demonstrate that 1H MRS-detectable glutamate/glutamine/ GLX can be used as early biomarkers of TMZ response in mutant IDH1 models, and 13C MRS-detectable production of hyperpolarized [1-13C]glutamate and [5-13C]glutamate from hyperpolarized [1-13C]α-KG and [2-13C]pyruvate, respectively, have potential as complementary approaches to monitor response. Collectively, these metabolic imaging biomarkers could help improve currently available imaging methods and provide an early indication of response to TMZ treatment in lower-grade mutant IDH1 glioma.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Findings show that glutamate can be used as a non-invasive, imageable metabolic marker for early assessment of tumor response to temozolomide, with the potential to improve treatment strategies for mutant IDH1 patients.
ACKNOWLEDGEMENTS
This work was supported by NIH R01CA172845, NIH R01CA1972254 and UCSF Brain Tumor Center Loglio Collective. The authors also acknowledge use of the P41EB013598 NIH-supported center.
Footnotes
Competing financial interests: Authors have no potential conflict of interest.
REFERENCES
- 1.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro-oncology 2019;21:v1–v100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nature medicine 2011;17:291–3 [DOI] [PubMed] [Google Scholar]
- 3.Thakkar JP, Dolecek TA, Horbinski C, Ostrom QT, Lightner DD, Barnholtz-Sloan JS, et al. Epidemiologic and molecular prognostic review of glioblastoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2014;23:1985–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine 2015;372:2481–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tom MC, Cahill DP, Buckner JC, Dietrich J, Parsons MW, Yu JS. Management for Different Glioma Subtypes: Are All Low-Grade Gliomas Created Equal? Am Soc Clin Oncol Educ Book 2019;39:133–45 [DOI] [PubMed] [Google Scholar]
- 6.Oberheim Bush NA, Chang S. Treatment Strategies for Low-Grade Glioma in Adults. J Oncol Pract 2016;12:1235–41 [DOI] [PubMed] [Google Scholar]
- 7.Darlix A, Mandonnet E, Freyschlag CF, Pinggera D, Forster MT, Voss M, et al. Chemotherapy and diffuse low-grade gliomas: a survey within the European Low-Grade Glioma Network. Neurooncol Pract 2019;6:264–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hafazalla K, Sahgal A, Jaja B, Perry JR, Das S. Procarbazine, CCNU and vincristine (PCV) versus temozolomide chemotherapy for patients with low-grade glioma: a systematic review. Oncotarget 2018;9:33623–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson SJ. Assessment of therapeutic response and treatment planning for brain tumors using metabolic and physiological MRI. NMR in biomedicine 2011;24:734–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Usinskiene J, Ulyte A, Bjornerud A, Venius J, Katsaros VK, Rynkeviciene R, et al. Optimal differentiation of high- and low-grade glioma and metastasis: a meta-analysis of perfusion, diffusion, and spectroscopy metrics. Neuroradiology 2016;58:339–50 [DOI] [PubMed] [Google Scholar]
- 11.van den Bent MJ, Wefel JS, Schiff D, Taphoorn MJ, Jaeckle K, Junck L, et al. Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. The Lancet Oncology 2011;12:583–93 [DOI] [PubMed] [Google Scholar]
- 12.Sagiyama K, Mashimo T, Togao O, Vemireddy V, Hatanpaa KJ, Maher EA, et al. In vivo chemical exchange saturation transfer imaging allows early detection of a therapeutic response in glioblastoma. Proceedings of the National Academy of Sciences of the United States of America 2014;111:4542–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceccon G, Lohmann P, Tscherpel C, Dunkl V, Rapp M, Stoffels G, et al. NIMG-79. Early treatment response assessment using o-(2-(18)f-fluoroethyl)-l-tyrosine (FET) PET compared to MRI in malignant gliomas treated with adjuvant temozolomide chemotherapy. Neuro-oncology 2018;20:vi193-vi [Google Scholar]
- 14.Murphy PS, Viviers L, Abson C, Rowland IJ, Brada M, Leach MO, et al. Monitoring temozolomide treatment of low-grade glioma with proton magnetic resonance spectroscopy. Br J Cancer 2004;90:781–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillevin R, Menuel C, Taillibert S, Capelle L, Costalat R, Abud L, et al. Predicting the outcome of grade II glioma treated with temozolomide using proton magnetic resonance spectroscopy. Br J Cancer 2011;104:1854–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izquierdo-Garcia JL, Viswanath P, Eriksson P, Chaumeil MM, Pieper RO, Phillips JJ, et al. Metabolic reprogramming in mutant IDH1 glioma cells. PloS one 2015;10:e0118781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, Bankson JA, Brindle K, Cunningham CH, et al. Hyperpolarized (13)C MRI: Path to Clinical Translation in Oncology. Neoplasia (New York, NY) 2019;21:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung BT, Chen HY, Gordon J, Mammoli D, Sriram R, Autry AW, et al. First hyperpolarized [2-(13)C]pyruvate MR studies of human brain metabolism. Journal of magnetic resonance (San Diego, Calif : 1997) 2019;309:106617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaumeil MM, Larson PE, Yoshihara HA, Danforth OM, Vigneron DB, Nelson SJ, et al. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat Commun 2013;4:2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izquierdo-Garcia JL, Viswanath P, Eriksson P, Cai L, Radoul M, Chaumeil MM, et al. IDH1 Mutation Induces Reprogramming of Pyruvate Metabolism. Cancer research 2015;75:2999–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mancini A, Xavier-Magalhaes A, Woods WS, Nguyen KT, Amen AM, Hayes JL, et al. Disruption of the beta1L Isoform of GABP Reverses Glioblastoma Replicative Immortality in a TERT Promoter Mutation-Dependent Manner. Cancer cell 2018;34:513–28.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luchman HA, Stechishin OD, Dang NH, Blough MD, Chesnelong C, Kelly JJ, et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro-oncology 2012;14:184–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luchman HA, Chesnelong C, Cairncross JG, Weiss S. Spontaneous loss of heterozygosity leading to homozygous R132H in a patient-derived IDH1 mutant cell line. Neuro-oncology 2013;15:979–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oldrini B, Vaquero-Siguero N, Mu Q, Kroon P, Zhang Y, Galán-Ganga M, et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. bioRxiv 2020:2020.03.10.985226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tateishi K, Higuchi F, Miller JJ, Koerner MVA, Lelic N, Shankar GM, et al. The Alkylating Chemotherapeutic Temozolomide Induces Metabolic Stress in IDH1-Mutant Cancers and Potentiates NAD(+) Depletion-Mediated Cytotoxicity. Cancer research 2017;77:4102–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savorani F, Tomasi G, Engelsen SB. icoshift: A versatile tool for the rapid alignment of 1D NMR spectra. Journal of magnetic resonance (San Diego, Calif : 1997) 2010;202:190–202 [DOI] [PubMed] [Google Scholar]
- 27.Subramani E, Jothiramajayam M, Dutta M, Chakravorty D, Joshi M, Srivastava S, et al. NMR-based metabonomics for understanding the influence of dormant female genital tuberculosis on metabolism of the human endometrium. Human reproduction (Oxford, England) 2016;31:854–65 [DOI] [PubMed] [Google Scholar]
- 28.Radoul M, Chaumeil MM, Eriksson P, Wang AS, Phillips JJ, Ronen SM. MR Studies of Glioblastoma Models Treated with Dual PI3K/mTOR Inhibitor and Temozolomide:Metabolic Changes Are Associated with Enhanced Survival. Molecular cancer therapeutics 2016;15:1113–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batsios G, Viswanath P, Subramani E, Najac C, Gillespie AM, Santos RD, et al. PI3K/mTOR inhibition of IDH1 mutant glioma leads to reduced 2HG production that is associated with increased survival. Scientific reports 2019;9:10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaumeil MM, Radoul M, Najac C, Eriksson P, Viswanath P, Blough MD, et al. Hyperpolarized (13)C MR imaging detects no lactate production in mutant IDH1 gliomas: Implications for diagnosis and response monitoring. NeuroImage Clinical 2016;12:180–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magnetic resonance in medicine 1993;30:672–9 [DOI] [PubMed] [Google Scholar]
- 32.Chaumeil MM, Larson PE, Woods SM, Cai L, Eriksson P, Robinson AE, et al. Hyperpolarized [1–13C] glutamate: a metabolic imaging biomarker of IDH1 mutational status in glioma. Cancer research 2014;74:4247–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalpathy-Cramer J, Gerstner ER, Emblem KE, Andronesi O, Rosen B. Advanced magnetic resonance imaging of the physical processes in human glioblastoma. Cancer research 2014;74:4622–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dworkin M, Mehan W, Niemierko A, Kamran SC, Lamba N, Dietrich J, et al. Increase of pseudoprogression and other treatment related effects in low-grade glioma patients treated with proton radiation and temozolomide. Journal of neuro-oncology 2019;142:69–77 [DOI] [PubMed] [Google Scholar]
- 35.Shim H, Holder CA, Olson JJ. Magnetic resonance spectroscopic imaging in the era of pseudoprogression and pseudoresponse in glioblastoma patient management. CNS Oncol 2013;2:393–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park I, Bok R, Ozawa T, Phillips JJ, James CD, Vigneron DB, et al. Detection of early response to temozolomide treatment in brain tumors using hyperpolarized 13C MR metabolic imaging. Journal of magnetic resonance imaging : JMRI 2011;33:1284–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molloy AR, Najac C, Viswanath P, Lakhani A, Subramani E, Batsios G, et al. MR-detectable metabolic biomarkers of response to mutant IDH inhibition in low-grade glioma. Theranostics 2020;10:8757–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zacharias NM, Baran N, Shanmugavelandy SS, Lee J, Lujan JV, Dutta P, et al. Assessing Metabolic Intervention with a Glutaminase Inhibitor in Real-Time by Hyperpolarized Magnetic Resonance in Acute Myeloid Leukemia. Molecular cancer therapeutics 2019;18:1937–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radoul M, Najac C, Viswanath P, Mukherjee J, Kelly M, Gillespie AM, et al. HDAC inhibition in glioblastoma monitored by hyperpolarized (13) C MRSI. NMR in biomedicine 2019;32:e4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johannessen TA, Mukherjee J, Viswanath P, Ohba S, Ronen SM, Bjerkvig R, et al. Rapid Conversion of Mutant IDH1 from Driver to Passenger in a Model of Human Gliomagenesis. Mol Cancer Res 2016;14:976–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chesnelong C, Chaumeil MM, Blough MD, Al-Najjar M, Stechishin OD, Chan JA, et al. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro-oncology 2014;16:686–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazor T, Chesnelong C, Pankov A, Jalbert LE, Hong C, Hayes J, et al. Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proceedings of the National Academy of Sciences of the United States of America 2017;114:10743–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandith AA, Qasim I, Zahoor W, Shah P, Bhat AR, Sanadhya D, et al. Concordant association validates MGMT methylation and protein expression as favorable prognostic factors in glioma patients on alkylating chemotherapy (Temozolomide). Scientific reports 2018;8:6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.St-Coeur PD, Poitras JJ, Cuperlovic-Culf M, Touaibia M, Morin P, Jr. Investigating a signature of temozolomide resistance in GBM cell lines using metabolomics. Journal of neuro-oncology 2015;125:91–102 [DOI] [PubMed] [Google Scholar]
- 45.Bartha R, Megyesi JF, Watling CJ. Low-grade glioma: correlation of short echo time 1H-MR spectroscopy with 23Na MR imaging. AJNR Am J Neuroradiol 2008;29:464–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steidl E, Pilatus U, Hattingen E, Steinbach JP, Zanella F, Ronellenfitsch MW, et al. Myoinositol as a Biomarker in Recurrent Glioblastoma Treated with Bevacizumab: A 1H-Magnetic Resonance Spectroscopy Study. PloS one 2016;11:e0168113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chronaiou I, Stensjøen AL, Sjøbakk TE, Esmaeili M, Bathen TF. Impacts of MR spectroscopic imaging on glioma patient management. Acta Oncol 2014;53:580–9 [DOI] [PubMed] [Google Scholar]
- 48.Ganji SK, An Z, Tiwari V, McNeil S, Pinho MC, Pan E, et al. In vivo detection of 2-hydroxyglutarate in brain tumors by optimized point-resolved spectroscopy (PRESS) at 7T. Magnetic resonance in medicine 2017;77:936–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park I, Mukherjee J, Ito M, Chaumeil MM, Jalbert LE, Gaensler K, et al. Changes in pyruvate metabolism detected by magnetic resonance imaging are linked to DNA damage and serve as a sensor of temozolomide response in glioblastoma cells. Cancer research 2014;74:7115–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pereira MSL, Klamt F, Thomé CC, Worm PV, de Oliveira DL. Metabotropic glutamate receptors as a new therapeutic target for malignant gliomas. Oncotarget 2017;8:22279–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.