SUMMARY
Cell differentiation and function are regulated across multiple layers of gene regulation, including modulation of gene expression by changes in chromatin accessibility. However, differentiation is an asynchronous process precluding a temporal understanding of regulatory events leading to cell fate commitment. Here we developed simultaneous high-throughput ATAC and RNA expression with sequencing (SHARE-seq), a highly scalable approach for measurement of chromatin accessibility and gene expression in the same single cell, applicable to different tissues. Using 34,774 joint profiles from mouse skin, we develop a computational strategy to identify cis-regulatory interactions and define domains of regulatory chromatin (DORCs) that significantly overlap with super-enhancers. During lineage commitment, chromatin accessibility at DORCs precedes gene expression, suggesting that changes in chromatin accessibility may prime cells for lineage commitment. We computationally infer chromatin potential as a quantitative measure of chromatin lineage priming and use it to predict cell fate outcomes. SHARE-seq is an extensible platform to study regulatory circuitry across diverse cells in tissues.
In Brief
SHARE-seq, a highly scalable approach to measure chromatin accessibility and gene expression in the same single cell, links distal regulatory elements to key lineage-specifying genes and finds that chromatin accessibility foreshadows gene expression.
Graphical Abstract

INTRODUCTION
Regulation of chromatin structure and gene expression underlies key developmental transitions in cell lineages (Novershtern et al., 2011; Shema et al., 2019; Spitz and Furlong, 2012). In recent years, genome-wide profiling of gene expression and chromatin has helped uncover mechanisms of chromatin change at key points of multi-lineage cell fate decisions (Shema et al., 2019; Spitz and Furlong, 2012). Prior studies comparing profiles of purified populations have observed that changes in histone modifications and binding of lineage-associated transcription factors (TFs) may precede and foreshadow changes in gene expression, creating poised or primed chromatin states that bias genes for activation or repression to alter lineage outcomes (Bernstein et al., 2006; Lara-Astiaso et al., 2014; Rada-Iglesias et al., 2011). For example, deposition of the histone modification H3K4me1 has been shown to prime regulatory elements, biasing cells for differentiation (Lara-Astiaso et al., 2014; Rada-Iglesias et al., 2011) or immune cell activation (Heinz et al., 2010; Ostuni et al., 2013). However, approaches to analyze primed chromatin states rely on bulk measurements of histone modifications, largely restricting analysis to well-defined chromatin states and synchronous cell culture models or stem cell systems with well-defined markers for fluorescence-activated cell sorting (FACS) isolation. We therefore reasoned that an experimental approach to measure chromatin accessibility and gene expression in the same single cell may enable identification of primed versus active accessible chromatin, providing a means to identify new mechanisms of chromatin-mediated lineage-priming in new cellular contexts at single-cell resolution.
Methods of combining measurements of different layers of gene regulation in single cells may serve to determine regulators of cell differentiation and can function as sensitive markers of cell identity and cell potential (Kelsey et al., 2017; Shema et al., 2019). Computational methods (Rusk, 2019) have had some success in integrating single-cell epigenome, transcriptome, and protein measurements profiled separately; however, these methods assume that these distinct measurements reflect a common cell identity and may not correctly recover changes unique to one layer, such as chromatin accessibility-mediated lineage priming. Emerging single-cell “multi-omics” technologies offer a direct means to determine the coordination between layers of gene regulation. Prior studies have sought to correlate gene expression with regulatory element accessibility (Cao et al., 2018; Chen et al., 2019; Zhu et al., 2019). However, these approaches had limited throughput or sensitivity, restricting their ability to recover fine but biologically important differences between chromatin accessibility and gene expression.
Here we investigate the dynamics of the epigenomic and transcriptomic basis of cellular differentiation by developing simultaneous high-throughput ATAC (Buenrostro et al., 2013) and RNA expression with sequencing (SHARE-seq) for individual or joint measures of single-cell chromatin accessibility and gene expression at low cost and on a massive scale. Using SHARE-seq, we profiled 84,426 cells across 4 different cell lines and 3 tissue types, including mouse lung, brain, and skin. Applying SHARE-seq to mouse skin shows that cell type definitions are correlated between chromatin accessibility and gene expression, with notable exceptions including cell cycle genes. We leverage the heterogeneity across single cells to infer chromatin-expression relationships and identify 63,110 peak-gene associations in adult mouse skin. High-density peak-gene-associated regions, to which we refer as domains of regulatory chromatin (DORCs), are enriched for lineage-determining genes and overlap with known super-enhancers. Strikingly, during hair follicle differentiation, chromatin at DORC-regulated genes becomes accessible before induction of the corresponding gene’s expression, identifying a role of chromatin accessibility in priming active chromatin states. Building on this finding, we develop an analytical framework, called “chromatin potential,” to infer cell fate choices de novo. We describe an experimental and analytical basis for integrated measurements of the epigenome and transcriptome, opening new avenues to uncover principles of gene regulation and cell fate specification across single cells in diverse systems.
RESULTS
SHARE-Seq for Joint Profiling of Chromatin Accessibility and Gene Expression at Scale
To create a chromatin accessibility and mRNA expression co-profiling approach that is scalable and sensitive, we built on SPLiT-seq and Paired-seq (Rosenberg et al., 2018; Zhu et al., 2019) to develop SHARE-seq, which uses multiple rounds of hybridization blocking to uniquely and simultaneously label mRNA and chromatin fragments in the same single cell (Figure 1A; Figures S1A and S1B; STAR Methods). Briefly, in SHARE-seq, (1) fixed and permeabilized cells or nuclei are transposed by Tn5 transposase to mark regions of open chromatin; (2) mRNA is reverse transcribed using a poly(T) primer containing a unique molecular identifier (UMI) and a biotin tag; (3) permeabilized cells or nuclei are distributed in a 96-well plate to hybridize well-specific barcoded oligonucleotides to transposed chromatin fragments and poly(T) cDNA; (4) hybridization is repeated three times, expanding the barcoding space to approximately 106 (963) barcode combinations (Figure S1B; Table S1), and, following hybridization, cell barcodes are ligated simultaneously to cDNA and chromatin fragments; (5) reverse crosslinking is performed to release barcoded molecules; (6) cDNA is specifically separated from chromatin using streptavidin beads, and each library is prepared for sequencing; and (7) paired profiles are identified using the common combination of well-specific barcodes (Figure S1A). This barcoding strategy may be extended to even larger experiments by using additional rounds of hybridization (Figure S1B).
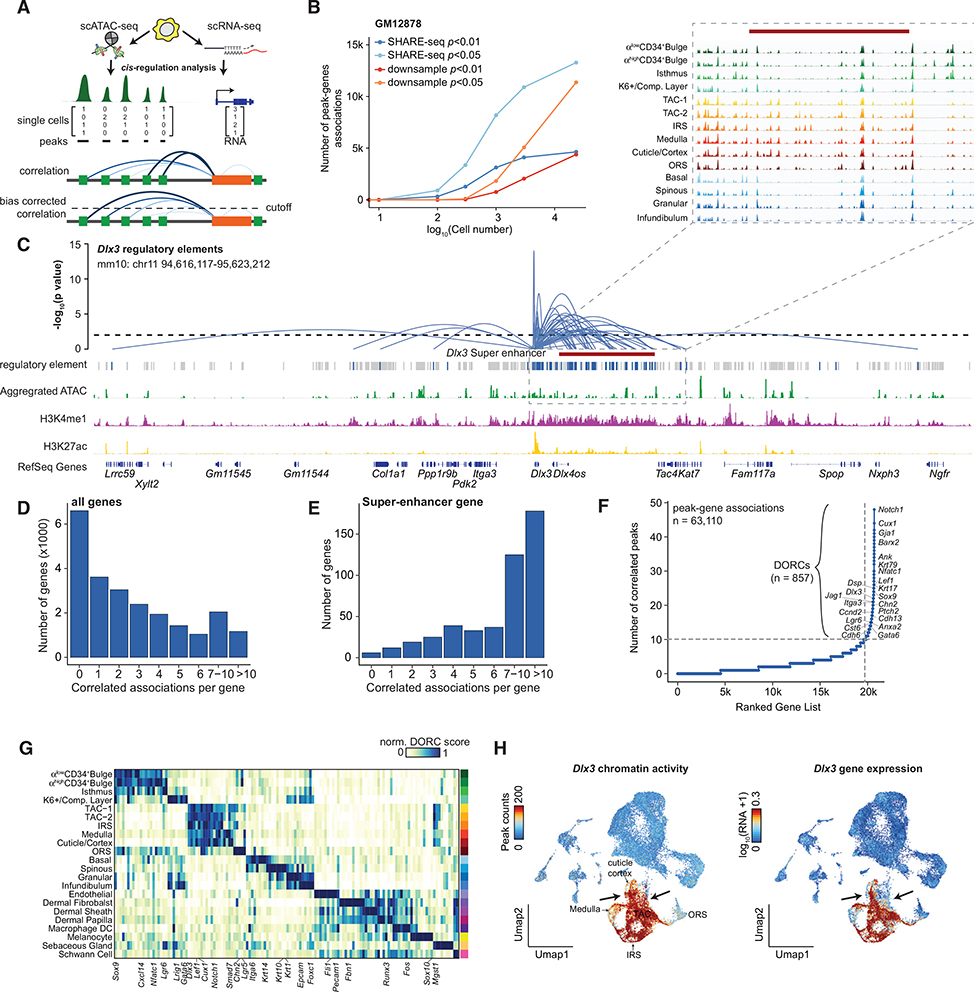
Figure 1. SHARE-Seq Provides an Accurate Co-measure of Chromatin Accessibility and Gene Expression.
(A) Workflow for measuring scATAC and scRNA from the same cell using SHARE-seq.
(B and C) Unique ATAC fragments (B) and RNA UMIs (C) aligning to the human or mouse genome. The experiment is performed using a mix of human (GM12878) and mouse (NIH/3T3) cell lines.
(D) The percentage of ATAC or RNA reads aligning to the human genome relative to all reads mapping uniquely to the human or mouse genomes.
(E) Number of ATAC fragments in peaks or RNA UMIs for SHARE-seq (this study), sci-CAR (Cao et al., 2018), SNARE-seq (Chen et al., 2019), or Paired-seq (Zhu et al., 2019). Boxplots denote the medians and the quartile ranges (25% and 75%), and the length of whiskers represents 1.5× interquartile ranges (IQRs).
(F) Aggregated single-cell chromatin accessibility and gene expression profiles in GM12878 cells (top), individual cell profiles (bottom), and single-cell average (right). Single-cells are sorted by the normalized ATAC-seq yield of the depicted NFkB1 locus.
SHARE-Seq Generates High-Quality Chromatin and Expression Profiles across Diverse Cell Lines and Tissues
To validate specificity and data quality, we first performed SHARE-seq on a mixture of human (GM12878) and mouse (NIH 3T3) cell lines. Human and mouse reads were well separated on chromatin and transcriptome profiles, resulting in 903 human and 1,341 mouse cells passing filters of 2,000 expected cells (Figures 1B–1D). We identified only one cell doublet, representing a remarkably low 0.04% collision rate (consistent with the expected rate of 0.052%; Figure S1C), a benefit of the large SHARE-seq barcoding space. Cells passing filter (STAR Methods) had, on average, 2,545 RNA UMIs (9,660 estimated UMI library size) and 8,252 unique ATAC-seq fragments (19,723 estimated library size with 65.5% fragments in peaks) (Figures S1D and S1E).
SHARE-seq had similar performance across replicates and additional cell lines (Figures S1F–S1M) and showed high concordance with previously published scATAC-seq datasets (STAR Methods; Figure S1J). SHARE-seq also consistently outperformed previously published joint ATAC-RNA approaches (Cao et al., 2018; Chen et al., 2019; Zhu et al., 2019; Figure 1E). Notably, SHARE-seq RNA reads (starting with cells or nuclei) are enriched for intronic regions, similar to single-nucleus RNA sequencing (snRNA-seq) (Habib et al., 2016; Figure S1N), which may be due to cell membrane lysis and serial washes. Intronic RNA is enriched for nascent RNA, which can be used not only to identify cell types (Habib et al., 2017) but also to investigate temporal processes in single cells (La Manno et al., 2018). Finally, chromatin accessibility at the NFkB1 locus and NFkB1 gene expression significantly co-varied across single cells (Spearman ρ = 0.31, p < 10−6, Z test), validating our expectation that increased chromatin accessibility results in higher gene expression and that SHARE-seq may be used to query chromatin-gene expression relationships (Figure 1F).
SHARE-seq performed well with cells or nuclei from a broad range of tissues, including mouse skin, brain, and lung (Figures 2A–2C; Figure S2). SHARE-seq performed comparably with scATAC-only approaches (Lareau et al., 2019; Mezger et al., 2018) applied to adult mouse lung (STAR Methods; Figure 2B) and snRNA-seq and scRNA-seq (Saunders et al., 2018; Zeisel et al., 2018; Habib et al., 2017) of adult mouse brain (STAR Methods; Figure 2C; Figures S2C and S2D). SHARE-seq shows similar data quality when starting with cells or nuclei (Figures S2E–S2G), with the expected increase in the portion of intronic RNA in nuclei. Importantly, SHARE-seq also enabled experiments to be performed at a substantially lower cost than prior methods. Altogether, these points validate the accuracy and utility of SHARE-seq for integrated measures of chromatin accessibility and gene expression in cell lines or primary tissues.
Figure 2. SHARE-Seq Enables Joint Profiling of Chromatin Accessibility and Gene Expression in Tissues.
(A) A schematic of tissues profiled with SHARE-seq, highlighting the cellular diversity within mouse skin.
(B and C) Comparison of library size estimates of SHARE-seq and other single-cell or nucleus-based approaches for scATAC-seq (B) and scRNA-seq (C) approaches. Boxplots denote the medians and the quartile ranges (25% and 75%), and the length of whiskers represents 1.5× IQRs.
(D) SHARE-seq uniform manifold approximation and projection (UMAP) visualization of single cells derived from mouse skin showing UMAP coordinates defined by RNA. Points colored by clusters are defined by RNA clustering, and cell types are assigned to clusters on the basis of marker genes, TF motifs, and chromatin accessibility peaks. Computational pairing (Stuart et al., 2019) of scATAC-seq to scRNA-seq (right) is colored by predicted cell type. The IRS cluster is highlighted.
(E) SHARE-seq UMAP visualization of single cells derived from mouse skin showing UMAP coordinates defined by ATAC.
(F) Heatmap showing the proportion of cells in the RNA cluster that overlaps with chromatin-defined clusters.
(G) Marker gene expression and TF motif scores for each cluster.
(H) Aggregated scATAC-seq tracks denoting marker chromatin accessibility peaks for each cluster.
(I) The cluster-cluster correlation (Spearman correlation coefficient) of scATAC-seq (top right) and scRNA-seq (bottom left). The scATAC-seq correlation was calculated based on the average peak counts per cluster. The scRNA-seq correlation was calculated based on average gene expression per cluster.
(J) Cells colored by the activity of cell cycle genes (left panel). An RNA cluster marked by high expression of cell cycle genes is highlighted in scRNA UMAP (top right panel) and scATAC UMAP space (bottom right panel).
Broad Congruence between Chromatin and RNA Defined Cell Types from SHARE-Seq
To utilize SHARE-seq to query the relationship between chromatin accessibility and gene expression, we focused on mouse skin. Skin is enriched for cell types from diverse lineages—some are highly proliferative, whereas others are dormant or cycling slowly—with multiple populations of stem cells giving rise to well-defined cell types (Adam et al., 2015; Cohen et al., 2018; Fan et al., 2018; Hsu et al., 2014; Joost et al., 2018, 2020; Lien et al., 2011; Salzer et al., 2018).
Leveraging the increased throughput and resolution of SHARE-seq, we assessed the congruence between the epigenome and transcriptome across an atlas of 34,774 high-quality profiles from adult mouse skin (Figure 2D; Figures S2H and S2I). To define cell subsets, we clustered the RNA portion of SHARE-seq data (Table S2). We then projected the cells based on ATAC-seq and RNA-seq independently to a low dimensional space (STAR Methods) and found that both projections separated these scRNA-seq-defined clusters (Figures 2D and 2E). SHARE-seq not only resolved cell types from distinct lineages but could also distinguish between cells of closely related types (for example, αhigh CD34+ bulge versus αlow CD34+ bulge; Blanpain et al., 2004). Moreover, cell membership in scATAC-seq clusters was highly congruent with membership in scRNA-seq clusters (Figure 2F), and both measures revealed the same major cell types, such as transit-amplifying cells (TACs), inner root sheath (IRS), outer root sheath (ORS), and hair shaft cells (Figures 2D–2F).
Cells in the RNA-based clusters can also be distinguished by chromatin accessibility features, further confirming their identity (Figures 2G and 2H). We annotated clusters by the activity of lineage-determining TFs, inferred from the scATAC-seq data (Figure 2G; Schep et al., 2017), and their correlation with TF RNA expression levels (Figures S2I and S2K; STAR Methods). This analysis revealed the global transcriptional activators Dlx3 and Sox9 and repressors Zeb1 and Sox5 (Adam et al., 2015; Huang et al., 2008; Spaderna et al., 2008), among many others (Figure S2K). Thus, SHARE-seq provides insights into cell identity at multiple scales, including chromatin regulation by key lineage-determining TFs.
Nevertheless, some cell states may be identified at higher resolution by chromatin or gene expression features. Specifically, grouping clusters by their aggregate (pseudo-bulk) profiles revealed more distinctive chromatin accessibility differences between the permanent portion (clusters 1–4) and regenerative portion (clusters 5–9) of the hair follicle. Conversely, cells corresponding to the granular layer are easier to distinguish as a unique cluster at the gene expression level (Figure 2I). Moreover, a subset of actively proliferating basal cells strongly expressing cell cycle genes (Figure 2J), which formed a single group by RNA, was not identified as a coherent cluster by chromatin accessibility (with one of four different dimensionality reduction approaches; Figure 2J; Figure S2L; STAR Methods). On the other hand, the TAC populations, which also strongly expressed cell cycle genes, were identified as a unique cluster by chromatin accessibility (Figure S2M). In the TAC populations, expression of cell cycle genes is also coupled to changes in lineage identity factors (Figure 2G). This suggests that the cell cycle is more associated with changes in gene expression and less predominant in chromatin accessibility profiles.
We reasoned that SHARE-seq can be used to directly test the accuracy of computational approaches (Stuart et al., 2019) that pair data from scATAC-seq and scRNA-seq from separately measured cells. Such methods typically assume congruence and may miss asynchrony or distinctions between these features of cellular identity. We thus tested a canonical correlation analysis (CCA)-based method (Stuart et al., 2019) by providing the ATAC-seq and RNA-seq portions of the SHARE-seq measurements separately and comparing inferred pairing with the correct (measured) coupling. Profiles from the same cell were assigned properly (defined as membership in the same cluster) with variable accuracy (74.9% in skin and 36.7% in mouse brain) (Figures S2N–S2S), with most mis-assignments between clusters representing similar cell types (e.g., IRS to TACs; Figure 2D; Figures S2Q and S2S). By down-sampling, we find that computational errors are exaggerated when sequencing depth or cell numbers are limited (Figure S2T). This suggests that SHARE-seq may help train computational pairing approaches across tissues or test their performance and help with further improvements.
Paired Measurements Associate Chromatin Peaks and Target Genes in cis
Cells exhibit significant variations in gene expression and the underlying regulation of chromatin because of intrinsic (e.g., bursts of expression; Larsson et al., 2019) and extrinsic (e.g., cell size, level of regulatory proteins; Lin and Amir, 2018) factors. SHARE-seq may allow us to infer the relationship between chromatin and gene expression. To test this, we developed an analytical framework to link distal peaks to genes in cis, based on the co-variation in chromatin accessibility and gene expression across cells, while controlling for technical biases in chromatin accessibility measurements (Figure 3A; Figure S3; STAR Methods). We first applied this approach to a dataset of 23,278 GM12878 cells and identified 13,277 significant peak-gene associations (Figures S3B and S3C; Table S3; p < 0.05, false discovery rate [FDR] = 0.11). To appropriately determine the probability of interaction between our peaks and the identified target gene, we first sought to normalize for ATAC-seq peak density surrounding gene promoters; ATAC-seq peaks are commonly located near gene promoters (in these data, 61.3% of ATAC-seq peaks are within 2 kb of transcription start sites [TSSs]). First normalizing for peak density near genes, we determined the half-life of these peak-gene associations to be 24.4 ± 3.6 kb (95% confidence interval), resulting in a finding surprisingly similar to a recent report assessing the effect of cis-regulatory regions on gene expression using CRISPR-based perturbation (24.1 kb) (Gasperini et al., 2019). Further validating our approach, peak-gene associations are significantly enriched in loops identified with H3K27ac HiChIP (p < 10−608, hypergeometric test) and depleted in the repressive histone mark H3K27me3 (Figure S3D). Determining peak-gene associations without correction of technical biases (peak intensity and GC content) resulted in a much smaller half-life (2.6 kb; Figure S3A). Notably, down-sampling of cell numbers or reads dramatically reduces the ability to discover peak-gene associations (Figure 3B). This demonstrates that the computational and experimental improvements reported here are essential for accurately determining peak-gene associations.
Figure 3. cis Regulation Determines DORCs.
(A) Schematic depicting an analytical framework for analysis of distal regulatory elements and expression of genes.
(B) Number of peak-gene associations after down-sampling the number of cells or reads in the GM12878 SHARE-seq dataset. Reads are down-sampled to match the number of reads recovered to match those obtained by sci-CAR (Cao et al., 2018).
(C) Loops denote the p value of chromatin accessibility of each peak and Dlx3 RNA expression (± 500 kb from TSSs). Loop height represents the significance of the correlation. H3K4me1 and H3K27ac ChIP-seq tracks and super-enhancer annotation were generated from an isolated TAC population (Adam et al., 2015). Grey bars denote scATAC-seq peaks. Blue bars denote peaks that are significantly associated with the Dlx3 gene. The magnified tracks show the aggregated ATAC-seq data of each cluster.
(D and E) The number of significant peak-gene associations for all genes (D) and previously defined (Adam et al., 2015) super-enhancer genes (E).
(F) The number of significantly correlated peaks (p < 0.05) for each gene (± 50 kb from TSSs). Known super-enhancer-regulated genes are highlighted.
(G) Representative DORCs for each cluster; values are normalized by the min and max activity.
(H) The peak counts of all Dlx3 correlated peaks (left) and Dlx3 gene expression (right) colored in UMAP. The arrows point to regions with differential signals.
Applying this framework to the mouse skin dataset, we identified 63,110 significant peak-gene associations (within ± 50 kb around TSSs, p < 0.05, FDR = 0.1; Table S4; Figures S3H and S3I). These peak-gene associations were enriched proximally to the TSSs (Figure S3G; p < 2.2 × 10−16, KS test), and most of the associations regulate a single gene (83.9%; Figure S3J). Interestingly, in rare cases, individual peaks associated with 4 or more genes (0.14% of peaks), including well known gene clusters (Histone, Hox, and Keratin), suggesting that these peaks may coincide with regulatory hubs controlling the expression of gene clusters (Figure S3K). Although interesting, these occurrences were rare and may also reflect technical artifacts; we therefore only considered the most significant peak-gene association when associating target genes with peaks. Finally, most of the chromatin peaks (82%; Figure S3L) were not correlated with expression of any gene, a finding that supports a previous observation that perturbations in only a small portion of candidate enhancers significantly alter the expression of genes (Gasperini et al., 2019).
A subset of genes, including key fate determination genes, was associated with a large number of peaks (p < 2.2 × 10−16, permutation test). For example, 22 peaks were significantly associated (within ± 50 kb around TSSs, p < 0.05) with Dlx3, highly expressed in TACs (Figure 3C; Adam et al., 2015). These results are reminiscent of previous observations describing regulatory locus complexity at key lineage genes (González et al., 2015). Further, regions with a high density of peak-gene associations significantly overlapped known super-enhancers (Adam et al., 2015; Figure 3C; Table S5; 2.1-fold enrichment, p = 10−238, hypergeometric test)—enhancer regions that are cell-type-specific and highly enriched in histone H3K27 acetylation (Whyte et al., 2013). This relationship was not simply driven by super-enhancer length (Spearman ρ = 0.04; Figure S3M) or the total number of peaks surrounding a gene (Figure S3N). Furthermore, super-enhancer-regulated genes are associated with more peaks compared with all genes (10.9 versus 4.4 associated peaks per gene, on average; p< 2.2 × 10−16, KS test; Figures 3D and 3E; Figure S3O). Finally, most annotated cell cycle genes (n = 97) had lower-than-expected numbers of peak-gene associations (on average, 3.4 associations for cell cycle genes versus 4.4 associations for all genes; p = 0.026, t test), further supporting a limited contribution of chromatin accessibility to cell cycle-associated gene expression changes and suggesting that variable expression is not sufficient for determining peak-gene associations.
DORCs Identify Key Lineage-Determining Genes De Novo
We define the 857 regions with an exceptionally large (>10) number of significant peak-gene associations as DORCs, identified as those exceeding an inflection point (“elbow”) when ranking genes by the number of significant associations (Figure 3F). We quantified the activity of DORCs as the sum of accessibility at peaks significantly associated with the DORC-regulated gene. The DORCs identified in sub-populations strongly overlap with DORCs identified with all cells (p = 10−201, hypergeometric test; Figure S3P). Moreover, DORCs were strongly enriched for known key regulators of lineage commitment across the expected lineages (Figure 3G; Figure S3Q). Notably, only 34.4% of DORCs active in hair follicle stem cells (HFSCs) or TACs overlap with super-enhancers identified in these same cell types (Adam et al., 2015), suggesting that DORCs may encompass other mechanisms promoting formation of enhancer clusters (Hnisz et al., 2017; Li et al., 2002). Finally, there were significant differences in DORCs even between closely related populations (Figures S3R and S3S), suggesting that DORCs are highly cell type specific.
Interestingly, gain of chromatin accessibility does not always equate productive transcription. For example, although Dlx3 DORC and Dlx3 gene expression were active in TAC/IRS/medulla cells, this was not the case in cuticle/cortex cells, where the Dlx3 DORC is active and the Dlx3 gene is not highly expressed (Figure 3H; Figure S3T). Thus, DORCs provide an unsupervised, readily accessible approach to simultaneously identify key lineage-determining genes and their regulatory regions at single-cell resolution without the need to know the cell type identity in advance, isolate cell subsets, and conduct challenging chromatin immunoprecipitation sequencing (ChIP-seq) experiments from primary samples.
Lineage Priming at Enhancers Precedes Gene Expression in DORCs
The hair follicle is a highly regenerative epithelial tissue that cycles between growth (anagen), degeneration (catagen), and rest (telogen). At anagen onset, hair follicle stem cells located at the bulge and hair germ proliferate transiently to produce short-lived TACs. These TACs are some of the most proliferative cells in adult mammals; they divide rapidly to produce multiple morphologically and molecularly distinct downstream differentiated cell types that constitute the mature hair follicle, including the companion layer, IRS, and hair shaft (hair shaft cuticle, cortex, and medulla) (Zhang and Hsu, 2017; Zhang et al., 2016).
We readily recovered three hair follicle differentiation trajectories (IRS, medulla, and cuticle/cortex, differentiated from TACs) from chromatin accessibility (Figure 4A). Systematically analyzing the cuticle/cortex trajectory revealed that DORCs generally become accessible prior to onset of their associated gene’s expression, consistent with lineage priming. For example, Wnt3 RNA became detectable at the late stage of hair shaft differentiation (Millar et al., 1999); however, accessibility in the Wnt3 DORC activated at TACs prior to gene expression before lineage commitment (Figure 4B), which we quantified by computing “residuals” (defined as the difference of chromatin accessibility and expression of the gene; STAR Methods). Despite peak-gene associations being defined by high correlation, we found that residuals were typically positive across most of the DORC-regulated genes (92%) and lineages (Figure 4C; Figures S4A and S4B). Thus, sufficiently high RNA expression is only detectable in a subset of DORC-active cells, likely reflecting a requisite gain of accessible chromatin at that gene’s locus.
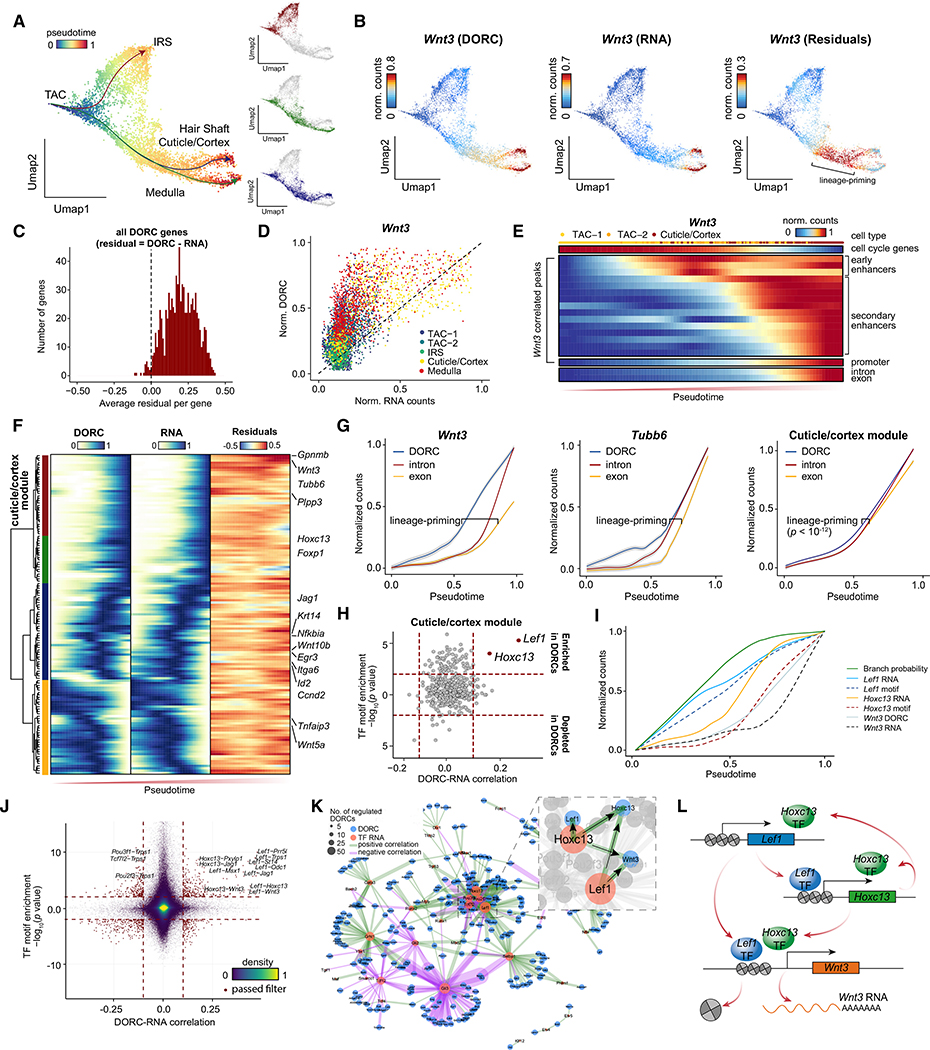
Figure 4. Lineage Dynamics of Chromatin and Expression Defines Lineage Priming.
(A) Pseudotime for three cell fate decisions shown on scATAC UMAP coordinates.
(B) Difference (residuals) for Wnt3 between chromatin accessibility and gene expression for the regenerative portion of the hair follicle.
(C) Histogram of the average difference (residuals) for each gene between chromatin accessibility and gene expression.
(D) Scatterplot of the Wnt3 DORC score and Wnt3 gene expression.
(E) Dynamics of gene expression (intron and exon) and individual chromatin accessibility peaks for the cuticle/cortex lineage.
(F) Hierarchical clustering of chromatin accessibility, expression of DORC-regulated genes, and the difference between chromatin accessibility and gene expression (residuals) for the cuticle/cortex lineage. Cells are ordered by pseudotime.
(G) Lineage dynamics for individual DORC-regulated genes, highlighting lineage-priming in Wnt3 (left), Tubb6 (center), and the mean of the cuticle/cortex module (red cluster in F). The shaded region presents a 0.99 confidence interval.
(H) TF motif enrichment in lineage-priming DORCs plotted against Spearman correlation of the cuticle/cortex module DORC score and TF gene expression.
(I) Lineage dynamics of Lef1 and Hoxc13 motif scores and gene expression precede Wnt3 DORC activation in the hair shaft lineage. A green line shows the branch probability of the hair shaft lineage.
(J) Pairwise correlation of DORC to expression of DORC-regulated genes and enrichment of the TF motif in the DORC. The red dashed lines indicate the cutoff to determine meaningful regulations.
(K) TF regulatory network showing the driver TF for each DORC. The width of an edge indicates the significance (−log10(FDR)) of correlation between TF RNA expression and DORC score. The magnified image shows TFs and DORCs that are directly connected to the Lef1 TF.
(L) Schematic of the stepwise model of Lef1, Hoxc13, and Wnt3 activation in the hair shaft lineage.
To further understand the gene-regulatory mechanisms underlying these residuals, we tracked the accessibility changes at individual peaks near the Wnt3 locus along differentiation pseudotime from TACs to cuticle/cortex cells (Figure 4D; Figure S4C). We found sequential activation of peaks in the Wnt3 DORC, with individual enhancer peaks activating much earlier than the Wnt3 promoter, followed by activation of nascent RNA expression (estimated by intron counts) and, finally, mature RNA expression (estimated by exon counts) (Figure 4E). We posit that activation of the promoter was the limiting factor for induced expression of Wnt3. For better visualization of DORC-associated residuals, we clustered DORCs into 4 groups (Figure 4F; Figure S4G), including a late-activating group to which we refer as the “cuticle/cortex module.” There was a lag of pseudotime between the onset of accessibility and the corresponding RNA expression (Figure 4G). Notably, some TACs at late pseudotime are still proliferative (as estimated by cell cycle gene expression); however, they already show activated enhancer peaks, suggesting a switch between proliferation and differentiation transitioning (Figure 4E). Importantly, lineage priming is not restricted to the cuticle/cortex lineage but also exists in IRS and medulla lineages (Figure S4H). These analyses support the long-standing hypothesis that enhancer activation foreshadows expression of target genes (Lara-Astiaso et al., 2014; Rada-Iglesias et al., 2011) and implicates chromatin accessibility as a marker for lineage priming.
We further investigated the mechanisms leading to chromatin accessibility-primed chromatin states and hypothesized that TFs that prime are distinct from TFs that activate enhancers. Indeed, we found that binding sites for Lef1 and Hoxc13 TFs were strongly enriched (p < 10−4, KS test; Figure 4H) in cuticle/cortex module DORCs (including the Wnt3 DORC). By pseudo-temporal ordering of RNA expression and TF motif activity (inferred from ATAC), we found clear ordering of Lef1 RNA onset first, followed by Lef1 motif accessibility, Hox13 RNA, Hoxc13 motif accessibility along with Wnt3 DORC activity, and, finally, Wnt3 RNA. This implicates Lef1 as the lineage-priming TF (Merrill et al., 2001). Hoxc13 expression following this first wave of chromatin accessibility likely further induces Wnt3 DORC accessibility, which finally promotes expression of Wnt3 (Figure 4I; Figure S4I). Interestingly, the pseudotime-determined cuticle/cortex branch probability tracks closely with Lef1 gene expression, particularly at early stages of differentiation, providing a genome-wide measure supporting Lef1 as the TF regulating lineage choice. This supports a model where distinct modes of regulation exist to prime chromatin accessibility and foreshadow lineage choice.
Leveraging DORCs and the TFs that regulate them, we constructed a TF-regulatory network that underlies hair follicle differentiation (STAR Methods), relating each TF to each DORC (Figures 4J and 4K; https://buenrostrolab.shinyapps.io/skinnetwork/). Consistent with our stepwise model of regulatory events leading to lineage commitment, Lef1 and Hoxc13 TFs are central components of the network driving the activity of cuticle/cortex genes, including Wnt3 and Trps1 (Figure 4L). Interestingly, we also identified highly connected transcriptional repressors (TFs that negatively correlate with DORC activity levels), including Gli3 and Tcf12, associated with the Sonic hedgehog (Shh) signaling pathway (Park et al., 2000; Mill et al., 2003). Thus, SHARE-seq, together with our computational framework, can measure lineage priming and predict novel regulators.
Chromatin Accessibility Priming Coincides with Multilineage Fate Bias and Histone Modifications
We further sought to find out how early during differentiation we could identify markers of lineage commitment. To investigate this, we identified DORCs that were differentially active between cuticle/cortex and medulla cells preceding the lineage decision, including Notch1, Cux1, and Lef1 (Figure S5A). Notch1, highly expressed in hair shaft cells, is critical in controlling hair follicle differentiation (Pan et al., 2004). When we partitioned the lineage-priming region into 3 sub-regions by the DORCs’ accessibility (Figures 5A and 5B), Notch1+ and Notch1− regions showed distinct chromatin patterns with coordinated changes in gene expression (Figures S5A and S5B), whereas Notch1+ cells were not distinctly identified by their gene expression pattern alone (Figure 5B). We observed clear chromatin differences across Notch1+ and Notch1− cells in the lineage priming-associated regions (Figures 5C and 5D). This further demonstrates that genome-wide changes in chromatin accessibility reflect lineage-primed cell states and highlights Notch1- and Tchh-specific chromatin changes priming gene expression activation.
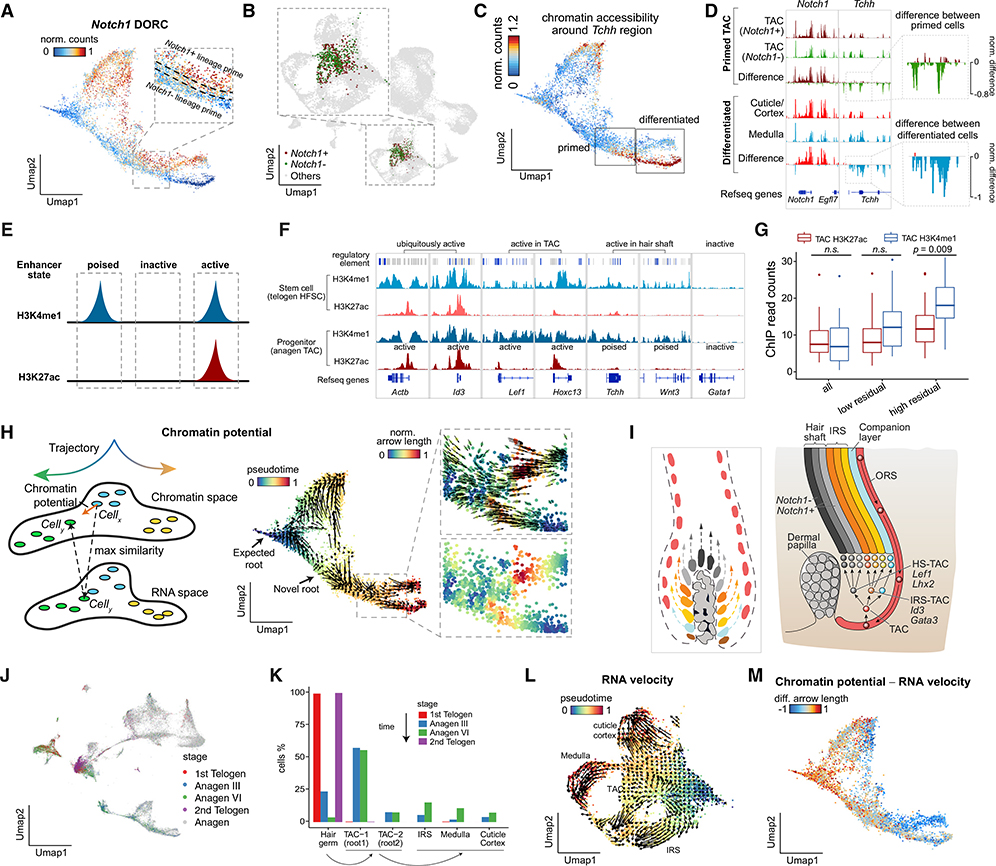
Figure 5. Chromatin Potential Describes Chromatin-to-Gene Expression Dynamics during Differentiation.
(A) Chromatin accessibility of the Notch1 DORC, highlighting the lineage-priming region.
(B) Distribution of Notch1+ and Notch1− lineage-primed cells in the scRNA UMAP.
(C) UMAP visualization of the chromatin accessibility profiles around the Tchh region in (D).
(D) Aggregated chromatin accessibility profiles of lineage-primed (Notch1+/−) progenitor cells (TACs) and differentiated cells (cuticle/cortex and medulla).
(E–G) The chromatin accessibility-primed chromatin state is reflected by the histone modification state. The ChIP-seq data were downloaded from Adam et al. (2015) and Lien et al. (2011).
(E) Schematic showing different categories of enhancers.
(F) ChIP-seq genome tracks showing lineage-priming genes that are identified by SHARE-seq and are active in TACs. Genes expressed in differentiated cells are poised in TACs.
(G) The ChIP-seq signal enrichment in lineage-specific genes that are defined by genes with higher expression in the hair shaft than in TACs. The lineage-specific genes are further classified by their residuals (DORC-RNA expression). Low-residual DORCs and high-residual DORCs refer to the bottom 10 percentile and top 10 percentile of genes ranked by residuals, respectively.
(H) Schematic of the conceptual workflow for determining chromatin potential (left). Chromatin potential is visualized on the scATAC UMAP space, and arrows denote the extrapolated gene expression state of the cell (right).
(I) Schematic of TAC heterogeneity. Left: heterogeneous progenitor cells vertically differentiate to fat-committed cells. Right: TACs differentiate to IRS-TACs and HS-TACs first and further differentiate to HS and IRS.
(J) UMAP visualization of the ATAC portion of SHARE-seq data on different hair follicle stages, projected to the data in Figure 2E.
(K) The cell type compositions of different hair follicle stages.
(L) RNA velocity visualized on scRNA UMAP coordinates.
(M) The difference between the neighborhood predicted by chromatin potential and RNA velocity.
Further analyses of chromatin accessibility-primed states revealed that lineage-primed loci also reflect primed histone modification states. Enhancers can be categorized as poised (H3K4me1high and H3K27aclow), active (H3K4me1high and H3K27achigh), and inactive (H3K4me1low and H3K27aclow) (Lara-Astiaso et al., 2014; Rada-Iglesias et al., 2011; Figure 5E). We found that Lef1 and Hoxc13 loci are poised in telogen HFSCs located at the bulge and hair germ (Rompolas et al., 2012, 2013) and then become active when HFSCs differentiate to TACs (Figure 5F). Furthermore, Wnt3 and Tchh loci are poised in TACs and then become active when TACs differentiate to hair shaft cells (Figure 5F). Extending this analysis to all DORCs, we found that DORCs with higher residuals coincided with a stronger signature of a poised chromatin state (p = 0.009; Figure 5G). We demonstrate that low levels of chromatin accessibility at DORCs are a marker of poised chromatin correlating with lineage fate outcomes across single cells.
Chromatin Potential Describes Chromatin-to-Gene Expression Dynamics during Differentiation
Empowered by our findings, we hypothesized that lineage priming by chromatin accessibility may foreshadow gene expression and may be used to predict lineage choice prior to lineage commitment. Focusing on DORC-regulated genes, we devised an approach to calculate “chromatin potential,” defined as the future RNA state most compatible with a cell’s current chromatin state (STAR Methods). We computed RNA-chromatin neighbors (k-NN, k = 10) and found, for each cell (cell x, chromatin neighborhood), 10 cells (cell y, RNA neighborhood) whose RNA expression of DORC-regulated genes is most correlated with the current chromatin state. Chromatin potential (arrow) is the direction and distance between each cell (cell x, chromatin neighborhood) and the 10 nearest cells (cell y, RNA neighborhood) in chromatin low-dimensional space (Figure 5H; Figures S5C and S5D). Its arrow length is a measure of how different the chromatin state is from the “future” RNA state. Notably, this analysis does not rely on the inferred pseudotime. Chromatin potential relates a potential “future” RNA state (observed in another cell) that is best predicted by the chromatin state of a given cell.
In general, chromatin potential flows from progenitor cells (TACs) to differentiated cells (IRS/hair shaft) (Figure 5H). Chromatin potential arrows are longer at key multi-lineage defining transitions, including the branchpoint that defines the cuticle/cortex and medulla lineages. However, some long arrows reflecting rare cells may be due to noise in the assay, errors in embedding the cells into lower dimensions, or technical biases. Chromatin potential identified a distinct root-like position in addition to the original pseudotime-identified root (Figure 5H). TACs are molecularly and spatially heterogeneous (Genander et al., 2014; Joost et al., 2020; Legué and Nicolas, 2005; Xin et al., 2018; Yang et al., 2017). These progenitors divide perpendicularly relative to the basement membrane and are set up by epithelial-mesenchymal niches in the regenerating hair follicle (Figure 5I; Figure S5E). At anagen III, lineage-primed and spatially restricted TACs emerge and are largely unipotent progenitors. These TACs are distinct and emerge from multi-lineage progenitor TACs seen in earlier anagen stages. The identified two roots show distinct molecular profiles consistent with previous reports of lineage-biased TACs (Joost et al., 2020; Yang et al., 2017; Figure S5F). The novel root is supported by RNA velocity (Figure S5G) as well as pseudotime inferred from the scRNA-seq data alone (Figure S5H), further supporting a model where both roots are associated with cells in an undifferentiated state.
To further validate the hypothesis that the novel root reflects lineage-biased progenitors, we performed SHARE-seq on different hair follicle stages (first telogen, anagen III, anagen VI, and second telogen) and evaluated the cell type compositions (Figures 5J and 5K; Figures S5I–S5K). Consistent with prior studies (Joost et al., 2020; Yang et al., 2017), we found a similar proportion of TAC-1 (the pseudotime-defined root) and TAC-2 (the additional root determined by chromatin potential) cells in anagen III and anagen VI stages and a significantly higher portion of stem cells in first telogen (hair germ) and differentiated cells (IRS/medulla/hair shaft) in anagen VI. This suggests that TAC-1 and TAC-2 are the roots in differentiation (Figure S5K). Chromatin potential allows us to relate the chromatin state of one cell to future RNA states, identifying likely paths cells may follow during developmental transitions.
Chromatin potential exceeds our ability to predict future RNA states from the cell’s current RNA state by its mRNA or nascent RNA (as shown by RNA velocity; La Manno et al., 2018), emphasizing the longer timescales foreshadowed by chromatin states (Figures S5L–S5N). RNA velocity-derived vectors provided little resolution of cell fate dynamics within TACs (Figure 5L; Figure S5G). The discrepancy between RNA velocity and chromatin potential is most prominent in TACs (Figure 5M). Interestingly, chromatin potential has a longer reach (prediction timescales) at early stages, whereas RNA velocity has a farther reach at late pseudotimes (p < 2.2 × 10−16, KS test; Figure S5P). RNA velocity, which relies on differences between unspliced and spliced mRNAs (Rabani et al., 2011), predicts the future of individual cells on a timescale of hours (La Manno et al., 2018). In contrast, the timescales of changes associated with chromatin states are less well defined (Kelsey et al., 2017); however, they are largely established prior to gene expression (Bernstein et al., 2006; Rada-Iglesias et al., 2011; Shema et al., 2019). We therefore reasonably expect that chromatin potential may predict the future cell state on a timescale greater than that seen by RNA velocity, especially during differentiation.
DISCUSSION
To infer transcriptional regulation and recover key regulatory regions in differentiation, SHARE-seq provides a means to infer DORCs reflecting key lineage-determining genes and the TFs that regulate them. Leveraging SHARE-seq, it should now be possible to identify key regulatory regions, including developmental super-enhancers, and their associated target genes without isolating specific cell subsets or ChIP-seq experiments, which can be challenging for in vivo samples or may not even be known a priori. Inclusion of more layers of measurements and improved computational methods to illustrate the differences between chromatin regulators and gene expression should enable a more robust approach to defining chromatin-gene dynamics in complex tissues. This is important in developmental biology, cancer research, and especially human genetics, where genetic variants associated with complex human diseases are found in non-coding regions, which can be challenging to relate to specific cell types and target genes.
We define chromatin potential to describe the difference between chromatin and expression upon hair follicle differentiation. Recently, RNA velocity approaches have predicted a cell’s future state from the differences between nascent and mature RNA (categorized by intronic and exonic reads). In contrast, chromatin potential enables analysis of genes irrespective of whether they have introns (e.g., the TFs Jun, Sox2, and Foxq1 in our analysis have no introns) and prediction of lineage fates prior to nascent transcription. Chromatin changes may also reflect lineage bias rather than lineage choice. Primed chromatin states can be altered or reversed (Bernstein et al., 2006; Lara-Astiaso et al., 2014; Ostuni et al., 2013; Weiner et al., 2016). Therefore, unlike RNA velocity, we consider chromatin potential a measure of what the cell will likely do rather than a measure of what the cell has committed to do. A fascinating direction for future research will be to determine differences in chromatin potential, RNA velocity, and lineage choice (through lineage tracing). Furthermore, building computational tools to integrate chromatin potential with RNA velocity may enable multi-state vectors aimed to predict the continuous trajectory a cell may follow.
SHARE-seq provides a generalizable platform and opportunity to include additional layers of information per cell. With further development, we expect to integrate other scRNA-seq-compatible measurements (Stuart et al., 2019), such as protein measurements (Stoeckius et al., 2017), genotyping, and lineage barcoding. Furthermore, powered by the massive scalability of this approach, SHARE-seq may be adapted to identify RNA barcodes, particularly useful for CRISPR-based perturbation screens (Dixit et al., 2016). As we move toward a cell atlas, we anticipate that SHARE-seq will likely play a key role in determining the full diversity of cell types and cell states, the regulators that define them, and the effect of common genetic variants on molecular processes in specific cell types.
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jason Buenrostro (jason_buenrostro@harvard.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Gene Expression Omnibus: SHARE-seq data are deposited under accession number GEO: GSE140203, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140203
The R Shiny-based web application for data visualization is accessible here: https://buenrostrolab.shinyapps.io/skinnetwork/
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
GM12878 cells were cultured in RPMI 1640 medium (11875–093, ThermoFisher) supplemented with 15% FBS (16000044, ThermoFisher) and 1% penicillin-streptomycin (15140122, ThermoFisher). NIH/3T3 and RAW 264.7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, 11965092, ThermoFisher) with the addition of 10% FBS and 1% of penicillin-streptomycin. Cells were incubated at 37°C in 5% CO2 and maintained at the exponential phase. NIH/3T3 and RAW 264.7 cells were digested with accutase for preparing single-cell suspension.
Mice
Mice were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) approved animal facility at Harvard University and MIT. Procedures were approved by the Institutional Animal Care and Use Committee of all institutions (institutional animal welfare assurance no. A-3125–01, 14–03-194 and 14–07-209). Normal lung, skin and brain were collected from wild-type mice from C57BL/6 mice aged to 6–8 weeks. Mice of both sexes were used for experiments.
Mouse skin
Female C57BL/6J mouse dorsal skins were collected at late anagen (P32). The hair cycle stages were confirmed using cryosectioning. To generate whole skin a single cell suspension, skin samples were incubated in 0.25% collagenase in HBSS at 37°C for 35–45 minutes on an orbital shaker. Samples were gently scraped from the dermal side and the single-cell suspension was collected by filtering through a 70μm filter followed by a 4μm filter. The epidermal portion of the skin samples were incubated in 0.25% trypsin-EDTA at 37°C for 35–45 minutes on the shaker and cells were gently scraped from the epidermal side. Single-cell suspensions were combined and centrifuged for 5 minutes at 4°C, resuspended in 0.25% FBS in PBS, and stained with DAPI (0.05 μg/ml). Live cells were enriched by FACS. To enrich epidermal populations, CD140a negative populations were purified by FACS and combined with whole skin cells in a ratio of 1:1.
Mouse brain
An adult mouse brain was dissected, snap-frozen on dry ice, and stored at −80°C. A single nucleus suspension was prepared following the OMNI-ATAC protocol (Corces et al., 2017). Frozen brain tissue was placed into a pre-chilled 2 mL Dounce homogenizer with 2 mL of 1 × homogenization buffer (320 mM sucrose, 0.1 mM EDTA, 0.1% NP40, 5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM Tris pH 7.8, 1% Protease Inhibitor Cocktail), and 1mM DTT). Tissue was homogenized with 10 strokes with the pestle A, followed by 20 strokes with the pestle B. The sample was centrifuged at 100 g for 1 min to remove large debris. 400 μL of the supernatant was transferred to a pre-chilled 2 mL round bottom tube. 400 μL of a 50% iodixanol solution (50% iodixanol in 1 × homogenization buffer) was added and mixed. 600 μL of a 29% iodixanol solution (29% iodixanol and 480 mM sucrose in 1 × homogenization buffer) was layered underneath the 25% iodixanol mixture. 600 μL of a 35% iodixanol solution (35% iodixanol and 480 mM sucrose in 1 × homogenization buffer) was layered underneath the 29% iodixanol solution. In a swinging-bucket centrifuge, nuclei were centrifuged for 20 min at 3,000 g. Nuclei were resuspended in PBSI (0.1U/μl Enzymatics RNase Inhibitor, Y9240L, QIAGEN; 0.05U/μl SUPERase inhibitor, AM2696, ThermoFisher; 0.04% Bovine Serum Albumin, BSA, 15260037, ThermoFisher in PBS) and proceed to fixation.
Mouse lung
Mouse lung was dissociated with fine scissors followed by proteolytic digestion using the Lung Dissociation kit (Miltenyi Biotech) following the manufacturer’s instructions. Dissociated cells were then incubated at 37°C for 20 minutes with rotation, then filtered using a 100 μm strainer. Red blood cells were lysed using the ACK buffer (A1049201, ThermoFisher).
METHOD DETAILS
Transposome preparation
Tn5 was produced in-house by following a published protocol with minor modifications (Picelli et al., 2014). The pTXB1-Tn5 expression vector was transformed into C3013 cells (NEB) following the manufacturer’s protocol. Each colony was incubated in a 5ml LB medium at 37°C for overnight shaking at 200rpm. That culture was used to start a 1L LB culture with 100μg/ml ampicillin and incubated on a shaker until it reached O.D. ~0.6 (~3 hours). Then the culture was chilled on ice for 30min. Fresh IPTG was added to 0.25mM to induce expression, and the culture was incubated at 18°C on a shaker at 200rpm overnight. The culture was collected by centrifugation at 6,000rpm, 4°Cfor 15min. The bacterial pellet was frozen and stored at −80°C for at least 30min. The frozen pellet was resuspended in 40ml chilled HEGX Buffer (20mM HEPES-KOH at pH 7.2, 0.8M NaCl, 1mM EDTA, 10% glycerol, 0.2% Triton X-100) including 1 × Roche Complete EDTA-free protease inhibitor tablets and 10 μL Benzonase nuclease (Sigma E1014). The lysate was sonicated on the Bioruptor until a large fraction of cells were lysed. The sonicated lysate was centrifuged at 30,000 g at 4°C for 20min. A 2ml aliquot of chitin slurry resin (NEB, S6651S) was packed into a disposable column (Bio-rad 7321010). Columns were washed with 30ml of a HEGX buffer. The soluble fraction was added to the chitin resin slowly, then incubated on a rotator at 4°C 8 hours or overnight. The unbound soluble fraction was drained, and the columns were washed thoroughly with a 40ml HEGX buffer. The chitin slurry was eluted with 10ml of elution buffer (10ml HEGX with 100mM DTT) on rotator at 4°C for ~48h. The eluate was collected and dialyzed twice in 500 ml of Tn5 Dialysis Buffer (100 mM HEPES-KOH pH 7.2, 0.2M NaCl, 0.2mM EDTA, 2mM DTT, 0.2% Triton X-100, 20%Glycerol). The dialyzed protein solution was concentrated using an Amicon Ultra-4 Centrifugal Filter Units 30 K (Millipore UFC803024), and sterile glycerol was added to make a final 50% glycerol stock of the purified protein. If an extra lower weight band was observed when running the product on a protein gel, the product was further purified using a gel filtration column.
Transposome activity quantification
To evaluate the activity of the homemade Tn5, we compared the efficiency of homemade transposome with Nextera TDE1 transposome. We performed standard bulk ATAC-seq experiment (Buenrostro et al., 2013) using Nextera TDE1 or homemade Tn5 diluted with the dilution buffer (50mM Tris, 100mM NaCl, 0.1mM EDTA, 1mM DTT, 0.1% NP-40, and 50% glycerol) at different ratios. The tagmention was performed on 50ng purified genomic DNA instead of cells. We quantified the number of required cycles to reach 1/3 of the plateau fluorescence by qPCR (Buenrostro et al., 2013) and determined the final dilution factor of homemade Tn5 that showed the most similar number of cycles as Nextera TDE1.
SHARE-seq
Preparing oligonucleotides for ligations
There are three barcoding rounds of hybridization reactions in SHARE-seq, with a different 96-well barcoding plate for each round (Table S1). Hybridization oligos have a universal linker sequence that is partially complementary to well-specific barcode sequences. These strands were annealed prior to cellular barcoding to create a DNA molecule with three distinct functional domains: a 5′ overhang that is complementary to the 5′ overhang present on the cDNA molecule or transposed DNA molecules (may originate from RT primer, transposition adaptor or previous round of barcoding), a unique well-specific barcode sequence, and another 5′ overhang complementary to the overhang present on the DNA molecule to be subsequently ligated. Linker strands and barcode strands for the hybridization rounds were added to RNase-free 96-well plates to a total volume of 10 μl/well with the following concentrations: round 1 plates contain 9 μM round 1 linker strand and 10 μM barcodes, round 2 plates contain 11 μM round 2 linker strand and 12 μM barcodes, and round 3 plates contain 13 μM round 3 linker strand and 14 μM barcodes. The oligos are dissolved in STE buffer (10mM Tris pH 8.0, 50mM NaCl, and 1mM EDTA). Oligos are annealed by heating plates to 95°C for 2 minutes and cooling down to 20°C at a rate of −1°C per minute.
Blocking strands are complementary to the 5′ overhang present on the DNA barcodes used during hybridization barcoding rounds. Blocking occurs after well-specific barcodes have hybridized to cDNA molecules, but before all cells are pooled back together. The blocking step minimizes the possibility that unbound DNA barcodes mislabel cells in future barcoding rounds. 10 μL of each blocking strand solution was added to each of the 96 wells after the first, second, and third round of hybridization of DNA barcodes, respectively. Blocking strand solutions were prepared at a concentration of 22 μM for round 1, 26.4 μM for round 2, and 23 μM for round 3. Blocking strands for the first two rounds were in a 2 × T4 DNA Ligase buffer (NEB) while the third round was in 0.1% Triton X-100. Both ligation reaction and blocking reaction were incubated with cells for 30 minutes at room temperature with gentle shaking (300rpm). All the oligos are thawed to room temperature before using.
Fixation
For simplicity, cells and nuclei, which were processed identically for the following steps, are both referred to as cells. Cells were centrifuged at 300 g for 5 minutes and resuspended to 1 million cells/ml in PBSI. Cells were fixed by adding formaldehyde (28906, ThermoFisher, final concentration of 0.1% for cell lines or 0.2% for primary tissues) and incubated at room temperature for 5 minutes. The amount of fixation affects both sequencing library complexity and ambient RNA contamination. We chose 0.1%–0.2% FA fixation to improve library complexity. If a significant amount of ambient RNA contamination is observed, more stringent fixation conditions (up to 1%) could be used. The fixation was stopped by adding 56.1 μL of 2.5M glycine, 50 μL of 1M Tris-HCl pH 8.0, and 13.3 μL of 7.5% BSA on ice. The sample was incubated at room temperature for 5 minutes and then centrifuged at 500 g for 5 minutes to remove supernatant. All centrifugations were performed on a swing bucket centrifuge. The cell pellet was washed twice with 1ml of PBSI, and centrifuged at 500 g for 5 minutes between washings. The cells were resuspended in PBS with 0.1U/μl Enzymatics RNase Inhibitor and aliquoted for transposition.
Transposition
All the oligos used in this protocol can be found in Table S1. The 100 μM Read1 and phosphorylated Read2 oligos were annealed with an equal amount of 100 μM blocked ME-complement oligo by heating at 85°C for 2 minutes and slowly cooling down to 20°C at a ramp rate of −1°C/minute. The annealed oligos were mixed with an equal volume of cold glycerol and stored at −80°C until use. Inhouse produced Tn510 was mixed with an equal volume of dilution buffer (50mM Tris, 100mM NaCl, 0.1mM EDTA, 1mM DTT, 0.1% NP-40, and 50% glycerol). Diluted Tn5 was then mixed with an equal volume of annealed oligos and incubated at room temperature for 30 minutes before transposition.
For each transposition reaction, 5 μL of cells (10,000–20,000 cells in PBSI) and 42.5 μL of transposition buffer (38.8mM Tris-acetate, 77.6mM K-acetate, 11.8mM Mg-acetate, 18.8% DMF, 0.12% NP-40, 0.47% Protease Inhibitor Cocktail, and 0.8U/μl Enzymatics RNase Inhibitor) were mixed and incubated at room temperature for 10 minutes. 2.5 μL of assembled Tn5 was added to the transposition reaction. Depending on the target number of cells to be recovered, the number of transposition reactions can be scaled up. In general, we prepare 10–40 reactions, which is equivalent to 100,000–800,000 cells. The transposition was carried out at 37°C for 30 minutes with shaking at 500rpm. The sample was centrifuged at 1,000 g for 3 minutes and then washed with 1ml Nuclei Isolation Buffer (NIB) (10mM Tris buffer pH 7.5, 10mM NaCl, 3mM MgCl2, 0.1% NP-40, freshly added 0.1U/μl Enzymatics RNase Inhibitor, and 0.05U/μl SUPERase RI). The sample was then resuspended to 60 μL of NIB and before proceeding to reverse transcription.
Reverse transcription
Transposed cells (60 μl) were mixed with 240 μL of RT mix (final concentration of 1 × RT buffer, 0.4U/μl Enzymatics RNase Inhibitor, 500 μM dNTP, 10 μM RT primer with an affinity tag, 15% PEG 6000, and 25U/μl Maxima H Minus Reverse Transcriptase). The RT primer contains a poly-T tail, a Unique Molecular Identifier (UMI), a universal ligation overhang, and a biotin molecule. The sample was heated at 50°C for 10 minutes, then went through 3 thermal cycles (8°C for 12 s, 15°C for 45 s, 20°C for 45 s, 30°C for 30 s, 42°Cfor 120 s and 50°C for 180 s), and finally incubated at 50°C for 5 minutes. After reverse transcription, 300 μL of NIB was added and the sample was centrifuged at 1,000 g for 3 minutes to remove supernatant. Cell pellet was washed with 0.5ml of NIB and centrifuged at 1,000 g for 3 minutes. Cells were resuspended in 4,608 μL of hybridization mix (1x T4 ligation buffer, 0.32 U/μl Enzymatics RNase Inhibitor, 0.05 U/μl SUPERase RI, 0.1% Triton X-100, and 0.25 × NIB).
Hybridization and ligation
Cells in ligation mix (40 μl) were added to each of the 96 wells in the first-round barcoding plate. Each well already contained 10 μL of the appropriate DNA barcodes. The round 1 barcoding plate was incubated for 30 minutes at room temperature with gentle shaking (300rpm) to allow hybridization to occur before adding blocking strands. 10 μL of round 1 blocking oligo was added and the plate was incubated for 30 minutes at room temperature with gentle shaking (300rpm). Cells from all 96 wells were combined into a single multichannel basin. Subsequent steps in round 2 and round 3 were identical to round 1, except that 50 μL and 60 μL of pooled cells were split and added to barcodes in round 2 (total volume of 60 μl/well) and round 3 (total volume of 70 μl/well), respectively. After adding the round 3 blocking oligo, cells from all wells were combined and centrifuged at 1,000 g for 3 minutes to remove supernatant. The cell pellet was washed twice with 1ml of NIB, and centrifuged at 1,000 g for 3 minutes between washings. Cells were resuspended in the ligation mix (1x T4 ligation buffer, 0.32U/μl Enzymatics RNase Inhibitor, 20U/μl T4 DNA ligase (M0202L, NEB), 0.1% Triton X-100, 0.2 × NIB) and incubated for 30 minutes at room temperature with gentle shaking (300rpm). Cells were washed with 0.5ml NIB and resuspended in 100 μL of NIB, counted and aliquoted to 0.2ml PCR tubes with 1,000–20,000 cells per tube. SHARE-seq allows preparation of libraries from large numbers of cells and easily enables sequencing from subsets of barcoded cells allowing for easy QC of new samples and for reducing sequencing costs, useful features when performing large scale experiments.
Reverse crosslinking and affinity pull-down
NIB was added to each sample to bring the volume to 50 μL in total. 50 μL of 2 × reverse crosslinking buffer (100mM Tris pH 8.0, 100mM NaCl, and 0.04% SDS), 2 μL of 20mg/ml proteinase K, and 1 μL of SUPERase RI were mixed with each sample and incubated at 55°C for 1 hour. 5 μL of 100mM PMSF was added to the reverse crosslinked sample to inactivate proteinase K and incubated at room temperature for 10 minutes. For each sample, 10 μL of MyOne C1 Dynabeads were washed twice with 1 × B&W-T buffer (5mM Tris pH 8.0, 1M NaCl, 0.5mM EDTA, and 0.05% Tween 20) and once with 1 × B&W-T buffer supplemented with 2U/μl SUPERase RI. After washing, the beads were resuspended in 100 μL of 2 × B&W buffer (10mM Tris pH 8.0, 2M NaCl, 1mM EDTA, and 4U/μl SUPERase RI) and mixed with the sample. The mixture was rotated on an end-to-end rotator at 10rpm for 60 minutes at room temperature. The lysate was put on a magnetic stand to separate supernatant and beads.
scATAC-seq library preparation
The supernatant that contained the transposed DNA fragments was purified with QIAGEN Minelute PCR clean up kit and eluted to 20 μL of Tris buffer (pH 8.0). Fragments were amplified in 50 μL PCR reaction (1 × NENnext, 0.5 μM library-specific Ad1 primer, and 0.5 μM P7 primer. The PCR reaction was carried out at the following conditions: 72°C for 5 minutes, 98°C for 30 s, and then 5 cycles at 98°C for 10 s, 65°C for 30 s and 72°C for 1 minute. After running 5 cycles of PCR, we took a 25 μL sample, add 10 μL of PCR cocktail with 0.6 × SYBRgreen, and ran qPCR. The qPCR reactions were amplified to saturation to determine the number of cycles required for the remaining samples on the plate. The number of extra cycles was determined as the number of qPCR cycles to reach 1/3 of saturated signal. The qPCR reaction was carried out at the following conditions: 95°C for 3 minutes, and then 20 thermal cycles at 98°C for 30 s, 65°C for 20 s and 72°C for 3 minutes.
cDNA library preparation
Beads were washed three times with 1 × B&W-T buffer and once with STE (10mM Tris pH 8.0, 50mM NaCl, and 1mM EDTA) both supplemented with 1U/μl SUPERase inhibitor. Beads were resuspended in 50 μL of template switch mix (15% PEG 6000,1 × Maxima RT buffer, 4% Ficoll PM-400, 1mM dNTPs, 4U/μl NxGen RNase Inhibitor, 2.5 μM TSO, and 10U/μl Maxima H Minus Reverse Transcriptase). Beads were rotated on an end-to-end rotator at 10rpm for 30 minutes at room temperature, and then shaken at 300rpm for 90 minutes at 42°C. Beads were resuspended by pipetting every 30 minutes during agitation. After template switching, 100 μL of STE were added to each tube to dilute the sample. The supernatant was removed by placing the sample on a magnetic stand. Beads were washed with 200 μL of STE without disturbing the bead pellet. Beads were then resuspended in 55 μL of PCR mix (1 × Kapa HiFi PCR mix, 400nM P7 primer, and 400nM RNA PCR primer). The PCR reaction was carried out at the following conditions: 95°C for 3 minutes, and then 5 cycles at 98°C for 30 s, 65°C for 45 s and 72°C for 3 minutes. We then took a 2.5 μL sample, added 7.5 μL of PCR cocktail with 1 × EvaGreen (Biotium), and ran qPCR. The qPCR reactions were amplified to saturation to determine the number of cycles required for the remaining samples on the plate. The number of extra cycles was determined as the number of qPCR cycles to reach 1/3 of saturated signal. The qPCR reaction was carried out at the following conditions: 95°C for 3 minutes, and then 20 thermal cycles at 98°C for 30 s, 65°C for 20 s and 72°C for 3 minutes. Amplified cDNA was purified by 0.8 × (for cell line) or 0.6 × (for primary tissue) AMPure beads and eluted to 10 μL of Tris pH 8.0 buffer. The amount of cDNA was quantified by Qubit (ThermoFisher).
Tagmentation and scRNA-seq library preparation
100 μM Read1 oligo was annealed with an equal amount of 100 μM blocked ME-complement oligo and assembled with Tn5 as described above. For each sample, 50ng cDNA was fragmented in a 50 μL tagmentation mix (1 × TD buffer from Illumina Nextera kit (10mM Tris HCl pH 7.5, 5mM MgCl2,10% DMF final concentration), and 5 μL assembled Tn5) at 55°C for 5 minutes. Fragmented cDNA was purified with the DNA Clean and Concentrator kit (Zymo) and eluted to 10 μL of Tris pH 8.0 buffer. Purified cDNA was then mixed with tagmentation PCR mix (25 μL of NEBNext High-Fidelity 2 × PCR Master Mix, 1 μL of 25 μM P7 primer and 1 μL of 25 μM Ad1 primer with sample barcodes). PCR was carried out at the following conditions: 72°C for 5 minutes, 98°C for 30 s, and then 7 cycles at 98°C for 10 s, 65°C for 30 s and 72°C for 1 minute. The amplified library was purified by 0.7 × AMpure beads and eluted to 10 μL of Tris buffer (pH 8.0).
Quantification and sequencing
Both scATAC-seq and scRNA-seq libraries were quantified with the KAPA Library Quantification Kit and pooled for sequencing. Libraries were sequenced on the Next-seq platform (Illumina) using a 150-cycle High-Output Kit (Read 1: 30 cycles, Index 1: 99 cycles, Index 2: 8 cycles, Read 2: 30 cycles) or the Nova-seq platform (Illumina) using a 200-cycle S1 kit (Read 1: 50 cycles, Index 1: 99 cycles, Index 2: 8 cycles, Read 2: 50 cycles).
SHARE-ATAC-seq pre-processing
Raw sequencing reads were trimmed with a custom python script. Reads were aligned to the hg19 or mm10 genome using bowtie2 (Langmead and Salzberg, 2012) with (-X2000) option. For each read, there are four sets of barcodes (eight bases each) in the indexing reads. The data were demultiplexed, tolerating one mismatched base in each 8-base barcode. Reads with alignment quality < Q30, improperly paired, mapped to the unmapped contigs, chrY, and mitochondria, were discarded. Duplicates were removed using Picard tools (http://broadinstitute.github.io/picard/). Open chromatin region peaks were called on individual samples using MACS2 peak caller (Zhang et al., 2008) with the following parameters:–nomodel –nolambda –keep-dup -call-summits. Peaks from all samples were merged and peaks overlapping with ENCODE blacklisted regions (https://sites.google.com/site/anshulkundaje/projects/blacklists) were filtered out. Peak summits were extended by 150bp on each side and defined as accessible regions. Peaks were annotated to genes using Homer (Heinz et al., 2010). The fragment counts in peaks and TF scores were calculated using chromVAR (Schep et al., 2017).
SHARE-RNA-seq pre-processing
Base calls were converted to the fastq format using bcl2fastq. Reads were trimmed with a custom python script. We removed reads that do not have TTTTTT at the 11–16 bases of Read 2 allowing one mismatch. Reads were aligned to the mouse genome (version mm10) using STAR (Dobin et al., 2013) (STAR–chimOutType WithinBAM–outFilterMultimapNmax 20–outFilterMismatchNoverLmax 0.06–limitOutSJcollapsed 2000000). For species mixing experiments, reads were aligned to a combined human (hg19) and mouse (mm10) genome and only primary alignments were considered. Data were demultiplexed, tolerating one mismatched base in each 8-base barcode. Aligned reads were annotated to both exons and introns using featurecounts (Liao et al., 2014). To speed up processing, only barcode combinations with > 100 reads were retained. UMI-Tools (Smith et al., 2017) was used to collapse UMIs of aligned reads that were within 1nt mismatch of another UMI. UMIs that were only associated with one read were removed as potential ambient RNA contamination. A matrix of gene counts by cell was created with UMI-Tools. For cell line data, cells that expressed > 7,500 genes, < 300 genes, or > 1% mitochondrial reads were removed. For tissue data, cells that expressed > 10,000 genes, < 100 genes, or > 2% mitochondrial reads were removed. Expression counts (number of transcripts) for a given gene in a given cell were determined by counting unique UMIs and compiling a Digital Gene Expression (DGE) matrix. Mitochondrial genes are removed. Seurat V3 (Stuart et al., 2019) was used to scale the DGE matrix by total UMI counts, multiplied by the mean number of transcripts, and values were log transformed. To visualize data, the top 3,000 variable genes were projected into 2D space by UMAP (McInnes et al., 2018).
Peak-gene cis-association and DORC identification
To calculate peak-gene associations in cis, we considered all ATAC peaks that are located in the ± 50 kb or ± 500 kb window around each annotated TSS. We used peak counts and gene expression values to calculate the observed Spearman correlation (obs) of each peak-gene pair. To estimate the background, we used chromVAR to generate 100 background peaks for each peak by matching accessibility and GC content, and calculated the Spearman correlation coefficient between those background peaks and the gene, resulting in a null peak-gene Spearman correlation distribution that is independent of peak-gene proximity. We calculated the expected population mean (pop.mean) and expected population standard deviation (pop.sd) from expected Spearman correlations. The Z score is calculated by z = (obs-pop.mean)/pop.sd. We observed a small portion of peaks are negatively correlated with gene expression (1.2% of total peaks). For simplicity, we used a one-sided z-test to determine p-values. For peaks associated with multiple genes, we only kept peak-gene associations with the smallest p-value. Of note, when background peak correction is not performed, the peak-gene associations show strong bias toward higher accessibility regions resulting in a strong bias toward promoter associated interactions.
To define DORCs (a set of nearby peaks per gene), we rank genes by the number of significantly associated peaks (± 50kb around TSSs, p < 0.05). We used 10 and 5 peaks per gene as cutoffs for skin data and GM12878 data, respectively. We then re-calculate peak-gene association by expanding the window to ± 500kb around TSSs. Prior to calculating DORC scores, we first normalized peak counts by the total number of unique fragments in peaks per cell. Following normalization, we defined the DORC score for a gene as the sum of counts in all significantly correlated peaks per gene to obtain a cell x DORC score matrix. To calculate the DORC score per cell type, we simply compute the average across all cells per cell type.
TF regulatory network
To define the TFs that regulate DORCs, we set two criteria. First, we calculated the −log10(p value) of the Spearman correlation between mean normalized DORC gene expression and DORC score. Second, we calculated the TF motifs that are enriched in the DORC. To do this, we performed PCA on DORC scores and found the k-nearest neighbors (k-NN, k= 50) of each DORC in PC space. We then obtained a PWM score matrix (peak x TF motif) using matchMotifs (out = “scores,” p.cutoff = 0.05) function in the chromVAR package (Schep et al., 2017). The enrichment of TF motifs is defined by the −log10(p value) (KS test) of the PWM scores in peaks encompassed in the k-NN of the DORCs compared to PWM scores in GC- and accessibility- matched peaks. The GC and accessibility matched peaks were derived using the getBackgroundPeaks function in chromVAR. Based on the distribution of −log10(p value) of the Spearman correlation and TF motif enrichment, we manually set cutoffs to select TFs that regulate DORCs.
HiChIP
Processed HiChIP fragments file was downloaded from Mumbach et al. (2017) and converted to bedpe using hicpropairs2bedpe function in cLoops package (Cao et al., 2020). The loops are called per chromosome using cLoops -f bedpe.gz -o dir -w -j -s -m 4. To compare with peak-gene associations, we filtered HiChIP loops that have at least one end anchored at promoters (+2kb and −200bp of TSSs).
Comparison to other technologies
We compared the performance of SHARE-seq to sci-CAR (Cao et al., 2018), SNARE-seq (Chen et al., 2019) and Paired-seq (Zhu et al., 2019) using cell line data. We used deeply sequenced GM12878 data for SHARE-seq, published A549 cell line data for sci-CAR (Cao et al., 2018) and published cell line mixture data for SNARE-seq (Chen et al., 2019) and Paired-seq (Zhu et al., 2019). We used the authors’ count matrices, which were obtained on libraries that were sequenced to saturation. For each assay, we determine the cutoff by ranking the number of unique molecules per cell barcode. We set cutoff at the steep drop-off which indicates separation between the cell-associated barcodes and the barcodes associated with debris.
To compare SHARE-seq with other high-throughput scATAC-seq methods using cell line data, we used the approach described in previous paper (Lareau et al., 2019), and compared with published datasets, including Cusanovich et al. (2015) (GSE67446), Pliner et al. (2018) (GSE109828), Preissl et al. (2018) (GSE1000333), Lareau et al. (2019) (GSE123581), and Buenrostro et al. (2015) (GSE65360).
To compare scATAC-seq technologies in primary tissue, we generated sci-ATAC, SHARE-seq, and 10x Genomics scATAC-seq datasets on an adult mouse lung using the same sample processing method (above).
To compare SHARE-seq with other high-throughput scRNA-seq/snRNA-seq methods, we processed four adult mouse brain datasets the same way as SHARE-seq. We downloaded count matrix for nuclei (https://support.10xgenomics.com/single-cell-gene-expression/datasets/2.1.0/nuclei_2k) and cells (Zeisel et al., 2018) processed by 10x Genomics (P60 cortex, SRP135960), cells processed by Drop-seq (Saunders et al., 2018) (P60 Cortex, GSE116470), and nuclei processed by DroNc-seq (Habib et al., 2017) (PFC, GSE71585).
Cell cycle signature
To calculate the cell cycle signature, we used our previously published cell cycle gene list (Tirosh et al., 2016) and summed up the normalized cell cycle gene counts per cell. We did not regress out the cell cycle signature, because it is one of the most important signatures in TACs.
Computational pairing
To confirm if computational pairing methods correctly predict cell type in scATAC-seq based on a scRNA-seq profile, we used Seurat v3.0 (Stuart et al., 2019) to calculate gene activity scores from scATAC-seq. Next, we identified anchors between the scATAC-seq and scRNA-seq datasets using CCA (Stuart et al., 2019) and used these anchors to transfer cell-type labels from scRNA-seq to scATAC-seq. We calculated the percent of mismatch between the predicted cell type to the actual cell type.
Brain data analysis
For the brain sample, we aggregated scATAC-seq data generated using SureCell (Lareau et al., 2019) as pseudo-bulk samples, then extracted a small number of principal components (PCs) from the normalized pseudo-bulk count matrix. We next projected the scATAC-seq data to the space spanned by the PCs. The projected data was then visualized using tSNE and UMAP. To jointly cluster on ATAC and RNA signals, we used Similarity NEtwork FUSION (Wang et al., 2014) to combine the distance matrix in chromatin space and RNA space. After generating the fused distance matrix, we then calculated the k-nearest neighbor graph and found clusters using the Louvain community detection algorithm. The clusters were assigned based on both marker genes and scATAC-seq signals.
Skin scATAC-seq peak count matrix
To ensure our peak set in skin includes ATAC peaks from rare populations, we performed two rounds of peak calling. We first called peaks on filtered reads from all cells and generated a 1st-round cell-peak count matrix. We then filtered cells based on both ATAC and RNA profiles and identified clusters based on RNA profiles. We next called peaks again on aggregated pseudo bulk samples from each cluster and merged all peak summits, to generate a 2nd-round cell-peak count matrix.
Skin scATAC-seq dimension reduction
To reduce the dimension of ATAC-seq data, we tested cisTopic (Bravo González-Blas et al., 2019), chromVAR motif score and Kmer (Schep et al., 2017), and snapATAC (Preissl et al., 2018) approaches using default parameters.
Pseudotime inference
To calculate pseudotime based on scATAC-seq data, we analyzed the cells from TACs, IRS and Hair Shaft populations. We provided 10 normalized topics from cisTopic (Bravo González-Blas et al., 2019) and scATAC UMAP coordinates as inputs to Palantir (Setty et al., 2019) to construct a diffusion map (palantir.utils.run_diffusion_maps(pca_projections, n_components = 10)). The pseudotime and branch probabilities were inferred using the following parameters in Palantir (num_waypoints = 1000, knn = 30). We then defined lineages by manually examining the distribution of branch probability and selecting cells above a certain cutoff.
We calculated pseudotime based on scRNA-seq data similarly to scATAC-seq data. We provided 10 normalized principal components (PCs) from Seurat (Stuart et al., 2019) and scRNA UMAP coordinates as inputs to Palantir (Setty et al., 2019). The pseudotime and branch probabilities were inferred using the following parameters in Palantir (num_waypoints = 1000, knn = 30). We then defined lineages by manually examining the distribution of branch probability and selecting cells above a certain cutoff.
Residual analysis
Both DORC scores and gene expression were smoothed over pseudotime with local polynomial regression fitting (loess) separately, then min-max normalized. The residual for each gene was calculated by subtracting normalized gene expression from normalized DORC scores.
Chromatin potential
To calculate chromatin potential, we first smoothed DORC scores (chromatin space) and corresponding gene expression (RNA space) over a k-nearest neighbor graph (k-NN, k = 50), calculated using normalized ATAC topics from cisTopic. Next, we calculated another k-NN (k = 10), between the smoothed chromatin profile of a given cell (Catac, i), and the smoothed gene expression profile of each cell (Crna, i, j). We then calculated the distance (Di, j) between the Catac, i and the average of Crna, j in chromatin space. The arrow length is defined by normalizing Di, j. For visualization, we smoothed arrows with the 15 k-NNs in low dimensional space. For grid view, we divided the UMAP space into a 40 × 40 grid, then averaged the arrows for all the cells within each grid.
RNA velocity
RNA velocity was calculated using Velocyto (La Manno et al., 2018) with default settings. For visualization, we smoothed arrows with the 15 RNA k-NNs. For grid view, we divided the UMAP space into a 40 × 40 grid, then averaged the arrows for all the cells within each grid.
Cost
SHARE-seq significantly reduces the amount of consumed enzyme by performing all reactions (including ligation, transposition, reverse transcription, and tagmentation) in bulk (about 10,000 cells per reaction), which dramatically reduces cost. The library preparation cost for SHARE-seq in our hands is only about $433 for 100,000 cells, including approximately $50 oligos, $50 enzymes (Tn5, ligase, etc.), $121 RNase inhibitors and other consumables. By comparison, the cost of sci-CAR scales with the number of cells to be recovered. For each experiment, 96 RT reactions, 96 transposition reactions, one tagmentation reaction per 25 nuclei and 2 PCR reactions per 25 nuclei are required. It would cost more than $30,000 to prepare a sequencing library for 100,000 cells for sci-CAR.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Methods
All of the statistical details for experiments can be found in the figure legends as well as the Method Details section. For all comparisons of independent observations between two groups, two-tailed t tests were performed, with p values unless otherwise specified. Z-tests were used to describe variance across groups.
Reads in Peaks Counts for ATAC-seq Data
To generate peak count matrices for scATAC-seq data, the number of reads overlapping a given peak window in the determined peak set was calculated for each unique cell barcode. FRIP was computed as the fraction of the number of sequenced reads per cell that fall in peaks and total unique nuclear reads per cell.
TF motif score
We used TF motif scores and gene expression values to calculate the observed Spearman correlation (obs) of each TF-gene pair. The TF motif scores were derived from chromVAR and were described under the Method Details sections and figure legends. TF motif scores were root-mean-square normalized and gene expression values were normalized using the SCtransform function in Seurat. Z scores and p-values were calculated in the same way in the cis-analysis.
Collision rate estimation
We estimate the collision by implementing a solution that was used in the birthday paradox (http://matt.might.net/articles/counting-hash-collisions/). In SHARE-seq, we introduce three rounds of barcoding during hybridization and ligation with 96 barcodes for each round. The cells are aliquoted to sub-libraries and another round of barcodes is added to each sub-library during the PCR step. In each sub-library, the total number of barcode combinations D = 96 3 96 3 96 = 884,736.
The expected number of collisions for N cells in a sub-library is
Assuming we have 20,000 cells recovered per sub-library, the number of expected collisions is
The expected collision rate is 224/20000 ≈1%. The number of cells that could fit in one SHARE-seq run is 20,000 3 96 ≈2 million with about 1% collision rate.
Library size estimation
The number of unique molecules in scRNA and scATAC (library size) was estimated per cell based on the Lander-Waterman equation that is implemented in Picard tools.
where X is the number of distinct molecules in the library, N is the number of read pairs, and C is the number of distinct fragments observed in read pairs (UMIs in the case of scRNA).
Supplementary Material
Figure S1. The Principle of SHARE-Seq and Data Quality Control on Cell Line Datasets, Related to Figure 1
(A) The structure of scATAC-seq and scRNA-seq sequencing library.
(B) The expected number of barcode combinations exponentially scales with the rounds of barcoding.
(C) Expected barcode collision happens with a large number of cells (> 105).
(D) Aggregate single-cell accessibility and gene expression profiles in GM12878 cells.
(E) Scatterplot of the portion of reads in peaks (FRIP) of GM12878 ATAC-seq data.
(F) The enrichment of ATAC-seq reads around TSSs.
(G) The insert size distribution of ATAC-seq fragments.
(H and I) The SHARE-seq reproducibility between biological replicates on ATAC-seq (H) and RNA-seq (I).
(J) Aggregated ATAC-seq portion of SHARE-seq profile compares to Cusanovich et al. (2015), Pliner et al. (2018), Preissl et al. (2018), SureCell (Lareau et al., 2019), sci-ATAC-seq (LaFave et al., 2020), Flugidm C1 dataset (Buenrostro et al., 2015), and DNase-seq (ENCODE).
(K and L) The estimated library size (the unique molecules could be recovered by sequencing to saturation, estimated based on the duplication rate and recovered unique molecule) in SHARE-ATAC-seq (K) and SHARE-RNA-seq (L).
(M) The aggregated single-cell SHARE-seq accessibility profiles across different cell lines.
(N) The RNA read distribution of SHARE-seq in the genome.
Figure S2. SHARE-Seq Generates High-Quality Libraries on Multiple Tissues and Reveals Misassignment of Cell Types in Computational Paring of ATAC-RNA, Related to Figure 2
(A) The enrichment of ATAC-seq reads around TSSs.
(B) The insert size distribution of ATAC-seq fragments.
(C) Comparison of SHARE-RNA-seq to previously deposited 3′ single cell/nuclei adult mouse brain datasets (STAR Methods) in terms of the number of genes detected.
(D) ATAC UMAP and RNA UMAP colored by the cell type assigned by the joint clustering of ATAC-seq and RNA-seq data in the mouse brain.
(E–G) Comparison of SHARE-seq starting with brain nuclei or brain cells. (E) The RNA read distribution of SHARE-seq in the genome. (F) The fraction of reads in peaks (frip) of ATAC-seq fragments. (G) The insert size distribution of ATAC-seq fragments.
(H) The hair follicle cell types shift during hair follicle cycles.
(I) Schematic of a computational pipeline to process SHARE-seq data on adult mouse skin.
(J and K) The TF motif scores to gene expression correlation in GM12878 cells (J), skin cells (K). The dots color denotes the significance of the correlation.
(L) ATAC UMAP visualization with Seurat LSI, chromVAR Kmer, and snapATAC approaches (STAR Methods). Points are colored by clusters labels.
(M) Cells colored by the activity of cell cycle genes (left panel). An RNA cluster marked by high expression of cell cycle genes is highlighted in scRNA UMAP (top right panel) and scATAC UMAP space (bottom right panel).
(N) ATAC UMAP colored by computationally inferred cell type in the mouse brain. The computational pairing was performed by transferring the assigned cluster label to the ATAC cluster using Seurat (Stuart et al., 2019).
(O) Heatmap showing the proportion of cells in the joint cluster that overlaps in ATAC clusters in the mouse brain.
(P) Marker genes for each assigned cell type in the mouse brain.
(Q) Histogram showing the percentage of cells that are correctly computationally assigned for each cell type in the mouse brain.
(R) UMAP visualization of computationally inferred cell type in mouse skin. The cell type labels are transferred from RNA-seq to ATAC-seq using Seurat (Stuart et al., 2019).
(S) Histogram showing the percentage of cells that are correctly computationally assigned for each cell type in mouse skin.
(T) The percentage of cells that are correctly computationally assigned for each cell type in mouse skin using Seurat (STAR Methods). The RNA reads, ATAC reads or both RNA and ATAC reads are down-sampled to 50% or 25% of the original number of reads. The full data (35k cells) or randomly selected 5k cells are used for computation.
Figure S3. cis Associations Overlap with Known Super-Enhancers and Are Gene and Cell Stage Specific, Related to Figure 3
(A) Distribution of peak-gene associations relative to TSSs in GM12878 cell line data. The distribution is normalized to the distribution of all the ATAC-seq peaks.
(B) The number of significant peak-gene associations for each gene in GM12878 cell line data.
(C) Representative peak-gene associations in GM12878 cell line data. Loops denote the correlation of peak accessibility and RNA expression at the SH3RF3 locus, loop height represents the significance of the correlation.
(D) Fold enrichment of histone modifications in peak-gene association in GM12878 cell line data. The bars are colored by significance of the enrichment. Downloaded ENCODE histone modification ChIP-seq data were intersected with ATAC-seq peak and compared to randomly selected genomic regions.
(E) The number of genes associated with each significant peak.
(F) Histogram of the number of significant peak-gene associations per gene for all the genes (left) and super-enhancer related genes (right) in GM12878 cell line data.
(G) The distance of each significant peak-gene association (p < 0.05) to the TSS of each gene.
(H–T) Cis-regulation analysis in the mouse skin dataset.
(H and I) The distribution (H) and p value (I) of peak-gene correlation in the mouse skin dataset.
(J) The number of genes associated with each significant peak for all genes and super-enhancer related genes.
(K) The number of significant peak-gene associations for each peak.
(L) The portion of peaks associated with genes varies with chromatin accessibility level.
(M) The scatterplot showing the length of super-enhancer is not correlated with the number of associated peaks.
(N) The scatterplot showing the number of peaks around a gene and the number of associated peaks to a gene within 50 kp windows of TSSs.
(O) A cumulative distribution function plot of peak-gene associations for each gene.
(P) The overlapping DORCs identified in TAC/IRS/Hair shaft and in all cells.
(Q) DORC activity for each defined cluster, values are normalized by the min and max activity.
(R and S) Differential DORC score between medulla and cuticle/cortex (R) and between medulla and IRS (S).
(T) Scatterplot of the Dlx3 DORC score and Dlx3 gene expression.
Figure S4. Lineage Priming Validation and Characterization, Related to Figure 4
(A) The scatterplot of the Tubb6 DORC score and Tubb6 gene expression.
(B) Distribution of residual and Spearman correlation of DORC to expression of DORC-regulated genes across cells in the hair shaft lineage.
(C) Change of aggregated chromatin accessibility profiles and aggregated RNA profiles over pseudotime. Loops denote the p-value of chromatin accessibility of each peak and Wnt3 RNA expression. Loop height represents-the significance of the correlation. Grey bars denote scATAC-seq peaks. Blue bars denote peaks that are significantly associated with the Wnt3 gene. The inset shows a zoom-in image of aggregated chromatin accessibility around Wnt3 locus.
(D) Hierarchical clustering of chromatin accessibility peak and expression of associated genes for the hair shaft lineage. Cells are ordered by pseudotime.
(E and F) Histogram of the average difference (residuals) for each gene between chromatin accessibility and gene expression with (E) and without (F) bias correction.
(G) Normalized residuals between chromatin accessibility and gene expression for hair-shaft lineages.
(H) UMAP visualization of DORC and RNA expression in medulla lineage and IRS lineage.
(I) ATAC UMAP visualization of gene expression (top) and motif score (bottom) inferred from ATAC-seq.
(J) Schematic showing the workflow of calculating the TF regulatory network.
Figure S5. Characterization of Chromatin Potential, Related to Figure 5
(A) Volcano plot of differentially enriched DORCs between Notch1+ and Notch1− lineage-prime cells.
(B) Volcano plot of differentially enriched DORC-regulated genes between Notch1 + and Notch1−lineage-priming cells.
(C) The raw chromatin potential. The arrow denotes the distance between a cell in chromatin accessibility space to its most similar cell in RNA space.
(D) Raw chromatin potential was smoothed by averaging 15 k-nearest neighbors for each given cell.
(E) UMAP colored by normalized DORC score, RNA expression and residual (DORC-RNA) of Lef1, which is a known marker of HS and HS-TAC.
(F) Differential gene expression between the two roots identified by pseudotime and chromatin potential respectively. Marker genes identified in previous reports (Joost et al., 2020; Yang et al., 2017) are labeled with an asterisk (*)
(G) RNA velocity visualized on scATAC UMAP coordinates. Umap colored by pseudotime. The big arrows point to the roots identified by chromatin potential.
(H) Pseudotime inferred using scRNA-seq from SHARE-seq for cell fate decisions shown on scRNA UMAP coordinates.
(I–K) SHARE-seq on different hair follicle stages. The cell types are identified by projecting on SHARE-seq data in Figure 2E. (I,J) UMAP visualization of ATAC portion of SHARE-seq data on different hair follicle stages. (K) UMAP visualization of the distribution of Anagen III cells. The number was normalized to the total numbers of Anagen III and Anagen VI cells and smoothed in the UMAP space.
(L) The arrows denote the potential “future” RNA state (observed in another cell) which is best predicted by the current RNA state. The arrows show the most correlated neighbor in RNA space for a given cell in RNA space.
(M) The arrows denote the potential “future” chromatin state (observed in another cell) which is best predicted by the current chromatin state. The arrows show the most correlated neighbor in chromatin space for a given cell in chromatin space.
(N) Comparison of the arrow lengths between chromatin potential, RNA-RNA prediction and chromatin-chromatin prediction.
(O) The Pearson correlations between chromatin state of a cell and the potential “future” RNA state of the given cell, predicted by either chromatin potential (left) or RNA velocity (right).
(P) A scatterplot shows the differences in arrow length between chromatin potential or RNA velocity. The dot color denotes pseudotime.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD140a | eBioscience | 13-1401-82 |
| Bacterial and Virus Strains | ||
| C3013 | NEB | C3013I |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RNase Inhibitor | QIAGEN Enzymatics | Y9240L |
| SUPERase·In RNase Inhibitor | Thermo Fisher Scientific | AM2696 |
| NxGen RNase Inhibitor | Lucigen | 30281-2 |
| 16% Formaldehyde (w/v) | Thermo Fisher Scientific | 28906 |
| Glycine | Sigma Aldrich | 50049 |
| 1M Tris HCl pH 7.5 | Thermo Fisher Scientific | 15567027 |
| 1M Tris HCl pH 8.0 | Thermo Fisher Scientific | 15568025 |
| 5M NaCl | Thermo Fisher Scientific | AM9760G |
| 1M MgCl2 | Sigma Aldrich | 63069 |
| 10% NP-40 Surfact-Amps | Thermo Fisher Scientific | 28324 |
| Buffer EB | QIAGEN | 19086 |
| DNA Clean & Concentrator-5 | Zymo | D4014 |
| PEG 6000 | Sigma Aldrich | 528877 |
| Maxima H Minus Reverse Transcriptase (200 U/μL) | Thermo Fisher Scientific | EP0753 |
| Deoxynucleotide (dNTP) Solution Mix | NEB | N0447L |
| T4 DNA ligase | NEB | M0202L |
| T4 DNA Ligase Reaction Buffer | NEB | B0202S |
| Proteinase K from Tritirachium album | Sigma Aldrich | P2308-100MG |
| Sodium Dodecyl Sulfate 20% (SDS) Solution | VWR | 97062-440 |
| Phenylmethanesulfonyl fluoride (PMSF) | Sigma Aldrich | P7626 |
| 2-Propanol | Sigma Aldrich | I9516 |
| 0.5M EDTA | Thermo Fisher Scientific | AM9260G |
| TWEEN20 | Sigma-Aldrich | P9416-100ML |
| Ficoll PM-400 (20%) | Sigma-Aldrich | F5415-25ML |
| KAPA HiFi HotStart ReadyMix | Fisher Scientific | NC0295239 |
| AMPure XP | Beckman Coulter | A63880 |
| Ethanol | Sigma-Aldrich | 8.18760.2500 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32854 |
| FlashGel DNA Cassettes | Lonza | 57031 |
| Dithiothreitol (DTT), 0.1M Solution | Thermo Fisher Scientific | 707265ML |
| NEBNext High-Fidelity 2X PCR Master Mix | NEB | M0541L |
| Glycerol | Thermo Fisher Scientific | 15514011 |
| Protease Inhibitor Cocktail | Sigma-Aldrich | P8340 |
| TRIS-Acetate Buffer 0.2M, pH 7.8 | Bioworld | 40120265-2 |
| Potassium acetate | Sigma-Aldrich | 95843-100ML-F |
| Magnesium acetate | Sigma-Aldrich | 63052-100ML |
| SYBR Green I Nucleic Acid Gel Stain | Thermo Fisher Scientific | S7563 |
| Dimethylformamide (DMF) | Thermo Fisher Scientific | 20673A |
| Quick-Load® Purple 100 bp DNA Ladder | NEB | N0551S |
| Gel Loading Dye, Purple (6X) | NEB | B7024S |
| PBS | Thermo Fisher Scientific | 10010049 |
| Deposited Data | ||
| SHARE-seq data | This manuscript | GEO: GSE140203 |
| Visualization of SHARE-seq skin data | This manuscript | https://buenrostrolab.shinyapps.io/skinnetwork/ |
| sci-ATAC data | Cusanovich et al., 2015 | GEO: GSE67446 |
| sci-ATAC data | Pliner et al., 2018 | GEO: GSE109828 |
| sci-ATAC data | Preissl et al., 2018 | GEO: GSE1000333 |
| SureCell scATAC data | Lareau et al., 2019 | GEO: GSE123581 |
| Flugidm scATAC data | Buenrostro et al., 2015 | GEO: GSE65360 |
| sci-ATAC data | LaFave et al., 2020 | GEO: GSE134812 |
| 10x snRNA brain data | 10x genomics | https://support.10xgenomics.com/single-cell-gene-expression/datasets/2.1.0/nuclei_2k |
| 10x scRNA brain data | Zeisel et al., 2018 | NCBI SRA: SRP135960 |
| Drop-seq brain data | Saunders et al., 2018 | GEO: GSE116470 |
| DroNc-seq brain data | Habib et al., 2017 | GEO: GSE71585 |
| HiChIP data | Mumbach et al., 2017 | GEO: GSM2705041 |
| sci-CAR A549 cell line data | Cao et al., 2018 | GEO: GSE117089 |
| SNARE-seq cell line mixture data | Chen et al., 2019 | GEO: GSE126074 |
| Paired-seq cell line mixture data | Zhu et al., 2019 | GEO: GSE130399 |
| Skin ChIP data | Adam et al., 2015 | GEO: GSE61316 |
| Skin ChIP data | Lien et al., 2011 | GEO: GSE31239 |
| Experimental Models: Cell Lines | ||
| GM12878 | Coriell Institute | GM12878 |
| NIH/3T3 | ATCC | CRL-1658 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | Jackson Labs | stock 000664 |
| Oligonucleotides | ||
| Oligo seqeuences | see Table S1 | N/A |
| Software and Algorithms | ||
| R (v3.5.3) | R Development Core Team, 2019 | https://www.R-project.org |
| chromVAR R package (v0.2.0) | Schep et al., 2017 | https://github.com/GreenleafLab/chromVAR |
| bowtie2 (v2.3.3.1) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS2 (v2.1.2) | Zhang et al., 2008 | https://github.com/macs3-project/MACS |
| samtools (v1.9) | Li et al., 2009 | http://samtools.sourceforge.net |
| Picard toolkit (2.14.1-SNAPSHOT) | NA | http://broadinstitute.github.io/picard |
| cLoops (v0.93) | Cao et al., 2018 | https://github.com/YaqiangCao/cLoops |
| STAR (v2.7.5) | Dobin et al., 2012 | https://github.com/alexdobin/STAR |
| UMI-Tools | Smith et al., 2017 | https://github.com/CGATOxford/UMI-tools |
| featurecounts | Liao et al., 2014 | http://subread.sourceforge.net |
| Seurat (v3) | Stuart et al., 2019 | https://satijalab.org/seurat/ |
| Similarity NEtwork FUSION | Wang et al., 2014 | http://compbio.cs.toronto.edu/SNF/SNF/Software.html |
| cisTopic (v2) | Bravo González-Blas et al., 2019 | https://github.com/aertslab/cisTopic |
| snapATAC | Fang et al., 2019 | https://github.com/r3fang/snATAC |
| Palantir (v0.2.6) | Setty et al., 2019 | https://github.com/dpeerlab/Palantir |
| Velocyto | La Manno et al., 2018 | http://velocyto.org |
Highlights.
Cell states marked by chromatin and gene expression are correlated but distinct
Lineage-determining genes are marked by domains of regulatory chromatin (DORCs)
DORCs are accessible prior to gene expression, foreshadowing lineage choice
Chromatin accessibility lineage priming predicts cell fate decisions
ACKNOWLEDGMENTS
We thank members of the Regev and Buenrostro labs for critical reading of the manuscript and helpful discussions. We are grateful to Jonathan Strecker for providing Tn5 and the Bauer Core at Harvard for providing sequencing services. J.D.B. and the Buenrostro lab acknowledge support from the Allen Distinguished Investigator Program through the Paul G. Allen Frontiers Group, the Chan Zuckerberg Initiative, and the NIH New Innovator Award (DP2). A.R. is an Investigator of the Howard Hughes Medical Institute. Work was supported by the NHGRI Center for Cell Circuits (to A.R.), the Klarman Cell Observatory (to A.R.), a grant from the BRAIN Initiative (to A.R.), the Smith Family Foundation Odyssey Award (to Y.-C.H.), and NIH R01-AR070825 (to Y.-C.H.). Y.-C.H. is a Pew Scholar and a NYSCF – Robertson Investigator. B.Z. is an awardee of the Charles A. King Trust postdoctoral research fellowship.
Footnotes
DECLARATION OF INTERESTS
A.R. is a founder of and equity holder in Celsius Therapeutics, an equity holder in Immunitas, and an SAB member of Thermo Fisher Scientific, Syros Pharmaceutical, Asimov, and Neogene Therapeutics. J.D.B. holds patents related to ATAC-seq and is an SAB member of Camp4 and seqWell. J.D.B., A.R., and S.M. submitted a provisional patent application based on this work.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.09.056.
REFERENCES
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, and Fuchs E (2015). Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, and Fuchs E (2004). Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648. [DOI] [PubMed] [Google Scholar]
- Bravo González-Blas C, Minnoye L, Papasokrati D, Aibar S, Hulselmans G, Christiaens V, Davie K, Wouters J, and Aerts S (2019). cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 16, 397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, Daza RM, McFaline-Figueroa JL, Packer JS, Christiansen L, et al. (2018). Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Chen Z, Chen X, Ai D, Chen G, McDermott J, Huang Y, Guo X, and Han JJ (2020). Accurate loop calling for 3D genomic data with cLoops. Bioinformatics 36, 666–675. [DOI] [PubMed] [Google Scholar]
- Chen S, Lake BB, and Zhang K (2019). High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 37, 1452–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I, Zhao D, Bar C, Valdes VJ, Dauber-Decker KL, Nguyen MB, Nakayama M, Rendl M, Bickmore WA, Koseki H, et al. (2018). PRC1 Fine-tunes Gene Repression and Activation to Safeguard Skin Development and Stem Cell Specification. Cell Stem Cell 22, 726–739.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, and Shendure J (2015). Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Wang D, Burgmaier JE, Teng Y, Romano R-A, Sinha S, and Yi R (2018). Single Cell and Open Chromatin Analysis Reveals Molecular Origin of Epidermal Cells of the Skin. Dev. Cell 47, 133. [DOI] [PubMed] [Google Scholar]
- Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS, et al. (2019). A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 176, 1516. [DOI] [PubMed] [Google Scholar]
- Genander M, Cook PJ, Ramsköld D, Keyes BE, Mertz AF, Sandberg R, and Fuchs E (2014). BMP signaling and its pSMAD1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell 15, 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González AJ, Setty M, and Leslie CS (2015). Early enhancer establishment and regulatory locus complexity shape transcriptional programs in hematopoietic differentiation. Nat. Genet 47, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, and Regev A (2016). Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, and Sharp PA (2017). A Phase Separation Model for Transcriptional Control. Cell 169, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-C, Li L, and Fuchs E (2014). Emerging interactions between skin stem cells and their niches. Nat. Med 20, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D-Y, Lin Y-T, Jan P-S, Hwang Y-C, Liang S-T, Peng Y, Huang C-Y, Wu H-C, and Lin C-T (2008). Transcription factor SOX-5 enhances nasopharyngeal carcinoma progression by down-regulating SPARC gene expression. J. Pathol 214, 445–455. [DOI] [PubMed] [Google Scholar]
- Joost S, Jacob T, Sun X, Annusver K, La Manno G, Sur I, and Kasper M (2018). Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing. Cell Rep. 25, 585–597.e7. [DOI] [PubMed] [Google Scholar]
- Joost S, Annusver K, Jacob T, Sun X, Dalessandri T, Sivan U, Sequeira I, Sandberg R, and Kasper M (2020). The Molecular Anatomy of Mouse Skin during Hair Growth and Rest. Cell Stem Cell 26, 441–457.e7. [DOI] [PubMed] [Google Scholar]
- Kelsey G, Stegle O, and Reik W (2017). Single-cell epigenomics: Recording the past and predicting the future. Science 358, 69–75. [DOI] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. (2014). Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, Pokholok D, Aryee MJ, Steemers FJ, Lebofsky R, and Buenrostro JD (2019). Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol 37, 916–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson AJM, Johnsson P, Hagemann-Jensen M, Hartmanis L, Faridani OR, Reinius B, Segerstolpe Å, Rivera CM, Ren B, and Sandberg R (2019). Genomic encoding of transcriptional burst kinetics. Nature 565, 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legué E, and Nicolas J-F (2005). Hair follicle renewal: organization of stem cells in the matrix and the role of stereotyped lineages and behaviors. Development 132, 4143–154. [DOI] [PubMed] [Google Scholar]
- Li Q, Peterson KR, Fang X, and Stamatoyannopoulos G (2002). Locus control regions. Blood 100, 3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). Feature Counts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lien W-H, Guo X, Polak L, Lawton LN, Young RA, Zheng D, and Fuchs E (2011). Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 9, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, and Amir A (2018). Homeostasis of protein and mRNA concentrations in growing cells. Nat. Commun 9, 4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Saul N, and Großtberger L (2018). UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw 3, 861. [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, and Fuchs E (2001). Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 15, 1688–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, Farmer A, Fordyce P, Linnarsson S, and Greenleaf W (2018). High-throughput chromatin accessibility profiling at single-cell resolution. Nat. Commun 9, 3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill P, Mo R, Fu H, Grachtchouk M, Kim PCW, Dlugosz AA, and Hui C-C (2003). Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 17, 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar SE, Willert K, Salinas PC, Roelink H, Nusse R, Sussman DJ, and Barsh GS (1999). WNT signaling in the control of hair growth and structure. Dev. Biol. 207, 133–149. [DOI] [PubMed] [Google Scholar]
- Mumbach MR, Satpathy AT, Boyle EA, Dai C, Gowen BG, Cho SW, Nguyen ML, Rubin AJ, Granja JM, Kazane KR, et al. (2017). Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet 49, 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. (2011). Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, and Natoli G (2013). Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171. [DOI] [PubMed] [Google Scholar]
- Pan Y, Lin M-H, Tian X, Cheng H-T, Gridley T, Shen J, and Kopan R (2004). γ-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev. Cell 7, 731–743. [DOI] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, and Joyner AL (2000). Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593–1605. [DOI] [PubMed] [Google Scholar]
- Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, and Sandberg R (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 24, 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. (2018). Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol. Cell 71, 858–871.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, et al. (2018). Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci 21, 432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2019). R: A language and environment for statistical computing (R Foundation for Statistical Computing; ). [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. (2011). Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol 29, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, and Wysocka J (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haber-man AM, and Greco V (2012). Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487, 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, and Greco V (2013). Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, et al. (2018). Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusk N (2019). Multi-omics single-cell analysis. Nat. Methods 16, 679. [DOI] [PubMed] [Google Scholar]
- Salzer MC, Lafzi A, Berenguer-Llergo A, Youssif C, Castellanos A, Solanas G, Peixoto FO, Stephan-Otto Attolini C, Prats N, Aguilera M, et al. (2018). Identity Noise and Adipogenic Traits Characterize Dermal Fibroblast Aging. Cell 175, 1575–1590.e22. [DOI] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, and Greenleaf WJ (2017). chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, and Pe’er D (2019). Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol 37, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Bernstein BE, and Buenrostro JD (2019). Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet 51, 19–25. [DOI] [PubMed] [Google Scholar]
- Smith T, Heger A, and Sudbery I (2017). UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, et al. (2008). The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 68, 537–544. [DOI] [PubMed] [Google Scholar]
- Spitz F, and Furlong EEM (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet 13, 613–626. [DOI] [PubMed] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, and Smibert P (2017). Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. (2016). Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, and Goldenberg A (2014). Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 11, 333–337. [DOI] [PubMed] [Google Scholar]
- Weiner A, Lara-Astiaso D, Krupalnik V, Gafni O, David E, Winter DR, Hanna JH, and Amit I (2016). Co-ChIP enables genome-wide mapping of histone mark co-occurrence at single-molecule resolution. Nat. Biotechnol 34, 953–961. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin T, Gonzalez D, Rompolas P, and Greco V (2018). Flexible fate determination ensures robust differentiation in the hair follicle. Nat. Cell Biol 20, 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Adam RC, Ge Y, Hua ZL, and Fuchs E (2017). Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices. Cell 169, 483–496.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, and Hsu Y-C (2017). Emerging roles of transit-amplifying cells in tissue regeneration and cancer. Wiley Interdiscip. Rev. Dev. Biol 6 10.1002/wdev.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Tsai P-C, Gonzalez-Celeiro M, Chung O, Boumard B, Perdigoto CN, Ezhkova E, and Hsu Y-C (2016). Hair follicles’ transit-amplifying cells govern concurrent dermal adipocyte production through Sonic Hedgehog. Genes Dev. 30, 2325–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Yu M, Huang H, Juric I, Abnousi A, Hu R, Lucero J, Behrens MM, Hu M, and Ren B (2019). An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol 26, 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The Principle of SHARE-Seq and Data Quality Control on Cell Line Datasets, Related to Figure 1
(A) The structure of scATAC-seq and scRNA-seq sequencing library.
(B) The expected number of barcode combinations exponentially scales with the rounds of barcoding.
(C) Expected barcode collision happens with a large number of cells (> 105).
(D) Aggregate single-cell accessibility and gene expression profiles in GM12878 cells.
(E) Scatterplot of the portion of reads in peaks (FRIP) of GM12878 ATAC-seq data.
(F) The enrichment of ATAC-seq reads around TSSs.
(G) The insert size distribution of ATAC-seq fragments.
(H and I) The SHARE-seq reproducibility between biological replicates on ATAC-seq (H) and RNA-seq (I).
(J) Aggregated ATAC-seq portion of SHARE-seq profile compares to Cusanovich et al. (2015), Pliner et al. (2018), Preissl et al. (2018), SureCell (Lareau et al., 2019), sci-ATAC-seq (LaFave et al., 2020), Flugidm C1 dataset (Buenrostro et al., 2015), and DNase-seq (ENCODE).
(K and L) The estimated library size (the unique molecules could be recovered by sequencing to saturation, estimated based on the duplication rate and recovered unique molecule) in SHARE-ATAC-seq (K) and SHARE-RNA-seq (L).
(M) The aggregated single-cell SHARE-seq accessibility profiles across different cell lines.
(N) The RNA read distribution of SHARE-seq in the genome.
Figure S2. SHARE-Seq Generates High-Quality Libraries on Multiple Tissues and Reveals Misassignment of Cell Types in Computational Paring of ATAC-RNA, Related to Figure 2
(A) The enrichment of ATAC-seq reads around TSSs.
(B) The insert size distribution of ATAC-seq fragments.
(C) Comparison of SHARE-RNA-seq to previously deposited 3′ single cell/nuclei adult mouse brain datasets (STAR Methods) in terms of the number of genes detected.
(D) ATAC UMAP and RNA UMAP colored by the cell type assigned by the joint clustering of ATAC-seq and RNA-seq data in the mouse brain.
(E–G) Comparison of SHARE-seq starting with brain nuclei or brain cells. (E) The RNA read distribution of SHARE-seq in the genome. (F) The fraction of reads in peaks (frip) of ATAC-seq fragments. (G) The insert size distribution of ATAC-seq fragments.
(H) The hair follicle cell types shift during hair follicle cycles.
(I) Schematic of a computational pipeline to process SHARE-seq data on adult mouse skin.
(J and K) The TF motif scores to gene expression correlation in GM12878 cells (J), skin cells (K). The dots color denotes the significance of the correlation.
(L) ATAC UMAP visualization with Seurat LSI, chromVAR Kmer, and snapATAC approaches (STAR Methods). Points are colored by clusters labels.
(M) Cells colored by the activity of cell cycle genes (left panel). An RNA cluster marked by high expression of cell cycle genes is highlighted in scRNA UMAP (top right panel) and scATAC UMAP space (bottom right panel).
(N) ATAC UMAP colored by computationally inferred cell type in the mouse brain. The computational pairing was performed by transferring the assigned cluster label to the ATAC cluster using Seurat (Stuart et al., 2019).
(O) Heatmap showing the proportion of cells in the joint cluster that overlaps in ATAC clusters in the mouse brain.
(P) Marker genes for each assigned cell type in the mouse brain.
(Q) Histogram showing the percentage of cells that are correctly computationally assigned for each cell type in the mouse brain.
(R) UMAP visualization of computationally inferred cell type in mouse skin. The cell type labels are transferred from RNA-seq to ATAC-seq using Seurat (Stuart et al., 2019).
(S) Histogram showing the percentage of cells that are correctly computationally assigned for each cell type in mouse skin.
(T) The percentage of cells that are correctly computationally assigned for each cell type in mouse skin using Seurat (STAR Methods). The RNA reads, ATAC reads or both RNA and ATAC reads are down-sampled to 50% or 25% of the original number of reads. The full data (35k cells) or randomly selected 5k cells are used for computation.
Figure S3. cis Associations Overlap with Known Super-Enhancers and Are Gene and Cell Stage Specific, Related to Figure 3
(A) Distribution of peak-gene associations relative to TSSs in GM12878 cell line data. The distribution is normalized to the distribution of all the ATAC-seq peaks.
(B) The number of significant peak-gene associations for each gene in GM12878 cell line data.
(C) Representative peak-gene associations in GM12878 cell line data. Loops denote the correlation of peak accessibility and RNA expression at the SH3RF3 locus, loop height represents the significance of the correlation.
(D) Fold enrichment of histone modifications in peak-gene association in GM12878 cell line data. The bars are colored by significance of the enrichment. Downloaded ENCODE histone modification ChIP-seq data were intersected with ATAC-seq peak and compared to randomly selected genomic regions.
(E) The number of genes associated with each significant peak.
(F) Histogram of the number of significant peak-gene associations per gene for all the genes (left) and super-enhancer related genes (right) in GM12878 cell line data.
(G) The distance of each significant peak-gene association (p < 0.05) to the TSS of each gene.
(H–T) Cis-regulation analysis in the mouse skin dataset.
(H and I) The distribution (H) and p value (I) of peak-gene correlation in the mouse skin dataset.
(J) The number of genes associated with each significant peak for all genes and super-enhancer related genes.
(K) The number of significant peak-gene associations for each peak.
(L) The portion of peaks associated with genes varies with chromatin accessibility level.
(M) The scatterplot showing the length of super-enhancer is not correlated with the number of associated peaks.
(N) The scatterplot showing the number of peaks around a gene and the number of associated peaks to a gene within 50 kp windows of TSSs.
(O) A cumulative distribution function plot of peak-gene associations for each gene.
(P) The overlapping DORCs identified in TAC/IRS/Hair shaft and in all cells.
(Q) DORC activity for each defined cluster, values are normalized by the min and max activity.
(R and S) Differential DORC score between medulla and cuticle/cortex (R) and between medulla and IRS (S).
(T) Scatterplot of the Dlx3 DORC score and Dlx3 gene expression.
Figure S4. Lineage Priming Validation and Characterization, Related to Figure 4
(A) The scatterplot of the Tubb6 DORC score and Tubb6 gene expression.
(B) Distribution of residual and Spearman correlation of DORC to expression of DORC-regulated genes across cells in the hair shaft lineage.
(C) Change of aggregated chromatin accessibility profiles and aggregated RNA profiles over pseudotime. Loops denote the p-value of chromatin accessibility of each peak and Wnt3 RNA expression. Loop height represents-the significance of the correlation. Grey bars denote scATAC-seq peaks. Blue bars denote peaks that are significantly associated with the Wnt3 gene. The inset shows a zoom-in image of aggregated chromatin accessibility around Wnt3 locus.
(D) Hierarchical clustering of chromatin accessibility peak and expression of associated genes for the hair shaft lineage. Cells are ordered by pseudotime.
(E and F) Histogram of the average difference (residuals) for each gene between chromatin accessibility and gene expression with (E) and without (F) bias correction.
(G) Normalized residuals between chromatin accessibility and gene expression for hair-shaft lineages.
(H) UMAP visualization of DORC and RNA expression in medulla lineage and IRS lineage.
(I) ATAC UMAP visualization of gene expression (top) and motif score (bottom) inferred from ATAC-seq.
(J) Schematic showing the workflow of calculating the TF regulatory network.
Figure S5. Characterization of Chromatin Potential, Related to Figure 5
(A) Volcano plot of differentially enriched DORCs between Notch1+ and Notch1− lineage-prime cells.
(B) Volcano plot of differentially enriched DORC-regulated genes between Notch1 + and Notch1−lineage-priming cells.
(C) The raw chromatin potential. The arrow denotes the distance between a cell in chromatin accessibility space to its most similar cell in RNA space.
(D) Raw chromatin potential was smoothed by averaging 15 k-nearest neighbors for each given cell.
(E) UMAP colored by normalized DORC score, RNA expression and residual (DORC-RNA) of Lef1, which is a known marker of HS and HS-TAC.
(F) Differential gene expression between the two roots identified by pseudotime and chromatin potential respectively. Marker genes identified in previous reports (Joost et al., 2020; Yang et al., 2017) are labeled with an asterisk (*)
(G) RNA velocity visualized on scATAC UMAP coordinates. Umap colored by pseudotime. The big arrows point to the roots identified by chromatin potential.
(H) Pseudotime inferred using scRNA-seq from SHARE-seq for cell fate decisions shown on scRNA UMAP coordinates.
(I–K) SHARE-seq on different hair follicle stages. The cell types are identified by projecting on SHARE-seq data in Figure 2E. (I,J) UMAP visualization of ATAC portion of SHARE-seq data on different hair follicle stages. (K) UMAP visualization of the distribution of Anagen III cells. The number was normalized to the total numbers of Anagen III and Anagen VI cells and smoothed in the UMAP space.
(L) The arrows denote the potential “future” RNA state (observed in another cell) which is best predicted by the current RNA state. The arrows show the most correlated neighbor in RNA space for a given cell in RNA space.
(M) The arrows denote the potential “future” chromatin state (observed in another cell) which is best predicted by the current chromatin state. The arrows show the most correlated neighbor in chromatin space for a given cell in chromatin space.
(N) Comparison of the arrow lengths between chromatin potential, RNA-RNA prediction and chromatin-chromatin prediction.
(O) The Pearson correlations between chromatin state of a cell and the potential “future” RNA state of the given cell, predicted by either chromatin potential (left) or RNA velocity (right).
(P) A scatterplot shows the differences in arrow length between chromatin potential or RNA velocity. The dot color denotes pseudotime.
Data Availability Statement
Gene Expression Omnibus: SHARE-seq data are deposited under accession number GEO: GSE140203, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140203
The R Shiny-based web application for data visualization is accessible here: https://buenrostrolab.shinyapps.io/skinnetwork/