Abstract
Intellectual abilities and their clinical presentations are extremely heterogeneous in autism spectrum disorder (ASD). The main causes of ASD remain unclear. ASD is frequently associated with sleep disorders. Biologic rhythms are complex systems interacting with the environment and controlling several physiological pathways, including brain development and behavioral processes. Recent findings have shown that the deregulation of the core clock neurodevelopmental signaling is correlated with ASD clinical presentation. One of the main pathways involved in developmental cognitive disorders is the canonical WNT/β-catenin pathway. Circadian clocks have a main role in some tissues by driving circadian expression of genes involved in physiologic and metabolic functions. In ASD, the increase of the canonical WNT/β-catenin pathway is enhancing by the dysregulation of circadian rhythms. ASD progression is associated with a major metabolic reprogramming, initiated by aberrant WNT/β-catenin pathway, the aerobic glycolysis. This review focuses on the interest of circadian rhythms dysregulation in metabolic reprogramming in ASD through the aberrant upregulation of the canonical WNT/β-catenin pathway.
Subject terms: Molecular neuroscience, Autism spectrum disorders
Introduction
Intellectual abilities and their clinical presentations are extremely heterogeneous in autism spectrum disorder (ASD). ASD includes Asperger, autism and pervasive developmental disorder-not otherwise specified (PDD-NOS). ASD is characterized by numerous and complex etiologies, including inflammation, metabolic, environmental or genetic determinants. Nevertheless, the main causes of ASD remain unclear. ASD is diagnosed within the first 3 years of life. Social interaction impairment, repetitive and restrictive behaviors or stereotypic patterns of behaviors characterize the different symptoms of ASD1. Early diagnosis is major issue for better prognosis and therapeutic care2,3.
ASD is frequently associated with sleep disorders4. Brain structures and functions development are considered as dynamic processes, dependent on genetic and social/physical environmental cues, involving homeostasis processes at each time, and maturation of new functions in time. Biologic rhythms are complex systems interacting with the environment and controlling several physiological pathways, including brain development5 and behavioral processes6.
Recent studies have shown that the deregulation of the core neurodevelopmental signaling is correlated with ASD clinical presentation. One of the main pathways implicated in developmental cognitive disorders is the canonical WNT/β-catenin pathway7. Several genetic mutations observed in ASD are linked with the dysregulation of the WNT/β-catenin pathway through interactions between chromodomain helicase DNA binding protein 8 (CHD8) and CTNNB1 (β-catenin)8. In many ASD findings, the WNT/β-catenin pathway is increased7,9–11.
Circadian clocks have a main role in some tissues by driving circadian expression of genes involved in physiologic and metabolic functions12. One of the key integrator of these complex mechanisms is the canonical WNT/β-catenin pathway13,14. ASD progression is associated with a major metabolic reprogramming, initiated by aberrant WNT/β-catenin pathway, the aerobic glycolysis11. In parallel, the dysregulation of circadian rhythms (CRs) upregulates the WNT/β-catenin pathway15, which in turn participates to the ASD initiation. This review focuses on the interest of CRs dysregulation in metabolic reprogramming in ASD through the aberrant upregulation of the canonical WNT/β-catenin pathway.
Circadian rhythms
CRs are major biological phenomena found in all universal processes. Their endogenous characteristic is an innate oscillation associated with a period of over 1 day. All the studied organisms show this oscillatory process. Numerous cell functions present temporal variations driven by these oscillatory and circadian ways, including gene expression, metabolic reprogramming, and molecular and cellular pathways. Different integration levels allow the study of the CRs, as endocrinal, physiological, neuronal cell behaviors. Although the coordination and the modulation of CRs are organized by specific pacemaker structures, the primary circadian oscillations are controlled at the cell level. These oscillations are determined by numerous clock genes16. The control of the circadian clock is based on an intracellular temporal tracking system that allows anterior organisms to change direction and thus adapt their behavior and the physiology of their life span17. It is well known that in many animals species, circadian clock is formed by a specific set of transcription factors which constitutes its molecular architecture. These determinants are both used in positive and negative feedback, which are modulated through a cell-autonomous manner18.
Endogenous oscillations generate a freewheeling period, which is close to 24 h, to maintain for the organism constant ambient conditions. These oscillators, at the molecular level, are based on the products of clock regulator genes organized in a transcriptional feedback loop. Circadian oscillations are the product of these post-transcriptional modifications of proteins19. A complex loop operates with clock gene transcriptional activators and in turn clock genes with a negative feedback role inhibiting their expression by disrupting the activity of their activators20. Several input pathways involve environmental information which interacts with the different compounds of the oscillators. The oscillators are synchronized with the 24 h solar day. The input pathways generate a day-time to transpose it by the oscillators to the output pathways. These output pathways control and regulate the expression of circadian clock genes to generate the rhythmicity. Moreover, the output pathways are predicted to be rhythmic and then controlled by the clock gene transcription factors. These compounds, in turn, regulate downstream the circadian clock genes in a time-of-day-specific manner21. This system can synchronize with its environmental time by its internal clock. To respect the environment, the input pathways are vital to maintain this timing for oscillators. A the process named entrainment, the input pathways can reset the activity of the oscillators to stay in a conform 24-h period of the environment21. Environmental cues can be detected by input pathways which in turn can modulate many mechanisms to control the activity or level of compounds of oscillators to keep a correct time-of-day expression. This phenomenon is observed in several environmental cues, including nutrition, social interactions and temperature22,23. Furthermore, the clock allows a strategy named gating to restrict responses to environmental cues at some day times. Diurnal mammals are insensitive to a light pulse during the day. Nevertheless, during the night, a light pulse can advance or delay the clock to synchronize diurnal mammals with the environment18. Environmental signals can interact with molecular oscillators in some cells in complex multicellular organisms. In unicellular organisms, each cell is modulated by oscillators in response to light24. However, in multicellular organisms, only a part of the cells has sensory capabilities leading to clock oscillators. The oscillators, and thus the overall rhythmicity of organisms, are concentrated into compounds including a master pacemaker and peripheral oscillators25. Face to these sensory inputs, the organism presents some nervous systems which possess environmental cue abilities as a central oscillators or pacemakers rather than to individual cells. In humans, the sensory clock inputs are localized in the brain, where signals from the master pacemaker leads to oscillators in some tissues of the organism.
Nonvisual retinal ganglion cells receive and perceive the light, and transmit this information to the master pacemaker (localized in the hypothalamus) through neural connections. The central pacemaker synchronizes oscillators to the other tissues by using circadian input pathways from the nervous system to peripheral cell systems. Moreover, to maintain the entrainment of these peripheral oscillators by the environment, this central system ensures that cellular oscillations within tissues are properly in phase to provide resonance between individual cellular rhythms6. Melatonin operates as a major synchronizer in humans and provides temporal feedback to oscillators within the nervous system to control the circadian phase and the stability of the rhythm26. In humans, as in other mammals, melatonin is considered as the main influencer of CRs through its action on receptors in the nervous system27.
The sleep–wake pattern is controlled by both CRs and homeostasis. Sleep pressure has been enhanced during the phase of waking and then decreases during the phase of sleeping. The sleep–wake pattern is controlled by the cycle of light–darkness28. Through a feedback, sleep–wake pattern can also control the CRs. For many studies, this pattern can be defined as an interface between environmental information (social, mood and cognition) and CRs29. Moreover, these two patters are influenced by melatonin4. Melatonin level is modulated by the light–dark exposure. Melatonin is associated with sleep initiation. Mutations in Clock, Bmal1, Cry1, Cry2 genes lead to the initiation of some alterations in sleep time and sleep fragmentation30,31.
Circadian clock
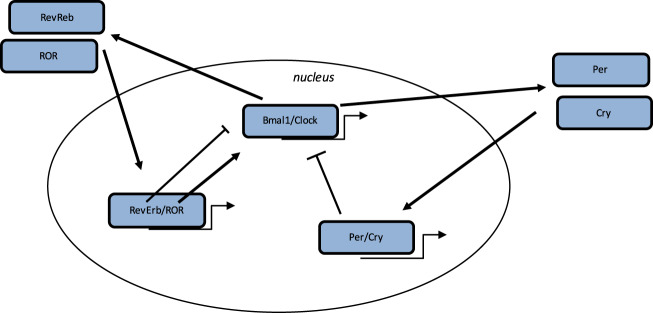
Some biological mechanisms in humans are controlled by the circadian “clock” (circadian locomotors output cycles kaput) (Fig. 1). The circadian clock is localized in the hypothalamic suprachiasmatic nucleus (SCN). CRs are endogenous and entrainable free-running periods that last ~24 h. Several transcription factors can control and modulate CRs. These factors are named circadian locomotor output cycles kaput (Clock), brain and muscle aryl-hydrocarbon receptor nuclear translocator-like 1 (Bmal1), Period 1 (Per1), Period 2 (Per2), Period 3 (Per3) and Cryptochrome (Cry1 and Cry2)32,33. They are modulated by positive and negative self-loop-regulation mediated by CRs18,34. Clock and Bmal1 heterodimerize and lead to the transcription of Per1, Per2, Cry1 and Cry2 (ref. 35). The Per/Cry heterodimer inhibits its activation by a negative feedback. It translocates back to the nucleus to directly downregulate the Clock/Bmal1 complex and then inhibits its own transcription35. The Clock/Bmal1 heterodimer activates the transcription of retinoic acid-related orphan nuclear receptors, Rev-Erbs and retinoid-related orphan receptors (RORs). Through a positive feedback loop, RORs activate the transcription of Bmal1, whereas through a negative feedback loop, Rev-Erbs downregulate their transcription35.
Fig. 1. Circadian clock genes.
The clock process is a stimulatory circle, involving the Bmal1/Clock heterodimer which activates the transcription of Per and Cry genes, and the inhibitory feedback circle with the Per/Cry heterodimer which translocates to the nucleus and which represses the transcription of the Clock and Bmal1 genes. An additional circle implicates the RORs and Rev-Erbs factors with a positive feedback by RORs and a negative feedback by Rev-Erbs.
Circadian clocks in ASD
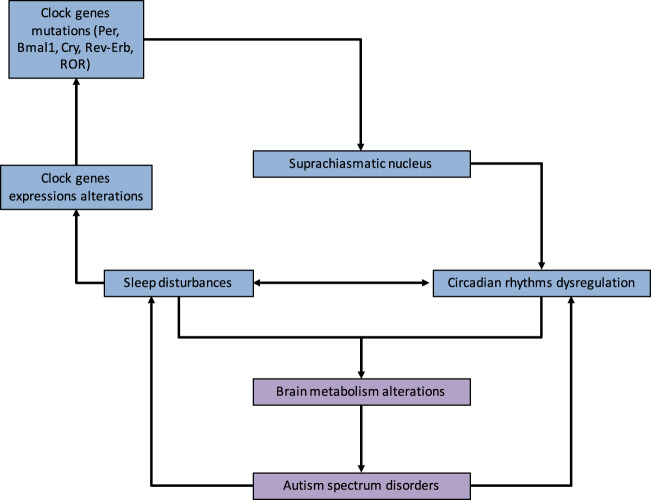
Sleep disorders are frequently associated with ASD (Fig. 2)36. Patients suffering from ASD are more correlated with falling asleep anxiety, sleep disorders and CRs disturbance36. Moreover, the prevalence of sleep disturbance is associated with cognitive impairments36–38. In parallel, melatonin levels have been dysregulated in ASD patients4. Melatonin diurnal levels are decreased as well as melatonin nocturnal levels, showing a global decrease in melatonin production in ASD children and a dysregulation of day–night rhythm39–42. Deregulation in the production of melatonin in ASD patients has been observed due to HIOMT (hydroxiindole O-methyltransferase) deficiency43. Recent studies have shown a potential interest in melatonin therapy in the treatment of sleep onset disorder for ASD patients44. Significant allelic correlation has been observed for clock genes, including Per1, and other nucleotids45–47. Genes involved in ASD pathogenesis are part of pathway networks enhanced in synapse formation, including Neuroxin-Neuroligin genes and in the alteration of the balance excitation-inhibition48. Parvalbumin expressing interneurons play a main role in ASD initiation. The knockout of Parvalbumin is associated with core symptoms of ASD patients49. CRs dysregulation may downregulate the maturation of Parvalbumin cells and then the timing of critical period of plasticity50. The dysregulation of CRs could impact the temporal organization of brain maturation and could have a cascade effect on several brain functions. Negative environmental conditions (sleep deprivation, stress…) could impact the CRs and thus redox homeostasis and transcriptional control of Parvalbumin genes involved in synapse formation and maturation of brain functions.
Fig. 2. Circadian rhythms and autism spectrum disorder.
Relationship between ASD, circadian rhythms and sleep disturbance. Alterations in clock genes and melatonin pathway contribute to the dysregulation of circadian sleep rhythmicity. Circadian rhythms deregulation leads to brain metabolism alterations contributing to ASD. In a negative feedback, ASD symptomatology reinforces circadian rhythms and sleep disturbances creating a self-reinforcing circle.
Melatonin and ASD
Melatonin plays a main role in neurodevelopment51. Disturbances in sleep–wake rhythm in ASD could be due to the dysregulation of melatonin52. Sleep latency and nocturnal and early awakenings have been reported in ASD41. In ASD, the nocturnal secretion of melatonin is low53. Intellectual disabilities are associated with melatonin abnormalities54. However, in the Down syndrome, the production of melatonin is normal whereas melatonin production is increased in the Fragile X patients55. Several therapeutic studies have focused on the interest of melatonin strategy in ASD56. Melatonin use in ASD children is associated with improvement of communication57, stereotyped behaviors58, anxiety59 and social withdrawal58. Nevertheless, melatonin is influenced by age and pubertal stage60, and in these studies the autistic behavioral impairment was generally enough described57.
Aerobic glycolysis
In mammalian cells, glucose is the main source of energy. All tissues need ATP to function normally. Cells produce ATP by a careful drop in oxidation state from energy-rich molecules like glucose, through the cell respiration process, down to product CO2 at the end. This process occurs in an aerobic or anaerobic manner, depending on whether oxygen is available. Glycolysis is the first step in glucose metabolism signaling and occurs in the cytosol of all cells. The presence of O2 is important because of oxidation of glucose under aerobic conditions results in ~32 molecules of ATP per molecule of glucose. Under anaerobic conditions, only two molecules of ATP can be produced. Aerobic glycolysis can occur in these two stages. The first occurs in the cytosol and involves the conversion of glucose to pyruvate resulting in NADH production and generating two molecules of ATP. In normal conditions, when oxygen is available, the energy contained in NADH is further released via re-oxidization of the mitochondrial chain and leads to the release of 30 molecules of ATP per molecule of glucose. Pyruvate is reduced to lactate, instead of re-oxidized, under aerobic glycolysis61. Thus, glucose is metabolized in order to produce ATP, by a cytosolic glycolysis and oxygen-dependent mitochondrial physiological mechanism. Glucose entry into the tricarboxylic acid (TCA) cycle is modulated by pyruvate dehydrogenase complex (PDH)62. Pyruvate is oxidized to acetyl-coA in mitochondria by the PDH. Acetyl-coA translocates to the TCA cycle for oxidation. Under aerobic glycolysis, pyruvate is converted into lactate in the cytosol by lactate dehydrogenase A (LDH-A). Moreover, aerobic glycolysis is caused by the involvement of hexokinase 2 (HK2) instead of HK1 and pyruvate kinase M2 (PKM2) instead of PKM1 (ref. 63). This phenomenon is called aerobic glycolysis or the Warburg effect.
Aerobic glycolysis in ASD
Few studies have investigated the potential impact of aerobic glycolysis in ASD, and thus the expression of the glycolytic enzymes (Fig. 2)11. However, numerous findings have highlighted increased lactate levels in ASD patients64–71.
Pyruvate production is stimulated65,67, but with an increased ratio of lactate-to-pyruvate65,68 showing a high glucose metabolism and LDH-A activity in ASD patients. Moreover, LDH-A expression has been increased in ASD patients71. A recent study has presented a decrease level of pH correlated with high levels of lactate in ASD patients72. These observations could suggest an increase of aerobic glycolysis in ASD since the dysregulation of this balance has been proposed as a candidate cause of ASD73.
CRs and aerobic glycolysis
Few studies have focused on the relationship between CRs and aerobic glycolysis (Fig. 2). Nevertheless, this relation could be mainly interesting in the development of tumors74. In the same way, melatonin expression and modulation by CRs in cancers is associated with the disruption of the aerobic glycolysis75–77. Thermodynamic and energy reprogramming highlight this relation in fibrosis78, neurodegenerative diseases79,80 and cancers81. The importance of 24-h fluctuations in the aerobic glycolysis and the availability of nicotinamide adenine dinucleotide phosphate hydrogen (NADPH) in cancer has been shown through the consideration of the redox influence on NADPH82.
The canonical WNT/β-catenin pathway
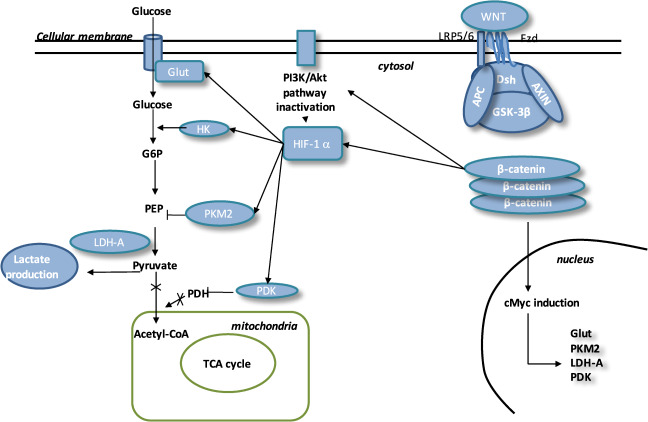
The Wingless/Int (WNT) pathway is a family of secreted lipid-modified glycoproteins (Fig. 3)83. Several nervous molecular mechanisms are modulated by the WNT/β-catenin pathway, including development of synapses in the central nervous system84,85, synaptogenesis86,87 and control of synaptic formation84,88. Numerous pathophysiologic signalings are mediated by the dysregulation of this pathway, including cancers89,90, fibrosis91, neurodegenerative diseases80 and angiogenesis13,92. Stimulation of β-catenin signaling needs the presence of the complex LRP5 /LRP693. LRP5 has a main role while LRP6 presents a minor role in the retinal vascularization94,95. Disheveled (Dsh) forms a complex with Axin, and this prevents the phosphorylation of β-catenin by glycogen synthase kinase-3β (GSK-3β). Then, β-catenin accumulation in the cytosol is observed and translocates to the nucleus to bind T-cell factor/lymphoid enhancer factor (TCF/LEF) co-transcription factors. This nuclear binding allows the transcription of WNT-responsive genes, such as cyclin D1, c-Myc, PDK1, MCT-1 (refs. 96,97).
Fig. 3. The canonical WNT/β-catenin pathway.
Inactivated WNT: Under physiologic circumstances, the cytoplasmic β-catenin is linked to its destruction complex, consisting of APC, AXIN and GSK-3β. β-catenin is phosphorylated by GSK-3β. Thus, phosphorylated β-catenin is destroyed in the proteasome. Then, the cytoplasmic level of β-catenin is kept low in the non-presence of WNT ligands. If β-catenin is not accumulated in the nucleus, the TCF/LEF complex does not stimulate the target genes. DKK1 inhibits the WNT/β-catenin pathway through binding to WNT ligands or LRP5/6. Activated WNT: When WNT ligands activate both FZD and LRP5/6, DSH is stimulated and phosphorylated by FZD. Phosphorylated DSH in turn activates AXIN, which comes off the β-catenin destruction complex. Thus, β-catenin escapes from phosphorylation and then accumulates in the cytoplasm. The accumulated cytosolic β-catenin moves into the nucleus, where it interacts with TCF/LEF and stimulates the transcription of target genes.
WNT ligands’ absence is associated with cytosolic β-catenin phosphorylation by GSK-3β.
A destruction complex is composed by tumor suppressor adenomatous polyposis coli (APC), Axin, GSK-3β and β-catenin. Then, phosphorylated β-catenin is destroyed in the proteasome. WNT inhibitors, including DKKs and SFRPs, control the WNT/β-catenin pathway by preventing its ligand–receptor interactions98.
GSK-3β, a neuron-specific intracellular serin-threonin kinase, is the major inhibitor of the WNT pathway99. GSK-3β regulates numerous pathophysiological pathways (cell membrane signaling, neuronal polarity and inflammation)100–102. GSK-3β downregulates β-catenin cytosolic accumulation and then its nuclear translocation100. GSK-3β diminishes β-catenin, mTOR pathway, HIF-1α and VEGF expression103.
The canonical WNT/β-catenin pathway in ASD
Some studies have observed the potential role of over-increased WNT/β-catenin pathway in ASD patients104–106. Furthermore, many gene compounds are associated with the development of ASD, including WNT2 ligand107, the WNT target MET (hepatocyte growth factor receptor)108,109, CHD8 (chromo-helicase domain protein 8) and DYRK1 (refs. 110–112). Other components of the WNT pathway have been involved in ASD pathogenesis, including WNT1 (ref. 113), WNT2 (ref. 107), WNT3 (ref. 114), WNT7A115, APC116–118, β-catenin8,119, TCF4 (refs. 120,121) and TCF7 (ref. 122). The CTNNB1 gene, a β-catenin encoder, is a major controller of the WNT pathway. Its expression is associated with sporadic ASD. Alterations of CTNNB1 correspond to intellectual disability and thus, ASD8. In mice, the expression of CTNNB1 expression is correlated with brain development123. Knockout of CTNNB1 in the Parvalbumin interneurons leads to the impairment of social interactions and enhancement of repetitive behaviors.
Phosphatase and tensin homolog protein (PTEN) inhibition is associated with the increase of the WNT pathway and high risk of ASD development124–126. In Purkinje cells, PTEN inhibition impairs social relation, behavior and deficits in motor learning127,128. PTEN and β-catenin control each other to normal brain growth and development129.
CRs and WNT/β-catenin pathway
RORs are upstream effectors of the WNT/β-catenin pathway130. By this interaction, circadian genes can modulate the cell cycle progression131. A Bmal1 knockdown can downregulate the WNT/β-catenin pathway132. In wild-type mice, the levels of WNT-related genes are higher than those observed in Bmal1 knockdown mice133,134. The proliferation and progression of cell cycle are controlled by Bmal1 by activating the WNT/β-catenin pathway135. Bmal1 involves the β-catenin transcription, diminishes the β-catenin degradation and then inhibits GSK-3β activity136. In the intestinal mucosa of ApcMin/+ mice, the degradation of Per2 leads to β-catenin increase by circadian disruption137.
In normal conditions, the core circadian genes operate in accurate feedback loops and keep the molecular clockworks in the SCN. They allow the control of peripheral clocks18,34. Per1 and Per2 maintain cell CRs and modulate cell-related gene activity, such as c-Myc, so as to sustain the physiologic cell cycle138,139.
Aerobic glycolysis and WNT/β-catenin pathway
Some reports have highlighted that the WNT/β-catenin pathway is closely associated and a main effector of the aerobic glycolysis (Fig. 4)11,78,140–142. The PI3K/Akt pathway stimulates the glucose metabolism to enhance protein and lipid synthesis143. Moreover, PI3K/Akt pathway increases the glucose metabolism to protect cells against reactive oxygen species (ROS) stress induced by activated HIF-1α and decreasing the glucose entry into the TCA cycle144. HIF-1α stimulates pyruvate dehydrogenase kinase (PDK) to phosphorylate PDH and inactivates it, leading to cytosolic pyruvate being shunted into lactate by LDH-A145. HIF-1α is transcriptionally activated by PI3K/Akt/mTOR pathway through 4E-BP1 and STAT3 (refs. 146–151).
Fig. 4. Interactions between WNT pathway and energy metabolism in ASD.
In ASD, the WNT pathway is activated. In the presence of WNT ligands, cytosolic β-catenin is accumulated in cytosol and GSK-3β is inhibited. APC and Axin combine with GSK-3β and DSH to form a complex with LRP 5/6 and FZD. β-catenin translocates to the nucleus and binds to TCF/LEF co-transcription factor. WNT target genes, such as cMyc, are activated. β-catenin accumulation increases the level of PI3K/Akt pathway and results in the activation of HIF-1α. Activated HIF-1α stimulates Glut, HK, PKM2, LDH-A and PDK1. Activation of HIF-1α involves PKM2 translocation to the nucleus. PKM2 activates PEP cascade and the formation of pyruvate. PKM2 binds to β-catenin and induces cMyc-mediated expression of glycolytic enzymes (Glut, LDH-A, PDK1). Activation of Glut and HK involves glucose hyper-metabolism with increase in glucose transport and phosphorylation rates. PDK1 inhibits PDH to downregulate the pyruvate entrance into mitochondria. Lactate production is activated by LDH-A. This is aerobic glycolysis.
Numerous studies have observed that the WNT/β-catenin pathway can downregulate the pyruvate oxidation in the TCA cycle140,152. WNT/β-catenin pathway, by activating both the PI3K/Akt/mTOR pathway and HIF-1α, can lead to aerobic glycolysis140,152,153. PI3K/Akt pathway can also control the β-catenin accumulation and then the expression of the downstream genes154. c-Myc directly stimulates the HIF-1α155, PDK and lactate transporter (MCT-1) expressions152. The stimulation of HIF-1α leads to the overexpression of glucose transporters (Glut), hexokinase (HK), pyruvate kinase (PK), PDK1 and LDH-A156–159.
Conclusion
Changes in energy metabolism are modulated by abnormal CRs in ASD patients. In ASD, the canonical WNT/β-catenin pathway is increased. Energy behaviors of metabolic enzymes in ASD are modified by this upregulation of the WNT/β-catenin pathway leading to the enhancement of aerobic glycolysis and thus the production of lactate. This explains the glucose hyper-metabolism observed in ASD. WNT pathway is driven by the CRs and operates under a circadian regime evolving to changes in energy metabolism. CRs directly contribute to the regulation of the molecular pathways WNT/β-catenin pathway involved in the reprogramming of cellular energy metabolism enabling ASD.
Author contributions
All authors listed have contributed to the work, and approved it for submitting to publication.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Posar A, Resca F, Visconti P. Autism according to diagnostic and statistical manual of mental disorders 5(th) edition: the need for further improvements. J. Pediatr. Neurosci. 2015;10:146–148. doi: 10.4103/1817-1745.159195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ospina MB, et al. Behavioural and developmental interventions for autism spectrum disorder: a clinical systematic review. PLoS ONE. 2008;3:e3755. doi: 10.1371/journal.pone.0003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altemeier WA, Altemeier LE. How can early, intensive training help a genetic disorder? Pediatr. Ann. 2009;38:167–170. doi: 10.3928/00904481-20090301-01. [DOI] [PubMed] [Google Scholar]
- 4.Geoffray M-M, Nicolas A, Speranza M, Georgieff N. Are circadian rhythms new pathways to understand Autism Spectrum Disorder? J. Physiol. Paris. 2016;110:434–438. doi: 10.1016/j.jphysparis.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi Y, Ye Z, Hensch TK. Clock genes control cortical critical period timing. Neuron. 2015;86:264–275. doi: 10.1016/j.neuron.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 7.Kwan V, Unda BK, Singh KK. Wnt signaling networks in autism spectrum disorder and intellectual disability. J. Neurodev. Disord. 2016;8:45. doi: 10.1186/s11689-016-9176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caracci MO, Ávila ME, De, Ferrari GV. Synaptic Wnt/GSK3β signaling hub in autism. Neural Plast. 2016;2016:9603751. doi: 10.1155/2016/9603751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulligan KA, Cheyette BNR. Neurodevelopmental perspectives on Wnt signaling in psychiatry. Mol. Neuropsychiatry. 2017;2:219–246. doi: 10.1159/000453266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vallée A, Vallée J-N. Warburg effect hypothesis in autism spectrum disorders. Mol. Brain. 2018;11:1. doi: 10.1186/s13041-017-0343-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards J, Gumz ML. Advances in understanding the peripheral circadian clocks. FASEB J. 2012;26:3602–3613. doi: 10.1096/fj.12-203554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 14.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Matsu-Ura T, Moore SR, Hong CI. WNT takes two to tango: molecular links between the circadian clock and the cell cycle in adult stem cells. J. Biol. Rhythms. 2018;33:5–14. doi: 10.1177/0748730417745913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackey SR, Golden SS, Ditty JL. The itty-bitty time machine genetics of the cyanobacterial circadian clock. Adv. Genet. 2011;74:13–53. doi: 10.1016/B978-0-12-387690-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/S0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 18.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 19.Hastings, M. H., Maywood, E. S. & Brancaccio, M. The mammalian circadian timing system and the suprachiasmatic nucleus as its pacemaker. Biology8, 13 (2019). [DOI] [PMC free article] [PubMed]
- 20.Atger F, Mauvoisin D, Weger B, Gobet C, Gachon F. Regulation of mammalian physiology by interconnected circadian and feeding rhythms. Front. Endocrinol. 2017;8:42. doi: 10.3389/fendo.2017.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson CH, Elliott JA, Foster R. Entrainment of circadian programs. Chronobiol. Int. 2003;20:741–774. doi: 10.1081/CBI-120024211. [DOI] [PubMed] [Google Scholar]
- 22.Carneiro BTS, Araujo JF. Food entrainment: major and recent findings. Front. Behav. Neurosci. 2012;6:83. doi: 10.3389/fnbeh.2012.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloch G, Herzog ED, Levine JD, Schwartz WJ. Socially synchronized circadian oscillators. Proc. Biol. Sci. 2013;280:20130035. doi: 10.1098/rspb.2013.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cassone VM. Effects of melatonin on vertebrate circadian systems. Trends Neurosci. 1990;13:457–464. doi: 10.1016/0166-2236(90)90099-V. [DOI] [PubMed] [Google Scholar]
- 27.Weaver DR, Stehle JH, Stopa EG, Reppert SM. Melatonin receptors in human hypothalamus and pituitary: implications for circadian and reproductive responses to melatonin. J. Clin. Endocrinol. Metab. 1993;76:295–301. doi: 10.1210/jcem.76.2.8381796. [DOI] [PubMed] [Google Scholar]
- 28.Wirz-Justice A, Terman M. Chronotherapeutics (light and wake therapy) as a class of interventions for affective disorders. Handb. Clin. Neurol. 2012;106:697–713. doi: 10.1016/B978-0-444-52002-9.00042-5. [DOI] [PubMed] [Google Scholar]
- 29.Wulff K, Gatti S, Wettstein JG, Foster RG. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat. Rev. Neurosci. 2010;11:589–599. doi: 10.1038/nrn2868. [DOI] [PubMed] [Google Scholar]
- 30.Naylor E, et al. The circadian clock mutation alters sleep homeostasis in the mouse. J. Neurosci. 2000;20:8138–8143. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laposky A, et al. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28:395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 32.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl Acad. Sci. USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 34.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/S0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 35.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006;15:R271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 36.Kotagal S, Broomall E. Sleep in children with autism spectrum disorder. Pediatr. Neurol. 2012;47:242–251. doi: 10.1016/j.pediatrneurol.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Allik H, Larsson J-O, Smedje H. Sleep patterns in school-age children with Asperger syndrome or high-functioning autism: a follow-up study. J. Autism Dev. Disord. 2008;38:1625–1633. doi: 10.1007/s10803-008-0543-0. [DOI] [PubMed] [Google Scholar]
- 38.Richdale AL, Schreck KA. Sleep problems in autism spectrum disorders: prevalence, nature, & possible biopsychosocial aetiologies. Sleep. Med. Rev. 2009;13:403–411. doi: 10.1016/j.smrv.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Nir I, et al. Brief report: circadian melatonin, thyroid-stimulating hormone, prolactin, and cortisol levels in serum of young adults with autism. J. Autism Dev. Disord. 1995;25:641–654. doi: 10.1007/BF02178193. [DOI] [PubMed] [Google Scholar]
- 40.Bourgeron T. The possible interplay of synaptic and clock genes in autism spectrum disorders. Cold Spring Harb. Symp. Quant. Biol. 2007;72:645–654. doi: 10.1101/sqb.2007.72.020. [DOI] [PubMed] [Google Scholar]
- 41.Tordjman S, et al. Advances in the research of melatonin in autism spectrum disorders: literature review and new perspectives. Int. J. Mol. Sci. 2013;14:20508–20542. doi: 10.3390/ijms141020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tordjman S, et al. Autism as a disorder of biological and behavioral rhythms: toward new therapeutic perspectives. Front. Pediatr. 2015;3:1. doi: 10.3389/fped.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melke J, et al. Abnormal melatonin synthesis in autism spectrum disorders. Mol. Psychiatry. 2008;13:90–98. doi: 10.1038/sj.mp.4002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guénolé F, et al. Melatonin for disordered sleep in individuals with autism spectrum disorders: systematic review and discussion. Sleep. Med. Rev. 2011;15:379–387. doi: 10.1016/j.smrv.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Nicholas B, et al. Association of Per1 and Npas2 with autistic disorder: support for the clock genes/social timing hypothesis. Mol. Psychiatry. 2007;12:581–592. doi: 10.1038/sj.mp.4001953. [DOI] [PubMed] [Google Scholar]
- 46.Yang Z, et al. Circadian-relevant genes are highly polymorphic in autism spectrum disorder patients. Brain Dev. 2016;38:91–99. doi: 10.1016/j.braindev.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Goto M, et al. Role of a circadian-relevant gene NR1D1 in brain development: possible involvement in the pathophysiology of autism spectrum disorders. Sci. Rep. 2017;7:43945. doi: 10.1038/srep43945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger RH, Miller AL, Seifer R, Cares SR, LeBourgeois MK. Acute sleep restriction effects on emotion responses in 30- to 36-month-old children. J. Sleep. Res. 2012;21:235–246. doi: 10.1111/j.1365-2869.2011.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wöhr M, et al. Lack of parvalbumin in mice leads to behavioral deficits relevant to all human autism core symptoms and related neural morphofunctional abnormalities. Transl. Psychiatry. 2015;5:e525. doi: 10.1038/tp.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hensch TK. Critical period plasticity in local cortical circuits. Nat. Rev. Neurosci. 2005;6:877–888. doi: 10.1038/nrn1787. [DOI] [PubMed] [Google Scholar]
- 51.de Faria Poloni J, Feltes BC, Bonatto D. Melatonin as a central molecule connecting neural development and calcium signaling. Funct. Integr. Genomics. 2011;11:383–388. doi: 10.1007/s10142-011-0221-8. [DOI] [PubMed] [Google Scholar]
- 52.Glickman G. Circadian rhythms and sleep in children with autism. Neurosci. Biobehav. Rev. 2010;34:755–768. doi: 10.1016/j.neubiorev.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 53.Tordjman S, Anderson GM, Pichard N, Charbuy H, Touitou Y. Nocturnal excretion of 6-sulphatoxymelatonin in children and adolescents with autistic disorder. Biol. Psychiatry. 2005;57:134–138. doi: 10.1016/j.biopsych.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 54.Yamashita Y, Matsuishi T, Murakami Y, Kato H. Sleep disorder in Rett syndrome and melatonin treatment. Brain Dev. 1999;21:570. doi: 10.1016/S0387-7604(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 55.Reiter RJ, Tan DX, Manchester LC, Qi W. Biochemical reactivity of melatonin with reactive oxygen and nitrogen species: a review of the evidence. Cell Biochem. Biophys. 2001;34:237–256. doi: 10.1385/CBB:34:2:237. [DOI] [PubMed] [Google Scholar]
- 56.Lord C. What is melatonin? Is it a useful treatment for sleep problems in autism? J. Autism Dev. Disord. 1998;28:345–346. doi: 10.1023/A:1026072922104. [DOI] [PubMed] [Google Scholar]
- 57.Wright B, et al. Melatonin versus placebo in children with autism spectrum conditions and severe sleep problems not amenable to behaviour management strategies: a randomised controlled crossover trial. J. Autism Dev. Disord. 2011;41:175–184. doi: 10.1007/s10803-010-1036-5. [DOI] [PubMed] [Google Scholar]
- 58.Malow B, et al. Melatonin for sleep in children with autism: a controlled trial examining dose, tolerability, and outcomes. J. Autism Dev. Disord. 2012;42:1729–1737. doi: 10.1007/s10803-011-1418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wasdell MB, et al. A randomized, placebo-controlled trial of controlled release melatonin treatment of delayed sleep phase syndrome and impaired sleep maintenance in children with neurodevelopmental disabilities. J. Pineal Res. 2008;44:57–64. doi: 10.1111/j.1600-079X.2007.00528.x. [DOI] [PubMed] [Google Scholar]
- 60.Touitou Y. Human aging and melatonin. Clinical relevance. Exp. Gerontol. 2001;36:1083–1100. doi: 10.1016/S0531-5565(01)00120-6. [DOI] [PubMed] [Google Scholar]
- 61.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 62.Bender T, Martinou J-C. The mitochondrial pyruvate carrier in health and disease: to carry or not to carry? Biochim. Biophys. Acta. 2016;1863:2436–2442. doi: 10.1016/j.bbamcr.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 63.L‚veillard T, Sahel J-A. Metabolic and redox signaling in the retina. Cell. Mol. Life Sci. 2017;74:3649–3665. doi: 10.1007/s00018-016-2318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moreno H, et al. [Clinical heterogeneity of the autistic syndrome: a study of 60 families] Invest. Clin. 1992;33:13–31. [PubMed] [Google Scholar]
- 65.László A, Horváth E, Eck E, Fekete M. Serum serotonin, lactate and pyruvate levels in infantile autistic children. Clin. Chim. Acta. 1994;229:205–207. doi: 10.1016/0009-8981(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 66.Oliveira G, et al. Mitochondrial dysfunction in autism spectrum disorders: a population-based study. Dev. Med. Child Neurol. 2005;47:185–189. doi: 10.1017/S0012162205000332. [DOI] [PubMed] [Google Scholar]
- 67.Germanò E, et al. [Neurobiology of autism: study of a sample of autistic children] Minerva Pediatr. 2006;58:109–120. [PubMed] [Google Scholar]
- 68.Correia C, et al. Brief report: high frequency of biochemical markers for mitochondrial dysfunction in autism: no association with the mitochondrial aspartate/glutamate carrier SLC25A12 gene. J. Autism Dev. Disord. 2006;36:1137–1140. doi: 10.1007/s10803-006-0138-6. [DOI] [PubMed] [Google Scholar]
- 69.Weissman JR, et al. Mitochondrial disease in autism spectrum disorder patients: a cohort analysis. PLoS ONE. 2008;3:e3815. doi: 10.1371/journal.pone.0003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol. Psychiatry. 2012;17:290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khemakhem, A. M., Frye, R. E., El-Ansary, A., Al-Ayadhi, L. & Bacha, A. B. Novel biomarkers of metabolic dysfunction is autism spectrum disorder: potential for biological diagnostic markers. Metab. Brain Dis.32, 1983–1997 (2017). [DOI] [PubMed]
- 72.Hagihara, H. et al. Decreased brain pH as a shared endophenotype of psychiatric disorders. Neuropsychopharmacology10.1038/npp.2017.167 (2017). [DOI] [PMC free article] [PubMed]
- 73.Marín O. Interneuron dysfunction in psychiatric disorders. Nat. Rev. Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 74.Cao Y, Wang R-H. Associations among metabolism, circadian rhythm and age-associated diseases. Aging Dis. 2017;8:314–333. doi: 10.14336/AD.2016.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blask DE, et al. Light exposure at night disrupts host/cancer circadian regulatory dynamics: impact on the Warburg effect, lipid signaling and tumor growth prevention. PLoS ONE. 2014;9:e102776. doi: 10.1371/journal.pone.0102776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dauchy RT, et al. Daytime blue light enhances the nighttime circadian melatonin inhibition of human prostate cancer growth. Comp. Med. 2015;65:473–485. [PMC free article] [PubMed] [Google Scholar]
- 77.Mao L, et al. Melatonin suppression of aerobic glycolysis (Warburg effect), survival signalling and metastasis in human leiomyosarcoma. J. Pineal Res. 2016;60:167–177. doi: 10.1111/jpi.12298. [DOI] [PubMed] [Google Scholar]
- 78.Vallée, A., Lecarpentier, Y. & Vallée, J. -N. Thermodynamic aspects and reprogramming cellular energy metabolism during the fibrosis process. Int. J. Mol. Sci. 18, 2537 (2017). [DOI] [PMC free article] [PubMed]
- 79.Vallée A, Lecarpentier Y, Guillevin R, Vallée J-N. Reprogramming energetic metabolism in Alzheimer’s disease. Life Sci. 2018;193:141–152. doi: 10.1016/j.lfs.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 80.Vallée A, Lecarpentier Y, Guillevin R, Vallée J-N. Thermodynamics in neurodegenerative diseases: interplay between canonical WNT/beta-catenin pathway-PPAR gamma, energy metabolism and circadian rhythms. Neuromolecular Med. 2018;20:174–204. doi: 10.1007/s12017-018-8486-x. [DOI] [PubMed] [Google Scholar]
- 81.Vallée A, Lecarpentier Y, Guillevin R, Vallée J-N. Thermodynamics in gliomas: interactions between the canonical WNT/beta-catenin pathway and PPAR gamma. Front. Physiol. 2017;8:352. doi: 10.3389/fphys.2017.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Méndez I, Vázquez-Martínez O, Hernández-Muñoz R, Valente-Godínez H, Díaz-Muñoz M. Redox regulation and pro-oxidant reactions in the physiology of circadian systems. Biochimie. 2016;124:178–186. doi: 10.1016/j.biochi.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 83.Al-Harthi L. Wnt/β-catenin and its diverse physiological cell signaling pathways in neurodegenerative and neuropsychiatric disorders. J. Neuroimmune Pharmacol. 2012;7:725–730. doi: 10.1007/s11481-012-9412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahmad-Annuar A, et al. Signaling across the synapse: a role for Wnt and Dishevelled in presynaptic assembly and neurotransmitter release. J. Cell Biol. 2006;174:127–139. doi: 10.1083/jcb.200511054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- 86.Itasaki N, et al. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development. 2003;130:4295–4305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- 87.Caricasole A, et al. Functional characterization of WNT7A signaling in PC12 cells: interaction with A FZD5 x LRP6 receptor complex and modulation by Dickkopf proteins. J. Biol. Chem. 2003;278:37024–37031. doi: 10.1074/jbc.M300191200. [DOI] [PubMed] [Google Scholar]
- 88.Sharma K, et al. High-throughput genetic screen for synaptogenic factors: identification of LRP6 as critical for excitatory synapse development. Cell Rep. 2013;5:1330–1341. doi: 10.1016/j.celrep.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vallée, A., Lecarpentier, Y., Guillevin, R. & Vallée, J.-N. Opposite interplay between the canonical WNT/β-catenin pathway and PPAR gamma: a potential therapeutic target in gliomas. Neurosci. Bull. 34, 573–588 (2018). [DOI] [PMC free article] [PubMed]
- 90.Vallée A, Guillevin R, Vallée J-N. Vasculogenesis and angiogenesis initiation under normoxic conditions through Wnt/β-catenin pathway in gliomas. Rev. Neurosci. 2018;29:71–91. doi: 10.1515/revneuro-2017-0032. [DOI] [PubMed] [Google Scholar]
- 91.Vallée A, Lecarpentier Y, Guillevin R, Vallée J-N. Interactions between TGF-β1, canonical WNT/β-catenin pathway and PPAR γ in radiation-induced fibrosis. Oncotarget. 2017;8:90579–90604. doi: 10.18632/oncotarget.21234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fuhrmann S. Wnt signaling in eye organogenesis. Organogenesis. 2008;4:60–67. doi: 10.4161/org.4.2.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye X, et al. Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell. 2009;139:285–298. doi: 10.1016/j.cell.2009.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou Y, et al. Canonical WNT signaling components in vascular development and barrier formation. J. Clin. Invest. 2014;124:3825–3846. doi: 10.1172/JCI76431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang W, et al. Critical endothelial regulation by LRP5 during retinal vascular development. PLoS ONE. 2016;11:e0152833. doi: 10.1371/journal.pone.0152833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shtutman M, et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Angers, S. & Moon, R. T. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol.10, 468–477 (2009). [DOI] [PubMed]
- 98.Cruciat C-M, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb. Perspect. Biol. 2013;5:a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-catenin is a target for the ubiquitin–proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu D, Pan W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem. Sci. 2010;35:161–168. doi: 10.1016/j.tibs.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hur E-M, Zhou F-Q. GSK3 signalling in neural development. Nat. Rev. Neurosci. 2010;11:539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ambacher KK, et al. The JNK- and AKT/GSK3β- signaling pathways converge to regulate puma induction and neuronal apoptosis induced by trophic factor deprivation. PLoS ONE. 2012;7:e46885. doi: 10.1371/journal.pone.0046885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yokosako K, et al. Glycolysis in patients with age-related macular degeneration. Open Ophthalmol. J. 2014;8:39–47. doi: 10.2174/1874364101408010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.De Ferrari GV, Moon RT. The ups and downs of Wnt signaling in prevalent neurological disorders. Oncogene. 2006;25:7545–7553. doi: 10.1038/sj.onc.1210064. [DOI] [PubMed] [Google Scholar]
- 105.Okerlund ND, Cheyette BNR. Synaptic Wnt signaling-a contributor to major psychiatric disorders? J. Neurodev. Disord. 2011;3:162–174. doi: 10.1007/s11689-011-9083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kalkman HO. A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol. Autism. 2012;3:10. doi: 10.1186/2040-2392-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wassink TH, et al. Evidence supporting WNT2 as an autism susceptibility gene. Am. J. Med. Genet. 2001;105:406–413. doi: 10.1002/ajmg.1401. [DOI] [PubMed] [Google Scholar]
- 108.Boon EMJ, van der Neut R, van de Wetering M, Clevers H, Pals ST. Wnt signaling regulates expression of the receptor tyrosine kinase met in colorectal cancer. Cancer Res. 2002;62:5126–5128. [PubMed] [Google Scholar]
- 109.Tuynman JB, et al. Cyclooxygenase-2 inhibition inhibits c-Met kinase activity and Wnt activity in colon cancer. Cancer Res. 2008;68:1213–1220. doi: 10.1158/0008-5472.CAN-07-5172. [DOI] [PubMed] [Google Scholar]
- 110.Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol. Cell Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hong JY, et al. Down’s-syndrome-related kinase Dyrk1A modulates the p120-catenin-Kaiso trajectory of the Wnt signaling pathway. J. Cell Sci. 2012;125:561–569. doi: 10.1242/jcs.086173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Martin P-M, et al. A rare WNT1 missense variant overrepresented in ASD leads to increased Wnt signal pathway activation. Transl. Psychiatry. 2013;3:e301. doi: 10.1038/tp.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Turner TN, et al. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barber JC, et al. Adenomatous polyposis coli and a cytogenetic deletion of chromosome 5 resulting from a maternal intrachromosomal insertion. J. Med. Genet. 1994;31:312–316. doi: 10.1136/jmg.31.4.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou X-L, et al. Association of adenomatous polyposis coli (APC) gene polymorphisms with autism spectrum disorder (ASD) Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:351–354. doi: 10.1002/ajmg.b.30415. [DOI] [PubMed] [Google Scholar]
- 118.Mohn JL, et al. Adenomatous polyposis coli protein deletion leads to cognitive and autism-like disabilities. Mol. Psychiatry. 2014;19:1133–1142. doi: 10.1038/mp.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Talkowski ME, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lotan A, et al. Neuroinformatic analyses of common and distinct genetic components associated with major neuropsychiatric disorders. Front. Neurosci. 2014;8:331. doi: 10.3389/fnins.2014.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dong F, et al. Deletion of CTNNB1 in inhibitory circuitry contributes to autism-associated behavioral defects. Hum. Mol. Genet. 2016;25:2738–2751. doi: 10.1093/hmg/ddw131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Spinelli L, Black FM, Berg JN, Eickholt BJ, Leslie NR. Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. J. Med. Genet. 2015;52:128–134. doi: 10.1136/jmedgenet-2014-102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Frazier TW, et al. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol. Psychiatry. 2015;20:1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kwon C-H, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lugo JN, et al. Deletion of PTEN produces autism-like behavioral deficits and alterations in synaptic proteins. Front. Mol. Neurosci. 2014;7:27. doi: 10.3389/fnmol.2014.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen Y, Huang W-C, Séjourné J, Clipperton-Allen AE, Page DT. Pten mutations alter brain growth trajectory and allocation of cell types through elevated β-catenin signaling. J. Neurosci. 2015;35:10252–10267. doi: 10.1523/JNEUROSCI.5272-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen TL. Inhibition of growth and differentiation of osteoprogenitors in mouse bone marrow stromal cell cultures by increased donor age and glucocorticoid treatment. Bone. 2004;35:83–95. doi: 10.1016/j.bone.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 131.Soták M, Sumová A, Pácha J. Cross-talk between the circadian clock and the cell cycle in cancer. Ann. Med. 2014;46:221–232. doi: 10.3109/07853890.2014.892296. [DOI] [PubMed] [Google Scholar]
- 132.Guo B, et al. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012;26:3453–3463. doi: 10.1096/fj.12-205781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yasuniwa Y, et al. Circadian disruption accelerates tumor growth and angio/stromagenesis through a Wnt signaling pathway. PLoS ONE. 2010;5:e15330. doi: 10.1371/journal.pone.0015330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Janich P, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 135.Lin F, Chen Y, Li X, Zhao Q, Tan Z. Over-expression of circadian clock gene Bmal1 affects proliferation and the canonical Wnt pathway in NIH-3T3 cells. Cell Biochem. Funct. 2013;31:166–172. doi: 10.1002/cbf.2871. [DOI] [PubMed] [Google Scholar]
- 136.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 137.Yang X, et al. Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J. Biochem. 2009;145:289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 138.Duffield GE, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol. 2002;12:551–557. doi: 10.1016/S0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 139.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 140.Thompson CB. Wnt meets Warburg: another piece in the puzzle? EMBO J. 2014;33:1420–1422. doi: 10.15252/embj.201488785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vallée A, Lecarpentier Y, Guillevin R, Vallée J-N. Aerobic glycolysis hypothesis through WNT/beta-catenin pathway in exudative age-related macular degeneration. J. Mol. Neurosci. 2017;62:368–379. doi: 10.1007/s12031-017-0947-4. [DOI] [PubMed] [Google Scholar]
- 142.Vallée A, Lecarpentier Y, Vallée J-N. Circadian rhythms and energy metabolism reprogramming in Parkinson’s disease. Curr. Issues Mol. Biol. 2019;31:21–44. doi: 10.21775/cimb.031.021. [DOI] [PubMed] [Google Scholar]
- 143.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lum JJ, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21:1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 146.Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG. TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4:147–158. doi: 10.1016/S1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 147.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jung JE, et al. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. FASEB J. 2005;19:1296–1298. doi: 10.1096/fj.04-3099fje. [DOI] [PubMed] [Google Scholar]
- 149.Land SC, Tee AR. Hypoxia-inducible factor 1alpha is regulated by the mammalian target of rapamycin (mTOR) via an mTOR signaling motif. J. Biol. Chem. 2007;282:20534–20543. doi: 10.1074/jbc.M611782200. [DOI] [PubMed] [Google Scholar]
- 150.Toschi A, Lee E, Gadir N, Ohh M, Foster DA. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J. Biol. Chem. 2008;283:34495–34499. doi: 10.1074/jbc.C800170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu Q, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24:5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 152.Pate KT, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J. 2014;33:1454–1473. doi: 10.15252/embj.201488598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Esen E, et al. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab. 2013;17:745–755. doi: 10.1016/j.cmet.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sami A, Karsy M. Targeting the PI3K/AKT/mTOR signaling pathway in glioblastoma: novel therapeutic agents and advances in understanding. Tumour Biol. 2013;34:1991–2002. doi: 10.1007/s13277-013-0800-5. [DOI] [PubMed] [Google Scholar]
- 155.Kim J, Gao P, Liu Y-C, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shibuya K, et al. Targeting the facilitative glucose transporter GLUT1 inhibits the self-renewal and tumor-initiating capacity of cancer stem cells. Oncotarget. 2015;6:651–661. doi: 10.18632/oncotarget.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sun Q, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc. Natl Acad. Sci. USA. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yeung SJ, Pan J, Lee M-H. Roles of p53, MYC and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell. Mol. Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]