Abstract
Craniofacial osteosarcoma is rare (2–10% of all osteosarcomas). Most low grade fibroblastic osteosarcomas of the long bones are characterized by amplification of chromosome12q including MDM2 and CDK4 genes. This study aims to investigate the utility of MDM2 and CDK4 immunostains as well as MDM2 FISH in craniofacial osteosarcomas as a means of distinguishing them from benign fibro-osseous lesions. Cases of primary osteosarcoma and benign fibro-osseous lesions of the craniofacial bones were identified in the diagnostic pathology archives. MDM2 (SMP14 and/or IF2) and CDK4 (D9G3E and/or DCS-31) immunostains were performed on a representative block from each osteosarcoma and benign case. Fluorescence in situ hybridization (FISH) for MDM2 was performed on non-decalcified osteosarcomas. In osteosarcomas, the rate of expression of either MDM2 IF2, MDM2 SMP14, CDK4 DCS-31, or CDK4 D9G3E was 72.7% (8/11 cases), usually focal and weak. Using the MDM2 IF2 clone and the CDK4 DCS-31 clone, MDM2 and CDK4 were negative in lesional cells in all 14 benign fibro-osseous lesions. Using the IF2 and SMP14 clones, MDM2 nuclear expression was present in associated osteoclast-like giant cells in both benign and malignant cases. Of 4 successful cases, 1 high grade osteosarcoma was positive for MDM2 amplification. MDM2 or CDK4 expression or MDM2 amplification may aid in a diagnosis of head and neck osteosarcoma. However, when absent, sarcoma is not excluded. Due to focal weak expression of MDM2 in tumor cells in conjunction with nuclear expression in associated giant cells, caution should be exercised when interpreting positive stains.
Keywords: Osteosarcoma; Craniofacial abnormalities; MDM2 protein, human; CDK4 protein, human; Immunohistochemistry; In-situ hybridization, fluorescence
Introduction
Osteosarcoma of the craniofacial bones is rare, representing 2–10% of all osteosarcomas [1–3]. Osteosarcoma is more common in long bones, in which 5–7% are low grade [4]. Compared to the long bones, craniofacial osteosarcomas are more frequently chondroblastic, occur in an intermediate age range (median age fourth decade), demonstrate lower recurrence rates with complete resection (regardless of grade) as well as lower response rates to chemotherapy, have lower rates of distant metastasis, and suffer mortality primarily due to local recurrence [1, 5–7].
The majority of low grade fibroblastic osteosarcomas of the long bones harbor a supernumerary ring chromosome with amplification of the 12q13-15 region including the MDM2 and CDK4 genes [8]. Up to one third of low grade osteosarcomas can dedifferentiate in to high grade osteosarcoma in long bones, and, if present, they retain chromosome 12q amplification [8]. High grade osteosarcomas (not arising from low grade) have complex karyotypes different from low grade osteosarcomas [8]. In long bones, MDM2 and CDK4 immunohistochemical (IHC) stains are useful in distinguishing low grade osteosarcoma from other benign mimics [4, 9, 10]. In addition to MDM2 expression by IHC, amplification of MDM2 by FISH has also been demonstrated [11].
In the craniofacial bones, distinction of low grade osteosarcoma from benign mimics (such as fibrous dysplasia and ossifying fibroma) may be difficult on small biopsy [9, 12]. Other studies have evaluated the utility of MDM2 and CDK4 IHC to distinguish osteosarcoma from benign fibro-osseous lesions in the head and neck, and have found MDM2 and CDK4 positivity in 8–100% and 33–84%, respectively, of head and neck osteosarcomas [5, 6, 8, 10]. All evaluated benign head and neck fibro-osseous lesions have been negative [8, 9]. The utility of MDM2 FISH in craniofacial osteosarcomas has not been investigated, and only a single reported high grade maxillary osteosarcoma demonstrated MDM2 amplification by FISH [13]. This study aims to investigate the utility of MDM2 and CDK4 immunostains as well as MDM2 FISH in craniofacial osteosarcomas as a means of distinguishing them from benign fibro-osseous lesions.
Materials and Methods
Cases of primary osteosarcoma and benign fibro-osseous lesions of the craniofacial bones were identified in the diagnostic pathology archives of The University of Chicago and of the Oral and Maxillofacial Pathology Biopsy Service (R.M.). The slides were reviewed by two head & neck pathologists (A.L.L. and N.A.C.) to confirm the diagnosis and to select blocks for immunostaining and FISH. Patient demographics, radiographic features, and clinical follow-up were gathered from the electronic medical record.
MDM2 and CDK4 immunostains were performed on a representative block from each osteosarcoma and benign case: Immunostain for MDM2 was performed using the 1F2 clone from Millipore at 1:200 titer on Bond using Define kit. Immunostain for CDK4 was performed using the DCS-31 clone from Invitrogen at 1:100 Casian titer on the Bond using Refine kit. MDM2 or CDK4 were considered positive if any amount of nuclear expression was seen in tumor cells.
Additional MDM2 and CDK4 immunostains were performed on a representative block from 7 select osteosarcoma cases, in order to evaluate possible differences in antibodies: Immunostain for MDM2 was performed using mouse MAb, clone SMP14 from Santa Cruz Biotechnology, at dilution 1:200 (conc 1 ug/ml), and for CDK4 using rabbit MAb, clone D9G3E from Cell Signalling Technology, at dilution 1:200. Immunostain for both antibodies was performed on automated IHC/ISH platform Bond 3 (Leica Microsystems. Buffalo Grive, IL), following on platform Heat Epitope antigen retrieval ER2 for 20 min, and Refine detection kit. Immunohistochemistry protocol included 25 min incubation with the primary anti-CDK4 RMab (D9G3E) or 25 and 50 min with anti MDM2 Mab (SMP14), 15 min post-primary step, and 25 min incubation with polymer HPR, followed 10 min peroxidase development with DAB. MDM2 or CDK4 were considered positive if any amount of nuclear expression was seen in tumor cells.
Fluorescence in situ hybridization (FISH) for MDM2 was attempted on 6 non-decalcified osteosarcoma cases. (All blocks selected for evaluation by FISH were undecalcified based on the gross descriptions. However, decalcification may have been performed but not accounted for).
A bacterial artificial clone (RP11-77H17) containing sequences specific to MDM2 was used to generate custom FISH probes by standard nick translation methods. Extracted DNA was labeled with fluorescently labeled dUTPs (Spectrum Orange; Abbott) using a Nick translation kit (Abbott, Chicago, IL). Probe performance and specificity was assessed using metaphase preparations from a pooled source of normal male controls. A dual-color FISH probe set was generated by mixing labeled probe in corresponding proportions with probe targeting the centromeric region (CEP) of Chromosome 12 (PBR12). Hybridization of slides was performed using standard methods. Slides were baked overnight at 70 °C followed by deparaffinization with Citrisolve (Fisher, Waltham, MA) and dehydration with ethanol series. Slides were incubated in 1 M NaSCN at 80 °C followed by 0.2 N HCI at room temperature for 30 min. Pepsin Protease solution was used for proteolytic pre-treatment of slides and fixed in 10% formaldehyde. Slides were manually denatured in 70% formamide at 80 °C and hybridized overnight at 37 °C in a humidity chamber. Slides were washed in SSC/NP40 the next day, and counterstained with DAPI (Sigma-Aldrich, St. Louis, MO).
Using standard FISH scoring techniques, a minimum of 30 tumor nuclei were manually enumerated for each target from all cores using fluorescent microscopy (Zeiss Axio Imager.Z2; Zeiss, Jena, Germany). Representative images were captured using CytoVysion analysis software, v.7.4 (Leica, Buffalo Grove, IL). Cutoff values were calculated using the beta inverse statistic (Excel; Microsoft, Redmond, WA) from data collected from 9 cases of liposarcoma. Cases were interpreted as positive for an amplified cell signal with ratio MDM2/CEP12 above 2.0.
Results
Malignant cases included 11 osteosarcomas from 11 patients (8 males, 3 females) with an average age of 34 years (range 14–70 years). Two patients had LiFraumeni syndrome, 1 had Soto syndrome, 1 had medulloblastoma and desmoid tumors suggestive of a possible syndrome, and 1 had a previous colon carcinoma. Anatomic locations included the mandible (6), skull (3), and maxilla (2). Four cases were predominantly chondroblastic, 4 fibroblastic, and 3 osteoblastic. Six cases were low grade, 2 intermediate grade, and 3 high grade. Three cases were decalcified using 10% Hydrochloric Acid (Table 1).
Table 1.
Clinicopathologic features of head & neck osteosarcoma patients
| Patient # | Age | Sex | Location | Histologic subtype | Histologic grade | Decalcified | MDM2 IF2 IHC | MDM2 SMP14 IHC | CDK4 DCS- 31 IHC | CDK4 D9G3E IHC | MDM2 FISH | Type of treatment | Status | Time of F/U (years) | Time of F/U (months) | Other history |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 25 | F | Mandible | Osteoblastic | Intermediate | Yes | Neg | NP | Neg | NP | NP | Adjuvant chemotherapy | NEO local disease; new lung adenocarcinoma; new melanoma, new iliac/sacral osteosarcoma with metastasis to T11 | 6.0 | 73.3 | Li Fraumeni syndrome |
| 2 | 62 | M | Mandible | Osteoblastic | High | No | Neg | Pos, focal | Neg | Pos | Pos: multiple cells amplified | Neoadjuvant chemotherapy, adjuvant chemoradiotherpy | Died of myocarditis | 0.9 | 11.0 | Prior colon cancer status post resection |
| 3 | 70 | M | Skull (frontal/parietal bone) | Fibroblastic | Low | Yes | Neg | Neg | Neg | Neg | NP | Adjuvant chemoradiotherapy | LTF | |||
| 4 | 15 | M | Skull (frontal bone) | Osteoblastic | High | No | Neg | NP | Pos, focal | NP | Neg: 1 cell amplified | Surgery only | Died of sepsis | 0.1 | 0.7 | Soto syndrome, pineoblastoma |
| 5 | 14 | M | Mandible | Fibroblastic > Osteoblastic | Low | Yes | Neg | NP | Neg | NP | NP | UNK | LTF | |||
| 6 | 34 | M | Mandible | Osteoblastic > Chondroblastic | Low | No | Pos, focal | Pos | Neg | Pos | Neg | UNK | LTF | |||
| 7 | 41 | F | Maxilla | Fibroblastic | Low | No | pos, focal | NP | neg | NP | Failed | Adjuvant radiation | NEO local disease | 5.0 | 60.7 | |
| 8 | UNK | M | Maxilla | Chondroblastic | Low | No | Pos, focal | Pos | Neg | Pos | Neg: 1 cell amplified | UNK | LTF | |||
| 9 | 18 | M | Skull | Fibroblastic > Osteoblastic | Intermediate | No | Pos, focal | Pos, focal | Pos, focal | Pos, focal | Failed | Adjuvant chemotherapy | Metastasis to T5 | 3.1 | 37.7 | Medulloblastoma, recurrent desmoid tumors |
| 10 | 19 | M | Mandible | Osteoblastic > Chondroblastic | High | No | Neg | Pos | Neg | pos, focal | NP | Adjuvant chemotherapy with growth, therefore re-resection | Nodal metastasis at primary resection; Local recurrence plus pulmonary metastasis | 0.1 | 1.4 | Li Fraumeni syndrome |
| 11 | 41 | F | Mandible | Chondroblastic | Low | No | Neg | Pos | Neg | Pos | NP | Surgery only | NEO local disease | 0.2 | 2.5 |
UNK unknown, neg negative, pos positive, NP not performed, NEO no evidence of, LTF lost to follow up, F/U follow up
Benign cases included 14 lesions from 14 patients (5 males, 9 females) with an average age of 36 years (range 6–86 years). Diagnoses included 5 fibrous dysplasias, 3 cemento-ossifying fibromas, 2 peripheral ossifying fibromas, 2 central giant cell lesions, 1 endosteal osteoma, and 1 odontogenic fibromyxoma. Anatomic locations included the mandible (10), gingiva (2), maxilla (1), and sphenoid (1).
Of all 11 osteosarcomas, 4 (36.4%) were positive for MDM2 IF2 clone and 2 (18.2%) were positive for CDK4 DCS-31 clone, often focally: 1 expressed combined MDM2/CDK4, 3 isolated MDM2, and 1 isolated CDK4. The rate of expression of either MDM2 IF2 or CDK4 DCS-31 was 45.5%. Of 7 available osteosarcomas, 6 (85.7%) were positive for both MDM2 SMP14 clone and CDK4 D9G3E clone, also often focally (Fig. 1, Table 1). Comparing the clones: 4 cases stained the same for MDM2 IF2 and MDM2 SMP14 (3 positive and 1 negative) and 3 were positive only for MDM2 SMP14; 2 cases stained the same for CDK4 DCS-31 and CDK4 D9G3E (1 positive and 1 negative) and 5 were positive only for CDK4 D9G3E. The rate of expression of either MDM2 IF2, MDM2 SMP14, CDK4 DCS-31, or CDK4 D9G3E was 72.7% (8/11 cases). Using the MDM2 IF2 clone and the CDK4 DCS-31 clone, MDM2 and CDK4 were negative in lesional cells in all 14 benign fibro-osseous lesions. With both the IF2 and SMP14 clones, MDM2 nuclear expression was present in associated osteoclast-like giant cells in both benign and malignant cases (Fig. 2).
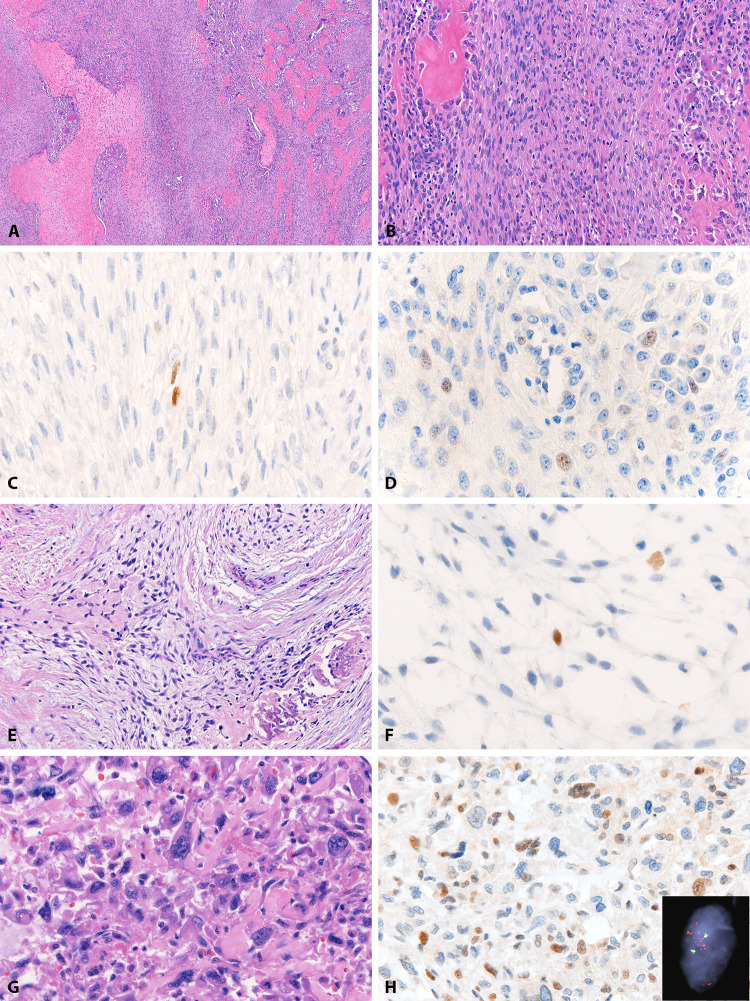
Fig. 1.
MDM2 in osteosarcomas. An intermediate grade predominantly fibroblastic osteosarcoma (patient 9) (a, b) with focal nuclear expression of MDM2 IF2 in spindled tumor cells (c) and CDK4 DCS-31 in plump tumor cells (d). A low grade osteoblastic/chondroblastic osteosarcoma (patient 6) (e) in which rare stellate-shaped tumor cells express MDM2 IF2 (f). A high grade osteoblastic osteosarcoma (patient 2) (g) with positive MDM2 SMP14 immunostain (h) and multiple cells showing MDM2 amplification by FISH (H inset)
Fig. 2.
MDM2 in tumor-associated giant cells. A low grade fibroblastic osteosarcoma (patient 5) (a) in which paratrabecular osteoclast-like giant cells (b) show nuclear MDM2 IF2 expression (c). Similarly, a high grade osteoblastic osteosarcoma (patient 4) (d) with MDM2 IF2 expression in tumor-associated giant cells (e). MDM2 IF2 is also expressed in osteoclast-like giant cells in benign lesions, such as this central giant cell lesion (f, g). In all cases, background lesional cells are negative
FISH for MDM2 was attempted in 6 non-decalcified osteosarcomas. Hybridization failed in 2 cases. In the remaining 4 successful cases: one high grade osteosarcoma was positive for MDM2 amplification in multiple cells, two had only a single cell with amplification and were considered negative, one was not amplified. The single MDM2 amplified case was positive for MDM2 SMP14 and CDK4 D9G3E, but negative for MDM2 IF2 and CDK4 DCS-31 by IHC (Fig. 1, Table 1). FISH was not performed on benign fibro-osseous lesions.
Preoperative radiographic data from 6 of the 11 patients (54.5%) was available. In 4 (66.7%), the features were compatible with malignancy: Patient 2 (a high grade osteoblastic osteosarcoma) had a panorex and CT scan showing a large (not measured) necrotic mass in the left masticator space with erosion of the left mandible as well as cervical lymphadenopathy; Patient 4 (a high grade osteoblastic osteosarcoma) had MR and CT scans showing a 3.5 cm mass in the frontal bone that eroded through and caused destruction of the frontal bone; Patient 7 (a low grade fibroblastic osteosarcoma) had CT scans showing a 7 cm mass with bone matrix originating from the right maxilla, eroding the anterior wall of the sinus, filling the right nasal cavity, extending through the nasal choana to the posterior nasopharynx, destroying the floor of the orbit, and deviating the right globe; Patient 10 (a high grade osteoblastic/chondroblastic osteosarcoma) had MR and CT scans showing a sclerotic, enhancing 3.6 cm mass in the right mandibular body with buccal and lingual soft tissue extension as well as cortical thinning and widening of the mental foramen. In 2 patients (33.3%), imaging was not specific for malignancy: Patient 5 (a low grade fibroblastic/osteoblastic osteosarcoma) had a CT showing a focal lesion between teeth 18 and 19 without comment on the appearance; Patient 11 (a low grade chondroblastic osteosarcoma) had MR and CT scans showing ill-defined sclerosis/enhancement in the left mandibular body marrow with lingual cortex periosteal reaction and mild ill-defined soft-tissue enhancement without a discrete soft tissue mass.
Of the 11 patients with osteosarcoma, all underwent local resection; 1 received neoadjuvant chemotherapy and adjuvant chemoradiotherapy; 5 received adjuvant therapy (3 chemotherapy, 1 radiotherapy, 1 chemoradiotherapy); 2 were treated with surgery alone; postsurgical treatment was not known in 3 patients. Four patients were lost to follow-up. Two patients died: one of sepsis one month after surgery and another of myocarditis one year after surgery. Two patients developed metastases: one to bone 3 years after surgery and one to regional lymph nodes at the time of surgery followed by lung 2 months after surgery. The remaining 3 patients were alive with no evidence of osteosarcoma after 3 months, 5 years, and 6 years (Table 1).
Discussion/Conclusions
In this study, we found that MDM2 or CDK4 protein expression by IHC or MDM2 gene amplification by FISH were occasionally present in head and neck osteosarcomas, while all benign fibro-osseous lesions were negative for MDM2 and CDK4 IHC. Immunoprofile varied by clone used. Osteosarcomas were more frequently positive using MDM2 SMP14 and CDK4 D9G3E (85.7% for both clones) compared to MDM2 IF2 (36.4%) and CDK4 DCS-31 (18.2%). One high grade osteosarcoma had MDM2 amplification by FISH and was positive for MDM2 SMP14 and CDK4 D9G3E.
Osteosarcoma of the craniofacial bones is rare and can be difficult to distinguish from benign lesions on small biopsy [9, 12]. Radiographically and histologically, low grade osteosarcoma may mimic benign chondroid, fibrous, or fibro-osseous lesions, such as fibrous dysplasia, ossifying/cemento-ossifying fibroma, periapical cemento-osseous dysplasia, and giant cell rich lesions [9, 12]. Of the 4 cases in which pre-operative imaging suggested malignancy (3 high and 1 low grade), all were positive for one of the MDM2 or CDK4 IHC clones. Of the 2 cases in which pre-operative imaging was not diagnostic of malignancy (both low grade), 1 was positive. Of the 5 cases in which pre-operative imaging was not available (3 low and 2 intermediate grade), 3 were positive. Therefore, in the absence of radiographic features of malignancy or on a challenging biopsy, focal positivity can be suggestive of osteosarcoma. Of the 8 low or intermediate grade sarcomas, 5 (62.5%) were positive by IHC. All 3 high grade tumors were positive.
In the long bones, low grade fibroblastic osteosarcomas are characterized by supernumerary ring chromosomes composed of 12q13-15 clusters containing MDM2 and CDK4 genes [11, 14, 15]. MDM2 (murine double minute-2) encodes a nuclear phosphoprotein that binds and inactivates p53 [15]. CDK4 (cyclin dependent kinase 4) phosphorylates and inactivates the retinoblastoma tumor suppressor protein during the G1 phase of the cell cycle, leading to cell cycle progression [15]. Gene amplification and overexpression of MDM2 and CDK4 can lead to dysregulation of the cell cycle and tumor progression [6]. Many studies have shown MDM2 and CDK4 expression by IHC to have good sensitivity and specificity in differentiating low grade osteosarcoma from benign mimics in long bones, with up to 97.5% sensitivity and 100% specificity [4, 7, 9, 10, 16, 17]. The amount of nuclear expression varies greatly between studies, with 22–70% of tumor cells expressing MDM2 and 66–92% expressing CDK4 [5, 6, 10, 16, 18].
Craniofacial osteosarcomas are clinically and pathologically different from those of the long bones, and may possess different genetic drivers [1, 5–7, 9]. Craniofacial osteosarcomas are more frequently chondroblastic, occur in an intermediate age range (median age fourth decade), demonstrate lower recurrence rates with complete resection (regardless of grade) as well as lower response rates to chemotherapy, have lower rates of distant metastasis, and suffer mortality primarily due to local recurrence [19–23].
Other studies have evaluated MDM2 and CDK4 IHC in craniofacial osteosarcoma as well as gene amplification by PCR (Table 2). Amplification of MDM2 by FISH has not been systematically evaluated in craniofacial osteosarcoma, with the exception of a single case report [13]. A limitation of the current study is the relatively small number of osteosarcomas tested. In light of the rarity of this disease, a review of literature was undertaken. In 2001, Lopes et al. found high rates of CDK4 (8/9, 89%) and MDM2 (5/9, 56%) expression in craniofacial osteosarcomas of all histologic grades. Positive expression correlated with amplification of either MDM2 or CDK4 DNA as evaluated by PCR in all but 2 cases (positive by IHC but not amplified by PCR) [6]. In 2003, Junior et al. evaluated 25 intermediate and high grade craniofacial osteosarcomas and found MDM2 and CDK4 expression in 6 (24%) and 21 (84%), respectively [5]. In 2010, Yoshida et al. evaluated low grade fibroblastic osteosarcomas of all sites, 3 of which were in the head and neck. All 3 (100%) expressed MDM2, and 1 (33%) expressed CDK4 [10]. Most recently, Guerin et al. examined 36 high grade mandibular osteosarcomas, of which 3 (8%) expressed MDM2. Amplification of MDM2 by PCR was present in 5/14 (36%) cases, 3 of which were negative by IHC [8]. There is wide variation in the amount and intensity of immunostaining among these studies. Differences in staining patterns could be due to a number of factors including differences in formalin fixation, decalcification, antibody dilution, and antigen retrieval [9].
Table 2.
Literature review of MDM2 and CDK4 in head & neck osteosarcoma
| Article | Craniofacial osteosarcomas | Benign lesions | |||||
|---|---|---|---|---|---|---|---|
| MDM2 IHC | CDK4 IHC | MDM2 amp | CDK4 amp | Grade evaluated | Subtype evaluated | MDM2 & CDK4 IHC | |
| Lopes et al. [6] | 8/9 (89%) 4 strong | 5/9 (56%) all strong | 6/9 (67%) | 6/9 (67%) | All | Chondroblastic osteoblastic parosteal | NP |
| Junior et al. [5] | 6/25 (24%) 2 moderate 4 weak | 21/25 (84%) 10 strong 9 moderate 2 weak | NP | NP | Intermediate high | Chondroblastic osteoblastic | NP |
| Yoshida et al. [10] | 3/3 (100%) 1 strong diffuse 1 moderate diffuse 1 moderate focal | 1/3 (33%) strong diffuse | NP | NP | Low | Fibroblastic | 1/40 EC (3%) ulnar BPOP pos |
| Guerin et al. [8] | 3/36 (8%) UNK pattern | NP | 5/14 (36%) | NP | High | Chondroblastic osteoblastic fibroblastic | 0/25 CF (0%) MDM2 only UNK histology |
| Hirose et al. [13] | 1/1 | 1/1 | 1/1 | 1/1 | High | Osteoblastic, giant cell rich | NP |
| Dujardin et al. [9] | NP | NP | NP | NP | NP | NP | 0/97 EC (0%) 0/10 CF (0%) 6 COF, 2 FD, 1 OD, 1 MF |
| Lott Limbach (current) | 7/11 (63%) all focal weak | 7/11 (63%) all focal weak | 1/4 (25%) | NP | All | Chondroblastic osteoblastic fibroblastic |
0/14 CF (0%) 6 FD, 3 COF, 2 POF, 2 CGCL, 1 EO, 1 OFM |
| Total | 28/85 (33%) | 35/49 (71%) | 13/28 (46%) | 7/10 (70%) | All | All | 1/186 (< 1%) |
IHC immunohistochemistry, amp amplification, NP not performed, UNK unknown, EC extracranial, BPOP bizarre parosteal osteochondromatous proliferation, CF craniofacial, COF cemento-ossifying fibroma, FD fibrous dysplasia, OD osseous dysplasia, MF myofibroma, POF peripheral ossifying fibroma, CGCL central giant cell lesion, EO endosteal osteoma, OFM odontogenic fibromyxoma
In prototypical MDM2 or CDK4-amplified sarcomas (namely, well-differentiated/dedifferentiated liposarcoma or low grade fibroblastic osteosarcoma of long bones), MDM2 and CDK4 are expressed at high rates (70–100% of cases) [10, 24–27]. However, in other lipomatous tumors, expression rates can vary: 9–39% express MDM2 and 3–13% express CDK4 [24–27]. Similarly, other non-MDM2 amplified sarcomas can also have expression: 0–35% MDM2 and 3–43% CDK4, ranging from focal weak to diffuse strong [24–27]. Therefore, it is not entirely unexpected for non-MDM2 amplified sarcomas (such as some in this study) to focally express MDM2 or CDK4. In these cases, the reason for MDM2 or CDK4 overexpression is not entirely understood, may be due to other neoplastic cell-cycle perturbations outside gene amplification.
Only two prior studies have evaluated MDM2 and CDK4 expression in benign fibro-osseous lesions of the head and neck, and both found all cases to be negative (n = 35) [8, 9]. An additional 97 benign extracranial fibro-osseous lesions were also negative [9]. One study evaluated benign bony lesions of all extracranial sites and found 1 ulnar bizarre parosteal osteochondromatous proliferation to be positive in the spindle cells [9] (Table 2).
In this series of craniofacial osteosarcomas, MDM2 or CDK4 were expressed in a total of 8 (72.7%) sarcomas using either combination of clones. Sarcomas of all grades (low, intermediate, and high) and all morphologies (chondroblastic, osteoblastic, fibroblastic) had expression. Many cases had focal or multifocal expression. None had strong diffuse expression. All 14 benign fibro-osseous lesions of the head and neck were negative by MDM2 and CDK4 IHC. FISH for MDM2 amplification was successfully performed in 4 head and neck osteosarcomas, 1 (25%) of which was positive. Of the 7 other cases with MDM2 or CDK4 expression, 3 were not amplified by FISH, hybridization failed in 2, and FISH was not attempted in 2.
The utility of MDM2 IHC compared to MDM2 FISH has only been evaluated in soft tissue tumors [28, 29]. Sirvent et al. found FISH had a sensitivity of 95% and specificity of 100%, while IHC had a sensitivity of 80% and specificity of 93%. The concordance rate of IHC with FISH and Q-PCR was 40% [29]. A more recent study by Kimura et al. found sensitivity and specificity of FISH to be 100% and 95%, respectively. Sensitivity and specificity of IHC was 100% and 87%, respectively [28, 29]. Both studies suggest that IHC is a good initial test, but FISH is more specific. Our results differ in that IHC-positive cases were unlikely to be amplified by FISH.
One pitfall in the use of MDM2 immunostain is the presence of nuclear MDM2 expression in tumor-associated osteoclast-like giant cells in both benign lesions and osteosarcomas. This finding was also reported by Yoshida et al. [4]. Close examination of the background counterstain and morphology of the positive cells can assist in identification of cell type. In high grade sarcomas, nuclei of giant cells should be smaller than tumor cells and clustered within a cell. Nuclear size of low grade sarcomas may be similar to giant cells, however, clustering should still be present in giant cells.
An additional interesting finding was the prevalence of a syndrome or features suggestive of a syndrome in 4 (36%) patients: 2 LiFraumeni, 1 Soto, and 1 suspected tumor syndrome. Sporadic tumors occurred in the remaining 7 (64%). Paget’s disease, fibrous dysplasia, and radiation are known predisposing factors for osteosarcoma [19]. Tumorigenesis in long bones is hypothesized to be more frequently related to rapid bone growth during adolescence, rather than predisposing disease [19]. Our findings support a high prevalence of predisposing conditions in craniofacial osteosarcomas.
When present, MDM2 or CDK4 expression or MDM2 amplification may aid in a diagnosis of head and neck osteosarcoma. However, when absent, sarcoma is not excluded. Variability in expression by different antibody clones is a potential pitfall. Additionally, due to focal weak expression of MDM2 in tumor cells in conjunction with nuclear expression in associated giant cells in both benign and malignant lesions, caution should be exercised when interpreting positive stains. As osteosarcomas of craniofacial bones are rare, additional studies are needed to further elucidate the role of MDM2 and CDK4 in these tumors.
Funding
No funding obtained.
Compliance with Ethical Standards
Conflict of interest
All authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee RJ, Arshi A, Schwartz HC, Christensen RE. Characteristics and prognostic factors of osteosarcoma of the jaws: a retrospective cohort study. JAMA Otolaryngol Head Neck Surg. 2015;141(5):470–477. doi: 10.1001/jamaoto.2015.0340. [DOI] [PubMed] [Google Scholar]
- 2.Baumhoer D, Brunner P, Eppenberger-Castori S, Smida J, Nathrath M, Jundt G. Osteosarcomas of the jaws differ from their peripheral counterparts and require a distinct treatment approach. Experiences from the DOESAK Registry. Oral Oncol. 2014;50(2):147–153. doi: 10.1016/j.oraloncology.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhary M, Chaudhary SD. Osteosarcoma of jaws. J Oral Maxillofac Pathol. 2012;16(2):233–238. doi: 10.4103/0973-029X.99075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida A, Ushiku T, Motoi T, Beppu Y, Fukayama M, Tsuda H, et al. MDM2 and CDK4 immunohistochemical coexpression in high-grade osteosarcoma: correlation with a dedifferentiated subtype. Am J Surg Pathol. 2012;36(3):423–431. doi: 10.1097/PAS.0b013e31824230d0. [DOI] [PubMed] [Google Scholar]
- 5.Junior AT, de Abreu AF, Pinto CA, Carvalho AL, Kowalski LP, Lopes MA. Clinicopathological and immunohistochemical analysis of twenty-five head and neck osteosarcomas. Oral Oncol. 2003;39(5):521–530. doi: 10.1016/S1368-8375(03)00017-4. [DOI] [PubMed] [Google Scholar]
- 6.Lopes MA, Nikitakis NG, Ord RA, Sauk J., Jr Amplification and protein expression of chromosome 12q13-15 genes in osteosarcomas of the jaws. Oral Oncol. 2001;37(7):566–571. doi: 10.1016/S1368-8375(00)00130-5. [DOI] [PubMed] [Google Scholar]
- 7.van den Berg H, Schreuder WH, de Lange J. Osteosarcoma: a comparison of jaw versus nonjaw localizations and review of the literature. Sarcoma. 2013;2013:316123. doi: 10.1155/2013/316123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerin M, Thariat J, Ouali M, Bouvier C, Decouvelaere AV, Cassagnau E, et al. A new subtype of high-grade mandibular osteosarcoma with RASAL1/MDM2 amplification. Hum Pathol. 2016;50:70–78. doi: 10.1016/j.humpath.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Dujardin F, Binh MB, Bouvier C, Gomez-Brouchet A, Larousserie F, Muret A, et al. MDM2 and CDK4 immunohistochemistry is a valuable tool in the differential diagnosis of low-grade osteosarcomas and other primary fibro-osseous lesions of the bone. Mod Pathol. 2011;24(5):624–637. doi: 10.1038/modpathol.2010.229. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida A, Ushiku T, Motoi T, Shibata T, Beppu Y, Fukayama M, et al. Immunohistochemical analysis of MDM2 and CDK4 distinguishes low-grade osteosarcoma from benign mimics. Mod Pathol. 2010;23(9):1279–1288. doi: 10.1038/modpathol.2010.124. [DOI] [PubMed] [Google Scholar]
- 11.Gisselsson D, Palsson E, Hoglund M, Domanski H, Mertens F, Pandis N, et al. Differentially amplified chromosome 12 sequences in low- and high-grade osteosarcoma. Genes Chromosom Cancer. 2002;33(2):133–140. doi: 10.1002/gcc.1219. [DOI] [PubMed] [Google Scholar]
- 12.Koury ME, Regezi JA, Perrott DH, Kaban LB. "Atypical" fibro-osseous lesions: diagnostic challenges and treatment concepts. Int J Oral Maxillofac Surg. 1995;24(2):162–169. doi: 10.1016/S0901-5027(06)80094-9. [DOI] [PubMed] [Google Scholar]
- 13.Hirose K, Okura M, Sato S, Murakami S, Ikeda JI, Noda Y, et al. Gnathic giant-cell-rich conventional osteosarcoma with MDM2 and CDK4 gene amplification. Histopathology. 2017;70(7):1171–1173. doi: 10.1111/his.13141. [DOI] [PubMed] [Google Scholar]
- 14.Szymanska J, Mandahl N, Mertens F, Tarkkanen M, Karaharju E, Knuutila S. Ring chromosomes in parosteal osteosarcoma contain sequences from 12q13-15: a combined cytogenetic and comparative genomic hybridization study. Genes Chromosom Cancer. 1996;16(1):31–34. doi: 10.1002/(SICI)1098-2264(199605)16:1<31::AID-GCC4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Wunder JS, Eppert K, Burrow SR, Gokgoz N, Bell RS, Andrulis IL. Co-amplification and overexpression of CDK4, SAS and MDM2 occurs frequently in human parosteal osteosarcomas. Oncogene. 1999;18(3):783–788. doi: 10.1038/sj.onc.1202346. [DOI] [PubMed] [Google Scholar]
- 16.Park HR, Jung WW, Bertoni F, Bacchini P, Park JH, Kim YW, et al. Molecular analysis of p53, MDM2 and H-ras genes in low-grade central osteosarcoma. Pathol Res Pract. 2004;200(6):439–445. doi: 10.1016/j.prp.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Jeon DG, Koh JS, Cho WH, Song WS, Kong CB, Cho SH, et al. Clinical outcome of low-grade central osteosarcoma and role of CDK4 and MDM2 immunohistochemistry as a diagnostic adjunct. J Orthop Sci. 2015;20(3):529–537. doi: 10.1007/s00776-015-0701-0. [DOI] [PubMed] [Google Scholar]
- 18.Gamberi G, Ragazzini P, Benassi MS, Ferrari C, Sollazzo MR, Molendini L, et al. Analysis of 12q13-15 genes in parosteal osteosarcoma. Clin Orthop Relat Res. 2000;377(377):195–204. doi: 10.1097/00003086-200008000-00026. [DOI] [PubMed] [Google Scholar]
- 19.Mardinger O, Givol N, Talmi YP, Taicher S. Osteosarcoma of the jaw. The Chaim Sheba Medical Center experience. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;91(4):445–451. doi: 10.1067/moe.2001.112330. [DOI] [PubMed] [Google Scholar]
- 20.Fernandes R, Nikitakis NG, Pazoki A, Ord RA. Osteogenic sarcoma of the jaw: a 10-year experience. J Oral Maxillofac Surg. 2007;65(7):1286–1291. doi: 10.1016/j.joms.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Demicco EG, Deshpande V, Nielsen GP, Kattapuram SV, Rosenberg AE. Well-differentiated osteosarcoma of the jaw bones: a clinicopathologic study of 15 cases. Am J Surg Pathol. 2010;34(11):1647–1655. doi: 10.1097/PAS.0b013e3181f7dac6. [DOI] [PubMed] [Google Scholar]
- 22.Clark JL, Unni KK, Dahlin DC, Devine KD. Osteosarcoma of the jaw. Cancer. 1983;51(12):2311–2316. doi: 10.1002/1097-0142(19830615)51:12<2311::AID-CNCR2820511224>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 23.Bertoni F, Bacchini P, Fabbri N, Mercuri M, Picci P, Ruggieri P, et al. Osteosarcoma. Low-grade intraosseous-type osteosarcoma, histologically resembling parosteal osteosarcoma, fibrous dysplasia, and desmoplastic fibroma. Cancer. 1993;71(2):338–345. doi: 10.1002/1097-0142(19930115)71:2<338::AID-CNCR2820710212>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 24.Thway K, Flora R, Shah C, Olmos D, Fisher C. Diagnostic utility of p16, CDK4, and MDM2 as an immunohistochemical panel in distinguishing well-differentiated and dedifferentiated liposarcomas from other adipocytic tumors. Am J Surg Pathol. 2012;36(3):462–469. doi: 10.1097/PAS.0b013e3182417330. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi A, Sakuma T, Fujimoto M, Jimbo N, Hirose T. Diagnostic utility and limitations of immunohistochemistry of p16, CDK4, and MDM2 and automated dual-color in situ hybridization of MDM2 for the diagnosis of challenging cases of dedifferentiated liposarcoma. Appl Immunohistochem Mol Morphol. 2019;27(10):758–763. doi: 10.1097/PAI.0000000000000677. [DOI] [PubMed] [Google Scholar]
- 26.Binh MB, Sastre-Garau X, Guillou L, de Pinieux G, Terrier P, Lagace R, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol. 2005;29(10):1340–1347. doi: 10.1097/01.pas.0000170343.09562.39. [DOI] [PubMed] [Google Scholar]
- 27.Aleixo PB, Hartmann AA, Menezes IC, Meurer RT, Oliveira AM. Can MDM2 and CDK4 make the diagnosis of well differentiated/dedifferentiated liposarcoma? An immunohistochemical study on 129 soft tissue tumours. J Clin Pathol. 2009;62(12):1127–1135. doi: 10.1136/jcp.2009.070201. [DOI] [PubMed] [Google Scholar]
- 28.Kimura H, Dobashi Y, Nojima T, Nakamura H, Yamamoto N, Tsuchiya H, et al. Utility of fluorescence in situ hybridization to detect MDM2 amplification in liposarcomas and their morphological mimics. Int J Clin Exp Pathol. 2013;6(7):1306–1316. [PMC free article] [PubMed] [Google Scholar]
- 29.Sirvent N, Coindre JM, Maire G, Hostein I, Keslair F, Guillou L, et al. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am J Surg Pathol. 2007;31(10):1476–1489. doi: 10.1097/PAS.0b013e3180581fff. [DOI] [PubMed] [Google Scholar]