Abstract
Breast cancer is the most common cancer type in women worldwide and its early detection is crucial to curing the disease. Tissue biopsy, currently the method of choice to obtain tumour molecular information, is invasive and might be affected by tumour heterogeneity rendering it incapable to portray the complete molecular picture. Liquid biopsy permits to study disease features in a more comprehensive manner by sampling biofluids and extracting tumour components such as circulating-tumour DNA (ctDNA), circulating-tumour cells (CTCs), and/or circulating-tumour RNA (ctRNA) amongst others in a monitoring-compatible manner. In this review, we describe the recent progress in the utilization of the circulating tumour components using early breast cancer samples. We review the most important analytes and technologies employed for their study.
Keywords: Liquid biopsy, Localized breast cancer, Circulating-tumour DNA, Circulating-tumour cells, Circulating-tumour RNA
1. Liquid biopsy and breast cancer
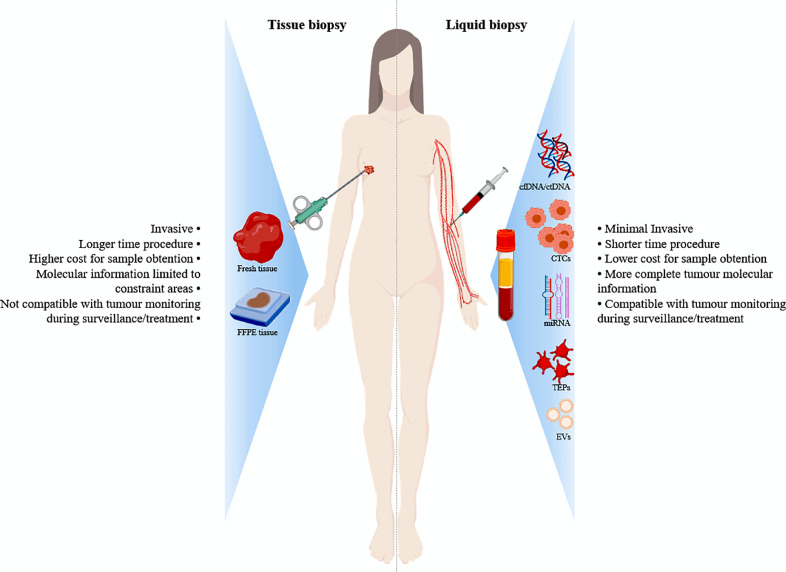
Solid biopsies are the current standard-of-care in clinical cancer management with an unquestionable utility in the oncologic field. They provide information about tumour histology, standard biomarkers for subtyping and treatment planning and molecular profiles for predictive and prognostic signatures with optimal cost-effectiveness ratio. Nevertheless, they inherently show several limitations. On one hand, tumour primary and/or metastatic tissue is not always accessible and processing turnaround times are often tedious. On the other hand, the tumour molecular and genetic information is limited to the biopsy area, possibly misleading the interpretation due to the tumour heterogeneity. On top of that, solid biopsies are incompatible with longitudinal monitoring and require an invasive procedure that may be painful and potentially risky to the patient [1]. Therefore, novel approaches to complement the above-mentioned caveats had to be further explored (Fig. 1).
Fig. 1.
Diagram summarizing the main components in blood liquid biopsies as well as the advantages over the conventional ones.
Abbreviations: FFPE, Formalin-Fixed Paraffin-Embedded; cfDNA, circulating-free DNA; CTCs, circulating-tumour cells; miRNA, microRNA; TEPs, tumour-educated blood platelets; EVs, extracellular vesicles.
The history of liquid biopsy started in 1869, when the existence of circulating tumour cells (CTCs) was first observed in metastatic cancer patients [2]. Later on in 1948, circulating nucleic acids were detected and quantified in blood [3] followed by the demonstration of the existence of cell-free DNA (cfDNA) in lupus patients in 1960 [4]. In oncology, the presence of cfDNA was noted towards the beginning the 80 s [5,6]. Since then, research using cfDNA had been on the rise firstly culminating in the identification of specific mutations in cfDNA in 1994 [7]. Nowadays, liquid biopsy permits the collection and study of multitude of circulating components released into the bloodstream by necrosis, apoptosis or actively by tumour cells [8]. These components, termed tumour circulome, can be divided into ctDNA, CTCs, circulating cell-free RNA (cfRNA), tumour-educated platelets (TEPs), extracellular vesicles (EVs) and a wide spectrum of proteins and metabolites amongst others [9] (Fig. 1).
Breast cancer is the most common neoplasm affecting women worldwide accounting for 23% of total oncologic diagnoses and 14% of cancer-related deaths [10]. Around 95% of breast cancer patients are detected in early stage without macroscopic evidence of metastases using existing screening techniques [11]. Despite the lack of evidences for a distant relapse, approximately 20% of patients will recur over the disease course with incurable frank-metastatic disease, suggesting a non-detectable tumour spread could already be at play from diagnosis [12]. In this regard, liquid biopsy combined with highly sensitive molecular technologies and advance bioinformatics protocols can substantially help to improve diagnosis and monitoring at this stage. These methods allow to obtain detailed tumour molecular information in a non-invasive manner. The potential of tumour circulome has been demonstrated in numerous studies, not only to diagnose breast cancer but also in treatment response monitoring during adjuvant and neoadjuvant therapies, minimal residual disease (MRD) identification and early relapse detection in localized and locally advance breast cancer. The so-far published studies addressing these questions mainly employed ctDNA and CTCs detection, oftentimes underscoring the ultra-low presence of these components in bodily fluids. This is especially notable for MRD detection as well as during and after neoadjuvant treatments, where multitude of ultra-sensitive technologies are under development to overcome this challenge.
In this review, we describe the most noteworthy research results highlighting the most commonly studied circulating tumour components in the early breast cancer setting.
2. Circulating-tumour DNA in early breast cancer
cfDNA refers to the bulk fragmented DNA released into the bloodstream by all cells of human body whereas circulating-tumour DNA (ctDNA) represents only the part of cfDNA that originates exclusively from the tumour tissue [13]. ctDNA can be released into the bloodstream mostly by apoptosis but also by necrosis or actively by tumour cells [14]. As a consequence of its apoptotic caspase-dependant cleavage origin, ctDNA is generally around 166 bp long [13]. The ctDNA fraction fluctuates according to the stage and cancer type. In patients with advanced or metastatic tumours, the ctDNA fraction represents up to 10%, in contrast to ~1% in the case of non-metastatic advanced cancer patients and <0•1% in patients with early localized tumours [15]. Consequently, ctDNA detection in early cancer stages is the most challenging and requires ultra-sensitive technologies. In this setting, ctDNA is undetectable in 90% of patients undertaking neoadjuvant therapy (NAT), which further decreases ctDNA levels [15], and no correlation exists between ctDNA detection and complete pathological response (pCR) [16]. On top of that, MRD is hardly measurable after surgery using current technologies.
Nevertheless, ctDNA-derived information has already served to detect cancer in its early stages [17], [18], [19], to monitor treatment response [20], therapeutic resistance [21], MRD after primary treatments, and/or risk-of-relapse (ROR) determination [22,23]. ctDNA is generally analysed either by digital-droplet PCR (ddPCR) or by next generation sequencing (NGS)-based procedures. ddPCR is currently the gold-standard for ctDNA assessment due to its excellent sensitivity, simplicity, and an interesting price/sample ratio. It allows longitudinal tracking of somatic mutations and a highly precise calculation of the total number of ctDNA molecules in a given sample. This technology was instrumental in a crucial study published by García-Murillas et al. [24], where the ddPCR system was used to detect MRD and predict the risk of relapse in breast cancer patients treated with NAT. The authors tracked mutations previously characterized in the primary tumour to detect ctDNA in post-surgery and in longitudinal follow-up plasma samples. They observed a strong positive correlation between ctDNA detection and future relapse. Importantly, they calculated a lead time of 7•9 months from ctDNA detection over clinical relapse. They also characterized the mutational profiles of ctDNA in the follow-up plasma samples to decipher the genetics of the therapy-resistant tumour clones using massive-parallel sequencing (MPS). Additionally, expanding the study set in a more recent paper the authors corroborated the high capacity of the methodology to monitor early breast cancer disease with an improved lead time of ctDNA detection over clinical relapse of 10•7 months [25]. In a different study developed by Rothé et al. [26] ddPCR was also employed in the neoadjuvant setting. Therein, the authors showed the feasibility of using ctDNA as biomarker for tumour response to NAT.
Another PCR-based technology is the so-called BEAMing, which combines magnetic beads, amplification steps and microfluidics to identify and quantify somatic mutations in biofluids. The most important study using this platform was published by Bettegowda et al. [27], where they investigated the technology capacity to detect ctDNA in early and advance cancers. Importantly, they included localized breast tumours where they detected ctDNA in a modest 50% of the total breast cancer cases.
On the other hand, the NGS-based approaches additionally permit the description of the cancer mutational profiles. They can be based on fixed panels composed of genes involved in cancer development and/or on patient-specific panels. With respect to fixed panels, it is important to mention the development of the so-called cancer personalized profiling by deep sequencing (CAPP-Seq). This is an ultrasensitive method for quantifying ctDNA using the most common genetic alterations of a given cancer and complex wet-lab methodologies together with customized bioinformatic analyses to build up a high sensitive ctDNA detection strategy [28]. Another example of fixed panels, which additionally incorporates current breast cancer surveillance procedures, was published by Zhang et al. [29]. In detail, the authors developed a novel methodology to detect ctDNA, designing a panel based on COSMIC data covering 136 genes, which was integrated with the Breast Imaging Reporting and Data System classification (BI-RADS). The predictive value of this combination increased up to 92% and ctDNA detection also served as a predictor of worse prognosis. These results indicate that the combination of ctDNA detection with current imaging techniques might be used to avoid post-surgery overtreatment. In another study, the utilization of fixed panels permitted the development of a novel technology called Targeted Error Correction Sequencing (TEC-Seq) to early detect different cancer types, including breast cancer, using MPS [18]. Therein, 55 commonly mutated driver genes were analysed using an amplicon-based technology. Unexpectedly, the ctDNA detection rate was 67% in stage I breast cancer, while this percentage decreased in stage II and III to 59% and 46%, respectively. These surprising results were probably caused by the low number of patients included in the study. In a recent study, Jimenez-Rodriguez et al. [30] compared the mutational profiles of solid tumours and plasma samples from early breast cancer patients using the amplicon-based SafeSEQ (Sysmex Inostics) technology and a fixed-gene panel for plasma sequencing. Interestingly, the authors observed that plasma DNA sequencing permitted the identification of clonal mutations not detected in tumour biopsy sequencing. On top of that, they showed correlation between ctDNA detection with age, tumour grade and size, immunohistochemical subtype, BI-RADS category, and lymph node positivity. This study highlights the importance to test plasma DNA for somatic mutation detection and improve the clinical management in localized breast cancer. Finally, Chen et al., [31] utilized the commercially available Ion Ampliseq Oncomine Research Panel to detect somatic mutations in primary tumour, blood and plasma DNA in pre and post-surgery early breast cancer samples. They showed very limited sensitivity (31%) in detecting ctDNA in relapsed patients. This is potentially due to the low capability of the employed technology to detect ctDNA at ultra-low allele frequencies required in this setting.
Recent efforts have also focused on the development of methodologies combining fixed NGS gene panels for multi-cancer ctDNA detection together with other circulating biomarkers. The CancerSEEK is a method for early detection of eight different cancer types, including breast cancer, as well as for the location of the organ-of-origin [17]. This test combines protein analysis with ctDNA detection, using driver mutations to detect tumours in early stages. The technology uses PCR amplification with molecular barcodes and NGS. The sensitivity for cancer detection ranges from 69 to 98% in five cancers with a specificity of 99%. However, the sensitivity to detect breast cancer in early stages is rather limited (33%), highlighting the restriction in using fixed panels in this cancer type considering the low mutagenicity rate observed in these tumours [32].
In a recent study Wan et al. developed a novel methodology for ctDNA detection, called INtegration of VAriant Reads (INVAR), and applied it to several cancer types including breast, both in the early and advanced stage. INVAR scans up to a thousand loci for mutations by error-suppression methods and signal-enrichment procedures. As little as one mutant molecule per 100,000 can be detected, thus significantly increasing the ctDNA detection sensitivity. In localized breast cancers, ctDNA was found in 62•5% of the cases with a specificity of 90%. The median integrated mutant allele fraction (IMAF) obtained in early breast cancer patients was 5•2 parts-per-million (ppm), substantially lower compared with the 15,000 ppm obtained in advanced melanoma, highlighting again the difficulties to detect ctDNA in localized breast tumours [33].
Other NGS-based procedures, including whole-exome or genome sequencing, can also be employed to delimit broad mutational profiles and copy number variations (CNVs) in ctDNA [13] and construct patient-specific panels. In this respect, Coombes et al. [22] designed personalized panels composed of 16 patient-specific variants, selected from primary tumour whole-exome sequencing data and ultradeep sequencing, for recurrence detection in localized breast cancer. ctDNA was detected in 89% of the patients who relapsed and not detected in 100% of the non-relapsed ones. Molecular relapse was detected up to two years before clinical or radiological relapse. Another example of a clever use of NGS technologies to design patient-specific panels in the neoadjuvant setting is the technology developed by McDonald et al. [34]. In detail, TARDIS uses patient-specific panels composed of multiple tumour mutations as ctDNA biomarkers for disease monitoring in the pre-surgery setting as well as to detect MRD. This method combines a targeted linear pre-amplification, followed by unique molecular identifiers (UMIs) ligation, targeted exponential PCR, and ultra-deep sequencing. Applying this technology, the authors observed a ctDNA allele frequency (AF) of 0•11% before and 0•003% and 0•017% after NAT in patients with pCR and residual disease, respectively. TARDIS demonstrated the ctDNA clinical relevance as a biomarker for NAT treatment response and MRD monitoring in early breast cancer patients, showing excellent sensitivity with a 100-fold improvement beyond current ctDNA detection methods.
Evidently, both PCR and NGS-based techniques have shown promising results for early breast cancer patient management (Table 1), although neither of them is currently incorporated into the clinical practise. Since ctDNA detection in localized tumours continues to be challenging, more and more studies and techniques are under development to reconfirm its diagnostic, prognostic and predictive potential.
Table 1.
Liquid biopsy studies in early breast cancer.
| Study | Analyte | Method | Findings | References |
|---|---|---|---|---|
| Bettegowda et al., 2014 | ctDNA | BEAMing |
|
[27] |
| García-Murillas et al., 2015 | ddPCR |
|
[24] | |
| Riva et al., 2017 | ddPCR |
|
[16] | |
| Phallen et al., 2017 | TEC-Seq |
|
[18] | |
| Chen et al., 2017 | Oncomine panel |
|
[31] | |
| Cohen et al., 2018 | CancerSEEK |
|
[17] | |
| McDonald et al., 2019 | TARDIS |
|
[34] | |
| García-Murillas et al., 2019 | ddPCR |
|
[25] | |
| Coombes et al., 2019 | Personalized panels and ultra-deep sequencing |
|
[22] | |
| Rothé et al., 2019 | ddPCR |
|
[26] | |
| Zhang et al., 2019 | Large NGS panels |
|
[29] | |
| Jimenez-Rodriguez et al., 2019 | SafeSEQ |
|
[30] | |
| Wan et al., 2020 | INVAR |
|
[33] | |
| Pierga et al., 2008 | CTCs | CellSearch® |
|
[56] |
| Bidard et al., 2010 | CellSearch® |
|
[49] | |
| Lucci et al., 2012 | CellSearch® |
|
[58] | |
| Bidard et al., 2013 | CellSearch® |
|
[50] | |
| Strati et al., 2013 | Adnatest, RT-qPCR |
|
[62] | |
| Pierga et al., 2015 | CellSearch® |
|
[53] | |
| Hall et al., 2015 | CellSearch® |
|
[51] | |
| Kasimir-Bauer et al., 2016 | AdnaTest® |
|
[61] | |
| Khosravi et al., 2016 | Nanotube-CTC—Chip |
|
[66] | |
| Pierga et al., 2017 | CellSearch® |
|
[57] | |
| Riethdorf, 2017 | CellSearch® |
|
[55] | |
| Politaki et al., 2017 | CellSearch®, RT-qPCR, dIF |
|
[63] | |
| Bidard et al., 2018 | CellSearch® |
|
[35] | |
| Sparano et al., 2018 | CellSearch® |
|
[52] | |
| Kwan et al., 2018 | CTC-iChip, RNAseq, microarray |
|
[64] | |
| Goodman et al., 2018 | CellSearch® |
|
[59] | |
| Loeian et al., 2019 | Nanotube-CTC—Chip |
|
[67] | |
| Trapp et al., 2019 | CellSearch® |
|
[60] | |
| Radovich et al., 2020 | ctDNA CTCs |
ctDNA: NGS fixed panel CTCs: microfluidic device (Ep-CAM positive selection) |
|
[65] |
| Roth et al., 2010 | cfmiRNAs | RT-qPCR |
|
[76] |
| Asaga et al., 2011 | RT-qPCR-DS |
|
[77] | |
| Kodahl et al., 2014 | RT-qPCR |
|
[78] | |
| Matamala et al., 2015 | Microarray, qRT-PCR |
|
[79] | |
| Kleivi Sahlberg et al., 2015 | RT-qPCR |
|
[82] | |
| Shimomura et al., 2016 | Microarray, qRT-PCR |
|
[80] | |
| Hamam et al., 2016 | Microarray, qRT-PCR |
|
[81] | |
| Papadaki et al., 2018 | RT-qPCR |
|
[83] | |
| Rodríguez-Martínez et al., 2019 | RT-qPCR |
|
[84] |
Abbreviations: ctDNA, circulating-tumour DNA; ddPCR, digital droplet PCR; MRD, minimal residual disease; NAT, neoadjuvant therapy; VAF, variant allele frequency; pathCR, pathological complete response; NGS, next generation sequencing; RT-qPCR, quantitative reverse transcription PCR; BI-RADS, Breast Imaging Reporting and Data System; IMAF, integrated mutant allele fraction; CTCs, circulating-tumour cells; PFS, progression-free survival; OS, overall survival; DFS, disease-free survival; DDFS, distant disease-free survival; DMFS, distant metastasis-free survival IF, Immunofluorescence; dIF, double immunofluorescence; LRFS, local recurrence-free survival; cfmiRNAs, circulating-free miRNAs; miRNA, micro RNA; RT-qPCR-DS, RT-qPCR applied directly in serum; ER, oestrogen receptor; TNBC, triple-negative breast cancer.
3. Circulating-tumour cells in early breast cancer
CTCs are released from the bulk tumour and migrate into the blood vessels by trans-endothelial transition to enter the body circulation. Such cells that are additionally capable to adapt and survive in different tissues can originate new tumours or metastases [35]. CTCs can be released from the primary tumour and/or the metastases at any stage of the tumorigenesis, even including localized tumours [36]. CTCs population is very heterogenous with the majority of cells being highly differentiated, while others have stem-cell like properties (CSCs) including increased self-renewal and marked resistance to therapies [37]. CTCs characteristics additionally reflect the cancer type and stage, e.g. in early breast cancer CTCs do not present increased “mitotic features” compared to the metastatic disease [38]. Important phenotypic changes are induced in CTCs by the epithelial-to-mesenchymal transition (EMT), adapting them to survive in the bloodstream and colonize other tissues [39]. Interactions with numerous blood components such as platelets are crucial for promoting their metastatic potential [40], conferring them protection from the host´s immune system [41,42]. CTCs can travel as individual cells through the bloodstream but it was recently discovered that they can also associate in clusters, which increases 23–50 times their metastatic capacity [43]. CTCs half-life is established in between one to three hours, with only 0•1% surviving for more than 24 h in the bloodstream, and 0•01% having the ability to metastasize [44,45].
CTCs are substantially less abundant in the blood of patients with early tumours, however there is evidence in the literature about the detection of CTCs in all breast cancer stages and their correlation with unfavourable prognosis, lack of treatment efficacy and tumour progression [46,47]. CTCs detection and characterization require to firstly perform an enrichment step considering CTCs rareness in the circulation, especially in the early cancer setting. This is implemented using molecular markers on the surface of these cells such as the epithelial cell adhesion molecule (EpCAM). This enrichment step is classified depending on whether the target cells or non-target cells are bound by the antibodies (positive or negative selection, respectively). Other procedures based on cell density or size are also gaining importance because of their capacity to process large volumes of samples in a more cost-effective manner [48].
There is a wide variety of technologies for CTCs detection and characterization developed by the biotech industry, which is growing exponentially at the same time as the CTCs’ clinical impact is being highlighted. The methodology developed by Menarini Silicon Biosystems called CellSearch® is the gold standard and the only technique approved by the FDA for the isolation and detection of CTCs in metastatic breast, prostate and colon cancer. This method is based on CTCs immunoisolation by positive selection targeting EpCAM. However, CTCs that are not expressing EpCAM because of the EMT or stem-cell like properties will not be detected. Nevertheless, this methodology is the most employed in studies involving early and advanced breast cancer samples. Numerous investigations confirmed that a count of ≥1 CTCs in 7•5 ml of blood with CellSearch® system at different time-points before and after surgery of localized breast tumours is associated with inferior progression-free survival (PFS), overall survival (OS) [49], [50], [51],and late recurrence [51,52] In the Beverly study [53,54], those patients with detectable CTCs at baseline had shorter disease-free survival (DFS) and OS. Similar results were found in the GeparQuattro trial [55], which included patients treated with NAT. In two different studies, Pierga et al. [56,57] investigated the existence of CTCs in pre- and post-neoadjuvant blood samples in 118 and 137 non-metastatic breast cancer patients, respectively. They also observed that patients with ≥1 CTCs showed worse DFS and OS. Another example is the study published by Lucci et al. [58] in 2012 showing the correlation between one or more CTCs in 7•5 ml of blood with worse PFS and OS. A meta-analysis published by Bidard et al. [35] in 2018 analysed the presence of CTCs from different studies in a total of 1574 samples from early stage patients before NAT treatment and 1200 samples from patients before surgery using this system. They observed that one or more CTCs were detected in 25•2% of pre-NAT patients and this was associated with tumour size. Additionally, the number of detected CTCs inversely correlated with the OS, DFS, and locoregional relapse-free interval, but not with the pCR. Patients with more than one CTC prior to NAT showed increased risk of death. Furthermore, in 861 patients where CTCs screening was performed before NAT, an increase in the prognostic ability for OS, distant DFS, and locoregional relapse-free interval was addressed. CTCs counts were an independent quantitative prognostic factor in patients with early breast cancer treated with NAT and they could complement current prognostic models based on tumour characteristics and response to treatment. Additionally, in one study published by Goodman et al. [59], the association between CTCs detection and the benefit of radiotherapy (RT) was assessed in 3213 patients from the NCDB and SUCCESS trials. CTCs were detected in 23•5% and 19•4% patients, respectively. In both cohorts, RT was associated with better OS in patients with positive CTCs counts, but not in the negative ones. In addition, in the SUCCESS cohort, RT was associated with longer DFS and local recurrence-free survival (LRFS). Finally, Trapp et al. [60] recently investigated the prognostic potential of CTCs detection in 1087 early breast cancer patients using CellSearch. A multivariable analysis showed that CTCs status at baseline and two years after chemotherapy was statistically significantly associated with OS and DFS in almost all breast cancer subtypes.
Another commercial method for CTCs analysis is the Adnatest (QIAGEN®). This procedure uses a combination of antibodies conjugated with magnetic beads for selecting tumour and epithelial markers and an RT-PCR for detecting breast cancer mRNAs biomarkers. Kasimir-Bauer et al. [61] employed this methodology to isolate CTCs in the breast cancer neoadjuvant setting. According to the results, the survival and detection of resistant and stem cell-like resistant CTCs after NAT was associated with worse prognosis in these patients. Adnatest was also applied in a comparative study using three different methods for detection and molecular characterization of CTCs in early and metastatic breast cancer patients [62]. CTCs were isolated from the peripheral blood of 254 early breast cancer patients. Molecular characteristics were assessed using a singleplex RT-qPCR for CK19, a multiplex RT-qPCR, and the AdnaTest BreastCancer™ and compared. CTCs positivity was observed in 14•2%, 22•8% and 16•5% of the cases, respectively. In the early setting, the concordance between the AdnaTest and CK-19 RTqPCR was 72•4% while the AdnaTest and multiplex RT-qPCR showed a concordance of 64•6%. This low correlation was attributable to CTCs’ heterogeneity and the diversity of genes included in the different methodologies. In another comparative study, Politaki et al. tested three different methods for CTCs detection in 200 early breast cancer patients. Using the CellSearch® System, CTCs (≥1 and ≥2) were detected in 23 ml of blood in 37% and 16•5% of the early breast cancer patients, respectively. On the other hand, 18•0% showed positivity by RT-qPCR and 16•9% by immunofluorescence. Importantly, no agreement was observed between methods [63].
The prognostic value of CTCs detection has also been demonstrated in a recent study published by Kwan et al. [64]. In this research, a digital RNA signature and a new technology called CTC-iChip were employed for CTCs isolation and detection in early and metastatic breast cancer patients. To develop the RNA expression signature, RNASeq and microarray expression data from breast cancer, blood and normal breast tissue were analysed. A 17-gene signature confirmed by single cell RNA sequencing and ddPCR was developed. In the localized breast cancer cohort (n = 54), no association between CTC-score and tumour grade or nodal status was found. However, a high baseline CTC-score was associated with residual disease. Furthermore, an elevated CTC-score during NAT (≥3 cycles) was associated with residual disease at the time of surgery. Therefore, the authors supported the prognostic value of CTCs detection in localized breast cancer patients. A recent study correlated ctDNA and CTCs detection with recurrence in triple-negative breast cancer (TNBC) patients after NAT. The results showed that the detection of either ctDNA or CTCs is independently associated with recurrence. Both DFS and distant disease-free survival (DDFS) were inferior for patients with detectable ctDNA and CTCs in contrast to those with undetectable circulating analytes [65].
An additional noteworthy novel methodology to detect CTCs in early breast cancer patients is the so-called nanotube-CTC-chip [66,67]. This system relies on label-free nanotube-antibody microarrays applying breast cancer-specific antibodies such as anti-EpCAM and anti-her2 amongst others. The authors not only demonstrated that this new technology is able to capture a mean of 62% of tumour cells in spike-in experiments, but they also showed the capability of the system to identify CTCs in the 100% of the studied breast cancer peripheral blood samples.
In conclusion, CTCs detection in the early breast cancer setting remains challenging, partially due to the high heterogeneity observed amongst CTCs, which makes the concordance and inter-method variability between studies an important caveat to conquer (Table 1). However, the use of CTCs identification and characterisation possess a great potential to further increase the clinical value of these cells in breast cancer management.
4. Circulating-tumour RNAs in early breast cancer
In tumour tissue, it is already known that RNA, especially the non-coding counterpart, plays important roles in the deregulation of cell homoeostasis and cancer development. RNA can also be released from tumour cells into the circulation of oncologic patients and thus be a potential analyte for liquid biopsy-based approaches. However, its study in liquid biopsies, correlation with cancer stages, clinical presentations and/or treatment responses are severely affected by its limited stability and the variability in the methodologies employed [68]. There is still little evidence regarding the circulating tumour RNA's utility to depict the tumour molecular profile or to use it as a biomarker in cancer patients, especially for localized tumours where its amounts are under the limit of detection for most current technologies. Nevertheless, several types of circulating tumour RNAs could actually be studied. The interest in the circulating non-coding RNAs (ncRNAs) is exponentially growing since their important implication related to cancer biology was described. They represent 80% of the total circulating RNA and are involved in the regulation of transduction pathways, acting also as cancer drivers [69]. Long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), small nuclear and nucleolar RNAs (snRNAs and snoRNAs), and PIWI-interacting RNAs (piRNAs) are ncRNAs molecules of great interest with important roles in cancer biology [70], [71], [72], [73]. However, little is known about their presence and utility as liquid biopsy biomarkers, requiring further studies to determine their clinical interest.
Conversely, miRNAs are ncRNAs small in size (~22 nt), capable to regulate gene expression at the post-transcriptional level with important implications in the development of a plethora of tumours [74]. They are present in blood and in other biological fluids and they are the most abundant cfRNAs [75]. miRNAs can circulate as free particles or associated with exosomes, either in their lumen or on their surface, which affects their stability. miRNAs are the most studied RNA species in tissues from localized breast cancers but also in the bloodstream, where several studies demonstrated their clinical potential (Table 1).
Circulating miRNA profiles were already employed in breast cancer patients across several studies. Different methods were applied to analyse them including RT-qPCR, digital PCR (dPCR), microarrays, and NGS. Currently, RT-qPCR and microarrays are mostly employed. In this regard, Roth et al. [76] investigated the expression of four miRNAs in the serum of primary and metastatic breast cancer patients. Their results demonstrated that the total amount of circulating RNAs and the serum miR-155 concentration could differentiate between patients with primary breast cancer and healthy individuals, while miR-10b and miR-34a levels were increased in metastatic patients and allowed their identification. Furthermore, they observed that advanced tumour stages were associated with an increased amount of total circulating RNAs and specifically with the overexpression of miR-34a. In another example, Asaga et al. [77] demonstrated that miR-21 was upregulated in serum of patients with any breast cancer stage, including localized tumours, compared to serum from healthy controls. Kodahl et al. [78] additionally identified a signature composed of nine miRNAs to discriminate between early breast cancer patients and healthy controls. Matamala et al. [79] studied the presence of four miRNAs in blood and found them to be overexpressed in pre-treated patients compared to controls and treated individuals. Shimomura et al. [80] analysed the serum of 1280 patients with early breast cancer and showed that the combination of five miRNAs was capable to detect early breast cancer with 98•0% sensitivity. Finally, Hamam et al. detected 18 upregulated miRNAs in patients with stage I, II and III, compared to stage IV, indicating that miRNAs showed potential to stratify tumours by stage [81].
The miRNAs were also investigated in early breast cancer as prognostic biomarkers. Kleivi Sahlberg et al. [82] demonstrated that the expression of four miRNAs predicted tumour relapse and OS. High-risk patients tended to express these four miRNAs more often and had lower OS than low-risk patients. Papadaki et al. [83] studied the expression levels of a different set of four miRNAs in a series of samples from localized breast cancer patients, observing an increase in the expression of miR-21, miR-23b, and miR-200c accompanied by a decrease in miR-190 in relapsed patients compared to the non-relapsed ones. They concluded that the combined expression of these four miRNAs as well as the miR-200c expression by itself could predict relapse and thus help to better stratify patients.
On top of that, miRNAs can also be utilized to monitor treatment response in the early breast cancer setting. Rodríguez-Martínez et al. [84] studied the role of exosomal miRNAs as a complementary tool for treatment response prediction in patients treated with NAT. miRNAs expression was measured before and after NAT, observing that miR-21 and miR-105 were overexpressed in metastatic patients compared to non-metastatic ones as well as controls.
Considering all the above-mentioned, RNAs in circulation have an interesting potential to improve the clinical management of breast cancer patients using an easy-to-obtain blood sample.
5. Outstanding questions
Liquid biopsy is a non-invasive methodology which serves to obtain key tumour information and it is rapidly transforming the cancer patient's clinical management. The assessment of the tumour circulating components in the early and advanced setting is very well documented and it is also reflected in the multitude of commercial assays that use ctDNA to determine the tumour genetic profiles, the treatment response including resistance as well as a biomarker for disease monitoring [65,85]. Indeed, the clinical validity of liquid biopsy in the early breast cancer setting is more than evident; multitude of clinical trials, aiming to demonstrate that the clinical decision-making based on these circulating components could serve to increase patient´s survival, are on-going or already finished. It is indisputable that liquid biopsies show great promise based on the data presented by several observational studies, albeit with limited number of patients. However, its clinical utility is still to be fully established and requires performing multicentre clinical studies including large series of breast cancer patients.
The detection and characterization of circulating tumour materials in the early setting present much more difficulties due to their low amounts in the biofluids from the patients. Important advances were performed using ctDNA, where ultrasensitive cutting-edge NGS technologies and patient-specific panels have served to characterize the tumours, to assess responses to neoadjuvant chemotherapies as well as to detect MRD after surgery. This revolution in methodology development gave rise to several NGS procedures employing different pipelines with the same objective - to detect ultra-low diluted ctDNA. However, there are still important caveats to their clinical extrapolation such as their technical complexity, especially at the bioinformatic level where non-specialized user-friendly software needs to be developed.
On the other hand, additional research is needed to validate the utility of CTCs, where so far only CTCs enumeration shows prognostic capacities in early breast cancer. The use of the CellSearch® System demonstrated validity in CTCs’ detection for outcome prediction in observational studies including early breast cancer patients (Table 1). However, interventional clinical trials addressing the clinical relevance of this technology in detecting CTCs, stratifying patients and applying personalized treatments are still missing.
Considering the difficulty to isolate CTCs from early cancer patients using standard volumes of blood draws, possibly single cell molecular characterization is the natural next step to uncover the full potential of CTCs in this setting. Studying CTCs at single-cell level could provide insights about the genetic characteristics of potential occult metastasis and their heterogeneity [86]. Such molecular characterization could permit to track disease evolution in follow-up blood samples using genetic aberrations as biomarkers and/or to treat patients and prevent disease relapses based on targetable driver mutations, as was previously demonstrated by detecting ESR1 resistance mutations in metastatic breast tumours [87].
Finally, the study of RNAs in circulation is showing great interest since more and more publications are presenting interesting results. Most of these articles employed miRNAs for molecular tumour stratification, treatment response and/or relapse prediction. However, the characterization of other circulating RNA types, such as the lncRNAs or the circRNAs that demonstrated importance in other cancer types [88,89], will likely open new avenues to uncover the real capacity of these analytes in breast cancer patients.
Overall, more research needs to be performed in the standardization of sample extraction procedures, circulating-tumour material isolation and the very diverse methodologies employed, aiming to extrapolate the use of the different circulating molecules into the “real world” in clinics. On top of that, the “only one analyte” barrier should be crossed, focusing more on multi-omics approaches to finally unleash the full clinical utility of liquid biopsy in cancer patients.
6. Search strategy and selection criteria
Data for this Review were identified by searches in MEDLINE, Current Contents, PubMed, and references from relevant articles using the terms “ctDNA”, “CTCs”, “early breast cancer”, “liquid biopsy”, and “cfRNA”. Only articles published in English and French were included with preference for those published in the last ten years.
Contributors
ICM and EA conceptualized and supervised the conception of the manuscript. ICM, EA and AAB coordinated and performed the literature search with RLV, MEDR, BJR, MIQO contribution. AAB and ICM designed and performed figures and tables. ICM and AAB wrote the manuscript with important inputs from all authors. All authors reviewed and agree with the content of the manuscript. All authors have read and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
ICM´s contract is funded by the Spanish Association Against Cancer (AECC). AAB is contracted by the “Garantía Juvenil en I+D+i Subprograma Estatal de Incorporación” by the Ministry of Science and Innovation (Spanish Government). MIQO contract is supported by the “Miguel Servet Type II” program (CPI13/00003), ISCIII, Spain and co-funded by the "Fondo Europeo de Desarrollo Regional-(FEDER)", and by the “Nicolas Monardes” research program from the "Consejería de Salud" (C-0030-2018), Andalusian Gobernment, Spain.
We thank the “Consejería de Salud de la Junta de Andalucía” and the “Fundación Unicaja” for their support to ICM´s projects.
We thank Veronika Mancikova for editing the paper.
The above-mentioned funders have no role in this review article.
Contributor Information
Emilio Alba, Email: ealbac@uma.es.
Iñaki Comino-Méndez, Email: inaki.comino@ibima.eu.
References
- 1.Kilgour E., Rothwell D.G., Brady G., Dive C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell. 2020;37(4):485–495. doi: 10.1016/j.ccell.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Ashworth T. A case of cancer in which cells similar to those in the tumors were seen in blood after death. Med J Aust. 1869;14:145–147. [Google Scholar]
- 3.Mandel P., Metais P.C.R. Les acides nucléiques du plasma sanguin chez l'homme. (Article in French) C R Seances Soc Biol Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 4.Chen J.A., Meister S., Urbonaviciute V., Rodel F., Wilhelm S., Kalden J.R. Sensitive detection of plasma/serum DNA in patients with systemic lupus erythematosus. Autoimmunity. 2007;40(4):307–310. doi: 10.1080/08916930701356317. [DOI] [PubMed] [Google Scholar]
- 5.Leon S., Shapiro B., Sklaroff D., Yaros M. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- 6.Stroun M., Anker P., Maurice P., Lyautey J., Lederrey C., Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46(5):318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 7.Sorenson G., Pribish D., Valone F., Memoli V., Bzik D., Yao S. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3(1):67–71. [PubMed] [Google Scholar]
- 8.Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F.O., Hesch R.D. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 9.Bardelli A., Pantel K. Liquid Biopsies, What We Do Not Know (Yet) Cancer Cell. 2017;31(2):172–179. doi: 10.1016/j.ccell.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 11.Howlader N., Altekruse S.F., Li C.I., Chen V.W., Clarke C.A., Ries L.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst. 2014;106(5) doi: 10.1093/jnci/dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry D.A., Cronin K.A., Plevritis S.K., Fryback D.G., Clarke L., Zelen M. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353(17):1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 13.Alimirzaie S., Bagherzadeh M., Akbari M.R. Liquid biopsy in breast cancer: a comprehensive review. Clin Genet. 2019;95(6):643–660. doi: 10.1111/cge.13514. [DOI] [PubMed] [Google Scholar]
- 14.Thierry A.R., El Messaoudi S., Gahan P.B., Anker P., Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35(3):347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riva F., Bidard F.C., Houy A., Saliou A., Madic J., Rampanou A. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin Chem. 2017;63(3):691–699. doi: 10.1373/clinchem.2016.262337. [DOI] [PubMed] [Google Scholar]
- 17.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J., et al. Direct detection of early-stage cancers using circulating tumor DNA. 2017;9(403):eaan2415. [DOI] [PMC free article] [PubMed]
- 19.Beaver J.A., Jelovac D., Balukrishna S., Cochran R.L., Croessmann S., Zabransky D.J., et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. 2014;20(10):2643–50. [DOI] [PMC free article] [PubMed]
- 20.Thierry A.R., Mouliere F., El Messaoudi S., Mollevi C., Lopez-Crapez E., Rolet F. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20(4):430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 21.Misale S., Yaeger R., Hobor S., Scala E., Janakiraman M., Liska D. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coombes R.C., Page K., Salari R., Hastings R.K., Armstrong A., Ahmed S., et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. 2019;25(14):4255–63. [DOI] [PubMed]
- 23.Garcia-Murillas I., Chopra N., Comino-Mendez I., Beaney M., Tovey H., Cutts R.J. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. 2019 doi: 10.1001/jamaoncol.2019.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Murillas I., Schiavon G., Weigelt B., Ng C., Hrebien S., Cutts R.J. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133. doi: 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Murillas I., Turner N.C. Clinical benefit of circulating tumor DNA analysis in follow-up of patients with early-stage breast cancer-reply. JAMA Oncol. 2019 doi: 10.1001/jamaoncol.2019.5682. [DOI] [PubMed] [Google Scholar]
- 26.Rothé F., Silva M.J., Venet D., Campbell C., Bradburry I., Rouas G., et al. Circulating tumor DNA in HER2-amplified breast cancer: a translational research substudy of the NeoALTTO phase III Trial. 2019;25(12):3581–8. [DOI] [PubMed]
- 27.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C., Modlin L.A. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Zhao W., Wei W., You Z., Ou X., Sun M. Parallel analyses of somatic mutations in plasma circulating tumor DNA (ctDNA) and matched tumor tissues in early-stage breast cancer. Clin Cancer Res. 2019;25(21):6546–6553. doi: 10.1158/1078-0432.CCR-18-4055. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez B.J., Cordoba G.D., Aranda A.G., Alvarez M., Vicioso L., Perez C.L. Detection of TP53 and PIK3CA mutations in circulating tumor DNA using next-generation sequencing in the screening process for early breast cancer diagnosis. J Clin Med. 2019;8(8) doi: 10.3390/jcm8081183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y.H., Hancock B.A., Solzak J.P., Brinza D., Scafe C., Miller K.D. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer. 2017;3:24. doi: 10.1038/s41523-017-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martincorena I., Campbell P.J. Somatic mutation in cancer and normal cells. Science. 2015;349(6255):1483–1489. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 33.Wan J.C.M., Heider K., Gale D., Murphy S., Fisher E., Mouliere F., et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. 2020;12(548):eaaz8084. [DOI] [PubMed]
- 34.McDonald B.R., Contente-Cuomo T., Sammut S.-.J., Odenheimer-Bergman A., Ernst B., Perdigones N. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med. 2019;11(504):eaax7392. doi: 10.1126/scitranslmed.aax7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bidard F.C., Michiels S., Riethdorf S., Mueller V., Esserman L.J., Lucci A. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J Natl Cancer Inst. 2018;110(6):560–567. doi: 10.1093/jnci/djy018. [DOI] [PubMed] [Google Scholar]
- 36.Rhim A.D., Mirek E.T., Aiello N.M., Maitra A., Bailey J.M., McAllister F. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Micalizzi D.S., Maheswaran S., Haber D.A. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31(18):1827–1840. doi: 10.1101/gad.305805.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams D.L., Adams D.K., Stefansson S., Haudenschild C., Martin S.S., Charpentier M. Mitosis in circulating tumor cells stratifies highly aggressive breast carcinomas. Breast Cancer Res. 2016;18(1):44. doi: 10.1186/s13058-016-0706-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu M., Bardia A., Wittner B.S., Stott S.L., Smas M.E., Ting D.T., et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. 2013;339(6119):580–4. [DOI] [PMC free article] [PubMed]
- 40.Labelle M., Begum S., Hynes R.O. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20(5):576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohme M., Riethdorf S., Pantel K. Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14(3):155–167. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 42.Kitamura T., Qian B.Z., Pollard J.W. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15(2):73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gkountela S., Castro-Giner F., Szczerba B.M., Vetter M., Landin J., Scherrer R. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176(1–2):98–112.e14. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molloy T.J., Bosma A.J., Baumbusch L.O., Synnestvedt M., Borgen E., Russnes H.G. The prognostic significance of tumour cell detection in the peripheral blood versus the bone marrow in 733 early-stage breast cancer patients. Breast Cancer Res. 2011;13(3):R61. doi: 10.1186/bcr2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braun M., Markiewicz A., Kordek R., Sądej R., Romańska H. Profiling of invasive breast carcinoma circulating tumour cells-are we ready for the 'liquid' revolution? Cancers (Basel) 2019;11(2) doi: 10.3390/cancers11020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rack B., Schindlbeck C., Jückstock J., Andergassen U., Hepp P., Zwingers T. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J Natl Cancer Inst. 2014;106(5) doi: 10.1093/jnci/dju066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L., Riethdorf S., Wu G., Wang T., Yang K., Peng G. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res. 2012;18(20):5701–5710. doi: 10.1158/1078-0432.CCR-12-1587. [DOI] [PubMed] [Google Scholar]
- 48.Martos T., Casadevall D., Albanell J. Circulating tumor cells: applications for early breast cancer. In: Piñeiro R, editor. Circulating tumor cells in breast cancer metastatic disease. Springer International Publishing; Cham: 2020. pp. 135–146. editor. [DOI] [PubMed] [Google Scholar]
- 49.Bidard F.C., Mathiot C., Delaloge S., Brain E., Giachetti S., de Cremoux P. Single circulating tumor cell detection and overall survival in nonmetastatic breast cancer. Ann Oncol. 2010;21(4):729–733. doi: 10.1093/annonc/mdp391. [DOI] [PubMed] [Google Scholar]
- 50.Bidard F.C., Belin L., Delaloge S., Lerebours F., Ngo C., Reyal F. Time-dependent prognostic impact of circulating tumor cells detection in non-metastatic breast cancer: 70-month analysis of the REMAGUS02 Study. Int J Breast Cancer. 2013;2013 doi: 10.1155/2013/130470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall C., Karhade M., Laubacher B., Anderson A., Kuerer H., DeSynder S. Circulating tumor cells after neoadjuvant chemotherapy in stage I-III triple-negative breast cancer. Ann Surg Oncol. 2015;22(Suppl 3):S552–S558. doi: 10.1245/s10434-015-4600-6. [DOI] [PubMed] [Google Scholar]
- 52.Sparano J., O'Neill A., Alpaugh K., Wolff A.C., Northfelt D.W., Dang C.T. Association of circulating tumor cells with late recurrence of estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 2018;4(12):1700–1706. doi: 10.1001/jamaoncol.2018.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pierga J.Y., Petit T., Lévy C., Ferrero J.M., Campone M., Gligorov J. Pathological response and circulating tumor cell count identifies treated HER2+ inflammatory breast cancer patients with excellent prognosis: BEVERLY-2 survival data. Clin Cancer Res. 2015;21(6):1298–1304. doi: 10.1158/1078-0432.CCR-14-1705. [DOI] [PubMed] [Google Scholar]
- 54.Bertucci F., Fekih M., Autret A., Petit T., Dalenc F., Levy C. Bevacizumab plus neoadjuvant chemotherapy in patients with HER2-negative inflammatory breast cancer (BEVERLY-1): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2016;17(5):600–611. doi: 10.1016/S1470-2045(16)00011-5. [DOI] [PubMed] [Google Scholar]
- 55.Riethdorf S., Müller V., Loibl S., Nekljudova V., Weber K., Huober J. Prognostic impact of circulating tumor cells for breast cancer patients treated in the neoadjuvant "Geparquattro" trial. Clin Cancer Res. 2017;23(18):5384–5393. doi: 10.1158/1078-0432.CCR-17-0255. [DOI] [PubMed] [Google Scholar]
- 56.Pierga J.Y., Bidard F.C., Mathiot C., Brain E., Delaloge S., Giachetti S. Circulating tumor cell detection predicts early metastatic relapse after neoadjuvant chemotherapy in large operable and locally advanced breast cancer in a phase II randomized trial. Clin Cancer Res. 2008;14(21):7004–7010. doi: 10.1158/1078-0432.CCR-08-0030. [DOI] [PubMed] [Google Scholar]
- 57.Pierga J.Y., Bidard F.C., Autret A., Petit T., Andre F., Dalenc F. Circulating tumour cells and pathological complete response: independent prognostic factors in inflammatory breast cancer in a pooled analysis of two multicentre phase II trials (BEVERLY-1 and -2) of neoadjuvant chemotherapy combined with bevacizumab. Ann Oncol. 2017;28(1):103–109. doi: 10.1093/annonc/mdw535. [DOI] [PubMed] [Google Scholar]
- 58.Lucci A., Hall C.S., Lodhi A.K., Bhattacharyya A., Anderson A.E., Xiao L. Circulating tumour cells in non-metastatic breast cancer: a prospective study. Lancet Oncol. 2012;13(7):688–695. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 59.Goodman C.R., Seagle B.-.L.L., Friedl T.W.P., Rack B., Lato K., Fink V. Association of circulating tumor cell status with benefit of radiotherapy and survival in early-stage breast cancer. JAMA Oncol. 2018;4(8) doi: 10.1001/jamaoncol.2018.0163. e180163-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapp E., Janni W., Schindlbeck C., Jückstock J., Andergassen U., de Gregorio A. Presence of circulating tumor cells in high-risk early breast cancer during follow-up and prognosis. J Natl Cancer Inst. 2019;111(4):380–387. doi: 10.1093/jnci/djy152. [DOI] [PubMed] [Google Scholar]
- 61.Kasimir-Bauer S., Bittner A.K., Konig L., Reiter K., Keller T., Kimmig R. Does primary neoadjuvant systemic therapy eradicate minimal residual disease? Analysis of disseminated and circulating tumor cells before and after therapy. Breast Cancer Res. 2016;18(1):20. doi: 10.1186/s13058-016-0679-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strati A., Kasimir-Bauer S., Markou A., Parisi C., Lianidou E.S. Comparison of three molecular assays for the detection and molecular characterization of circulating tumor cells in breast cancer. Breast Cancer Res. 2013;15(2):R20. doi: 10.1186/bcr3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Politaki E., Agelaki S., Apostolaki S., Hatzidaki D., Strati A., Koinis F. A comparison of three methods for the detection of circulating tumor cells in patients with early and metastatic breast cancer. Cell Physiol Biochem. 2017;44(2):594–606. doi: 10.1159/000485115. [DOI] [PubMed] [Google Scholar]
- 64.Kwan T.T., Bardia A., Spring L.M., Giobbie-Hurder A., Kalinich M., Dubash T. A digital RNA signature of circulating tumor cells predicting early therapeutic response in localized and metastatic breast cancer. Cancer Discov. 2018;8(10):1286–1299. doi: 10.1158/2159-8290.CD-18-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radovich M., Jiang G., Hancock B.A., Chitambar C., Nanda R., Falkson C. Association of circulating tumor DNA and circulating tumor cells after neoadjuvant chemotherapy with disease recurrence in patients with triple-negative breast cancer: preplanned secondary analysis of the BRE12-158 randomized clinical trial. JAMA Oncol. 2020 doi: 10.1001/jamaoncol.2020.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khosravi F., Trainor P.J., Lambert C., Kloecker G., Wickstrom E., Rai S.N. Static micro-array isolation, dynamic time series classification, capture and enumeration of spiked breast cancer cells in blood: the nanotube-CTC chip. Nanotechnology. 2016;27(44):44lt03. doi: 10.1088/0957-4484/27/44/44LT03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loeian M.S., Mehdi Aghaei S., Farhadi F., Rai V., Yang H.W., Johnson M.D. Liquid biopsy using the nanotube-CTC-chip: capture of invasive CTCs with high purity using preferential adherence in breast cancer patients. Lab Chip. 2019;19(11):1899–1915. doi: 10.1039/c9lc00274j. [DOI] [PubMed] [Google Scholar]
- 68.Yuan T., Huang X., Woodcock M., Du M., Dittmar R., Wang Y. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413. doi: 10.1038/srep19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lanzós A., Carlevaro-Fita J., Mularoni L., Reverter F., Palumbo E., Guigó R. Discovery of cancer driver long noncoding RNAs across 1112 tumour genomes: new candidates and distinguishing features. Sci Rep. 2017;7:41544. doi: 10.1038/srep41544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 71.Dvinge H., Guenthoer J., Porter P.L., Bradley R.K. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res. 2019;29(10):1591–1604. doi: 10.1101/gr.246678.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Y., Dou M., Song X., Dong Y., Liu S., Liu H. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18(1):123. doi: 10.1186/s12943-019-1052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marchese F.P., Huarte M. Long non-coding RNAs and chromatin modifiers: their place in the epigenetic code. Epigenetics. 2014;9(1):21–26. doi: 10.4161/epi.27472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rupaimoole R., Calin G.A., Lopez-Berestein G., Sood A.K. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016;6(3):235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roth C., Rack B., Müller V., Janni W., Pantel K., Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12(6):R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Asaga S., Kuo C., Nguyen T., Terpenning M., Giuliano A.E., Hoon D.S. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57(1):84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 78.Kodahl A.R., Lyng M.B., Binder H., Cold S., Gravgaard K., Knoop A.S. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol Oncol. 2014;8(5):874–883. doi: 10.1016/j.molonc.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matamala N., Vargas M.T., González-Cámpora R., Miñambres R., Arias J.I., Menéndez P. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61(8):1098–1106. doi: 10.1373/clinchem.2015.238691. [DOI] [PubMed] [Google Scholar]
- 80.Shimomura A., Shiino S., Kawauchi J., Takizawa S., Sakamoto H., Matsuzaki J. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107(3):326–334. doi: 10.1111/cas.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hamam R., Ali A.M., Alsaleh K.A., Kassem M., Alfayez M., Aldahmash A. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci Rep. 2016;6:25997. doi: 10.1038/srep25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kleivi Sahlberg K., Bottai G., Naume B., Burwinkel B., Calin G.A., Børresen-Dale A.L. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res. 2015;21(5):1207–1214. doi: 10.1158/1078-0432.CCR-14-2011. [DOI] [PubMed] [Google Scholar]
- 83.Papadaki C., Stratigos M., Markakis G., Spiliotaki M., Mastrostamatis G., Nikolaou C. Circulating microRNAs in the early prediction of disease recurrence in primary breast cancer. Breast Cancer Res. 2018;20(1):72. doi: 10.1186/s13058-018-1001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodríguez-Martínez A., de Miguel-Pérez D., Ortega F.G., García-Puche J.L., Robles-Fernández I., Exposito J. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019;21(1):21. doi: 10.1186/s13058-019-1109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vidula N., Dubash T., Lawrence M.S., Simoneau A., Niemierko A., Blouch E. Identification of somatically acquired BRCA1/2 mutations by cfDNA analysis in patients with metastatic breast cancer. Clin Cancer Res. 2020;26(18):4852–4862. doi: 10.1158/1078-0432.CCR-20-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rossi T., Gallerani G., Angeli D., Cocchi C., Bandini E., Fici P. Single-cell NGS-based analysis of copy number alterations reveals new insights in circulating tumor cells persistence in early-stage breast cancer. Cancers (Basel) 2020;12(9) doi: 10.3390/cancers12092490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paolillo C., Mu Z., Rossi G., Schiewer M.J., Nguyen T., Austin L. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells. Clin Cancer Res. 2017;23(20):6086–6093. doi: 10.1158/1078-0432.CCR-17-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang S., Du L., Wang L., Jiang X., Zhan Y., Li J. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23(2):1396–1405. doi: 10.1111/jcmm.14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pan B., Qin J., Liu X., He B., Wang X., Pan Y. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. doi: 10.3389/fgene.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]