Abstract
Background
Diagnosis of rib fractures plays an important role in identifying trauma severity. However, quickly and precisely identifying the rib fractures in a large number of CT images with increasing number of patients is a tough task, which is also subject to the qualification of radiologist. We aim at a clinically applicable automatic system for rib fracture detection and segmentation from CT scans.
Methods
A total of 7,473 annotated traumatic rib fractures from 900 patients in a single center were enrolled into our dataset, named RibFrac Dataset, which were annotated with a human-in-the-loop labeling procedure. We developed a deep learning model, named FracNet, to detect and segment rib fractures. 720, 60 and 120 patients were randomly split as training cohort, tuning cohort and test cohort, respectively. Free-Response ROC (FROC) analysis was used to evaluate the sensitivity and false positives of the detection performance, and Intersection-over-Union (IoU) and Dice Coefficient (Dice) were used to evaluate the segmentation performance of predicted rib fractures. Observer studies, including independent human-only study and human-collaboration study, were used to benchmark the FracNet with human performance and evaluate its clinical applicability. A annotated subset of RibFrac Dataset, including 420 for training, 60 for tuning and 120 for test, as well as our code for model training and evaluation, was open to research community to facilitate both clinical and engineering research.
Findings
Our method achieved a detection sensitivity of 92.9% with 5.27 false positives per scan and a segmentation Dice of 71.5%on the test cohort. Human experts achieved much lower false positives per scan, while underperforming the deep neural networks in terms of detection sensitivities with longer time in diagnosis. With human-computer collobration, human experts achieved higher detection sensitivities than human-only or computer-only diagnosis.
Interpretation
The proposed FracNet provided increasing detection sensitivity of rib fractures with significantly decreased clinical time consumed, which established a clinically applicable method to assist the radiologist in clinical practice.
Funding
A full list of funding bodies that contributed to this study can be found in the Acknowledgements section. The funding sources played no role in the study design; collection, analysis, and interpretation of data; writing of the report; or decision to submit the article for publication .
Keywords: Rib fracture, Deep learning, Detection and segmentation
Research in context.
Evidence before this study
Quickly and precisely identifying the rib fractures in a large number of CT images is a tough and important task, which plays an important role in identifying trauma severity. Deep leanring has achieved a great success in medical image analysis. In this study, we aimed at a clinically applicable deep learning system to automatically detect and segment rib fractures.
Added value of this study
We present a deep learning system, named FracNet, for automatic detection and segmentation of the rib fractures. The proposed FracNet achieved high detection sensitivity, acceptable false positive per scan and segmentation overlap, which was proven to improve the human detection sensitivity with reduced clinical time comsued in our observer study. Besides, a subset of our dataset was open-source to research community, which is the first open large-scale dataset in this application.
Implications of all the available evidence
The proposed FracNet could help the radiologists in the diagnosis of rib fractures, to increase the efficiency of the clinical workflow, without decreasing the diagnostic accuracy at the same time.
Alt-text: Unlabelled box
1. Introduction
Recent advances in artificial intelligence and computer vision lead to a rapid development of deep learning technology [1] in medical image analysis and digital medicine [2], [3], [4], [5], [6], [7]. With end-to-end learning of deep representation, deep supervised learning, as a unified methodology, achieved remarkable success in numerous 2D and 3D medical image tasks, e.g., classification [8], detection [9], segmentation [10]. With the rise of deep learning, infrastructures, algorithms and data (with annotations) are known to be the keys to its success. Computer-aided diagnosis with a high-performance deep learning is expected to save human labor, improve diagnosis consistency and accuracy, personalize patient treatment, and improve patient–doctor relationship. [11]
Rib fracture represents an important indicator of trauma severity; the number of fractured ribs increases morbidity and mortality [12]. Multidetector computed tomography (CT) provides a more accurate assessment to evaluate for the presence of rib fractures when standard posteroanterior (PA) chest radiograph is specific but insensitive [12], [13], [14], [15]. Definite diagnosis (counting) of the number of rib fractures is also an important indicator in forensic examination for degree of disability [14,16,17]. However, the identification of rib fracture in CT images using conventional axial thin (1–1.5 mm) images is a difficult and labor-intensive task. Each rib has a complex shape with a diagonal course across numerous CT sections [18], which leads to missing rib fracture diagnosis (detection) in clinical practice. For instance, buckle fractures are the most frequently missing type of fracture reported in 2012 [19,20], due to the confusing appearance; nondisplaced rib fractures could be missing when parallel to the scan plane of the CT images. Besides, diagnosing subtle fractures is tedious and time-consuming for a large number of CT slices, which must be evaluated sequentially, rib-by-rib and side-by-side [18].
In this study, we aim at a clinically applicable automatic system for rib fracture detection and segmentation from CT scans. Few prior studies explore the development and validation of deep learning algorithms in this application. We proposed an automatic system named FracNet based on 3D UNet [21], trained and evaluated with a large-scale dataset, named RibFrac Dataset, consisting of 7,473 voxel-level rib fracture segmentation from 900 chest-abdomen CT scans (332,483 CT slices). The annotation of RibFrac Dataset followed a human-in-the-loop labeling procedure, which ensures a high standard of annotation quality. On RibFrac test cohort, the proposed FracNet system achieved a detection sensitivity of 92.9% (with 5.27 false positives per scan) and a segmentation Dice Coefficient of 71.5%, which outperformed counterpart methods based on 3D variants of FCN [22] and DeepLab v3+ [23] with a 3D ResNet-18 backbone [24,25]. Furthermore, observer studies with two experienced radiologists, including independent human-only study and human-collobration study, were designed to validate the clinical value of the proposed system. Our system achieved higher detection sensitivities than human experts. Importantly, human-computer collaboration significantly improved detection sensitivities over computer-only and human-only diagnosis, with reduced clinical time compared to human-only diagnosis.
As the first open research in this application, a subset of the annotated RibFrac Dataset (600 CT scans, 221,308 CT slices) and our code for model training and evaluation will be open-source. We believe this large-scale dataset could facilitate both clinical research for automatic rib fracture diagnosis and engineering research for 3D computer vision.
2. Materials and methods
2.1. RibFrac dataset
2.1.1. Ethics
This retrospective study was approved by the ethics committee of Huadong Hospital affiliated to Fudan University (NO.2019K146), which waived the requirement for informed consent.
2.1.2. Inclusion Criteria
From January 2017 to December 2018, a search of the electronic medical records and the radiology information systems of the hospital for patients with traumatic rib fractures identified on chest-abdomen CT scans (1–1.25 mm) was performed by one author. A total of 7,473 traumatic rib fractures from 900 patients [mean age, 55.1 years 11.82 (standard deviation); range, 21–94 years] were enrolled in the study. There were 580 men [63.8%] and 329 women [36.2%]. Traumatic abdomen-thorax CT was performed by using the following two CT scanners: 16 cm wide coverage detector CT (Revolution CT, GE Healthcare, WI, USA); second-generation dual-source CT scanner (Somatom Definition Flash, Siemens Healthcare, Forchheim, Germany) with following parameters: 120 kVp; 100–200 mAs; pitch, 0.75–1.5; and collimation, 1–1.25 mm, respectively. All imaging data were reconstructed by using a bone or medium sharp reconstruction algorithm with a thickness of 1–1.25 mm.
As detailed in Fig. 1 (a), the inclusion criteria are as follows: (1) Traumatic patients with thin-slice chest-abdomen CT images (1–1.25 mm) containing all ribs, and (2) Thin-slice CT images without breathing artifact debasing diagnostic accuracy.
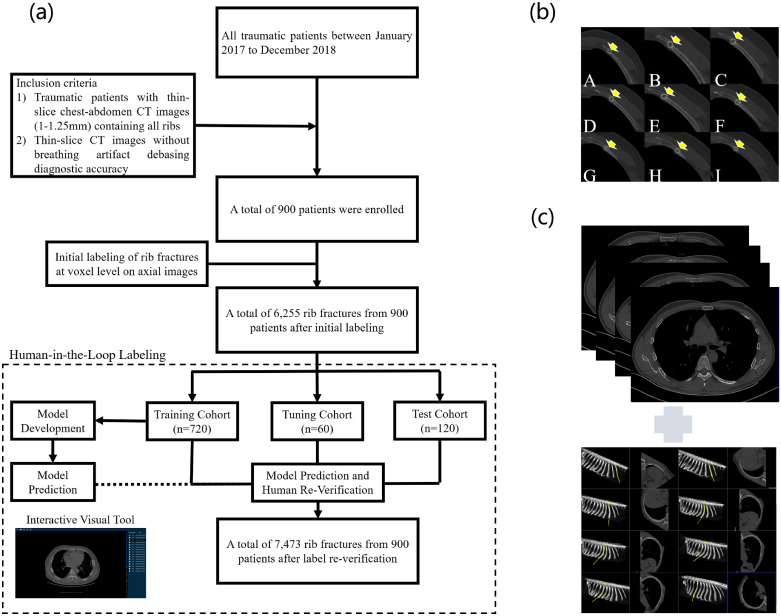
Fig. 1.
(a) Flowchart of RibFrac Dataset setup, including human-in-the-loop labeling of rib fractures. (b) Illustration of manual rib fracture labeling. (c) Verification of manual labeling with axial images (top) and manually curve planar reformation images (bottom).
2.1.3. Human-in-the-loop labeling of rib fractures
In the whole labeling procedure, there were 5 radiologists involved: A (3-5 years), B (10-20 years), C (5 years), D (5 years), E (20 years); numbers in the brackets denote the years of experience in chest CT interpretation.
All enrolled CT scans were first randomly diagnosed by two radiologists A and B in radiology department after the CT examinations in 48 hours, who did not participate in this study. Two junior radiologists C and D manually delineated the volume of interest (VOI) of the traumatic rib fractures with diagnosed CT reports at voxel level on axial CT images with the help of the diagnosis reports (by the radiologists A or B) and a medical image processing and navigation software 3D Slicer (version 4.8.1, Brigham and Women's Hospital). The broken ends of fractured bone were included as much as possible for the volume of the fractures as Fig. 1 (b); Besides, as illustrated in Fig. 1 (c), axial images combining manually curve planar reformation images were used together to insure the accuracy of labeling the real fractures [14], as rib fractures can be variable and inconspicuous if the fracture line is not present or parallels the detection plane [18]. After labeling by C and D, the VOIs were then confirmed by another senior radiologist E.
An initial deep learning model following a same pipeline as FracNet (Section 2.2) was developed on the RibFrac training cohort (Section 2.1.3). The initial system was used to predict fractures on the RibFrac training, tuning and test cohorts. We excluded all predicted fractures with high overlap between any initial label; all remaining predictions were feedback to the radiologist E to verify (reduce false positives). This procedure was assisted by an interactive visual tool (see Supplementary Materials). Around 20% annotations were missing from initial labeling and added with the human-in-the-loop labeling. The verified annotations were used for the development and validation of the deep learning system. Please note that there was no data leakage issue in the human-in-the-loop labeling procedure and the following development and validation, since our deep learning system was only trained on the training cohort.
2.1.4. Dataset pretreatment
The chest-abdomen CT DICOM (Digital Imaging and Communications in Medicine) format images were imported into the software for delineating, and the images with VOI information were then extracted with NII or NIFTI (Neuroimaging Informatics Technology Initiative) format for next-step analysis.
As depicted in Table 1, we randomly split the whole RibFrac Dataset (900 cases, 332,483 CT slices in total) into 3 cohorts: training (720 cases, to train the deep learning system), tuning (60 cases, to tune hyper-parameters of the deep learning system) and test (120 cases, to evaluate the model and human performance). In standard machine learning terminology, tuning is regarded as “validation”; in standard medical terminology, test in regarded as “validation”.
Table 1.
RibFrac Dataset Overview.
| Cohorts | Availability | No. Patients / CT Scans | No. CT Slices | No. Fractures |
|---|---|---|---|---|
| Training | Total | 720 | 265,302 | 6,156 |
| Public | 420 | 154,127 | 3,987 | |
| In-House | 300 | 111,175 | 2,169 | |
| Tuning | Public | 60 | 22,562 | 435 |
| Test | Public | 120 | 44,619 | 882 |
Considering several practical issues, we open source a subset of 600 cases (221,308 CT slices in total), with 420 cases for training, 60 cases for tuning and 120 for test. To our knowledge, it is the first open research dataset in this application. On the open-source subset of RibFrac Dataset, a deep learning system with same architecture of FracNet could be developed and validated with an acceptable performance. Please refer to Supplementary Materials for details.
2.2. Development of deep learning system
2.2.1. Model pipeline
Our algorithm follows a data-driven approach: it relies on the human annotations of rib fractures and learns to directly predict the voxel-level segmentation of fractures. Notably, the proposed FracNet does not rely on the extraction of rib centerlines in typical rib analysis algorithms [27]. As illustrated in Fig. 2 (a), our model pipeline consists of three stages: (a) pre-processing, (b) sliding-window prediction, and (c) post-processing.
Fig. 2.
(a) The pipeline for detecting rib fractures from CT scans. A 3D convolutional neural network, named FracNet, was developed to segment the fractures in a sliding window fashion. Pseudo-color in the figure is used for better visualizing binary images of bones and segmentation results. (b) Neural network architecture of FracNet based on 3D UNet [21]. Our code in PyTorch [26] for model training and evaluation will be soon open source.
(a) Pre-processing: To speed up the detection, we extracted the bone areas through a series of morphological operations (e.g., thresholding and filtering). The original spacing was preserved since only thin-section CT scans were included in our dataset. The intensity of input voxels was clipped to the bone window (level=450, width=1100) and normalized to [-1,1].
(b) Sliding-window prediction: Considering the elongated shape of rib fractures, standard labeling with bounding boxes could be missing much details. Therefore, we formulated the rib fracture detection as a 3D segmentation task. A customized 3D UNet, named FracNet (Section 2.2.2.), was developed to perform segmentation in a sliding-window fashion. Since a whole-volume CT scan could be too large to fit in a regular GPU memory, we cropped patches in a sliding-window fashion with a stride of 48 and feed them to our network. A raw segmentation was obtained by assembling patches of prediction. Maximum values were kept in the overlapping regions of multiple predictions.
(c) Post-processing: To efficiently reduce the false positive in our predictions, predictions of small sizes (smaller than 200 voxels) were filtered out. We also removed the spine regions according to their coordinates on the raw segmentation. To generate detection proposal, we first binarized the post-processed segmentation results with a low threshold of 0.1, and then computed connected components on the binary segmentation. Each connected component was regarded as a detection proposal, with a probability calculated by averaging raw segmentation scores over all voxels within the connected component.
2.2.2. Network architecture of FracNet and counterparts
To capture both local and global contexts, we proposed a customized 3D UNet [21] architecture, named FracNet, following an encoder-decoder architecture in Fig. 2 (b). The encoder was a series of down-sampling stage, each of which is composed of 3D convolution, batch-normalization [28], non-linearity and max pooling. The resolution of feature maps was halved after each down-sampling stage, while the number of channels was doubled. In the decoder, the feature map resolution was gradually restored through a series of transposed convolution. Features from the encoder were reused through feature concatenation from the same levels of the encoder and decoder. After the feature maps were recovered to the original size, we used a convolution layer to shrink the output channel to 1. Activated with a sigmoid function, the output denoted background=0 and lesions=1.
To benchmark our method, we also designed 3D variants of FCN and DeepLab v3+ 3for 3D segmentation. In both models, we used a 3D backbone, named 3D ResNet18-HR based on ResNet [2] to encode the 3D representation. Compared to standard ResNet achitecture (3D ResNet18-LR), the initial convolution layer with a stride of 2 followed by a down-sampling max pooling was modified into a single convolution layer with a stride of 1, thus the resolution of initial feature map from 3D ResNet18-HR is 4 times large as that of 3D ResNet18-LR. For 3D DeepLab, we added a 3D variant of atrous spatial pyramid pooling (ASPP) [3] between the encoder and decoder of 3D FCN to refine the output features. The neural network architectures of 3D FCN and 3D DeepLab is illustrated in Supplementary Materials.
2.2.3. Model training
Since rib fracture annotations were very sparse in whole CT volumes, during model training, we adopted a sampling strategy to alleviate the imbalance between positive and negative samples. Positive samples of size 64 × 64 × 64 were randomly cropped from a 96 × 96 × 96 region centered at the rib fracture, while negative samples were extracted within bone regions without fractures. During training, each batch consisted of 12 positive and 12 negative samples. Data augmentation of random plane flipping was applied. We used a combination of soft Dice loss and binary cross-entropy (BCE) to train our network:
where denote the ground truth and prediction of rib fracture segmentation, respectively, and denotes the batch size. We trained the network using Adam optimizer [29] with a warm-up training strategy. The learning rate linearly increased from 0.00001 to 0.1 during the first epoch, and then linearly decreased to 0.001 in 100 epochs. The RibFrac tuning cohort was used for tuning the hyper-parameters, including choosing the best model snapshot to be evaluated on the test cohort.
2.3. Model evaluation and statistical analysis
2.3.1. Metrics
Our method followed a segmentation methodology to perform a detection task, therefore both segmentation and detection metrics were critical to evaluate the model performance. For segmentation, we reported Dice Coefficient (Dice) and Intersection-over-Union (IoU),
Note that both Dice and IoU are positively correlated, where Dice is the most popular metric for medical image segmentation.
The evaluation of detection performance was based on Free-Response Receiver Operating Characteristic (FROC) analysis, an evaluation approach balancing both sensitivity and false positives. The FROC analysis was reported with sensitivities at various false positive (FP) levels, typically FP. We also reported their average as the overview metric for FROC analysis. Besides the FROC analysis, the overall detection sensitivity and average false positives per scan were also reported, which denoted the maximum sensitivity at maximum FP level in FROC analysis.
For each detection proposal, it was regarded as a hit when overlapped with IoU between any rib fracture annotation. Please note that for objects with elongated shape, the IoU tended to vary, which was the reason why we chose IoU as the detection hit criterion. See Section 3.1 for more explanation on this issue.
2.3.2. Observer study
To benchmark the proposed deep learning system with human experts, two radiologists R1 (a junior radiologist with more than 3 years of experience in chest CT interpretation) and R2 (a senior radiologist with 10 years of experience in chest CT interpretation) were required to participate in an independent human-only observer study. R1 and R2 were shown the RibFrac test cohort with randomized order to independently detect and segment each rib fracture, blinded to the fracture results and patient information. We then computed the detection and segmentation metrics with the ground truth labels with a human-in-the-loop annotation procedure (Section 2.1.2). The standard of reference for the diagnosis of rib fractures was the accurate location of the fractured rib and positive rib fracture [14].
Besides the independent observer study, a human-computer collaboration study was conducted to simulate the real clinical scenario.
2.4. Role of funding source
The funding sources played no role in the study design; collection, analysis, and interpretation of data; writing of the report; or decision to submit the article for publication.
3. Results
3.1. FracNet performs consistently on RibFrac cohorts
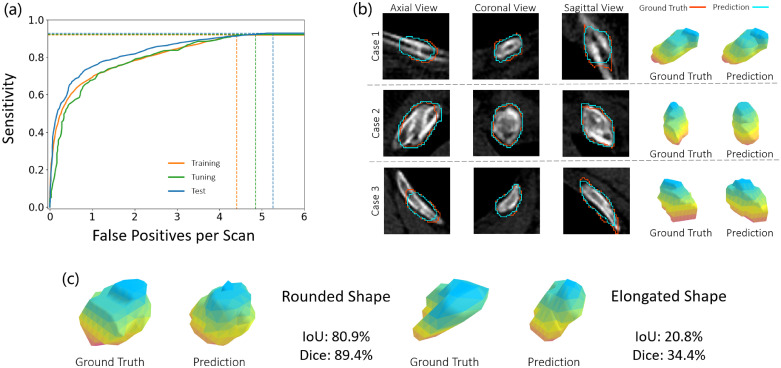
We first reported the performance of the proposed FracNet on our RibFrac training, tuning and test cohorts. As illustrated in Fig 3 (a) and Table 2, our method achieved detection sensitivities of around with average false positives per scan on the three cohorts consistently. Besides, our method achieved an acceptable segmentation performance, Diceon the training, tuning and test cohorts, respectively. Illustration of the predicted segmentation by FracNet was depicted in Fig. 3 (b). There was overfitting observed in the segmentation tasks, as segmentation was the proxy task for training the FracNet system; however, no overfitting was observed on the detection task. Please note that numbers of lesions in the rib fracture task were associated with elongated shapes, while object segmentation with elongated shape tended to be associated with low segmentation metrics (IoU and Dice). In Fig. 3 (c), we demonstrated 2 cases with rounded and elongated shape. Both cases were predicted with visually similarly segmentation to ground truth, while the segmentation metrics (IoU and Dice) of elongated shape were dramatically lower than those of rounded shape. It also explained why we choosed IoU as the detection hit criterion.
Fig. 3.
(a) FROC curves of FracNet detection performance on the RibFrac training, tuning and test cohorts. (b) Illustration of predicted segmentation on RibFrac test cohorts. (c) A comparison of segmentation metrics (IoU and Dice) for rounded and elongated shape. In (b) and (c), the pseudo-color in the 3D shape is only for visualization purpose.
Table 2.
FracNet performance on RibFrac training, tuning and test cohorts, in terms of detection and segmentation performance. FP: false positives per scan. IoU: Intersection-over-Union. Dice: Dice Coefficient.
| Cohorts | Detection Sensitivities @ FP Levels |
Detection |
Segmentation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.5 | 1 | 2 | 4 | 8 | Avg | Sensitivity | Avg FP | IoU | Dice | |
| Training | 60.3% | 69.3% | 78.3% | 90.0% | 91.9% | 77.9% | 91.9% | 4.41 | 77.5% | 87.3% |
| Tuning | 55.6% | 67.8% | 78.9% | 89.7% | 92.2% | 76.8% | 92.2% | 4.85 | 58.7% | 74.0% |
| Test | 66.0% | 75.0% | 81.7% | 90.5% | 92.9% | 81.2% | 92.9% | 5.27 | 55.6% | 71.5% |
3.2. Benchmarking FracNet with counterparts and experts
To validate the effectiveness of the proposed FracNet system, we compared the model performance with several deep neural network counterparts and human experts in Table 3. As demonstrated, the FracNet outperformed 3D FCN and 3D DeepLab by large margins, which verified the effectiveness of network design in the proposed FracNet. Please note that the model size of FracNet was smaller than these of 3D FCN and 3D DeepLab. Moreover, we conducted observer studies with two radiologists (R1 and R2, details in Section 2.3.2). Remarkably, though human experts achieved much lower false positives per scan, they underperformed the deep neural networks in terms of detection sensitivities. As for segmentation performance, FracNet underperformed R1 while outperformed R2. We also evaluated the performance of human collaboration with a simple union of human annotations (R1 R2); the union improved detection sensitivities with a cost of additional false positives introduced.
Table 3.
A comparison of detection and segmentation performance on RibFrac Test Set, of FracNet, two deep neural network counterparts (3D FCN and 3D DeepLab), two radiologists (R1 and R2) and their union.
| Methods | Detection Sensitivities @ FP Levels |
Detection |
Segmentation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.5 | 1 | 2 | 4 | 8 | Avg | Sensitivity | Avg FP | IoU | Dice | |
| FracNet | 66.0% | 75.0% | 81.7% | 90.5% | 92.9% | 81.2% | 92.9% | 5.27 | 55.6% | 71.5% |
| 3D FCN | 59.9% | 69.7% | 76.1% | 84.4% | 87.8% | 75.6% | 87.8% | 7.02 | 49.1% | 66.2% |
| 3D DeepLab | 63.7% | 72.5% | 79.2% | 88.2% | 91.3% | 79.0% | 91.3% | 6.11 | 50.3% | 68.7% |
| R1 | / | / | / | / | / | / | 79.1% | 1.34 | 47.4% | 64.3% |
| R2 | / | / | / | / | / | / | 75.9% | 0.92 | 36.7% | 53.1% |
| R1 R2 | / | / | / | / | / | / | 83.1% | 1.80 | 47.8% | 64.7% |
| FracNet R1 | / | / | 83.9% | 90.4% | 93.8% | 82.6% | 93.8% | 5.99 | 54.9% | 70.9% |
| FracNet R2 | / | / | 85.8% | 92.6% | 95.7% | 84.4% | 95.7% | 5.83 | 52.5% | 68.9% |
We further evaluate the performance of human-computer unions (FracNet R1 and FracNet R2). The detection probabilities of human were set to 1, therefore sensitivities with low FP level 0.5 and 1 were missing. Excitingly, dramatical improvement in detection sensitivities was observed, which was the foundation of human-computer collaboration (Section 3.3).
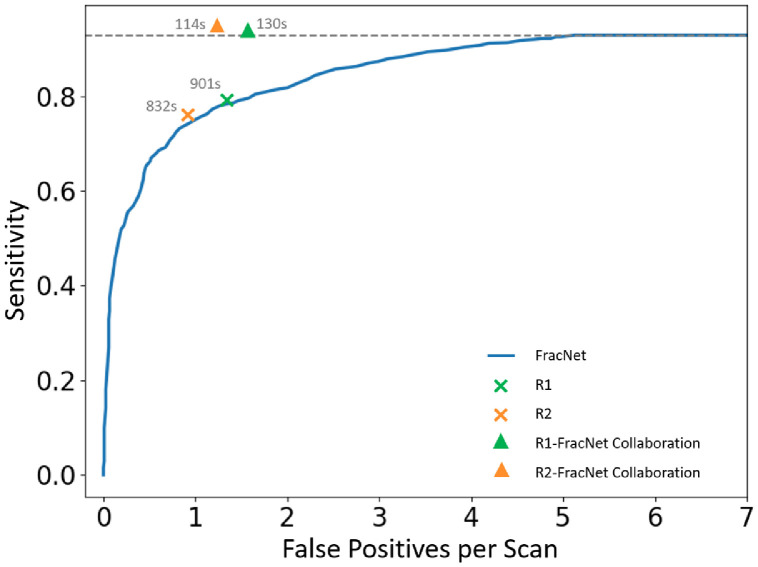
3.3. Human-computer collaboration
In this section, we validated the human-computer collaboration performance (Section 2.3.3) in Table 4 and Fig. 4. Average clinical time for detecting and segmenting all rib fractures was also reported. The average model time was measured with an implementation of PyTorch 1.3.1 and Python 3.7, on a machine with a single NVIDIA GTX 1080Ti with Intel Xeon E5-2650 and 128 G memory. The human-only diagnosis outperformed FracNet with given false positive levels. However, the human-computer collaboration could further improve their performance with reduced clinical time. Basically, the human-computer collaboration followed the workflow of the FracNet system in clinical scenario: (a) model prediction (Model), (b) manual false positive reduction and verification (FPR), and (c) missing lesion detection and segmentation (Segmentation). Compared to conventional manual diagnosis by human experts (R1 and R2), the human-computer collaboration significantly improved the detection sensitivities by large margins, with a sight cost in increasing false positives. Nevertheless, the computer-aided diagnosis with FracNet reduced the clinical time for rib fracture detection and segmentation. In real clinical practice, the clinicians are not asked to segmentation the rib fractures, where only diagnosis time should be counted. Even in such cases, human-computer collaboration could reduce clinical time with even better diagnosis performance.
Table 4.
A comparison of detection performance and clinical time on RibFrac Test Set. FPR: Manual False Positive Reduction with our interactive visual tool. Co.: collaboration.
| Detection Performance |
Clinical Time |
|||
|---|---|---|---|---|
| Sensitivity | Avg FP | Workflow | Average Time | |
| FracNet | 92.9% | 5.27 | Model (31s) | 31s |
| R1 | 79.1% | 1.34 | Diagnosis (322s) + Segmentation (579s) | 901s |
| R2 | 75.9% | 0.92 | Diagnosis (282s) + Segmentation (550s) | 832s |
| R1-FracNet Co. | 93.4% | 1.58 | Model (31s) + FPR (79s) + Segmentation (20s) | 130s |
| R2-FracNet Co. | 94.4% | 1.21 | Model (31s) + FPR (58s) + Segmentation (25s) | 114s |
Fig. 4.
A comparison of human-only and human-computer collaboration detection performance, where the clinical time used on average are also depicted on the figure.
4. Discussion
This study proposed a deep learning system, named FracNet, to detect and segment the rib fractures from CT scans. In rib fracture detection, our model performed high sensitivity (92.9%) and average FPs (5.27); as a comparison, human experts achieve 79.1%, 1.34 and 75.9%, 0.92. Besides, our deep learning system showed acceptable performance on rib fracture segmentation (IoU: 55.6%; Dice: 71.5%), which had never been reported in prior studies . Collaborated with the deep learning system, sensitivity of rib fractures increased (up to 94.4%) with acceptable false positives and reduced clinical time consuming (approximate 86% clinical time decreased).
Through the observer study, the junior radiologist had higher sensitivity (79.1%) of rib fractures detection with increased FPs (1.34) than the senior radiologist (75.9%, 0.9), indicating that the radiologists have their own interpretation in rib fractures. Although the junior radiologist achieved 3.2% higher sensitivity of rib fractures, the FPs also increased about 31%. The human-computer collaboration improved both the sensitivity and FPs compared with human-only or computer-only diagnosis, indicating the existence of model-detected rib fractures that were missed by radiologists, and vice versa. The inspiring results achieved by human-computer collaboration were consistent with a pervious study [4] in chest radiograph interpretation. When collaborated with human experts, FracNet achieved higher sensitivities with significantly reduced false positives. Moreover, deep-learning-assisted diagnosis significantly decreased about 86.3% and 85.6% clinical time with comparable or even better diagnostic accuracy (higher sensitivities and FPs).
Before our study, there were two related recent studies using deep learning to detect the rib fractures from CT images [30,31]. Both studies formalized the task as 2D detection, however our study formalized it as 3D segmentation. As discussed in Section 3.1, the rib fractures were generally associated with elongated shapes; The formalization with segmentation masks in our study was expected to be more accurate than that with detection bounding boxes in these related studies. To our knowledge, it is the first study for rib fracture segmentation. Besides, the data and annotation were of higher standard in our study. High-quality thin-slice CT scans with thickness of 1–1.25 mm were used in our study, compared to 1.5 mm [30] and partially 5 mm [679 of 974 patients (about 69.7%)] [31]. It was reported that thin-slice images could be helpful for the diagnosis of bone fractures and incidental findings [32]. On the other hand, we adapted a human-in-the-loop labeling procedure (Section 2.1.2), five radiologists were envovled to ensure the high quality of our annotations, which could help to reduce the risk of overestimating model performance [4,[33], [34], [35]]. For these reasons, our model achieved a significantly higher detection sensitivity with less time-comsuing as time is crucial for trauma patients in the emergency setting throughout the whole diagnostic and therapeutic management process [36]. The model performance was consistent on our external training, tuning and test cohorts. More importantly, we open source the first large scale dataset for rib fracture detection and segmentaiton with voxel-level annotations, to improve research reproducibility and facilitate further research.
There are limitations in this study. Although developed and validated on a large-scale dataset, this is a single-center study. In our site, the performance of diagnostic performance between junior and senior human experts was similar, this may benefit from the expertise of our radiologists in rib fracture diagnosis. However, in our experience, the diagnostic performance of radiologists with different expertise from different sites may vary significantl. Besides, even the annotations were verified with a human-in-the-loop labeling procedure, there could still be false positive or false negative annotation. Moreover, the landscape of deep neural networks was not fully explored. In further studies, we are investigating model generalization of our method on multi-center datasets, with more rounds of human-in-the-loop labeling procedure. It is also interesting to explore segmentation loss for elongated objects[37], [38] or leverage the pretraining from natural / medical images [39,40]. Apart from automatic rib fracture detection and segmentation, we are also developing datasets and models to automatically classify the fracture types. We will also introduce recent advances in 3D deep learning to improve the model performance.
In conclusion, our deep learning model collaborated with human experts could help to increase the diagnostic effectiveness and efficiency in the diagnosis of rib fractures, which implied the great potential of deep-learning-assisted diagnosis in clinical practice.
Contributors
Conception and design: L. Jin, J. Yang, M. Li
Development of methodology: L. Jin, J. Yang, Y. Sun, Y. Gao, B. Ni, M. Li
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): L. Jin, J. Yang, Y. Sun, W. Ma, M. Tan, P. Gao, M. Li
Analysis and interpretation of data (e.g., statistical analysis, computational analysis): L. Jin, J. Yang, Y. Sun, K. Kuang, H. Kang, J. Chen, M. Li
Writing, review, and/or revision of the manuscript: L. Jin, J. Yang, Y. Sun, B. Ni, M. Li
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): L. Jin, K. Kuang, H. Kang, J. Chen, M. Li
Study supervision: L. Jin, J. Yang, B. Ni, M. Li
Algorithm and software development: J. Yang, K. Kuang, H. Kang, J. Chen
All authors read and approved the final version of the manuscript.
Data sharing section
The data and code from Huadong Hospital affiliated to Fudan University used in this study are available at https://m3dv.github.io/FracNet/ to users who agree with our data license (CC BY-NC 4.0) and code license (Apache-2.0 License).
Declaration of Competing interest
All authors declare that they have no conflict of interests.
Acknowledgments
This study has received funding from the Medical Imaging Key Program of Wise Information Technology of 120-Health Commission of Shanghai 2018ZHYL0103 (Ming Li), the National Natural Science Foundation of China 61976238 (Ming Li) and "Future Star" of famous doctors' training plan of Fudan University (Ming Li). This study has received funding from Shanghai Youth Medical Talents Training Funding Scheme AB83030002019004 (Liang Jin). This study was also supported by National Science Foundation of China 61976137, U1611461 (Bingbing Ni). The funding sources played no role in the study design; collection, analysis, and interpretation of data; writing of the report; or decision to submit the article for publication.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103106.
Appendix. Supplementary materials
Supplementary Video. Illustration of manual false positive reduction and label verification on model prediction with our interactive visualization tool. This tool is used in the human-in-the-loop labeling (Sec. 2.1.2) and human-computer collaboration study (Sec. 2.3.2).
References
- 1.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 2.Zhao W, Yang J, Sun Y. 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res. 2018;78(24):6881–6889. doi: 10.1158/0008-5472.CAN-18-0696. [DOI] [PubMed] [Google Scholar]
- 3.Zhang HT, Zhang JS, Zhang HH. Automated detection and quantification of COVID-19 pneumonia: CT imaging analysis by a deep learning-based software. Eur J Nucl Med Mol Imaging. 2020;47(11):2525–2532. doi: 10.1007/s00259-020-04953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majkowska A, Mittal S, Steiner DF. Chest radiograph interpretation with deep learning models: assessment with radiologist-adjudicated reference standards and population-adjusted evaluation. Radiology. 2020;294(2):421–431. doi: 10.1148/radiol.2019191293. [DOI] [PubMed] [Google Scholar]
- 5.Chilamkurthy S, Ghosh R, Tanamala S. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet. 2018;392(10162):2388–2396. doi: 10.1016/S0140-6736(18)31645-3. [DOI] [PubMed] [Google Scholar]
- 6.Lindsey R, Daluiski A, Chopra S. Deep neural network improves fracture detection by clinicians. Proc Natl Acad Sci USA. 2018;115(45):11591–11596. doi: 10.1073/pnas.1806905115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Gu Y, Dvornek N, Staib LH, Ventola P, Duncan JS. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results. Med Image Anal. 2020;65 doi: 10.1016/j.media.2020.101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bien N, Rajpurkar P, Ball RL. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLoS Med. 2018;15(11) doi: 10.1371/journal.pmed.1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, Zhou F, Liu B, Fu D, Bai X. Efficient multiple organ localization in CT image using 3D region proposal network. IEEE Trans Med Imaging. 2019 doi: 10.1109/TMI.2019.2894854. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, Xie P, Wang M. Deep learning-based fully automated detection and segmentation of lymph nodes on multiparametric-mri for rectal cancer: a multicentre study. EBioMedicine. 2020;56 doi: 10.1016/j.ebiom.2020.102780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 12.Talbot BS, Gange CP, Jr., Chaturvedi A, Klionsky N, Hobbs SK, Chaturvedi A. Traumatic rib injury: patterns, imaging pitfalls, complications, and treatment. Radiographics. 2017;37(2):628–651. doi: 10.1148/rg.2017160100. [DOI] [PubMed] [Google Scholar]
- 13.Urbaneja A, De Verbizier J, Formery AS. Automatic rib cage unfolding with CT cylindrical projection reformat in polytraumatized patients for rib fracture detection and characterization: Feasibility and clinical application. Eur J Radiol. 2019;110:121–127. doi: 10.1016/j.ejrad.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Jin L, Ge X, Lu F. Low-dose CT examination for rib fracture evaluation: a pilot study. Medicine (Baltimore) 2018;97(30):e11624. doi: 10.1097/MD.0000000000011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SJ, Bista AB, Min YG. Usefulness of low dose chest CT for initial evaluation of blunt chest trauma. Medicine (Baltimore) 2017;96(2):e5888. doi: 10.1097/MD.0000000000005888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolopp M, Douis N, Urbaneja A. Automatic rib unfolding in postmortem computed tomography: diagnostic evaluation of the OpenRib software compared with the autopsy in the detection of rib fractures. Int J Legal Med. 2020;134(1):339–346. doi: 10.1007/s00414-019-02195-x. [DOI] [PubMed] [Google Scholar]
- 17.Glemser PA, Pfleiderer M, Heger A. New bone post-processing tools in forensic imaging: a multi-reader feasibility study to evaluate detection time and diagnostic accuracy in rib fracture assessment. Int J Legal Med. 2017;131(2):489–496. doi: 10.1007/s00414-016-1412-6. [DOI] [PubMed] [Google Scholar]
- 18.Ringl H, Lazar M, Topker M. The ribs unfolded - a CT visualization algorithm for fast detection of rib fractures: effect on sensitivity and specificity in trauma patients. Eur Radiol. 2015;25(7):1865–1874. doi: 10.1007/s00330-015-3598-2. [DOI] [PubMed] [Google Scholar]
- 19.Dankerl P, Seuss H, Ellmann S, Cavallaro A, Uder M, Hammon M. Evaluation of Rib fractures on a single-in-plane image reformation of the rib cage in CT examinations. Acad Radiol. 2017;24(2):153–159. doi: 10.1016/j.acra.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 20.Cho SH, Sung YM, Kim MS. Missed rib fractures on evaluation of initial chest CT for trauma patients: pattern analysis and diagnostic value of coronal multiplanar reconstruction images with multidetector row CT. Br J Radiol. 2012;85(1018):e845–e850. doi: 10.1259/bjr/28575455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falk T, Mai D, Bensch R. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16(1):67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 22.Shelhamer E, Long J, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(4):640–651. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 23.Chen L-C, Zhu Y, Papandreou G, Schroff F, Adam H. ECCV; 2018. Encoder-decoder with atrous separable convolution for semantic image segmentation. [Google Scholar]
- 24.Hara K, Kataoka H, Satoh Y. CVPR; 2018. Can spatiotemporal 3d cnns retrace the history of 2d cnns and imagenet? [Google Scholar]
- 25.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. CVPR. 2016 [Google Scholar]
- 26.Paszke A, Gross S, Massa F. NeurIPS; 2019. PyTorch: an imperative style, high-performance deep learning library. [Google Scholar]
- 27.Lenga M, Klinder T, Bürger C, Berg JV, Franz A, Lorenz C. MSKI@MICCAI; 2018. Deep learning based rib centerline extraction and labeling. [Google Scholar]
- 28.Ioffe S, Szegedy C. ICML; 2015. Batch normalization: accelerating deep network training by reducing internal covariate shift. [Google Scholar]
- 29.Kingma DP, Ba J. ICLR; 2015. Adam: a method for stochastic optimization. [Google Scholar]
- 30.Weikert T, Noordtzij LA, Bremerich J. Assessment of a deep learning algorithm for the detection of rib fractures on whole-body trauma computed tomography. Korean J Radiol. 2020;21(7):891–899. doi: 10.3348/kjr.2019.0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou QQ, Wang J, Tang W. Automatic detection and classification of rib fractures on thoracic CT using convolutional neural network: accuracy and feasibility. Korean J Radiol. 2020;21(7):869–879. doi: 10.3348/kjr.2019.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guchlerner L, Wichmann JL, Tischendorf P. Comparison of thick- and thin-slice images in thoracoabdominal trauma CT: a retrospective analysis. Eur J Trauma Emerg Surg. 2020;46(1):187–195. doi: 10.1007/s00068-018-1021-9. [DOI] [PubMed] [Google Scholar]
- 33.Taylor AG, Mielke C, Mongan J. Automated detection of moderate and large pneumothorax on frontal chest X-rays using deep convolutional neural networks: a retrospective study. PLoS Med. 2018;15(11) doi: 10.1371/journal.pmed.1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajpurkar P, Irvin J, Ball RL. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018;15(11) doi: 10.1371/journal.pmed.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang EJ, Park S, Jin KN. Development and validation of a deep learning-based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw Open. 2019;2(3) doi: 10.1001/jamanetworkopen.2019.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khung S, Masset P, Duhamel A. Automated 3D rendering of ribs in 110 polytrauma patients: strengths and limitations. Acad Radiol. 2017;24(2):146–152. doi: 10.1016/j.acra.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, Ma Y, He S, Zhu J. 3D-GIoU: 3D generalized intersection over union for object detection in point cloud. Sensors (Basel) 2019;19(19) doi: 10.3390/s19194093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordström M, Bao H, Löfman F, Hult H, Maki A, Sugiyama M. Springer International Publishing; Cham: 2020. MICCAI; pp. 269–278. (Calibrated surrogate maximization of dice). [Google Scholar]
- 39.Yang J, He Y, Huang X. AlignShift: bridging the gap of imaging thickness in 3D anisotropic volumes. MICCAI. 2020 [Google Scholar]
- 40.Yang J, Huang X, Ni B, Xu J, Yang C, Xu G. 2019. Reinventing 2D Convolutions for 3D medical images. arXiv preprint arXiv:191110477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video. Illustration of manual false positive reduction and label verification on model prediction with our interactive visualization tool. This tool is used in the human-in-the-loop labeling (Sec. 2.1.2) and human-computer collaboration study (Sec. 2.3.2).