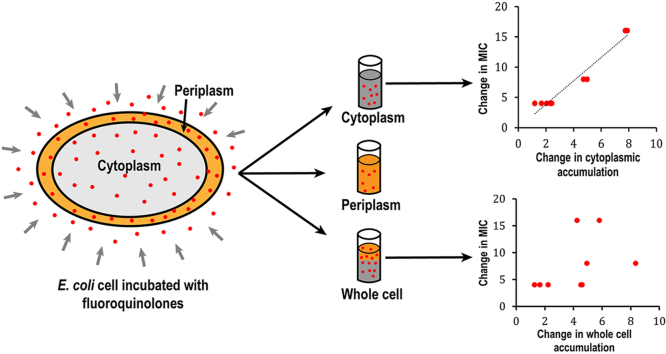
Abstract
The double-layered cell envelope of Gram-negative bacteria and active drug efflux present a formidable barrier for antimicrobial compounds to penetrate. Fluoroquinolones are among the few classes of antimicrobials that are clinically useful in the treatment of Gram-negative bacterial infection. Previous studies on fluoroquinolone accumulation measured total bacteria associated compounds, rather than the cytoplasmic accumulation. Fluoroquinolones target the type II topoisomerases in the cytoplasm. Thus, the cytoplasmic accumulation is expected to be more relevant to the potency of the drugs. Here, we fractionated and measured the concentration of nine fluoroquinolone compounds in the periplasm and the cytoplasm of two strains of E. coli cells, a parent strain and its isogenic efflux-deficient tolC knockout strain. The potency of the drugs was determined using the minimum inhibitory concentration (MIC) assay. We found that all fluoroquinolones tested accumulated at much higher concentrations in the periplasm than in the cytoplasm. The periplasmic concentrations were 2–15 folds higher than the cytoplasmic concentration, while the actual distribution ratio varies drastically among the compounds tested. Good correlation between the MIC and the cytoplasmic accumulation, but not whole cell accumulation, was observed using a pair of isogenic wild type and drug-efflux deficient strains.
Keywords: Gram-negative bacteria, Periplasm accumulation, Fluoroquinolone, Minimum inhibitory concentration
Graphical abstract
Highlights
-
•
Fluoroquinolones accumulate to high concentration in the periplasm of E. coli
-
•
Periplasmic concentrations are higher than the cytoplasmic and exterior media concentration.
-
•
Good correlation was observed for the cytoplasmic drug concentration and the antimicrobial potency.
-
•
Disruption of active efflux has a larger impact on the periplasmic than the cytoplasmic accumulation.
1. Introduction
Antibiotic resistance is becoming a serious global problem of the current era. According to an interagency coordination group report to the Secretary-General of the United Nations in April 2019, approximately 700,000 deaths occur every year globally due to multidrug resistant infections and this figure could reach 10 million deaths per year by 2050 [1]. Most of the antibiotics that are used currently were developed during the golden era of antibiotics in the 1940s–1960s and are gradually becoming less effective due to the development of bacterial resistance [[2], [3], [4], [5]]. A clinically useful antimicrobial needs to be selectively toxic towards bacteria, with excellent potency, and effectively penetrating to the target site inside the bacteria cell. The latter is a major barrier in the development of drugs for Gram-negative bacteria due to their double-membrane cell envelope structure. The outer membrane contains lipopolysaccharide (LPS) in the outer leaflet and phospholipids in the inner leaflet. LPS forms a hydrophilic barrier on the surface of the bacteria. The outer membrane contains protein channels called porins, which allow the passage of selected compounds [[6], [7], [8], [9], [10]]. The inner membrane is a phospholipid bilayer, which is a hydrophobic barrier [2]. In addition, Gram-negative bacteria have an array of drug efflux pumps that remove toxic compounds from the cells [11]. Because of these obstacles many antimicrobials effective against Gram-positive bacteria do not work for Gram-negative bacteria [2,12,13].
The importance of cellular accumulation to any pharmaceuticals is apparent. Several studies have been conducted to determine the correlation between antibiotic accumulation in bacteria and its MIC [[14], [15], [16], [17], [18], [19]]. Bazile et al. determined the cellular accumulation of 11 fluoroquinolones and their DNA gyrase inhibition activity [18]. No correlation was found between MIC and accumulation, but the correlation between the MIC and the minimal effective dose improved on considering accumulation as a factor. In addition, they observed that the hydrophobicity of compounds correlated positively with accumulation in S. aureus and negatively with accumulation in E. coli and P. aeruginosa [18]. Inspired by this pioneering work, Piddock and her group conducted a series of studies on the accumulation of compounds in bacteria. They found that the highly hydrophobic rifampicin accumulated much less in E. coli compared to S. aureus, and speculated that the presence of the outer membrane in E. coli was the major reason for the reduced accumulation, while efflux also played a minor role [19]. In the study of fluoroquinolones accumulation in Mycobacterium tuberculosis [16], no correlation between MIC and accumulated concentration of fluoroquinolones was found. Instead, a clear negative correlation between the molecular size and accumulation, and positive correlation between hydrophobicity and accumulation were observed [16]. In the study of fluoroquinolone accumulation in S. aureus, no correlation of accumulation with hydrophobicity, but positive correlation with molecular weight, was observed. However, the steady state concentration of accumulated compounds did not correlate with their effectiveness as revealed by MIC [14]. In the study of accumulation of 10 fluoroquinolones in Streptococcus pneumoniae, hydrophobicity and molecular weight of the compounds were observed to correlate negatively with the steady state accumulation concentration of fluoroquinolones. Surprisingly, MIC of the fluoroquinolones was found to correlate negatively with steady state concentration with more effective antibiotics accumulating less [15].
In a recent study, Iyer et al. determined accumulation of fluoroquinolones and a collection of other DNA ligase inhibitors in E. coli using mass spectroscopy and observed that there was no correlation between accumulation of fluoroquinolones versus their effectiveness in inhibiting bacteria cell growth [17]. No correlation was observed between accumulation concentration and the ratio of IC50 to MIC of the DNA ligases. It was suggested that the lack of correlation could be because the accumulation data represented the accumulation of drugs in entire cell rather than the amount available for target inhibition [17]. When we first started this project, no publications were available to our knowledge that studied the accumulation of antibiotics in subcellular fractions of Gram-negative bacteria. A very recent report described fractionation and quantification of the antibiotic accumulation using mass spectroscopy [20]. The accumulation of four antibiotics from different classes were investigated. But the drastic differences between the structure and mechanism of the drugs made comparison among them difficult.
To further explore the penetration barrier presented by Gram-negative bacteria, in this study we examined the accumulation of nine fluoroquinolones in subcellular compartments of an E. coli strain and its isogenic tolC knock out strain. The potency of the compounds as a growth inhibitor was determined using the minimum inhibitory concentration (MIC) assay. Relative distribution of fluoroquinolones in the periplasm and cytoplasm, and their correlation with antimicrobial activity were studied to reveal new insight in our understanding about how antimicrobials work.
2. Materials and methods
2.1. Drug accumulation assay
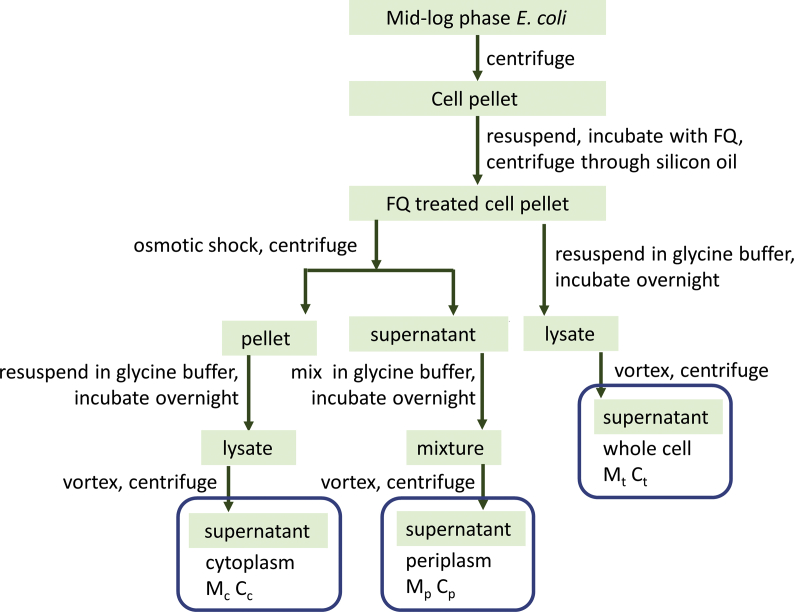
Two strains of E. coli, both obtained from the Yale Coli Genetic Stock Center, were used in the measurement of accumulation, BW25113 and BW25113 ΔtolC (referred to below as WT and ΔtolC). Bacteria were cultured in LB broth to mid-log phase and harvested by centrifugation at 3,000 g for 15 min at room temperature. All following operations were conducted at room temperature unless otherwise noted. The cell pellet was resuspended in a NaPi-Mg buffer (50 mM sodium phosphate buffer, 0.25% MgCl2, pH 7.0) to a final concentration of optical density at 600 nm (OD600) of 6.4. The bacterial density was approximately 6.4 × 109 colony-forming units per ml. For drug treatment, the indicated fluoroquinolone was added to the bacteria suspension to a final drug concentration of 2.0 μg/ml. The mixture was incubated for 15 min with constant shaking at 250 rpm. After incubation, 700 μl of the drug-treated bacterial cells was layered carefully on top of 700 μl of silicon oil in a microcentrifuge tube. The sample was centrifuged at 13,000 g for 1 min. The supernatant was discarded, and the inner wall of the centrifuge tubes cleaned carefully using a paper rod made out of Kimwipe. For the “whole cell” samples, 1.0 ml of glycine-HCl buffer (0.1 M glycine-HCl, pH 3.0) was added directly to lyse the cell as described [21]. The pellet was resuspended and incubated overnight. The next morning, cell debris and membrane were removed through centrifugation at 15,000 g for 15 min. The supernatant was collected and diluted 1:1 using the glycine-HCl buffer before fluorescence emission spectra were measured. For the fractionated samples, the periplasm and cytoplasm were obtained as described below, and 1.0 ml of glycine-HCl buffer was added into each sample followed by mixing and overnight incubation. In the next morning, all samples were centrifuged after a brief vortex, and the supernatant was collected and diluted before measurement as described above. The peak intensity at the corresponding excitation and emission wavelength was used to determine the concentration of each fluoroquinolone according to its standard calibration curve (see below). Fig. 1 is a flow chart of the procedure. All measurements were done in triplicate and the data were presented as the average ± standard deviation.
Fig. 1.
Flow chart showing the sample preparation procedure.
To evaluate the effect of wash, cells were incubation with drugs and centrifuged through silicon oil as described above. The cell pellet was quickly resuspended in 300 μl of 70 mM Tris buffer (pH 8.0) and centrifuged again at 13,000 g for 1 min. The supernatant was collected as the “wash solution”. Glycine-HCl buffer (1.0 ml) was added and the concentration of fluoroquinolone in the wash solution was determined similarly as in the other samples.
2.2. Separation of the periplasm and cytoplasm
An osmotic shock method was used to extract the periplasm with modifications to minimize the incubation time [22]. Briefly, cell pellet was resuspended in 100 μl of periplasm preparation buffer (200 mM Tris-Cl, 1 mM EDTA, 20% sucrose, 1 mg/ml lysozyme, pH 8.0) and incubated for 5 min. Next, 200 μl of ice-cold deionized water was added and the sample was mixed by tapping. The mixture was incubated on ice for 2 min and then centrifuged at 13,000 g for 1 min. The supernatant was collected, which contains the periplasmic component. The pellet contains the cytoplasm and cell membranes.
To assess see potential leakage of cytosolic protein, BW25113 transformed with plasmid pBAD-sfGFP (a gift from Ryan Mehl, Addgene plasmid # 85,482) [23] was cultured to OD600 0.5, and induced using 0.2% arabinose for 0.5 h. Cell pellet was collected by centrifugation through silicon oil and subjected to the osmotic shock procedure exactly as described above. After the supernatant containing the periplasm was collected, the spheroplast was resuspended in 300 μl tris buffer, lysed through sonication and centrifuged to collect the solution component (cytoplasm). In parallel, a duplicate cell pellet was resuspended in 300 μl tris buffer, sonicated and centrifuged to collect the supernatant (whole cell). The sfGFP concentration in all three samples were determined using the intrinsic fluorescence of sfGFP at excitation and emission wavelengths of 495 nm/510 nm.
To monitor the leakage of ATP from the cytoplasm, the periplasm, cytoplasm, and whole cell samples were prepared from BW25113 cell pellet as described above for the drug accumulation assay, and the ATP concentration was determined following well established protocols [24]. Briefly, cell pellet was collected by centrifugation through silicon oil and subjected to the osmotic shock procedure exactly as described above. After the supernatant containing the periplasm was collected, the spheroplast was resuspended in 450 μl ice cold 0.4 M perchloric acid and vortexed for 10 s. The mixture was incubated on ice for 15 min and spun down at 13,000×g for 5 min. To neutralize the acid, 200 μl of the supernatant was transferred to a fresh tube and mixed with 100 μl of a solution containing 0.72 M KOH and 0.16 M KHCO3. The neutralized mixture was then centrifuged at 13,000×g for 5 min and the supernatant was transferred to a fresh tube for use in the ATP assay. Similarly, 150 μl of ice cold 1.2 M perchloric acid was added into 300 μl of the periplasm sample (to a final concentration of 0.4 M perchloric acid) and vortexed for 10 s, followed by the same procedure to prepare samples for ATP analysis. In parallel, a duplicate cell pellet was resuspended in 450 μl ice cold 0.4 M perchloric acid and processed exactly as described for the spheroplast sample. ATP concentration in the samples were determined using a luciferase – based assay (Staybrite™ Highly stable ATP bio luminescence kit, BioVision Incorporated.). The ATP level was determined by measuring luminescence levels using BioTek Synergy HT multimode plate reader.
2.3. Preparation of the calibration curves
Cell culture was prepared similarly as described above except that NaPi-Mg buffer, instead of the fluoroquinolone solution, was used in the “drug treatment” step. Cells were aliquoted and pelleted through silicon oil similarly and the whole cell, periplasm, and cytoplasm extracts were obtained separately as described above. These solutions were used as the background to obtain the respective calibration curves. For each fluoroquinolone used in this study, standard solutions were prepared by spiking the extracts with known concentration of each compound. All samples were vortexed and centrifuged for 15 min at 15,000 g and the supernatant was used to measure the fluorescence after 1:1 dilution using the glycine-HCl buffer.
The emission spectra of the samples were obtained to determine the peak intensity. Graph was plotted with the emission peak intensity in Y-axis and the concentration of drug in X-axis. Every sample was done in triplicate and plotted as the average with error bars representing the standard deviation. Standard calibration curves for all compounds used in this study can be found in supplementary materials (Fig. S1). Structure of these compounds are shown in Fig. S2. Excitation/emission wavelength (nm) of compounds used in this study: ciprofloxacin (Cipro, 280/447), norfloxacin (Nor, 278/444), enrofloxacin (Enro, 278/445), levofloxacin (Levo, 293/502), lomefloxacin (Lome, 286/450), ofloxacin (Oflo, 293/501), fleroxacin (Flero, 286/453), marbofloxacin (Marbo, 298/510), moxifloxacin (Moxi, 296/509).
2.4. Quantification of accumulated drug
For each sample, the measured fluorescence intensity was converted into sample concentration using the respective calibration curve. The product of the sample concentration and sample volume yielded the mass of the compounds associated with the whole cell pellet (Mt), periplasm (Mp) or cytoplasm (Mc) from 0.7 ml of OD600 6.4 bacteria, or ~4.5 × 109 cells. As described above, total volume of the periplasm, cytoplasm, and whole cell samples are approximately 1.3, 1.0, and 1.0 ml, respectively. Similarly, the mass of the compounds in the wash solution (Mw) was determined from multiplying the wash solution concentration and the wash sample volume (1.3 ml).
To calculate the total concentration of each compound in the cell (Ct), since the actual cell volume of the pellet was 16.1 μl (see below):
Concentration of drug in the periplasm (Cp) is determined assuming the periplasm volume to be 10% of the total cell volume:
Similarly, the total volume of the cytoplasm was approximately 13.8 μl, thus the concentration in the spheroplast Cc:
2.5. Volume of cellular compartments
The reported volume of the periplasmic space of Gram-negative bacteria ranges from 7% to 40% [20,[25], [26], [27], [28], [29], [30], [31]]. A seminal study by Stock et al. reported that the periplasmic space in E. coli and S. typhimurium constitute of approximately 20–40% of the total cell volume [25]. The volume was determined experimentally through exploiting the selective permeability of the inner and outer membranes. More recent studies used electron microscopy to measure the gap between the outer membrane and inner membrane, and the reported thickness of the periplasmic space ranged from 10 nm to 33 nm [[26], [27], [28]]. More recently, Pilizota et al. estimated the periplasmic space to be approximately 16% of the total cell volume based on fluorescence imaging of E. coli cells expressing fluorescent proteins [29]. Prochnow et al. took the cellular dimensions from different published sources and calculated the volume of the periplasm of E. coli to be ~7% of the total cell volume [20]. Based on these discussions, in our study we assumed the periplasmic volume to be 10% of the total cell volume.
The total volume of E. coli in 1 ml of OD600 1.0 culture has been reported to be approximately 3.6 μl [32]. This value was used as the cellular volume for the calculation of drug concentration in “whole cell” samples. The cell density in the drug treatment mixture is OD600 6.4. Each pellet contained cells from 700 μl of OD600 6.4 culture, which yielded a total cell volume of 3.6 × 6.4 × 0.7 = 16.1 μl. Assuming 10% of the total volume to be the periplasm, volume of the periplasm was 1.6 μl. Cell membranes has been estimated to account for 4% of the overall volume, and thus the cytoplasm was 13.8 μl [20].
2.6. Determination of the minimum inhibitory concentration (MIC)
BW25113 wild type and tolC knockout strains were cultured overnight in LB broth at 37 °C with shaking at 250 rpm. The overnight culture was diluted into fresh LB broth at a final concentration of 5 × 105 CFU/ml. The diluted cells were then aliquoted into a 48-well plate, the indicated drug was added with 2-fold dilutions to create a series of concentrations. Plates were then incubated at 37 °C with shaking at 160 rpm overnight. The concentration of drug at which no visible growth was observed was reported as the MIC.
3. Results and discussion
3.1. Cellular accumulation of fluoroquinolones
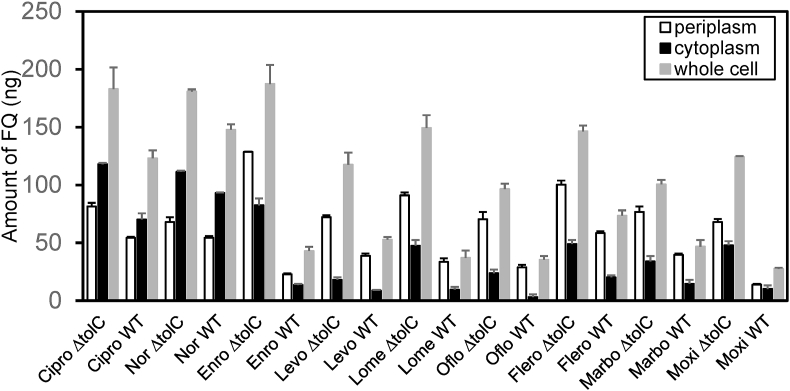
We analyzed nine fluoroquinolone family compounds that are commercially available and have high fluorescence intensity. Accumulation studies were performed as described in Materials and Methods. After incubation with fluoroquinolones and cell fractionation, fluorescence emission spectra were collected for each sample. The intensity at the corresponding wavelength was used to determine compounds concentration using the calibration curves. Calculated concentrations Mp, Mc, and Mt were shown in Fig. 2 and Table 1.
Fig. 2.
Accumulation of fluoroquinolones (FQs) in the periplasm (Mp), cytoplasm (Mc), and whole cell (Mt) in BW25113 (WT) and BW25113ΔtolC (ΔtolC) cells.
Table 1.
Accumulation of fluoroquinolones (FQs) in two strains, BW25113 (WT) and BW25113Δtolc (ΔC). Mt, Mp, and Mc are mass of accumulated compounds in the whole cell, periplasm, and cytoplasm, respectively. The difference (Δ) between the sum of the measured periplasmic and cytoplasmic accumulation to that of the whole cell accumulation was calculated. Cp, Cc and Ct refer to the concentration of drug in the periplasm, cytoplasm, and whole cell respectively.
| FQs/strain | Mp (ng) | Mc (ng) | Mt (ng) | Δ (ng) | Cp (μg/ml) | Cc (μg/ml) | Ct (μg/ml) |
|---|---|---|---|---|---|---|---|
| Ciprofloxacin ΔC | 81.4 ± 3.1 | 118.0 ± 1.1 | 183.0 ± 18.8 | 16.5 | 50.9 ± 1.9 | 8.6 ± 0.1 | 11.4 ± 1.2 |
| Ciprofloxacin WT | 54.2 ± 1.0 | 70.2 ± 5.3 | 123.1 ± 6.8 | 1.4 | 33.9 ± 0.6 | 5.1 ± 0.4 | 7.6 ± 0.4 |
| Norfloxacin ΔC | 67.9 ± 4.1 | 111.4 ± 0.9 | 180.8 ± 1.9 | −1.4 | 42.5 ± 2.6 | 8.1 ± 0.1 | 11.2 ± 0.1 |
| Norfloxacin WT | 54.4 ± 1.6 | 93.0 ± 0.5 | 147.7 ± 4.7 | −0.3 | 34.0 ± 1.0 | 6.7 ± 0.1 | 9.2 ± 0.3 |
| Enrofloxacin ΔC | 128.6 ± 0.3 | 82.5 ± 5.8 | 187.4 ± 16.6 | 23.7 | 80.4 ± 0.2 | 6.0 ± 0.4 | 11.6 ± 1.0 |
| Enrofloxacin WT | 22.8 ± 0.9 | 13.5 ± 1.0 | 42.9 ± 3.7 | −6.6 | 14.2 ± 0.5 | 1.0 ± 0.1 | 2.7 ± 0.2 |
| Levofloxacin ΔC | 72.2 ± 1.6 | 17.9 ± 2.2 | 117.5 ± 10.4 | −27.4 | 45.1 ± 1.0 | 1.3 ± 0.2 | 7.3 ± 0.6 |
| Levofloxacin WT | 38.8 ± 1.9 | 8.7 ± 0.5 | 52.7 ± 2.4 | −5.2 | 24.2 ± 1.2 | 0.6 ± 0.1 | 3.3 ± 0.1 |
| Lomefloxacin ΔC | 91.1 ± 2.4 | 47.4 ± 5.0 | 149.3 ± 11.1 | −10.7 | 56.9 ± 1.5 | 3.4 ± 0.3 | 9.3 ± 0.7 |
| Lomefloxacin WT | 33.5 ± 3.1 | 9.5 ± 6.5 | 37.0 ± 6.4 | 6.0 | 20.9 ± 1.9 | 0.7 ± 0.4 | 2.3 ± 0.4 |
| Ofloxacin ΔC | 70.4 ± 6.2 | 23.7 ± 3.1 | 96.5 ± 4.6 | −2.4 | 44.0 ± 3.9 | 1.7 ± 0.2 | 6.0 ± 0.3 |
| Ofloxacin WT | 28.8 ± 2.1 | 3.0 ± 2.3 | 35.3 ± 3.2 | −3.5 | 18.0 ± 1.3 | 0.2 ± 0.2 | 2.2 ± 0.2 |
| Fleroxacin ΔC | 100.3 ± 3.5 | 49.0 ± 3.4 | 146.4 ± 5.1 | 3.0 | 62.7 ± 2.2 | 3.6 ± 0.2 | 9.1 ± 0.3 |
| Fleroxacin WT | 58.4 ± 1.6 | 20.2 ± 1.7 | 73.4 ± 4.7 | 5.2 | 36.5 ± 1.0 | 1.5 ± 0.1 | 4.6 ± 0.3 |
| Marbofloxacin ΔC | 76.7 ± 4.8 | 34.0 ± 4.7 | 100.5 ± 3.8 | 10.2 | 47.9 ± 3.0 | 2.5 ± 0.3 | 6.2 ± 0.2 |
| Marbofloxacin WT | 39.6 ± 1.1 | 14.5 ± 3.5 | 46.7 ± 5.6 | 7.4 | 24.7 ± 0.7 | 1.1 ± 0.2 | 2.9 ± 0.3 |
| Moxifloxacin ΔC | 68.1 ± 2.6 | 47.8 ± 0.5 | 124.2 ± 0.9 | −8.3 | 42.5 ± 1.6 | 3.5 ± 0.1 | 7.7 ± 0.1 |
| Moxifloxacin WT | 13.9 ± 0.6 | 10.1 ± 3.1 | 27.5 ± 1.0 | −3.5 | 8.7 ± 0.4 | 0.7 ± 0.2 | 1.7 ± 0.1 |
Gram negative bacteria have four subcellular compartments, the outer membrane (OM), the inner membrane (IM), the periplasm, and the cytoplasm. It is impossible to separate the IM and OM quantitatively in a timely manner, thus our original plan was to fractionate cells into three components, the periplasm, the cytoplasm, and the membrane. However, in practice we realized that fractionating the membrane component while keeping the original compound distribution was technically impossible. To separate the membrane component, cells need to be disrupted vigorously to burst the cells and fragment the cell membrane, followed by prolonged ultra-centrifugation to collect the membrane vesicles as a pellet, as described in the recent publication by Prochnow et al. [20]. The entire process takes longer than an hour. During the process, drug redistribution among lysed cell fragments and the soluble component would certainly occur. Thus, the value determined may not faithfully reflect the actual amount of compound in each component. In this study we will focus on the quantification of accumulation in the two aqueous compartments, the periplasm and cytoplasm.
3.2. All compounds accumulated more in BW25113Δtolc than in BW25113
As expected, for all compounds, the accumulation levels in the wild type strain is lower than that in the tolC knockout strain. This is an additional evidence validating the experimental method and measurements in this study. The ratio of difference varies drastically among compounds used in this study, potentially reflecting the difference in efflux efficiency of the AcrAB-TolC system for different fluoroquinolones. Ciprofloxacin, norfloxacin, and enrofloxacin are the top three accumulated drugs in the whole-cell and the cytoplasm, while enrofloxacin, levofloxacin, and marbofloxacin are the top three accumulators in the periplasm. The accumulation in the whole cell should equal to the sum of accumulations in the cytoplasm, periplasm and the membranes. The difference between the combined accumulation measured in the aqueous compartments and the whole cell accumulation was calculated: Δ=(Mp + Mc)-Mt (Table 1). Overall the Δ values are reasonably small, validating the accuracy of the measurements. The accumulation in the cell membrane, which was not measured in this study, could also contribute to a negative value to the difference.
3.3. Intracellular concentration of fluoroquinolones
To further evaluate the concentration of fluoroquinolones in the periplasm, cytoplasm, and the whole cell, we divided the mass of the accumulated compounds by the volume of each compartment. The calculated concentration can be found in Table 1. Interestingly, we found the periplasmic concentrations of the compounds (Cp) were much higher than the concentrations in the cytoplasm (Cc) and the whole cell (Ct).
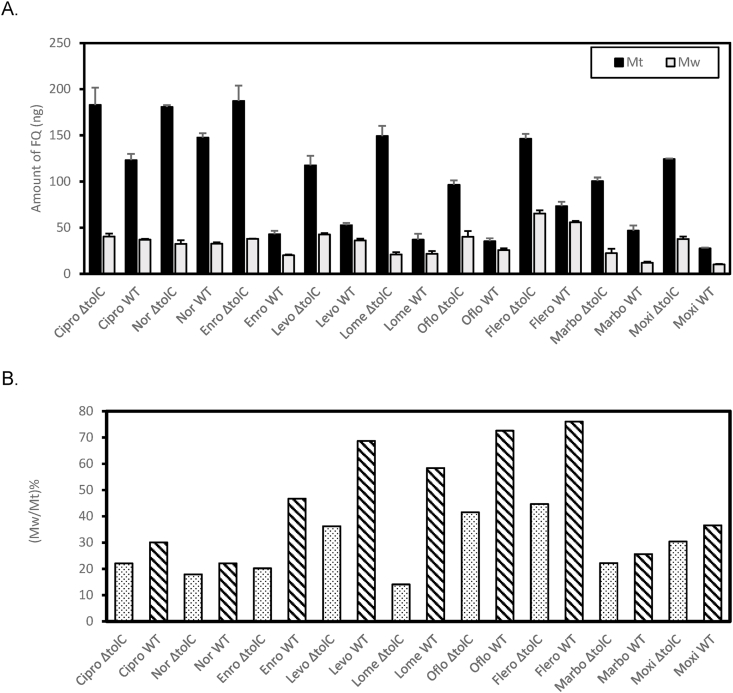
While the logic of washing the cell pellet to remove compounds loosely attached to the exterior of the bacteria is apparent, there is also concern of losing compound, especially from the periplasm, during the washing step. To avoid the wash step, a method was developed to centrifuge cells through a silicon oil layer [17,33,34]. We adopted the silicon oil method in this study, but the very high drug concentration measured in the periplasm made us reconsider the procedure. Inefficient separation of exterior compounds is likely to lead to a larger false positive impact for the periplasmic accumulation, as these compounds could be washed off during the osmotic shock procedure and contribute to the measured periplasmic accumulation.
To evaluate the potential impact of a wash step to the measurement, we quantified compounds that were released from the drug-treated cell pellet during a quick wash step after centrifugation of the bacteria through the silicon oil layer. Fig. 3A and B showed the Mw values for all fluoroquinolones in the two strains, as well as the respective Mt for comparison. Even after centrifugation through the silicon oil layer, the washing process still led to a significant reduction of the associated compounds for all fluoroquinolones. The level of reduction varies among the compounds. Compounds “washed off” could either be loosely associated to the exterior of the cell surface or flushed out from the cell. Interestingly, for every compound, the wild type (WT) strain BW25113 (diagonal lines) lost a higher percentage of associated drug during the wash step than the efflux-deficient strain (dotted), clearly indicating the involvement of active efflux and thus loss of compound from the inside of the bacteria during the quick wash procedure (Fig. 3B).
Fig. 3.
Loss of compounds during wash. A. Measured Mw values. The corresponding Mt was also shown for comparison. B. Relative Mw to Mt for BW25113 (diagonal lines) and BW25113ΔtolC (dotted).
The accumulation data of the periplasm and whole cell accumulation, after wash correction, were shown in Table 2. Since the cytoplasmic accumulation data were collected from samples after the removal of the periplasm, there is no need to apply wash correction to the cytoplasmic accumulation data. Even with the wash correction, the periplasmic concentrations remain to be higher than both the cytoplasmic and external media concentrations.
Table 2.
Accumulation of fluoroquinolones (FQs) in two strains, BW25113 (WT) and BW25113Δtolc (ΔC) after correction for wash. Mtw and Mpw are accumulation numbers in the whole cell and periplasm and cytoplasm, respectively. Cpw and Ctw refer to the concentration of drug in the periplasm and whole cell respectively. Accumulated fluoroquinolones were at higher concentrations than the external media.
| FQs/strain | Mw (ng) | Mpw (ng) | Mtw (ng) | Cpw (μg/ml) | Ctw (μg/ml) |
|---|---|---|---|---|---|
| Ciprofloxacin ΔC | 40.4 ± 7.3 | 41.0 ± 7.9 | 142.6 ± 20.1 | 25.6 ± 5.0 | 8.9 ± 1.2 |
| Ciprofloxacin WT | 37.0 ± 5.6 | 17.3 ± 5.7 | 86.1 ± 8.8 | 10.9 ± 3.5 | 5.3 ± 0.5 |
| Norfloxacin ΔC | 32.4 ± 1.1 | 35.6 ± 4.3 | 148.4 ± 2.2 | 22.3 ± 2.7 | 9.2 ± 0.1 |
| Norfloxacin WT | 32.6 ± 1.4 | 21.8 ± 2.1 | 115.0 ± 4.9 | 13.6 ± 1.3 | 7.1 ± 0.3 |
| Enrofloxacin ΔC | 37.9 ± 3.9 | 90.7 ± 3.9 | 149.5 ± 17.0 | 56.7 ± 2.4 | 9.3 ± 1.1 |
| Enrofloxacin WT | 20.2 ± 3.4 | 2.7 ± 3.5 | 22.8 ± 5.0 | 1.7 ± 2.2 | 1.4 ± 0.3 |
| Levofloxacin ΔC | 42.6 ± 0.6 | 29.6 ± 1.7 | 74.9 ± 10.5 | 18.5 ± 1.1 | 4.7 ± 0.6 |
| Levofloxacin WT | 36.2 ± 3.1 | 2.6 ± 3.7 | 16.5 ± 3.9 | 1.6 ± 2.3 | 1.0 ± 0.2 |
| Lomefloxacin ΔC | 21.0 ± 5.5 | 70.1 ± 6.0 | 128.0 ± 12.4 | 43.8 ± 3.7 | 8.0 ± 0.8 |
| Lomefloxacin WT | 21.6 ± 5.9 | 11.9 ± 6.6 | 15.4 ± 8.7 | 7.4 ± 4.2 | 1.0 ± 0.5 |
| Ofloxacin ΔC | 40.1 ± 5.0 | 30.3 ± 8.0 | 56.4 ± 6.8 | 19.0 ± 5.0 | 3.5 ± 0.4 |
| Ofloxacin WT | 25.7 ± 0.7 | 3.2 ± 2.2 | 9.7 ± 3.2 | 2.0 ± 1.4 | 0.6 ± 0.2 |
| Fleroxacin ΔC | 65.4 ± 6.4 | 34.9 ± 7.3 | 81.0 ± 8.1 | 21.8 ± 4.5 | 5.0 ± 0.5 |
| Fleroxacin WT | 55.8 ± 2.0 | 2.6 ± 2.5 | 17.6 ± 5.0 | 1.7 ± 1.6 | 1.1 ± 0.3 |
| Marbofloxacin ΔC | 22.3 ± 3.5 | 54.4 ± 5.9 | 78.1 ± 5.2 | 34.0 ± 3.7 | 4.9 ± 0.3 |
| Marbofloxacin WT | 12.0 ± 1.6 | 27.6 ± 1.9 | 34.8 ± 5.8 | 17.2 ± 1.2 | 2.2 ± 0.4 |
| Moxifloxacin ΔC | 37.7 ± 4.3 | 30.3 ± 5.0 | 86.5 ± 4.3 | 19.0 ± 3.1 | 5.4 ± 0.3 |
| Moxifloxacin WT | 10.1 ± 2.1 | 3.9 ± 2.2 | 17.4 ± 2.3 | 2.4 ± 1.4 | 1.1 ± 0.1 |
3.4. Accumulated fluoroquinolone were at higher concentration than the external media
In the efflux deficient strain ΔtolC, the concentrations of compound accumulated in the cells were always higher than the external concentration of compound used in the study (2.0 μg/ml). The observation of higher drug concentration collected inside the cell than the external concentration has been reported in several previous studies, however, whole cell accumulations were measured in these cases [[35], [36], [37], [38]]. We found that the accumulated concentrations in the periplasm were higher than concentrations both the in the cytoplasm and the external media. Two potential artifacts could lead to evaluated number of the measured Cp: first, nonspecific binding of compounds to the cell surface, which was later detached from the membrane during the osmotic shock step; second, leakage during the osmotic shock step, which may lead to the release of compounds from the cytoplasm to the osmotic shock solution. To minimized potential effect of the first possibility, we evaluated the effect of washing as described above. Even with the wash correction, which likely leads to an underestimate on the measured Cpw due to loss of compound from the periplasm during wash, the periplasmic concentrations for most cases remained higher than 2.0 μg/ml. To address the second possible source of error, we used a very quick osmotic shock procedure (less than 10 min) during our study. To examine the integrity of the inner membrane during the process, we conducted two assays. First, we used BW25113 expressing sfGFP. As described in materials and methods, the cells were subjected to identical treatment as cells used in the drug accumulation assay to obtain the periplasmic, cytoplasm, and whole cell extractions. Measurement of sfGFP fluorescence in the three samples revealed that the signal in the periplasmic sample was less than 2% of the whole cell signal. We acknowledge that sfGFP is much larger than the fluoroquinolones, however the lack of sfGFP leakage suggests that the procedure did not lead to a large-scale membrane disruption. To further confirm that the high fluoroquinolone concentration in the periplasm was not a result of leakage from the cytoplasm, we fractionated BW25113 as described and measured the ATP concentration in the periplasm, cytoplasm, and the whole cell samples. Since ATP is only present in the cytoplasm, the detection of ATP in the periplasm would indicate that leakage occurred during the osmotic shock procedure. We found that the relative ATP concentration in the periplasm was ~6% of the concentration determined for the whole cell sample, indicating that the leakage of small molecules such at ATP from the cytoplasm was minimum during the osmotic shock procedure (Fig. S3 in Supplementary materials).
3.5. Correlation between MIC ratio versus accumulation ratio in two isogenic strains
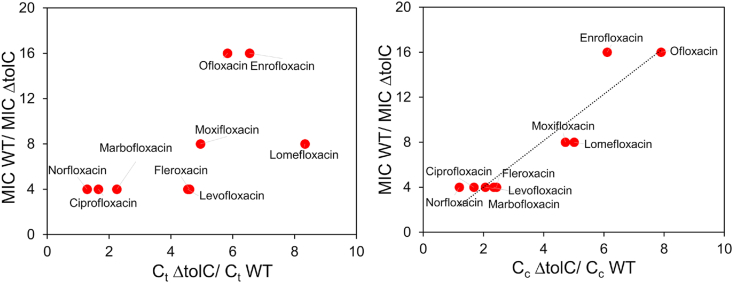
MIC of all fluoroquinolones were determined for both BW25113 and BW25113ΔtolC (Table 3). Compounds with higher MIC are less effective against a certain bacterium. While higher accumulation inside the cell makes a compound a better antibiotic, accumulation alone never correlated well with the effectiveness of an antibiotic as reported in previous studies [[14], [15], [16], [17], [18], [19]]. Although the efficacy of an antibiotic against a bacterium depends on several factors, if other parameters remain consistent except for accumulation, we expect that the accumulation at the target site to correlate directly with the efficacy of the antibiotic. We measured the accumulation of the nine fluoroquinolones in a pair of isogenic strains, BW25113 and BW25113ΔtolC. The only difference between the two strains is the deficiency of efflux in the knockout strain, thus we expect the MIC ratio between the two strains to correlate negatively with the cytosolic accumulation ratio of these compounds. As expected, the MIC ratio correlated very well with the ratio of cytoplasmic accumulation Cc with a Pearson's correlation coefficient of 0.94 (Fig. 4). However, no correlation was observed when the MIC ratio was plotted against the ratio of the whole cell accumulation Ct.
Table 3.
MIC of fluoroquinolones in two E. coli strains, BW25113 (WT) and BW25113 ΔtolC.
| Fluoroquinolones | MIC (μg/ml) | RMIC | |
| WT | ΔtolC | ||
| Ciprofloxacin | 0.016 | 0.004 | 4 |
| Norfloxacin | 0.064 | 0.016 | 4 |
| Enrofloxacin | 0.032 | 0.002 | 16 |
| Levofloxacin | 0.032 | 0.008 | 4 |
| Lomefloxacin | 0.125 | 0.016 | 8 |
| Ofloxacin | 0.125 | 0.008 | 16 |
| Fleroxacin | 0.064 | 0.016 | 4 |
| Marbofloxacin | 0.032 | 0.008 | 4 |
| Moxifloxacin | 0.064 | 0.008 | 8 |
Fig. 4.
Evaluation of correlation between MIC and accumulation. MIC ratio in the wild type and efflux deficient strain plotted against the ratio in whole cell accumulation (left) or spheroplast accumulation (right).
3.6. Effect of hydrophobicity in partition across the inner membrane
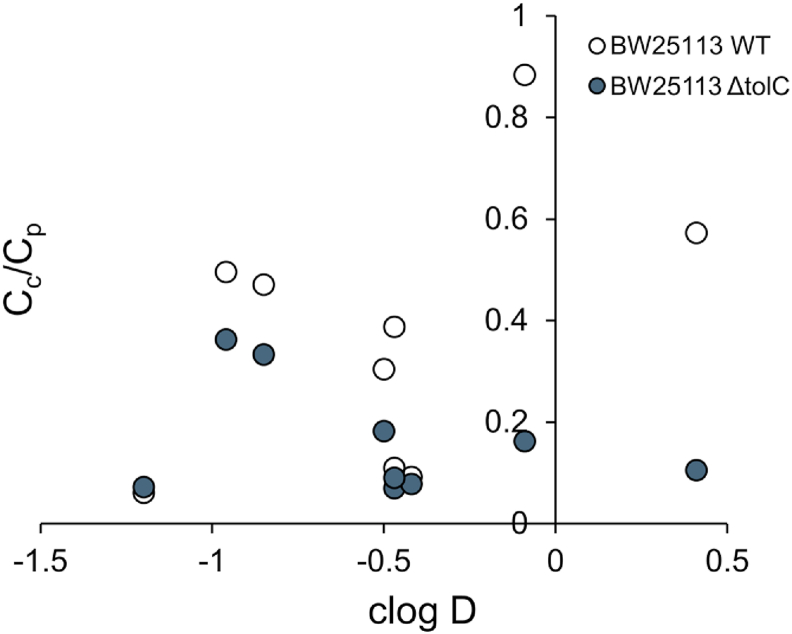
As mentioned in the introduction, the correlation between hydrophobicity of a compound with its accumulation has been investigated by several studies [[14], [15], [16],18,19]. There is no consistent conclusion about how hydrophobicity affects the overall accumulation in Gram-negative bacteria. Here, the cytoplasmic and periplasmic accumulation data offered us an opportunity to evaluate the role of the inner membrane as a hydrophobic barrier. The ratio of the cytoplasmic to periplasmic accumulation (Cc/Cp) were plotted against the hydrophobicity of the compounds (Fig. 5). Higher hydrophobicity, as represented by the clogD values, did not correlate with a higher percentage of compounds accumulated into the cytoplasm.
Fig. 5.
Cc/Cp plotted against clogD. clogD values were calculated using Chemaxon, MarvinSketch version 18.10 at pH 7.4, with electrolyte concentration of 0.1 mol/dm3 Cl−, 0.1 mol/dm3 Na+/K+.
4. Discussion
The main purpose of this study was to characterize antimicrobials accumulation and distribution inside the periplasm and cytoplasm of Gram-negative bacterial cell and examine the impact of this distribution on the efficacy of the drug. The separation of the whole cell accumulation into the periplasmic and cytoplasmic components offered us the unique opportunity to make several interesting observations.
First, we observed that inside the Gram-negative bacterium E. coli, the accumulated fluoroquinolone concentration was very different in different subcellular compartments. For all nine compounds in both strains, we observed a much higher concentration of fluoroquinolones accumulated in the periplasm compared to the cytoplasm. In most cases the periplasmic fluoroquinolone concentration is even higher than the concentration in the exterior media. Prochnow et al. also observed that the concentration of ciprofloxacin in the periplasm of E. coli was higher than the external concentration used in the accumulation assay [20]. While no active uptake has been identified so far for fluoroquinolones, the negative inside Donnan potential across the outer membrane has been considered to be a major contributing factor that lead to the accumulation of these compounds inside the Gram negative bacteria [25,39]. Another possibility is the binding of fluoroquinolones to yet unknown periplasmic components, which may have reduced the concentration of free drug in the periplasm to be lower than the exterior concentration.
Another interesting observation is the lack of correlation between the hydrophobicity of the compounds and their ability to accumulate in the cytoplasm. The inner membrane is clearly not just a hydrophobic barrier in this case. Other factors that could potentially affect the Cc/Cp ratio are the structure of the compounds, and presence of binding partners in the cytoplasm and/or periplasm. Binding to a partner will switch a molecule from the free state to a bound state, which may affect the equilibrium across the inner membrane.
To understand the structural features that promote a compound's ability to penetrate the Gram-negative cellular envelope, several studies were performed recently to investigate the accumulation of compounds in Gram-negative bacteria [7,13,[40], [41], [42], [43]]. O'shea and Moser observed that antimicrobials effective for Gram-negative bacteria were normally small (less than 600 Da) and much more polar in general than other pharmaceuticals [41]. Silver et al. compared the antimicrobials effective against Gram-negative bacteria with the ones effective against Gram-positive bacteria and reported that the former group is smaller in size and less polar. Among the ones active against Gram-negative bacteria, those that enter through passive diffusion are less polar compared to the those entering through active transport [42]. Richter et al. measured accumulation of a large group of compounds to distill principles favoring accumulation in Gram-negative bacteria. They concluded that good accumulators tend to have amine functional groups, be amphiphilic and rigid, and with low globularity [13]. More importantly, these rules were applied to convert deoxynybomycin from a Gram-positive-only antibiotic into an antibiotic that is active against a wide range of multi-drug resistant bacteria including Gram-negative ones [13]. Acosta-Gutierrez et al. focused on porins to study the constricted region of the channel, the electrostatic interactions, and size limit to understand the mechanisms of penetration through these channels. They concluded that positively charged groups are important to promote penetration, which is consistent with the mechanisms of the rules proposed by Richter et el [13,40]. In all these studies, whole cell accumulation was measured. We argue that quantification of the accumulation at different subcellular compartments will lead to valuable insight into the mechanism of effective penetration. Collecting data on the accumulation of a large group of compounds in sub-cellular compartments is a cumbersome and labor-intensive, yet necessary effort. Here we reported the preliminary result on a group of nine fluoroquinolones. Additional studies on diverse sets of compounds would be critically necessary to fully understand the structural features that favor the accumulation of antimicrobials in bacterial cells.
Author contribution
YW and AP designed the project. AP, OA, YC conducted experiments and analyzed data. YW and AP wrote the manuscript.
Acknowledgement
We thank Dr. Alice Erwin for critical reading and suggestions of this manuscript. We thank Dr. Harry LeVine for his assistance in the ATP assay. This work is supported by NIH grant number 1R56AI137020, 1R21AI142063-01, and NSF grant number CHE-1709381.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrep.2020.100849.
Appendix A. Supplementary data
References
- 1.IACG . Report To the Secretary General Of the United Nations. 2019. No time to wait–securing the future from drug-resistant infections. [Google Scholar]
- 2.Lewis K. New approaches to antimicrobial discovery. Biochem. Pharmacol. 2017;134:87–98. doi: 10.1016/j.bcp.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013;12:371. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 4.Vranakis I., Goniotakis I., Psaroulaki A., Sandalakis V., Tselentis Y., Gevaert K., Tsiotis G. Proteome studies of bacterial antibiotic resistance mechanisms. Journal of Proteomics. 2014;97:88–99. doi: 10.1016/j.jprot.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Bello C. Antibiotic adjuvants–A strategy to unlock bacterial resistance to antibiotics. Bioorg. Med. Chem. Lett. 2017;27:4221–4228. doi: 10.1016/j.bmcl.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Choi U., Lee C.R. Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front. Microbiol. 2019;10:953. doi: 10.3389/fmicb.2019.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zgurskaya H.I., Lopez C.A., Gnanakaran S. Permeability barrier of gram-negative cell envelopes and approaches to bypass it. ACS Infect. Dis. 2015;1:512–522. doi: 10.1021/acsinfecdis.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delcour A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pages J.M., James C.E., Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi A., Nakashima R., Sakurai K. Structural basis of RND-type multidrug exporters. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epand R.M., Walker C., Epand R.F., Magarvey N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta Biomembr. 2016;1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Richter M.F., Drown B.S., Riley A.P., Garcia A., Shirai T., Svec R.L., Hergenrother P.J. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. 2017;545:299. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piddock L.J.V., Jin Y.F., Griggs D.J. Effect of hydrophobicity and molecular mass on the accumulation of fluoroquinolones by Staphylococcus aureus. J. Antimicrob. Chemother. 2001;47:261–270. doi: 10.1093/jac/47.3.261. [DOI] [PubMed] [Google Scholar]
- 15.Piddock L.J.V., Johnson M.M. Accumulation of 10 fluoroquinolones by wild-type or efflux mutant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2002;46:813–820. doi: 10.1128/AAC.46.3.813-820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piddock L.J.V., Ricci V. Accumulation of five fluoroquinolones by Mycobacterium tuberculosis H37Rv. J. Antimicrob. Chemother. 2001;48:787–791. doi: 10.1093/jac/48.6.787. [DOI] [PubMed] [Google Scholar]
- 17.Iyer R., Ye Z., Ferrari A., Duncan L., Tanudra M.A., Tsao H., Wang T., Gao H., Brummel C.L., Erwin A.L. Evaluating LC–MS/MS to measure accumulation of compounds within bacteria. ACS Infect. Dis. 2018;4:1336–1345. doi: 10.1021/acsinfecdis.8b00083. [DOI] [PubMed] [Google Scholar]
- 18.Bazile S., Moreau N., Bouzard D., Essiz M. Relationships among antibacterial activity, inhibition of DNA gyrase, and intracellular accumulation of 11 fluoroquinolones. Antimicrob. Agents Chemother. 1992;36:2622–2627. doi: 10.1128/aac.36.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams K.J., Piddock L.J. Accumulation of rifampicin by Escherichia coli and Staphylococcus aureus. J. Antimicrob. Chemother. 1998;42:597–603. doi: 10.1093/jac/42.5.597. [DOI] [PubMed] [Google Scholar]
- 20.Prochnow H., Fetz V., Hotop S.-K., García-Rivera M.A., Heumann A., Brönstrup M. Subcellular quantification of uptake in gram-negative bacteria. Anal. Chem. 2019;91:1863–1872. doi: 10.1021/acs.analchem.8b03586. [DOI] [PubMed] [Google Scholar]
- 21.Piddock L.J.V., Jin Y.-F., Ricci V., Asuquo A.E. Quinolone accumulation by Pseudomonas aeruginosa, Staphylococcus aureus and Escherichia coli. J. Antimicrob. Chemother. 1999;43:61–70. doi: 10.1093/jac/43.1.61. [DOI] [PubMed] [Google Scholar]
- 22.Davidson V.L., Sun D. Lysozyme-osmotic shock methods for localization of periplasmic redox proteins in bacteria. Methods Enzymol. 2002;353:121–130. doi: 10.1016/s0076-6879(02)53042-1. [DOI] [PubMed] [Google Scholar]
- 23.Miyake-Stoner S.J., Refakis C.A., Hammill J.T., Lusic H., Hazen J.L., Deiters A., Mehl R.A. Generating permissive site-specific unnatural aminoacyl-tRNA synthetases. Biochemistry. 2010;49:1667–1677. doi: 10.1021/bi901947r. [DOI] [PubMed] [Google Scholar]
- 24.Bagnara A.S., Finch L.R. Quantitative extraction and estimation of intracellular nucleoside triphosphates of Escherichia coli. Anal. Biochem. 1972;45:24–34. doi: 10.1016/0003-2697(72)90004-8. [DOI] [PubMed] [Google Scholar]
- 25.Stock J.B., Rauch B., Roseman S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 1977;252:7850–7861. [PubMed] [Google Scholar]
- 26.Van Wielink J.E., Duine J.A. How big is the periplasmic space? Trends Biochem. Sci. 1990;15:136. doi: 10.1016/0968-0004(90)90208-s. [DOI] [PubMed] [Google Scholar]
- 27.Graham L., Beveridge T., Nanninga N. Periplasmic space and the concept of the periplasm. Trends Biochem. Sci. 1991;16:328–329. doi: 10.1016/0968-0004(91)90135-i. [DOI] [PubMed] [Google Scholar]
- 28.Seltmann G., Holst O. The Bacterial Cell Wall. Springer; 2002. Periplasmic space and rigid layer; pp. 103–132. [Google Scholar]
- 29.Pilizota T., Shaevitz J.W. Fast, multiphase volume adaptation to hyperosmotic shock by Escherichia coli. PloS One. 2012;7 doi: 10.1371/journal.pone.0035205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasupuleti S., Sule N., Cohn W.B., MacKenzie D.S., Jayaraman A., Manson M.D. Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3, 4-dihydroxymandelic acid. J. Bacteriol. 2014;196:3992–4000. doi: 10.1128/JB.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller S.I., Salama N.R. The gram-negative bacterial periplasm: size matters. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volkmer B., Heinemann M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PloS One. 2011;6 doi: 10.1371/journal.pone.0023126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cory T.J., Winchester L.C., Robbins B.L., Fletcher C.V. A rapid spin through oil results in higher cell-associated concentrations of antiretrovirals compared with conventional cell washing. Bioanalysis. 2015;7:1447–1455. doi: 10.4155/bio.15.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X.Z., Livermore D.M., Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline. chloramphenicol, and norfloxacin, Antimicrobial Agents and Chemotherapy. 1994;38:1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams K.J., Chung G.A.C., Piddock L.J.V. Accumulation of Norfloxacin by Mycobacterium aurum and Mycobacterium smegmatis. Antimicrob. Agents Chemother. 1998;42:795–800. doi: 10.1128/aac.42.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phetsang W., Pelingon R., Butler M.S., Kc S., Pitt M.E., Kaeslin G., Cooper M.A., Blaskovich M.A.T. Fluorescent trimethoprim conjugate probes to assess drug accumulation in wild type and mutant Escherichia coli. ACS Infect. Dis. 2016;2:688–701. doi: 10.1021/acsinfecdis.6b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y., Joubran C., Miller-Vedam L., Isabella V., Nayar A., Tentarelli S., Miller A. Thinking outside the “bug”: a unique assay to measure intracellular drug penetration in gram-negative bacteria. Anal. Chem. 2015;87:3579–3584. doi: 10.1021/ac504880r. [DOI] [PubMed] [Google Scholar]
- 38.Davis T.D., Gerry C.J., Tan D.S. General platform for systematic quantitative evaluation of small-molecule permeability in bacteria. ACS Chem. Biol. 2014;9:2535–2544. doi: 10.1021/cb5003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikaido H., Thanassi D.G. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob. Agents Chemother. 1993;37:1393–1399. doi: 10.1128/aac.37.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acosta-Gutiérrez S., Ferrara L., Pathania M., Masi M., Wang J., Bodrenko I., Zahn M., Winterhalter M., Stavenger R.A., Pagès J.-M., Naismith J.H., van den Berg B., Page M.G.P., Ceccarelli M. Getting drugs into gram-negative bacteria: rational rules for permeation through general porins. ACS Infect. Dis. 2018;4:1487–1498. doi: 10.1021/acsinfecdis.8b00108. [DOI] [PubMed] [Google Scholar]
- 41.O'Shea R., Moser H.E. Physicochemical properties of antibacterial compounds: implications for drug discovery. J. Med. Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 42.Silver L.L. A Gestalt approach to Gram-negative entry. Bioorg. Med. Chem. 2016;24:6379–6389. doi: 10.1016/j.bmc.2016.06.044. [DOI] [PubMed] [Google Scholar]
- 43.Brown D.G., May-Dracka T.L., Gagnon M.M., Tommasi R. Trends and exceptions of physical properties on antibacterial activity for Gram-positive and Gram-negative pathogens. J. Med. Chem. 2014;57:10144–10161. doi: 10.1021/jm501552x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.