Abstract
Background
Calcium signaling and membrane fusion play key roles in exocytosis of drug-containing vesicles through the blood-brain barrier (BBB). Identifying the role of synaptotagmin-like protein4-a (Slp4-a) in the presence of Ca2+ ions, at the pre-fusion stage of a vesicle with the basolateral membrane of endothelial cell, can reveal brain drug transportation across BBB.
Methods
We utilized molecular dynamics (MD) simulations with a coarse-grained PACE force field to investigate the behaviors of Slp4-a with vesicular and endothelial membranes at the pre-fusion stage of exocytosis since all-atom MD simulation or experiments are more time-consuming and expensive to capture these behaviors.
Results
The Slp4-a pulls lipid membranes (vesicular and endothelial) into close proximity and disorganizes lipid arrangement at contact points, which are predictors for initiation of fusion. Our MD results also indicate that Slp4-a needs Ca2+ to bind with weakly-charged POPE lipids (phosphatidylethanolamine).
Conclusions
Slp4-a is an important trigger for membrane fusion in BBB exocytosis. It binds to lipid membranes at multiple binding sites and triggers membrane disruption for fusion in calcium-dependent case.
General significance
Understanding the prefusion process of the vesicle will help to design better drug delivery mechanisms to the brain through formidable BBB.
Keywords: Synaptotagmin-like protein4-a (Slp4-a), Blood-brain barrier (BBB), Receptor-mediated pre-fusion, Brain drug delivery, Molecular dynamics
Highlights
-
•
Role of Ca2+ on Slp4-a is studied for vesicle pre-fusion in EC to initiate exocytosis.
-
•
Coarse-grained MD is used to study large scale conformation change of Slp-4a.
-
•
Interaction between C2A domain and lipids is much stronger than that of C2B.
-
•
Slp4-a can bind to bilayer membrane in Ca2+-bound case to close membrane gap.
1. Introduction
Treatment of neurodegenerative diseases requires delivery of drugs across the blood-brain barrier (BBB), a specialized tight junction formed by microvascular endothelial cells (ECs) [1,2]. Successful delivery of drugs through BBB requires both endocytosis and exocytosis. Although there are many studies for endocytosis of nanocarriers such as clathrin-mediated endocytosis, caveolar endocytosis [[3], [4], [5], [6]], the mechanism of exocytosis is poorly understood, especially for the BBB system. However, the exocytosis mechanism is well studied in the synapses of neurons for neurotransmitter delivery [7,8]. Here we adopt a similar exocytosis mechanism for vesicle exocytosis from the endothelial cell using functional proteins found in the Weibel-Palade body (WPB) exocytosis in the endothelial cell [9,10]. Table 1 lists the necessary functional proteins in synaptic exocytosis and their equivalent counterpart for vesicle transport based on WPB exocytosis from an endothelial cell.
Table 1.
List of equivalent functional proteins and processes in synaptic and BBB exocytosis.
| Equivalent protein or process | Synaptic exocytosis [7,8] | BBB exocytosis [9,10] |
|---|---|---|
| Synaptosomal Associated Protein (SNAP) | SNAP25 | SNAP23 |
| Vesicle Associated Membrane Protein (VAMP) | VAMP2 | VAMP8 |
| Syntaxin Protein | Syntaxin1 | Syntaxin2 |
| Calcium Sensor Protein | Synaptotagmin 1 (Syt1) | Synaptotagmin-like protein4-a (Slp4-a) |
| Syntaxin-binding Protein | Mammalian Uncoordinated 18–1 (Munc18-1) | Munc18-1 |
| First protein bridge for vesicle docking with the cell membrane | Munc13-1 connects to lipids | Munc13-4 connects to Annexin complex |
| Second protein bridge for vesicle docking with the cell membrane | Connection between Syt1, CAPS (Calcyphosin) and Munc18-1 | Connection between Slp4-a and Munc18-1 |
| SNAREs (SNAP REceptor) complex formed at the priming stage | Consist of SNAP25, VAMP2, and Syntaxin1 | Consist of SNAP23, VAMP8 and Syntaxin2 |
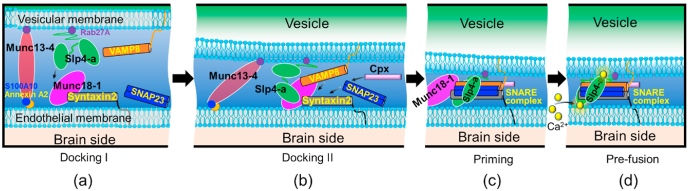
In a synaptic system, the exocytosis of neurotransmitters is proceeded by docking, priming, and pre-fusion. During the docking stage of synaptic exocytosis [9], Munc13-1 protein uses its C2 domains to link the vesicular membrane to the cell membrane to form the first docking bridge. This bridge not only helps vesicular protein VAMP2 to bind to Munc18-1 (a syntaxin binding protein), but also facilitates the formation of the second docking bridge by connecting Munc18-1, Syt1 (synaptotagmin 1) and CAPS (calcyphosin) together. Similarly, in the BBB system, the first docking bridge (Fig. 1a) is proposed to form by Munc13-4, where it binds to Rab27A and annexin A2-S100A10 complex to link the vesicle to endothelial membrane like in WPB [10]. This link helps Slp4-a to form the second docking bridge by connecting with Munc18-1, which consequently closes the gap between the vesicle and endothelial membrane [10,11] (Fig. 1b). One difference between Slp4-a and Syt1 is that Slp4-a binds to the vesicular membrane through Rab27A (Ras-related protein 27A) [12], while Syt1 connects directly to the vesicular membrane by its transmembrane domain [8].
Fig. 1.
Schematic of vesicle docking at the pre-fusion stage in BBB exocytosis. (a) Vesicle connects to the endothelial membrane through the first protein bridge (Docking I). Important proteins involved in this bridge are Munc13-4, Rab27A, S100A10, and Annexin A2. (b) The two membranes get closer when Slp4-a binds to Munc18-1 as well as syntaxin2 to form the second docking bridge (Docking II). (c) SNAREs complex formed by the merging of Syntaxin2, VAMP8, and SNAP23 into 4 helix bundle for priming. In this stage, Slp4-a starts to connect with SNAREs and complexin. (d) Calcium ion-dependent pre-fusion, where the protein system could trigger the membrane fusion.
In the synaptic system, the vesicle associated membrane protein (VAMP2) connects with Munc18-1 at the docking II stage as a pre-requisite for the formation of SNAREs [8]. Similarly, VAMP8 is assumed to connect with Munc18-1 (Syntaxin2 binding protein) in the BBB system at the end of the docking stage II as shown in Fig. 1b.
During the priming stage, Munc18-1 initiates the assembly of the SNARE four-helix bundle (in the synaptic system) by incorporation of SNAP-25 [8]. Through circular dichroism spectroscopy, Zhou et al. [13] reported that complexin 1 (cpx1) also binds with SNAREs complex in the priming stage. The priming stage information does not exist for the BBB system. However, owing to the presence of the similar proteins in the BBB system, we presented an analogous priming mechanism for the vesicle and the endothelial cells where SNAP23 binds to Munc18-1 (as well as VAMP8 and Syntaxin2) to form the 4 helix bundle SNAREs complex (Fig. 1c).
In the synaptic system, the pre-fusion stage starts with the increase of calcium ion concentration in the neuron. Experimental studies suggest that the membrane fusion is actuated by increasing levels of free Ca2+ ions in the cytosol [8]. Ca2+sensor proteins (Syt1) act as the fusion initiator because the Ca2+ bindings trigger the general movement and structural changes of the two domains (C2A and C2B) of these proteins, leading to an initial lipid binding [8]. However, the pre-fusion step is quite unclear in the BBB system. We thus propose a similar pre-fusion stage for BBB based on the synaptic pre-fusion stage, as shown in Fig. 2a, where Slp4-a is located next to SNAREs and complexin. Similar to Syt1 in the synaptic system, which triggers the fusion process in the presence of calcium ions, the Slp4-a is a calcium sensor [14]. Even though the Syt1 protein of the synaptic system has been extensively studied [15,16], the role of Slp4-a is not known at the pre-fusion stage of BBB exocytosis. Through an experiment work, Lyakhova et al. [17] demonstrated a strong interaction between Slp4-a (especially its C2A domain) and PIP2 plasma membrane for vesicle-docking and fusion through anionic binding. Nevertheless, molecular details of how Slp4-a facilitates the lipid interaction or triggers membrane fusion are not clear.
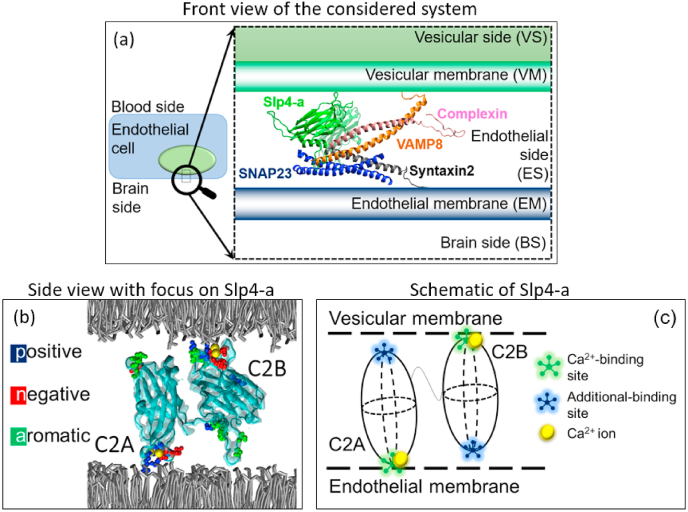
Fig. 2.
Structure of the protein system at the pre-fusion stage of a vesicle in BBB. (a) The zoomed-in view of key components: Vesicular membrane, endothelial cell membrane in the brain side, and associate proteins involved in the pre-fusion stage. Protein complex involves in this process includes VAMP8 (orange), Complexin (pink), Syntaxin2 (gray), and SNAP23 (blue), and Slp4-a (green). (b) The new cartoon model for Slp4-a (cyan-colored), which consists of C2A and C2B domains. Each C2 domain shows both the Ca2+ binding site and additional binding sites with some important amino acids. Calcium ions (yellow) are shown in the calcium binding site. The additional binding site is located on the opposite side of the Ca2+ binding site in each domain. Lipids are shown as gray layers located at the top and bottom sides of the Slp4-a. (c) Sketch of the Slp4-a model with four important regions: two Ca2+ binding sites in green and two additional binding regions in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The structure of Slp4-a has been presented by Wang et al. [18] (PDB: 3FDW). As shown in Fig. 2b, the Ca2+ binding site of the C2A (called C2A-bottom) is mainly controlled by negatively-charged Aspartic (D) residues such as D387 and D418. The number of D residues in the C2A-bottom of Slp4-a is less than that of Syt1-C2A. Similarly, the Ca2+ binding site of the C2B (called C2B-top) of Slp4-a is mainly formed by a group of negatively-charged Aspartic residues including D548, D603, and D611, which is similar to Syt1-C2B [18]. Besides, other groups of heavily-charged amino acids are found in both C2A (top side) and C2B (bottom side) domain of Slp4-a and they act as additional lipid binding sites (see Fig. 2c). These residues include negatively charged E434, D466, and D471 at the top of the C2A domain (called C2A-top) and E590, D591, D634, and E638 at the bottom of C2B domain (called C2B-bottom). All binding sites can also have both positively charged and hydrophobic amino acid residues (Fig. 2b) [18]. The number of Ca2+ ions in each calcium binding pocket of Slp4-a and how differently those pockets work compared with synaptic ones are not known.
To understand the role of Slp4-a in the prefusion stage, we will use MD simulations. MD simulations have been used to investigate behaviors of many nanoscale systems, including large scale conformational changes of synaptotagmin in the synaptic system [19,20], but it has not been used for Slp4-a in the endothelial cells (ECs) of BBB. In the BBB system, available MD studies are only limited to penetration of drug-like molecules to the lipid membrane of ECs through receptor-mediated endocytosis [21,22]. Thus, it is important to understand the molecular details of how Slp4-a triggers the pre-fusion for the exocytosis process. In this work, we conducted MD simulations using the hybrid PACE (protein in atomistic details coupled with a coarse-grained environment) force field [[23], [24], [25]] to examine the Ca2+-dependent response of Slp4-a. POPE is used as lipid membranes to study the interaction between Slp4-a and lipid membranes. We are particularly interested in studying the large scale conformational changes of Slp4-a in the presence of calcium ions, which might trigger the pre-fusion.
2. Models and methods
The general scope of this research is at the pre-fusion stage for exocytosis in the BBB system as shown in Fig. 2a. During the pre-fusion stage, the vesicle membrane is in close proximity to the plasma membrane, and the SNARE complex is already formed by the combination of various functional proteins such as Syntaxin2 (SX2), VAMP8, and SNAP23. Based on the synaptic system, it is assumed that Slp4-a and complexin remain very close to each other through helix domain interactions, and they have non-bonded interactions with SNAREs complex, especially VAMP8 and syntaxin2 [13].
Steered MD simulations have been reported for SNARE complex in the synaptic system to study membrane fusion using a coarse-grained MARTINI force field [26,27]. In this study, the PACE force field is used to study the pre-fusion stage since it could capture conformational change of proteins, which is difficult for all-atom MD simulations [24,28]. In PACE, proteins are modeled with a united-atom-based model, while coarse-grained MARTINI (four heavy molecules represent one site) is used for water and lipid [26,27]. In the united-atom-based protein model, each heavy atom with bound hydrogens is modeled as one bead, however, important hydrogens on the backbone or side chain could be modeled as separate beads [29]. The 12-6 Lennard–Jones (LJ) potential is utilized for non-bonded interactions between water and protein & water and water. Modified LJ potentials are used for hydrogen bonds; Coulomb potentials are used to handle electrostatic interactions between polar sites of proteins [30].
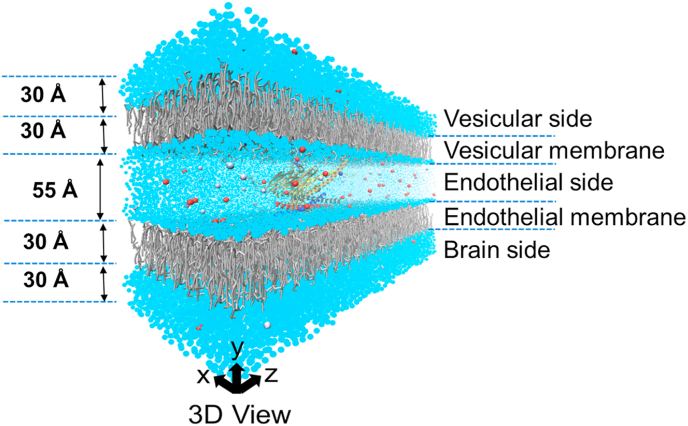
We performed all simulations in NAMD 2.10 [31]. The initial configuration was built using VMD [32] and CHARMM-GUI [33,34], and it consists of water, SNAREs, Slp4-a, complexin, and two bilayers (POPE) lipid membranes as shown in Fig. 3. Our simulation system contains three separate segments: the vesicles medium at the top, the endothelium medium with all functional proteins at the middle, and the brain side medium at the bottom. In our simulation system, Syntaxin2 and VAMP8 are attached to the bottom and top lipid membrane, respectively by the transmembrane domains to keep the initial membrane distance at 55 Å [13]. The size of the simulation system is ~160 × 175 × 300 Å3, which contains 37,800 coarse-grain water molecules. In addition, 150 mM NaCl was added to the system as the neutralizer (see Fig. 3). The simulation systems were equilibrated systematically using the standard equilibration process [35].
Fig. 3.
Coarse-grained (CG) simulation model for the pre-fusion of vesicle initiated by Slp4-a. The model contains ~77,679 atoms with an overall dimension of 160 × 175 × 300 Å3. The center of mass of the whole protein system is located at the center of the model system. CG waters are shown in bright blue color, while CG lipids are presented in gray color. The initial distance between the two membranes is 55 Å. NaCl is added as a neutralizer for the system and Ca2+ is added in a smaller concentration than NaCl. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In simulations, the periodic boundary conditions were applied in all three directions. The van der Waals interactions were calculated using LJ potential with a cutoff of 12 Å. Production simulations were carried out for ~450 ns using NPT (T = 300 K, P = 1 atm) ensembles. A time-step of 5 fs was chosen for simulation, which is similar to the all-atom simulation, but the total number of atoms is significantly reduced compared to all-atom simulations.
3. Results and discussion
To study the large-scale conformation change of the system under the influence of Ca2+, we created a model system, named Ca2+-bound, where two Ca2+ ions located in each calcium binding pocket of Slp4-a (Fig. 2c). For comparison purposes, we also created another model system without Ca2+ ions in these calcium binding pockets (called Ca2+-unbound). Three independent realizations were conducted for each model for statistical consistency. Since all three realizations show similar results, here we only show one realization for illustration. We have studied the molecular interactions between Slp4-a and lipid membranes in terms of radial distribution functions and the interaction potential energy (PE). Furthermore, we studied the membrane gap changes under different Ca2+ binding within Slp4-a.
3.1. Interaction between Slp4-a and lipids
3.1.1. Radial distribution function of lipids around calcium binding sites of Slp4-a
Radial distribution function (RDF), g(r), is the number of molecules or particles within a spherical region around a reference point, and it is used to quantify the relative abundance of lipids around a protein [36]. In this study, we calculated the RDF of phospholipid at different times around the center of mass of the following binding sites of Slp4-a :
-
•
C2B-top (Ca2+ binding site at the top side of C2B),
-
•
C2B-bottom (additional binding site at the bottom side of C2B),
-
•
C2A-bottom (Ca2+ binding site at the bottom side of C2A), and
-
•
C2A-top (additional binding region at the top side of C2A)
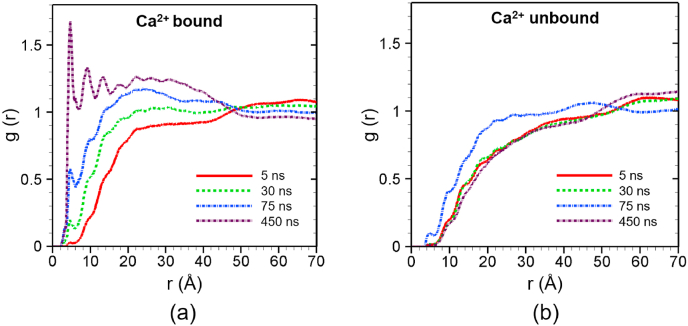
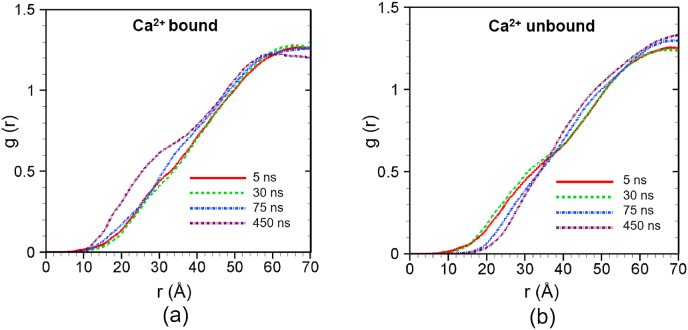
Fig. 4 shows the RDF of lipids around the C2B-top during the simulations. In the Ca2+-bound case of C2B-top, the first g(r) peak is gradually rising up from almost 0 at 5 ns to over 1.5 after 450 ns (Fig. 4a). The first peak is found at the scanning radius (r) of 4 Å, which is equivalent to the closest van der Waals distance between nonbonded particles in our simulations [30]. This result reveals that there are direct non-bonded contacts between lipid and the C2B-top site, and these non-bonded contacts enhanced with time as demonstrated by the rise of the first peak. The RDF data at 450 ns shows multiple peaks after the first one, and these subsequent peaks indicate that more amino acids of the C2B-top site are getting in contact with lipids. In contrast, in the absence of Ca2+, C2B-top does not demonstrate any interaction with lipids within 4 Å of the protein site as shown in Fig. 4b. These results clearly demonstrate the roles of Ca2+ in the interactions between C2B top residues and lipids. In the former case (Fig. 4a), Ca2+ is able to strengthen the interaction between the protein site and lipids, and thus supporting the fact that amino acids can dive deeper into lipid membranes.
Fig. 4.
Radial distribution function (g(r)) of lipid around the Ca2+ binding site of C2B (C2B top) at different times for (a) Ca2+-bound and (b) Ca2+ -unbound scenarios. The profiles are obtained by averaging over 5 ns around the specific time frames.
The RDF is also calculated for lipids around the C2B-bottom, as shown in Fig. 5, for both Ca2+ - bound and unbound cases. Our simulation results show that there is no peak for the g(r) at the C2B-bottom side even though there are calcium ions on its binding sites. The g(r) value is 0 within the 10 Å scanning radius around the C2B-bottom site (Fig. 5a) indicating that there is no direct contact between lipids (endothelial membrane) and the C2B-bottom. Thus, Ca2+ binding at the C2B-top does not support the ability of the C2B-bottom domain in binding with surrounding lipids. Similarly, at the corresponding Ca2+-unbound case, the radial distribution function (Fig. 5b) does not show the existence of lipids within the 10 Å scanning radius. This similarity with its corresponding Ca2+-bound case implies no direct contact formed between the protein site and endothelial cell membrane lipids. These results reveal that Ca2+ binding could initiate a strong contact between C2B-top and vesicular lipids either through C2B-top penetration to lipid membrane or via drop-down of membrane surface for protein binding, but they do not support movement of C2B-bottom towards the endothelial membrane.
Fig. 5.
Radial distribution function of lipid around the additional binding site of C2B (C2B bottom) at different times for (a) Ca2+-bound and (b) Ca2+ -unbound scenarios. The profiles are obtained by averaging over 5 ns around the specific time frames.
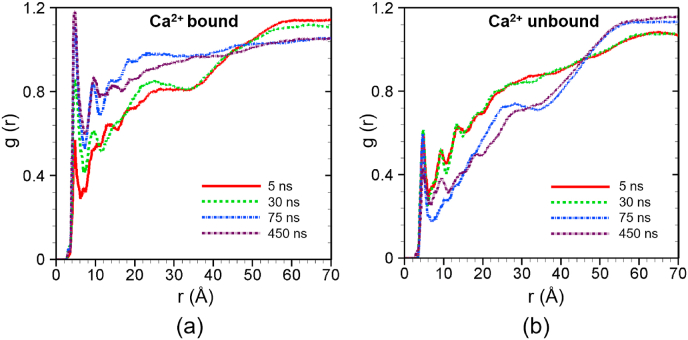
Next, we study the interactions for the C2A domain. Fig. 6a presents the radial distribution function of lipids around the C2A-bottom site for Ca2+-bound case. In this case, the first peak is evident within a scanning radius of 4 Å for all simulations. The magnitude of the first peak is increasing with time indicating that the C2A bottom is penetrating deep into the endothelial membrane. This is due to the fact that calcium binding at the corresponding C2A binding site can make it more positive, which enhances the electrostatic attraction between the C2A bottom site and endothelial cell lipids.
Fig. 6.
Radial distribution function of lipid around the Ca2+ binding site of C2A (C2A bottom) at different times for (a) Ca2+-bound and (b) Ca2+ -unbound scenarios. The profiles are obtained by averaging over 5 ns around the specific time frames.
Unlike the C2B, for the Ca2+-unbound case, there is still significant interaction between C2A-bottom and surrounding lipids from the endothelial cell as shown in Fig. 6b. Here the first peak appears within the 4 Å for all cases. However, the magnitude of the peak does not change with time and the peak value is much lower than that of Ca2+-bound case. This might be due to the presence of positive residues such as Arg 392, Arg 451, Arg 454, etc. around the calcium binding site in the C2A-bottom.
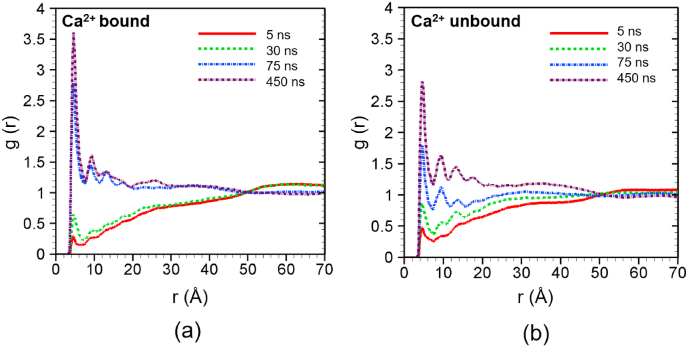
Fig. 7 interestingly shows high interaction between C2A-top and surrounding lipids in both Ca2+-bound (Fig. 7a) and Ca2+-unbound (Fig. 7b) cases as demonstrated by the existence of the first peak within 4 Å scanning radius. In Fig. 7a, the magnitude of the first peak rises from 0.25 at 5 ns to 3.6 at 450 ns indicating that the lipid membrane and the C2A top site are coming closer. In the absence of any Ca2+ binding at C2A-top, the enhanced penetration of the C2A into vesicular membrane might be facilitated by aromatic (hydrophobic) amino acids such as Leu 433, Leu 436, Ala 438, Leu 470, etc. at the C2A-top. These amino acids are well known for their hydrophobic property that causes them to hide into lipid bilayer to avoid hydration environment [20]. Similar lipid contacts are also found in the Ca2+-unbound case at the C2A-top as evident from the first peak of g(r) (Fig. 7b). Here the magnitude of the first peak increases from 0.5 at 5 ns to 2.8 at 450 ns. The reason for this interaction is the same as in the Ca2+-bound case; it is primarily caused by the aromatic residues at the top. However, the strength of the interaction is weaker for the Ca2+-unbound case indicating that calcium ions have some effects even on the distal site. Although calcium ions bind to the corresponding binding sites at the bottom part of the C2A, these calcium bindings could stabilize the (perpendicular) orientation of C2A with respect to both membranes. This might support the interactions of aromatic residues at the C2A-top with vesicular membrane for Ca2+-bound case. On the other hand, the Ca2+-unbound C2A domain could freely move or rotate and the C2A-top can tip out of facing-lipid position, which consequently weakens its lipid binding.
Fig. 7.
Radial distribution function of lipid around the additional binding site of C2A (C2A top) at different times for (a) Ca2+-bound and (b) Ca2+ -unbound scenarios. The profiles are obtained by averaging over 5 ns around the specific time frames.
It is also interesting to note that the lipid interaction behavior is different at C2A (top) and C2B (bottom) sites. This is due to the presence of flexible loops as well as multiple hydrophobic amino acids at C2A-top, which approach and penetrate into lipid membrane. On the other hand, C2B-bottom consists of more rigid helices and it has much fewer hydrophobic amino acids compared to C2A-top site. These results are consistent with the reported mechanism that the hydrophobic amino acids at the additional binding sites (C2A-top) optimally dive into the lipid bilayers [37,38].
In summary, each domain of C2A and C2B induces different RDFs from their different interactions with lipids, however, all results indicate that Ca2+-activated Slp4-a is likely to initiate fusion actions in BBB exocytosis by enhancing membrane connection. This is similar to what have been found in Ca2+-activated Syt1 [19,20]. Moreover, the results showed that the C2A domain is more effective in binding with lipids than the C2B domain, even in a Ca2+-unbound case. This supports the conclusion from the experiment of Lyakhova et al. where Slp4-a is found strongly binds with PIP2 lipids in a Ca2+ independent manner mainly with the C2A domain [17]. However, in a weakly charged lipids such as POPE, as used in our work, Slp4-a connects with lipids strongly in a Ca2+ dependent manner, especially at the C2A-bottom and C2B-top. Besides, Wang et al. [39] proposed a multiple binding site mechanism for synaptic Syt1 where the proteins can bind with one or many types of lipids in a membrane through different regions of functional amino acids. Similar to Syt1 in the synaptic system, it is possible that Slp4-a protein could concurrently bind with high-affinity lipids (such as PIP2) through its strongly charged or aromatic amino acid groups, in one location and weakly charged lipids (such as POPE) in other locations of a target membrane through calcium binding sites to trigger membrane fusion.
3.2. Potential energy (PE) between Slp4-a and lipids
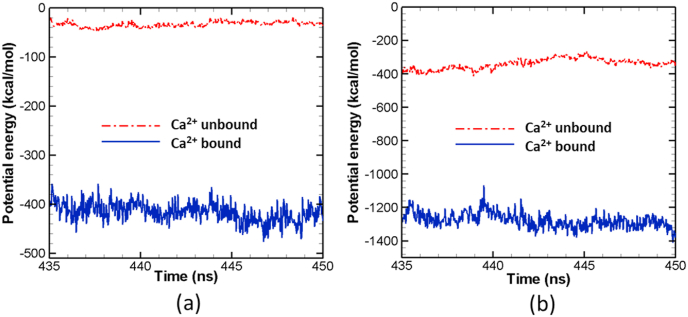
To further understand the interaction between Slp4-a and the lipid membranes, we measure the potential energy of all charged amino acids of each Ca2+ binding sites with lipid molecules. The energy is the sum of van der Waals and electronic potential energy between charged carbon atoms and the surrounding charged phosphate head in lipid molecules (PO4). For C2B top calcium binding sites we considered Asp 548, Asp 603, Asp 611, Glu 605, Arg 604, and Lys 542, while for C2A bottom calcium ion binding site Asp 387, Glu 388, Arg 392, Arg 451, Arg 454, Lys 390, and Lys 391 are used. Fig. 8 shows the potential energy for both Ca2+ bound and unbound cases. The potential energy is stronger for Ca2+-bound case compare to the unbound one. In the C2B domain, once calcium binds at their respective binding site it turns more positive, which initiates stronger interaction for calcium binding cases. On the other hand, in the absence of the calcium ions, there are still positively charged residues such as Arginine 604 and Lysine 542 at the C2B top, which provides some interaction with surrounding lipids, albeit at much lower strength due to the mild positive charge of those amino acids. In other words, the interaction between the C2B domain and the surrounding lipids strongly depends on the calcium ion binding.
Fig. 8.
Potential Energy (PE) between calcium binding sites of Slp4-a and surrounding lipids for (a) C2B and (b) C2A domain.
Similar calcium ion dependent interaction results are observed for the C2A domain as shown in Fig. 8b. For the C2A bottom domain, the potential energy is much stronger in the Ca2+-bound case compare to the Ca2+-unbound case. Similar to C2B, once Ca2+ ion is occupied in the binding pocket of C2A, it strongly enhances the positive charge of the pocket and thus facilitates a stronger binding between the pocket and lipids. For the Ca2+-bound case, the magnitude of interaction is much higher for the C2A (bot) domain compare to C2B (top) even though their radial distribution functions (Fig. 4, Fig. 6b) are very similar. This might be due to the structure of these calcium binding pockets. For instance, there are many positively charged amino acids such as Lys 390, Lys 391, Arg 392 in C2A (bot) calcium ion binding site. On the other hand, there are only two positively charged amino acids (Lys 542 and Arg 604) in the corresponding binding site of C2B. Moreover, it is important to note that the radial distribution function in C2B (top) is influenced by more hydrophobic residues such as Alanine 540, Alanine 541, Valine 601, and Tryptophan 602 at lipid contact, and these residues are not counted in the potential energy. Nevertheless, the interaction between bound Ca2+ ions and lipids constrains C2A as well as C2B domain at or around perpendicular orientation with respect to lipid membranes and thus facilitates more interaction between Slp4-a and lipids.
Fig. 8b shows that, for Ca2+-unbound cases, the potential energy is also higher for C2A (bot) with respect to the C2B (top) domain. This behavior can be explained with radial distribution function results of lipid around calcium binding pockets of C2B and C2A in Ca2+-unbound cases as shown in Fig. 4, Fig. 6b, respectively. Due to the presence of a large number of positively charged residues in C2A (bot), it is more interactive with lipids than C2B in a calcium independent manner.
We also observed some differences in interaction behavior between synaptotagmin (Syt1) and synaptotagmin like protein (Slp4-a). For calcium binding sites in Slp4-a, we placed two Ca2+ ions in each calcium binding pockets of the Slp4-a, but only one Ca2+ ion stably remains at each site. The corresponding number in the Syt1 case is 2 for each C2 domain [19]. Indeed, the less aspartic (Asp) acids (negatively charged residues) in the binding pocket, the weaker calcium holding. For instance, five aspartic acids in each binding pocket are responsible to hold Ca2+ in Syt1 [19] compare to only three aspartic acids in each Ca2+ binding pocket of Slp4-a [18]. Our modeling results confirm that the Ca2+ dependent binding ability of Slp4-a is, nonetheless, weaker than Syt1.
3.3. Membrane responses
Several studies suggest that the interaction of synaptotagmin with lipid membrane can cause bending of the membrane, which might initiate the vesicle-cell fusion [20,40]. The detailed study of the fusion process is beyond the current computational capability of molecular simulations due to the large time scale. Thus, we here examined the changes of lipid-gap due to the presence of synaptotagmin like protein (Slp4-a). The distance between two lipid (vesicle and endothelial) membranes were measured with (as well as without) calcium ion binding at the respective C2A and C2B domains of the Slp4-a.
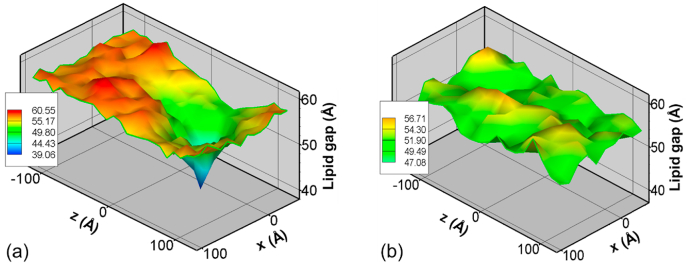
Fig. 9 shows the lipid gap for Ca2+-bound and unbound cases due to the interaction of Slp4-a at 450 ns.The lipid gap (XZ plane) profile is calculated by measuring the distance between the bottom layer of PO4 lipid head of the vesicular membrane and the top layer of PO4 of the endothelial membrane. The distance is measured at each split cubic bin with a bin size of 5 Å. For Ca2+-bound case, as shown in Fig. 9a, there is a clear drop in the gap between lipid membranes at Slp4-a locations indicating that the binding of Ca2+ in C2 domains is responsible for this change. In addition to the Ca2+ binding, the additional binding regions of each C2 domain can penetrate to lipid membranes and locally disturb the lipids’ order at the contacts. This result is similar to previous molecular dynamics study in a synaptic system where Sty1 can bend the lipid membranes in Ca2+-bound case. As the membrane gap decreases due to the bending of the membrane, initiated by the Slp4-a, the lipid membranes can be pulled by SNAREs to form the membrane fusion [20] (Fig. 9a).
Fig. 9.
Slp4-a induced lipid membrane gap (between the vesicular membrane and endothelial cell membrane) changes in a BBB system for (a) Ca2+-bound and (b) Ca2+-unbound cases at 450 ns.
To demonstrate the crucial role of Ca2+, the lipid gap data from the corresponding Ca2+-unbound case is shown in Fig. 9b. Unlike the earlier case, no noticeable drop in the membrane gap is observed for the Ca2+-unbound case except for some thermal fluctuations. Our simulation results show that even though the C2A domains itself could actively interact with lipid membranes (at the aromatic amino acid group - C2A top), there is no significant change in membrane gap without support from Ca2+ ions. With Ca2+ binding, the lipid gap around Slp4-a drops nearly 30% after 450 ns compared with the Ca2+-unbound case. Our results prove the previous hypothesis (McMahon et al., 2010 [40], Wu and Schulten. 2014 [20] and Rizo 2018 [8]) that the C2 domains insert into the target membrane in a Ca2+-dependent manner, resulting in membrane buckling. The lipid disordering regions can potentially stimulate membrane fusion.
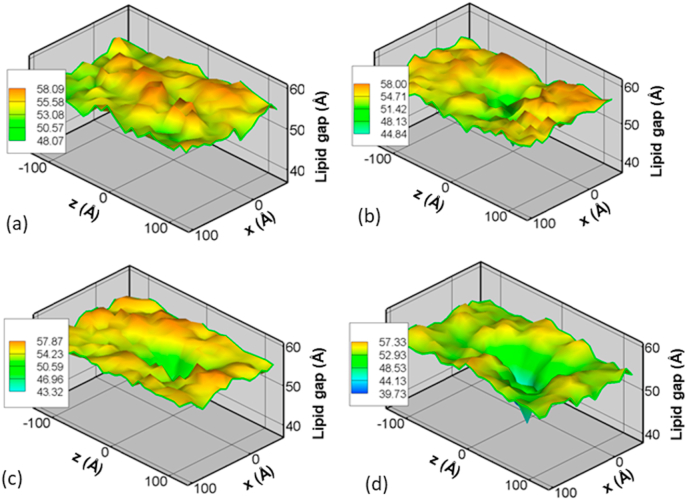
Until now all results are presented under the assumption that the calcium binding site of C2B domain of Slp4-a is located very close to the vesicular membrane, while the calcium-binding site of the C2A domain of the same protein is in close proximity of the endothelial membrane as shown in Fig. 2 (b&c). However, based on the observed orientation of synaptotagmin (Syt1) in the synaptic system [8], it is likely to have other orientations for the Slp4-a during the pre-fusion stage. To investigate the effect of initial Slp4-a configurations, we have studied the membrane dynamics by slightly rotating the Slp4-a from its original orientation (Configuration 1) shown in Fig. 2. In the new configuration, the additional binding sites, located on the opposite side of the calcium-binding site, of the both C2B and C2A domains are in close proximity to membranes. In other words, the additional binding site of the C2A is located next to the top (vesicular) membrane and the additional binding site of the C2B is very close to the bottom (endothelial) membrane in configuration 2. In this new configuration, the two calcium binding sites of Slp4-a are located further away from lipid membranes compared with Configuration 1. We are particularly interested in the role of calcium ion in changing the membrane gap since the calcium binding site is relatively away from the membrane. As shown in Fig. 10, no significant changes in the distance between the vesicular membrane and endothelial cell membrane are observed within the first 30 ns. However, the membrane gap starts dropping afterward. At the 450 ns, the membrane gap approaches value similar to the ones obtained for Configuration 1 (see Fig. 9a). Both configurations 1 and 2 show a 30% drop in the membrane gap in the Ca2+-bound case in less than 0.5 μs at Slp4-a location. However, the location of the membrane gap change shifts slightly due to the shift of the binding site locations. Nevertheless, the calcium ions are responsible for the decrease in membrane gap for any arbitrary initial configuration of Slp4-a, which could trigger the membrane pre-fusion for exocytosis of vesicles.
Fig. 10.
Effect of Ca2+ in Slp4-a induced membrane gap (between the vesicular membrane and endothelial cell membrane) changes for Configuration 2 in a BBB system at (a) 5 ns, (b) 30 ns, (c) 75 ns and (d) 450 ns.
4. Conclusions
We have conducted MD simulations to investigate the behavior of Slp4-a protein during the vesicle-endothelial pre-fusion stage to further understand the exocytosis of vesicle from endothelial cells in BBB. We have developed a model analogous to the synaptic exocytosis using existing proteins in endothelial cells. We measured the radial distribution function of lipids around binding sites of Slp4-a and found that Ca2+ plays a significant role in the interactions between Slp4-a and lipids. In the Ca2+-bound case, Ca2+ is able to strengthen the interaction between Slp4-a and lipids by enhancing the electrostatic attraction between them. Our numerical results also indicate that the hydrophobic amino acids at the additional binding sites of C2A could dive into the lipid bilayers. These interactions through additional binding sites confirm that the C2A domain could strongly interact with lipids even in the absence of calcium. Moreover, based on recent experimental observations and our molecular simulations, it can be concluded that Slp4-a protein could concurrently bind with high-affinity lipids (such as PIP2) through its strongly charged or aromatic amino acid groups, in one location and weakly charged lipids (such as POPE in our work) in other locations of a target membrane through calcium binding sites to trigger membrane fusion.
We also measured the potential energy between calcium binding sites of Slp4-a and lipids of the neighboring membrane. Our numerical results show that the interaction between these protein sites and the surrounding lipids strongly depends on the calcium ion binding. However, the potential energy magnitude is different for the C2A and C2B domains. Due to the presence of a large number of positively charged residues in the calcium binding site of C2A, it is more interactive with lipids than that of C2B.
Finally, we measured the change in the membrane gap between the vesicular membrane and endothelial membrane to find the influence of Slp4-a as well as calcium ions. We observed a significant drop in the gap between lipid membranes at Slp4-a locations for Ca2+-bound case, but not much change in membrane gap without the support from Ca2+ ions. Changes in the membrane gap are also explored for other configurations of the Slp4-a, where the protein is rotated slightly to provide a gap between the calcium ion binding site and the neighboring lipids. The new configuration also shows significant changes in the membrane gap in Ca2+ bound case. In other words, calcium ions are responsible for the decrease in membrane gap for any arbitrary initial configuration of Slp4-a, which could trigger the membrane pre-fusion for exocytosis of vesicles.
CRediT authorship contribution statement
Quyen Van Dinh: Methodology, Coarse-grained molecular dynamics, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Jin Liu: Writing - review & editing. Prashanta Dutta: Conceptualization, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM122081. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Computational resources were provided in part by the Extreme Science and Engineering Discovery Environment (XSEDE) under grant No. MCB170012.
References
- 1.Abbott N.J., Patabendige A.A.K., Dolman D.E.M., Yusof S.R., Begley D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010;37:13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 2.Pardridge W.M. Molecular Trojan horses for blood-brain barrier drug delivery. Curr. Opin. Pharmacol. 2006;6:494–500. doi: 10.1016/j.coph.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Khan A.I., Liu J., Dutta P. Iron transport kinetics through blood-brain barrier endothelial cells. Biochemica Et Biophysica Acta-Gen. Subj. 2018;1862:1168–1179. doi: 10.1016/j.bbagen.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng H., Dutta P., Liu J. Stochastic simulations of nanoparticle internalization through transferrin receptor dependent clathrin-mediated endocytosis. Biochemica Et Biophysica Acta-Gen. Subj. 2018;1862:2104–2111. doi: 10.1016/j.bbagen.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan A.I., Lu Q., Du D., Lin Y.H., Dutta P. Quantification of kinetic rate constants for transcytosis of polymeric nanoparticle through blood-brain barrier. Biochemica Et Biophysica Acta-Gen. Subj. 2018;1862:2779–2787. doi: 10.1016/j.bbagen.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan A.I., Liu J., Dutta P. Bayesian inference for parameter estimation in lactoferrin-mediated iron transport across blood-brain barrier. Biochemica Et Biophysica Acta-Gen. Subj. 2020;1864:129459. doi: 10.1016/j.bbagen.2019.129459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreutzberger A.J.B., Kiessling V., Liang B.Y., Seelheim P., Jakhanwal S., Jahn R., Castle J.D., Tamm L.K. Reconstitution of calcium-mediated exocytosis of dense-core vesicles. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1603208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizo J. Mechanism of neurotransmitter release coming into focus. Protein Sci. 2018;27:1364–1391. doi: 10.1002/pro.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mourik M., Eikenboom J. Lifecycle of Weibel-Palade bodies. Hämostaseologie. 2017;37:13–24. doi: 10.5482/HAMO-16-07-0021. [DOI] [PubMed] [Google Scholar]
- 10.Schillemans M., Karampini E., Kat M., Bierings R. Exocytosis of Weibel-Palade bodies: how to unpack a vascular emergency kit. J. Thromb. Haemostasis. 2019;17:6–18. doi: 10.1111/jth.14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda M., Imai A., Nashida T., Shimomura H. Slp4-a/granuphilin-a interacts with syntaxin-2/3 in a Munc18-2-dependent manner. J. Biol. Chem. 2005;280:39175–39184. doi: 10.1074/jbc.M505759200. [DOI] [PubMed] [Google Scholar]
- 12.van Breevoort D., Snijders A.P., Hellen N., Weckhuysen S., van Hooren K., Eikenboom J., Valentijn K., Fernandez-Borja M., Ceulemans B., De Jonghe P., Voorberg J., Hannah M., Carter T., Bierings R. STXBP1 promotes Weibel-Palade body exocytosis through its interaction with the Rab27A effector Slp4-a. Blood. 2014;123:3185–3194. doi: 10.1182/blood-2013-10-535831. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Q.J., Zhou P., Wang A.L., Wu D., Zhao M.L., Sudhof T.C., Brunger A.T. The primed SNARE-complexin-synaptotagmin complex for neuronal exocytosis. Nature. 2017;548:420–+. doi: 10.1038/nature23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharda A., Flaumenhaft R. 2018. The Life Cycle of Platelet Granules. F1000Research 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi U.B., Strop P., Vrljic M., Chu S., Brunger A.T., Weninger K.R. Single-molecule FRET-derived model of the synaptotagmin 1-SNARE fusion complex. Nat. Struct. Biol. 2010;17:318–U384. doi: 10.1038/nsmb.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang H., Cafiso D.S. Conformation and membrane position of the region linking the two C2 domains in synaptotagmin 1 by site-directed spin labeling. Biochemistry. 2008;47:12380–12388. doi: 10.1021/bi801470m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyakhova T.A., Knight J.D. The C2 domains of granuphilin are high-affinity sensors for plasma membrane lipids. Chem. Phys. Lipids. 2014;182:29–37. doi: 10.1016/j.chemphyslip.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Takeuchi T., Yokota H., Izumi T. Novel rabphilin-3-like protein associates with insulin-containing granules in pancreatic beta cells. J. Biol. Chem. 1999;274:28542–28548. doi: 10.1074/jbc.274.40.28542. [DOI] [PubMed] [Google Scholar]
- 19.Bykhovskaia M. Calcium binding promotes conformational flexibility of the neuronal Ca2+ sensor synaptotagmin. Biophys. J. 2015;108:2507–2520. doi: 10.1016/j.bpj.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z., Schulten K. Synaptotagmin's role in neurotransmitter release likely involves Ca2+-induced conformational transition. Biophys. J. 2014;107:1156–1166. doi: 10.1016/j.bpj.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shamloo A., Pedram M.Z., Heidari H., Alasty A. Computing the blood brain barrier (BBB) diffusion coefficient: a molecular dynamics approach. J. Magn. Magn Mater. 2016;410:187–197. [Google Scholar]
- 22.Miranda A., Cova T., Sousa J., Vitorino C., Pais A. Computational modeling in glioblastoma: from the prediction of blood-brain barrier permeability to the simulation of tumor behavior. Future Med. Chem. 2018;10:121–131. doi: 10.4155/fmc-2017-0128. [DOI] [PubMed] [Google Scholar]
- 23.Jewel Y., Liu J., Dutta P. Coarse-grained simulations of conformational changes in the multidrug efflux transporter AcrB. Mol. Biosyst. 2017;13:2006–2014. doi: 10.1039/c7mb00276a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han W., Schulten K. Further optimization of a hybrid united-atom and coarse-grained force field for folding simulations: improved backbone hydration and interactions between charged side chains. J. Chem. Theor. Comput. 2012;8:4413–4424. doi: 10.1021/ct300696c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jewel Y., Van Dinh Q., Liu J., Dutta P. Substrate dependent transport mechanism in AcrB of multidrug resistance bacteria. Proteins. 2020;88:853–864. doi: 10.1002/prot.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marrink S.J., De Vries A.H., Mark A.E. Coarse grained model for semiquantitative lipid simulations. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
- 27.Marrink S.J., Risselada H.J., Yefimov S., Tieleman D.P., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 28.Han W., Wu Y.D. Coarse-grained protein model coupled with a coarse-grained water model: molecular dynamics study of polyalanine-based peptides. J. Chem. Theor. Comput. 2007;3:2146–2161. doi: 10.1021/ct700151x. [DOI] [PubMed] [Google Scholar]
- 29.Han W., Wan C.K., Jiang F., Wu Y.D. PACE Force field for protein simulations. 1. Full parameterization of version 1 and verification. J. Chem. Theor. Comput. 2010;6:3373–3389. doi: 10.1021/ct1003127. [DOI] [PubMed] [Google Scholar]
- 30.Wan C.K., Han W., Wu Y.D. Parameterization of PACE force field for membrane environment and simulation of helical peptides and helix-helix association. J. Chem. Theor. Comput. 2012;8:300–313. doi: 10.1021/ct2004275. [DOI] [PubMed] [Google Scholar]
- 31.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 33.Jo S., Kim T., Iyer V.G., Im W. Software news and updates - CHARNIM-GUI: a web-based grraphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 34.Qi Y.F., Cheng X., Han W., Jo S., Schulten K., Im W. CHARMM-GUI PACE CG builder for solution, micelle, and bilayer coarse-grained simulations. J. Chem. Inf. Model. 2014;54:1003–1009. doi: 10.1021/ci500007n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jo S., Lim J.B., Klauda J.B., Im W. CHARMM-GUI Membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma S., Kim B.N., Stansfeld P.J., Sansom M.S.P., Lindau M. A coarse grained model for a lipid membrane with physiological composition and leaflet asymmetry. PloS One. 2015;10 doi: 10.1371/journal.pone.0144814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang J., Adamian L., Jackups R. The membrane-water interface region of membrane proteins: structural bias and the anti-snorkeling effect. Trends Biochem. Sci. 2005;30:355–357. doi: 10.1016/j.tibs.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Shityakov S., Roewer N., Broscheit J.-A., Förster C. In silico models for nanotoxicity evaluation and prediction at the blood-brain barrier level: a mini-review. Computational Toxicology. 2017;2:20–27. [Google Scholar]
- 39.Wang S., Li Y., Ma C. Synaptotagmin-1 C2B domain interacts simultaneously with SNAREs and membranes to promote membrane fusion. Elife. 2016;5 doi: 10.7554/eLife.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMahon H.T., Kozlov M.M., Martens S. Membrane curvature in synaptic vesicle fusion and beyond. Cell. 2010;140:601–605. doi: 10.1016/j.cell.2010.02.017. [DOI] [PubMed] [Google Scholar]