Abstract
To tackle the problem of antimicrobial resistance (AMR) surveillance programmes are in place within Europe applying phenotypic methods, but there are plans for implementing whole genome sequencing (WGS). We tested the benefits of WGS using Escherichia coli collected from pig surveillance performed between 2013 to 2017. WGS was performed on 498 E. coli producing ESBL and AmpC enzymes, recovered from pig caeca on MacConkey + cefotaxime (McC + CTX) agar, as recommended by the European Commission, or ESBL agar, used additionally by United Kingdom. Our results indicated WGS was extremely useful for monitoring trends for specific ESBL genes, as well as a plethora of AMR genotypes, helping to establish their prevalence and co-linkage to certain plasmids. Recovery of isolates with multi-drug resistance (MDR) genotypes was lower from McC + CTX than ESBL agar. The most widespread ESBL genes belonged to the blaCTX-M family. blaCTX-M-1 dominated all years, and was common in two highly stable IncI1 MDR plasmids harbouring (blaCTX-M-1,sul2, tetA) or (blaCTX-M-1, aadA5, sul2, dfrA17), in isolates which were phylogenetically dissimilar, suggesting plasmid transmission. Therefore, WGS provided a wealth of data on prevalence of AMR genotypes and plasmid persistence absent from phenotypic data and, also, demonstrated the importance of culture media for detecting ESBL E. coli.
Subject terms: Computational biology and bioinformatics, Microbiology, Molecular biology, Diseases, Pathogenesis
Introduction
Increasing levels of antimicrobial resistance (AMR) is a global concern to both human and animal health. The O’Neill report estimated 10 million lives will be at risk per year due to resistant infections by 2050 if policies to stop the spread of AMR are not implemented1. The One Health concept requires a holistic approach to looking at AMR in humans, animals and the environment. Therefore, gaining information on AMR isolates in indicator species, such as commensal E. coli from livestock, is invaluable to characterise the resistome and track any emerging trends through the food chain. As WGS has become a more widely utilised resource, EU Agencies such as EFSA and ECDC have proposed that it is used by EU member states in surveillance activities whether it be harmonised AMR surveillance2, or for outbreak investigations3.
The WHO has identified and ranked antimicrobials by those deemed critically important for human medicine4. This list includes third and higher generation cephalosporins, which are targeted by bacterial production of extended spectrum beta-lactamase (ESBL) and AmpC enzymes, and E. coli harbouring these genes are the subject of specific surveillance by EU members, which is monitored by EFSA. These antibiotics have been used in human medicine since the early 1980s with resistance to extended-spectrum cephalosporins (ESCs) first noted in Klebsiella pneumoniae and Serratia marcesens in 1983, rising rapidly in the intervening years5,6. One of the primary drivers behind this increase in prevalence of resistant isolates is the occurrence of promiscuous plasmids harbouring blaCTX-M genes, which are often co-located with other resistance genes, found in Enterobacteriaceae5. Plasmid types harbouring ESBL genes vary, but frequently include plasmids of incompatibility groups I1, and F (IncI1 and IncF). ESBL-producing E.coli have been isolated from multiple sources including healthy humans, livestock and companion animals, in addition to food-stuffs, and are the most common bacterial hosts of acquired blaCTX-M7–13. ESBL harbouring E. coli can belong to different STs with ST131 most commonly associated with infection in humans and which has rarely been identified in livestock; others such as ST10 and ST88 have been found in both humans and animals7,8,11,14.
The aim of this study was to perform molecular characterisation using WGS to determine the benefits of using this approach for surveillance. We expected to identify AMR genes and circulating plasmids in E. coli isolated on selective agars containing ESCs. These were collected from porcine caecal samples from healthy pigs from a UK nationwide pig study in 2013, and EU mandatory monitoring of healthy pigs at slaughter in the UK in 2015 and 2017; each caecal content was cultured on two different ESC containing agar plates. Although the isolates from 2013 had previously been analysed using phenotypic methods15, they had not been genotyped and so are included here for comparison with datasets from 2015 and 2017. A secondary aim of the study was to compare isolates from the EU monitoring agar, MacConkey + 1 mg/L cefotaxime with an additional ESBL agar, to determine any benefits from using an additional agar. AMR genotypes of isolates were characterised in detail through the APHA SeqFinder WGS pipeline that we have validated for use in AMR surveillance16. Selected plasmids carrying AMR genes, including MDR plasmids, were characterised by long read sequencing, and compared to those from NCBI and other studies to look at transmission and persistence.
Results
Antimicrobial resistance identified in E. coli isolates from EU and national surveillance
Genotypic profiles were established through WGS of E. coli producing ESBL or AmpC enzymes (Supplementary Table S1) isolated in 2015 and 2017 from EU surveillance on two agar plates, one specified by EFSA (McC + CTX) and an additional plate (ESBL) supplemented by UK for national surveillance activities in livestock, which are reported through VARSS17,18. Moreover, WGS of E. coli collected through national surveillance of pigs in 2013 selected on ESBL agars have also been included for investigation of trends over five years. From the McC + CTX isolates, overall there was a ~ 97% correlation between the phenotype (based on ECOFFs) and the resistance genotype, with the majority of antimicrobial classes showing excellent to good correlations (Supplementary Table S3). The discordance was primarily attributed to azithromycin over-reporting, because the presence of mphA does not always result in an azithromycin MIC above the cut-off19. In addition, the presence of a tet(A) variant that was initially not in our database resulted in non-concordance between the genotype and phenotype of five isolates. Once this gene was added there was a 100% correlation for tetracycline phenotypic and genotypic resistance, increasing the overall correlation to ~ 98%.
AMR genotypes in isolates recovered on McC + CTX agar from 2015 and 2017 EU surveillance
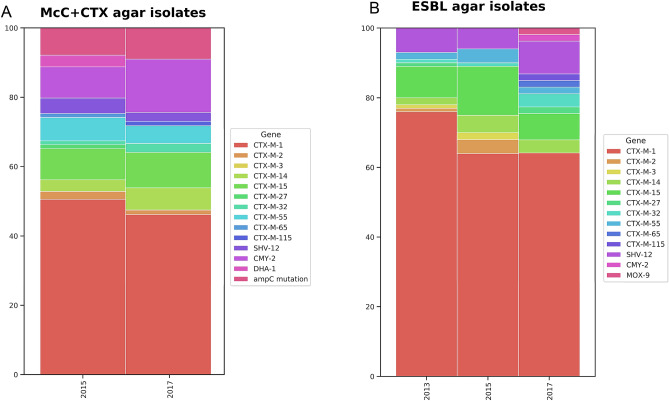
Eighty-nine isolates were recovered from 313 caecal samples on McC + CTX agar in 2015, and 75 from 347 caecal samples in 2017; as expected all isolates harboured resistance genes to ESCs, and to other antimicrobials, including the ten others in the EFSA panel for E. coli outlined in Decision 2013/652/EU. A greater proportion of isolates in 2015 were identified as ESBLs compared to those in 2017, as more allelic variants of blaCTX-M were present in 2015 (Fig. 1, Supplementary Table S2). blaCMY-2 and AmpC promoter mutations were found in isolates from both years from these plates and comprised 15/89 (17%) and 20/75 (27%) of the isolates recovered in 2015 and 2017 respectively; three isolates from 2015 contained blaDHA-1, the only time point this gene was identified in this study. Genotypic resistance of isolates to the non-beta lactam antimicrobials present in the EFSA panel (Supplementary Table S3), as a proportion of the total isolates examined, was lower in 2017, except for trimethoprim; but this reduction was not statistically significant. Genotypic resistance to the highest non-beta-lactam antimicrobials, sulfamethoxazole, tetracycline and trimethoprim, in both years was most often attributed to the presence of sul2, tet(A) and dfrA17. Genes conferring resistance to five antimicrobials not in the EFSA panel were present in both years, with spectinomycin/streptomycin most frequently detected, primarily due to presence of aadA variants, ant3-1a, strA and strB (Table S3). Genotypic resistance to the critically important antimicrobial fosfomycin, conferred by fosA3, was identified in one isolate from 2015, and two isolates from 2017. The most common resistance gene combination in 2015 from the McC + CTX agar was blaCTX-M-1, sul2 and tet(A) (4%), but in 2017 isolates AmpC promoter mutations were the most common (4%; Table 1).
Figure 1.
Differing proportions of AmpC/ESBL genes were present in isolates from McC + CTX (A) or ESBL (B) agar collected in 2013, 2015 and 2017.
Table 1.
Most common genes conferring genotypic resistance per set of isolates and the proportion of isolates with that resistance gene pattern.
| Year and agar type | Most common resistance gene allelic combination and proportion |
|---|---|
| 2013 ESBL | aadA5, blaCTX-M-1, sul2, dfrA17 (8%) |
| 2015 ESBL | blaCTX-M-1, sul2, tet(A) (7%) |
| 2015 McC + CTX | blaCTX-M-1, sul2, tet(A) (4%) |
| 2017 ESBL | ant3-1a, blaCTX-M-1, sat2A, sul2, tet(A) (8%) |
| 2017 McC + CTX | AmpC promoter mutation (4%) |
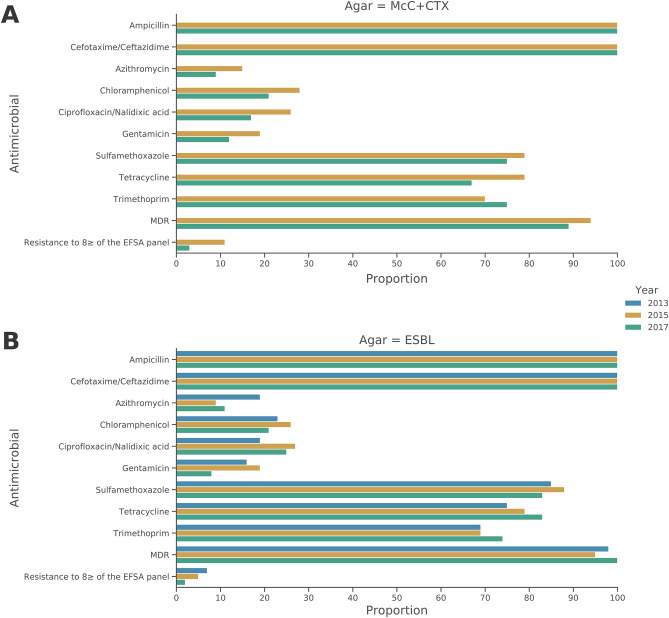
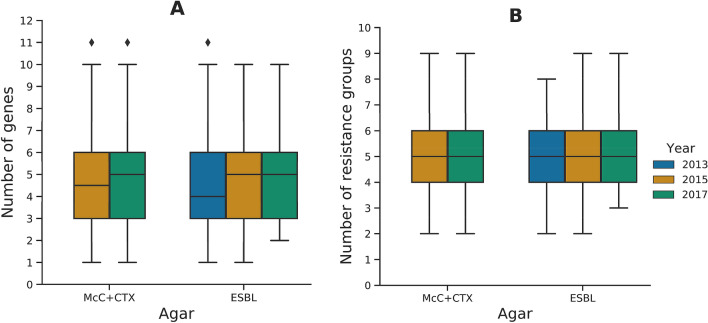
The proportion of isolates with genotypic MDR i.e. presence of resistance genes to three or more AMR classes, was slightly higher in isolates from 2015 (94%), than for isolates from 2017 (89%; Fig. 2A, Supplementary Table S3) and these figures were the same as those calculated from the MIC phenotypic data available (not shown). Significantly (P = < 0.05), the percentage of isolates that were resistant genotypically to eight or more of the EFSA panel were substantially reduced from 11 to 3%, in this period. The median number of resistance genes per isolate was five in 2017, compared to 4.5 in 2015 (Fig. 3), but the number of EFSA antimicrobial classes these genes conferred resistance to remained at five. Under EFSA MDR rules, ampicillin and cefotaxime/ceftazidime are treated as independent classes. However, because ESC genes confer resistance to both of these, an isolate harbouring a single ESC gene will count as having resistance to two classes therefore, sometimes elevating the median number of antimicrobial classes the isolate is resistant to beyond the number of resistance genes it harbours. blaCTX-M-1 was the most common ESC gene present in isolates from the 2015 and 2017 McC + CTX plate (Fig. 1A, Supplementary Table S2).
Figure 2.
Proportion of resistance to antimicrobials present in the EFSA E. coli monitoring panel in isolates originating from the MacConkey + cefotaxime agar in 2015 and 2017 (A) and ESBL agars in 2013 (Chromagar CTX, ESBL Brilliance), 2015 (ESBL Brilliance) and 2017 (Chromagar ESBL; B).
Figure 3.
Boxplot of the number of acquired resistance genes per isolate conferring genotypic resistance to antimicrobials in the EFSA panel for the different agar types and years (A), and number of resistance groups in the EFSA panel per isolate (B).
AMR genotypes in isolates recovered on ESBL agar from a 2013 national survey, and 2015, 2017 EU surveillance
More ESBL isolates were collected from the national survey (n = 185/637) in 2013, than in 2015 (n = 96/313) and 2017 (n = 53/347) from ESBL agar plates. Isolates were most likely to harbour blaCTX-M-1, which accounted for the highest proportion of ESBL genes detected in 2013 (76%, Fig. 1B, Supplementary Table S2). Overall, nine other blaCTX-M variants were recovered from ESBL agar; 14% of isolates harboured blaCTX-M-15 in 2015; the only other non-blaCTX-M variant ESBL gene identified was blaSHV-12 (Supplementary Table S2). Unexpectedly, blaCMY-2 and blaMOX-9 without an ESBL gene were present in one isolate each from 2017, indicating possible disruption of porins thus allowing the sample to grow on CHROMagar ESBL20. Mutations in the AmpC promoter region were identified in one isolate from each year; these isolates also harboured an ESBL gene. As with isolates recovered from McC + CTX agar, sul2, tet(A) and dfrA17 were the most frequently detected non-beta-lactamases. Azithromycin resistance was highest in the 2013 ESBL isolates, and there were also a high proportion of isolates with resistance to the non-EFSA antimicrobial streptomycin/spectinomycin, conferred by aadA variants, strA, strB and ant3-1a, just as for the McC + CTX isolates. Genotypic fosfomycin resistance, attributed to fosA3, was identified in three isolates from 2013 and an isolate from 2015 ESBL agar. The most common gene combination in 2013 was aadA5, blaCTX-M-1, sul2 and dfrA17, found in 8% of isolates. Both 2015 and 2017 isolates harboured blaCTX-M-1, sul2 and tet(A), but the most common genotype in 2017 also included ant3-1a and sat2A (Table 1). The median number of resistance genes in isolates from 2013 (n = 4) was slightly lower compared to 2015 and 2017 (n = 5); however the median number of resistances classes was five for all years. The reason for the number of genes being lower than the number of classes is due to ESBL and AmpC genes confer resistance to two EFSA classes: ampicillin and cefotaxime or ceftazidime. This is similar to the McC + CTX isolates where a level of gene redundancy was also observed (Fig. 3B).
The proportion of isolates demonstrating genotypic MDR in 2013 was 98%, which was higher than the 2015 ESBL isolates (94%) but lower than the 2017 ESBL isolates, where 100% of isolates were genotypically MDR. The 2013 and 2017 MDR proportions were higher than recovered from the McC + CTX agar, however, the 2015 ESBL and 2015 McC + CTX proportions were the same (Fig. 2B, Supplementary Table S3). Despite the fluctuation in genotypic MDR from 2013 to 2017, the ratio of isolates recovered from ESBL plates resistant to eight or more of the EFSA antimicrobial panel decreased from 7% in 2013 to 2% in 2017, however this was not a statistically significant reduction (Fig. 2B).
Characterisation of plasmid genomes
Phenotypic characterisation provides no detail on plasmids most likely responsible for AMR transmission, which is an advantage of WGS. To aid their characterisation, long and short read WGS data were used to circularise eight plasmids from six isolates. Isolate selection was based upon either incomplete short-read plasmid data, or the ESBL/AmpC gene they possessed, so the most complete information on the plasmid host background could be obtained. Of the 498 ESBL/AmpC isolates included in this study with short-read WGS data available, 67% of isolates did not contain the same genes as those present on the circularised ESBL/ampC gene-harbouring plasmids with > 99% sequence similarity, indicating the immense genetic diversity of these plasmids.
The blaCTX-M-1 genes were primarily identified on IncI1-type ST3 plasmids, which formed two groups, similar to pPE13096 or pESBL138 (Table 1). pESBL138 blaCTX-M-1 IncI1-ST3 plasmids contained sul2 and tetA, whereas pPE13096 blaCTX-M-1 was co-located with aadA5, sul2 and dfrA17 (Table 2; Supplementary Fig. S1a). The 108 kb pESBL138 was most similar to plasmids isolated from broilers in France in 2010–2012 (Supplementary Fig. S1b)21; and the 110 kb pPE13096, showed highest similarity to two plasmids of unknown French origin (GenBank accession: LT985235.1 and LT985286.1, Supplementary Fig. S1b). The two plasmid types were not related to a particular host E. coli ST. Using the resolved plasmid genomes as reference we noted 42 isolates to harbour > 99% average nucleotide identity to the genes of pPE13096, and 131 isolates to pESBL138, indicating likely presence of similar plasmids (Table 2).
Table 2.
Plasmids that were circularised from hybrid short and long read sequencing with their Incompatibility type, associated AMR genes, size and number of isolates that shared > 99% similarity with the circularised plasmids.
| Plasmid ID | Year host E. coli was isolated | Plasmid Inc-type | Associated AMR genes | Size (kb) | No. of isolates from all years with > 99% identity |
|---|---|---|---|---|---|
| pPE13096 | 2013 | I1 | aadA5, blaCTX-M-1, sul2, dfrA17 | 110 | 42 |
| pPO125 | 2015 | Y | blaCTX-M-15, dfrA14, qnrS1, strA, strB, sul2, blaTEM-1b, tetA | 98 | 5 |
| pPO189_X1 | 2015 | X1 | dfrA14, lnuF, sul3, blaTEM-1b, tetA | 33 | 3 |
| pPO189_F | F | dfrA8, blaTEM-1b, strA, strB, sul2 | 74 | 5 | |
| pESBL138 | 2015 | I1 | blaCTX-M-1, sul2, tetA | 108 | 131 |
| p3682_I1 | 2017 | I1 | blaCMY-2 | 91 | 1 |
| p3682_FQ | F/Q | ant3-Ia, catA1, dfrA1, mphB, blaTEM-1b, tetA, strA, strB, sul2, sul3 | 169 | 1 | |
| pPO116 | 2017 | X3 | blaSHV-12, qnrS1 | 42 | 11 |
p3682_I1, a 91 kb IncI1 plasmid that harboured blaCMY-2 was only present in isolate 3682 (Table 2). The backbone of this plasmid was most similar to a plasmid isolated from a bovine Salmonella in USA which lacked blaCMY-2, and also from a Scottish canine E. coli with blaCMY-2 (Supplementary Fig. S2)22. A 169 kb MDR plasmid, p3682_FQ that contained both IncF and IncQ replicons was also identified in isolate 3682 (Table 2), indicating presence of a possible plasmid hybrid. Similar plasmids were not identified in NCBI, but it was comparable to other plasmids from ongoing APHA projects (data not shown), suggesting wider distribution.
The blaCTX-M-15 plasmid sequenced was pPO125 with an IncY replicon and multiple resistance genes (Table 2). This plasmid showed some similarity to two plasmids from the USA, including one that was isolated from a hospital plumbing system (Supplementary Fig. S3)23; and only showed sequence similarity with five other isolates in our collection. pP0116, a 42 kb IncX3 plasmid that contained blaSHV-12 and qnrS1, matched to 11 isolates from our study and showed high sequence similarity to other IncX3 plasmids from a human urinary-tract infection and chicken faeces isolated in the Netherlands in 2009 and 2014, respectively (Supplementary Fig. S4)24.
Hybrid assembly of isolate PO189 containing blaCTX-M-55, qnrS1 and aac3-IVa indicated that these genes had integrated into the same chromosomal region and the chromosomal location of blaCTX-M-55 and qnrS1 was identified in ten isolates. Although isolate PO189 did not harbour an ESBL gene on a plasmid, it harboured two MDR plasmids (pPO189_X1 and pPO189_F), which were further identified in three and five isolates, respectively (Table 2). Both plasmids contained genes that conferred resistance to trimethoprim, sulphonamide and ampicillin, showing a level of redundancy in the host (Table 2).
Diversity of E. coli genotypes
Another advantage of WGS is the ability to characterise the host genome harbouring AMR genes. In silico MLST performed on the isolates assigned 114 STs, in addition to 20 STs that were novel, indicating much diversity in the host. The most abundant STs were ST88 (n = 38), ST10 (n = 35) and ST101 (n = 35), but the dominant ST for each year fluctuated. Two isolates from 2017 were assigned to ST131, an E. coli ST associated with human infections. However, one isolate contained blaCTX-M-1 and the other contained blaCTX-M-27, rather than blaCTX-M-15 typically associated with the epidemic clone of ST131 in humans. Although the blaCTX-M-1 isolate harboured the fimH22 allele, the blaCTX-M-27 isolate did harbour the fimH30 allele, fluroquinolone resistance and the prophage region associated with the C1-M27 subclade associated with human pathogenic ST131 strains25.
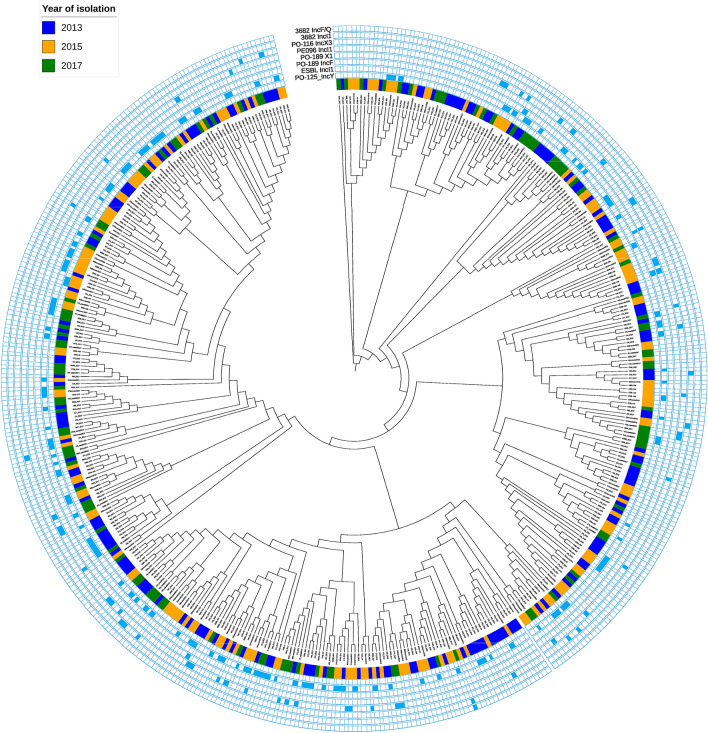
To establish any links between isolates from different years, a maximum-likelihood phylogenetic tree of the core-genome SNPs was constructed. The tree reiterated that the isolates recovered from each year were not clonal and did not cluster by year (Fig. 4). The isolates with > 99% match to the circularised plasmids were also mapped to the tree to determine their distribution. Isolates that contained either IncI1 blaCTX-M-1 plasmid types (pPE13096 or pESBL138), which was the most common plasmid type, were interspersed throughout the tree, indicating these plasmids were not restricted to a narrow E. coli host range.
Figure 4.
Phylogenetic tee of E. coli isolates from 2013, 2015 and 2017. The inner circle colours dictate the year the E. coli were isolated and the other rings show the presence of plasmids with high similarity to those listed in Table 2.
Discussion
The monitoring of isolates by WGS provides a wealth of data, as we have shown previously16, and in this paper there was very good correlation between resistance genotypes and resistance phenotypes determined by MICs and applying ECOFFs, which improved further on addition of a gene variant to our APHA SeqFinder AMR database. We believe this criterion applies widely, and demonstrates the need for continual assessment, and iterative improvement of WGS pipelines. Furthermore, as demonstrated here and previously16, for a small number of antimicrobial classes, gene presence was not always associated with a phenotype. Nevertheless, WGS is extremely useful for monitoring the trends of specific resistances and their underlying mechanism, in addition to tracking the host E. coli and their resistance plasmids, and therefore is a great improvement on phenotyping methods currently used for reporting AMR trends across Europe by EU agencies. The use of hybrid sequencing contributed further to this characterisation because it provided a greater chance of circularising plasmids more accurately than short-read data alone, identifying the diversity within these mobile-genetic elements, to help monitor their stability and propensity to transfer between different hosts. The reduction of sequencing costs, improved turn-around-time, objectivity and ease of sharing data beyond just AMR profiles has led to the use of WGS for monitoring of key organisms considered by EFSA, with optional sequencing and submission of results in 2021, and mandatory WGS monitoring by 2026. However, it can be important to continue MIC alongside the WGS, at least for a proportion of isolates, because it allows the training of AMR reference databases and inclusion of any gene omissions. Furthermore, discrepancies between the two methods can aid the discovery of new genes or novel AMR genes, and the development of more accurate rules for gene presence.
Although different commercial ESBL agars were used in this study, we believe any variation in selection of isolates between the ESBL agars should be less than between ESBL and McC + CTX agar. This is because McC + CTX allows for the growth of organisms that harbour AmpC, ESBL or carbapenemases, however the ESBL agars are intended to select for only ESBL producers and not AmpC producers. This was demonstrated by the genotypes of the respective isolates from each agar with the main differences between isolates recovered being the increased recovery of isolates from McC + CTX agar with blaCMY-2 or AmpC mutations instead of an ESBL genotype and a lower proportion of MDR genotypes. In contrast, isolates from ESBL agars mostly harboured an ESBL genotype and had a greater probability of genetic linkage with other AMR genes, and thus were MDR. These results highlight the influence of selective agars on the isolates cultured from the same sample and the diversity of resistance genes they harbour, and we therefore recommend the use of both for surveillance. A further benefit of including both agar types, is that a greater proportion of the bacterial population is likely to be sampled, as suggested by the phylogenetic tree, so important isolates such as the ESBL harbouring ST131 C1-M27 pathogen, which was recovered from the additional ESBL agar not mandated by the EU25, are not missed.
Genes belonging to the blaCTX-M family were the most common ESBL genes from both agar types (McC + CTX or commercial ESBL agar); blaSHV-12, was the only other ESBL gene present in a handful of isolates from each year. This concurs with the findings of a Dutch study of E. coli from livestock from 2007–2017, which found blaCTX-M genes were most common and blaSHV-12 was present at a lower frequency26. Unsurprisingly, blaCTX-M-1, commonly associated with livestock27, was dominant in all years of the study; it’s prevalence from a range of sources has been noted previously7,26. IncI1 replicon bearing plasmids were most often associated with blaCTX-M-1 genes, with 43% of blaCTX-M-1 isolates harbouring genes showing > 99% identity with pESBL138. The presence of this plasmid in isolates recovered over a five-year period demonstrated its stability, which was unusual compared to the majority of plasmids circularised in this study, that were only found in a handful of isolates, with exception of another blaCTX-M-1 plasmid, pPE13096. Plasmid stability has been noted in studies of broilers, so this may not be a novel observation26,28. However, due to short-read WGS data being used for the majority of isolates in this study, a full plasmid comparison was difficult and it requires further long-read sequencing to determine the variability at minutiae of the plasmids recovered; which was too costly for us to undertake currently. Excluding beta-lactams, isolates from all years showed the highest proportion of genotypic co-resistance to the same three antimicrobials: sulfamethoxazole, tetracycline and trimethoprim, which since 2013 are some of the most sold antimicrobials for animal therapeutics (UK-VARSS 2017, 2018). Resistance to these antimicrobials persisted although results from monitoring showed a decrease in 2017 compared to 2015 in E. coli isolated from pigs at slaughter (UK-VARSS 2017, 2018). The co-resistance, resulting in MDR, is likely driven by co-location of sul, tet and dfrA gene variants on plasmids such as IncI1 carrying blaCTX-M-1 type ESBL genes, as shown in this study. There was a slight decrease in the proportion of MDR isolates in the McC + CTX agar from 2015 to 2017, which coincided with a 50% reduction in antibiotic sales approved for usage in pigs and poultry, and 48% reduction in antibiotics approved for pigs only, from 2013 to 201717. However, there was an increase in the level of MDR from the ESBL isolates from 2013 to 2017. This could be attributed to the MDR rules that are stipulated by EFSA when analysing ESBL and AmpC producing isolates. As ESBL and AmpC producing isolates are also resistant to aminopenicillins (ampicillin), they automatically fulfil two of the three groups required for MDR designation, and as ESBL isolates are more likely to harbour additional resistance genes on plasmids, these isolates are more prone to MDR designation. However, if ampicillin was combined with cefotaxime and ceftazidime to form a single group, a 3–11% reduction in the proportion of MDR would result. Notably, resistance genes to the highest priority-critically important antimicrobials (HP-CIAs) colistin, meropenem and tigecycline not recommended for use in food-producing animals were not detected in any isolates. However, genes conferring resistance to other important antimicrobials such as gentamicin, rifampicin, azithromycin, fosfomycin and ciprofloxacin were detected in multiple isolates. Some of these resistances are not novel, for example, gentamicin and ciprofloxacin resistance have previously been reported in E. coli from livestock in Europe29,30. Although, identification of fosA3, a gene rare in Europe, in five isolates from all years sampled was surprising as this gene is more commonly associated with Asia31. fosA3 was first reported in Europe in 2013 from a migratory bird in Germany, where the gene was located on a MDR IncA/C plasmid containing blaNDM-132. Resistance to antibiotics less commonly used in human medicine but used in treating livestock, such as spectinomycin, was common in all years.
One limitation of this study is that it focussed only on E. coli isolates collected from selective media and not on E. coli collected from non-selective media from EU surveillance. This only allowed investigation into trends of E. coli that are ESBL/AmpC producers and their co-resistances, and not the general resistance trends of all E. coli that may be present in livestock and probably harbour other antimicrobial groups. So isolates that may harbour resistances to other HP-CIAs but are not ESCs may be have been missed, as demonstrated during the retrospective mcr-1 investigations30,33,34.
In conclusion, this study demonstrated how the availability of WGS data from both short and long read sequences in bacteria collected from national surveillance activities allowed in-depth scrutiny of resistance trends and dissemination of plasmids, which generally harbour AMR genes. Such analysis if incorporated into surveillance activities would provide a wealth of data to help monitor long-term national AMR trends more accurately, including rapid retrospective analysis of a particular gene, ST or AMR profile. Furthermore, having this genomic information provides enormous value for rapid retrospective molecular characterisation to determine prevalence and transmission, and facilitates comparison with new or current datasets in the event on an outbreak.
Methods
DNA extraction of E. coli isolated from pig caecal contents
Archived E. coli isolated from pig caeca in 2013 using selective agar (CHROMagar CTX or Oxoid Brilliance ESBL) originated from a nationwide pig study by Randall et al. 201415 DNA from these were isolated using the QuickGene system (Fujifilm, Japan), and an MP FastPrep (MP Biomedicals, USA), for additional mechanical lysis. The 2015 and 2017 isolates originated from archived E. coli previously isolated from the EU mandatory monitoring of AMR in zoonotic and commensal bacteria, outlined by the Commission Implementing Decision 2013/652/EU and the MICs of these isolates to 16 antimicrobials recommended by EFSA were obtained by following EU monitoring protocols2. They were grown on either MacConkey supplemented with 1 mg/L cefotaxime (McC + CTX) or CHROMagar ESBL, depending on which the E. coli had originally been isolated on. DNA was extracted from the 2015 isolates using a crude boilate as described previously30,33. For the 2017 isolates, a single colony was used to inoculate a 3 mL overnight culture in LB. The overnight culture was the starting material for the MagMax core nucleic acid purification kit and extracted with a Kingfisher Flex system (Thermofisher, USA) using the heated script protocol. For minION Nanopore sequencing, DNA was extracted using a Qiagen genomic 100/G tip, barcoded using a rapid barcoding kit, used as per manufacturer’s instructions.
WGS and analysis
The 2013 (n = 185) isolates were sequenced with an Illumina HiSeq, whilst the 2015 (n = 188) and 2017 (n = 128) isolates were sequenced using an Ilumina MiSeq or NextSeq. A subset of six isolates were sequenced on an Oxford Nanopore minION. SPAdes 3.11.1 was used for assembling the short-read data; Unicycler for the short and long-read hybrid data; and Prokka 1.11 for annotation35–37. MLST of the isolates was completed with SRST2, and presence of AMR genes and plasmids determined with APHA SeqFinder and Abricate30,38,39. The isolates were aligned to E. coli K12 with Snippy, Gubbins was used to remove recombination regions, and RAxML was used to build a phylogeny (trees were annotated with iTOL), and Kraken was used to for taxonomic classification40–43. Circularised plasmid sequences were compared to the genome assemblies of all other isolates using fastANI44 and blastN for comparing to those held by NCBI. pMLST was used to determine the plasmid ST45. An isolate was considered MDR if it contained genes conferring resistance to three or more different groups of antimicrobials listed in the scientific report published by EFSA in 2018 46. Following the EFSA MDR stipulations, cefotaxime and ceftazidime were treated as a single group, as were ciprofloxacin and nalidixic acid. Genotypic resistance was compared to previously generated phenotypic MIC data for the 201518 and 201717 McC + CTX isolates using methods outlined in Stubberfield et al.16.
Supplementary information
Acknowledgements
We would like to thank Dr Richard Ellis for the 2015–2017 Illumina sequencing and Charlotte Bickers for her help with Nanopore sequencing. We thank the Veterinary Medicines Directorate (VMD) for funding the project under grants FZ2200 to CT and VM0529 to MFA, and 2013 isolates under grant VM0506 to CT. This project has also received funding to MFA from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 773830, in the ARDIG project within the One Health European Joint Programme. The 2013 sequencing was funded by the National Institute for Health Research (NIHR) Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance at University of Oxford in partnership with Public Health England (PHE) (HPRU-2012-10041). DC is NIHR Senior Investigator.
Author contributions
N.D. performed the DNA extractions for 2015 and 2017 isolates, the Nanopore sequencing, and analysed the genomic data. M.A. performed the DNA extractions for the 2013 samples. L.R., J.R., R.H. and F.L. performed the bacterial isolation from the caecal samples; D.C. sequenced the 2013 isolates; M.F.A. and N.D. coordinated the study; M.F.A. and C.T. provided scientific guidance and oversight; N.D. and M.F.A. drafted the manuscript.
Data availability
All raw Illumina and Nanopore data produced in this study have been deposited under PRJEB34493. Plasmid sequences have been deposited under accession numbers MW077910-MW077917.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-76877-7.
References
- 1.O’Neill J. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014;20:1–20. [Google Scholar]
- 2.Authority, E. F. S. et al. (2019) Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food‐producing animals and food. EFSA J.17, e05709. [DOI] [PMC free article] [PubMed]
- 3.Control, E. C. f. D. P. a. ECDC strategic framework for the integration of molecular and genomic typing into European surveillance and multi-country outbreak investigations 2019–2021. (Stockholm, 2019).
- 4.WHO. Critically important antimicrobials for human medicine: Ranking of antimicrobial agents for risk management of anti microbial resistance due to non-human use. Report No. 9241512229, (World Health Organization, 2017).
- 5.Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 7.Day MJ, et al. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J. Antimicrob. Chemother. 2016;71:1178–1182. doi: 10.1093/jac/dkv485. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh H, et al. bla(CTX-M-27)-Encoding Escherichia coli sequence type 131 lineage C1–M27 clone in clinical isolates, Germany. Emerg. Infect. Dis. 2017;23:1754–1756. doi: 10.3201/eid2310.170938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopkins KL, Batchelor MJ, Anjum M, Davies RH, Threlfall EJ. Comparison of antimicrobial resistance genes in nontyphoidal salmonellae of serotypes Enteritidis, Hadar, and Virchow from humans and food-producing animals in England and Wales. Microb. Drug Resist. 2007;13:281–288. doi: 10.1089/mdr.2007.779. [DOI] [PubMed] [Google Scholar]
- 10.Kirchner M, Lemma F, Randall L, Anjum M. Loop-mediated isothermal amplification for extended spectrum β-lactamase gene detection in poultry carcases. Vet. Rec. 2017;181:119–119. doi: 10.1136/vr.104150. [DOI] [PubMed] [Google Scholar]
- 11.Abuoun M, et al. Characterising antimicrobial resistant Escherichia coli and associated risk factors in a cross-sectional study of pig farms in Great Britain. Front. Microbiol. 2020;11:861. doi: 10.3389/fmicb.2020.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Card RM, et al. Impact of ciprofloxacin and clindamycin administration on gram-negative bacteria isolated from healthy volunteers and characterization of the resistance genes they harbor. Antimicrob. Agents Chemother. 2015;59:4410–4416. doi: 10.1128/AAC.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchner M, et al. Antimicrobial resistance characteristics and fitness of Gram-negative fecal bacteria from volunteers treated with minocycline or amoxicillin. Front. Microbiol. 2014;5:722. doi: 10.3389/fmicb.2014.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoesser N, et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio. 2016;7:e02162–e12115. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randall LP, et al. Prevalence of extended-spectrum-β-lactamase-producing Escherichia coli from pigs at slaughter in the UK in 2013. J. Antimicrob. Chemother. 2014;69:2947–2950. doi: 10.1093/jac/dku258. [DOI] [PubMed] [Google Scholar]
- 16.Stubberfield E, et al. Use of whole genome sequencing of commensal Escherichia coli in pigs for antimicrobial resistance surveillance, United Kingdom, 2018. Eurosurveillance. 2019;24:50. doi: 10.2807/1560-7917.ES.2019.24.50.1900136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UK-VARSS. UK Veterinary Antibiotic Resistance and Sales Surveillance Report, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/915743/_1691664-v1-VARSS_2017_Watermark_FINALx-accessible.pdf (2017).
- 18.UK-VARSS. UK Veterinary Antibiotic Resistance and Sales Surveillance Report, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/582341/1051728-v53-UK-VARSS_2015.pdf (2015).
- 19.Gomes C, Ruiz-Roldán L, Mateu J, Ochoa TJ, Ruiz J. Azithromycin resistance levels and mechanisms in Escherichia coli. Sci. Rep. 2019;9:6089–6089. doi: 10.1038/s41598-019-42423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagès J-M, James CE, Winterhalter M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008;6:893. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 21.Touzain F, et al. Characterization of plasmids harboring blaCTX-M and blaCMY genes in E. coli from French broilers. PLoS ONE. 2018;13:e0188768. doi: 10.1371/journal.pone.0188768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner S, Lupolova N, Gally DL, Argyle SA. Convergence of plasmid architectures drives emergence of multi-drug resistance in a clonally diverse Escherichia coli population from a veterinary clinical care setting. Vet. Microbiol. 2017;211:6–14. doi: 10.1016/j.vetmic.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weingarten RA, et al. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio. 2018;9:e02011–02017. doi: 10.1128/mBio.02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liakopoulos A, et al. Genomic and functional characterisation of IncX3 plasmids encoding blaSHV-12 in Escherichia coli from human and animal origin. Sci. Rep. 2018;8:7674. doi: 10.1038/s41598-018-26073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duggett N, et al. Detection in livestock of the human pandemic Escherichia coli ST131 fimH30(R) clone carrying blaCTX-M-27. J. Antimicrob. Chemother. 2020 doi: 10.1093/jac/dkaa407. [DOI] [PubMed] [Google Scholar]
- 26.Ceccarelli, D. et al. Diversity of plasmids and genes encoding resistance to extended spectrum cephalosporins in commensal Escherichia coli from dutch livestock in 2007–2017. Front. Microbiol.10, (2019). [DOI] [PMC free article] [PubMed]
- 27.Fischer J, et al. blaCTX-M-15-carrying Escherichia coli and Salmonella isolates from livestock and food in Germany. J. Antimicrob. Chemother. 2014;69:2951–2958. doi: 10.1093/jac/dku270. [DOI] [PubMed] [Google Scholar]
- 28.Dierikx C, et al. Extended-spectrum-β-lactamase-and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J. Antimicrob. Chemother. 2012;68:60–67. doi: 10.1093/jac/dks349. [DOI] [PubMed] [Google Scholar]
- 29.Szmolka A, Anjum MF, La Ragione RM, Kaszanyitzky ÉJ, Nagy B. Microarray based comparative genotyping of gentamicin resistant Escherichia coli strains from food animals and humans. Vet. Microbiol. 2012;156:110–118. doi: 10.1016/j.vetmic.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 30.Anjum MF, et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J. Antimicrob. Chemother. 2016;71:2306–2313. doi: 10.1093/jac/dkw149. [DOI] [PubMed] [Google Scholar]
- 31.Ana CM, et al. Importation of fosfomycin resistance fosA3 Gene to Europe. Emerg. Infect. Dis. J. 2016;22:346. doi: 10.3201/eid2202.151301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villa L, et al. IncA/C plasmid carrying bla(NDM-1), bla(CMY-16), and fosA3 in a Salmonella enterica serovar corvallis strain isolated from a migratory wild bird in Germany. Antimicrob. Agents Chemother. 2015;59:6597–6600. doi: 10.1128/AAC.00944-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duggett NA, et al. Molecular epidemiology of isolates with multiple MCR plasmids from a pig farm in Great Britain: The effects of colistin withdrawal in the short and long term. J. Antimicrob. Chemother. 2018;73:3025–3033. doi: 10.1093/jac/dky292. [DOI] [PubMed] [Google Scholar]
- 34.Duggett NA, et al. Occurrence and characterization of mcr-1-harbouring Escherichia coli isolated from pigs in Great Britain from 2013 to 2015. J. Antimicrob. Chemother. 2017;72:691–695. doi: 10.1093/jac/dkw477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 37.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inouye M, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seemann, T. ABRicate, https://github.com/tseemann/abricate (2016).
- 40.Seemann, T. Snippy, https://github.com/tseemann/snippy (2014).
- 41.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15–e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Authority, E. F. S., Prevention, E. C. f. D. & Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J.16, e05182, (2018). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw Illumina and Nanopore data produced in this study have been deposited under PRJEB34493. Plasmid sequences have been deposited under accession numbers MW077910-MW077917.