Abstract
The intestine interacts with many factors, including dietary components and ethanol (EtOH), which can impact intestinal health. Previous studies showed that different types of dietary fats can modulate EtOH-induced changes in the intestine; however, mechanisms underlying these effects are not completely understood. Here, we examined intestinal transcriptional responses to EtOH in WT and transgenic fat-1 mice (which endogenously convert n6 to n3 polyunsaturated fatty acids [PUFAs]) to identify novel genes and pathways involved in EtOH-associated gut pathology and discern the impact of n3 PUFA enrichment. WT and fat-1 mice were chronically fed EtOH, and ileum RNA-seq and bioinformatic analyses were performed. EtOH consumption led to a marked down-regulation of genes encoding digestive and xenobiotic-metabolizing enzymes, and transcription factors involved in developmental processes and tissue regeneration. Compared to WT, fat-1 mice exhibited a markedly plastic transcriptome response to EtOH. Cell death, inflammation, and tuft cell markers were downregulated in fat-1 mice in response to EtOH, while defense responses and PPAR signaling were upregulated. This transcriptional reprogramming may contribute to the beneficial effects of n3 PUFAs on EtOH-induced intestinal pathology. In summary, our study provides a reference dataset of the intestinal mucosa transcriptional responses to chronic EtOH exposure for future hypothesis-driven mechanistic studies.
Subject terms: Alcoholic liver disease, Nutrition, Fatty acids
Introduction
The gastrointestinal (GI) tract interacts dynamically with a variety of environmental factors, including different dietary components and ethanol (EtOH). Dietary fats are among the most important factors contributing to intestinal homeostasis and basic functions, and are tightly regulated in order to maintain intestinal health and overall well-being1,2. Dietary nutrients can also shape the GI microbiota and intestinal immunity3. Therefore, a well-balanced diet is crucial for good health. However, in the Western diet, n6 polyunsaturated fatty acid (PUFA) consumption has become progressively higher than n3 PUFA consumption, and this unbalanced diet may have a negative impact on health and multiple pathological conditions. An increase in the n6/n3 PUFA ratio has been shown to be detrimental in multiple chronic diseases4,5. It is well recognized that dietary PUFAs play an important role in intestinal health. Reduced intestinal inflammation by n3 PUFA supplementation has been shown both in patients and in animal models, suggesting anti-inflammatory and pro-resolving properties of n3 PUFAs and their derivatives in the gut (reviewed in6). EtOH consumption, in general, negatively affects the GI tract causing damaging effects on intestinal health in experimental animals and in humans. These effects include pathological changes in numerous metabolic pathways and functions such as loss of gut barrier integrity, increased inflammation, and microbial dysbiosis, among others7–9. Notably, composition of dietary fats can modulate (either exacerbate or attenuate) EtOH-induced changes in the intestine. It has been demonstrated that EtOH and a diet enriched in corn oil (containing a high amount of the n6 PUFA linoleic acid), but not a diet enriched in medium chain fatty acids (MCFAs), results in increased gut permeability, alterations in the intestinal mucus layer and antimicrobial defense, and an increased intestinal pro-inflammatory response8,10. Supplementation with saturated long-chain fatty acids (LCFAs) positively affected EtOH-mediated alterations in the intestinal barrier and gut microbiota dysbiosis11. It has been shown that tributyrin, a triglyceride found in butter and margarine, prevents alcohol-induced tight-junction (TJ) disruption, which in turn protects against intestinal hyperpermeability12. Various dietary fatty acids also differentially modulate EtOH-induced changes in the gut microbiome. For example, EtOH in combination with n6 PUFAs, but not MCFAs, causes an increase in Proteobacteria and Actinobacteria13.
A recent study from our group demonstrated that the increase in endogenous n3 PUFAs and subsequent decrease in the n6/n3 PUFA ratio in transgenic fat-1 mice (which endogenously convert n6 to n3 PUFAs) attenuated EtOH-mediated alterations in the gut-liver axis14. Specifically, we found protection against EtOH-mediated downregulation of intestinal TJ proteins ex vivo in organoid cultures and in vivo in fat-1 mice. Further, although EtOH feeding did not cause marked structural damage to the ileum in either WT or fat-1 mice, expression of markers of intestinal stem cells growth and proliferation were higher in ileal organoids obtained from fat-1 vs WT animals. In addition, compared to WT, fat-1 mice developed less intestinal inflammation in response to EtOH and lipopolysaccharide (LPS) challenge. Thus, the types of dietary fat that reach the intestine and the endogenous tissue levels of n3 and n6 PUFAs are critical modulators of EtOH-associated gut abnormalities. Although recent evidence supports the beneficial effects of n3 PUFAs on EtOH-mediated alterations in intestinal homeostasis14, the molecular mechanisms underlying these effects are not completely understood. The current study is an extension of our previous work analyzing the intestinal transcriptional responses to chronic EtOH administration that addresses the following goals: (1) to identify novel genes and pathways of intestinal dysregulation that contribute to alcohol-associated gut pathology; and (2) to examine gene expression signatures underlying the beneficial effects of n3 PUFA-mediated intestinal protection against EtOH-induced damage. Understanding changes in the intestinal transcriptome and how these changes influence host metabolism/health in vivo may identify novel therapeutic targets for the treatment of alcohol-induced intestinal pathology.
Results
Characteristics of the animal model and overview of intestinal RNAseq analysis
To examine the impact of chronic EtOH administration and elevated levels of endogenous n3 PUFAs on the intestinal transcriptome, we performed RNAseq analysis and compared the transcriptomes of intestinal epithelial tissue obtained from WT and fat-1 mice subjected to control (pair-fed mice, PF) or EtOH-containing diets (EtOH-fed mice, see Supplemental Fig. S1a for study design). Animal daily food consumption and body weights can be found in Supplemental Fig. S1b,c, respectively. While there were no differences in food consumption between genotypes, as well as between PF and EtOH-fed animals, WT EtOH, WT PF, and fat-1 PF mice gained body weight by the end of the experiment, whereas fat-1 EtOH mice did not. Compared to WT, fat-1 animals exhibited elevated intestinal tissue n3 PUFA levels in both PF and EtOH-fed mice (see ref.14 and Supplemental Fig. S1d). There were no differences in n6 PUFAs between WT and fat-1 control PF mice; however, elevated intestinal n6 PUFA levels were observed in response to EtOH in WT but not in fat-1 littermates. In addition, the ratio of n6/n3 PUFAs was significantly lower in fat-1 compared to WT mice in both PF and EtOH-fed groups.
Intestinal RNAseq analysis performed on these mice revealed that both the genotype and EtOH administration caused alterations in the intestinal transcriptome; the full list of significant differentially expressed genes (DEGs) can be found in Supplemental Materials Excel File S1. We next applied a cutoff of ≥ two-fold change for the DEGs used in Volcano Plots, GO Processes, and STRING Cluster Analyses. This threshold was used to identify robust gene expression changes and the accompanying biological processes. There were 630 DEGs between fat-1 PF and WT PF animals (420 up and 210 down). Of those genes, 276 DEGs between fat-1 PF and WT PF animals met the two-fold change threshold for further analysis (193 up and 83 down). EtOH administration resulted in 1144 DEGs (481 up and 663 down) in the WT EtOH vs WT PF group, and of these genes, 439 met the two-fold change threshold (163 up and 276 down). While 2107 genes were differentially expressed in fat-1 EtOH vs fat-1 PF mice (1045 up and 1062 down), 1501 met the two-fold change threshold for further analyses (812 up and 689 down). 94 genes were significantly different (80 up and 14 down) in the WT EtOH group when compared to fat-1 EtOH mice (all of these DEGs met the two-fold change threshold).
Differentially expressed genes between fat-1 and WT control pair-fed animals
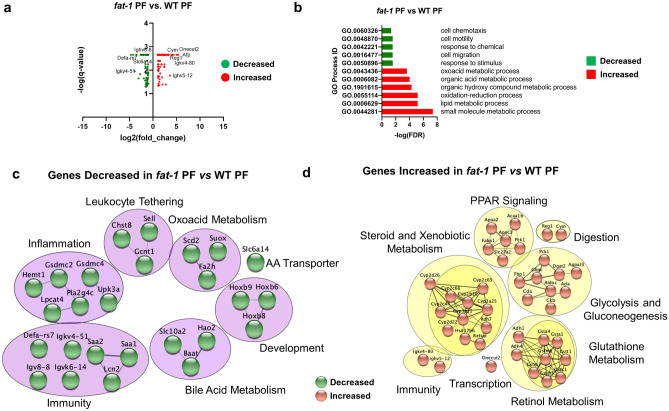
The most prominent changes among genes differentially expressed in fat-1 PF compared to WT PF mice (≥ two-fold change, Fig. 1a and Supplementary Table S1) were observed for Onecut2, Reg1, Igkv4-80, Cym, Ighv5-12, and Afp (increased expression), and Defa-rs7, Igk4-51, Slc6a14, and Ighv8-8, (decreased expression). In order to best classify fat-1 PF vs WT PF mice with respect to all DEGs that met the two-fold threshold, we performed Gene Ontology (GO) pathway analysis in CytoScape. The top enriched and diminished pathways are shown in Fig. 1b. Next, gene clusters identified by STRING were used to visualize gene interactions and identify processes affected by the endogenous increase in n3 PUFAs in fat-1 compared to WT mice. The gene and protein interactions inferred from differentially expressed transcripts between fat-1 PF and WT PF included down-regulated gene networks (Fig. 1c) for leukocyte tethering (Chst8, Gcnt1, Sell), inflammation (Gsdmc2, Gsdmc4, Pla2g4c), immunity (Saa2, Saa1, Lcn2), bile acid metabolism (Slc10a2, Hao2, Baat), development (Hoxb9, Hoxb6, Hoxb8), oxoacid metabolism (Scd2, Suox2, Fa2h), and amino acid transport (Slc6a14). Up-regulated gene networks (Fig. 1d) included metabolism of steroids/xenobiotics (Cyp2c65, Cyp2b10, Cyp2b66), glycolysis and gluconeogenesis (Fbp1, G6pc, Pck1), glutathione metabolism (Gstt1, Gstm3, Gsta1), peroxisome proliferator-activated receptor (PPAR) signaling (Apoa2, Fabp1, Acaa1b), immunity (Igkv4-80, Ighv5-12), digestion (Reg1, Cym), and transcription (Onecut2).
Figure 1.
Differentially expressed genes between fat-1 and WT control pair-fed animals. (a) Volcano plot analysis of significant genes from the fat-1 PF vs WT PF comparison. Genes are plotted as log2(fold change) vs antilog(p-value). (b) GO Processes representative of decreased gene expression (green) and increased gene expression (red) for the fat-1 PF vs WT PF comparison. (c) Gene cluster analysis of genes decreased in fat-1 PF vs WT PF comparison (purple circles, green nodes). (d) Gene cluster analysis of genes increased in fat-1 PF vs WT PF comparison (yellow circles, copper nodes). Significant DEGs were selected based on a q-value < 0.05. Significant DEGs with fold-change greater than or equal to two-fold were used for analyses in (a–d). n = 3–5 mice per group.
EtOH-mediated alterations in the intestinal transcriptome: similarity in transcriptional responses between WT and fat-1 mice
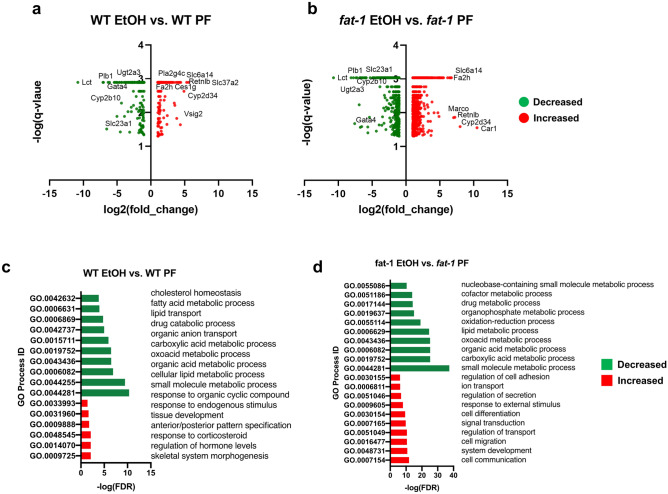
EtOH administration resulted in substantial alterations in intestinal gene expression. Notably, there were almost 2 times more genes differentially expressed in fat-1 mice compared to WT littermates in response to EtOH (2107 DEGs vs 1144 DEGs, respectively), suggesting that the alterations in the associated lipid profile and, possibly, lipid homeostasis in fat-1 mice resulted in elevated gene transcription changes in response to EtOH challenge. Further analysis revealed a set of 835 genes in WT and fat-1 mice with similarly changed expression in response to EtOH (321 ≥ two-fold change), while 1272 and 309 genes were exclusively changed by EtOH in fat-1 and WT mice, respectively (180 and 118 ≥ two-fold change). Volcano plot analysis of DEGs that met the two-fold threshold in WT-EtOH relative to WT-PF (Fig. 2a) and in fat-1 EtOH relative to fat-1 PF (Fig. 2b) demonstrated that the top down-regulated genes shared by both genotypes included Lct, Cyp2b10, Ugt2a3, Gata4, and Plb1. Among the genes up-regulated by EtOH in both WT and fat-1 mice were Cyp2d34, Retnlb, Slc6a14, and Fa2h. The top DEGs that changed their expression in response to EtOH in WT and fat-1 mice, as well as shared genes between WT and fat-1 mice in response to EtOH are listed in Supplementary Tables S2, S3, and S4, respectively. Cytoscape GO analysis identified downregulated processes in WT-EtOH vs WT-PF mice including lipid metabolism, oxoacid metabolism, anion transport, fatty acid metabolism, and cholesterol homeostasis; while upregulated processes included development and response to hormones (Fig. 2c). GO processes diminished in fat-1 EtOH vs fat-1 PF mice included oxoacid metabolism, lipid metabolism, drug metabolism, and nucleobase metabolism, while cell communication, cell migration, signal transduction, transport, cell adhesion, and cell differentiation were enriched (Fig. 2d).
Figure 2.
Changes in intestinal transcriptional responses to EtOH in WT and fat-1 mice. (a) Volcano plot analysis of significant genes from the WT EtOH vs WT PF comparison plotted as log2(fold change) vs antilog(p-value) for individual genes. (b) Volcano plot analysis for the fat-1 EtOH vs fat-1 PF comparison plotted as log2(fold change) vs antilog(p-value) for individual genes. (c) GO Process analysis representative of genes decreased in expression (green) and genes increased in expression (red) for the WT EtOH vs WT PF comparison. (d) GO Process analysis representative of genes decreased in expression (green) and genes increased in expression (red) for the fat-1 EtOH vs fat-1 PF comparison. Significant DEGs that met a two-fold threshold were used for the analyses in (a–d). n = 3–5 mice per group.
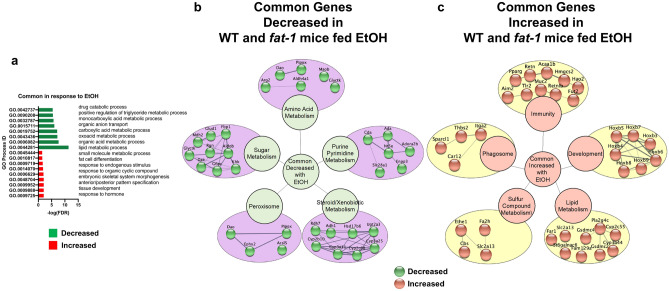
GO processes representative of the decreased genes in response to EtOH shared between genotypes included lipid metabolism, carboxylic acid metabolism, drug metabolism, and anion transport. The elevated shared genes due to EtOH included hormone and developmental processes (Fig. 3a). Gene clusters identified by STRING were used to visualize gene interactions and pathways among DEGs commonly affected by EtOH in both genotypes. Common pathway clusters decreased by EtOH in WT and fat-1 mice included amino acid metabolism (Arg2, Dao, Pipox), sugar metabolism (Aldob, G6pc, Fbp1), purine and pyrimidine metabolism (Ada, Cda, Nt5e), steroid and xenobiotic metabolism (Rdh7, Adh1, Cyp2b10), and the peroxisome (Ephx2, Dao, Acsl5) (Fig. 3b). Pathway clusters representative of common genes elevated by EtOH in WT and fat-1 mice included immunity (Pparγ, Muc2, Tlr2), development (Hoxb4, Hoxb8, Hoxb5), lipid metabolism (Cyp2c55, Cyp3a44, Far1), sulfur compound metabolism (Cbs, Ethe1, Fa2h), and the phagosome (Itga2, Thbs2, Sparcl1) (Fig. 3c).
Figure 3.
Similarity in transcriptional responses of ileum tissue to EtOH in WT and fat-1 mice. (a) GO Process analysis representative of genes decreased in expression (green) and genes increased in expression (red) that were shared between genotypes in response to EtOH. (b) Gene cluster analysis of genes shared between genotypes that were decreased in response to EtOH (purple circles, green nodes). (c) Gene cluster analysis of genes shared between genotypes that were increased in response to EtOH (yellow circles, copper nodes). Significant DEGs that met a two-fold change threshold were used for the analyses in (a–c). n = 3–5 mice per group.
EtOH-mediated alterations in the intestinal transcriptome: unique transcriptional responses to EtOH in WT or fat-1 mice
Next, we identified the specific transcriptional responses to EtOH in the intestinal mucosa of WT and fat-1 mice. There were a set of genes most prominently up and down-regulated in response to EtOH treatment exclusively in WT but not in fat-1 mice including Slc37a2, Ighv14-4, Ighv1-63, Abca12, and Rn7sk (~ 47-, 15-, 14-, 13-, and 11- fold increases, respectively), and Fabp1, Ighv2-3, Igkv4-51, Gip, and Ighv8-8 (~ 70-, 16-, 15-, 14-, and 8- fold decreases, respectively, Supplementary Table S5). The genes most highly changed in response to EtOH treatment exclusively in fat-1 but not in WT mice included: Car1, Marco, Nov, Plet1, and Cdhr1 (~ 1454-, 151-, 97-, 31-, and 30-fold increases, respectively), and Bbox1, Slc4a5, Dnah2, Rec8, and Afp (~ 59-, 25-, 20-, 19-, and 16-fold decreases, respectively, Supplementary Table S6).
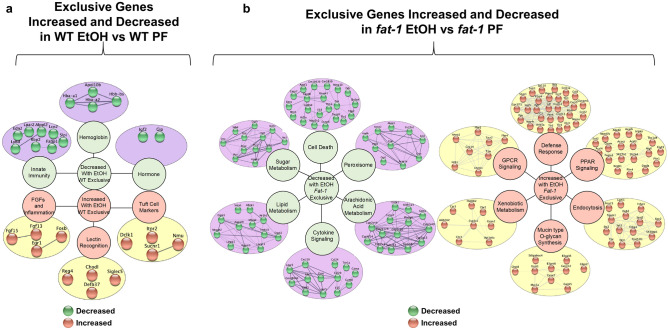
Cytoscape cluster analysis identified genotype-specific pathway alterations in response to EtOH. Pathways that were downregulated in response to EtOH in WT mice exclusively included hormones (Igf2, Gip), hemoglobin (Hba-a1, Hba-a2, Hbb-bs), and innate immunity (Lcn2, Slpi, Ly6d). Pathway clusters upregulated in response to EtOH in WT mice only included tuft cell markers (Itpr2, Dclk1, Sucnr1), lectin recognition (Chodl, Siglec5, Reg4), and fibroblast growth factors (FGFs) (Fgf13, Fgf15) and inflammation (Fosb, Egr1) (Fig. 4a). Clustered pathways decreased in response to EtOH in fat-1 mice exclusively include cell death (Gsdmd, Il18, Casp7), the peroxisome (Hadh, Crat, Acaa1a), arachidonic acid metabolism (Cyp2c66, Cyp2j6, Cyp2u1), cytokine signaling (Il15, Cxcl16, Gzma), lipid metabolism (Nr1h3, Dgat1, Dgat2), and sugar metabolism (Pklr, Eno1, Mdh1) (Fig. 4b). Pathway clusters increased in response to EtOH in fat-1 mice only included defense response (Marco, Il1rn, Tlr4), PPAR signaling (Pnpla3, Plin4, Plin1), endocytosis (Syt1, Dab2, Epn2), mucin O-glycan synthesis (B3gnt6, Gcnt1, Gaint12), xenobiotic metabolism (Gstm2, Cbr2, Cbr3), and GPCR signaling (Ffar2, Ffar4, Tacr2) (Fig. 4b).
Figure 4.
Genotype exclusive transcriptional responses to EtOH in WT and fat-1 mice. (a) Cluster analysis of genes exclusively decreased (purple circles, green nodes) or increased in response to EtOH in WT mice (yellow circles, copper nodes). (b) Cluster analysis of genes exclusively decreased (purple circles, green nodes) or increased in response to EtOH in fat-1 mice (yellow circle, copper nodes). Significant DEGs that met a two-fold change threshold were used for the analyses in (a,b). n = 3–5 mice per group.
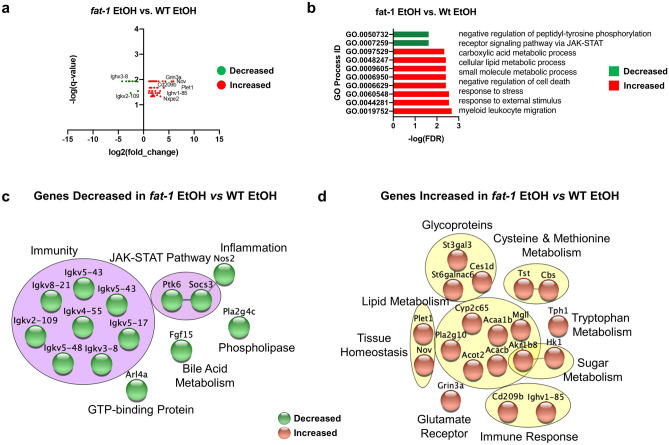
Differential transcriptional responses to EtOH between fat-1 and WT mice
The differences described above in transcriptional responses to EtOH in fat-1 and WT mice resulted in a total of 94 genes that were differentially regulated between fat-1 EtOH and WT EtOH animals. Among 80 up-regulated DEGs in fat-1 EtOH vs WT EtOH, the greatest increase was observed for Nov, Plet1, Cd209b, Ighv1-85, Grin3a, Nxpe2 (~ 61-, 46-, 20-, 16-, 14-, and tenfold increases, respectively, Fig. 5a, Supplementary Table S7). Immunoglobulins, including Ighv3-8 and Igkv2-109 were among the 14 most decreased DEGs in fat1-1 EtOH vs WT EtOH mice. Processes representative of genes decreased in fat-1 EtOH mice included negative regulation of tyrosine phosphorylation and JAK-STAT signaling, while leukocyte migration, stress response, negative regulation of cell death, and lipid metabolism were increased (Fig. 5b). Gene clusters identified by STRING analysis and the pathways they represent were used to visualize ileum DEGs between fat-1 EtOH and WT EtOH mice. Clustered pathways downregulated in fat-1 EtOH vs WT EtOH mice included immunity (Igkv8-21, Igkv2-109, Igkv5-17), the JAK-STAT pathway (Ptk6, Socs3), inflammation (Nos2), GTP-binding protein (Arl4a), bile acid metabolism (Fgf15), and phospholipase (Pla2g4c) (Fig. 5c). Pathway clusters upregulated in fat-1 EtOH vs WT EtOH mice included glycoproteins (St3gal3, St6galnac6, Ces1d), lipid metabolism (Acaa1b, Cyp2c65, Acacb), cysteine-methionine metabolism (Tst, Cbs), tryptophan metabolism (Tph1), sugar metabolism (Hk1, Akr1b8), immune response (Cd209b, Ighv1-85), glutamate receptor (Grin3a), and tissue homeostasis (Plet1, Nov) (Fig. 5d).
Figure 5.
Differential transcriptional responses in fat-1 EtOH vs WT EtOH mice. (a) Volcano plot analysis of significant genes from the fat-1 EtOH vs WT EtOH comparison plotted as log2(fold change) vs. log(p-value) for individual genes. (b) GO Process analysis representative of genes decreased in expression (green) and genes increased in expression (red) from the fat-1 EtOH vs WT EtOH comparison. (c) Gene cluster analysis of genes decreased in the fat-1 EtOH vs WT EtOH comparison (purple circles, green nodes). (d) Gene cluster analysis of genes increased in the fat-1 EtOH vs WT EtOH comparison (yellow circles, copper nodes). Significant DEGs that met a two-fold change threshold were used for the analyses in (a–d). n = 3–5 mice per group.
Targeted analysis of selected intestinal metabolic pathways relevant to diet and EtOH-mediated alterations of gut homeostasis and overall well-being
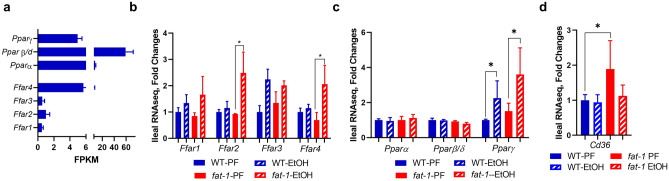
Effects of EtOH on the expression of free fatty acid receptors in WT and fat-1 mice
Nutrient sensing in the gut epithelium is fundamental for intestinal health. Since dietary fatty acids (FAs) can impact intestinal homeostasis, and n3 and n6 PUFAs act as signaling molecules, we were interested in evaluating the expression of receptors recognizing these molecules, including the FFAR and PPAR families. We observed that Pparβ/δ was the most abundantly expressed among all FA receptors (Fig. 6a). Ffar4, which recognizes MCFAs and LCFAs15,16, had the highest expression among the FFARs in the intestinal mucosa (Fig. 6a). There were no evident differences in FA receptors between WT PF and fat-1 PF mice. EtOH administration resulted in elevated levels of Ffar2 and Ffar4 in fat-1 but not WT mice, and an increase in Pparγ, which recognizes LCFAs and LCFA metabolites, in both WT and fat-1 EtOH-fed animals (Fig. 6b,c). The expression of Cd36, which mainly recognizes LCFAs was significantly elevated in fat-1 vs WT PF but not EtOH-fed animals (Fig. 6d).
Figure 6.
Expression of free fatty acid receptors in fat-1 vs WT mice. (a) Relative gene expression of FFA receptors (Ppars and Ffars) in ileal mucosa from WT PF mice. (b) Gene expression of Ffars in ileal mucosa among all treatment groups. (c) Gene expression of Ppars in ileal mucosa among all treatment groups. (d) Gene expression of Cd36 in ileal mucosa among all treatment groups. Data are presented as mean ± SEM for FPKM (a) and fold changes (b–d). *p < 0.05. n = 3–5 mice per group.
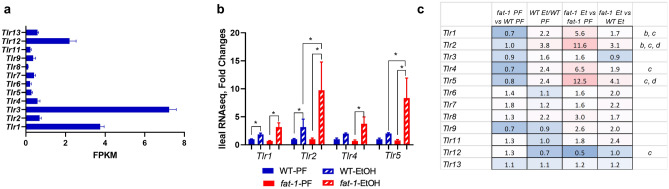
Changes in the expression of microbial sensing genes in WT and fat-1 mice exposed to EtOH
The intestinal epithelium expresses a range of pattern recognition receptors (PRRs) that sense and respond to a variety of microbial signals to maintain mucosal homeostasis17. Among the PRRs, toll-like receptors (TLRs) are key players in microbe and microbial-product recognition. TLRs are also significantly involved in host defense and tissue repair responses18, and play a critical role in EtOH-mediated immune response19. In addition, saturated and unsaturated FAs may exert their effects via activation of TLRs like TLR2 and TLR420. In our study, we observed that Tlr3 was the highest expressed TLR in the intestinal mucosa, followed by Tlr1 and Tlr12 (Fig. 7a). Further, there was a significant increase in Tlr1 and Tlr2 expression in both fat-1 and WT EtOH-fed mice compared to control PF animals, while Tlr4 and Tlr5 were significantly up-regulated by EtOH only in fat-1 mice. Interestingly, the expression of Tlr2 and Tlr5 were significantly higher in fat-1 EtOH compared to WT EtOH (Fig. 7b). Tlr12 was the only family member decreased by EtOH in fat-1 mice (Fig. 7c). There were no significant differences between any experimental groups in the expression of other Tlrs or other PRR families, e.g., nucleotide-binding oligomerization domain-like receptors (Nlrs, data not shown).
Figure 7.
Changes in the expression of microbial sensing genes in WT and fat-1 mice exposed to EtOH. (a) Relative gene expression of Tlr genes in ileal mucosa from WT PF mice. Data are presented as mean ± SEM for FPKM. (b) Gene expression of Tlr genes in ileal mucosa from all treatment groups. Data are expressed as fold changes vs WT PF set as 1, * p < 0.05. (c) Heatmap analysis of ileal mucosa Tlr genes across multiple comparisons. Data are expressed as fold changes for indicated groups. Significance for the fat-1 PF vs WT PF comparison is denoted by (a), for the WT EtOH vs WT PF (b), for fat-1 EtOH vs fat-1 PF (c), and for fat-1 EtOH vs WT EtOH (d). n = 3–5 mice per group.
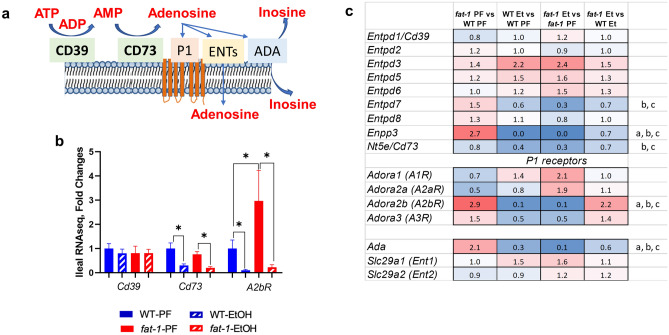
Impact of EtOH on the intestinal genes involved in the adenosine signaling pathway in WT and fat-1 mice
Adenosine signaling is recognized as an important endogenous anti-inflammatory pathway in various diseases, including intestinal injury and inflammation21,22. Adenosine triphosphate (ATP) is released by stressed, apoptotic, and necrotic intestinal epithelial cells (IECs) as well as bacteria during inflammation23. ATP is converted to adenosine by cell surface ectonucleotidases ectonucleoside triphosphate diphosphohydrolase-1 (CD39) and ecto-5′ nucleotidase (CD73). Adenosine signals via several receptors and is inactivated by equilibrative nucleoside transporters (ENTs) or by adenosine deaminase (ADA)24 (Fig. 8a). RNAseq analysis in our study revealed no significant differences in Cd39 due to EtOH consumption or genotype, while Cd73 was significantly down-regulated by EtOH in both WT and fat-1 mice (although the expression was not different between the fat-1-EtOH and WT EtOH groups) (Fig. 8b). Further, A2bR, a predominant adenosine receptor expressed in IECs21,25, was also significantly down-regulated by EtOH in both genotypes. Interestingly, A2bR expression was markedly higher in fat-1 vs WT PF but not EtOH-fed animals (Fig. 8b). There were no noticeable changes in the levels of Ent1 and Ent2 in response to EtOH administration or genotype (Fig. 8c). Ada expression was significantly increased in fat-1 PF vs WT PF mice but was down-regulated by EtOH in both genotypes (Fig. 8c). Overall, our results suggest that n3 PUFAs may enhance adenosine signaling initiating an anti-inflammatory signaling cascade in response to EtOH.
Figure 8.
Effects of EtOH on the expression of intestinal genes involved in adenosine signaling in WT and fat-1 mice. (a) Schematic of the adenosine signaling pathway. (b) Gene expression of Cd39, Cd73, and A2bR in ileal mucosa from all treatment groups. Data are expressed as fold changes vs WT PF set as 1, *p < 0.05. (c) Heatmap analysis of adenosine signaling genes in ileal mucosa across multiple comparisons. Data are expressed as fold changes for indicated groups. Significance for the fat-1 PF vs WT PF comparison is denoted by (a), for the WT EtOH vs WT PF (b), for fat-1 EtOH vs fat-1 PF (c), and for fat-1 EtOH vs WT EtOH (d). n = 3–5 mice per group.
Discussion
EtOH consumption can negatively affect intestinal health by altering intestinal homeostasis, gut microbiota, and gut barrier integrity, leading to enhanced translocation of bacteria and bacterial products which affect multiple host organs9,26. It has been shown that different types of dietary fats can modulate intestinal responses to chronic EtOH administration in mice10,13. However, the molecular mechanisms underlying these responses are not well understood. In this study we examined global intestinal transcriptome responses to chronic EtOH exposure and to increased endogenous n3 PUFA levels using WT and transgenic fat-1 mice which endogenously biosynthesize n3 PUFAs from n6 PUFAs. We observed that WT and fat-1 mice had both similarities and differences in intestinal transcriptional responses to EtOH suggesting that there were some conserved and unique mechanisms associated with EtOH exposure. The most prominently diminished gene caused by EtOH consumption in both genotypes was Lct (which was down-regulated more than 1500-fold). Lct is the gene encoding lactase, a digestive enzyme involved in lactose breakdown. Reduced lactase activity has been reported in humans (specifically those who drink alcohol)27 and in experimental animals (e.g. in rats after 3 months of EtOH consumption) likely due to direct effects of EtOH and its metabolites on the intestinal mucosa28. EtOH consumption also led to a pronounced down-regulation of a wide range of genes encoding drug- and xenobiotic-metabolizing enzymes (e.g., Cyp2b10, Ugt2a3, and the nuclear receptor Car [which regulates these genes]) in both WT and fat-1 mice. However, it should be also noted that Cyp2d34, another member of cytochrome P450 family, was the most up-regulated gene in both WT and fat-1 mice, demonstrating that EtOH significantly altered the intestinal biotransformation gene networks regardless of genotype. Among other genes that were elevated in both genotypes in response to EtOH were genes of the homeobox (Hoxb) family. Hoxb genes are transcription factors that are involved in various aspects of development and tissue regeneration, including patterning of the GI tract and differentiation of the epithelium29. The significance of the EtOH-induced up-regulation of Hoxb genes is not clear, but it may be an adaptive response to the damaging effects of EtOH on gut morphology and needs to be investigated further.
We identified gene expression patterns exclusively changed in WT or fat-1 mice in response to EtOH challenge. There were 309 genes altered by EtOH exclusively in WT but not in fat-1 mice. One such gene is Fabp1 (~ 70-fold decrease in WT EtOH vs WT PF mice). Fabp1 encodes fatty acid-binding protein 1, which is essential for proper lipid metabolism in differentiated enterocytes, and is required for enterocyte proliferation30, suggesting that downregulation of this gene may contribute to EtOH-induced alterations in intestinal homeostasis. We further observed that WT but not fat-1 mice subjected to EtOH feeding exhibited reduced expression of innate immunity genes in the intestine, including Slpi and Lcn2. Intestinal SLPI has antibacterial31 and anti-inflammatory32 functions. SLPI administration has been shown to be more effective than IL-10 and TGF-β at preventing intestinal inflammation in a colitis animal model32. LCN2 acts on intestinal macrophages to promote bacterial clearance leading to resolution of intestinal inflammation in colitis33. The host response to gut bacteria is critical, and tuft cells play an important role in that process. During bacterial infection, tuft cells proliferate to elicit a pro-inflammatory response34. Interestingly, the expression of tuft cell markers Sucnr1 and Dclk were elevated due to EtOH in WT but not fat-1 mice. Tuft cells are also chemosensory cells that coordinate immune responses to gut derived bacteria and bacterial products35. Tuft cell Sucnr1 is activated by bacterial succinate and elicits a type-2 inflammatory response in the intestine34. Since the expression of Sucnr1 was increased in response to EtOH in WT but not in fat-1 mice, the type-2 inflammatory response is likely reduced in fat-1 mice.
One of the important observations from the current study was that fat-1 mice exhibited greater transcriptional plasticity in response to EtOH in the intestinal mucosa. Transcriptional plasticity is considered a beneficial, adaptive response to stress36. Indeed, fat-1 EtOH-fed mice had 2107 DEGs relative to fat-1 PF mice in contrast to only 1144 DEGs observed for the WT EtOH vs WT PF mice. These diverse transcriptomic responses to EtOH in the intestines of fat-1 mice may explain the greater protection against EtOH-induced intestinal alterations, as previously reported14. Of note, there were 1272 DEGs exclusively changed in fat-1 EtOH vs fat-1 PF mice, including down-regulation of several genes involved in cell death and cytokine signaling and up-regulation of genes involved in defense response, xenobiotic metabolism, and PPAR signaling. Of particular interest, there were several transcripts encoding pyroptosis mediators, such as Gsdmd and Il18, that were decreased in fat-1 EtOH vs fat-1 PF mice but not in WT EtOH vs WT PF mice. Pyroptosis is a pro-inflammatory form of cell death that has been implicated in alcoholic-liver disease37, but not yet in alcohol-associated gut pathology. n3 PUFAs are precursors for specialized pro-resolving mediators (SPMs, e.g. RvD1, RvE1)38. Increased tissue n3 PUFA levels in fat-1 mice may lead to an increased pool of these SPMs39, which in turn may decrease the expression of pyroptosis genes in the intestinal mucosa as part of SPM-mediated resolution of inflammation. Previously, RvD1 treatment was shown to reduce pro-inflammatory cytokine levels in the ileum14. Fat-1 EtOH but not WT EtOH-fed mice also exhibited diminished expression of several genes encoding pro-inflammatory cytokines and chemokines that were previously implicated in multiple intestinal pathologies, including Cxcl1640, Il1541, and Ccl2042, suggesting that n3 PUFA enrichment may dampen the intestinal inflammatory response to EtOH.
We further identified a set of genes that were differentially regulated between fat-1 EtOH and WT EtOH-fed animals. Of interest, the expression of Socs3 and Ptk6 was downregulated in fat-1 EtOH mice relative to WT EtOH mice. Diminished expression of Socs3 is anti-inflammatory in Crohn’s disease43 and loss of Ptk6 expression enhances intestinal integrity44. Also, many immunoglobin genes (Igkv5-43, Igkv8-21, Igkv5-55, Igkv2-109, Igkv5-48, Igkv3-8, Igkv5-17) were downregulated in fat-1 EtOH relative to WT EtOH mice. The expression of genes at the Igkv loci determines the variable region of light chains on antibodies that bind to antigens (and therefore the specificity of antigen recognition)45. Decreased expression of Igkv genes may indicate that there are fewer antigens present in the intestine due to n3 PUFA enrichment. These results demonstrate that n3 PUFAs may negatively regulate transcription of these pro-inflammatory mediators and change the antibody repertoire after EtOH insult. In contrast, the expression of genes relating to intestinal lipid metabolism (Acaa1b and Acacb), sugar metabolism (Hk1, Akr1b8), and amino acid metabolism (Tph1, Tst, Cbs) were elevated in fat-1 EtOH relative to WT EtOH-fed mice. Preservation of metabolic gene expression may be one of the benefits of n3 PUFA enrichment after EtOH consumption.
n3 and n6 PUFAs may act as signaling molecules by activating several groups of receptors, including metabolite-sensing receptors of the FFAR and PPAR families15,16,46. In our study, we observed EtOH-induced expression of Ffar2 and Ffar4 in fat-1 but not WT mice. Since FFAR2 can positively regulate production of IL-22, a beneficial cytokine for intestinal health, as well as downstream antibacterial responses47, and FFAR4 activation is anti-inflammatory in IECs16, the up-regulation of these genes in fat-1 mice may contribute to the beneficial effects of a n3 PUFA-enriched environment in the intestine14. Pparγ, a nuclear receptor with anti-inflammatory functions in intestinal epithelial and immune cells48, and a critical regulatory role in mucosal defense49, was up-regulated by EtOH in both WT and fat-1 mice, most likely as an adaptive response to EtOH challenge.
Another interesting observation from our study was that the expression of several Tlrs, including Tlr1/2/4/5 were induced by EtOH in both WT and fat-1 mice. Moreover, fat-1 EtOH-fed mice had significantly increased Tlr2 and Tlr5 expression compared to WT EtOH-fed animals. Intestinal TLRs are important molecular regulators which protect the host from pathogens. They are involved in gut barrier function, tissue repair responses, and thus are critical for the maintenance of intestinal mucosal homeostasis. For example, it has been shown that activation of TLR2 enhanced intestinal epithelial barrier function and increased synthesis and release of IL-10, a critical anti-inflammatory cytokine50. Similarly, TLR5 activation by its agonist flagellin resulted in a transient transcription of genes encoding IL-17 and IL-22, which may be crucial for the early defenses against pathogen invasion of host tissues51. This would suggest that n3 PUFAs may enhance defense responses following an EtOH insult.
It is important to note that EtOH feeding differentially affected intestinal n6 and n3 PUFA levels in WT and fat-1 mice. A significant EtOH-mediated increase in n6 PUFAs (with a trend for n3 PUFAs) was observed in WT but not in fat-1 mice, suggesting that elevated levels of endogenous n6 PUFAs may also be responsible for EtOH-induced intestinal pathology in WT mice. This concept is supported by previous studies demonstrating that EtOH consumption and a diet enriched in n6 PUFA linoleic acid exacerbated intestinal inflammation and impaired gut barrier integrity and defense8,10. Elevated levels of various PUFAs after EtOH feeding in mice have been also reported in other organs such as the liver52. We do not have a definitive answer as to why there was an increase in n3 and n6-PUFAs in response to EtOH in our study, but it may be related to an overall disturbance in fatty acid metabolism, a hallmark of excessive alcohol consumption (e.g., decreased fatty acid oxidation53). Likely, fat-1 mice counteract the EtOH-mediated increase in n6-PUFAs through direct conversion to n3 PUFAs by the fat-1 desaturase enzyme.
Overall, our study provides a reference dataset of intestinal mucosa transcriptional responses to chronic EtOH exposure. These data will be instrumental in the formulation of specific hypotheses for follow-up studies potentially leading to new therapeutic interventions for intestinal pathological changes induced by EtOH. The study also demonstrated several favorable changes in the intestinal transcriptional repertoire in an n3 PUFA-enriched environment, which may ameliorate the deleterious effects of EtOH on intestinal homeostasis.
Methods
Study design
The study was approved by and performed in accordance with the guidelines of the University of Louisville Institutional Animal Care and Use Committee. The mice were handled and treated as previously described14. Male wild type (WT) C57Bl/6 mice and fat-1 littermates on a C57BL/6 background were used for this study. The fat-1 transgenic mouse line was obtained from Dr. J. X. Kang (Harvard Medical School) and has been described previously54. These mice express the n3 fatty acid desaturase gene fat-1, from C. elegans, in all tissues, leading to a direct conversion of n6 PUFAs to n3 PUFAs with a subsequent decrease in the n6/n3 PUFA ratio54. For the current study, the experimental mice were generated at the University of Louisville animal facility by breeding fat-1 heterozygotes to WT C57BL/6 mice to obtain fat-1 heterozygous and WT littermates. All mice were genotyped at weaning. Our pilot studies and reports from others54,55 confirmed that fat-1 mice exhibited elevated n3 PUFA levels and decreased n6/n3 PUFA ratio in various tissues. Expression of the fat-1 gene was validated by standard PCR (forward primer: 5′-CGGTTTCTGCGATGGATCCCAC-3′, reverse primer: 5′-CACAGGAACCGGGCAAAGAA-3′) on tail biopsies. The gene Gdf-5 was used as a positive control (forward primer: 5′-AAGCCCTCAGTCAGTTGTGC-3′, reverse primer: 5′-AAAACCATGAAAGGAGTGGG-3′). See Supplemental Fig. S2a,b for breeding scheme and representative results of genotype conformation by PCR, respectively. The 8–10 week old male WT and fat-1 littermates were placed on control or EtOH-containing Lieber-DeCarli liquid diets (BioServ, Flemington, NJ). The mice were fed for 6 weeks with a stepwise increase in EtOH concentration (0%, 1%, and 2% for two days each, 4% and 5% for one week each and then 6% for 3 weeks, see Supplemental Fig. S1a for experimental model). The control pair-fed mice (PF) received and consumed the same amount of isocaloric food (as determined by caloric intake with maltose dextrin substituting for EtOH) that EtOH-fed animals consumed in the previous day (pair-feeding protocol). Food consumption was recorded daily and body weights weekly. PF mice consumed the full amount of food provided each day (similar as EtOH-fed animals); the average daily food consumption by weeks is reported in Supplemental Fig. S1b. At the end of the feeding protocol, the mice were anesthetized with ketamine/xylazine (100/20 mg/kg) and euthanized by exsanguination and thoracotomy.
Intestinal tissue isolation and RNA preparation
After euthanasia, the intestines were removed, and the ileum was excised and thoroughly flushed with PBS. An approximately 1.5 cm segment of the terminal portion of the ileum was isolated and snap-frozen in liquid nitrogen, then stored at − 80 °C. See Supplemental Fig. S3 for the location of ileal segment used for RNA isolation. Total ileum RNA was isolated using Trizol reagent (Thermo Fisher, Waltham, MA) from 3–5 mice/group followed by removal of contaminating genomic DNA by digestion with DNase I (TURBO DNA-free kit, Thermo Fisher). RNA was further purified and concentrated using the GeneJET RNA cleanup and concentration micro kit (Thermo Fisher). RNA integrity was determined by the Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA). Only RNA samples with integrity values ranging from 7 to 9 were used for RNASeq analysis.
Intestinal tissue RNA-sequencing and bioinformatics
The transcriptome sequencing library preparation and sequencing was performed by the University of Louisville Center for Genetics in Molecular Medicine core facility using the TruSeq Stranded mRNA library prep kit (part no. 20020594). The full details for RNA Sequencing have been described previously14, but pertinent methodology is described below. Briefly, poly A purification and RNA fragmentation were performed on total RNA and Superscript II (Thermo-Fisher) was used to generate cDNA, with subsequent removal of the RNA template. Second strand synthesis was performed with dUTP to ensure stranded libraries, and double-stranded cDNA were purified with AMPure XP beads. 3′-Ends were adenylated, indexing adapters were ligated onto the ends, and libraries were once again purified with AMPure beads. Libraries were loaded onto an Agilent DNA 1000 chip and validated on an Agilent 2100 Bioanalyzer. Quantitation was performed with the Illumina Library Quantification Kit (Illumina Inc., San Diego, CA), ABI Prism qPCR Mix from Kapa Biosystems (Wilmington, MA). Three dilutions were tested in triplicate. Libraries were diluted to 10 nM, pooled, further diluted and denatured and analyzed on a NextSeq 500 v2 150 cycles High Output kit (cat#FC-404-2002) in a 2 × 75 bp PE.
RNA-seq bioinformatic and statistical analysis
Analysis was conducted by the NIH-funded Kentucky Biomedical Research Infrastructure Network Bioinformatics Core. The full details for this method have been published previously14, but pertinent details are described below. Sequences were directly aligned to the Mus musculus reference genome assembly (mm10.fa) using tophat2 (version 2.0.13)56 guided by Ensembl build 82 mouse transcripts. To confirm expression of the fat-1 transgene, we aligned each of the original reads to the C. elegans genome (ce10). A small fraction of the reads (less than 1%) mapped to C. elegans, as expected. We then extracted the reads from the region of the fat-1 gene and constructed a UCSC genome browser track for these regions. Differentially expressed genes (DEGs) between experimental groups were identified using the Tuxedo suite of programs including cuffdiff2 (version 2.2.1)57,58. A p-value cutoff ≤ 0.05, q-value cutoff ≤ 0.05 was used to determine differential expression. The full list of significant DEGs for each comparison can be found in Supplemental Materials Excel File S1. DEGs with a cutoff of ≥ two-fold change were used for further pairwise comparisons in Volcano Plots, GO Processes, and STRING Cluster Analysis. This threshold was used to obtain a higher magnitude of differences, and to identify more robust gene expression changes and the accompanying biological processes.
Cytoscape STRING plugin was used for visualizing molecular interactions between DEGs and identifying Gene Ontology:Biological Processes (GO:BPs) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways that include the DEGs59. Cytoscape analysis identified enriched GO:BPs (representative of DEGs increased) and diminished GO:BPs (representative of DEGs decreased) for their respective comparisons. The top ten enriched and diminished GO:BPs were reported (if 10 were identified) along with their associated false-discovery rate (FDR < 0.05). Similarly, Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) Analysis and gene clustering were conducted in CytoScape followed by KEGG Pathway enrichment (FDR < 0.05). Gene clusters and their associated KEGG Pathway were used for DEGs increased and decreased in the respective comparisons.
STRING cluster diagrams list gene clusters by biological pathways they represent. Genes with connections as determined by STRING analysis are within the same circle, and the associated process is listed near the cluster. Clusters with purple circles and green nodes were decreased for the respective comparison, while clusters with yellow circles and copper nodes were increased.
Measurement of n3 and n6 PUFAs
Fatty acids were measured in the intestines of the same mice as used in the RNAseq analysis. The assay was conducted at the Laboratory for Lipid Medicine and Technology at Massachusetts General Hospital and Harvard University according to a well-established protocol60 with slight modifications. Briefly, frozen ileum segments were ground under liquid nitrogen using a mortar and pestle. The samples were then processed for lipid extraction and methylation by adding 1:1 hexane and 14% boron trifluoride/methanol and heated at 100 °C for one hour. Fatty acid methyl esters were analyzed on a fully automated 6890 N Network Gas Chromatography Instrument equipped with a flame-ionization detector (Agilent Technologies, Palo Alto, CA). Individual fatty acid concentrations were determined by retention time using a reference standard, GLC461, and quantitated by a reference to an internal standard, C13:0 FAME (Nu-Chek Prep, Elysian, MN).
Supplementary information
Acknowledgements
The authors thank Marion McClain for manuscript editing. We also wish to acknowledge Dr. Shubha Gosh-Dastidar for her contributions to this project.
Author contributions
All authors have reviewed and approve this manuscript. Listed below are the fellow author contributions to this study. Conceptualization: I.K., C.M., D.W.; methodology: D.W., J.W., J.H., J.K.; formal analysis: J.H., J.W., D.W., Y.S., C.Y.C.; investigation: J.H., J.W., D.W., Y.S., C.Y.C.; resources: I.K., C.M., E.R.; data curation: E.R., I.K.; writing—original draft preparation: J.H., I.K.; writing—review and editing: J.W., D.W., I.K., C.M., E.R., C.Y.C., J.K.; visualization: J.H., I.K.; supervision: I.K., D.W.; project administration: I.K.; funding acquisition: I.K., C.M., E.R., J.H.
Data availability
The GEO accession number for the RNAseq data reported in this paper is GSE133253. All other data generated or analyzed during this study are included in this published article and its supplementary information file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Josiah E. Hardesty and Jeffrey B. Warner.
Supplementary information
is available for this paper at 10.1038/s41598-020-76959-6.
References
- 1.Francescangeli F, De Angelis ML, Zeuner A. Dietary factors in the control of gut homeostasis, intestinal stem cells, and colorectal cancer. Nutrients. 2019;11:2936. doi: 10.3390/nu11122936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugihara K, Morhardt TL, Kamada N. The role of dietary nutrients in inflammatory bowel disease. Front. Immunol. 2018;9:3183. doi: 10.3389/fimmu.2018.03183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spencer SP, Belkaid Y. Dietary and commensal derived nutrients: shaping mucosal and systemic immunity. Curr. Opin. Immunol. 2012;24:379–384. doi: 10.1016/j.coi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaliannan K, et al. Multi-omic analysis in transgenic mice implicates omega-6/omega-3 fatty acid imbalance as a risk factor for chronic disease. Commun. Biol. 2019;2:276. doi: 10.1038/s42003-019-0521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simopoulos AP. An increase in the omega-6/omega-3 fatty acid ratio increases the risk for obesity. Nutrients. 2016;8:128. doi: 10.3390/nu8030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ungaro F, Rubbino F, Danese S, D'Alessio S. Actors and factors in the resolution of intestinal inflammation: lipid mediators as a new approach to therapy in inflammatory bowel diseases. Front. Immunol. 2017;8:1331. doi: 10.3389/fimmu.2017.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirpich IA, et al. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol. 2008;42:675–682. doi: 10.1016/j.alcohol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirpich IA, et al. The type of dietary fat modulates intestinal tight junction integrity, gut permeability, and hepatic toll-like receptor expression in a mouse model of alcoholic liver disease. Alcohol Clin. Exp. Res. 2012;36:835–846. doi: 10.1111/j.1530-0277.2011.01673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leclercq S, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. USA. 2014;111:E4485–4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirpich IA, et al. Ethanol and dietary unsaturated fat (corn oil/linoleic acid enriched) cause intestinal inflammation and impaired intestinal barrier defense in mice chronically fed alcohol. Alcohol. 2013;47:257–264. doi: 10.1016/j.alcohol.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, P. et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology148, 203–214 e216. 10.1053/j.gastro.2014.09.014 (2015). [DOI] [PMC free article] [PubMed]
- 12.Donde H, et al. Tributyrin inhibits ethanol-induced epigenetic repression of CPT-1A and attenuates hepatic steatosis and injury. Cell Mol. Gastroenterol. Hepatol. 2019 doi: 10.1016/j.jcmgh.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirpich IA, et al. Saturated and unsaturated dietary fats differentially modulate ethanol-induced changes in gut microbiome and metabolome in a mouse model of alcoholic liver disease. Am. J. Pathol. 2016;186:765–776. doi: 10.1016/j.ajpath.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warner DR, et al. Decreased ω-6:ω-3 PUFA ratio attenuates ethanol-induced alterations in intestinal homeostasis, microbiota, and liver injury. J. Lipid Res. 2019;60:2034–2049. doi: 10.1194/jlr.RA119000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moniri NH. Free-fatty acid receptor-4 (GPR120): Cellular and molecular function and its role in metabolic disorders. Biochem. Pharmacol. 2016;110–111:1–15. doi: 10.1016/j.bcp.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anbazhagan AN, et al. A novel anti-inflammatory role of GPR120 in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2016;310:C612–C621. doi: 10.1152/ajpcell.00123.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukata M, Arditi M. The role of pattern recognition receptors in intestinal inflammation. Mucosal. Immunol. 2013;6:451–463. doi: 10.1038/mi.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otte JM, Cario E, Podolsky DK. Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology. 2004;126:1054–1070. doi: 10.1053/j.gastro.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 19.19Petrasek, J., Mandrekar, P. & Szabo, G. Toll-like receptors in the pathogenesis of alcoholic liver disease. Gastroenterol Res Pract2010. 10.1155/2010/710381 (2010). [DOI] [PMC free article] [PubMed]
- 20.Hwang DH, Kim JA, Lee JY. Mechanisms for the activation of Toll-like receptor 2/4 by saturated fatty acids and inhibition by docosahexaenoic acid. Eur. J. Pharmacol. 2016;785:24–35. doi: 10.1016/j.ejphar.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aherne CM, et al. Epithelial-specific A2B adenosine receptor signaling protects the colonic epithelial barrier during acute colitis. Mucosal. Immunol. 2015;8:1324–1338. doi: 10.1038/mi.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart ML, Jacobi B, Schittenhelm J, Henn M, Eltzschig HK. Cutting Edge: A2B Adenosine receptor signaling provides potent protection during intestinal ischemia/reperfusion injury. J. Immunol. 2009;182:3965–3968. doi: 10.4049/jimmunol.0802193. [DOI] [PubMed] [Google Scholar]
- 23.Cekic C, Linden J. Purinergic regulation of the immune system. Nat. Rev. Immunol. 2016;16:177–192. doi: 10.1038/nri.2016.4. [DOI] [PubMed] [Google Scholar]
- 24.Eltzschig HK, Sitkovsky MV, Robson SC. Purinergic signaling during inflammation. N. Engl. J. Med. 2012;367:2322–2333. doi: 10.1056/NEJMra1205750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strohmeier GR, Reppert SM, Lencer WI, Madara JL. The A2b adenosine receptor mediates cAMP responses to adenosine receptor agonists in human intestinal epithelia. J. Biol. Chem. 1995;270:2387–2394. doi: 10.1074/jbc.270.5.2387. [DOI] [PubMed] [Google Scholar]
- 26.Bujanda L. The effects of alcohol consumption upon the gastrointestinal tract. Am. J. Gastroenterol. 2000;95:3374–3382. doi: 10.1111/j.1572-0241.2000.03347.x. [DOI] [PubMed] [Google Scholar]
- 27.Perlow W, Baraona E, Lieber CS. Symptomatic intestinal disaccharidase deficiency in alcoholics. Gastroenterology. 1977;72:680–684. doi: 10.1016/S0016-5085(77)80154-6. [DOI] [PubMed] [Google Scholar]
- 28.Huang CC, et al. Effects of long-term ethanol consumption on jejunal lipase and disaccharidase activities in male and female rats. World J. Gastroenterol. 2005;11:2603–2608. doi: 10.3748/wjg.v11.i17.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richmond CA, Breault DT. Regulation of gene expression in the intestinal epithelium. Prog. Mol. Biol. Transl. Sci. 2010;96:207–229. doi: 10.1016/B978-0-12-381280-3.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez Sawicki, L. et al. FABP1 knockdown in human enterocytes impairs proliferation and alters lipid metabolism. Biochim Biophys Acta Mol Cell Biol Lipids1862, 1587–1594. 10.1016/j.bbalip.2017.09.006 (2017). [DOI] [PMC free article] [PubMed]
- 31.Si-Tahar M, Merlin D, Sitaraman S, Madara JL. Constitutive and regulated secretion of secretory leukocyte proteinase inhibitor by human intestinal epithelial cells. Gastroenterology. 2000;118:1061–1071. doi: 10.1016/s0016-5085(00)70359-3. [DOI] [PubMed] [Google Scholar]
- 32.Bermúdez-Humarán LG, et al. Serine protease inhibitors protect better than IL-10 and TGF-β anti-inflammatory cytokines against mouse colitis when delivered by recombinant lactococci. Microb. Cell Fact. 2015;14:26–26. doi: 10.1186/s12934-015-0198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyonaga T, et al. Lipocalin 2 prevents intestinal inflammation by enhancing phagocytic bacterial clearance in macrophages. Sci. Rep. 2016;6:35014–35014. doi: 10.1038/srep35014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadjsombati MS, et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity. 2018;49:33–41.e37. doi: 10.1016/j.immuni.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ting, H.-A. & von Moltke, J. The immune function of tuft cells at gut mucosal surfaces and beyond. J. Immunol. (Baltimore, Md.:1950)202, 1321–1329. 10.4049/jimmunol.1801069 (2019). [DOI] [PMC free article] [PubMed]
- 36.Li A, Li L, Wang W, Song K, Zhang G. Transcriptomics and fitness data reveal adaptive plasticity of thermal tolerance in oysters inhabiting different tidal zones. Front. Physiol. 2018;9:825. doi: 10.3389/fphys.2018.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khanova E, et al. Pyroptosis by caspase11/4-gasdermin-D pathway in alcoholic hepatitis in mice and patients. Hepatology. 2018;67:1737–1753. doi: 10.1002/hep.29645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serhan CN, Yacoubian S, Yang R. Anti-inflammatory and proresolving lipid mediators. Annu. Rev. Pathol. 2008;3:279–312. doi: 10.1146/annurev.pathmechdis.3.121806.151409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudert CA, et al. Transgenic mice rich in endogenous omega-3 fatty acids are protected from colitis. Proc. Natl. Acad. Sci. USA. 2006;103:11276–11281. doi: 10.1073/pnas.0601280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diegelmann J, et al. Expression and regulation of the chemokine CXCL16 in Crohn's disease and models of intestinal inflammation. Inflamm. Bowel Dis. 2010;16:1871–1881. doi: 10.1002/ibd.21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, Z. et al. IL-15 is highly expressed in inflammatory bowel disease and regulates local T cell-dependent cytokine production. J. Immunol. (Baltimore, Md.: 1950)164, 3608–3615. 10.4049/jimmunol.164.7.3608 (2000). [DOI] [PubMed]
- 42.Kaser A, et al. Increased expression of CCL20 in human inflammatory bowel disease. J. Clin. Immunol. 2004;24:74–85. doi: 10.1023/B:JOCI.0000018066.46279.6b. [DOI] [PubMed] [Google Scholar]
- 43.Cheng X, et al. miR-19b downregulates intestinal SOCS3 to reduce intestinal inflammation in Crohn's disease. Sci. Rep. 2015;5:10397–10397. doi: 10.1038/srep10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haines RJ, Beard RS, Jr, Eitner RA, Chen L, Wu MH. TNFα/IFNγ mediated intestinal epithelial barrier dysfunction is attenuated by MicroRNA-93 downregulation of PTK6 in mouse colonic epithelial cells. PLoS ONE. 2016;11:e0154351–e0154351. doi: 10.1371/journal.pone.0154351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins AM, Watson CT. Immunoglobulin light chain gene rearrangements, receptor editing and the development of a self-tolerant antibody repertoire. Front. Immunol. 2018;9:2249–2249. doi: 10.3389/fimmu.2018.02249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marion-Letellier R, Dechelotte P, Iacucci M, Ghosh S. Dietary modulation of peroxisome proliferator-activated receptor gamma. Gut. 2009;58:586–593. doi: 10.1136/gut.2008.162859. [DOI] [PubMed] [Google Scholar]
- 47.Chun E, et al. Metabolite-sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity. 2019;51:871–884.e876. doi: 10.1016/j.immuni.2019.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasan AU, Rahman A, Kobori H. Interactions between host PPARs and gut microbiota in health and disease. Int. J. Mol. Sci. 2019;20:387. doi: 10.3390/ijms20020387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peyrin-Biroulet L, et al. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc. Natl. Acad. Sci. USA. 2010;107:8772–8777. doi: 10.1073/pnas.0905745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Latorre E, et al. Toll-like receptors 2 and 4 modulate intestinal IL-10 differently in ileum and colon. United European Gastroenterol. J. 2018;6:446–453. doi: 10.1177/2050640617727180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Maele L, et al. TLR5 signaling stimulates the innate production of IL-17 and IL-22 by CD3(neg)CD127+ immune cells in spleen and mucosa. J. Immunol. 2010;185:1177–1185. doi: 10.4049/jimmunol.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puri P, et al. Alcohol produces distinct hepatic lipidome and eicosanoid signature in lean and obese. J. Lipid. Res. 2016;57:1017–1028. doi: 10.1194/jlr.M066175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You M, Arteel GE. Effect of ethanol on lipid metabolism. J. Hepatol. 2019;70:237–248. doi: 10.1016/j.jhep.2018.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang JX, Wang J, Wu L, Kang ZB. Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature. 2004;427:504. doi: 10.1038/427504a. [DOI] [PubMed] [Google Scholar]
- 55.Kaliannan K, Wang B, Li XY, Kim KJ, Kang JX. A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci. Rep. 2015;5:11276. doi: 10.1038/srep11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang JX, Wang J. A simplified method for analysis of polyunsaturated fatty acids. BMC Biochem. 2005;6:5. doi: 10.1186/1471-2091-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEO accession number for the RNAseq data reported in this paper is GSE133253. All other data generated or analyzed during this study are included in this published article and its supplementary information file.