Abstract
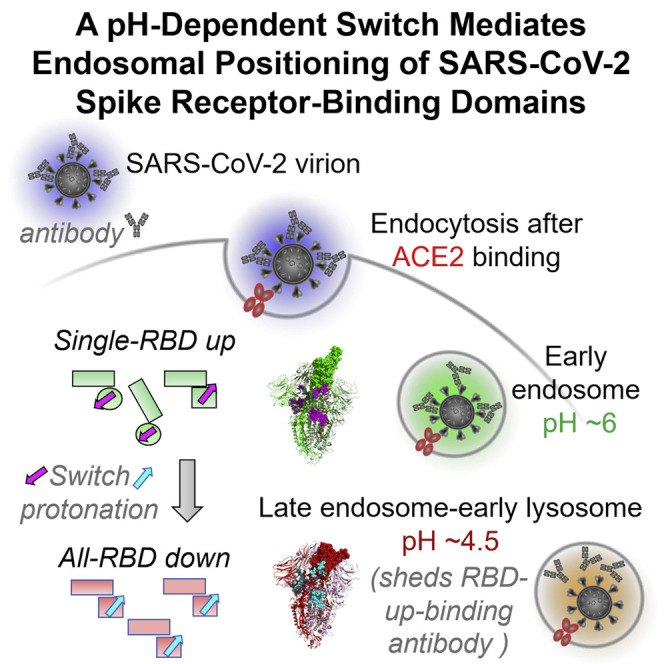
The SARS-CoV-2 spike employs mobile receptor-binding domains (RBDs) to engage the human ACE2 receptor and to facilitate virus entry, which can occur through low-pH-endosomal pathways. To understand how ACE2 binding and low pH affect spike conformation, we determined cryo-electron microscopy structures—at serological and endosomal pH—delineating spike recognition of up to three ACE2 molecules. RBDs freely adopted “up” conformations required for ACE2 interaction, primarily through RBD movement combined with smaller alterations in neighboring domains. In the absence of ACE2, single-RBD-up conformations dominated at pH 5.5, resolving into a solitary all-down conformation at lower pH. Notably, a pH-dependent refolding region (residues 824–858) at the spike-interdomain interface displayed dramatic structural rearrangements and mediated RBD positioning through coordinated movements of the entire trimer apex. These structures provide a foundation for understanding prefusion-spike mechanics governing endosomal entry; we suggest that the low pH all-down conformation potentially facilitates immune evasion from RBD-up binding antibody.
Keywords: ACE2 receptor, COVID-19, endosomal entry, pH-dependent switch, receptor-binding domain (RBD), structural rearrangement, type 1 fusion machine
Graphical Abstract
Highlights
-
•
Determine cryo-EM structures of SARS-CoV-2 spike along its endosomal entry pathway
-
•
Reveal structural basis by which a pH-dependent switch mediates RBD positioning
-
•
Show spike to exclusively adopt an all-RBD-down conformation at low pH
-
•
Suggest low-pH all-RBD-down conformation to provide a basis for immune evasion
Zhou et al. determine 12 structures of the SARS-CoV-2 spike, bound by ACE2 receptor and ligand free, that reveal a pH-dependent switch to mediate positioning of spike receptor-binding domains (RBDs). At low pH, the spike adopts an all-RBD-down conformation, which provides a potential means of immune evasion from RBD-up-recognizing antibody.
Introduction
Like other beta-coronaviruses, entry by SARS-CoV-2 involves its trimeric spike glycoprotein, a type 1 fusion machine that undergoes large-scale conformational changes between prefusion and postfusion conformations to facilitate fusion of viral and host cell membranes. The exact entry process for SARS-CoV-2 is still being defined but is known to involve spike interaction with the ACE2 receptor (Lan et al., 2020; Shang et al., 2020b; Wang et al., 2020; Zhou et al., 2020a). Entry can occur endosomally as well as at the cell surface, with inhibition of both the endosomal cathepsin L and the cell-surface TMPRSS2 required to inhibit fully SARS-CoV-2 entry (Hoffmann et al., 2020; Ou et al., 2020); cleavage of the spike by furin between S1 and S2 subunits can also occur, but furin cleavage does not appear to be essential for entry and occurs distal from the fusion peptide. Cryo-electron microscopy (cryo-EM) structures reveal two prevalent prefusion conformations for uncleaved and furin-cleaved SARS-CoV-2 spikes (Walls et al., 2020; Wrapp et al., 2020; Wrobel et al., 2020): a single-up conformation and an all-down conformation, related to the positioning of the receptor-binding domains (RBDs) in the S1 subunit. The “up” positioning of RBD is needed for interaction with ACE2 receptor and is also related to the epitope availability of RBD-directed antibodies. A postfusion structure (Cai et al., 2020), furthermore, confirms extensive structural rearrangements between prefusion and postfusion conformations. However, even within the prefusion state, the precise mechanism governing RBD positioning and the effect of ACE2 binding are still being delineated.
In addition to entry, accumulating data indicate the SARS-CoV-2 spike to be involved in evasion from the humoral immune response. Antibodies such as CR3022, with sub-nanomolar apparent affinity for a cryptic epitope on RBDs, fail to neutralize (Yuan et al., 2020); even among neutralizing antibodies, affinity to spike correlates only weakly with neutralization (Liu et al., 2020); moreover, neutralizing antibodies often show little somatic hypermutation, suggesting impaired maturation (Brouwer et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Seydoux et al., 2020). Although the high glycan density of the spike (Watanabe et al., 2020) might provide partial explanation for reduced immunogenicity, it is not clear why antibodies with high affinity for spike fail to neutralize.
To provide insight into receptor interactions and RBD positioning along the endosomal entry pathway, we determined 12 cryo-EM structures of the spike, alone or in complex with the ACE2 receptor, at serological and endosomal pHs. We used these structures to quantify domain movements and to delineate the interprotomer mechanism that mediates endosomal positioning of RBDs. We supplemented these structural studies with biochemical analyses that suggest a potential spike-based means of immune evasion. Overall, our findings provide structural definition for the binding of up to three ACE2 molecules per SARS-CoV-2 spike trimer and define a key structural transition between pH 5.5 and 4.5, highlighting the crucial role of a refolding region with multiple aspartic acid residues—a pH-dependent switch—that mediates positioning of RBDs, by twisting the relative orientation of disulfide-linked helices and coordinating the interprotomer movement of domains.
Results
Cryo-EM Structures of SARS-CoV-2 Spike with ACE2 at Serological and Endosomal pH
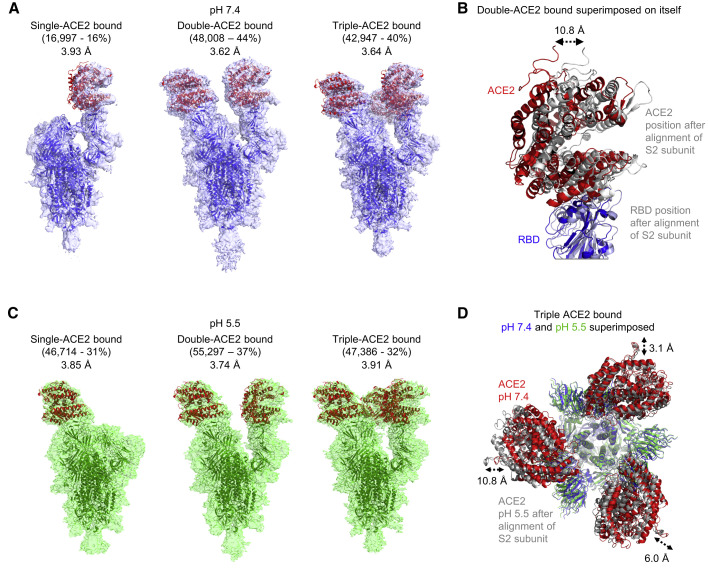
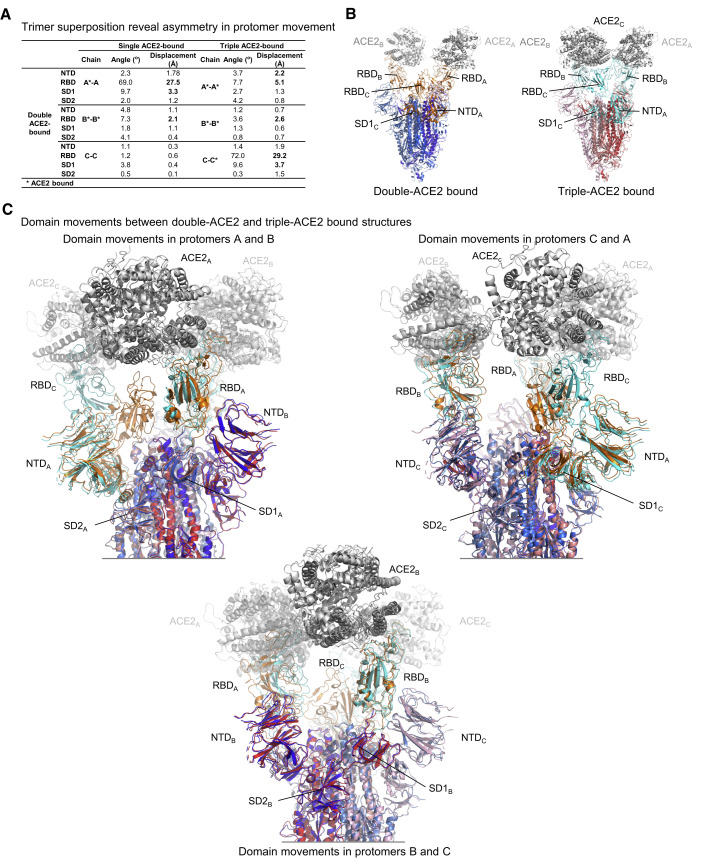
To provide structural insight into the recognition between ACE2 and prefusion spike trimer, we chose to focus on prefusion-stabilized spikes and analyzed the “2P”-prefusion stabilized soluble trimeric ectodomain (spike), which includes GSAS and PP mutations along with the T4 phage fibritin trimerization domain (Wrapp et al., 2020). We mixed soluble monomeric ACE2 with spike trimer at a 6:1 molar ratio at pH 7.4 and collected single-particle cryo-EM data on a Titan Krios. We obtained structures at 3.6–3.9 Å resolution and observed spike to bind ACE2 at stoichiometries of 1:1, 1:2, and 1:3, with prevalences of 16%, 44%, and 40%, respectively (Figures 1 A and S1; Table S1). ACE2 binding introduced trimer asymmetry. First, comparison of single-RBD-up conformations of the spike, for ACE2-bound and ligand-free structures, revealed recognition of ACE2 to induce a small movement of RBD (Figure S2). Second, although the membrane-proximal region of the spike in these complexes remained 3-fold symmetric, the ACE2-binding regions showed asymmetry with, for example, superposition of the double-ACE2-bound complex onto itself on the basis of membrane-proximal regions leading to displacement of ACE2 molecules by almost 11 Å (Figure 1B). The full complement of trimer superpositions (Figure S3A) revealed preferential ways to align trimers moving from single- to double- to triple-ACE2-bound conformations. Analysis of domain movements indicated the large movement of RBD (from “down” to “up” conformation) required to accommodate ACE2 binding to be accompanied by more subtle movements of neighboring domains (Figure 2 A), and we delineated the coordinated interprotomer domain movements that were involved in raising RBD (Figures 2B, 2C, and S3). However, the largest movement in S2 between single- and triple-ACE2-bound spikes occurred at the flexible C terminus of S2, with an overall root-mean square deviation (rmsd) for the S2 subunit of < 1 Å between single-, double-, and triple-ACE2-bound trimers (Figures S3A and S3B). Thus, ACE2-receptor engagement required RBD to be in the up position, but we could see no clear evidence for the binding of ACE2 priming S2 for substantial structural rearrangement, beyond the raising of RBD and a reduction of RBD interactions with S2.
Figure 1.
Cryo-EM Structures of SARS-CoV-2 Spike with ACE2 Show Similar Stoichiometries at Serological and Endosomal pH
(A) Cryo-EM structures of spike with single-, double-, or triple-bound ACE2 at serological pH.
(B) Structural comparison of the two ACE2-RBD in the double-ACE2-bound structure reveals different tilt angles resulting in as much as a 10.8 Å displacement as indicated.
(C) Cryo-EM structure of spike and ACE2 at endosomal pH.
(D) Comparison of triple-ACE2-bound spikes at serological and endosomal pH. Structures were aligned by S2-subunit superposition and are displayed with the trimer perpendicular to the page and with spike colored blue and green according to pH, and ACE2 colored red and gray for pH 7.4 and 5.5, respectively. Monomeric ACE2 was used as a ligand in these samples.
Figure 2.
Coordinated Inter-Protomer Domain Movements Assist in Raising RBD to Bind ACE2
(A) Domain movements between single-, double- and triple-bound ACE2.
(B and C) ACE2-induced conformational change. Protomers in the double-ACE2 bound spike are colored in shades of blue whereas those in the triple-ACE2-bound spike are colored in shades of red. Domains that move over 2 Å are colored orange and cyan for the double- and triple-ACE2-bound spikes, respectively.
See also Figures S2 and S3.
To provide insight into the effect of endosomal pH, we again mixed soluble monomeric ACE2 with spike trimer at a 6:1 molar ratio, but this time at pH 5.5, and determined the structure of the complex by using cryo-EM. Similar to serological pH, we obtained structures at 3.7–3.9 Å resolution and observed spike to bind ACE2 at stoichiometries of 1:1, 1:2, and 1:3 with prevalences of 31%, 37%, and 32%, respectively (Figures 1C and S1; Table S1). We superposed triple-ACE2-bound complexes determined at pH 7.4 and pH 5.5 and observed the membrane-proximal regions of the spike to align closely, whereas ACE2 molecules showed displacements of 3.1, 6.0, and 10.8 Å (Figure 1D). Overall, structures of the spike with ACE2 showed substantial populations of single-, double-, and triple-ACE2-bound spikes at both serological and endosomal pH. The observed distribution of spike bound to single-, double- and triple-ACE2s suggests—in the context of 2P-stabilization with S1-S2-cleavage site removed and in the presence of excess ACE2—the lack of a substantial energy barrier between different ACE2-spike stoichiometries.
Ligand-Free Cryo-EM Structures of SARS-CoV-2 Spike at Low pH
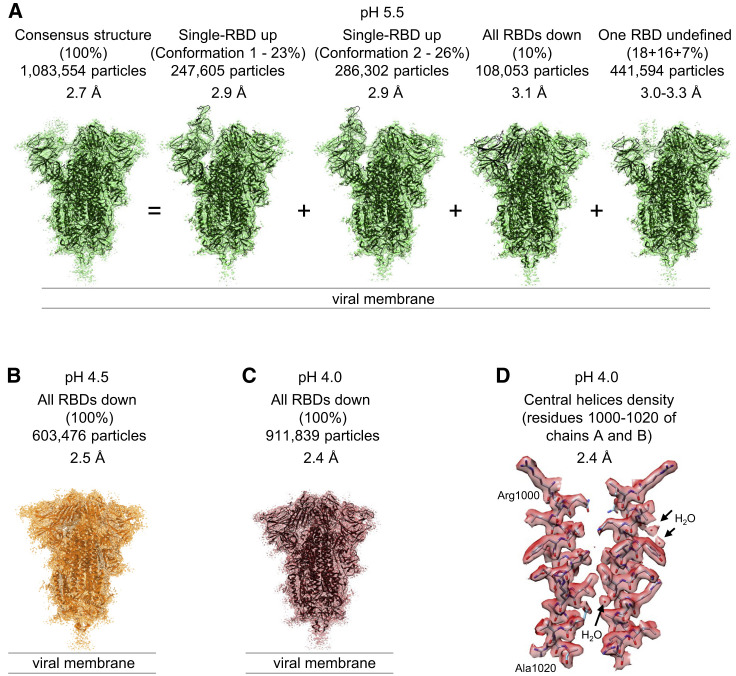
In light of the similarity of ACE2 complexes at pH 7.4 and 5.5 (Figure 1), we decided to analyze the structure of the ligand-free spike at pH 5.5 by single-particle cryo-EM. We determined a consensus structure from 1,083,554 particles at a resolution of 2.7 Å, in which most of the spike was well resolved, except for a lone RBD for which reconstruction density was poor (Figures 3 A and S4; Table S2). Analysis of structural heterogeneity in this region (Videos S1, S2, S3, and S4; Figure S4CA) produced 6 3D classes ranging in prevalence from 7% to 26% and describing 3 principal conformations, and the RBD was in the up or down position or without a defined position (Figure S4CB). Interestingly, unlike for ACE2-bound complexes, no double- or triple-RBD-up conformations were observed. Two classes with prevalences of 23% (Conformation 1, 2.9 Å resolution) and 26% (Conformation 2, 2.9 Å resolution) corresponded to two different single-RBD-up conformations. A third prevalent class representing 10% of the particles had all RBDs down. For all three of these prevalent classes, unlike the consensus structure, density for all RBD domains was well resolved (Figure S4CC), indicating multiple orientations of RBD in the spike at pH 5.5. In the remaining classes, the RBD did not assume a defined position, suggesting RBD mobility at pH 5.5.
Figure 3.
Cryo-EM Analyses Reveal Lower pH to Reduce Spike-Conformational Heterogeneity Culminating in an All RBD-Down Conformation at pH 4.0
(A) Structures at pH 5.5 with particle prevalence and resolution of determined structures.
(B) Structure of spike at pH 4.5.
(C) Structure of spike at pH 4.0.
(D) Example of reconstruction density. A region at the central helices of the pH 4.0 structure is shown with well-defined water molecules. The contour level is 0.015 (5.7 σ).
See also Figure S4, Tables S2 and S3, and Videos S1, S2, S3, and S4.
A ratcheting motion of one NTD domain results in increased mobility of the corresponding RBD (see Method Details for a detailed description). A corresponding top view is presented in Video S2.
The RBD is up and alternates between two positions (see Method Details for a detailed description). A corresponding side view is presented in Video S1.
A ratcheting motion of one NTD domain results in increased mobility of the corresponding RBD (see Method Details for a detailed description). A corresponding top view is presented in Video S4.
The RBD alternates between up and down positions (see Method Details for a detailed description). A corresponding side view is presented in Video S3.
To determine how even lower pH affected spike conformational heterogeneity, we sought to obtain a cryo-EM structure of the ligand-free spike at even lower pH. We collected cryo-EM datasets at both pH 4.5 and 4.0. Single-particle analysis of the pH 4.5 dataset comprising 603,476 particles resolved into an all-RBD-down conformation, and we refined this map to 2.5 Å resolution (Figures 3B and S4; Table S3); single-particle analysis of the pH 4.0 dataset comprising 911,839 particles resolved into a virtually identical all-RBD-down conformation (rmsd between the two structures of 0.9 Å) (Figures 3C and S4; Table S3). The similarity of the pH 4.5 and pH 4.0 structures indicated spike conformational heterogeneity to be reduced between pH 5.5 and 4.5, and then to remain unchanged as pH was reduced further. The pH 4.0 map was especially well-defined at 2.4 Å resolution (Table S3), enabling individual water molecules to be observed (Figure 3D), and we chose the pH 4.0 structure for comparative analysis.
Refolding at Spike Domain Interfaces Underlies Conformational Rearrangement
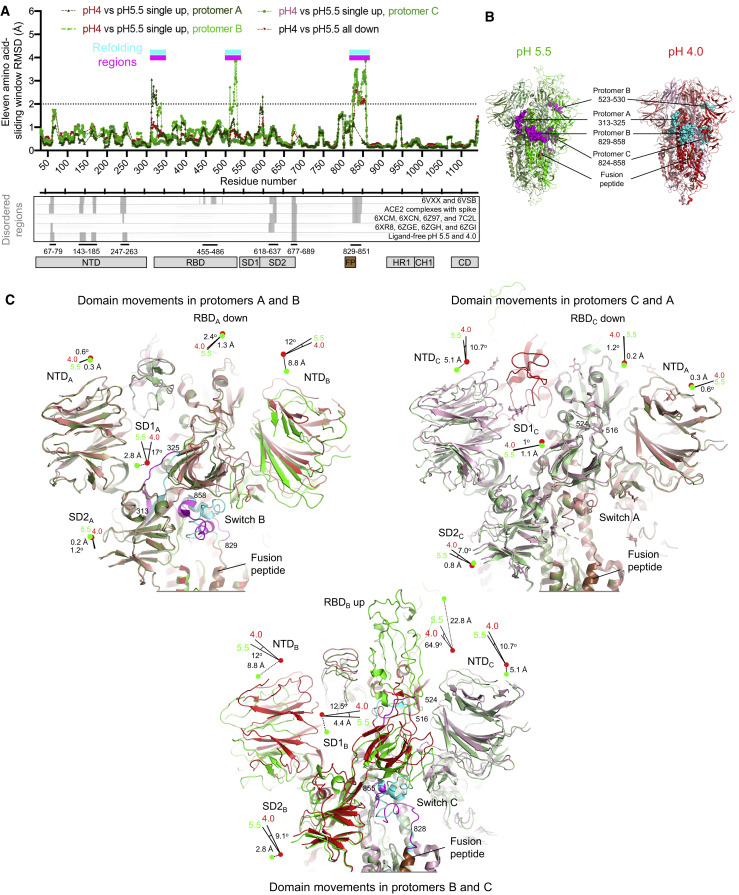
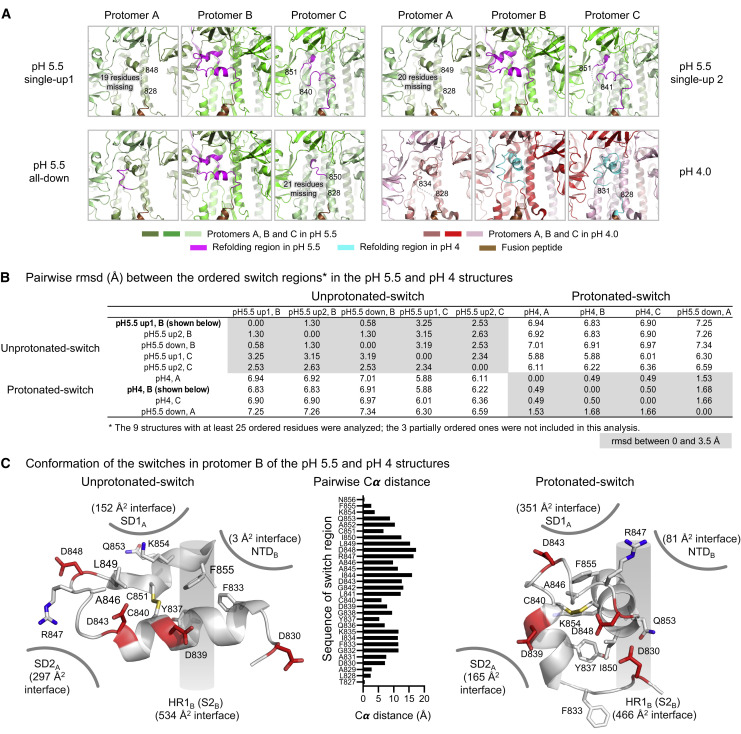
To identify critical components responsible for the reduction of conformational heterogeneity between pH 5.5 and lower pH and to shed light on the spike mechanism controlling the positioning of RBDs, we analyzed rmsds between the pH 5.5 structures and the all-down pH 4.0 conformation with an 11-residue sliding window to identify regions that refolded (Figures 4 A, top, and S5). Given that each protomer in the trimer displayed a different conformation in each of the pH 5.5 structures, we defined protomer B as the one with RBD in the up position in each of the single RBD-up conformations, and protomers A–C as appearing counter-clockwise when viewed along the trimer 3-fold axis toward the membrane. We observed significant rmsd peaks for short stretches around residue 320 in protomer A only and around residue 525 in protomer B only, and more substantially in a region comprising residues 824–858 (Figure S5B). This region, which for reasons described below we named the “switch” region, was fully defined in protomer B and partially resolved in protomer A (residues 824–828 and 848–858) and protomer C (residues 824–841 and 851–858). Notably, this region was almost entirely unresolved in our structures with ACE2 and in most published spike structures (Figure 4A, bottom), suggestive of substantial mobility.
Figure 4.
A Switch Domain Mediates RBD Position
(A) Identification of refolding regions through rmsd analysis with a 11-residue window (top) and comparison of disordered regions in cryo-EM structures (bottom).
(B) Refolding regions identified by sliding-window rmsd analysis are highlighted on the pH 5.5 single-up and pH 4.0 structures as spheres and are colored magenta and cyan, respectively. Protomers A, B, and C of the pH 5.5 structure are each colored smudge, green, or pale green, and the corresponding protomers in the pH 4.0 structure are colored salmon, red, or light pink, and fusion peptide is colored brown.
(C) Domain movements between pH 5.5 and 4.0. Three views are shown to depict the movements at the interfaces of protomers A-B, B-C, and C-A. Extent and direction of rotation and displacement are indicated for each domain with vectors and colored dots. Refolding regions are labeled and colored as in (B).
The asymmetry in distribution of refolding regions in the trimer between single-up and all-down structures (Figure 4B) suggested the up RBD to depend on concerted adjustments throughout the trimer. To delineate these, we determined angles and rigid-body translations between each of the subdomains (Table S4) for pH 5.5 single-RBD-up and 4.0 all-RBD-down structures. For clarity, we specify by subscript the protomer of each subunit or residue. Starting with the subdomain 1 of protomer A (SD1A) at the entrance loop of protomer A, and moving laterally around the trimer (Figure 4C, Video S5), we observed slight refolding in the 313–325A stretch, allowing a 17° rotation of SD1A to accommodate the switch region on the neighboring B protomer (switch B). At pH 5.5, switch B interacted with subdomain 2 of protomer A (SD2A) (buried surface area of ∼300 Å2), and this key inter-protomer contact coupled with SD1A rotation and 2.8 Å translation resulted in the 8.8 Å lateral displacement of N-terminal domain of protomer B (NTDB) toward the next RBD-switch (RBDB and switch C). The displaced NTDB induced consecutive shifts of SD2B and SD1B domains, which culminated in the 22.8 Å up translation (64.9° rotation) of RBDB versus its down equivalent.
The up positioning of RBDB was accommodated in part by a 5.1 Å mostly downward displacement of NTDC toward the viral membrane, which—continuing to the next RBD-switch (RBDC and switch A)—induced minor shifts of SD2C and SD1C domains and yielded RBDC and switch A in conformations that closely resembled those of the all-down pH 4.0 structure.
At pH 4.0, each of the RBD-switches closely resembled each other. The most dramatic refolding in relation to the switches at pH 5.5 occurred in switch B, where the guanidinium of residue Arg847B swivels over 25 Å from interacting in an inter-protomer manner with SD2A to interacting in an intra-protomer manner with NTDB of the same protomer. This swiveling breaks the coordinated displacements of domains across the protomer-protomer interface, reducing the SD2A interaction with switch B by half (buried surface area of ∼160 Å2).
Notably, refolding regions were observed to reside at critical inter-protomer contacts or at key joints between domains, especially the SD2 to SD1 joint, which cradles the switch of the neighboring protomer, and the SD1B joint with up-RBDB made up of refolding residues 523–530B.
A pH-Dependent Switch Domain Locks Spike in Down Position
The switch domain, which included aspartic acid residues at 830, 839, 843, and 848 and a disulfide linkage between Cys840 and Cys851, was located at the nexus of SD1 and SD2 from one protomer, and HR1 (in the S2 subunit) and NTD from the neighboring protomer. This region showed dramatic conformational changes (Figure 5 A). Pairwise rmsd comparisons (Figure 5B) indicated the cryo-EM-determined switch structures to segregate into two conformations: “unprotonated-switches” and “protonated-switches.”
Figure 5.
The pH-Switch Domain
(A) Switches in the pH 5.5 and pH 4.0 structures. The protomers and switches were colored as in Figure 4B. Disordered regions of the switches are shown as gray dashed lines and marked by flanking residue numbers.
(B) Pairwise rmsd between switch regions (residues 824–858) from different protomers. Of the 12 protomers determined in this study, only 9 had at least 25 ordered residues and were included in this pairwise-rmsd analysis; rmsds of less than 3.5 Å shaded gray. Switch regions for SARS-CoV-2 spike at higher pH were recently described (Cai et al., 2020; Wrobel et al., 2020), and these and switch regions from other coronaviruses are analyzed in Figure S7.
(C) Comparison of the unprotonated and protonated switches. Key residues are shown in stick representation, and Asp and Cys residues are colored red and yellow, respectively. Interactive surface areas with surrounding domains indicated. Pairwise Cα-distances between switch residues is shown in the middle.
See also Figures S5 and S7 and Tables S4 and S5.
Unprotonated-switches were exemplified by switches B and C at pH 5.5 and perhaps best by switch B in the pH 5.5 single-RBD-up structure (Figures 5A and 5C, left; Video S5). Continuing from fusion peptide (FPB), the N terminus of switch B formed several helical turns (833–842), extending laterally from HR1B to SD2A. A turn (843–848) provided extensive contacts with SD2A, before returning in helical turns (849–855) back to HR1B. Unprotonated-switches were stabilized by a hydrophobic core comprising the disulfide and residues Phe833, Tyr837, Ala846, Leu849, and Phe855 (Figure 5C, left). Notably, all four of the unprotonated-switch aspartic acids faced solvent and appeared to be negatively charged.
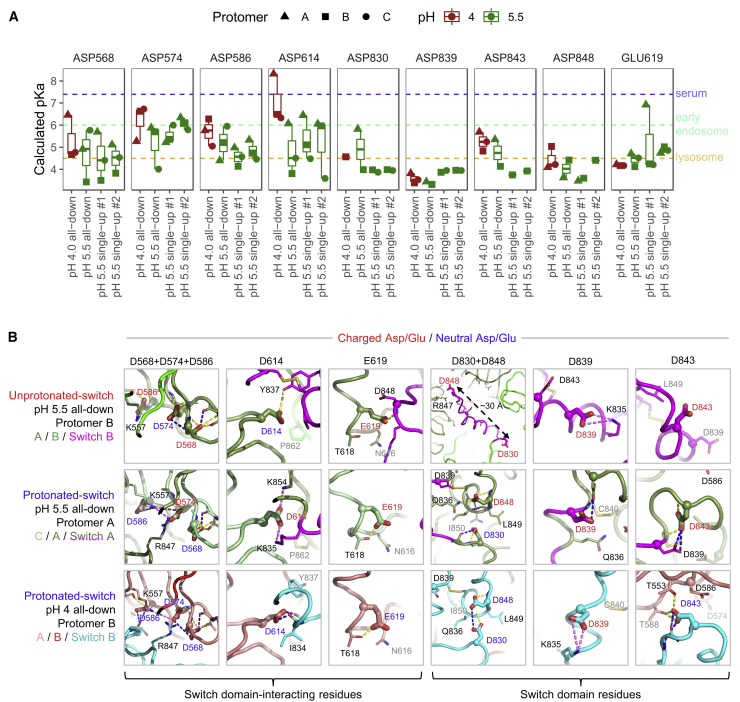
Protonated-switches were exemplified by switch A at pH 5.5 and by all switches at pH 4.0 (Figures 5A and 5C, right; Video S5). These switches reoriented their N-terminal helical turns to point toward SD1, swiveling the Cα-position of Arg847 over 15 Å to interact with NTD (Figure 5C and Table S5) before finishing the rest of the domain with a few helical turns (848–855). Protonated switches were stabilized by a hydrophobic core comprising Tyr837, Ile850, and aliphatic portions of the side chain from Lys854 on one side of the disulfide and Ala846 and Phe855 on the other. Notably, two of the switch domain Asp residues that appeared most likely to be protonated based on hydrogen-bonding patterns in the pH 4.0 structure (D830 and D843) also had higher calculated pKas than the unprotonated switch conformation, consistent with their observed hydrogen bonds and their apparent protonation at pH 5.5 (Figure 6 ). Additionally, three Asp residues from the neighboring protomer (D574, D586, and D614) had higher pKas in protonated-switch conformations than in unprotonated-switch conformations. In general, our pKa calculations and structural analyses both indicated increased Asp residue protonation in the protonated-switch conformation, and reflected the expected trend of increasing numbers of protonated Asp or Glu residues at lower pH.
Figure 6.
pKa Calculations for the pH-Switch Domain
(A) PROPKA-calculated pKas for pH-dependent switch domain residues in the pH 4.0 and 5.5 unliganded spike structures. pKas are plotted for titratable residues within and interacting with the 824–858 pH-dependent switch domain for in each structure, disordered regions excluded. Typical pH values for serum (7.4), early endosome (6.0), and late endosome (4.5) are indicated by dashed lines.
(B) Close-up views of Asp/Glu residues in (A) from the pH 4.0 and pH 5.5 structures depict changes in chemical environment for each residue between conformations. View angles with respect to superposed structures are the same within each residue column. Switch domain and surrounding protomers are colored as indicated at left. Highlighted residues are shown as thick sticks with labels colored based on pKa-based dominant protonation state at the structure pH: charged Asp/Glu in red, and neutral (protonated) Asp/Glu in blue. Residues within 4 Å are shown as thin sticks. Dashed lines indicate hydrogen bonds (yellow) or salt bridge interactions (violet), and hydrogen bonds requiring carboxylic acid group protonation are shown in blue. The pKa shifts between unprotonated- and protonated-switch conformations define a pH-dependent stability gradient that favors the protonated-switch form at lower pHs (Yang and Honig, 1993). However, other factors such as global conformational constraints might also play a role in favoring one conformation over another.
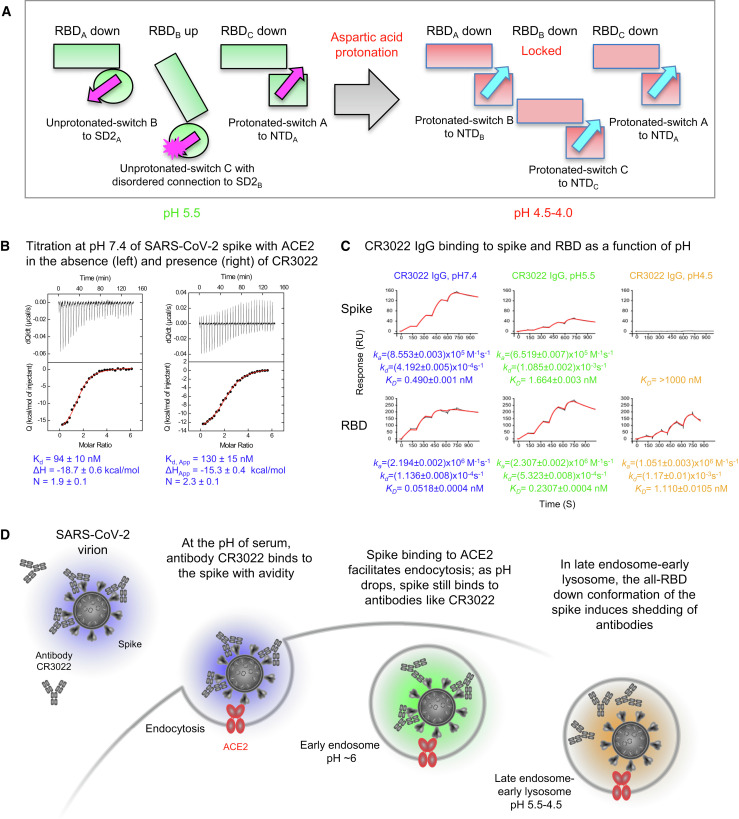
Analysis of switch domain conformations and RBD positions (Figure 7 A) indicated a concordance between switches interacting with NTD (breaking coordinated interprotomer interactions) and the locking of RBDs in the down position. Thus, at pH 5.5, the unprotonated-switches in protomers B and C interacted with the SD2 domain of the neighboring protomer to transmit lateral displacements of domains. At lower pH, the protonated-switches interrupt this interprotomer interaction, resulting in the locking of RBDs in the down position.
Figure 7.
SARS-CoV-2 Spike at Serological pH Binds ACE2 and CR3022 and at Lower pH Still Binds ACE2 but Not CR3022
(A) Schematic of the pH-switch locking of RBD in the down position.
(B) Isothermal titration calorimetry at pH 7.4 of ACE2 recognizing spike (left) or spike previously titrated with Fab CR3022 (right). Measurements were performed in duplicate; uncertainties are the average standard errors obtained by fitting to the two independent datasets.
(C) Apparent affinities of spike (top) and real affinities of RBD (bottom) to CR3022 IgG as a function of pH as measured by SPR.
(D) Schematic showing ACE2-dependent endosomal entry of SARS-CoV-2 and the pH-dependent shedding of antibodies like CR3022.
See also Figure S6.
Binding of ACE2 Receptor and CR3022 Antibody at Serological and Endosomal pH
We investigated the influence of these pH-dependent conformational changes on binding of antibody. For variation in pH over the course of virus entry to affect the binding of antibody, the antibody would need to allow spike recognition of the ACE2 receptor, thereby enabling the virus to initiate endosomal entry. We chose to focus on antibody CR3022, an antibody originally identified to neutralize SARS (ter Meulen et al., 2006), given that this antibody was shown to bind SARS-CoV-2 spike with sub-picomolar affinity (Yuan et al., 2020) and to induce spike disassembly (Huo et al., 2020), yet did not neutralize SARS-CoV-2 (Yuan et al., 2020).
CR3022 has been shown to not inhibit RBD binding to ACE2 (Yuan et al., 2020), but this had not been shown with spike. We used isothermal titration calorimetry (ITC) to determine whether binding of CR3022 to spike was compatible with ACE2 interaction at serological pH. We chose to use a monomeric version of ACE2 to test more sensitively the effect of antibody inhibition. First, we titrated ACE2 into soluble spike, and observed 1.9 ACE2 molecules to bind per spike trimer, with an affinity of 94 nM (Figure 7B, left). Next, we titrated soluble spike with the antigen-binding fragment (Fab) of CR3022 until saturation (Figure S6A) and further titrated ACE2 into the spike-CR3022 complex formed to observe 2.3 ACE2 molecules to bind each spike-CR3022 complex, with an affinity of 130 nM (Figure 7B, right). Thus, at serological pH, the SARS-CoV-2 spike appeared capable of recognizing ACE2 even in the presence of antibody CR3022, indicating that CR3022-bound spikes could initiate endosomal-based ACE2-dependent entry.
To gain insight into the effect of endosomal pH on ACE2 and CR3022 interactions with spike, we characterized their binding to both spike and RBD, expressed as a separate molecule. For these experiments, we chose to use dimeric ACE2 to more closely mimic native interactions with spike. Endosomes vary in pH from pH ∼6 (early endosomes) to pH ∼5 (late endosomes), with lysosomal pH as low as ∼4 (Benjaminsen et al., 2011; Turk and Turk, 2009). For endosomal pH, we chose to measure pH 5.5 and 4.5. At endosomal pH, surface plasmon resonance (SPR)-determined apparent ACE2 binding affinities to both spike and RBD were somewhat reduced from 0.82 nM at serological pH to 8.4 and 7.0 nM at pH 5.5 and 4.5, respectively, for spike, and from 1.0 nM at serological pH to 2.2 and 15 nM at pH 5.5 and 4.5, respectively, for RBD (Figure S6B). With CR3022 IgG, apparent affinities to spike and RBD were sub-nanomolar at serological pH, though with a 10-fold difference (0.49 and 0.052 nM to spike and RBD, respectively) (Figure 7C). At pH 5.5, this 10-fold difference was retained (1.7 and 0.23 nM, respectively). However, at pH 4.5, CR3022 still bound to RBD (1.1 nM), but its apparent affinity to spike was dramatically reduced with a KD > 1,000 nM—an apparent affinity difference we estimated to be >1,000-fold (Figures 7C and S6C). Because CR3022 still bound strongly to the isolated RBD, we attributed the dramatically reduced apparent affinity of CR3022 for spike at low pH to conformational constraints of the spike (Figure 7D).
Overall, the pH-induced retraction of RBDs through the spike adopting an all-down conformation can be described as a “conformational masking” energy barrier, which increased CR3022 affinity should be able to overcome. Relevant to this, a recent study has revealed a single mutation between SARS and SARS-CoV-2 spike in the CR3022 epitope that leads to increased CR3022 binding and neutralization for SARS, indicating the SARS-CoV-2 spike evasion from CR3022 neutralization to depend on the reduced affinity of CR3022 (Wu et al., 2020).
Discussion
Viral spikes are prime targets for neutralizing antibodies, and many have evolved mechanisms for immune evasion, some of which resemble aspects of the endosomal pH-dependent conformational masking described here. Receptor binding-site masking through endosomal cleavage, for example, occurs with the Ebola virus glycoprotein trimer (Kaletsky et al., 2007), and conformational masking has been previously described for the HIV-1 envelope trimer (Kwong et al., 2002), which is labile and elicits antibodies of little neutralization capacity. With the SARS-CoV-2 spike, we anticipate endosomal affinity reduction observed here for CR3022 to apply generally to RBD-up-recognizing antibodies, although this remains to be demonstrated explicitly; we note that the emerging D614G variant of SARS-CoV-2 (Korber et al., 2020; Plante et al., 2020) alters switch interactions, such that RBDs are not fully retracted (Yurkovetskiy et al., 2020); this variant appears to be more sensitive to neutralization by patient sera (Li et al., 2020) providing an estimate of the impact of this low-pH conformational masking on neutralization. In general, antibodies can neutralize SARS-CoV-2 through diverse mechanisms, including by competition for ACE2 binding or by stabilizing spike in the all-RBD-down state, and there are multiple ways these antibodies interact with spike (Barnes et al., 2020).
The functional purpose of the up-down positioning of RBD domains in coronaviruses has been a point of debate, given that structures with RBD-up and RBD-down have been determined (Beniac et al., 2006; Gui et al., 2017; Kirchdoerfer et al., 2016; Pallesen et al., 2017; Shang et al., 2020a; Shang et al., 2018; Song et al., 2018; Walls et al., 2016; Yuan et al., 2017). Do the waving RBDs of other coronavirus spikes elicit antibodies that are then shed through an endosomal mechanism along the lines that we outline for SARS-CoV-2? We note that the switch domains from bat RaTG13 and SARS-CoV-1 are virtually identical in sequence to that of SARS-CoV-2, and the aspartic acid residues of the switch domain are mostly conserved in MERS (Figure S7), potentially indicating the switch-based all-RBD-down locking strategy of immune evasion described here to enable other coronaviruses that utilize endosomal entry to avoid neutralization by RBD-up-recognizing antibodies.
Recent cryo-EM analyses of ACE2-bound structures (Benton et al., 2020), cryo-ET analysis of membrane-bound spikes (Ke et al., 2020; Turoňová et al., 2020), and smFRET analyses (Lu et al., 2020) provide additional contexts by which to interpret the structural results described here. Benton and colleagues suggest three-ACE2 to destabilize the prefusion spike, but in the context of ACE2-bound to 2P-stabilized spikes, no substantial changes in S2 conformation were induced by ACE2 binding. Meanwhile, the fascinating motions described by Ke and colleagues and by Turoňová and colleagues involve regions of spike that are below the ordered regions of S2 that we described here. Lastly, smFRET analysis suggests an on-path intermediate as the basis for the observed ACE2-induced trimer asymmetry; it will be fascinating to see whether smFRET analysis of soluble trimers and at endosomal pH can provide insight into the pH-induced alterations in spike conformation that we observe here.
We note that the critical switch region (residues 824–858) displays remarkable structural diversity within coronaviruses, segregating into three structural clusters (Figure S7). Each of the structures within these clusters generally comprises two helices, linked by a disulfide, in distinct orientations in relation to each other and to the surrounding domains. Overall, the structural diversity of the switch region, defined here for SARS-CoV-2 along its endosomal entry pathway and recently at higher pH (Cai et al., 2020; Wrobel et al., 2020), provides a further example of how type 1 fusion machines can use structural rearrangement not only to merge membranes (e.g., transitioning from prefusion to intermediate to postfusion states) but to evade potential neutralizing antibodies that recognize the prefusion state.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CR3022 | Yuan et al., 2020 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Superdex200 10/300GL column | GE Healthcare Life Sciences | Cat#28990944 |
| cOmplete His-tag Purification Resin | Roche | Cat#05893801001 |
| rmp Protein A Sepharose Fast Flow | GE Healthcare | Cat#17-5138-03 |
| 1X PBS | GIBCO | Cat#10010-031 |
| HEPES | Life Technologies | Cat#15630-080 |
| IgG elution buffer | Thermo Scientific | Cat#21009 |
| MES | Sigma | Cat#M8250 |
| Sodium acetate | Sigma | Cat#S7545 |
| Sodium chloride | Quality Biological, Inc | Cat#351-036-101 |
| Sodium citrate | Macron | Cat#0754-12 |
| Trizma base | Sigma | Cat#T1503 |
| Sodium tetraborate | Sigma | Cat#S9640 |
| SARS-CoV-2 spike protein | Wrapp et al., 2020 | N/A |
| Monomeric ACE2 | Zhou et al., 2020b | N/A |
| Dimeric ACE2 | Zhou et al., 2020b | N/A |
| Critical Commercial Assays | ||
| Turbo293™ Transfection Kit | ThermoFisher Scientific Inc. | Cat#A14525 |
| Deposited Data | ||
| Cryo-EM structure of SARS-CoV-2 spike in complex with single ACE2 at pH 7.4 | This study | EMDB: EMD-22941 PDB: 7KNB |
| Cryo-EM structure of SARS-CoV-2 spike in complex with double ACE2 at pH 7.4 | This study | EMDB: EMD-22932 PDB: 7KMZ |
| Cryo-EM structure of SARS-CoV-2 spike in complex with triple ACE2 at pH 7.4 | This study | EMDB: EMD-22927 PDB: 7KMS |
| Cryo-EM structure of Focused ACE2-RBD at pH 7.4, after C3 Symmetry Expansion | This study | EMDB: EMD-22922 PDB: 7KMB |
| Cryo-EM structure of SARS-CoV-2 spike in complex with single ACE2 at pH 5.5 | This study | EMDB: EMD-22943 PDB: 7KNE |
| Cryo-EM structure of SARS-CoV-2 spike in complex with double ACE2 at pH 5.5 | This study | EMDB: EMD-22949 PDB: 7KNH |
| Cryo-EM structure of SARS-CoV-2 spike in complex with triple ACE2 at pH 5.5 | This study | EMDB: EMD-22950 PDB 7KNI |
| Cryo-EM structure of SARS-CoV-2 spike at pH 5.5, consensus map | This study | EMDB: EMD-22253 PDB: 6XM0 |
| Cryo-EM structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | This study | EMDB: EMD-22254 PDB: 6XM3 |
| Cryo-EM structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | This study | EMDB: EMD-22255 PDB: 6XM4 |
| Cryo-EM structure of SARS-CoV-2 spike at pH 5.5, all RBD down | This study | EMDB: EMD-22256 PDB: 6XM5 |
| Cryo-EM structure of SARS-CoV-2 spike at pH 4.5 | This study | EMDB: EMD-22515 PDB: 7JWY |
| Cryo-EM structure of SARS-CoV-2 spike at pH 4.0 | This study | EMDB: EMD-22251 PDB: 6XLU |
| Experimental Models: Cell Lines | ||
| Expi293F cells | ThermoFisher Scientific Inc | Cat#A14527 |
| FreeStyle 293-F cells | ThermoFisher Scientific Inc | Cat#R79007 |
| Recombinant DNA | ||
| pVRC8400 vector | https://www.addgene.org | Cat#63160 |
| pVRC8400-CR3022 | Yuan et al., 2020 | N/A |
| pαH-SARS-CoV-2-S-2P | Wrapp et al., 2020 | N/A |
| Software and Algorithms | ||
| Coot | Emsley and Cowtan, 2004 | https://sbgrid.org/software/ |
| CTFFind4 | Rohou and Grigorieff, 2015 | http://grigoriefflab.janelia.org/ctffind4 |
| crYOLO | Wagner et al., 2019 | https://sphire.mpg.de/wiki/doku.php?id=pipeline:window:cryolo |
| cryoSPARC | Punjani et al., 2017 | https://cryosparc.com |
| GraphPad Prism Software | GraphPad Prism Software, Inc. | N/A |
| Leginon | Suloway et al., 2005 | https://sbgrid.org/software/titles/leginon |
| Phenix | Adams et al., 2010 | https://sbgrid.org/software/ |
| The PyMOL Molecular Graphics System | Schrödinger, LLC | https://pymol.org/2/ |
| RELION | Scheres, 2012 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| Scrubber 2.0 | BioLogic Software | http://www.biologic.com.au/scrubber.html |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter D. Kwong (pdkwong@nih.gov).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Cryo-EM structure coordinates and electron density maps for the SARS-CoV-2 spike ligand free and ACE2 complexes have been deposited with the Protein Data Bank and Electron Microscopy Data Bank (individual spike at pH 5.5, consensus structure: PDB 6XM0, EMD-22253; individual spike at pH 5.5 with single RBD in the up position (conformation up-1): PDB 6XM3, EMD-22254; individual spike at pH 5.5 with single RBD in the up position (conformation up-2): PDB 6XM4, EMD-22255; individual spike at pH 5.5 with all RBDs down: PDB 6XM5, EMD-22256; individual spike at pH 4.5: PDB 7JWY, EMD-22515; individual spike at pH 4.0: PDB 6XLU, EMD-22251; spike with single ACE2 at pH 7.4: PDB 7KNB, EMD-22941; spike with double ACE2 at pH 7.4: PDB 7KMZ, EMD-22932; spike with triple ACE2 at pH 7.4: PDB 7KMS, EMD-22927; Focused ACE2-RBD at pH 7.4, after C3 Symmetry Expansion: PDB 7KMB, EMD-22922; spike with single ACE2 at pH 5.5: PDB 7KNE, EMD-22943; spike with double ACE2 at pH 5.5: PDB 7KNH, EMD-22949; spike with triple ACE2 at pH 5.5: PDB 7KNI, EMD-22950). Additional supplemental items are available from Mendeley Data at https://doi.org/10.17632/y2j4k2mkd6.2.
Experimental Model and Subject Details
Cell Lines
FreeStyle 293-F (cat# R79007) and Expi293F cells (cat# A14528; RRID: CVCL_D615) were purchased from ThermoFisher Scientific Inc. FreeStyle 293-F cells were maintained in FreeStyle 293 Expression Medium, while Expi293F cells were maintained in Expi Expression Medium. The above cell lines were used directly from the commercial sources and cultured according to manufacturer suggestions.
Method Details
Production of Spike, ACE2 and Antibodies
SARS-CoV-2 spike (Wrapp et al., 2020) was expressed by transient transfection in 293 Freestyle cells. Briefly, 1 mg of DNA was transfected into 1L of cells using Turbo293 transfection reagent, and the cells were allowed to grow at 37°C for 6 days. Following expression, the supernatant was cleared by centrifugation and filtration, and then incubated with cOmplete His-Tag Purification resin. The resin was washed with PBS containing increasing concentrations of imidazole, and the protein eluted in 20 mM Tris pH8.0, 200 mM NaCl, 300 mM Imidazole. HRV3C protease was added at a 1:20 mass ratio and incubated overnight at 4°C to cleave the purification tags. The protein was then applied to a Superdex 200 column in PBS, after which the spike containing fractions were pooled and concentrated to 1 mg/mL. Single chain Fc tagged RBD and NTD domains were expressed in the same manner, and purified using capture by Protein A resin, followed by cleavage of the tag using HRV3C (Zhou et al., 2020b) and gel filtration.
Human ACE2 proteins were prepared in monomeric form (residues 1-620) and in dimeric form (residues 1-740). The expression plasmids were constructed and the protein purified as described previously (Zhou et al., 2020b). Briefly, DNA sequence encoding monomeric or dimeric ACE2 was synthesized and cloned into a plasmid with an HRV3C cleavage site, monomeric Fc tag and 8xHisTag at the 3′ end. The proteins were expressed by transient transfection of 293F cells and purified from a Protein A column. The tag was removed by overnight HRV3C digestion at 4°C. The proteins were further purified with a Superdex 200 16/60 column in 5 mM HEPES, pH7.5 and 150 mM NaCl.
For antibody preparation, DNA sequences of antibody CR3022 (Yuan et al., 2020) heavy and light chains were cloned into the pVRC8400 vector, as described previously (Wu et al., 2011), expressed and purified as described (Zhou et al., 2020b). The Fab fragments were generated by overnight digestion with Endoproteinase LysC (New England Biolabs) at 37°C and purified by protein A column to remove uncut IgG and Fc fragments.
Isothermal Titration Calorimetry
Calorimetric titration experiments were performed at 25°C using a VP-ITC microcalorimeter from MicroCal⁄Malvern Instruments (Northampton, MA, USA). The spike protein, ACE2 and Fab of CR3022 were prepared and exhaustively dialyzed against PBS, pH 7.4, prior to the experiments. Any dilution steps prior to the experiments were made using the dialysate to avoid any unnecessary heats of dilution associated with the injections. All reagents were thoroughly degassed prior to the experiments. For the direct determination of the binding to the spike protein, the solution containing either ACE2 or CR3022 (Fab) was added stepwise in 10 μL aliquots to the stirred calorimetric cell (∼1.4 ml) containing spike protein at 0.4 – 0.5 μM (expressed per trimer). The concentration of titrant in the syringe was 12 – 14 μM for both ACE2 and CR3022 (Fab). The effect of CR3022 on ACE2 binding to spike protein was studied by first titrating the spike protein with CR3022 until complete saturation was reached, and then performing a complete titration of the complex with ACE2. Despite the thorough dialysis, the heat of dilution/injection associated with the injection of ACE2 into the complex was considerable during the course of the titration and needed to be accounted for in the analysis. The heat evolved upon each injection was obtained from the integral of the calorimetric signal and the heat associated with binding was obtained after subtraction of the heat of dilution. The enthalpy change, ΔH, the association constant, K a, and the stoichiometry, N, were obtained by nonlinear regression of the data to a single-site binding model using Origin with a fitting function made inhouse. Gibbs energy, ΔG, was calculated from the binding affinity using ΔG = -RTlnK a, (R = 1.987 cal/(K × mol)) and T is the absolute temperature in kelvin). The entropy contribution to Gibbs energy, -TΔS, was calculated from the relation ΔG = ΔH -TΔS.
SPR Binding Experiments
SPR binding experiments were performed using a Biacore T200 biosensor, equipped with a Series S SA chip. The running buffer varied depending on the pH of the binding reaction; experiments at pH 7.4 were performed in a running buffer of 10 mM HEPES pH 7.4, 150 mM NaCl, 0.2 mg/mL BSA and 0.01% (v/v) Tween-20; at pH 5.5 experiments were performed in 10 mM sodium acetate pH 5.5, 150 mM NaCl, 0.2 mg/mL BSA and 0.01% (v/v) Tween-20; and at pH 4.5 in 10 mM sodium acetate pH 4.5, 150 mM NaCl, 0.2 mg/mL BSA and 0.01% (v/v) Tween-20. All measurements were performed at 25°C.
Biotinylated spike and RBD were captured over independent flow cells at 700-1000 RU and 150 RU respectively for both the CR3022 IgG and the dimeric ACE2 binding experiments. To avoid the difficulty in surface regeneration that arises with slow dissociation, we used single-cycle kinetics binding experiments. CR3022 IgG was tested at analyte concentrations 36-1.33 nM prepared in running buffer at each pH, using a three-fold dilution series. In addition, CR3022 IgG was tested over the spike at higher analyte concentrations ranging 108-4 nM, 360-13.33 nM and 1000-37.04 nM at pH 4.5, only to confirm the absence of binding to the spike at pH 4.5. Dimeric ACE2 was tested at 90-3.33 nM prepared in running buffer at each pH, using a three-fold dilution series. Binding over the spike or RBD surface as well as over a streptavidin reference surface was monitored for 120 s, followed by a dissociation phase of 120-900 s depending on the interaction at 50 μl/min. Four blank buffer single cycles were performed by injecting running buffer instead of Fab to remove systematic noise from the binding signal. The data were processed and fit to 1:1 single cycle model using Scrubber 2.0 (BioLogic Software).
Cryo-EM Structures of ACE2-Spike Complexes
SARS-CoV-2 spike was incubated with 3-fold molar excess of monomeric ACE2 receptor with a final trimer concentration of 1 mg/mL in either PBS, pH 7.4, or 10 mM sodium acetate, pH 5.5, with 150 mM NaCl. The samples (2 μl) were vitrified using a Leica EM GP and Vitrobot Mark IV plunge freezers on glow-discharged carbon-coated copper grid (protochip, CF 1.2/1.3). Data were collected on a 300 kV Titan Krios equipped with a Gatan K3-BioQuantum direct detection device using Leginon software (Suloway et al., 2005). The total dose was fractionated for 2 s over 40 raw frames. Motion correction, contrast transfer function (CTF) estimation, particle picking with topaz (Bepler et al., 2019) and extraction, 2D classification, ab initio model generation, 3D refinements and local resolution estimation were carried out in cryoSPARC 2.14 (Punjani et al., 2017). We note that some classes of unbound spike were also observed in both datasets; however, particle picking was optimized for complexes so the fraction was low. The 3D reconstructions were performed using C1 symmetry for all complexes as the ACE2-RBD region showed flexibility that prohibited typical symmetry operations in the triple-bound complexes. However, the RBD-ACE2 region was assessed in greater detail through focused refinement following particle expansion with C3 symmetry applied to the pH 7.4 triple bound reconstruction. This RBD-ACE2 model was then used as a reference structure for refinement of all other ACE2-bound models.
The coordinates of SARS CoV-2 spike ectodomain structures, PDB entries 6VXX and 6M0J (Walls et al., 2020), were employed as initial models for fitting the sharpened cryo-EM map of the ACE2-bound structures (Table S1). Manual and automated model building were iteratively performed using Coot (Emsley and Cowtan, 2004) and real space refinement in Phenix (Adams et al., 2010) to accurately fit the coordinates to the electron density map. Molprobity (Davis et al., 2004) was used to validate geometry and check structure quality. UCSF ChimeraX (Goddard et al., 2018) was used for map-fitting cross correlation calculation (Fit-in-Map tool) and for figure preparation.
Cryo-EM Structures of Ligand-Free Spikes
A sample of SARS-CoV-2 S in PBS with a protein concentration of 1 mg/mL was diluted to 0.5 mg/mL using 0.2 M sodium acetate, pH 4.0, pH 4.5, or pH 5.5 (final sodium acetate concentration: 0.1 M). Separate measurements with a pH meter confirmed that combining equal volumes of PBS and 0.2 M sodium acetate, pH 4.0, pH 4.5, or pH 5.5, produces solutions with pH 4.0, pH 4.5, and pH 5.5, respectively. Quantifoil R 2/2 gold grids were used for specimen preparation. The grids were glow-discharged using a PELCO easiGlow device (air pressure: 0.39 mBar, current: 20 mA, duration: 30 s) immediately before vitrification. Cryo-EM grids were prepared by plunge-freezing in liquid ethane using an FEI Vitrobot Mark IV plunger with the following settings: chamber temperature of 4°C, chamber humidity of 95%, blotting force of −5, blotting time of 5 s, and drop volume of 2.7 μl. Datasets were collected at the National CryoEM Facility (NCEF), National Cancer Institute, on a Thermo Scientific Titan Krios G3 electron microscope equipped with a Gatan Quantum GIF energy filter (slit width: 20 eV) and a Gatan K3 direct electron detector. Four movies per hole were recorded in the counting mode using Latitude software. The dose rate was 13.4 e−/s/pixel.
Each dataset was divided into subsets which were initially processed independently in parallel using Frederick Research Computing Environment (FRCE) computing cluster and later combined for the final refinement. Movie-frame alignment was performed using MotionCorr2 (Zheng et al., 2017). Ctffind4 was used to determine the parameters of CTF (Rohou and Grigorieff, 2015). The remaining processing steps were performed using Relion 3.0 (Scheres, 2012) unless otherwise stated. For spike at pH 4.0, a small particle set was selected manually and used to obtain 2D classes which were utilized as templates to select a larger set of particles. An initial 3D model was obtained using EMAN 2.1 (Tang et al., 2007) from the 2D classes generated from this extended particle set. This 3D model was then subjected to 3D auto-refinement, and the resulting map was used to generate low-pass filtered picking templates for the entire dataset. For spike at pH 5.5 and pH 4.5, particle picking was performed with cryOLO 1.5 (Wagner et al., 2019) using a general network model, and an initial 3D model was obtained with EMAN 2.1 from a subset of resulting 2D classes. The following steps included rounds of 3D classification, 3D auto-refinement, CTF refinement, and particle polishing. Map resolutions were calculated using the gold-standard approach (Henderson et al., 2012) at the FSC curve threshold of 0.143. ResMap 1.1.4 was used to asses local resolution (Kucukelbir et al., 2014). Map sharpening B factors are listed in Tables S2 and S3. Local map sharpening was performed using phenix.auto_sharpen (Terwilliger et al., 2018). SPIDER 22.1 was used for map conversion and re-sizing (Frank et al., 1996). Correlations between cryo-EM maps and atomic models were assessed using phenix.mtriage (Afonine et al., 2018). UCSF Chimera was used for docking and visualization (Pettersen et al., 2004). Despite the fact that C3 symmetry was imposed during the reconstruction of spike for the pH 4.0 dataset, the resulting map displayed some asymmetrical features in some regions, such as that around residue 830. Therefore, the three chains of the atomic model were built and refined individually. The coordinates of SARS CoV-2 spike ectodomain structures, PDB entries 6VXX and 6VYB, were used as initial models for fitting the cryo-EM map of the spike structures at pH 4.0 and pH 5.5 structures. The coordinates of SARS CoV-2 spike at pH 4.0 were used as initial model for the spike structure at pH 4.5. Iterative model building and real space refinement were carried out using Coot (Emsley and Cowtan, 2004) and Phenix to accurately fit the coordinates to the electron density map. Molprobity (Davis et al., 2004) was used to validate geometry and check structure quality.
3D Variability and RBD Conformation Analysis
For 3D variability analysis, a subset of 100,000 particles randomly selected from the final particle set at pH 5.5 was exported into cryoSPARC 2.15 (Punjani et al., 2017), and a homogeneous refinement was performed without imposing symmetry. The 3D variability analysis was set up to use three eigenvectors of the 3D covariance, and 20 frames were used for visualization of results. The eigenvectors describing movements of the RBD were identified via examining the resulting volume series and corresponding variability videos (Videos S1, S2, S3, and S4).
The structural heterogeneity of the consensus pH 5.5 map in the RBD region was analyzed using local 3D classification. To obtain an accurate mask encompassing the conformational space of the dynamic RBD, the four 3D variability volumes corresponding to the beginning and the end of the trajectories defined by eigenvectors 0 and 2 were first aligned to the consensus cryo-EM map. For each of the four volumes, the density corresponding to the dynamic RBD was isolated by performing volume segmentation in UCSF Chimera (Pettersen et al., 2004). These RBD sub-volumes were added together, and a soft mask was created from the resulting composite volume by low-pass filtering the density to 15 Å, extending the resulting volume by 2 pixels, and adding a soft edge of 5 pixels using relion_mask_create. Local 3D classification of the consensus dataset within this mask was performed without particle alignment in Relion 3 (Scheres, 2012), followed by global 3D refinement of each of the resulting six maps.
Identification of Spike Refolding Regions
We used a sliding window of 11 amino acids and 21 amino acids respectively to align and calculate backbone (C, Ca, O, N) rmsd values between the pH 4 structure (protomer B) and pH 5.5 single-RBD-up or pH 5.5 all-RBD-down structures, respectively, using PyMOL (Version 2.3.4). Calculation was omitted if the specified residue range had less than 22 backbone atoms. The average rmsd values of pH 5.5 single-RBD-up conformation 1 and conformation 2 were reported for pH 5.5 single-RBD-up analysis. The refolding regions were defined as residues with greater than 2-Å rmsd. Refolding regions with more than one consecutive residue were further considered, and single residue gaps were ignored when determining the residue ranges. Manual inspection revealed 11 amino acid-window to correspond better with domain movements. Therefore, the results from only 11 amino acid-window analysis were reported.
Clustering of Switch Region Structures
The coronavirus spike structures were obtained from PDB using sequence similarity search against SARS-CoV-2 spike protein with the default parameters. After manual examinations, structures that were not coronavirus spike trimers were excluded. For the rest of the structures, the sequences were aligned using ClustalW (Larkin et al., 2007) and chains with at least 70% of the residues determined of the switch region (residues 824-858, SARS-CoV-2 numbering) were further considered. The structures were clustered using the hclust function implemented in statistical package R based on the pairwise backbone rmsd distances calculated with the rms_cur function in PyMOL after the switch regions were aligned.
pKa Calculations
Individual residue pKas were calculated for the pH 4.0 all-down, pH 5.5 all-down, and pH 5.5 single-up (conformations 1 and 2) structures using PROPKA (Olsson et al., 2011; Søndergaard et al., 2011). For residues in the chain B 830-855 switch domains and titratable residues within 5Å of the switch domain, pKa data were analyzed and plotted using R (https://www.R-project.org/) in RStudio (http://www.rstudio.com/) with the ggplot2 library (Wickham, 2016) and structural figures were made using PyMOL.
Quantification and Statistical Analysis
SPR data were processed and fit using Scrubber 2.0 (BioLogic Software). Cryo-EM data were processed and analyzed using CryoSparc and Relion. Cryo-EM structural statistics were analyzed with Phenix and Molprobity. Statistical details of experiments are described in Method Details or figure legends, such as for isothermal titration calorimetry, where we state in the legend to Figure 7B that measurements were performed in duplicate, with uncertainties being the average standard errors obtained by fitting to the two independent datasets.
Other than standard approaches described in Method Details or figure legends such as splitting of cryo-EM data to allow for statistical analysis, no additional methods were used to determine whether the data met assumptions of the statistical approach.
Acknowledgments
We thank S. Goff for discussions on viral variants and entry mechanism; R. Grassucci, Y.-C. Chi, and Z. Zhang from the Cryo-EM Center at Columbia University for assistance with cryo-EM data collection; M.G. Joyce for antibody CR3022; J.S. McLellan for spike expression vector; E.H. Zhou for assistance with videos; and members of the Virology Laboratory and Vector Core, Vaccine Research Center, for discussions and comments on the manuscript. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), federal funds from the Frederick National Laboratory for Cancer Research under Contract HHSN261200800001E to A.S., T.S., and Y.T. Cryo-EM data for the spike-ACE2 complexes were collected at Columbia University Cryo-EM Center at the Zuckerman Institute, and at the National Center for CryoEM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and by grants from the Simons Foundation (SF349247) and NY State Assembly. Cryo-EM datasets for individual spike proteins were collected at the National CryoEM Facility (NCEF) of the National Cancer Institute. This research was, in part, supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E. We are especially grateful to U. Baxa, A. Wier, M. Hutchison, and T. Edwards of NCEF for collecting cryo-EM data and for technical assistance with cryo-EM data processing. Frederick Research Computing Environment (FRCE) high-performance computing cluster was used for processing cryo-EM datasets of individual spike proteins.
Author Contributions
T.Z. and Y.T. determined ligand-free spike structures at pH 5.5, 4.5, and 4.0; J.G., M.R., and G.C. determined spike-ACE2 structures; G.-Y.C. carried out informatics analyses; P.S.K. performed SPR; J.M.S. calculated pKa; A.S. performed ITC; J.B., W.S., I.T.T., and B.Z. provided reagents; J.C.B. analyzed switch mechanics; A.N. provided insight into pH effect on antibody binding; A.S.O. produced spike; M.S. produced spike expression vectors; T.S. prepared ligand-free cryo-EM specimens; J.S. assisted with entry mechanism; P.W. provided insight into neutralization; S.W. assisted with manuscript preparation; R.A.F. supervised pKa calculations; D.D.H. supervised P.W.; J.R.M. supervised reagents and analyses, L.S. supervised SPR and cryo-EM studies with ACE2; P.D.K. oversaw the project and—with T.Z., Y.T., A.S.O., J.G., P.S.K. A.N., A.S., P.W., W.S., B.Z., G.-Y.C., J.M.S., S.W., and L.S.—wrote the manuscript, with all authors providing revisions and comments.
Declaration of Interests
The authors declare no competing interests.
Published: November 17, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chom.2020.11.004.
Supporting Citation
The following references appear in the supplemental information: Xu et al., 2020
Supplemental Information
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Klaholz B.P., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., Adams P.D., Urzhumtsev A. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 2018;74:814–840. doi: 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020 doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjaminsen R.V., Sun H., Henriksen J.R., Christensen N.M., Almdal K., Andresen T.L. Evaluating nanoparticle sensor design for intracellular pH measurements. ACS Nano. 2011;5:5864–5873. doi: 10.1021/nn201643f. [DOI] [PubMed] [Google Scholar]
- Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020 doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A.J., Berger B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods. 2019;16:1153–1160. doi: 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020:eabd4251. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I.W., Murray L.W., Richardson J.S., Richardson D.C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615-9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Sali A., Baker M.L., Carragher B., Devkota B., Downing K.H., Egelman E.H., Feng Z., Frank J., Grigorieff N. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe. 2020;28:445–454. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R.L., Simmons G., Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020 doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Doyle M.L., Casper D.J., Cicala C., Leavitt S.A., Majeed S., Steenbeke T.D., Venturi M., Chaiken I., Fung M. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lu M., Uchil P.D., Li W., Zheng D., Terry D.S., Gorman J., Shi W., Zhang B., Zhou T., Ding S. Real-time Conformational Dynamics of SARS-CoV-2 Spikes on Virus Particles. Cell Host Microbe. 2020;28 doi: 10.1101/2020.09.10.286948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M.H., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J. Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxiv. 2020 doi: 10.1101/2020.05.12.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Luo C., Zhang W., Li F. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog. 2018;14:e1007009. doi: 10.1371/journal.ppat.1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Liu C., Yount B., Gully K., Yang Y., Auerbach A., Peng G., Baric R., Li F. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16:e1008392. doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søndergaard C.R., Olsson M.H., Rostkowski M., Jensen J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Sobolev O.V., Afonine P.V., Adams P.D. Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr. D Struct. Biol. 2018;74:545–559. doi: 10.1107/S2059798318004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk B., Turk V. Lysosomes as “suicide bags” in cell death: myth or reality? J. Biol. Chem. 2009;284:21783–21787. doi: 10.1074/jbc.R109.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bülow S., Gecht M., Bagola K., Hörner C. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., DiMaio F., Bosch B.J., Veesler D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag, New York; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N.S., Louder M., McKee K., NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Bangaru S., Huang D., Zhu X., Lee C.-C.D., Turner H.L., Peng L., Yang L., Nemazee D. A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody. bioRxiv. 2020 doi: 10.1101/2020.09.21.305441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., Wang Y., Hong Q., Wang S., Zhao Q. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. bioRxiv. 2020 doi: 10.1101/2020.06.30.177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A.S., Honig B. On the pH dependence of protein stability. J. Mol. Biol. 1993;231:459–474. doi: 10.1006/jmbi.1993.1294. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Teng I.-T., Olia A.S., Cerutti G., Gorman J., Nazzari A., Shi W., Tsybovsky Y., Wang L., Wang S. Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes. Cell Rep. 2020;33:108322. doi: 10.1016/j.celrep.2020.108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A ratcheting motion of one NTD domain results in increased mobility of the corresponding RBD (see Method Details for a detailed description). A corresponding top view is presented in Video S2.
The RBD is up and alternates between two positions (see Method Details for a detailed description). A corresponding side view is presented in Video S1.
A ratcheting motion of one NTD domain results in increased mobility of the corresponding RBD (see Method Details for a detailed description). A corresponding top view is presented in Video S4.
The RBD alternates between up and down positions (see Method Details for a detailed description). A corresponding side view is presented in Video S3.
Data Availability Statement
Cryo-EM structure coordinates and electron density maps for the SARS-CoV-2 spike ligand free and ACE2 complexes have been deposited with the Protein Data Bank and Electron Microscopy Data Bank (individual spike at pH 5.5, consensus structure: PDB 6XM0, EMD-22253; individual spike at pH 5.5 with single RBD in the up position (conformation up-1): PDB 6XM3, EMD-22254; individual spike at pH 5.5 with single RBD in the up position (conformation up-2): PDB 6XM4, EMD-22255; individual spike at pH 5.5 with all RBDs down: PDB 6XM5, EMD-22256; individual spike at pH 4.5: PDB 7JWY, EMD-22515; individual spike at pH 4.0: PDB 6XLU, EMD-22251; spike with single ACE2 at pH 7.4: PDB 7KNB, EMD-22941; spike with double ACE2 at pH 7.4: PDB 7KMZ, EMD-22932; spike with triple ACE2 at pH 7.4: PDB 7KMS, EMD-22927; Focused ACE2-RBD at pH 7.4, after C3 Symmetry Expansion: PDB 7KMB, EMD-22922; spike with single ACE2 at pH 5.5: PDB 7KNE, EMD-22943; spike with double ACE2 at pH 5.5: PDB 7KNH, EMD-22949; spike with triple ACE2 at pH 5.5: PDB 7KNI, EMD-22950). Additional supplemental items are available from Mendeley Data at https://doi.org/10.17632/y2j4k2mkd6.2.