Abstract
Access to rapid and accurate detection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA is essential for controlling the current global pandemic of coronavirus disease 2019. In this study, the use of oral rinses (ORs) and posterior oropharyngeal saliva as an alternative to swab collection methods from symptomatic and asymptomatic health care workers for the detection of SARS-CoV-2 RNA by RT-PCR was evaluated. For saliva samples, the overall agreement with oropharyngeal swabs was 93% (Ƙ = 0.84), with a sensitivity of 96.7% (95% CI, 83.3%–99.8%). The agreement between saliva and nasopharyngeal swabs was 97.7% (Ƙ = 0.93), with a sensitivity of 94.1% (95% CI, 73.0%–99.7%). ORs were compared with nasopharyngeal swabs only, with an overall agreement of 85.7% (Ƙ = 0.65), and a sensitivity of 63% (95% CI, 46.6%–77.8%). The agreement between a laboratory-developed test based on the CDC RT-PCR and two commercial assays, the Xpert Xpress SARS-CoV-2 and the Cobas SARS-CoV-2, was also evaluated. The overall agreement was >90%. Finally, SARS-CoV-2 RNA in saliva samples was shown to be stable, with no changes in viral loads over 24 hours at both room temperature and 4°C. Although the dilution of SARS-CoV-2 in ORs precluded its acceptability as a sample type, posterior oropharyngeal saliva was an acceptable alternative sample type for SARS-CoV-2 RNA detection.
Rapid and accurate viral detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is critical in controlling outbreaks in community and health care settings. The current test, track, and trace public health approach to surveillance relies heavily on testing to identify both symptomatic and asymptomatic patients. Nasopharyngeal swabs (NPSs) and oropharyngeal swabs (OPSs) are two of the most common upper respiratory tract specimen types recommended for SARS-CoV-2 diagnostic testing, with NPSs traditionally being the preferred sample type. However, the current pandemic has placed a significant supply strain on the availability and allocation of NPSs and OPSs. In addition, collection of swabs by health care workers (HCWs) poses a transmission risk and requires use of personal protective equipment. The limited inventory of protective gear and testing supplies has prompted evaluation of alternate sample types that can be obtained with minimized HCW exposure and which do not require specialized collection materials, including swabs and viral transport media. A few recent studies and case reports have evaluated oral rinses (ORs)1,2 and saliva samples in patients.3, 4, 5, 6 The goal of this study was to determine the reliability of self-collected ORs and posterior oropharyngeal saliva as alternative sample types for the detection of SARS-CoV-2 RNA in HCWs on three different molecular platforms.
Materials and Methods
Study Design
This study was conducted at Memorial Sloan Kettering Cancer Center (MSKCC) in New York City between April 4, 2020, and May 11, 2020, at the peak of the regional epidemic (https://www1.nyc.gov/site/doh/covid/covid-19-data.page, last accessed May 19, 2020). The cohort included MSKCC employees who completed an internally designed research electronic data capture screening tool designed to determine the need for coronavirus disease 2019 (COVID-19) testing based on either symptoms or exposures. Employees with a positive symptom screen (fever or chills, cough, shortness of breath, body aches, or new loss of sense of smell or taste) or exposure to a case of COVID-19 were scheduled for testing at a location for sample collection. For the purpose of this study, each enrolled employee had paired samples collected, either paired NPS and OR, paired NPS and posterior oropharyngeal saliva (saliva), or paired OPS and saliva samples. Verbal consent was obtained for collection of the alternative samples. The study was granted a Health Insurance Portability and Accountability Act waiver by the MSKCC Institutional Review Board (protocol 18-491).
Sample Collection
NPS and OPS samples were collected using flocked swabs (Copan Diagnostics, Murrieta, CA) and placed in viral transport media. For saliva specimens, HCWs were first asked to swallow and then bring up saliva from the back of the throat and spit at least 3.0 mL of saliva into an empty sterile container. For oral rinses, HCWs were asked to place 10 mL of sterile water in their mouth and with mouth closed, swish for 15 seconds, without gargling and spit the water in a sterile container. NPSs and OPSs were collected by dedicated nurses at test sites who were trained in the swab technique, whereas saliva and mouth wash samples were self-collected. All samples were stored at room temperature until transport to the laboratory within 2 hours.
COVID-19 Laboratory-Developed Real-Time RT-PCR
SARS-CoV-2 RNA was detected using a laboratory-developed real-time RT-PCR based on the CDC protocol, targeting two regions of the nucleocapsid gene (N1 and N2), with the following modifications:
Nucleic acid extraction (110 μL) was performed on either the NUCLISENS EasyMag (bioMérieux, Durham, NC) or the Chemagic 360 (PerkinElmer, Shelton, CT) following an off-board, vortexed, and prelysis step inside a biosafety cabinet 2 hood. For mucoid mouth washings or saliva samples, 1:1 dilution with phosphate-buffered saline was done before off-board lysis. Real-time reverse transcription PCR was performed on either an ABI 7500 Fast (Applied Biosystems, Foster City, CA) or the QuantStudio 5 (Applied Biosystems) system using 5 μL of extracted nucleic acids. Samples were considered positive if both analytic targets N1 and N2 were detected.
Limit of Detection
A limited limit of detection study was done to evaluate performance of the SARS-CoV-2 RT-PCR on saliva samples. The limit of detection of the saliva was estimated by spiking SARS-CoV-2 control material (AccuPlex SARS-CoV-2 Reference Material Kit; Sera Care Life Sciences, Milford, MA) into SARS-CoV-2 RNA-negative saliva samples and performing three 10-fold dilutions. Equivalent testing was performed in parallel in NPSs for comparison. Each dilution was tested in six replicates.
Stability Study
The stability of SARS-CoV-2 RNA in saliva between collection time and testing in the laboratory was evaluated. Three time points and two temperatures were evaluated to mimic the expected transport conditions at MSKCC. Five paired saliva samples and NPS samples with a range of viral loads (cycle threshold [CT] values ranging from 20 to 35) were stored in a transport cooler with (4°C) or without ice packs (room temperature). Nucleic acid extraction and PCR were performed on the same sample at time 0 and then after 8 and 24 hours of storage in the cooler with or without ice packs. At the 8- and 24-hour time points, samples were extracted and tested in triplicate, and the mean CT values at the three time points and two different storage temperatures were compared.
Commercial SARS-CoV-2 RT-PCR Performance on Saliva Samples
An analysis was performed to compare performance of two commercial Emergency Use Authorization tests and the modified CDC test on saliva samples. The Xpert SARS-CoV-2 test (Cepheid, Sunnyvale, CA) and the Cobas SARS-CoV-2 test (Roche Molecular Diagnostics, Pleasanton, CA) were used. Both tests are cleared by the US Food and Drug Administration under Emergency Use Authorization for nasopharyngeal and oropharyngeal swabs. Testing of saliva samples was performed following manufacturer's instructions for the approved sample types. Briefly, for the Xpert SARS-CoV-2 test, about 300 μL of specimen was transferred into the cartridge using a plastic Pasteur pipette provided with the kit. The cartridge was loaded on a GeneXpert instrument and run for 45 minutes, with results interpreted automatically by the instrument software. Results were considered positive if the N gene and/or the E gene was detected and presumptive positive if only the E gene was detected. For the Cobas SARS-CoV-2 test, 600 μL of specimen was transferred to a secondary tube that was loaded on the Cobas 6800 instrument. Results were available approximately after 3 hours and automatically interpreted by the instrument software as positive if the Open Reading Frame-1 gene a/b and/or E gene was detected and presumptive positive if only the E gene was detected. Mucoid samples were treated as described for the laboratory-developed test.
Statistical Analysis
Results were analyzed to determine the sensitivity and specificity of the detection of SARS-CoV-2 RNA in oral rinses or saliva compared with throat or nasopharyngeal swabs as the reference method. Percentage agreement between the laboratory-developed test and the two commercial SARS-CoV-2 Emergency Use Authorization tests was calculated. Data analysis was performed in GraphPad Prism 8.4.2. (GraphPad Software Inc., La Jolla, CA).
Results
Samples and Participants
Overall, 570 samples were collected from 285 unique HCW participants, including 98 paired oral rinses and NPSs, 100 paired saliva and OPS samples, and 87 paired saliva and NPS samples. A total of 224 HCWs were symptomatic, 35 were asymptomatic, and 26 had unknown symptom status at the time of sample collection. There was no difference in detection rate across samples types between symptomatic and asymptomatic participants (data not shown).
Comparison of ORs with NPSs
For the paired OR and NPS samples, the detection rate for SARS-CoV-2 RNA was 23.4% (n = 23/98) and 33.7% (n = 33/98), respectively. Overall agreement between the two sample types occurred in 85.7% (n = 84/98) of samples, including 21 positive pairs and 63 negative pairs (Table 1). SARS-CoV-2 RNA was detected in 12 additional NPSs only and in 2 oral rinses only. Using NPS as the reference standard, the sensitivity and specificity of SARS-CoV-2 RNA detection in oral rinses were 63.6% (95% CI, 46.6%–77.8%) and 96.9% (95% CI, 89.5%–99.5%), respectively.
Table 1.
Performance Characteristics of SARS-CoV-2 PCR on Oral Rinses Compared with NPSs
| Oral rinses | NPS specimens |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 21 | 2 | 23 |
| Negative | 12 | 63 | 75 |
| Total | 33 | 65 | 98 |
| Sensitivity, % | 63.6 (95% CI, 46.6–77.8) | ||
| Specificity, % | 96.9 (95% CI, 89.5–99.5) | ||
NPS, nasopharyngeal swab; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Comparison of Saliva with Swab Methods
For the paired OPS and saliva samples, the detection rate for SARS-CoV-2 RNA was 30% (n = 30/100) in OPSs and 35% (n = 35/100) in saliva samples. Overall agreement between the two sample types occurred in 93% (n = 93/100) of samples, including 29 positive pairs and 64 negative pairs (Table 2). For seven paired samples, SARS-CoV-2 RNA was detected only in OPSs for one pair and only in saliva for six pairs. Using detection rate in OPS as the gold standard, the sensitivity and specificity of SARS-CoV-2 RNA were 96.7% (95% CI, 83.3%–99.8%) and 91.4% (95% CI, 82.5%–96.0%), respectively. Finally, for the 87 paired NPS and saliva samples, the detection rate for SARS-CoV-2 RNA was 19.5% (n = 17/87). Overall agreement between the two sample types occurred in 98% (n = 85/87) of samples, including 16 positive pairs and 69 negative pairs (Table 3). For two paired samples, SARS-CoV-2 RNA was detected only in the nasopharyngeal swab for one pair and only in saliva for the other pair. Using detection rate in nasopharyngeal swabs as the gold standard, the sensitivity and specificity of SARS-CoV-2 RNA were 94.1% (95% CI, 70.2%–99.7%) and 98.6% (95% CI, 92.4%–99.9%), respectively.
Table 2.
Performance Characteristics of SARS-CoV-2 PCR on Saliva Compared with OPSs
| Saliva | OPS specimens |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 29 | 6 | 35 |
| Negative | 1 | 64 | 65 |
| Total | 30 | 70 | 100 |
| Sensitivity, % | 96.67 (95% CI, 83.3–99.8) | ||
| Specificity, % | 91.43 (95% CI, 82.5–96.01) | ||
OPS, oropharyngeal swab; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table 3.
Performance Characteristics of SARS-CoV-2 PCR on Saliva Compared with NPSs
| Saliva | NPS specimens |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 16 | 1 | 17 |
| Negative | 1 | 69 | 70 |
| Total | 17 | 70 | |
| Sensitivity, % | 94.1 (95% CI, 73.0–99.7) | 87 | |
| Specificity, % | 98.6 (95% CI, 92.3–99.9) | ||
NPS, nasopharyngeal swab; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Viral Load in Paired Samples
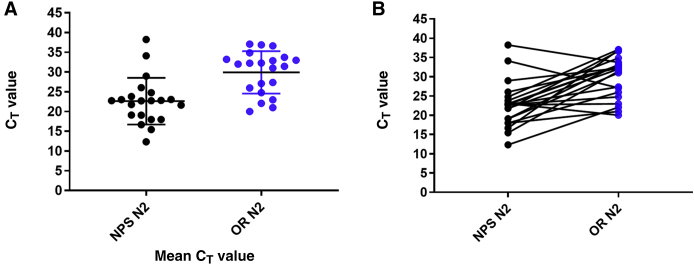
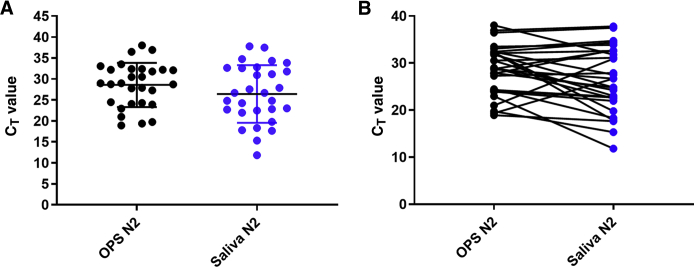
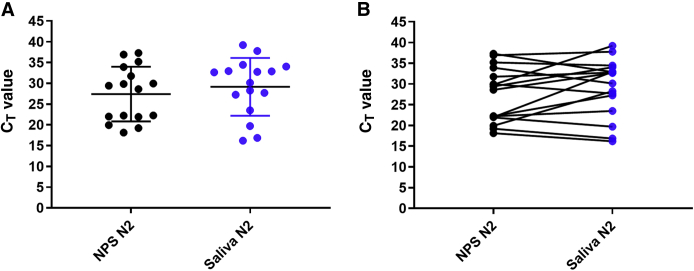
An estimate of the viral load in all paired samples was further evaluated by reviewing the PCR CT values for all positive samples. Given that the N1 and N2 CT values were mostly similar (ie, within ±1 to 2 CT; data not shown), comparison was focused on N2 CT values. The mean and median CT values for N2 in nasopharyngeal swabs were 22.6 (95% CI, 19.9–25.3) and 22.6 [interquartile range (IQR), 18.5 to 24.3], respectively, whereas the mean and median CT values in the corresponding oral rinses were 29.9 (95% CI, 29.7–34.1) and 32.0 (IQR, 25.4 to 33.5), respectively (Figure 1A). The mean and median CT values for N2 in oropharyngeal swabs were 28.6 (95% CI, 26.6–30.5) and 28.9 (IQR, 24.2 to 32.3), respectively, whereas the mean and median CT values in the saliva samples were 27.9 (95% CI, 25.5–30.5) and 27.9 (IQR, 22.2 to 36.6), respectively (Figure 2A). Finally, the mean and median CT values in the nasopharyngeal swabs were 28.1 (95% CI, 24.5–31.8) and 29.4 (IQR, 21.9 to 34.6), respectively, whereas the mean and median CT values in the corresponding saliva samples were 29.6 (95% CI, 26.0–33.2) and 32.7 (IQR, 25.4 to 34.23), respectively (Figure 3A). When looking at each individual pair, the sample type with the lower CT value (higher viral load) varied between each paired sample, with CT values being generally lower for NPSs when compared with ORs (90.4%) but with only 53.8% and 41.4% of samples having lower CT values in NPSs and OPSs, respectively, when compared with saliva (Figures 1B, 2B, and 3B). The differences between the CT values were statistically significant for NPSs versus ORs (P < 0.0001) and for OPSs versus saliva (P < 0.05) but not for NPSs versus saliva (P = 0.17).
Figure 1.
Comparison of severe acute respiratory syndrome coronavirus 2 cycle threshold (CT) values in oral rinses (ORs) and nasopharyngeal swabs (NPSs). Each circle represents the RT-PCR CT values for the N2 target in individual samples. Black circles are the CT values for NPSs. Blue circles represent CT values for ORs. A: Mean CT values in NPSs compared with mean CT values in ORs. B: Comparison of CT values in each NPS/OR paired sample.
Figure 2.
Comparison of severe acute respiratory syndrome coronavirus 2 cycle threshold (CT) values in saliva and oropharyngeal swabs (OPSs). Each circle represents the RT-PCR CT values for the N2 target in individual samples. Black circles are the CT values for OPSs. Blue circles represent CT values for saliva. A: Mean CT values in OPSs compared with mean CT values in saliva. B: Comparison of CT values in each OPS/saliva paired sample.
Figure 3.
Comparison of severe acute respiratory syndrome coronavirus 2 cycle threshold (CT) values in saliva and nasopharyngeal swabs (NPSs). Each circle represents the RT-PCR CT values for the N2 target in individual samples. Black circles are the CT values for NPSs. Blue circles represent CT values for saliva. A: Mean CT values in NPSs compared with mean CT values in saliva. B: Comparison of CT values in each NPS/saliva paired sample.
Limit of Detection
Each dilution was tested in six replicates, and the detection rate in both sample types was determined. The limit of detection was determined to be 500 copies/mL in both sample types (Supplemental Table S1).
Stability Study
Three time points and two storage conditions were used to evaluate the stability of SARS-CoV-2 in saliva samples between collection and receipt in the laboratory for testing. For samples stored both in a cooler with ice packs or at room temperature, there were no significant differences in CT values between time 0 and at 8 and 24 hours (Supplemental Table S2). Similar results were obtained with NPS samples.
Commercial SARS-CoV-2 RT-PCR Performance on Saliva Samples
A total of 24 of 48 positive saliva samples had enough remaining samples to evaluate on the Xpert SARS-CoV-2 test and the Cobas SARS-CoV-2 test (Table 4). On the Xpert, 2 of 24 had invalid results and not enough samples to repeat testing. The percentage agreement between the laboratory-developed test and Xpert on the remaining 21 samples was 91% (n = 20/22), with the Xpert missing one of the positive saliva samples. In addition, 10 randomly selected negative saliva samples were tested and confirmed negative by the Xpert test. On the Cobas, 2 of 24 had invalid results, with not enough samples to repeat testing. The percentage agreement between the laboratory-developed test and the Cobas on the remaining 22 samples was 100% (n = 22/22). In addition, 10 randomly selected negative saliva samples were tested and confirmed negative by the Cobas test.
Table 4.
Comparison of the Xpert Xpress SARS-CoV-2 and the Roche Cobas SARS-CoV-2 to the LDT COVID-19 RT-PCR
| Variable | Xpert Xpress SARS-CoV-2 | Cobas SARS-CoV-2 |
|---|---|---|
| Positive samples | 20/22 | 22/22 |
| Negative samples | 10/10 | 10/10 |
| Invalid samples | 2 | 2 |
| Positive agreement, % | 91 | 100 |
| 95% CI, % | 75.2–98.4 | 85.2–100 |
| Negative agreement, % | 100 | 100 |
| 95% CI, % | 72.3–100 | 72.3–100 |
COVID-19, coronavirus disease 2019; LDT, laboratory-developed test; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Discussion
This study provides data to support the use of saliva for the detection of SARS-CoV-2 RNA based on evaluation of samples from health care workers at risk for COVID-19. SARS-CoV-2 RNA detection from saliva was superior to oropharyngeal swabs and equivalent to nasopharyngeal swabs. Furthermore, these results were validated on two commercial SARS-CoV-2 PCR platforms, with 91% to 100% agreement. On the other hand, oral rinses were suboptimal for the diagnosis of SARS-CoV-2 when compared with nasopharyngeal swabs.
The performance characteristics of COVID-19 PCR on saliva and other less invasive sample types, including throat gargle (oral rinses or mouthwashes), has only recently been evaluated.1,2 Validation of these self-collection sample types holds immense promise in enabling broad testing strategies that mitigate risk and personal protective equipment resource utilization. For saliva samples, sensitivity for detection of SARS-CoV-2 RNA compared with NPS and/or OPS ranges from 84% to 97%.3, 4, 5, 6, 7, 8, 9 In one study from Hong Kong, saliva from 12 hospitalized patients with confirmed COVID-19 was tested, and 11 of 12 (91.7%) had COVID-19 RNA detected in the initial sample.3 However, no comparison to a paired NPS or sputum sample was made in the study. A study from Melbourne, Australia, compared paired saliva and NPS samples collected from 522 ambulatory patients.5 SARS-CoV-2 RNA was detected in 39 NPS samples, and 33 of 39 paired saliva samples were also positive, for a sensitivity of 84.6% (95% CI, 70.0%–93.1%). A sensitivity for saliva of 84.2% (95% CI, 60.4%–96.6%) was also reported in a study from Bangkok, Thailand, when the reference method was a combination of NPS and OPS.9 The lower sensitivity in these two studies, in contrast to the current findings on saliva and the dual reports by To et al,3,4 is probably explained by the dilution factor; samples in the current study were tested neat compared with a 1:1 dilution with Amies media for their study. The yield for these diluted samples was similar to those of the OR samples in the current study, suggesting that dilution of saliva samples could significantly impact detection rate. Notably, all of the studies mentioned above also report isolated cases where SARS-CoV-2 was only detected in saliva and missed in the corresponding NPS/OPS samples. This rare finding may be related to swab collection technique.
Comparison of data from published studies on saliva using different Emergency Use Authorization RT-PCR platforms is challenging because of the differences in sample collection and study design. However, validation across various platforms is essential before SARS-CoV-2 testing on saliva is broadly adopted. A comparison in a subset of samples in this study found high concordance among all three molecular tests, suggesting that testing saliva is feasible on these commercial assays. Two recent studies evaluated saliva samples on the Xpert SARS-CoV-27,8 test, with a sensitivity of 96% compared with NPS sample for undiluted saliva8 and 84% in the study where saliva samples were diluted.7
An interesting comparison that is often made in study with paired samples is comparing viral loads through comparison of CT values. In the current study, overall median CT in saliva and OPS samples was comparable but higher than NPS. Some variability in this observation was seen with higher salivary viral load, which is possibly related to the phase of illness at the time samples were collected. The symptom onset date was not systematically recorded, and further analysis on yield from saliva versus other samples from time of symptom onset could not be performed.
One of the main concerns related to saliva collection has been the stability of RNA in saliva samples, which has been extensively researched over the years.10 Samples were collected in 50-mL conical tubes that are readily available and inexpensive, and SARS-CoV-2 RNA viral load remained stable for up to 24 hours at room temperature or refrigerated, well within expected time to receipt in the laboratory (<24 hours) at MSKCC. It is possible that unlike host RNA that is readily accessible to salivary RNases, SARS-CoV-2 RNA is less susceptible to degradation.
The current study has several limitations. First, although almost 300 HCWs were enrolled, the declining incidence of COVID-19 cases over the study period limited the number of positive samples enrolled in the study. Second, limited self-reported clinical information was collected at the time of testing; date of symptom onset and type of symptoms were not recorded consistently throughout the study period. Given limited reagent availability, not all samples were tested using the Xpert and Cobas assays. It is possible that these two platforms may have detected additional positive samples, but only a limited subset of positive saliva samples were tested, limiting conclusions beyond additional proof of concept for these commercial platforms.
In summary, this report presents the most extensive data set on SARS-CoV-2 RNA detection in oral rinses to date and does not find it to be a suitable alternative to swab methods. In addition, undiluted saliva was shown to be a better sample than oral rinses for self-collection, on the basis of comparative performance of saliva and ORs with NPSs and OPSs. These results collectively suggest that saliva is an acceptable alternative to NPSs for SARS-CoV-2 RNA detection by RT-PCR, with the distinct advantages of being broadly deployable and obviating pervasive testing supply shortages.
Footnotes
Supported in part by National Cancer Institute Cancer Center support grant P30 CA008748.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.10.018.
Supplemental Data
References
- 1.Malecki M., Lusebrink J., Teves S., Wendel A.F. Pharynx gargle samples are suitable for SARS-CoV-2 diagnostic use and save personal protective equipment and swabs. Infect Control Hosp Epidemiol. 2020 doi: 10.1017/ice.2020.229. [Epub ahead of print] doi:10.1017/ice.2020.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito M., Adachi E., Yamayoshi S., Koga M., Iwatsuki-Horimoto K., Kawaoka Y., Yotsuyanagi H. Gargle lavage as a safe and sensitive alternative to swab samples to diagnose COVID-19: a case report in Japan. Clin Infect Dis. 2020;71:893–894. doi: 10.1093/cid/ciaa377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.To K.K., Tsang O.T., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J.M.C., Leung W.S., Chik T.S., Choi C.Y., Kandamby D.H., Lung D.C., Tam A.R., Poon R.W., Fung A.Y., Hung I.F., Cheng V.C., Chan J.F., Yuen K.Y. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58:300776-20. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyllie A.L., Fournier J., Casanovas-Massana J., Campbell M., Tokuyama M., Vijayakumar P. Saliva is more senstivie for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. N Engl J Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J.H., Yip C.C., Poon R.W., Chan K.H., Cheng V.C., Hung I.F., Chan J.F., Yuen K.Y., To K.K. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microbes Infect. 2020;9:1356–1359. doi: 10.1080/22221751.2020.1775133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCormick-Baw C., Morgan K., Gaffney D., Cazares Y., Jaworski K., Byrd A., Molberg K., Cavuoti D. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol. 2020;58:e01109–e01120. doi: 10.1128/JCM.01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Sungkanuparph S., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.05.001. [Epub ahead of print] doi:10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan R., Heavey S., Graham D.G., Wellman R., Khan S., Thrumurthy S., Simpson B.S., Baker T., Jevons S., Ariza J., Eneh V., Pye H., Luxton H., Hamoudi R., Whitaker H., Lovat L.B. An optimised saliva collection method to produce high-yield, high-quality RNA for translational research. PLoS One. 2020;15:e0229791. doi: 10.1371/journal.pone.0229791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.