Abstract
The world is engulfed by one of the most widespread and significant public health crises in decades as COVID-19 has become among the leading causes of death internationally. The novel SARS-CoV-2 coronavirus which causes COVID-19 has unified the scientific community in search of therapeutic and preventative solutions. The top priorities at the moment are twofold: first, to repurpose already-approved pharmacologic agents or develop novel therapies to reduce the morbidity and mortality associated with the ever-spreading virus. Secondly, the scientific and larger pharmaceutical community have been tasked with the development, testing, and production of a safe and effective vaccine as a longer-term solution to prevent further spread and recurrence throughout the populace. The purpose of this article is to review the most up-to-date published data regarding both the leading pharmacological therapies undergoing clinical trials and vaccine candidates in development to stem the threat of COVID-19.
Highlights
-
•
A pandemic caused by SARS-CoV-2 has become a leading cause of death globally.
-
•
The anti-viral agent remdesivir was effective in patients who were hospitalized with COVID-19 and had lower respiratory tract infection.
-
•
The results of a number of clinical trials repurposing immunomodulatory agents have been reported, with positive results for dexamethasone in severe COVID-19.
-
•
The development of an effective COVID-19 vaccine appears to be eminent.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive sense, enveloped RNA beta coronavirus that emerged in Wuhan, China, in December of 2019 [1]. It is the cause of the clinical disease known as COVID-19 that has resulted in more than 50 M infections and more than 1.25 M deaths according to the World Health Organization [2]. COVID-19 is the third respiratory pandemic or epidemic caused by infection with a novel coronavirus. The first, SARS, developed in Hong Kong in the early-2000s, presented an average 6 days after infection with fever, chills, headache, myalgia, and cough. The principal organs involved were the lungs, which with computerized tomography (CT) imaging demonstrated consolidations that evolved within 7–10 days into pulmonary infiltrates. A number of patients required mechanical ventilatory support, and by day 21 following initial onset of SARS-CoV, most patients had recovered, with mortality rate of approximately 9.6% [3,4]. The second clinical epidemic caused by a novel coronavirus was dubbed Middle East Respiratory Syndrome (MERS), and arose in 2012 in and near the Arabian Peninsula. This disease was associated primarily with fever, cough, and shortness of breath and it had a much higher 35% mortality rate [4,5]. Although SARS-CoV-2 shares sequence similarity with both SARS-CoV (79%) and MERS-CoV (50%), it has been most closely linked to two bat-derived SARS-like viruses (bat-SL-CoVZC45 and bat-SL-CoVZXC21, ~88% similarity) [1]. The novel SARS-CoV-2 virus has been officially classified into the subgenus Sarbecovirus of the Betacoronavirus genus. Although it shares many features with SARS, SARS-CoV-2 infection is unique in that viral particles are shed during the presymptomatic phase of infection [6], which has led to significant spread of the virus worldwide.
In this article, we will first offer a brief clinical overview of COVID-19, along with an introduction to the biology of the SARS-CoV-2 virus. Then, we will describe in detail the vaccine candidates and various therapeutic strategies, including pharmacologic therapies, convalescent plasma, and monoclonal antibodies, currently undergoing clinical trials.
2. Clinical overview
2.1. Symptoms
Patients with COVID-19 most commonly report fever, cough, myalgia, fatigue, dyspnea, anosmia, and ageusia [7,8]. In some cases, there is a presence of increased sputum production, headache, hemoptysis, diarrhea, and myalgia [[9], [10], [11], [12], [13], [14]], although roughly 20% percent of patients are thought to be truly asymptomatic (see “Disease Course” section below) [15].
2.2. Radiographic findings
Typical radiographic finding on chest roentgenogram or computerized tomography (CT) imaging demonstrates bilateral pulmonary involvement, commonly located in the posterior lung areas. Bilateral ground-glass opacifications are frequent (representing areas of active interstitial inflammation) in subsegmental areas of consolidation, which generally progress following clinical day five into lesions and mass shadows of high density [14,16]. Cavitations, discrete pulmonary nodules, pleural effusions, emphysema, and fibrosis are uncommon [17].
2.3. Laboratory studies
The most widely reported abnormal laboratory tests with COVID-19 include leucopenia, lymphopenia, and hypoalbuminemia [9,14]. As expected, the presence of elevated cytokines and inflammatory markers, including erythrocyte sedimentation rate, c-reactive protein, and d-dimer are present [11]. These occasionally signal the start of Cytokine Release Syndrome (CRS) in patients, which greatly increases the chances of both mortality and severe acute respiratory distress syndrome (ARDS) [18]. SARS-CoV-2 viral nucleic acid can be detected in the gastrointestinal tract, urine, and saliva [12], and it is not uncommon to encounter abnormal liver function tests [10] including elevated levels of alanine and aspartate aminotransferases (ALT, AST), creatine kinase, and lactate dehydrogenase [10,11,14].
A few laboratory markers have been noted to be predictive of severe illness. One is an increase in the neutrophil to lymphocyte ratio (NLR), demonstrated in patients who required intensive care and/or mechanical ventilation vs. patients with mild disease [19]. Additionally, recent studies have suggested a link between blood types and the severity of COVID-19 symptoms, with Rh-negative blood type being the most protective against severe disease outcomes [20].
2.4. Histopathological findings
Immunophenotyping of bronchoalveolar cells in COVID-19 patients has shown both M1 and M2- like macrophages in moderate to severe cases, along with elevations of inflammatory cytokines [21]. Direct measurement of circulating cytokines and chemokines in plasma showed higher levels of IL-1β, IL1RA, IL7, IL8, IL9, IL10 and most importantly IFN-γ and TNFα. Furthermore, FGF, CSF-3, CSF-2, CXCL10, CCL2, CCL3, CCL4, CCL8, CXCL2, CXCL8, CXCL9, CXCL16, PDGF, and VEGF were also elevated [9,14,22]. In addition to the serum elevations of IL-6 and TNFα, these were also found elevated in spleen and lymph node specimens which suggests that IL-6 may play a role in mediating lymphopenia in severe COVID-19 cases [23]. Furthermore single-cell analysis of bronchoalveolar immune cells revealed higher proportions of macrophages and neutrophils in severe cases of COVID-19 compared to moderate infections, while also showing increased proliferative and heterogenous phenotype of CD8+ T cells along with reduced clonal expansion. In addition, bronchiolar lavage fluid analysis from patients with severe disease also reported higher levels of IL-8, IL-6, and IL-1β [21].
2.5. Disease course
Upon infection, one of the first symptoms that appears is a fever that can persist for up to 12 days. Dyspnea and cough can develop soon thereafter and follow a similar duration; in one study, cough persisted in 45% of survivors even after hospital discharge [24,25]. Both viral nucleic acid and replication-competent virus are detectable at the onset of symptoms in nasal swabs. Titers of replication-competent virus decline and active viral infection stops after 10 to 15 days [26]. It is important to note that the first complications not directly attributable to viral infection, such as sepsis, occur around day 9. More serious lung disease, including acute respiratory distress syndrome (ARDS) and CRS tend to occur in the second week of infection [27] and may require mechanical ventilatory support. In a few initial reports it was found that up to 20% of patients admitted to the hospital had to be put on a mechanical ventilatory support, although this has thankfully declined with improvements in treatment [13,28]. Additionally, acute cardiac and kidney injuries can appear in a 10–20 day period [24,25]. In the US, according to the CDC there have been more than 300,000 excess deaths, two-thirds of which are caused by COVID-19 [29]. In February of 2020, when COVID-19 spread through the US, the primary “at risk” group was identified as individuals over the age of 45, particularly those with multiple preexisting chronic medical conditions. In the initial CDC reports, this group represented almost 80% of deaths in the US [30]. However, as the pandemic has progressed and larger numbers of individuals were infected, an increasing number of 25 to 44 year-old individuals have succumbed, likely owing to social factors such as quarantine fatigue [29].
Interestingly, recent findings suggest that despite recovery and discharge from the hospital, COVID-19 patients may have some long-term health sequelae, including the induction of diabetes [31]. Furthermore, cerebral micro-structural changes that occur post-recovery in part explain widespread reports of prolonged anosmia and “brain fog” or difficulty with various cognitive tasks [32]. As the pandemic wears on and more data are collected, no doubt additional long-term consequences of COVID-19 will be identified.
3. Viral biology
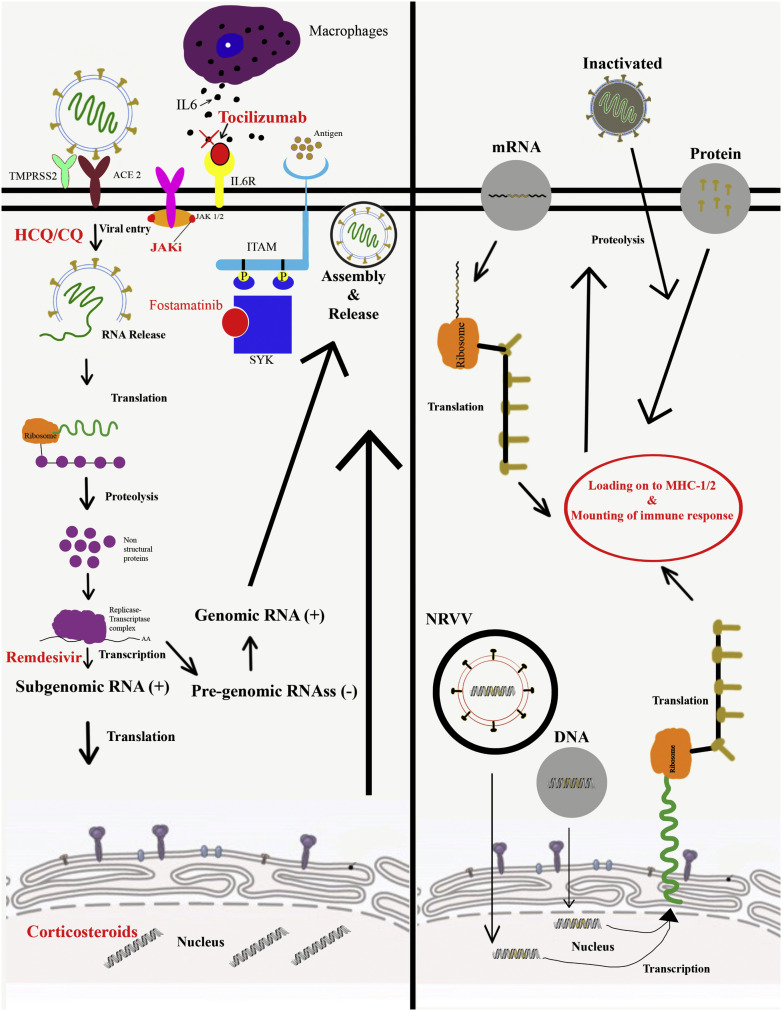
Although SARS-CoV-2 likely has more than one mechanism of cellular entry [33,34], the most closely examined is entry via the angiotensin-converting enzyme 2 (ACE2) protein [35] (Fig. 1 ). Viral entry into the cell is also highly dependent on transmembrane protease serine 2 (TMPRSS2). Other cleaving proteases such as cathepsins B and L (CatB/L) have also been shown to induce viral cleavage and facilitate cellular entry, but to a lesser extent [36,37]. Both ACE2 and TMPRSS2 have been detected in nasal and bronchial epithelium, while ACE2 gene expression has been identified in alveolar type II epithelial cells [36,38]. Additionally, ACE2 is found in the heart, cornea, esophagus, ileum, colon, liver, gallbladder, kidneys, and testis [36,39]. This broad ACE2 tissue expression likely drives SARS-CoV-2 viral entry, inflammatory reaction, and subsequent widespread damage across a variety of organs and organ systems [23,39,40]. The envelope-anchored spike protein (S protein) of SARS-CoV-2 binds to ACE2 to facilitate viral entry [[41], [42], [43]]. Cryo-electron microscopy suggests ACE2 is a homodimer that binds up to two S proteins of SARS-CoV-2 [44,45]. S protein is translated within the cell as an S1-S2 complex, with S2 being cleaved off (via the aforementioned TMPRSS2 and cathepsins) to facilitate the assembly of viable virions [37,44,45].
Fig. 1.
(A) Mechanisms of action of various therapeutic agents being deployed against SARS-CoV-2 and COVID-19; (B) An overview of the various vaccine candidate types and their mechanism of action.
It is still not fully understood how SARS-CoV-2 viral infection drives the pathology seen in the COVID-19 clinical disease, but certain factors have been suggested. Among these is interruption of the renin-angiotensin-aldosterone (RAS) pathway. ACE2 acts in the renin-angiotensin-aldosterone system (RAS) as an inhibitor of angiotensin II and its downstream effects. As a negative regulator of the pathway, ACE2 either converts angiotensin II into Ang-1-7, which acts on the MAS pathway, or angiotensin I into Ang-1-9, which works on the angiotensin II (ATII) receptor [46,47]. One of the functions of ACE2 in the lungs is to protect against ARDS and acute lung injury via reducing local inflammation [48]. A 2005 article by Kuba et al. demonstrated that SARS-CoV (the viral etiology of the first SARS pandemic) binds to the ACE2, elicits endocytosis, and reduces ACE2 protein levels on the cell membrane [49]. This type of viral entry ultimately leads to an increase in levels of angiotensin II and a decrease in ACE2, which could explain the pulmonary pro-inflammatory cytokine landscape and ARDS seen in COVID-19.
Although SARS-CoV-2 is very similar to SARS-CoV [1], key functional differences include receptor binding affinity and immunological evasion capacity. First, the receptor-binding domain (RBD) of SARS-CoV-2 Spike protein has a greater affinity to the ACE2 receptor than SARS-CoV [50,51]. Secondly, the SARS-CoV-2 S protein receptor binding domain has one “up” and two “down” conformations, equating to a less ‘exposed’ S protein and improved immune surveillance evasion [45]. This may ultimately prolong patient recovery time due to delays in the development of an appropriate antibody-mediated immune response in vivo [42,44,45,51].
4. Vaccine development
4.1. Non-replicating viral vectors (NRVV)
The most widely utilized virally vectored candidates for non-replicating SARS-CoV-2 vaccines are adenoviral based (Table 1 , Fig. 1). Adenoviruses are double-stranded DNA viruses that are typically rendered replication ineffective via deletion of their E1 region [52]. Upon infection of target cells, there are high levels of transgene expression and upregulation of costimulatory molecules that elicit the cytokine and chemokine responses [53], improving immunogenicity. Considering that SARS-CoV-2 uses S protein to gain entry into cells, all the vaccines currently in trials express either full-length S protein or S protein subunits [[54], [55], [56], [57]].
Table 1.
Summary list of vaccines against SARS-CoV-2 in human clinical trials of Phase ≥2.
| Type | Organization | Clinical phase | Name | Registry index |
|---|---|---|---|---|
| Inactivated | Sinovac | 3 | CoronaVac/PiCoVacc | NCT04456595NCT04582344 |
| Wuhan Institute of Biological Products/Sinopharm | 3 | Inactivated COVID-19 vaccine (Vero cells) | ChiCTR2000034780 | |
| Bejing Institute of Biological Products/Sinopharm | 3 | BBIBP-CorV | NCT04560881 ChiCTR2000034780 | |
| Institute of Medical Biology, Chinese Academy of Medical Sciences | 1/2 | Inactivated SARS-CoV-2 Vaccine | NCT04470609 | |
| Research Institute for Biological safety Problems, Republic of Kazahstan | 1/2 | QazCovid-in® | NCT04530357 | |
| Bharat Biotech (Whole Virion Inactivated) | 1/2 | BBV152A/B | NCT04471519 | |
| Non-Replicating Viral Vector | University of Oxford/AstraZeneca | 3 | AZD1222 (ChAdOx1 nCoV-19) | NCT04516746NCT04540393 |
| Cansino Biological Inc./Bejing Institute of Biotechnology | 3 | Ad5-nCoV | NCT04526990NCT04540419 | |
| Gamaleya Research Institute | 3 | Gam-COVID-Vac | NCT04530396NCT04564716 | |
| Janssen Pharmaceutical Companies (Johnson&Johnson) | 3 | Ad26.COV2·S | NCT04505722 | |
| mRNA | Moderna/NIAID | 3 | mRNA-1273 | NCT04470427 |
| BioNTech/Fosun Pharma/Pfizer | 3 | BNT162b2 and BNT162b1 | NCT04368728 | |
| Curevac | 1/2 | CVnCoV | NCT04515147 | |
| saRNA | Arcturus/Duke-NUS | 2 | ARCT-021 | NCT04480957 |
| Imperial College London | 1 | LNP-nCoVsaRNA | ISRCTN17072692 | |
| HDT Biocorp./University of Washington | N/A | LION/repRNA-CoV2S | doi: https://doi.org/10.1126/scitranslmed.abc9396 | |
| DNA | Inovio Pharmaceuticals/International Vaccine Institute | 1/2 | INO-4800 | NCT04447781 |
| Osaka University/AnGes/Takara Bio | 1/2 | AG0301-COVID19 and AG0302-COVID19 | NCT04463472NCT04527081 | |
| Cadila Healthcare Limited | 1/2 | nCov Vaccine | CTRI/2020/07/026352 | |
| Genexine Consortium | 1/2 | GX-19 | NCT04445389 | |
| Protein Subunit | Novavax | 3 | SARS-CoV-2 rS/Matrix-M1 Adjuvant (NVX-CoV2373) | 2020–004123–16 NCT04533399 |
| Anhui Zhigei Longcom Biopharmaceutical/Institute of Microbiology, Chinese Academy of Sciences | 2 | Recombinant new coronavirus vaccine (CHO cell) | NCT04466085 | |
| Kentucky Bioprocessing, Inc. | 1/2 | KBP-COVID-19 / KBP-201 | NCT04473690 | |
| Sanofi Pasteur/GSK | 1/2 | SARS-CoV-2 vaccine formulation 1/2 | NCT04537208 | |
| FBRI SRC VB VECTOR, Rospotrebnadsor, Koltsovo | 1/2 | EpiVacCorona | NCT04527575 |
One of the biggest disadvantages when using an adenoviral-vectored vaccine design is that most patients have pre-existing immunity against multiple adenovirus strains, which commonly circulate as upper respiratory infection pathogens, or they develop immunity soon after the first vaccine dose is administered [58]. A SARS-CoV-2 candidate made by CanSinoBiologics in collaboration with the Beijing Institute of Biotechnology uses adenovirus type 5 (Ad5). This design has proven to be effective; however, certain groups' pre-existing immunity led to a slight decrease in efficacy [56,59]. There have been reports that intranasal inoculation in animals is more effective than the intramuscular injection of Ad5 [57], thereby increasing Ad5 vectored vaccine efficacy. To avoid pre-existing adenoviral immunity, The University of Oxford, in collaboration with AstraZeneca, has designed a chimpanzee adenovirus-vectored vaccine that expresses the full S protein (AZD-1222, previously known as ChAdOx1-nCoV). The initial inoculation is followed by a booster dose 28 days later in order to ensure long-term immunity development. Overall, the vaccine has proven well tolerated and immunogenic [60,61]. To overcome Ad5 immunity concerns, the Gamaleya Research Institute has developed a vaccine candidate using recombinant Ad5 (rAd5) as the initial dose, followed by a booster dose using recombinant Adenovirus type 26 (Gam-COVID-Vac). Despite ChAdOx1 causing higher neutralizing antibody titers, Gam-COVID-Vac nonetheless demonstrated titers equivalent to recovered COVID-19 patients [55].
Clinical trial preliminary reports of nonreplicating viral vector vaccine candidates for SARS-CoV-2 have shown all these vaccines to be safe and immunogenic, where immunogenicity has been defined as the detection (using enzyme-linked immunosorbent assays, ELISA) of antibodies induced against the spike protein. Additionally, some groups have used measurements of interferon-gamma (IFNγ), IL-2, and TNFα to infer cellular immunity development. All of the aforementioned NRVV candidates induced anti-spike antibodies and induced a measurable cellular immune response. NRVV vaccines caused small local reactions after injection, such as swelling and redness, and systemic symptoms including malaise, fatigue, fever, and headache, all of which generally resolve within 96 h after vaccination.
4.2. Messenger RNA vaccine candidates
Although there have been to date no FDA-approved mRNA vaccines available for use in humans, the unprecedented times brought on by the COVID-19 pandemic require unusual solutions. Production of nucleic acid vaccine candidates is generally faster and cheaper than protein subunit vaccines [62] and, thus, there have been several mRNA-based SARS CoV-2 vaccine candidates developed and currently undergoing testing. Conventional mRNA vaccine design includes an open reading frame of the targeted antigen (in this case, spike protein) with a 3′ polyadenylated tail [63], which generally produces both humoral and cellular immune responses [64] (Fig. 1). A significant hurdle to mRNA vaccine development has been the propensity of mRNA to degrade; thus, stability and appropriate intracellular translation of mRNA are vital for the success of these vaccine candidates [65]. Various strategies have been developed to address these problems, including removal of double-stranded RNA [66] and embedding of mRNA in lipid nanoparticles [64]. These lipid nanoparticle delivery vehicles have also been leveraged as adjuvants, leading to increased T follicular helper- and germinal center B-cell responses [67].
The leading mRNA SARS CoV-2 vaccine candidate is being developed as a collaboration between the National Institute of Allergy and Infectious Diseases (NIAID) and Moderna. Their vaccine, mRNA-1273, encodes the spike-2 protein antigen, made up of SARS-CoV-2 glycoprotein with the transmembrane anchor and an intact S1-S2 cleavage site [68]. It was initially evaluated in nonhuman primates and has successfully induced a robust anti-SARS-CoV-2 neutralizing antibody response and rapid protection against pulmonary injury [69,70]. BioNTech and Pfizer have created four RNA-based vaccine candidates explored in early-stage clinical trials (Table 1), two of which proceeded to further testing. Their vaccines were also embedded in LNP and encode perfusion-stabilized, membrane-anchored SARS-CoV-2 full-length spike (BNT162b2) and secreted trimerized SARS-CoV-2 receptor-binding-domain (BNT162b1). Based on recently published preliminary results, BNT162b2 reduces systemic adverse reaction to the vaccine in all participants, especially in older adults [71]. Both BNT and mRNA-1273 require booster doses in order to ensure high neutralizing antibody titer and (presumably) long term immunogenicity. Despite the necessity of a second dose, the antibody response against the SARS-CoV-2 receptor-binding domain (RBD) of both vaccines showed significantly higher titers compared to patients who have recovered from COVID-19 [68,72,73]. Furthermore, the BNT162b1 vaccine demonstrated an anti-RBD IgG titer rising by nearly fifteen-fold on day 28 following first inoculation. Although these vaccines induced mild adverse symptoms following the initial dose, including mild fatigue, chills, headache, myalgia, and localized pain at the injection site, adverse reactions progressed with an increase in dosage, and booster doses in some patients caused moderate to severe local and systemic reactions [68,72].
4.3. Self-amplifying messenger RNA vaccine candidates
In addition to the above non-amplifying mRNA vaccines, self-amplifying RNA (saRNA) vaccine technology has been recently developed (Fig. 1). saRNA vaccines are synthesized using plasmids of Trinidad donkey Venezuelan equine encephalitis virus strains (VEEV). The VEEV structural coding regions are then replaced with pre-fusion Spike protein of SARS-CoV-2, while the self-amplifying coding region of VEEV alphavirus remains conserved [74,75]. These vaccines are promising because they have the potential to induce a more robust immunological response than a non-replicating mRNA vaccine. However, a major disadvantage of these vaccines is the length, given that they contain RNA sequence for replicon and the Spike protein. Some of the SARS CoV-2 saRNA vaccines currently under study include candidates by Imperial College in London [74], Arcturus/DUKE-NS, and the University of Washington [75] (Table 1). All of these utilize self-amplifying RNA constructs embedded in various forms of nanoparticles with adjuvant properties [[74], [75], [76]].
4.4. DNA vaccine candidates
As early as 1990, investigators have shown that direct injection of intact nucleic acids into muscles of mice was a potentially useful vaccination strategy [77]. There are several advantages to a DNA-based vaccination approach, including the dramatically increased stability of the DNA molecule compared to RNA and the potential of DNA constructs to produce a large number of mRNA molecules, thereby increasing the target antigen's immunologic exposure. Additionally, the thermal stability of DNA means that DNA-based vaccines have fewer refrigeration requirements than their mRNA-based counterparts.
DNA vaccines targeting both MERS and SARS-CoV-2 have both been developed. Inovio Pharmaceuticals had previously engineered MERS-CoV DNA vaccines, and now have designed a SARS-CoV-2 DNA-based vaccine candidate (INO-4800) (Table 1, Fig. 1). The vaccine was created based on a consensus SARS-CoV-2 spike glycoprotein sequence with an N-terminal IgE leader, added to enhance expression in target cells and increase immunogenicity [78]. In guinea pig testing, INO-4800 has shown humoral immunogenicity with anti-SARS-CoV-2 antibodies which inhibited viral binding to the ACE2 receptor. Furthermore, bronchoalveolar lavage fluid analysis revealed the presence of both cellular and humoral immune components after inoculation. There are three additional DNA vaccine candidates listed in the US clinicaltrials.gov database, but no additional preliminary data are available.
4.5. Inactivated whole-virus vaccine candidates
Vaccines that use inactivated pathogens to induce immunity have a longstanding history in pandemic response. Although this type of vaccination represents the vast majority of historically effective vaccines, their long production time has put them at a disadvantage in the current COVID-19 pandemic. The most promising inactivated SARS-CoV-2 vaccine candidates are currently in a phase 3 clinical trials in China (Table 1, Fig. 1).
This vaccine approach utilizes variants of the SARS-CoV-2 virion that are propagated via Vero (African Green Monkey) cell lines. Upon viral extraction, beta-propiolactone is used for inactivation with the viral particle then adsorbed onto an adjuvant (aluminum hydroxide) [79,80]. The present trials are investigating anti-viral immunity development at 14 and 28 days post-inoculation, with variations in timing and dose of booster vaccine, including an evaluation of two booster doses. When compared to other vaccine types, these inactivated viral vaccines appear to have reduced adverse effects. Most systemic adverse reactions were mild with no severe adverse reactions, while localized injection site redness and pain were common [79]. All adverse reactions have resolved 72 h after vaccine administration.
Comparing induced antiviral immunity of these inactivated viral vaccine candidates to the other vaccine types mentioned previously is difficult, as no comparisons were made between induced antiviral antibodies and those of recovered COVID-19 patient convalescent plasma. Nevertheless, the plaque reduction neutralization test (PRNT) analysis used in one of these studies mirrored that used in the mRNA-1273, BNT162b1, and ChAdOx1 studies. Inactivated vaccines showed similar titers to other approaches mentioned elsewhere in this article, and even higher titers than Ad5-vectored vaccines [80].
Despite this, there are several potential disadvantages of inactivated viral vaccines. Despite inactivated vaccines being considered one of the safer options in worldwide vaccination, the use of aluminum hydroxide as an adjuvant has been previously linked to vaccine-associated enhanced respiratory disease (VAERD), a driver of even more enhanced viral pulmonary pathology that has been reported since the 1960s as the complication in vaccine trials and studies of measles and respiratory syncytial virus [81,82]. No indications of VAERD have been noted in phase 1 or 2 of these trials published to date. More concerningly, the previously-developed inactivated viral vaccines against SARS-CoV (the causative agent of the initial SARS epidemic) showed that anti-viral IgG levels rapidly decline 16 months after inoculation, rendering them practically undetectable three years after inoculation [83], raising concerns for durable immunity in what is expected to be a multi-year pandemic.
4.6. Protein subunit vaccine candidates
An alternative method of vaccine construction, the synthetic-protein subunit approach lies between nucleic acid-based techniques and inactivated whole virus vaccines. This group of SARS-CoV-2 vaccine candidates contain a recombinant spike protein expressed in various (typically insect) cell lines. Similar to RNA-based approaches, peptides are often unstable in vivo and are typically packaged into nanoparticles adsorbed onto specific adjuvants structured to increase the uptake of protein cargo into host antigen presenting cells. The leading SARS-CoV-2 protein vaccine candidate contender is NVX-CoV2373, produced by Novavax, currently in phase 3 clinical trial. This vaccine candidate consists of a nanoparticle containing the full-length wild-type Spike glycoprotein that was engineered to be resistant to proteolytic cleavage and capable of binding ACE2 receptors with high affinity. Protein production was optimized in the established baculovirus Spodoptera frugiperda (Sf9) insect cell expression system, while the adjuvant used to increase the vaccine's immunogenicity is Novavax's Matrix-M1 [84]. In addition to the Sf9 expression system, another commonly used cell line for vaccine production is Chinese hamster ovary (CHO) [[85], [86], [87]]. The CHO cell line has been utilized in research settings for the production of MERS and SARS-CoV vaccines and has been used for COVID-19 serological testing [88].
Adverse effects from NVX-CoV2373 have been mild, including redness and swelling at the injection site, arthralgia, fatigue, headache, myalgia, nausea, and malaise dissipated after at least two days. No serious adverse effects have been noted. NVX-CoV2373 induces humoral and cellular immunity slightly stronger than that observed in convalescent plasma from recovered COVID19 patients.
5. Pharmacological therapies
5.1. Remdesivir
Remdesivir (GS-5734) is a broad-spectrum antiviral drug that is a 1′-substituted adenosine nucleotide analog. It has been proven an effective therapy against several RNA viruses, including Ebola, by competing with ATP for incorporation into RNA-dependent RNA polymerase [89]. During nascent viral RNA chain, RNA-dependent RNA polymerases are unable to process beyond the insertion of remdesivir, resulting in a chain elongation termination. Remdesivir has a conserved mode of action in a diverse group of RNA viruses [90], and was among the first antiviral agents to be tested for activity against SARS-CoV-2. Indeed, in initial in vitro testing, remdesivir inhibited SARS-CoV-2 infection in Vero E6 cells and human lung primary cell lines [91,92].
Consequently, many clinical trials are currently ongoing to test the drug's safety and efficacy for treatment against COVID-19 in human patients. A report from Beigel et al. showed that remdesivir successfully reduced the recovery time from 15 to 10 days on average in adults hospitalized with SARS-CoV-2 infection [93]. Additionally, the patients treated with remdesivir had almost a half reduced chance of mortality calculated by Kaplan-Meier estimates [93]. Even prior to the final report of the trial, the US Food and Drug Administration gave remdesivir Emergency Use Authorization (EUA) [94]; the drug label indicates a recommendation for use in patients 12 years of age and older and weighing at least 40 kg a 5-day course in mild to moderate infections and a 10-day course in severe infections requiring mechanical ventilation. Importantly, the maker of remdesivir (Gilead Sciences) has reported that parallel use of remdesivir and hydroxychloroquine might inhibit the effect of remdesivir and reduce both the rate and likelihood of recovery in patients receiving both drugs, a consideration given recent controversies surrounding the use of hydroxychloroquine in patients with COVID-19 [95].
5.2. Tocilizumab
Tocilizumab is a recombinant, fully humanized monoclonal antibody which targets both soluble and membrane-bound forms of the interleukin-6 receptor (IL-6R). It is used in clinical practice to treat adult patients with rheumatoid arthritis and pediatric patients with systemic juvenile idiopathic arthritis, although its effectiveness has recently been demonstrated in other systemic autoimmune and inflammatory conditions, including giant cell arteritis [96], multicentric Castleman's disease [97] and, importantly, was approved by the FDA in 2017 for the treatment of Cytokine Release Syndrome in chimeric antigen receptor (CAR) T-cell cancer therapy [98].
In COVID-19, elevated IL-6 levels have been correlated with increased mortality [24], sparking interest in the use of tocilizumab for COVID-19 therapy. There have been numerous case reports of tocilizumab improving oxygenation and reducing inflammatory biomarkers in hospitalized COVID-19 patients [99,100]. Although not yet associated with pathogenesis, an improvement in COVID-19-associated lymphopenia has also been reported following tocilizumab treatment [101]. A recent rapid meta-analysis of published case reports also demonstrated a reduction in mortality of 12% among COVID-19 patients treated with tocilizumab [102]. Despite these positive suggestions of efficacy, tocilizumab therapy for COVID-19 remains unproven; on August 27, 2020, the US National Institutes of Health (NIH) advised against the widespread use of anti-IL-6 receptor monoclonal antibodies (tocilizumab included) following conflicting published case reports and a lack of properly designed clinical trial data [103]. Very recently, a randomized double-blind placebo-controlled trial in hospitalized patients with severe COVID-19 showed that tocilizumab was ineffective in reducing mortality or the rate of intubation. Further, secondary end points including the need for supplemental oxygen and clinical worsening were not reduced by tocilizumab treatment compared to placebo [104].
5.3. Fostamatinib
Acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) exhibited by a subset of patients suffering from COVID-19 and other respiratory ailments have been linked to the elevated circulating levels of mucin-1 (KL-6/MUC1) and MUC5AC [105,106]. As a transmembrane protein of the mucosal epithelial cells, MUC1 plays a critical role in the lining of the airway lumen. Overexpression of this protein and excessive mucus production have been shown to increase the duration of infections as well as mortality from respiratory diseases [107]. In an attempt to target MUC1, a large pharmacological screening study done by Alimova et al. 2020 identified Fostamatinib as a potential antagonist of MUC1 [108].
Fostamatinib is an inhibitor of the spleen tyrosine kinase (SYK), an enzyme known for its role in the adaptive immune receptor signaling, cellular adhesion, innate immune recognition, and platelet function [109]. It has been used in the treatment of autoimmune diseases such as rheumatoid arthritis and immune thrombocytopenia (ITP) [110,111]. Moreover, the broad number of immune pathways involving SYK has been shown to play a role in the hyper-inflammatory response caused by anti-SARS-CoV-2-Spike IgG [112]. In particular, R406, an active component of Fostamatinib, has recently been confirmed to effectively block SYK and its downstream signaling and suggests potential efficacy of fostamatinib in the treatment of ALI and COVID-19 [108,112]. As it is already FDA approved, Fostamatinib has been accelerated into phase 2 clinical trials for COVID-19 (NCTNCT04579393 and NCT04581954).
5.4. Chloroquine/hydroxychloroquine (CQ/HCQ)
Quinine has been used as therapeutic agent for centuries. It was first introduced into the Western medical pharmacopeia in 1638 when the wife of the Spanish Viceroy of Peru, Countess Chincona, was given bark of the later-named Chincona tree by an Incan herbalist as a treatment for malaria. A derivative of quinine, chloroquine, saw significant use as an antimalarial drug during World War II. Its use to treat autoimmune disease, particularly systemic lupus erythematosus, began in the 1950s. Hydroxylation, leading to hydroxychloroquine, reduced systemic toxicities significantly and led to more widespread use in chronic diseases, including autoimmune diseases, particularly systemic lupus erythematosus and rheumatoid arthritis [[113], [114], [115]].
The possible mechanisms of action of chloroquine and its derivatives on coronavirus infection are several and not yet fully understood. Of relevance to coronavirus infection, these drugs increase endosomal pH, preventing acidification and reducing viral entry into cellular cytoplasm from endosomes [116]. Additionally, CQ inhibits glycosylation of the cellular ACE2 receptor, thereby interfering with SARS-CoV binding and infection [117]. As CQ/HCQ have been proven useful against SARS-CoV and other widely circulating human and bat coronaviruses (HCoV-229E and HCoV-O43) [91], it only seemed reasonable that it should work in a similar fashion with emerging SARS-CoV-2. Initial in vitro tests on Vero E6 cells showed promising viral inhibition, where both CQ and HCQ prevented the transport of virus from early endosomes to endolysosomes (suspected to be necessary for the release of viral genome) [116]. However, retrospective studies and clinical trial results show inconclusive evidence about the effect of these drugs against COVID-19. Initial reports from a multisite international randomized, double-blind, placebo-controlled trial showed that HCQ failed to produce a decrease in symptom presence and/or severity over a 14-day period [118]. Furthermore, additional observational and multi-center, randomized, controlled trial preliminary reports showed that HCQ did not significantly lower chances of required intubation or mortality rate [119,120]. Following these and other negative studies, treatment of COVID-19 with CQ and HCQ is not recommended by the NIH [121].
5.5. Janus kinase pathway inhibitors (JAKi)
The broad spectrum of inflammatory cytokines and chemokines present in COVID-19 patients suggest the activation of the Janus kinase-Signal Transducer and Activator of Transcription (JAK-STAT) pathway. The first approved JAK inhibitor was ruxolitinib [122]. Like tocilizumab, ruxolitinib has been reported to improve COVID-19 symptoms. The drug is well tolerated in patients and associated with very few adverse effects. A recent retrospective analysis showed that patients receiving ruxolitinib significantly reduced their COVID-19 inflammation scores with consistent clinical improvement in COVID-19 patients with evidence of cytokine release syndrome [123]. There are additional, ongoing clinical trials of other FDA-approved JAK inhibitors in COVID-19 patients, including tofacitinib and baracitinib, with some indications that combinations of JAK inhibitors (particularly, baracitinib) with the antiviral drug remdesivir may reduce time to recovery in hospitalized COVID-19 patients (ACTT-2 trial press release, Lilly corporation).
5.6. Corticosteroids
Considering that COVID-19 elicits a broad systemic inflammatory response, including inflammation-related damage of the lungs, corticosteroid therapy would seem an obvious therapeutic candidate. However, high doses and/or long-term use of corticosteroids are associated with a variety of deleterious effects, including metabolic derangements, an increased risk of infection, and bone abnormalities, among others [124]. Additionally, in MERS, corticosteroids have been linked to delayed viral clearance, which has also been reported in SARS-CoV-2 [125,126].
Despite this, several clinical trials are ongoing to evaluate the efficacy and appropriate timing of corticosteroid treatment for COVID-19. Perhaps the most influential of these, preliminary results of the RECOVERY trial published in mid-July 2020, reported a reduction in death rates associated with the administration of dexamethasone in hospitalized patients requiring supplemental oxygenation, but not among those receiving no respiratory support [127]. Interestingly, two previously-published retrospective analyses from Chinese cohorts had noted similar findings; the first study used methylprednisolone in severe COVID-19 cases presenting with acute respiratory distress syndrome [128], whereas the second study found a benefit in reducing length of hospitalization and improvement in chest imaging parameters in severe COVID-19 cases given methylprednisolone [129]. Additional studies report that low-dose, short-course corticosteroids might slow or prevent disease progression and reduce inflammation [130,131]. Based on current evidence, the NIH recommends the use of dexamethasone 6 mg per day for up to 10 days or until discharge for patients hospitalized with COVID-19 requiring supplemental oxygenation [132]. It should be noted that in milder COVID-19 there remains a possible risk that steroids might worsen the disease course, as steroids increase the risk of infection. Certainly, it should not be assumed that patients on chronic steroid therapy are immune from developing severe COVID-19 or that they are at a lesser risk of becoming infected by SARS-CoV-2.
6. Additional novel therapies
6.1. Complement inhibitors
The ability of antibodies and phagocytic cells to clear an infection is often enhanced by the complement system. The complement cascade is part of the innate immune system; dysregulation of complement has been linked to various autoimmune diseases such as C3 glomerulopathy, paroxysmal nocturnal hemoglobinuria, systemic lupus erythematosus, and many others [[133], [134], [135]]. Additionally, dysregulation of the complement system has been linked to coagulation pathways, thrombosis, and prolonged systemic inflammation [136]. Thrombosis plays a vital role in COVID-19 pathology [137] and is an important finding in the pulmonary disease seen with COVID-19, where histologic evaluation demonstrates deposition of complement components C5-9, C4d, and mannose lectin-associated serine protease 2 [138].
Complement-targeting therapeutics have garnered much research interest in the past decade owing to its role in autoimmune disease and are now being trialed in COVID-19. One of the early drugs that was tested was eculizumab, a monoclonal antibody targeting C5 protein convertase that recently showed effectiveness in improving recovery from COVID-19 [[139], [140], [141]]. In addition to C5 inhibition, the compstatin-based complement C3 inhibitory drug (AMY-101) has also shown some success [142]. When the two inhibitory drugs were compared, despite both eliciting a robust anti-inflammatory response, AMY-101 showed a greater decline in neutrophil counts and more effective lymphocyte recovery [143]. Further upstream, Conestat alfa (a human recombinant C1 esterase), was administered to a small group of COVID-19 patients resulting in near-immediate defervescence, improvement in inflammatory markers and stabilization or decrease in oxygen requirements [144]. Following these early successes, clinical trials are underway for many of the complement cascade inhibitors, including AMY-101 (NCT04395456), APL-9 (NCT04402060), Eculizumab (NCT04346797 and NCT04355494), Ravulizumab (NCT04369469 and NCT04390464), Zilucopan (NCT04382755), Avdoralimab (NCT04371367), Conestat Alfa (NCT04414631) and others.
6.2. Convalescent plasma (CP)
Although the mortality associated with COVID-19 is higher than other commonly circulating respiratory infections (e.g. influenza), the majority of individuals infected with SARS-CoV-2 have recovered. Convalescent plasma sourced from recovered COVID-19 patients contains naturally produced antibodies that, when administered to at-risk individuals, may provide temporary protection against the worst effects of the disease. There are several obstacles to the use of CP, perhaps the most serious being the lack of standardization among protocols for preparing and administering this heterogeneous product. Despite this, there have been several published reports describing reductions in ground-glass opacification in the lungs of COVID-19 patients (indicating a reduction in localized, pathogenic lung inflammation) and an increase in neutralizing antibody levels in patients treated with CP [145,146]. It is important to note that these studies have small sample sizes, limiting the firm conclusions that can be drawn from each. However, a report published in June of 2020 by Joyner et al. noted a low risk of adverse effects arising from CP treatment of COVID-19 patients in a large sample of roughly 20,000 patients in the US. The overall incidence of transfusion reaction was <1%, the highest frequency adverse effects were cardiac in nature, and judged to be related to the COVID-19 clinical disease rather than CP therapy itself [147]. Further examination is necessary to explore the efficacy of CP in widespread clinical administration, and further standardization of CP protocols would be desirable.
6.3. Anti-SARS-CoV-2 antibody cocktails
In contrast to the heterogeneity of CP, synthetic anti-SARS-CoV-2 antibody cocktails are highly enriched specific antibodies against the SARS-CoV-2 spike glycoprotein which bind to the virion and prevent cellular entry. The relatively rapid mutation rate of SARS-CoV-2 (as an RNA virus) raises the possibility of antibody-escaping mutants and the development of ‘resistance’ to antibody cocktails [[148], [149], [150]]. Therefore, to counteract the viral mutation, a cocktail of different variants against spike protein are generally administered together [150], thereby minimizing the likelihood of escape mutants. The development of these monoclonal antibodies began with the identification of humanized-mouse and human antibodies that have a high affinity for the receptor binding domain of the SARS-CoV-2 spike protein [151]. The first synthetic antibodies for clinical use have been developed by Regenron; the REGN-COV2 antibody cocktail is currently in clinical trials as part of the RECOVERY Collaborative Group Trials (NCT04381936) as well as Phase 1/2 trials (NCT04425629, NCT04426695). The cocktail consists of REGN10933 and REGN10987 antibodies, both of which bind differently to the spike protein and have previously shown promising results in non-human primates and hamsters [152].
7. Conclusion
Since the emergence of SARS-CoV-2, the scientific community has been working restlessly to find both short-term therapeutic approaches and a longer-term vaccine solution to reduce spread and curb COVID-19 morbidity and mortality. Despite significant progress and promising results from vaccine candidate studies, many obstacles remain, including the logistical difficulties surrounding mass production and delivery of millions or billions of doses to the worldwide population, which will likely represent the largest pipeline bottleneck. Unfortunately, some vaccine types, such as mRNAs, are quite unstable at room temperatures and may require freezers not commonly found in rural clinics and hospitals away from academic research centers; non-refrigerated vaccine types may prove a more viable solution for these locations. Despite the challenges posed by this novel and rapidly spreading viral infection, the world has seen an unprecedented level of scientific engagement and cooperation which no doubt will serve as a model for future pandemic responses.
Funding
The funding sources are NIH: NIAMS R61AR078075, NIH: NIAMS R01AR076440, NIH: NIAMS K08AR070891, DoD: CDMRP PRMRP PR191652.
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease (COVID-19) – World Health Organization, (n.d.). https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed November 9, 2020).
- 3.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C.B., To. K.F., Lui S.F., Szeto C.C., Chung S., Sung J.J.Y. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 4.Fani M., Teimoori A., Ghafari S. Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Futur. Virol. 2020 doi: 10.2217/fvl-2020-0050. [DOI] [Google Scholar]
- 5.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S., Kim T., Lee E., Lee C., Kim H., Rhee H., Park S.Y., Son H.-J., Yu S., Park J.W., Choo E.J., Park S., Loeb M., Kim T.H. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment Center in the Republic of Korea. JAMA Intern. Med. 2020;180:1447. doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornuss D., Lange B., Schröter N., Rieg S., Kern W.V., Wagner D. Anosmia in COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1426–1427. doi: 10.1016/j.cmi.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaira L.A., Salzano G., Deiana G., De Riu G. Anosmia and Ageusia: common findings in COVID-19 patients. Laryngoscope. 2020;130:1787. doi: 10.1002/lary.28692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J'an, Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D.S.C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-L., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S. China medical treatment expert group for covid-19, clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., the Northwell COVID-19 Research Consortium, Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA. 2020 doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buitrago-Garcia D., Egli-Gany D., Counotte M.J., Hossmann S., Imeri H., Ipekci A.M., Salanti G., Low N. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: a living systematic review and meta-analysis. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H., Ling Y., Jiang Y., Shi Y. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295:210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., Jacobi A., Li K., Li S., Shan H. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong M., Zhang H., Cao X., Mao X., Lu Z. Higher level of neutrophil-to-lymphocyte is associated with severe COVID-19. Epidemiol. Infect. 2020;148 doi: 10.1017/S0950268820001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zietz M., Tatonetti N.P. Testing the association between blood type and COVID-19 infection, intubation, and death. medRxiv. 2020 doi: 10.1101/2020.04.08.20058073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 22.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., ten Oever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.026. 1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Feng Z., Diao B., Wang R., Wang G., Wang C., Tan Y., Liu L., Wang C., Liu Y., Liu Y., Yuan Z., Ren L., Wu Y. 2020. The Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Directly Decimates Human Spleens and Lymph Nodes, Infectious Diseases (except HIV/AIDS) [DOI] [Google Scholar]
- 24.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., Ng O.-T., Marimuthu K., Ang L.W., Mak T.M., Lau S.K., Anderson D.E., Chan K.S., Tan T.Y., Ng T.Y., Cui L., Said Z., Kurupatham L., Chen M.I.-C., Chan M., Vasoo S., Wang L.-F., Tan B.H., Lin R.T.P., Lee V.J.M., Leo Y.-S., Lye D.C. Singapore 2019 novel coronavirus outbreak research team, epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020;323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N., van den Akker J.P.C., Endeman H., Gommers D.A.M.P.J., Cornelissen J.J., Hoek R.A.S., van der Eerden M.M., Hesselink D.A., Metselaar H.J., Verbon A., de Steenwinkel J.E.M., Aron G.I., van Gorp E.C.M., van Boheemen S., Voermans J.C., Boucher C.A.B., Molenkamp R., Koopmans M.P.G., Geurtsvankessel C., van der Eijk A.A. Infectious Diseases (except HIV/AIDS) 2020. Shedding of infectious virus in hospitalized patients with coronavirus disease-2019 (COVID-19): duration and key determinants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Ma X. Acute respiratory failure in COVID-19: is it “typical” ARDS? Crit. Care. 2020;24:198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karagiannidis C., Mostert C., Hentschker C., Voshaar T., Malzahn J., Schillinger G., Klauber J., Janssens U., Marx G., Weber-Carstens S., Kluge S., Pfeifer M., Grabenhenrich L., Welte T., Busse R. Case characteristics, resource use, and outcomes of 10 021 patients with COVID-19 admitted to 920 German hospitals: an observational study. Lancet Respir. Med. 2020;8:853–862. doi: 10.1016/S2213-2600(20)30316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossen L.M., Branum A.M., Ahmad F.B., Sutton P., Anderson R.N. Excess deaths associated with COVID-19, by age and race and ethnicity - United States, January 26-October 3, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:1522–1527. doi: 10.15585/mmwr.mm6942e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Severe Outcomes Among Patients With Coronavirus Disease 2019 (COVID-19) — United States, February 12–March 16, 2020. Vol. 68. 2020. https://stacks.cdc.gov/view/cdc/85951 (accessed October 27, 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubino F., Amiel S.A., Zimmet P., Alberti G., Bornstein S., Eckel R.H., Mingrone G., Boehm B., Cooper M.E., Chai Z., Del Prato S., Ji L., Hopkins D., Herman W.H., Khunti K., Mbanya J.-C., Renard E. New-onset diabetes in Covid-19. N. Engl. J. Med. 2020;383:789–790. doi: 10.1056/NEJMc2018688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y., Li X., Geng D., Mei N., Wu P.-Y., Huang C.-C., Jia T., Zhao Y., Wang D., Xiao A., Yin B. Cerebral micro-structural changes in COVID-19 patients - an MRI-based 3-month follow-up study. EClinicalMedicine. 2020;25:100484. doi: 10.1016/j.eclinm.2020.100484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J., Wang B., Sun X.-X., Wang C.-F., Yang X., Lin P., Deng Y.-Q., Wei D., Yang X.-M., Zhu Y.-M., Zhang K., Zheng Z.-H., Miao J.-L., Guo T., Shi Y., Zhang J., Fu L., Wang Q.-Y., Bian H., Zhu P., Chen Z.-N. 2020. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. 2020.03.14.988345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thépaut M., Luczkowiak J., Vivès C., Labiod N., Bally I., Lasala F., Grimoire Y., Fenel D., Sattin S., Thielens N., Schoehn G., Bernardi A., Delgado R., Fieschi F. 2020. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. 2020.08.09.242917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. HCA lung biological network, SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.-E., Barbry P., Leslie A., Kiem H.-P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J., HCA Lung Biological Network HCA lung biological network, SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.035. 1016–1035.e19, Electronic address: lung-network@humancellatlas.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L., Li X., Chen M., Feng Y., Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovasc. Res. 2020;116:1097–1100. doi: 10.1093/cvr/cvaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diao B., Wang C., Wang R., Feng Z., Tan Y., Wang H., Wang C., Liu L., Liu Y., Liu Y., Wang G., Yuan Z., Ren L., Wu Y., Chen Y. 2020. Human Kidney Is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection, Infectious Diseases (except HIV/AIDS) [DOI] [Google Scholar]
- 41.Ziegler C., Allon S.J., Nyquist S.K., Mbano I., Miao V.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., II, Kazer S., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Tzouanas C.N., Taliaferro F., Guo Z., Wang J.P., Dwyer D.F., Buchheit K.M., Boyce J., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Berger B., Leslie A., Flynn J.L., Fortune S.M., Finberg R.W., Kean L., Garber M., Schmidt A., Lingwood D., Shalek A.K., Ordovas-Montanes J., Lung Biological Network, HCA . 2020. SARS-CoV-2 Receptor ACE2 is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Enriched in Specific Cell Subsets Across Tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. 281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 47.Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 48.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.-C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894–904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benihoud K., Yeh P., Perricaudet M. Adenovirus vectors for gene delivery. Curr. Opin. Biotechnol. 1999;10:440–447. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- 53.Rea D., Schagen F.H., Hoeben R.C., Mehtali M., Havenga M.J., Toes R.E., Melief C.J., Offringa R. Adenoviruses activate human dendritic cells without polarization toward a T-helper type 1-inducing subset. J. Virol. 1999;73:10245–10253. doi: 10.1128/jvi.73.12.10245-10253.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., He X., Martinez D.R., Rutten L., Bos R., van Manen D., Vellinga J., Custers J., Langedijk J.P., Kwaks T., Bakkers M.J.G., Zuijdgeest D., Huber S.K.R., Atyeo C., Fischinger S., Burke J.S., Feldman J., Hauser B.M., Caradonna T.M., Bondzie E.A., Dagotto G., Gebre M.S., Hoffman E., Jacob-Dolan C., Kirilova M., Li Z., Lin Z., Mahrokhian S.H., Maxfield L.F., Nampanya F., Nityanandam R., Nkolola J.P., Patel S., Ventura J.D., Verrington K., Wan H., Pessaint L., Van Ry A., Blade K., Strasbaugh A., Cabus M., Brown R., Cook A., Zouantchangadou S., Teow E., Andersen H., Lewis M.G., Cai Y., Chen B., Schmidt A.G., Reeves R.K., Baric R.S., Lauffenburger D.A., Alter G., Stoffels P., Mammen M., Van Hoof J., Schuitemaker H., Barouch D.H. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020 doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., Grousova D.M., Erokhova A.S., Kovyrshina A.V., Botikov A.G., Izhaeva F.M., Popova O., Ozharovskaya T.A., Esmagambetov I.B., Favorskaya I.A., Zrelkin D.I., Voronina D.V., Shcherbinin D.N., Semikhin A.S., Simakova Y.V., Tokarskaya E.A., Lubenets N.L., Egorova D.A., Shmarov M.M., Nikitenko N.A., Morozova L.F., Smolyarchuk E.A., Kryukov E.V., Babira V.F., Borisevich S.V., Naroditsky B.S., Gintsburg A.L. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020 doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., Li J.-X., Yang B.-F., Wang L., Wang W.-J., Wu S.-P., Wang Z., Wu X.-H., Xu J.-J., Zhang Z., Jia S.-Y., Wang B.-S., Hu Y., Liu J.-J., Zhang J., Qian X.-A., Li Q., Pan H.-X., Jiang H.-D., Deng P., Gou J.-B., Wang X.-W., Wang X.-H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S., Zhong G., Zhang J., Shuai L., Zhang Z., Wen Z., Wang B., Zhao Z., Song X., Chen Y., Liu R., Fu L., Zhang J., Guo Q., Wang C., Yang Y., Fang T., Lv P., Wang J., Xu J., Li J., Yu C., Hou L., Bu Z., Chen W. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020;11:4081. doi: 10.1038/s41467-020-17972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang Z., Gao G., Reyes-Sandoval A., Cohen C.J., Li Y., Bergelson J.M., Wilson J.M., Ertl H.C.J. Novel, chimpanzee serotype 68-based adenoviral vaccine carrier for induction of antibodies to a transgene product. J. Virol. 2002;76:2667–2675. doi: 10.1128/JVI.76.6.2667-2675.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., Jia S.-Y., Jiang H.-D., Wang L., Jiang T., Hu Y., Gou J.-B., Xu S.-B., Xu J.-J., Wang X.-W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., Faust S.N., Finn A., Flaxman A.L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A.M., Pollock K.M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J. Oxford COVID Vaccine Trial Group, Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E.R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Pérez-Pérez L., Edwards N.J., Wright D., Bissett C., Gilbride C., Williamson B.N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P.W., Scott D., Saturday G., de Wit E., Gilbert S.C., Munster V.J. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020 doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graham B.S., Mascola J.R., Fauci A.S. Novel vaccine technologies: essential components of an adequate response to emerging viral diseases. JAMA. 2018;319:1431–1432. doi: 10.1001/jama.2018.0345. [DOI] [PubMed] [Google Scholar]
- 63.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.-J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., Mihai C., Lynn A., McFadyen I., Moore M.J., Senn J.J., Stanton M.G., Almarsson Ö., Ciaramella G., Brito L.A. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bashirullah A., Cooperstock R.L., Lipshitz H.D. Spatial and temporal control of RNA stability. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., LaBranche C.C., Montefiori D.C., Lobby J.L., Saunders K.O., Liao H.-X., Korber B.T., Sutherland L.L., Scearce R.M., Hraber P.T., Tombácz I., Muramatsu H., Ni H., Balikov D.A., Li C., Mui B.L., Tam Y.K., Krammer F., Karikó K., Polacino P., Eisenlohr L.C., Madden T.D., Hope M.J., Lewis M.G., Lee K.K., Hu S.-L., Hensley S.E., Cancro M.P., Haynes B.F., Weissman D. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., 2nd, Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H., mRNA-1273 Study Group An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., Dinnon K.H., Elbashir S.M., Shaw C.A., Woods A., Fritch E.J., Martinez D.R., Bock K.W., Minai M., Nagata B.M., Hutchinson G.B., Wu K., Henry C., Bahi K., Garcia-Dominguez D., Ma L., Renzi I., Kong W.-P., Schmidt S.D., Wang L., Zhang Y., Phung E., Chang L.A., Loomis R.J., Altaras N.E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M.K., Shi W., Leung K., Yang E.S., West A., Gully K.L., Stevens L.J., Wang N., Wrapp D., Doria-Rose N.A., Stewart-Jones G., Bennett H., Alvarado G.S., Nason M.C., Ruckwardt T.J., McLellan J.S., Denison M.R., Chappell J.D., Moore I.N., Morabito K.M., Mascola J.R., Baric R.S., Carfi A., Graham B.S. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020 doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., Nagata B.M., Andersen H., Martinez D.R., Noe A.T., Douek N., Donaldson M.M., Nji N.N., Alvarado G.S., Edwards D.K., Flebbe D.R., Lamb E., Doria-Rose N.A., Lin B.C., Louder M.K., O’Dell S., Schmidt S.D., Phung E., Chang L.A., Yap C., Todd J.-P.M., Pessaint L., Van Ry A., Browne S., Greenhouse J., Putman-Taylor T., Strasbaugh A., Campbell T.-A., Cook A., Dodson A., Steingrebe K., Shi W., Zhang Y., Abiona O.M., Wang L., Pegu A., Yang E.S., Leung K., Zhou T., Teng I.-T., Widge A., Gordon I., Novik L., Gillespie R.A., Loomis R.J., Moliva J.I., Stewart-Jones G., Himansu S., Kong W.-P., Nason M.C., Morabito K.M., Ruckwardt T.J., Ledgerwood J.E., Gaudinski M.R., Kwong P.D., Mascola J.R., Carfi A., Lewis M.G., Baric R.S., McDermott A., Moore I.N., Sullivan N.J., Roederer M., Seder R.A., Graham B.S. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walsh E.E., Frenck R., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Thompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Sahin U., Gruber W.C. RNA-based COVID-19 vaccine BNT162b2 selected for a pivotal efficacy study. medRxiv. 2020 doi: 10.1101/2020.08.17.20176651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Şahin U., Jansen K.U. Phase 1/2 study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020 doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 73.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O’Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., 2nd, Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H., mRNA-1273 Study Group Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Erasmus J.H., Khandhar A.P., Walls A.C., Hemann E.A., O’Connor M.A., Murapa P., Archer J., Leventhal S., Fuller J., Lewis T., Draves K.E., Randall S., Guerriero K.A., Duthie M.S., Carter D., Reed S.G., Hawman D.W., Feldmann H., Gale M., Jr., Veesler D., Berglund P., Fuller D.H. Single-dose replicating RNA vaccine induces neutralizing antibodies against SARS-CoV-2 in nonhuman primates. bioRxiv. 2020 doi: 10.1101/2020.05.28.121640. [DOI] [Google Scholar]
- 76.de Alwis R., Gan E.S., Chen S., Leong Y.S., Tan H.C., Zhang S.L., Yau C., Matsuda D., Allen E., Hartman P., Park J., Alayyoubi M., Bhaskaran H., Dukanovic A., Bao B., Clemente B., Vega J., Roberts S., Gonzalez J.A., Sablad M., Yelin R., Taylor W., Tachikawa K., Parker S., Karmali P., Davis J., Sullivan S.M., Hughes S.G., Chivukula P., Ooi E.E. 2020. A Single Dose of Self-Transcribing and Replicating RNA Based SARS-CoV-2 Vaccine Produces Protective Adaptive Immunity In Mice. 2020.09.03.280446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 78.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M., Xu Z., Walters J., Bhojnagarwala P., Yang M., Chokkalingam N., Pezzoli P., Parzych E., Reuschel E.L., Doan A., Tursi N., Vasquez M., Choi J., Tello-Ruiz E., Maricic I., Bah M.A., Wu Y., Amante D., Park D.H., Dia Y., Ali A.R., Zaidi F.I., Generotti A., Kim K.Y., Herring T.A., Reeder S., Andrade V.M., Buttigieg K., Zhao G., Wu J.-M., Li D., Bao L., Liu J., Deng W., Qin C., Brown A.S., Khoshnejad M., Wang N., Chu J., Wrapp D., McLellan J.S., Muthumani K., Wang B., Carroll M.W., Kim J.J., Boyer J., Kulp D.W., Humeau L.M.P.F., Weiner D.B., Broderick K.E. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y.-J., Zeng G., Pan H.-X., Li C.-G., Kan B., Hu Y.-L., Mao H.-Y., Xin Q.-Q., Chu K., Han W.-X., Chen Z., Tang R., Yin W.-D., Chen X., Gong X.-J., Qin C., Hu Y.-S., Liu X.-Y., Cui G.-L., Jiang C.-B., Zhang H.-M., Li J.-X., Yang M.-N., Lian X.-J., Song Y., Lu J.-X., Wang X.-X., Xu M., Gao Q., Zhu F.-C. Immunogenicity and safety of a SARS-CoV-2 inactivated vaccine in healthy adults aged 18–59 years: report of the randomized, double-blind, and placebo-controlled phase 2 clinical trial. Pub. Global Health. 2020 doi: 10.1101/2020.07.31.20161216. [DOI] [Google Scholar]
- 80.Xia S., Duan K., Zhang Y., Zhao D., Zhang H., Xie Z., Li X., Peng C., Zhang Y., Zhang W., Yang Y., Chen W., Gao X., You W., Wang X., Wang Z., Shi Z., Wang Y., Yang X., Zhang L., Huang L., Wang Q., Lu J., Yang Y., Guo J., Zhou W., Wan X., Wu C., Wang W., Huang S., Du J., Meng Z., Pan A., Yuan Z., Shen S., Guo W., Yang X. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. JAMA. 2020 doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fulginiti V.A., Eller J.J., Downie A.W., Henry Kempe C. Altered reactivity to measles virus: atypical measles in children previously immunized with inactivated measles virus vaccines. JAMA. 1967;202:1075–1080. doi: 10.1001/jama.202.12.1075. [DOI] [PubMed] [Google Scholar]
- 82.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., Parrott R.H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 83.Cao W.-C., Liu W., Zhang P.-H., Zhang F., Richardus J.H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 2007;357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 84.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J., McGrath M., Weston S., Piedra P.A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J.D., Griffin P., Wilkinson B., Glenn G.M. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Recombinant New Coronavirus Vaccine (CHO Cells) to Prevent SARS-CoV-2 Phase I Clinical Trial (≥60 Years Old), (n.d.). https://clinicaltrials.gov/ct2/show/NCT04550351?term=Recombinant+new+coronavirus+vaccine+%28CHO+cell%29&draw=2&rank=1 (accessed October 3, 2020).
- 86.Clinical Study of Recombinant Novel Coronavirus Vaccine, (n.d.). https://clinicaltrials.gov/ct2/show/NCT04466085?term=Recombinant+new+coronavirus+vaccine+%28CHO+cell%29&draw=2&rank=2 (accessed October 3, 2020).
- 87.Phase I Clinical Study of Recombinant Novel Coronavirus Vaccine, (n.d.). https://clinicaltrials.gov/ct2/show/NCT04445194?term=Recombinant+new+coronavirus+vaccine+%28CHO+cell%29&draw=2&rank=3 (accessed October 3, 2020).
- 88.Johari Y.B., Jaffé S.R.P., Scarrott J.M., Johnson A.O., Mozzanino T., Pohle T.H., Maisuria S., Bhayat-Cammack A., Brown A.J., Lan Tee K., Jackson P.J., Wong T.S., Dickman M.J., Sargur R., James D.C. 2020. Production of Trimeric SARS-CoV-2 Spike Protein by CHO Cells for Serological COVID-19 Testing, Infectious Diseases (except HIV/AIDS) [DOI] [PubMed] [Google Scholar]
- 89.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11 doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.-C., Tian G., Jiang H.-W., Tao S.-C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pruijssers A.J., George A.S., Schäfer A., Leist S.R., Gralinksi L.E., Dinnon K.H., III, Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., Gully K., Martinez D.R., Brown A.J., Graham R.L., Perry J.K., Du Pont V., Pitts J., Ma B., Babusis D., Murakami E., Feng J.Y., Bilello J.P., Porter D.P., Cihlar T., Baric R.S., Denison M.R., Sheahan T.P. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., de Castilla D. Lopez, Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.-D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., ACTT-1 Study Group Members Remdesivir for the treatment of covid-19 - preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.VEKLURY® (remdesivir), (n.d.). https://www.remdesivir.com/us/?gclid=CjwKCAjwiOv7BRBREiwAXHbv3J87tFSsxkuYa_BCJ7EKOz99HPLQf7up36YUbQp_pvlnUYJ5FEGKyRoC9EcQAvD_BwE&gclsrc=aw.ds (accessed October 5, 2020).
- 95.Gilead Presents Additional Data on Investigational Antiviral Remdesivir for the Treatment of COVID-19, (n.d.). https://www.gilead.com/news-and-press/press-room/press-releases/2020/7/gilead-presents-additional-data-on-investigational-antiviral-remdesivir-for-the-treatment-of-covid-19 (accessed October 5, 2020).
- 96.Villiger P.M., Adler S., Kuchen S., Wermelinger F., Dan D., Fiege V., Bütikofer L., Seitz M., Reichenbach S. Tocilizumab for induction and maintenance of remission in giant cell arteritis: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet. 2016;387:1921–1927. doi: 10.1016/S0140-6736(16)00560-2. [DOI] [PubMed] [Google Scholar]
- 97.Galeotti C., Boucheron A., Guillaume S., Koné-Paut I. Sustained remission of multicentric Castleman disease in children treated with tocilizumab, an anti-interleukin-6 receptor antibody. Mol. Cancer Ther. 2012;11:1623–1626. doi: 10.1158/1535-7163.MCT-11-0972. [DOI] [PubMed] [Google Scholar]
- 98.Le R.Q., Li L., Yuan W., Shord S.S., Nie L., Habtemariam B.A., Przepiorka D., Farrell A.T., Pazdur R. FDA approval summary: tocilizumab for treatment of chimeric antigen receptor T cell-induced severe or life-threatening cytokine release syndrome. Oncologist. 2018;23:943. doi: 10.1634/theoncologist.2018-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Price C.C., Altice F.L., Shyr Y., Koff A., Pischel L., Goshua G., Azar M.M., Mcmanus D., Chen S.-C., Gleeson S.E., Britto C.J., Azmy V., Kaman K., Gaston D.C., Davis M., Burrello T., Harris Z., Villanueva M.S., Aoun-Barakat L., Kang I., Seropian S., Chupp G., Bucala R., Kaminski N., Lee A.I., LoRusso P.M., Topal J.E., Dela Cruz C., Malinis M. Tocilizumab treatment for cytokine release syndrome in hospitalized COVID-19 patients: survival and clinical outcomes. Chest. 2020 doi: 10.1016/j.chest.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., Pan A., Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27 doi: 10.1016/j.chom.2020.04.009. 992–1000.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Malgie J., Schoones J.W., Pijls B.G. Decreased mortality in coronavirus disease 2019 patients treated with tocilizumab: a rapid systematic review and meta-analysis of observational studies. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.What's New, (n.d.). https://www.covid19treatmentguidelines.nih.gov/whats-new/ (accessed October 7, 2020).
- 104.Stone J.H., Frigault M.J., Serling-Boyd N.J., Fernandes A.D., Harvey L., Foulkes A.S., Horick N.K., Healy B.C., Shah R., Bensaci A.M., Woolley A.E., Nikiforow S., Lin N., Sagar M., Schrager H., Huckins D.S., Axelrod M., Pincus M.D., Fleisher J., Sacks C.A., Dougan M., North C.M., Halvorsen Y.-D., Thurber T.K., Dagher Z., Scherer A., Wallwork R.S., Kim A.Y., Schoenfeld S., Sen P., Neilan T.G., Perugino C.A., Unizony S.H., Collier D.S., Matza M.A., Yinh J.M., Bowman K.A., Meyerowitz E., Zafar A., Drobni Z.D., Bolster M.B., Kohler M., D’Silva K.M., Dau J., Lockwood M.M., Cubbison C., Weber B.N., Mansour M.K. BACC bay tocilizumab trial investigators, efficacy of tocilizumab in patients hospitalized with covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nakashima T., Yokoyama A., Ohnishi H., Hamada H., Ishikawa N., Haruta Y., Hattori N., Tanigawa K., Kohno N. Circulating KL-6/MUC1 as an independent predictor for disseminated intravascular coagulation in acute respiratory distress syndrome. J. Intern. Med. 2008;263:432–439. doi: 10.1111/j.1365-2796.2008.01929.x. [DOI] [PubMed] [Google Scholar]
- 106.Lu W., Liu X., Wang T., Liu F., Zhu A., Lin Y., Luo J., Ye F., He J., Zhao J., Li Y., Zhong N. Elevated MUC1 and MUC5AC mucin protein levels in airway mucus of critical ill COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.26406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yokoyama A., Kohno N., Hamada H., Sakatani M., Ueda E., Kondo K., Hirasawa Y., Hiwada K. Circulating KL-6 predicts the outcome of rapidly progressive idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 1998;158:1680–1684. doi: 10.1164/ajrccm.158.5.9803115. [DOI] [PubMed] [Google Scholar]
- 108.Alimova M., Sidhom E.-H., Satyam A., Dvela-Levitt M., Melanson M., Chamberlain B.T., Alper S.L., Santos J., Gutierrez J., Subramanian A., Grinkevich E., Bricio E.R., Kim C., Clark A., Watts A., Thompson R., Marshall J., Pablo J.L., Coraor J., Roignot J., Vernon K.A., Keller K., Campbell A., Emani M., Racette M., Bazua-Valenti S., Padovano V., Weins A., McAdoo S.P., Tam F.W.K., Ronco L., Wagner F., Tsokos G.C., Shaw J.L., Greka A. A high content screen for mucin-1-reducing compounds identifies fostamatinib as a candidate for rapid repurposing for acute lung injury during the COVID-19 pandemic. bioRxiv. 2020 doi: 10.1101/2020.06.30.180380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Geahlen R.L. Getting Syk: spleen tyrosine kinase as a therapeutic target. Trends Pharmacol. Sci. 2014;35:414–422. doi: 10.1016/j.tips.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mócsai A., Ruland J., Tybulewicz V.L.J. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat. Rev. Immunol. 2010;10:387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Weinblatt M.E., Kavanaugh A., Genovese M.C., Jones D.A., Musser T.K., Grossbard E.B., Magilavy D.B. Effects of fostamatinib (R788), an oral spleen tyrosine kinase inhibitor, on health-related quality of life in patients with active rheumatoid arthritis: analyses of patient-reported outcomes from a randomized, double-blind, placebo-controlled trial. J. Rheumatol. 2013;40:369–378. doi: 10.3899/jrheum.120923. [DOI] [PubMed] [Google Scholar]
- 112.Hoepel W., Chen H.-J., Allahverdiyeva S., Manz X., Aman J., A.U.C.-19 Biobank, Bonta P., Brouwer P., de Taeye S., Caniels T., van der Straten K., Golebski K., Griffith G., Jonkers R., Larsen M., Linty F., Neele A., Nouta J., van Baarle F., van Drunen C., Vlaar A., de Bree G., Sanders R., Willemsen L., Wuhrer M., Bogaard H.J., van Gils M., Vidarsson G., de Winther M., den Dunnen J. Anti-SARS-CoV-2 IgG from severely ill COVID-19 patients promotes macrophage hyper-inflammatory responses. Cold Spring Harbor Lab. 2020 doi: 10.1101/2020.07.13.190140. 2020.07.13.190140. [DOI] [Google Scholar]
- 113.Ben-Zvi I., Kivity S., Langevitz P., Shoenfeld Y. Hydroxychloroquine: from malaria to autoimmunity. Clin. Rev. Allergy Immunol. 2012;42:145–153. doi: 10.1007/s12016-010-8243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ruiz-Irastorza G., Ramos-Casals M., Brito-Zeron P., Khamashta M.A. Clinical efficacy and side effects of antimalarials in systemic lupus erythematosus: a systematic review. Ann. Rheum. Dis. 2010;69:20–28. doi: 10.1136/ard.2008.101766. [DOI] [PubMed] [Google Scholar]
- 115.M.J. Bergman, M.D. (5 P. Martin J. Bergman), The Nine Lives of Hydroxychloroquine, (n.d.). https://rheumnow.com/blog/nine-lives-hydroxychloroquine (accessed October 30, 2020).
- 116.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Skipper C.P., Pastick K.A., Engen N.W., Bangdiwala A.S., Abassi M., Lofgren S.M., Williams D.A., Okafor E.C., Pullen M.F., Nicol M.R., Nascene A.A., Hullsiek K.H., Cheng M.P., Luke D., Lother S.A., MacKenzie L.J., Drobot G., Kelly L.E., Schwartz I.S., Zarychanski R., McDonald E.G., Lee T.C., Rajasingham R., Boulware D.R. Hydroxychloroquine in nonhospitalized adults with early COVID-19: a randomized trial. Ann. Intern. Med. 2020 doi: 10.7326/M20-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D.K., Kubin C., Barr R.G., Sobieszczyk M.E., Schluger N.W. Observational study of hydroxychloroquine in hospitalized patients with covid-19. N. Engl. J. Med. 2020;382:2411–2418. doi: 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Horby P., Mafham M., Linsell L., Bell J.L., Staplin N., Emberson J.R., Wiselka M., Ustianowski A., Elmahi E., Prudon B. Effect of hydroxychloroquine in hospitalized patients with COVID-19: preliminary results from a multi-centre, randomized, controlled trial. MedRxiv. 2020 doi: 10.1101/2020.07.15.20151852. [DOI] [Google Scholar]
- 121.Chloroquine or Hydroxychloroquine, (n.d.). https://www.covid19treatmentguidelines.nih.gov/antiviral-therapy/chloroquine-or-hydroxychloroquine-with-or-without-azithromycin/ (accessed October 7, 2020).
- 122.Harrison C., Kiladjian J.-J., Al-Ali H.K., Gisslinger H., Waltzman R., Stalbovskaya V., McQuitty M., Hunter D.S., Levy R., Knoops L., Cervantes F., Vannucchi A.M., Barbui T., Barosi G. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N. Engl. J. Med. 2012;366:787–798. doi: 10.1056/NEJMoa1110556. [DOI] [PubMed] [Google Scholar]
- 123.La Rosée F., Bremer H.C., Gehrke I., Kehr A., Hochhaus A., Birndt S., Fellhauer M., Henkes M., Kumle B., Russo S.G., La Rosée P. The Janus kinase 1/2 inhibitor ruxolitinib in COVID-19 with severe systemic hyperinflammation. Leukemia. 2020;34:1805–1815. doi: 10.1038/s41375-020-0891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fardet L., Fève B. Systemic glucocorticoid therapy: a review of its metabolic and cardiovascular adverse events. Drugs. 2014;74:1731–1745. doi: 10.1007/s40265-014-0282-9. [DOI] [PubMed] [Google Scholar]
- 125.Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A., Jose J., Pinto R., Al-Omari A., Kharaba A., Almotairi A., Al Khatib K., Alraddadi B., Shalhoub S., Abdulmomen A., Qushmaq I., Mady A., Solaiman O., Al-Aithan A.M., Al-Raddadi R., Ragab A., Balkhy H.H., Al Harthy A., Deeb A.M., Al Mutairi H., Al-Dawood A., Merson L., Hayden F.G., Fowler R.A., Saudi Critical Care Trial Group Corticosteroid therapy for critically ill patients with middle east respiratory syndrome. Am. J. Respir. Crit. Care Med. 2018;197:757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 126.Huang R., Zhu C., Wang Jian, Xue L., Li C., Yan X., Huang S., Zhang B., Zhu L., Xu T., Ming F., Zhao Y., Cheng J., Shao H., Zhao X.-A., Sang D., Zhao H., Guan X., Chen X., Chen Y., Wei J., Issa R., Liu L., Yan X., Wu C. Corticosteroid therapy is associated with the delay of SARS-CoV-2 clearance in COVID-19 patients. Eur. J. Pharmacol. 2020;889:173556. doi: 10.1016/j.ejphar.2020.173556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.RECOVERY Collaborative Group. Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., Prudon B., Green C., Felton T., Chadwick D., Rege K., Fegan C., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Montgomery A., Rowan K., Juszczak E., Baillie J.K., Haynes R., Landray M.J. Dexamethasone in hospitalized patients with covid-19 - preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu C., Chen X., Cai Y., Xia J.'an, Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Y., Jiang W., He Q., Wang C., Wang B., Zhou P., Dong N., Tong Q. A retrospective cohort study of methylprednisolone therapy in severe patients with COVID-19 pneumonia. Signal Transduct Target Ther. 2020;5:57. doi: 10.1038/s41392-020-0158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bartoletti M., Marconi L., Scudeller L., Pancaldi L., Tedeschi S., Giannella M., Rinaldi M., Bussini L., Valentini I., Ferravante A.F., Potalivo A., Marchionni E., Fornaro G., Pascale R., Pasquini Z., Puoti M., Merli M., Barchiesi F., Volpato F., Rubin A., Saracino A., Tonetti T., Gaibani P., Ranieri V.M., Viale P., Cristini F., PREDICO Study Group Efficacy of corticosteroid treatment for hospitalized patients with severe COVID-19: a multicenter study. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hu Z., Lv Y., Xu C., Sun W., Chen W., Peng Z., Chen C., Cui X., Jiao D., Cheng C., Chi Y., Wei H., Hu C., Zeng Y., Zhang X., Yi Y. Clinical use of short-course and low-dose corticosteroids in patients with non-severe COVID-19 during pneumonia progression. Front. Public Health. 2020;8:355. doi: 10.3389/fpubh.2020.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Corticosteroids, (n.d.). https://www.covid19treatmentguidelines.nih.gov/immune-based-therapy/immunomodulators/corticosteroids/ (accessed October 7, 2020).
- 133.Malik T.H., Lavin P.J., Goicoechea de Jorge E., Vernon K.A., Rose K.L., Patel M.P., de Leeuw M., Neary J.J., Conlon P.J., Winn M.P., Pickering M.C. A hybrid CFHR3-1 gene causes familial C3 glomerulopathy. J. Am. Soc. Nephrol. 2012;23:1155–1160. doi: 10.1681/ASN.2012020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Jorge E.G., Macor P., Paixão-Cavalcante D., Rose K.L., Tedesco F., Cook H.T., Botto M., Pickering M.C. The development of atypical hemolytic uremic syndrome depends on complement C5. J. Am. Soc. Nephrol. 2011;22:137–145. doi: 10.1681/ASN.2010050451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hillmen P., Young N.S., Schubert J., Brodsky R.A., Socié G., Muus P., Röth A., Szer J., Elebute M.O., Nakamura R., Browne P., Risitano A.M., Hill A., Schrezenmeier H., Fu C.-L., Maciejewski J., Rollins S.A., Mojcik C.F., Rother R.P., Luzzatto L. The complement inhibitor eculizumab in paroxysmal nocturnal hemoglobinuria. N. Engl. J. Med. 2006;355:1233–1243. doi: 10.1056/NEJMoa061648. [DOI] [PubMed] [Google Scholar]
- 136.Satyam A., Graef E.R., Lapchak P.H., Tsokos M.G., Dalle Lucca J.J., Tsokos G.C. Complement and coagulation cascades in trauma. Acute Med. Surg. 2019;6:329–335. doi: 10.1002/ams2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., Li W.W., Li V.W., Mentzer S.J., Jonigk D. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mahajan R., Lipton M., Broglie L., Jain N.G., Uy N.S. Eculizumab treatment for renal failure in a pediatric patient with COVID-19. J. Nephrol. 2020 doi: 10.1007/s40620-020-00858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Diurno F., Numis F.G., Porta G., Cirillo F., Maddaluno S., Ragozzino A., De Negri P., Di Gennaro C., Pagano A., Allegorico E., Bressy L., Bosso G., Ferrara A., Serra C., Montisci A., D’Amico M., Morello S. Schiano Lo, Di Costanzo G., Tucci A.G., Marchetti P., Di Vincenzo U., Sorrentino I., Casciotta A., Fusco M., Buonerba C., Berretta M., Ceccarelli M., Nunnari G., Diessa Y., Cicala S., Facchini G. Eculizumab treatment in patients with COVID-19: preliminary results from real life ASL Napoli 2 Nord experience. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4040–4047. doi: 10.26355/eurrev_202004_20875. [DOI] [PubMed] [Google Scholar]
- 141.Laurence J., Mulvey J.J., Seshadri M., Racanelli A., Harp J., Schenck E.J., Zappetti D., Horn E.M., Magro C.M. Anti-complement C5 therapy with eculizumab in three cases of critical COVID-19. Clin. Immunol. 2020;219:108555. doi: 10.1016/j.clim.2020.108555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mastaglio S., Ruggeri A., Risitano A.M., Angelillo P., Yancopoulou D., Mastellos D.C., Huber-Lang M., Piemontese S., Assanelli A., Garlanda C., Lambris J.D., Ciceri F. The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clin. Immunol. 2020;215:108450. doi: 10.1016/j.clim.2020.108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mastellos D.C., Pires da Silva B.G.P., Fonseca B.A.L., Fonseca N.P., Auxiliadora-Martins M., Mastaglio S., Ruggeri A., Sironi M., Radermacher P., Chrysanthopoulou A., Skendros P., Ritis K., Manfra I., Iacobelli S., Huber-Lang M., Nilsson B., Yancopoulou D., Connolly E.S., Garlanda C., Ciceri F., Risitano A.M., Calado R.T., Lambris J.D. Complement C3 vs C5 inhibition in severe COVID-19: early clinical findings reveal differential biological efficacy. Clin. Immunol. 2020;220:108598. doi: 10.1016/j.clim.2020.108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Urwyler P., Moser S., Charitos P., Heijnen I.A.F.M., Rudin M., Sommer G., Giannetti B.M., Bassetti S., Sendi P., Trendelenburg M., Osthoff M. Treatment of COVID-19 with conestat alfa, a regulator of the complement, contact activation and Kallikrein-Kinin system. Front. Immunol. 2020;11:2072. doi: 10.3389/fimmu.2020.02072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ye M., Fu D., Ren Y., Wang F., Wang D., Zhang F., Xia X., Lv T. Treatment with convalescent plasma for COVID-19 patients in Wuhan, China. J. Med. Virol. 2020 doi: 10.1002/jmv.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Joyner M.J., Bruno K.A., Klassen S.A., Kunze K.L., Johnson P.W., Lesser E.R., Wiggins C.C., Senefeld J.W., Klompas A.M., Hodge D.O., Shepherd J.R.A., Rea R.F., Whelan E.R., Clayburn A.J., Spiegel M.R., Baker S.E., Larson K.F., Ripoll J.G., Andersen K.J., Buras M.R., Vogt M.N.P., Herasevich V., Dennis J.J., Regimbal R.J., Bauer P.R., Blair J.E., van Buskirk C.M., Winters J.L., Stubbs J.R., van Helmond N., Butterfield B.P., Sexton M.A., Soto J.C. Diaz, Paneth N.S., Verdun N.C., Marks P., Casadevall A., Fairweather D., Carter R.E., Wright R.S. Safety update: COVID-19 convalescent plasma in 20,000 hospitalized patients. Mayo Clin. Proc. 2020;95:1888–1897. doi: 10.1016/j.mayocp.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D., Guo L., Zhang G., Li H., Xu Y., Chen M., Gao Z., Wang J., Ren L., Li M. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin. Infect. Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., Chung K.M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W.-Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Hernandez A. Romero, Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D.C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., Atwal G.S., Oyejide A., Goez-Gazi Y., Dutton J., Clemmons E., Staples H.M., Bartley C., Klaffke B., Alfson K., Gazi M., Gonzalez O., Dick E., Jr., Carrion R., Jr., Pessaint L., Porto M., Cook A., Brown R., Ali V., Greenhouse J., Taylor T., Andersen H., Lewis M.G., Stahl N., Murphy A.J., Yancopoulos G.D., Kyratsous C.A. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020 doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]