Abstract
Unlike small molecule drugs, therapeutic proteins must maintain the proper higher order structure (HOS) in order to maintain safety and efficacy. Due to the sensitivity of many protein systems, even small changes due to differences in protein expression or formulation can alter HOS. Previous work has demonstrated how hydroxyl radical protein footprinting (HRPF) can sensitively detect changes in protein HOS by measuring the average topography of the protein monomers, as well as identify specific regions of the therapeutic protein impacted by the conformational changes. However, HRPF is very sensitive to the radical scavenging capacity of the buffer; addition of organic buffers and/or excipients can dramatically alter the HRPF footprint without affecting protein HOS. By compensating for the radical scavenging effects of different adalimumab biosimilar formulations using real-time adenine dosimetry, we identify that sodium citrate buffer causes a modest decrease in average solvent accessibility compared to sodium phosphate buffer at the same pH. We find that the addition of polysorbate-80 does not alter the conformation of the biosimilar in either buffer, but it does provide substantial protection from protein conformational perturbation during short periods of exposure to high temperature. Compensated HRPF measurements are validated and contextualized by dynamic light scattering (DLS), which suggests that changes in adalimumab biosimilar aggregation is a major driver in measured changes in protein topography. Overall, compensated HRPF accurately measured conformational changes in adalimumab biosimilar that occurred during formulation changes, and identified the effect of formulation changes on protection of HOS from temperature extremes.
Keywords: biosimilars, hydroxyl radical protein footprinting, mass spectrometry, protein conformations, therapeutic proteins
INTRODUCTION
Over the last thirty years, the global market for biopharmaceuticals has increased tremendously, reaching sales exceeding $176 billion in 2015 with robust growth projected (1). A major issue in therapeutic protein development is protein stability. Not only must the protein remain covalently stable (e.g. side chain oxidation, deamidation, disulfide bond isomerization, etc.), but the non-covalent higher order structure (HOS) must remain intact. Perturbation of the biotherapeutic HOS can lead to loss of efficacy due to precipitation, aggregation, or loss of conformational integrity (2). In some cases, HOS perturbation can not only cause a loss of efficacy but also can induce adverse drug reactions due to aberrant pharmacology and patient immune response (3, 4). These issues make biotherapeutic formulation a key step; the formulation is important for protecting the biotherapeutic from modification, but it simultaneously can alter the HOS and conformational stability in complex and often unpredictable ways. Methods for the analysis of protein therapeutic HOS are critical, but they must be able to handle not only the inherent covalent heterogeneity of many protein therapeutic samples (e.g. protein glycoforms, antibody-drug conjugate stoichiometry heterogeneity) but they must also be able to handle heterogeneity in HOS that can occur, especially in unstable formulations (e.g. polydisperse protein aggregates, dynamic conformations).
One tool that has emerged for HOS analysis of large, complex and heterogeneous proteins (as therapeutic proteins and protein aggregates often are) is hydroxyl radical protein footprinting (HRPF). In HRPF, proteins in aqueous solution are exposed to hydroxyl radicals diffusing freely in solution. These hydroxyl radicals diffuse to the analyte protein and can potentially react with any of the amino acid side chains in the protein. The rate of this reaction is a function of two factors: the chemical reactivity of the amino acid (which is dependent on the amino acid (5, 6) combined with the inductive effects of neighboring amino acids (7, 8)); and the accessibility of the amino acid side chain to the solvated hydroxyl radical. This apparent rate of side chain oxidation can be measured by liquid chromatography coupled to mass spectrometry (LC-MS), where the amount of oxidation of amino acids in a given proteolytic peptide can be determined based on the relative intensities of the oxidized and unoxidized versions of that peptide sequence(9–12). By comparing the extent of oxidation of the same amino acid sequence in two different conformational states (e.g. monomeric versus aggregate), the differences in solvent accessibility of each reactive amino acid side chain can be determined (8, 11, 13, 14).
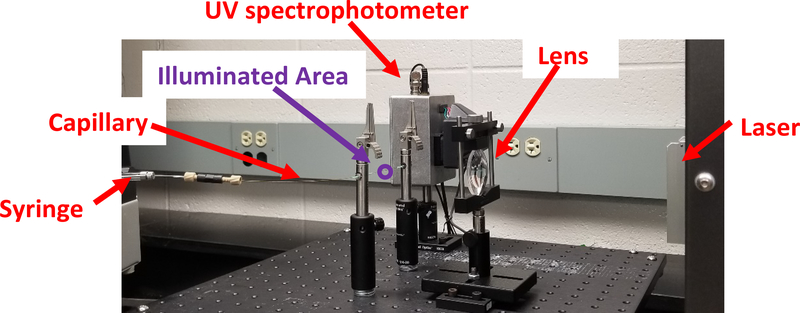
Many methods for generating hydroxyl radicals in situ with protein analytes have been developed (9, 10, 15–20). The most popular for the examination of therapeutic proteins is Fast Photochemical Oxidation of Proteins (FPOP, Figure 1) (19). In FPOP, proteins are mixed with hydrogen peroxide, along with a mild radical scavenger like glutamine. The mixture is flowed through a fused silica capillary, which passes through the path of a pulsed UV laser (usually a KrF excimer laser). The hydrogen peroxide absorbs the UV photon, photodissociating into two hydroxyl radicals. The glutamine radical scavenger limits the half-life of the hydroxyl radical, ensuring that the initial irreversible oxidation of the amino acid side chain by the hydroxyl radical is completed on the order of a microsecond or less (19). The illuminated sample flows out of the path of the UV laser before a second pulse is emitted, ensuring that no significant portion of the sample experiences more than a single hydroxyl radical exposure period, lasting less than a microsecond. Due to the short timescale of the reaction, proteins can be heavily modified by FPOP without labeling artifactual conformations induced by the FPOP process itself (8, 21). Unlike another popular mass spectrometry-based HOS analysis method, hydrogen-deuterium exchange, FPOP labeling is stable. This allows for much greater flexibility in post-labeling processing, clean-up and analysis.
Figure 1: FPOP optical bench for adalimumab biosimilar analysis.
Sample is mixed with H2O2, adenine radical dosimeter and glutamine scavenger and loaded into the syringe. Sample is pushed through the fused silica capillary through the focused beam path of a KrF excimer UV laser. The UV light photolyzes H2O2 into hydroxyl radicals, which oxidizes the protein and adenine dosimeter. The syringe flow pushes the illuminated sample out of the path of the laser prior to the next laser pulse, with an unilluminated exclusion volume between illuminated regions. Immediately after oxidation, the sample is passed through an inline UV spectrophotometer, which measures the UV absorbance of adenine at 265 nm. Sample is then deposited into a quench buffer to eliminate the remaining H2O2 and secondary oxidants.
Due to the ability to heavily label analyte proteins in a benchtop setup, FPOP has been used to examine therapeutic proteins and compare conformations between biosimilars and originator biotherapeutics (22), map antibody-antigen interaction interfaces (23–26), and examine structural consequences of gaps in the cold chain for therapeutic protein handling (22). However, a major difficulty in using HRPF to support formulation development is the radical scavenging effect of formulation components. Many formulation buffers and excipients are efficient hydroxyl radical scavengers, due to the ability of hydroxyl radicals to abstract hydrogens from C-H bonds and perform addition chemistry to aromatic systems with the corresponding organic radicals often reacting with molecular oxygen dissolved in solution (5). Comparing proteins in significantly different radical scavenging backgrounds results in a differential rate of labeling driven not by changes in conformation, but by differences in the effective hydroxyl radical concentration that can react with the analyte protein (27). Several methods have been published for measuring the effects of hydroxyl radical scavengers on the effective radical dose in FPOP experiments, a process known as hydroxyl radical dosimetry (28–30). One method published by our group is notable for its simple readout: adenine dosimetry (31, 32). Adenine absorbs UV light strongly at 265 nm; however, after reacting with hydroxyl radicals, the UV absorbance at 265 nm drops significantly. The loss of UV absorbance varies linearly with effective hydroxyl radical concentration, and similarly varies linearly with reducing protein or peptide oxidation in FPOP experiments (31).
Adenine dosimetry allows researchers to quickly measure if samples were measured under comparable radical scavenging backgrounds; however, the question remains how to correct for different radical scavenging backgrounds to allow for comparisons of HRPF data. Previous methods involved matching radical scavenging backgrounds through the addition of scavengers to solution (29). While this method works, it can alter the formulation properties and, in the presence of strong radical scavengers, can largely confound analysis by preventing most oxidation of the analyte system. Recently, we reported that altering the maximum hydroxyl radical concentration through manipulation of UV photon fluence and/or hydrogen peroxide concentration can compensate for different radical scavenging capacities of the analyte matrix in a technique we termed compensated FPOP. By measuring adenine dosimetry in real time with the addition of an inline UV detector immediately downstream of the FPOP illumination region (Figure 1), the effects of FPOP compensation can be measured in real time. Compensated FPOP was able to successfully provide the same HRPF footprint for two samples with the same conformation exposed under vastly different radical scavenging backgrounds (27). With this technology, we should be able to measure formulation-induced changes in biotherapeutic conformation and aggregation without compromising the amount of labeling of the analyte protein.
Here, we present the use of real time compensated FPOP to measure the effects of therapeutic protein formulation on protein conformation, aggregation, and stability to heat shock. Examining a heterologously expressed adalimumab biosimilar, we study the effects of changing buffer composition, as well as the addition of polysorbate-80 as a stabilizing excipient that is also a very potent hydroxyl radical scavenger. We also study the effects of buffer and polysorbate-80 on the conformational stability of the biotherapeutic after heat shock exposure. Using compensated FPOP, we were able to identify not only complex changes in aggregation properties, but also identify the regions of the protein that exhibited altered topography under these conditions. Data gathered were compared with dynamic light scattering (DLS), which were consistent with the observed compensated FPOP results and aided in interpretation of topography changes.
MATERIALS AND METHODS
Reagents
Sodium phosphate and sodium citrate, LC-MS grade water, acetonitrile, formic acid and hydrogen peroxide were purchased from Fisher Chemicals (Fair Lawn, NJ). Adenine and L-glutamine were purchased from Acros Biologicals. Catalase was purchased from Sigma (St. Louis, MO) Methionine amide was obtained from Bachem (Torrance, CA). Sequencing-grade modified trypsin was purchased from Promega Corp. (Madison, WI). All reagents were used without further purification. Purified water (18.2 MΩ) was obtained from a Synergy UV system (Millipore, Billerica, MA). Adalimumab was obtained from GlycoScientific (Athens, GA).
Compensated FPOP Labeling
A final adalimumab biosimilar concentration of 1mg/ml in 50 mM phosphate buffer, pH 6.0 or 50 mM sodium citrate buffer, pH 6.0 was incubated either at room temperature or at 55 °C for 1 hour. This mixture contained 2 mM adenine as a radical dosimeter to monitor the available radical dose in each sample (27, 31). 2 μL of freshly prepared 1 M hydrogen peroxide were added to the sample mixture immediately prior to laser irradiation. The total 20 μl of this sample mixture was irradiated by flow through the path of the pulsed ultraviolet laser beam. Briefly, a Compex Pro 102 KrF excimer laser (Coherent, Germany) was run at ~10.7 mJ/mm2/pulse, with a laser repetition rate of 15 Hz. The flow rate was adjusted to 11.7 μL/min to ensure a 15% exclusion volume between irradiated segments to help accounting for sample diffusion and laminar flow (33). The sample was passed through a Pioneer inline radical dosimeter (GenNext Technologies)for the real time monitoring of adenine readings. After illumination, each replicate was collected in a microcentrifuge tube containing 25 μl of quench mixture that contained 0.5 μg/μl H-Met-NH2 and 0.5 μg/μl catalase to eliminate secondary oxidants such as remaining hydrogen peroxide, protein peroxides, superoxide, etc. All experiments were performed in triplicate for statistical analysis. For performing compensation FPOP with sodium citrate buffer, the hydrogen peroxide concentration was increased to 130 mM in order to achieve the same ΔAbs265 as observed for the sample with sodium phosphate buffer.
FPOP Sample Processing and LC-MS
50 mM Tris, pH 8.0 containing 1 mM CaCl2 was added to the protein samples after FPOP. 5 mM of dithiothreitol was added and incubated the mixture at 95 °C for 15 minutes to denature and reduce the IgG1. After the mixture had been cooled to room temperature, a 1:20 trypsin weight ratio was added to the samples for overnight digestion at 37 °C with sample rotation. The trypsin reaction was stopped by adding 0.1% formic acid and the samples were stored at −20 °C until analysis was performed on nano-LC-MS/MS system.
The protein samples were analyzed on an Orbitrap Fusion instrument (Thermo Fisher Scientific) controlled with Xcalibur version 2.1.1 (Thermo Fisher, San Jose, CA). Samples were loaded onto an Acclaim PepMap 100 C18 nanocolumn (0.75 mm × 150 mm, 2 μm, Thermo Fisher Scientific). Separation of peptides on the chromatographic system was performed using mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) at a rate of 0.30 μL/min. The peptides were eluted with a gradient consisting of 2 to 10% solvent B over 5 min, then to 32% B in 26 min, ramped to 95 % B over 4 min, held for 4 min, and then returned to 2% B over 2 min and held for 8 min. Peptides were eluted directly into the nanospray source of an Orbitrap Fusion instrument using a conductive nanospray emitter obtained from Thermo Scientific. All data were acquired in positive ion mode. Collision-induced dissociation CID were used to fragment peptides, with an isolation width of 3 m/z units. The spray voltage was set to 2500 volts, and the temperature of the heated capillary was set to 300 °C. In CID mode, full MS scans were acquired from m/z 350 to 2000 followed by eight subsequent MS2 scans on the top eight most abundant peptide ions. The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
FPOP Data Analysis
Tryptic digests of adalimumab biosimilar were first identified using ByOnic version v2.10.5 (Protein Metrics) using its amino acid sequence. The enzyme specificity was set to cleave the protein after lysine and arginine. All possible major oxidation modifications (34) were included as variable modifications for the database searches. The peak intensities of the unoxidized peptides and their corresponding oxidation products observed in LC-MS were used to calculate the average oxidation events per peptide in the sample as previously reported (8). Briefly, peptide level oxidation was calculated by adding the ion intensities of all the oxidized peptides multiplied by the number of oxidation events required for the mass shift (e.g., one event for +16, two events for +32) and then divided by the sum of the ion intensities of all unoxidized and oxidized peptide masses as represented by equation 1.
| (1). |
where P denotes the oxidation events at the peptide level and I values are the peak intensities of oxidized and unoxidized peptides. For this system, no oxidation products other than the net addition of one or more oxygen atoms was detected at sufficient levels for quantification.
Dynamic Light Scattering
Adalimumab biosimilar at 1 mg/mL with either sodium phosphate or sodium citrate buffers in the presence or absence of 0.1% polysorbate-80 was incubated at room temperature or 55 °C for 1 hour. The hydrodynamic diameter of the protein samples was examined by dynamic light scattering on Zetasizer Nano ZS90 (Malvern Panalytical, Worcestershire, UK). The test was carried out at 25 °C using disposable polystyrene cuvettes. DLS was performed in triplicate, with representative spectra displayed herein.
RESULTS
Compensated FPOP Measures HOS Changes Driven by Buffer and Excipient
Using 50 mM sodium phosphate buffer, pH 6.0 as a reference state, we used FPOP to examine the changes in topography that occurred upon changing the buffer to 50 mM sodium citrate, pH 6.0. Without compensation, a strong increase in background radical scavenging was detected by adenine dosimetry upon switching to sodium citrate buffer. In phosphate buffer, adenine dosimetry gave a ΔAbs265 of 9.77±0.31 mAU; under identical experimental conditions, the same sample in citrate buffer gave a ΔAbs265 of 5.03±0.52 mAU. Examination of the FPOP footprint revealed a reduction in oxidation in all measured oxidized peptides (Figure S1, Supplementary Information), with all but two oxidized peptides showing statistically significant reductions in oxidation (α=0.05). However, the extent of this protection that is driven by topographical changes in the monoclonal antibody (mAb) versus protection due to excess hydroxyl radical scavenging is unknown.
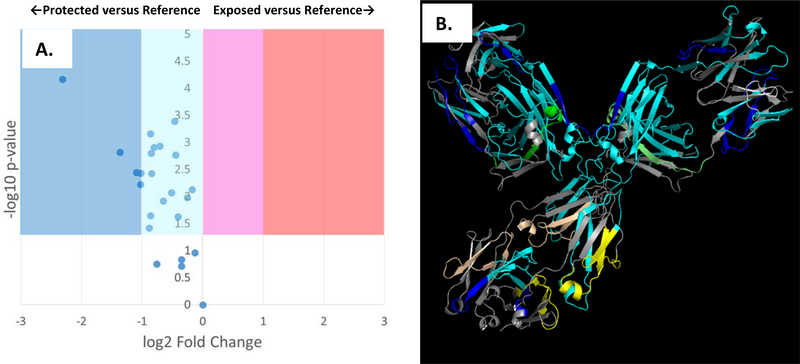
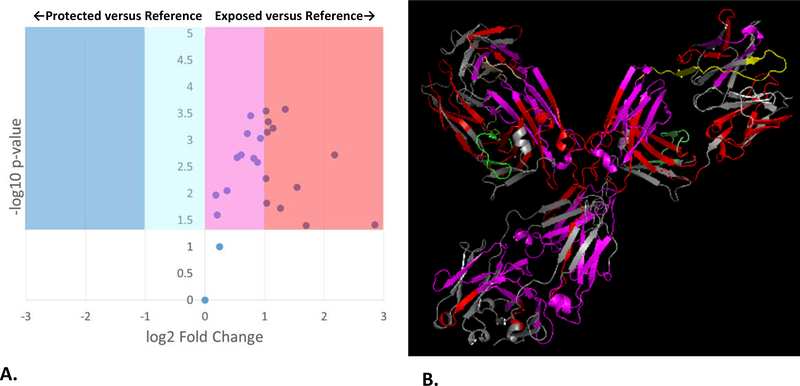
After compensation, adenine dosimetry of the adalimumab biosimilar in citrate buffer gave a ΔAbs265 of 9.43±1.27, within error of the dosimetry readings from the reference state. A comparison of the FPOP footprinting results is shown in Figure 2A. After compensation, the FPOP results reveal a significant decrease in solvent accessibility across almost all of the protein. The compensated FPOP data are plotted against a structural model of the adalimumab biosimilar in Figure 2B. Very high levels of protection are observed in the hinge region and the Fab region near the paratope. As FPOP reports on the weighted average of protein topography across all conformers in solution, these results are consistent with an increase in the aggregation of the mAb, through an increase in the proportion of antibody in an aggregated state and/or a marked increase in the average size of the aggregate. Dynamic light scattering of the adalimumab biosimilar support an interpretation of the data that combines an increase in aggregation with changes in conformation of the monomer. The Z-average of the adalimumab biosimilar in sodium phosphate buffer is 62.4±16.4 nm; in sodium citrate buffer, the Z-average increases to 105.3±6.4 nm. The DLS spectrum indicates the presence of a roughly equivalent amount of IgG1 (~0.1% of the total volume) in an aggregated state, with the size of both the aggregate and the monomer increasing in citrate buffer (Figures S3A and S4A, Supplementary Information). All DLS spectra shown are in intensity mode. Rayleigh scattering is proportional to the hydrodynamic diameter of the particle to the sixth power; the DLS spectra do not indicate the presence of high amounts of aggregate, but rather represent sensitive detection of low amounts of large aggregate. While the data generated clearly show that the buffer alters adalimumab higher order structure, it is unclear what mechanism is causing the buffer-induced changes in higher order structure. Both buffers have the same pH, the same buffer concentration, and use the same cation; however, the mechanism of differential structural effects of the phosphate anion versus the citrate anion are unclear.
Figure 2: FPOP comparison of adalimumab biosimilar in 50 mM sodium citrate buffer (pH 6.0) after dosimetry-based compensation versus 50 mM sodium phosphate buffer (pH 6.0) as a reference.
FPOP was performed with 15% exclusion volume, 100 mM H2O2, in 100 μm ID capillary, with the radical dosage altered to generate equivalent adenine dosimetry readings. (A.) Volcano plot of FPOP results, using oxidation in sodium phosphate as the reference sample. Each point represents one peptide from the biosimilar, with the X-axis representing the fold change in oxidation amount in citrate buffer as compared to phosphate buffer, and the Y-axis representing the p-value from a two-tailed Student’s t-test. The four shaded regions represent statistically significant changes in oxidation (α = 0.05), with the shade representing the color coding used in the structural visualization. (B.) Structural visualization of the differential FPOP data plotted in the volcano plot. Cyan: <2x decrease in oxidation; Blue: >2x decrease in oxidation; Magenta: <2x increase in oxidation; Red: >2x increase in oxidation; Grey: regions of the protein where no oxidized peptide was detected in any sample. Regions that showed no statistically significant changes in oxidation are colored green (light chain) or yellow (heavy chain).
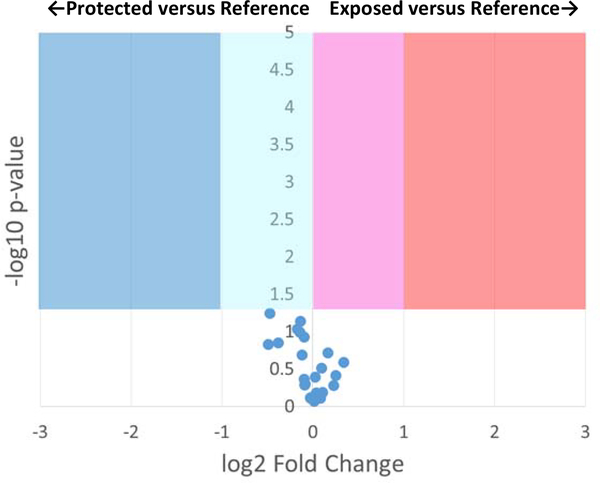
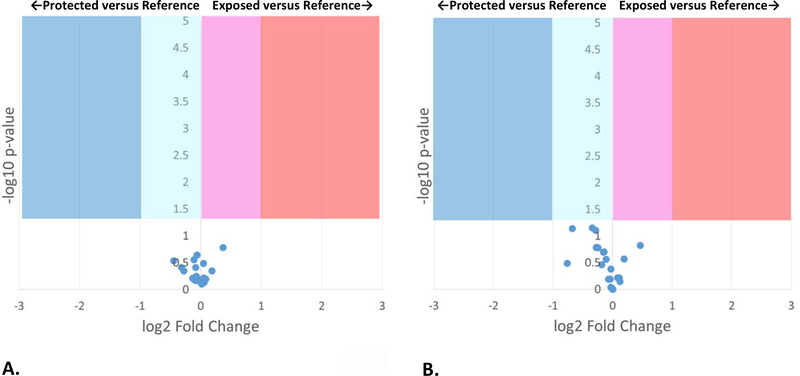
Compensated FPOP was also used to measure the effects of addition of 0.1% polysorbate-80 as a stabilizing excipient to adalimumab biosimilar in either phosphate or citrate buffer. Polysorbate-80 is one of the most commonly used excipients in biopharmaceuticals, well-known to protect proteins from aggregation (35). Adenine dosimetry readings for addition of polysorbate-80 to buffered adalimumab biosimilar were compensated to parity (sodium phosphate buffer: ΔAbs265 of 11.3±0.57 mAU without excipient, ΔAbs265 of 11.7±1.56 mAU with excipient; sodium citrate buffer: ΔAbs265 of 7.87±1.06 mAU without excipient, ΔAbs265 of 7.00±0.36 mAU with excipient). Compensated FPOP results showed no significant change in topography for any region of adalimumab biosimilar in phosphate buffer (Figure 3) or citrate buffer (Figure S2, Supplementary Information) upon addition of 0.1% polysorbate-80. Figure S5 shows that compensated FPOP is capable of generating sufficient radical to label equivalent amounts of the peptide in the presence of 0.1% polysorbate-80, generating sufficiently strong signal for both oxidized and unoxidized peptide for quantification. DLS also showed no substantial changes upon the addition of 0.1% polysorbate-80 (Figure S3B and S4B). Adalimumab biosimilar in phosphate buffer yielded a Z-average of 62.4±16.4 nm; upon addition of 0.1% polysorbate-80, the Z-average is 74.9±20.7 nm. In citrate buffer, the Z-average is 105.3±6.4 nm; upon addition of 0.1% polysorbate-80, the Z-average is 82.4±33.9 nm. The DLS spectra are visually unchanged upon addition of polysorbate-80 in either sodium phosphate buffer (Figure S3A and S3B) or sodium citrate buffer (Figure S4A and S4B).
Figure 3: Volcano plot of FPOP comparison of adalimumab biosimilar in 50 mM sodium phosphate buffer, 0.1% polysorbate-80 (pH 6.0) after dosimetry-based compensation versus 50 mM sodium phosphate buffer (pH 6.0) as a reference.
FPOP was performed with 15% exclusion volume, 100 mM H2O2, in 100 μm ID capillary, with the radical dosage altered to generate equivalent adenine dosimetry readings. Each point represents one peptide from the biosimilar, with the X-axis representing the fold change in oxidation amount in citrate buffer as compared to phosphate buffer, and the Y-axis representing the p-value from a two-tailed Student’s t-test. The four shaded regions represent statistically significant changes in oxidation (α = 0.05).
Effects of Buffer and Excipient on Heat Shock-Induced HOS Changes
FPOP topographical maps of adalimumab biosimilar were compared between mAb at room temperature, and mAb that was heated to 55 °C for one hour, and then cooled to room temperature. For adalimumab biosimilar in sodium phosphate buffer, heat shock caused a complex response. Most regions showed no change in topography or very slight decrease in solvent accessibility, while one region of the light chain showed a near two-fold increase in solvent accessibility (Figure 4A). Plotting these results in the context of the adalimumab structure indicates that the region of exposure is in the Fab near the hinge region, while the regions of very slight protection run along the central axis of the molecule through the hinge region (Figure 4B). Analysis of DLS results shows an average increase in particle size (Figure S3C), from a Z-average of 62.4±16.4 nm at room temperature to 107.3±68.5 nm after heat shock. However, more detailed examination of the DLS data shows a more complex response (Figure S3A and S3C). While heat shock increases the proportion of the mAb in the aggregate form (~33% intensity in the aggregate at room temperature to ~41% intensity after heat shock), the size of the aggregate decreases significantly (321.7 ± 76.9 nm at room temperature to 217.3 ± 47.1 nm after heat shock). No noticeable shift in monomer hydrodynamic diameter is detected in the DLS; however, conformational shifts that do not greatly change the hydrodynamic diameter would not be detectable by DLS. While the DLS data indicate that the change in topography measured by FPOP is driven at least in part by changes in aggregation, it is quite possible that changes in monomer conformation contribute as well.
Figure 4: FPOP comparison of adalimumab biosimilar in 50 mM sodium phosphate buffer (pH 6.0) after heat shock versus a room temperature reference.
FPOP was performed with 15% exclusion volume, 100 mM H2O2, in 100 μm ID capillary. (A.) Volcano plot of FPOP results, using oxidation at room temperature as the reference sample. Each point represents one peptide from the biosimilar, with the X-axis representing the fold change in oxidation amount after heat shock as compared to the reference, and the Y-axis representing the p-value from a two-tailed Student’s t-test. The four shaded regions represent statistically significant changes in oxidation (α = 0.05), with the shade representing the color coding used in the structural visualization. (B.) Structural visualization of the differential FPOP data plotted in the volcano plot. Cyan: <2x decrease in oxidation; Blue: >2x decrease in oxidation; Magenta: <2x increase in oxidation; Red: >2x increase in oxidation; Grey: regions of the protein where no oxidized peptide was detected in any sample. Regions that showed no statistically significant changes in oxidation are colored green (light chain) or yellow (heavy chain).
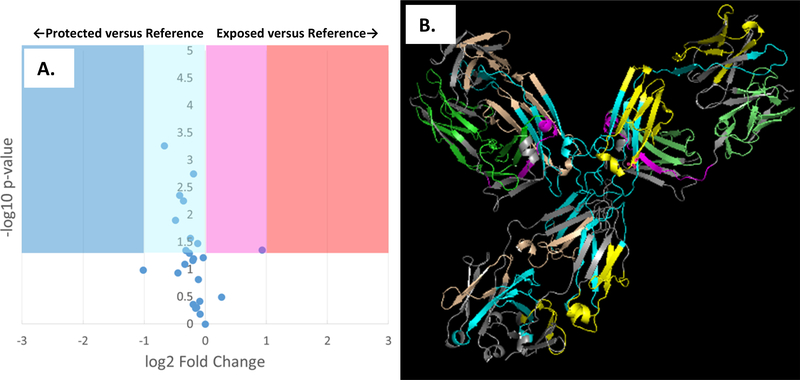
The response to heat shock in sodium citrate buffer is markedly different from that of sodium phosphate buffer. As shown in Figure 5A, heating of the adalimumab biosimilar in sodium citrate buffer shows an increase in average solvent accessibility of every oxidized peptide in the mAb, with all but one being statistically significant. Many of the increases are also large in magnitude, with one peptide showing a >7x increase in oxidation. Figure 5B places these topographical changes in a structural context. The results clearly show that, while the topographical exposure affects the entire molecule, the greatest exposure is found in the Fab and hinge region of the mAb. DLS data again show a significant change in aggregation (Figure S4C). The Z-average decreases from 105.3±6.4 nm at room temperature to 18.9±3.1 nm after heat shock. The fraction of the monomer increases significantly (from DLS signal intensity of ~65.9% to ~71.9%). The first aggregate not only decreases in intensity (from ~34.1% before heat shock to ~25.7% after heat shock), but also shown a significant and reproducible decrease in size (449.9±113.8 nm before heat shock; 178.7±48.83 nm after heat shock). We do also observe a new asymmetric, non-Gaussian DLS peak at 5289 nm after heat shock, with 2.4% DLS intensity. As this peak does not occur in the non-heat shock treated sample and persists across repeated DLS measurements, we do not believe this is an impurity; however, the unusual appearance of the peak warrants that it be interpreted with caution. Regardless, this large aggregate’s contribution by volume is very small due to the drastically increased DLS response for larger particles. Any changes in monomer conformation are too small to be detected by DLS, but again they may contribute to changes in topography measured by FPOP.
Figure 5: FPOP comparison of adalimumab biosimilar in 50 mM sodium citratete buffer (pH 6.0) after heat shock versus a room temperature reference.
FPOP was performed with 15% exclusion volume, 100 mM H2O2, in 100 μm ID capillary. (A.) Volcano plot of FPOP results, using oxidation at room temperature as the reference sample. Each point represents one peptide from the biosimilar, with the X-axis representing the fold change in oxidation amount after heat shock as compared to the reference, and the Y-axis representing the p-value from a two-tailed Student’s t-test. The four shaded regions represent statistically significant changes in oxidation (α = 0.05), with the shade representing the color coding used in the structural visualization. (B.) Structural visualization of the differential FPOP data plotted in the volcano plot. Cyan: <2x decrease in oxidation; Blue: >2x decrease in oxidation; Magenta: <2x increase in oxidation; Red: >2x increase in oxidation; Grey: regions of the protein where no oxidized peptide was detected in any sample. Regions that showed no statistically significant changes in oxidation are colored green (light chain) or yellow (heavy chain).
The addition of polysorbate-80 significantly changes the effect of heat shock on protein HOS, as measured both by FPOP and by DLS. As shown in Figure 6, after the addition of 0.1% polysorbate-80 no differences in adalimumab biosimilar topography was observed before and after heat shock for 50 mM sodium phosphate buffer (Figure 6A) or 50 mM sodium citrate buffer (Figure 6B). Comparisons of the FPOP footprint showed both low fold changes and not statistically significant differences, supporting no change in protein HOS after heat shock in the presence of 0.1% polysorbate-80. DLS data similarly support the ability of polysorbate-80 to preserve the higher order structure of adalimumab biosimilar from brief periods of elevated temperature as shown in Figures S3 and S4, Supplementary Information.
Figure 6: FPOP volcano plots of adalimumab biosimilar with 0.1% polysorbate-80 after heat shock versus a room temperature reference.
FPOP was performed with 15% exclusion volume, 100 mM H2O2, in 100 μm ID capillary. Each point represents one peptide from the biosimilar, with the X-axis representing the fold change in oxidation amount after heat shock as compared to the reference, and the Y-axis representing the p-value from a two-tailed Student’s t-test. The four shaded regions represent statistically significant changes in oxidation (α = 0.05). (A.) Adalimumab biosimilar in 50 mM sodium phosphate (pH 6.0); (B.) Adalimumab biosimilar in 50 mM sodium citrate (pH 6.0).
DISCUSSION
The idea of using hydroxyl radical dosimetry to compensate for differential radical scavenging properties of biopharmaceutical formulations was described in theory previously (22); here, we demonstrate it in practice using an inline adenine dosimeter to adjust effective hydroxyl radical doses. Using inline adenine dosimetry to compensate for differential radical scavenging properties as previously described (27), we were able to directly compare the protein HOS changes that occur due to altering buffers and the addition of excipients. As shown in a comparison of Figure 2 and Figure S1, hydroxyl radical scavengers decrease oxidation of all peptides, although not always to the same extent for each peptide. The general overall pattern of oxidation is maintained; however, without compensation, it is impossible to untangle the effects of scavenging from the effects of topographical protection. Peptides that are not topographically protected (or that are even topographically exposed) can appear protected due to scavenging. Additionally, the extent of topographical protection is obscured by the contributions of scavenging to the differences in HRPF labeling. Since the extent of HRPF protection/exposure is directly proportional to the extent of topographical protection/exposure (8, 36), the contribution of scavengers is essential to understand and correct for. We were able to perform dosimetry-based compensation despite the very high hydroxyl radical scavenging capacity of polysorbate-80 (5). The data that were generated using compensated FPOP were not only consistent with traditional DLS measurements, but complemented them. While DLS measurements gave information on particle size and dispersity, FPOP provided a molecular-level view. Together, the technologies gave a much more comprehensive view of biopharmaceutical HOS that is critical for biopharmaceutical development.
The benefits of compensated FPOP with inline dosimetry for biopharmaceutical development are widespread. The time for sample preparation and analysis are short: ~10 min for FPOP and quenching, followed by the proteolytic digestion and LC-MS analysis method of the user’s choice. With inline compensation of radical scavengers, FPOP is very flexible in accommodating almost any sample and aqueous formulation. Compensation with real-time inline dosimetry is fast and simple; each sample could be compensated in a few minutes each using different peroxide concentrations, with compensation by altering laser fluence being even faster. The method can accommodate a wide range of pH (5, 37). As illustrated by the work here, the method can detect changes not only in tertiary higher order structure, but also in aggregation profiles. FPOP reports on changes in the average solvent accessible surface area of the peptides measured (8, 36), making the analysis of polydisperse, heterogeneous and/or dynamic mixtures possible.
With the development and application of methods for compensating for differential radical scavenging capacities using inline radical dosimetry, two major hurdles remain in developing compensated FPOP as a technology for supporting biopharmaceutical development efforts. Currently, data analysis software tools that are suitable for the automated analysis of FPOP results are not widely acceptable. Published results almost invariably include manual or, at best, semi-automated quantification of peptide oxidation with significant manual user input. This manual analysis significantly limits the efficiency and throughput of the technology. Both academic and commercial efforts to address this problem are underway, but no reliable automated data analysis solutions for FPOP currently exist. Additionally, FPOP currently requires the use of custom-built systems using very high-powered lasers that have significant maintenance and workplace safety demands. Some alternative benchtop methods for hydroxyl radical footprinting have been published (9, 10, 17), but all are slow and run a significant risk of probing artifactual conformations induced by the labeling process. With the development of reliable software and an industry-friendly integrated method for rapid hydroxyl radical labeling of proteins, compensated FPOP offers considerable advantages as a method for biotherapeutic HOS analysis.
CONCLUSION
Here, we describe the application of a compensated FPOP method for the topographical analysis of an IgG1 biosimilar of adalimumab in four different formulations: sodium phosphate buffer; sodium citrate buffer; sodium phosphate buffer with 0.1% polysorbate-80; and sodium citrate buffer with 0.1% polysorbate-80. We are able to show that, at room temperature, significant and detectable changes in the IgG1 HOS occurs when changing buffer from phosphate to citrate. DLS indicates these changes are driven by an increase in the size of aggregation products in citrate buffer. We are also able to show that the buffer plays a large role in the response of the IgG1 mAb to heat shock. In citrate buffer, this change is represented in FPOP data as a widespread exposure of most of the surfaces of the protein. Using DLS, we can identify these changes in the average topography as being due to the disruption of most of the aggregate species present at room temperature. By contrast, heat shock in phosphate buffer results in a more complex response: a slight protection of certain regions of the IgG1 mostly concentrated in the Fc and hinge regions, and a slight exposure of one region in the light chain near the hinge. DLS confirms this complex response, with heat shock resulting in a larger proportion of the mAb in an aggregate form, but a smaller aggregate size than found in the room temperature sample. Finally, the addition of polysorbate-80 showed no perturbation of IgG1 HOS at room temperature for citrate or phosphate buffer. In heat shock experiments, polysorbate-80 was able to prevent any changes in protein topography and aggregation in both compensated FPOP and DLS experiments. These results indicate the power of compensated FPOP and the complementarity of topographical information with data generated from more traditional DLS analysis.
Supplementary Material
ACKNOWLEDGEMENTS
J.S.S., R.O. and S.R.W. acknowledge support from the National Institute of General Medical Sciences (R43GM125420) to support commercial development of a benchtop FPOP device. J.S.S and S.K.M acknowledge support from the National Institute of General Medical Sciences for the development of compensation protocols for high radical scavenging environments (R01GM127267).
Footnotes
FINANCIAL CONFLICT OF INTEREST DISCLOSURE
J.S.S., R.O. and S.R.W. disclose a significant financial interest in GenNext Technologies, Inc., an early-stage company seeking to commercialize technologies for protein higher order structure analysis. This manuscript and all data were reviewed by S.K.M., who has no financial conflict of interest, in accordance with University of Mississippi FCOI management practices.
REFERENCES
- 1.Global biopharmaceuticals market growth, trends and forecasts (2016–2021) Current trends in biopharmaceuticals market. Mordor Intelligence: Hyderabad, India; 2016. [Google Scholar]
- 2.Manning MC, Chou DK, Murphy BM, Payne RW, Katayama DS. Stability of protein pharmaceuticals: an update. Pharm Res. 2010;27(4):544–75. doi: 10.1007/s11095-009-0045-6. [DOI] [PubMed] [Google Scholar]
- 3.Giezen TJ, Mantel-Teeuwisse AK, Strauss S. Safety-related regulatory actions for biologicals approved in the United States and the Europena Union. Journal of the American Medical Society. 2008;300(16):1887–96. [DOI] [PubMed] [Google Scholar]
- 4.Giezen TJ, Schneider CK. Safety assessment of biosimilars in Europe: a regulatory perspective. Generics and Biosimilars Initiative Journal. 2014;September 2014:1–8. [Google Scholar]
- 5.Buxton GV, Greenstock CL, Helman WP, Ross AB. Critical-Review of Rate Constants for Reactions of Hydrated Electrons, Hydrogen-Atoms and Hydroxyl Radicals (.OH/.O-) in Aqueous-Solution. J Phys Chem Ref Data. 1988;17(2):513–886. [Google Scholar]
- 6.Xu G, Chance MR. Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Anal Chem. 2005;77(14):4549–55. [DOI] [PubMed] [Google Scholar]
- 7.Sharp JS, Tomer KB. Effects of anion proximity in peptide primary sequence on the rate and mechanism of leucine oxidation. Anal Chem. 2006;78(14):4885–93. [DOI] [PubMed] [Google Scholar]
- 8.Xie B, Sood A, Woods RJ, Sharp JS. Quantitative Protein Topography Measurements by High Resolution Hydroxyl Radical Protein Footprinting Enable Accurate Molecular Model Selection. Sci Rep. 2017;7(1):4552. doi: 10.1038/s41598-017-04689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp JS, Becker JM, Hettich RL. Protein surface mapping by chemical oxidation: structural analysis by mass spectrometry. Anal Biochem. 2003;313(2):216–25. [DOI] [PubMed] [Google Scholar]
- 10.Sharp JS, Becker JM, Hettich RL. Analysis of protein solvent accessible surfaces by photochemical oxidation and mass spectrometry. Anal Chem. 2004;76(3):672–83. [DOI] [PubMed] [Google Scholar]
- 11.Chance MR. Unfolding of apomyoglobin examined by synchrotron footprinting. Biochem Biophys Res Commun. 2001;287(3):614–21. [DOI] [PubMed] [Google Scholar]
- 12.Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR. Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. International Journal of Radiation Biology. 2002;78(2):101–14. [DOI] [PubMed] [Google Scholar]
- 13.Huang W, Ravikumar KM, Chance MR, Yang S. Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: a protection factor analysis. Biophys J. 2015;108(1):107–15. doi: 10.1016/j.bpj.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charvatova O, Foley BL, Bern MW, Sharp JS, Orlando R, Woods RJ. Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to galectin-1. J Am Soc Mass Spectrom. 2008;19(11):1692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maleknia SD, Chance MR, Downard KM. Electrospray-assisted modification of proteins: a radical probe of protein structure. Rapid Commun Mass Spectrom. 1999;13(23):2352–8. [DOI] [PubMed] [Google Scholar]
- 16.Goldsmith SC, Guan JQ, Almo S, Chance M. Synchrotron protein footprinting: a technique to investigate protein-protein interactions. J Biomol Struct Dyn. 2001;19(3):405–18. [DOI] [PubMed] [Google Scholar]
- 17.McClintock C, Kertesz V, Hettich RL. Development of an electrochemical oxidation method for probing higher order protein structure with mass spectrometry. Anal Chem. 2008;80(9):3304–17. [DOI] [PubMed] [Google Scholar]
- 18.Watson C, Janik I, Zhuang T, Charvatova O, Woods RJ, Sharp JS. Pulsed electron beam water radiolysis for submicrosecond hydroxyl radical protein footprinting. Anal Chem. 2009;81(7):2496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 2005;16(12):2057–63. [DOI] [PubMed] [Google Scholar]
- 20.Aye TT, Low TY, Sze SK. Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Anal Chem. 2005;77(18):5814–22. [DOI] [PubMed] [Google Scholar]
- 21.Gau BC, Sharp JS, Rempel DL, Gross ML. Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal Chem. 2009;81(16):6563–71. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson C, Sharp JS. Conformational analysis of therapeutic proteins by hydroxyl radical protein footprinting. AAPS J. 2012;14(2):206–17. doi: 10.1208/s12248-012-9336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin M, Krawitz D, Callahan MD, Deperalta G, Wecksler AT. Characterization of ELISA Antibody-Antigen Interaction using Footprinting-Mass Spectrometry and Negative Staining Transmission Electron Microscopy. J Am Soc Mass Spectrom. 2018;29(5):961–71. doi: 10.1007/s13361-017-1883-9. [DOI] [PubMed] [Google Scholar]
- 24.Storek KM, Auerbach MR, Shi H, Garcia NK, Sun D, Nickerson NN, et al. Monoclonal antibody targeting the beta-barrel assembly machine of Escherichia coli is bactericidal. Proc Natl Acad Sci U S A. 2018;115(14):3692–7. doi: 10.1073/pnas.1800043115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Wecksler AT, Molina P, Deperalta G, Gross ML. Mapping the Binding Interface of VEGF and a Monoclonal Antibody Fab-1 Fragment with Fast Photochemical Oxidation of Proteins (FPOP) and Mass Spectrometry. J Am Soc Mass Spectrom. 2017;28(5):850–8. doi: 10.1007/s13361-017-1601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Wei H, Krystek SR Jr., Bond D, Brender TM, Cohen D, et al. Mapping the Energetic Epitope of an Antibody/Interleukin-23 Interaction with Hydrogen/Deuterium Exchange, Fast Photochemical Oxidation of Proteins Mass Spectrometry, and Alanine Shave Mutagenesis. Anal Chem. 2017;89(4):2250–8. doi: 10.1021/acs.analchem.6b03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharp JS, Misra SK, Persoff JJ, Egan RW, Weinberger SR. Real Time Normalization of Fast Photochemical Oxidation of Proteins Experiments by Inline Adenine Radical Dosimetry. Anal Chem. 2018;90(21):12625–30. doi: 10.1021/acs.analchem.8b02787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Moniz H, Wang S, Ramiah A, Zhang F, Moremen KW, et al. High structural resolution hydroxyl radical protein footprinting reveals an extended robo1-heparin binding interface. J Biol Chem. 2015;290(17):10729–40. doi: 10.1074/jbc.M115.648410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu B, Mackness BC, Rempel DL, Zhang H, Cui W, Matthews CR, et al. Incorporation of a Reporter Peptide in FPOP Compensates for Adventitious Scavengers and Permits Time-Dependent Measurements. J Am Soc Mass Spectrom. 2017;28(2):389–92. doi: 10.1007/s13361-016-1552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niu B, Zhang H, Giblin D, Rempel DL, Gross ML. Dosimetry determines the initial OH radical concentration in fast photochemical oxidation of proteins (FPOP). J Am Soc Mass Spectrom. 2015;26(5):843–6. doi: 10.1007/s13361-015-1087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie B, Sharp JS. Hydroxyl Radical Dosimetry for High Flux Hydroxyl Radical Protein Footprinting Applications Using a Simple Optical Detection Method. Anal Chem. 2015;87(21):10719–23. doi: 10.1021/acs.analchem.5b02865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riaz M, Misra SK, Sharp JS. Towards high-throughput fast photochemical oxidation of proteins: Quantifying exposure in high fluence microtiter plate photolysis. Anal Biochem. 2018;561–562: 32–6. doi: 10.1016/j.ab.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konermann L, Stocks BB, Czarny T. Laminar Flow Effects During Laser-Induced Oxidative Labeling for Protein Structural Studies by Mass Spectrometry. Analytical Chemistry. 2010;82(15):6667–74. doi: Doi 10.1021/Ac101326f. [DOI] [PubMed] [Google Scholar]
- 34.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107(8):3514–43. [DOI] [PubMed] [Google Scholar]
- 35.Singh SM, Bandi S, Jones DNM, Mallela KMG. Effect of Polysorbate 20 and Polysorbate 80 on the Higher-Order Structure of a Monoclonal Antibody and Its Fab and Fc Fragments Probed Using 2D Nuclear Magnetic Resonance Spectroscopy. J Pharm Sci. 2017;106(12):3486–98. doi: 10.1016/j.xphs.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur P, Kiselar J, Yang S, Chance MR. Quantitative protein topography analysis and high-resolution structure prediction using hydroxyl radical labeling and tandem-ion mass spectrometry (MS). Mol Cell Proteomics. 2015;14(4):1159–68. doi: 10.1074/mcp.O114.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smedley JG, Sharp JS, Kuhn JF, Tomer KB. Probing the pH-dependent prepore to pore transition of Bacillus anthracis protective antigen with differential oxidative protein footprinting. Biochemistry. 2008;47(40):10694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.