Abstract
To identify drugs that could potentially be used to treat infection with SARS-CoV-2, a high throughput 384-well assay was developed to measure the binding of the receptor binding domain (RBD) of the viral S1 protein to its main receptor, angiotensin converting enzyme 2 (ACE2). The RBD was fused to both a HiBIT tag and an IL6 secretion signal to enable facile collection from the cell culture media. The addition of culture media containing this protein, termed HiBIT-RBD, to cells expressing ACE2 led to binding that was specific to ACE2 and both time and concentration dependant, Binding could be inhibited by both RBD expressed in E. coli and by a full length S1 - Fc fusion protein (Fc-fused S1) expressed in eukaryotic cells. The mutation of residues that are known to play a role in the interaction of RBD with ACE2 also reduced binding. This assay may be used to identify drugs which inhibit the viral uptake into cells mediated by binding to ACE2.
Keywords: SARS-CoV-2, ACE2, Binding assay, HiBIT tag, Mutation, Nano-luciferase
Graphical abstract
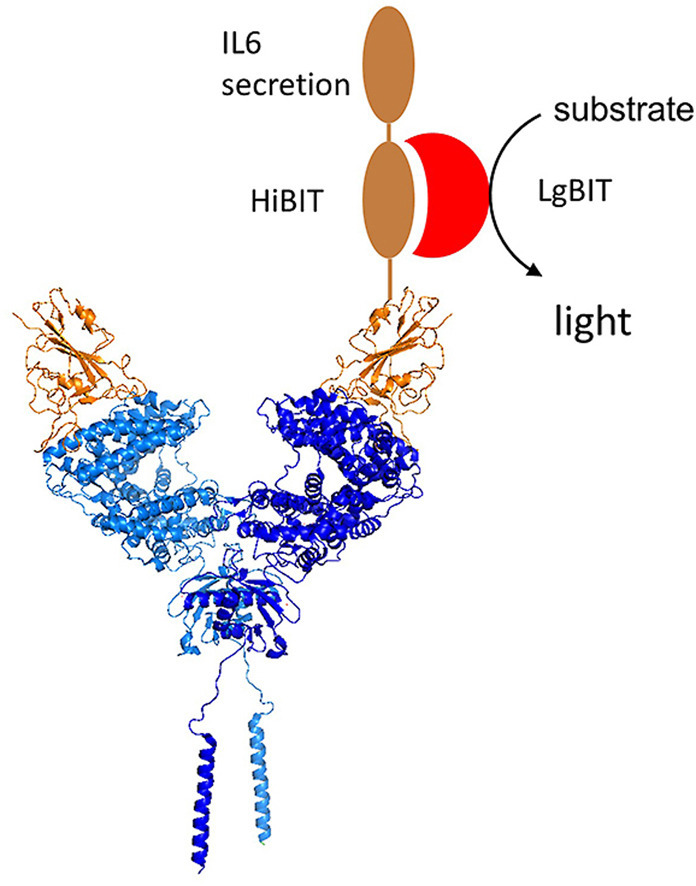
Abbreviations
- LgBIT
Large binary interaction technology
- HiBIT
High affinity binary interaction technology
- RBD
Receptor binding domain
- ACE2
Angiotensin converting enzyme 2
- SARS
Severe acute respiratory syndrome
1. Introduction
Zoonotic viruses make up the vast majority of emerging, human diseases owing to their ability to cross-spread between species from animals to humans [1]. SARS-CoV-2, the seventh coronavirus known to infect humans [2], hit the world rapidly and uncontrollably, resulting in the death of over 1 million people as of October 2020, alongside devastating social-economical burdens. To-date, no widely effective treatment or vaccines are available and a significant drive towards SARS-CoV-2 management relies on the effective track and trace of infected individuals in concert with restrictions on personal freedom. There is, therefore, an urgent need to identify effective therapies that ameliorate the significant health and socioeconomic burdens caused by SARS-CoV-2.
The viral tropism is determined by the interactions that occur between viral surface proteins and host cell receptors, and as such, these interactions are vital for effective cross-species and intra-species transmission [3]. Coronaviruses (CoVs) express a surface glycoprotein, termed spike, that binds to cell-surface receptors and mediate the entry of the virus into cells to initiate an infection [4]. Many host receptors for coronaviruses have been identified [4,5]; in the case of SARS-COV-2, the viral surface glycoprotein S1 contains a receptor binding domain (RBD) that interacts with angiotensin converting enzyme 2 (ACE2) and this is thought to mediate viral uptake into cells [6]. Thus, blocking this interaction could potentially limit viral infections. More recently, it has also been suggested that neuropilin can also mediate SARS-COV-2 uptake [7].
The current state of the pandemic demands a rapid response. Drug repurposing screens are a convenient and attractive route to the identification of drugs that could address unmet medical needs. This is particularly attractive in the case of SARS-COV-2 because drugs that are already licensed for the treatment of other diseases could be rapidly redeployed to treat the current pandemic [8]. Many marketed drugs are readily available in large quantities and their safety profiles are well understood. When compared to traditional drug discovery paradigms, drug repurposing significantly reduces the time required for the identification of drug candidates that may treat emerging viral infections. Nevertheless, suitable assays are required for the rapid identification of candidate SARS-COV-2 therapeutic drugs from the vast list of approved medicinal drugs. Computational screens provide an alternative strategy, although they lack a full consideration of the complex physiological environment [9] that dictates how molecules interact in vivo and, in any case, still require biochemical assays to validate the in silico results.
To develop an assay to measure the binding of SARS-COV-2 spike protein to ACE2, here the HiBIT system has been used. HiBIT is an 11 amino acid peptide that binds with high affinity to LgBIT, a fragment of an engineered form of luciferase termed nanoluc [10]. The binding of HiBIT to LgBIT renders the complex catalytically competent, creating a luminescence signal when an appropriate substrate is provided. The binding of HiBIT to LgBIT is of sufficiently high affinity that additional proteins are not required to colocalize the two proteins and create a bioactive complex. Thus, LgBIT can be utilised for the detection of a HiBIT-tagged protein. In this study, SARS-COV-2 RBD was fused inframe with a sequence encoding HiBIT to create a reagent that was anticipated to bind ACE2 and which could be detected by addition of LgBIT and the nanoluc substrate. An IL6 secretion signal was also included to allow HiBIT-tagged RBD secretion into cell culture medium of transfected cells. The resultant culture medium, containing recombinantly expressed HiBIT-tagged RBD, was subsequently added to COS cells ectopically expressing ACE 2 to measure RBD-ACE2 binding. This was used in a proof-of-concept screen of a small library of marketed drugs.
2. Methods
Materials. The plasmid pCDNA3 encoding ACE2 was obtained from Genescript (OHu 20260); the plasmid encoding prolactin was obtained from Sino biological (HG10275-CY). pRSETA encoding the SARS-CoV-2 S1 protein was obtained from GeneArt Gene Synthesis. Optimem and lipofectamine 2000 were obtained from Thermofisher. HiBIT detection reagent was obtained from Promega. The flexicloning transfer system (C8820), pFN39K and HiBIT detection reagent were obtained from Promega. The Q5 mutagenesis kit was obtained from New England Biolabs. SARS CoV-2 spike S1 protein was obtained from the Centre for AIDS Reagents, NIBSC, UK: Recombinant SARS CoV-2 spike S1 protein, consisting of a combination of the Fc fusion (25 kDa) and the SARS CoV-2 S1 domain (70 kDa) was obtained from Prof. Ian M Jones.
2.1. Molecular biology
RBD constructs were prepared using the flexicloning transfer system. The region encoding amino acids 328–536 of the SARS-CoV-2 spike S1 were amplified using Pfu polymerase and pRSETA SARS-CoV-2 S1 as the template. Forward (CCCGGCGATCGCCATGCGGTTCCCCAATATCACCAATCTG) and reverse (GTCGGTTTAAACGTTCTTCACGAGATTGGTGCTT) primers introduced Sgf1 and Pme1 restriction sites flanking the RBD. The product was gel purified, digested with flexiblend and ligated into pF4ACMV. After restriction analysis, selected clones were verified by sequencing. For site directed mutagenesis, mutations were introduced into the resulting pF4ACMV-RBD using a Q5 mutagenesis kit according to the manufacturer’s instructions. The desired clones were identified by sequencing. The wild-type and mutated pF4ACMV-RBD clones and pFN39K, which includes a HiBIT tag and an IL6 secretion signal, were subsequently digested with flexiblend, heat inactivated and ligated. pFN39K clones containing the RBD were selected using kanamycin and identified by restriction digest. Plasmids for transfection were prepared using a Qiagen plasmid purification kit.
2.2. Recombinant SARS-CoV2 spike S1 and SARS-CoV2 S1 receptor binding domain (RBD)
Residues 319–597 of the SARS-CoV-2 spike S1 RBD (GenBank: MN908947) were cloned upstream of an N-terminal 6XHisTag in the pRSET A expression vector and transformed into BL21(DE3) pLysS Competent Cells (Novagen, UK). Protein expression was carried out in MagicMedia™ E. coli Expression Media (Invitrogen, UK) at 37 °C for 24 h, 250 rpm. The bacterial pellet was suspended in 5 mL of lysis buffer (BugBuster Protein Extraction Reagent, Merck Millipore, UK; containing DNAse) and incubated at room temperature for 30 min. The protein was purified from inclusion bodies using IMAC chromatography under denaturing conditions [11]. Fractions were pooled and buffer-exchanged to phosphate-buffered saline (PBS; 140 mM NaCl, 5 mM NaH2PO4, 5 mM Na2HPO4, pH 7.4; Lonza, UK) using a Sephadex G-25 column (GE Healthcare, UK). Recombinant protein was stored at −20 °C until required.
2.3. Binding assay
COS cells were grown in DMEM supplemented with 10% (v/v) fetal calf serum and penicillin-streptomycin (50 U/ml). For each well of a 384-well plate, a transfection mix was prepared containing plasmids encoding ACE2 or prolactin (0.025 μg) in 6.25 μL optimem and mixed with an equal volume of 0.8% (v/v) lipofectamine 2000 in optimem. COS cells were collected by trypsinization, diluted in antibiotic free medium to 60,000 cells/ml and 25 μL per well added to the transfection mix. After mixing well, 25 μL were added per well of the 384-well plate. The outer two rows of the plate were not used but filled with medium to minimize edge effects from evaporation. The cells were incubated for 48 h before being used in the binding assay.
In parallel, a separate set of COS cells were transfected with pFN39K HiBIT RBD. 3 μg of DNA in 750 μL optimem were mixed with 750 μL of 0.8% (v/v) Lipofectamine 2000 in optimem. After 30 min, 3 mL of COS cells (60,000 cell/mL in antibiotic free medium) were added, the samples mixed and plated in a 6 well plate. After 16 h, the medium was aspirated and replaced with DMEM containing 2% (v/v) FCS. After a further 32 h, the cell culture medium was collected, cleared by centrifugation (150×g, 3 min) and supplemented with HEPES (10 mM) and stored on ice.
To perform the binding assay, the 384-well plate was placed on ice and washed twice (2 × 50 μL) with ice-cold DMEM containing 2% (v/v) FCS and 10 mM HEPES. If drugs were being tested, these were diluted in a 96 well plate in the HiBIT-RBD on ice. After shaking off the medium from the plate, 25 μL of the ice-cold culture medium containing HiBIT-RBD was added per well and the plate incubated on ice for 90 min. The plate was washed four times with 70 μL ice-cold DMEM containing 2% FCS and 10 mM HEPES and before filling wells with 10 μL of the same solution. The HiBIT detection reagent was prepared according to the manufacturer’s instruction and 10 μL added per well. The plate was shaken on ice in the dark for 5 min and then centrifuged briefly (150×g, 1 min, 0 °C) to remove bubbles. Luminescence was measured on a plate reader at intervals over 45 min returning the plate to ice each time; an integration time of 0.1 s per well was used to minimize plate warming.
2.4. Western blotting
Cells were transfected as described above with pFN39K HiBIT-RBD. Cells in a 12 well plate were lysed with a 250 μL modified RIPA as described [12], separated on a 4–12% SDS-polyacrylamide gel, transferred to PVDF and analysed by immunoblotting with anti ACE2 (ab15348, 1 μg/mL).
3. Results
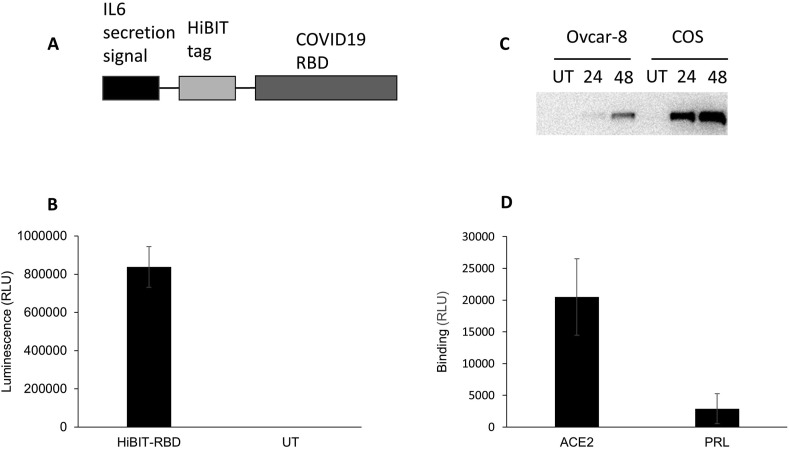
The receptor binding domain of SARS-CoV-2 was cloned into the pFN39K expression vector in frame with an IL6 secretion signal and the HiBIT tag (Fig. 1 a). This allowed the HiBIT-tagged protein (HiBIT-RBD) to be collected from the culture medium of cells transfected with the tag. The expression of the protein was verified by measuring luminescence after adding this culture medium to the HiBIT detection reagent (Fig. 1b). ACE2 was expressed by transient transfection of Ovcar-8 cells and COS cells. Western blotting (Fig. 1c) demonstrated higher levels of protein in COS cells, and subsequent experiments were performed with this cell line. HEK-293 cells were also tested, and although they allowed robust expression, these were considered not to be suitable for establishing a binding assay as the cells readily detached when washed with PBS, a step necessary to remove any unbound HiBIT-RBD. COS cells transiently expressing ACE2 or prolactin (PRL, negative control) were incubated with HiBIT-RBD cell culture supernatant, washed and the bound HiBIT-RBD measured by addition of the HiBIT detection reagent. In preliminary experiments, the incubation was conducted at either at 37 °C or at 0 °C to minimize the potential uptake of the receptor-ligand complex by endocytosis. After incubation at 0 °C, significantly more HiBIT-RBD was detected bound to cells expressing ACE2 than cells expressing PRL (Fig. 1d). In preliminary experiments, when the assay was conducted at 37 °C, less specific binding was observed (data not shown) so all further experiments were conducted at 0 °C.
Fig. 1.
Development of reagents to measure RBD binding to ACE2. A. pFN39K was engineered to express SARS-COV-2 RBD fused to the HiBIT tag. An inframe IL6 secretion signal was included to facilitate the collection of the reagent from cell culture media. B. COS cells were transfected transiently with pFN39K HiBIT-RBD and after 48 h, the expressed protein was detected in the cell culture supernatant by addition of HiBIT detection reagent (mean ± S. D, n = 4 replicates from one representative experiment). C. Ovcar-8 or COS cells were transfected with a plasmid expressing ACE2 and samples collected after 24 or 48 were analysed by immunoblotting. One experiment, representative of two is shown. D. COS cells transfected with plasmids encoding ACE2 or PRL were incubated on ice with cell culture media containing HiBIT-RBD, washed, and bound HiBIT-RBD detected by addition of HiBIT detection reagent.
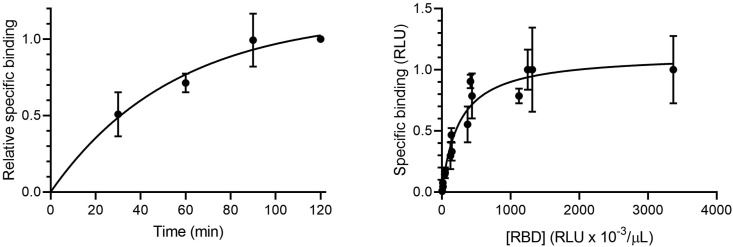
Next, the kinetics of HiBIT-RBD binding to ACE2 were investigated. COS cells expressing ACE2 or PRL were incubated with HiBIT-RBD at 0 °C for different periods and the specific binding, defined as binding to cells expressing ACE2 minus binding to cells expressing PRL, was determined (Fig. 2 a). Specific binding reached steady state after approximately 90 min. In all further experiments, an incubation period of 90 min at 0 °C was used. To evaluate the concentration dependency of HiBIT-RBD binding, COS cells expressing ACE2 or PRL were incubated with different dilutions of the HIBIT-RBD cell culture supernatant and after washing to remove unbound HiBIT-RBD, specific binding determined. As expected, the specific binding was saturable, consistent with HiBIT-RBD binding to a finite number of receptors.
Fig. 2.
HiBIT-RBD binds to ACE2 in a saturable and time dependant manner. A. RBD was collected from cell culture media from COS cells transiently expressing ACE2 for 48 h and incubated with COS cells transiently expressing ACE2 or PRL for different periods on ice. After washing, bound HiBIT-RBD was detected by addition of HiBIT detection reagent. Specific binding was defined as the binding measured in ACE2 expressing cells minus the binding measured in cells expressing PRL. The results (mean ± S.D, n = 3) are expressed as a fraction of the specific binding measured after 120 min. B. HiBIT-RBD was collected from cell culture media from COS cells transiently expressing ACE2 for 72 h (to achieve an adequate concentration of HiBIT-RBD to saturate the receptor). Separately, COS cells were transfected with plasmids encoding ACE2 or PRL. After 48 h, the cells were incubated with the serial dilutions of the cell culture medium (90 min, 0 °C), washed, and bound HiBIT-RBD measured by addition of HiBIT detection reagent. Specific binding was defined as the binding measured in ACE2 expressing cells minus the binding measured in cells expressing PRL. The results were expressed as a fraction of the maximum specific binding observed in each experiment. The results shown are the combined individual results from 3 separate experiments (each mean ± S.D.).
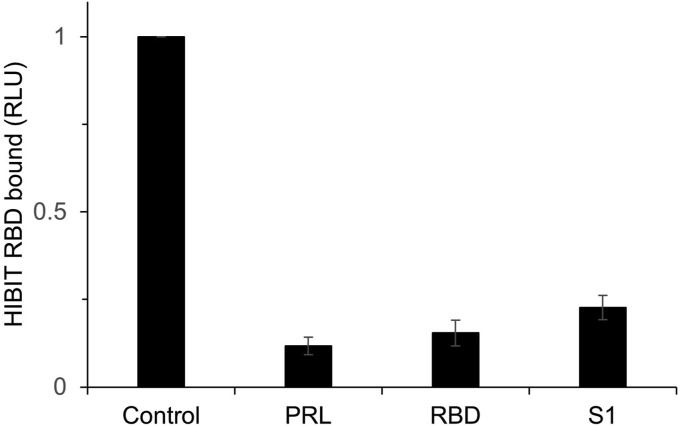
To provide evidence that the HiBIT-RBD provides a model of the viral spike protein binding to ACE-2, competition experiments were performed. In these experiments, SARS-CoV-2 S1 RBD and Fc-fused S1 (recombinantly produced in E. coli and eukaryotic cells, respectively) were used to give confidence that the binding mode was not unique to reagents prepared in COS cells. Cells transiently expressing ACE-2 were incubated with HiBIT-RBD in the presence of either RBD or with the full-length Fc-Fused S1. Both proteins were able to almost completely suppress the specific binding of HiBIT-RBD (Fig. 3 ).
Fig. 3.
Binding of HiBIT-RBD to cells expressing ACE2 is inhibited by both recombinant RBD and S1 protein. COS cells expressing ACE2 were incubated with cell culture supernatant containing HiBIT-RBD in the presence of PBS (vehicle) or purified RBD (residues 319–597) expressed in E. coli or a purified S1-Fc fusion protein expressed in eukaryotic cells (40 μg/mL). Alternatively, binding of HiBIT-RBD to cells expressing PRL was measured to define non-specific binding. The results are expressed as a fraction of the binding measured in cells expressing ACE2 (mean ± S.D, n = 3) and in the absence of a competing ligand.
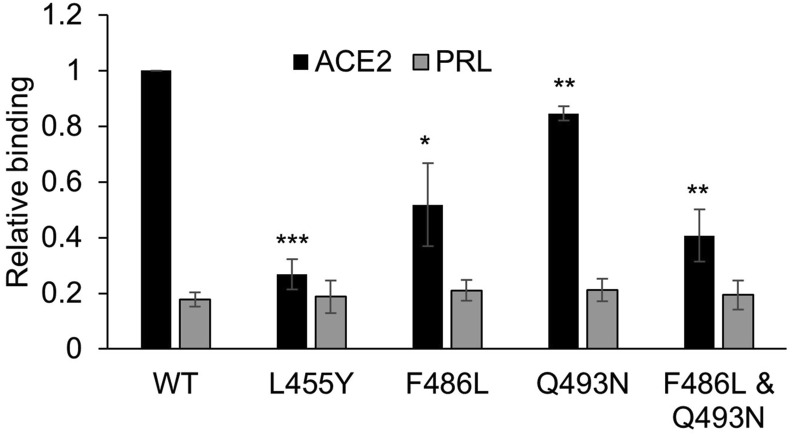
To provide further evidence that HiBIT-RBD bound ACE2 in a manner that reflected the virus binding to ACE2, we made use of reports [[13], [14], [15]] that identified crucial amino acids in the RBD that are involved in the RBD-ACE2 interaction. It has also been reported that mutation of L455, F486 and Q493 in the SARS-COV-2 RBD to the corresponding residues in SARS-COV-1 substantially reduces the binding of the SARS-COV-2 RBD to ACE2 when measured in an ELISA assay [16]. Consequently, mutations encoding L455Y, F486L and Q493N were engineered into the HiBIT-RBD construct. When these were tested in the HiBIT binding assay (Fig. 4 ), HiBIT-RBDL455Y and HiBIT-RBDF486L exhibited diminished binding to cells expressing ACE-2 and, in the case of HiBIT-RBDL455Y the specific binding was almost completely blocked. In the case of HiBIT-RBDQ493N, binding to ACE-2 expressing cells was reduced more modestly (although significantly). HiBIT-RBDF486L,Q493N, which contained both F486L and Q493 N mutations, also bound weakly to cells expressing ACE2, although not significantly more so than did HiBIT-RBDF486. There was no measurable effect of any of the mutations on the non-specific binding of the HiBIT-RBDs to control cells expressing prolactin.
Fig. 4.
Inhibition of HiBIT-RBD binding to ACE2 by site directed mutagenesis of the interaction domain. HiBIT-RBDs, containing mutations that have previously been shown to inhibit RBD to ACE2 in an ELISA assay, were transiently expressed in COS cell and the cell culture supernatant added to cells expressing either ACE2 or PRL. After incubation, binding was measured by addition of HiBIT detection reagent. The results are expressed as a proportion of the binding of wild-type RBD-HiBIT to ACE 2 measured in each experiment (mean ± S.D, n = 4) and are significantly different to this where shown (paired t-test; ∗, P < 0.05; ∗∗, P < 0.005; ∗∗∗, P < 0.0005).
4. Discussion
We have developed an assay that specifically measures the binding of SARS-COV2 to ACE2. The assay was implemented in a 384-well format allowing reasonable throughput. The assay can be conducted using widely available scientific instrumentation (a plate reader capable of measuring luminescence) and reagents, and it is envisaged that this technology could be deployed in many laboratory settings.
For this assay ACE2, the main SARS-COV-2 receptor, was transiently expressed in COS cells because these cells can be transfected with high efficiency. HEK-293 cells were also investigated, but owing to their propensity to become easily dislodged during washing it was deemed unlikely that reproducible data could be obtained with them. COS cells were also preferred over Ovcar-8 cells, which can also be transfected with relatively high efficiency, because of the higher level of expression observed in the former cells. In the future, it may be useful to develop a cell line stably expressing the receptor. However, since we hoped to identify compounds which could inhibit RBD binding to ACE2 as quickly as possible to assist in the SARS-COV-2 pandemic, that option was not pursued here.
Several lines of evidence suggest that the described assay faithfully measures RBD binding to ACE2. As expected, binding of HiBIT-RBD was both time dependant and saturable, consistent with binding to ACE2. Importantly, significantly more binding of HiBIT-RBD was measured in cells expressing ACE2 than to cells transfected with a negative control (PRL). This indicated that the measured binding was specific to ACE2. Furthermore, both RBD and full length S1 proteins obtained from distinct biological sources could compete with HiBIT-RBD for ACE2 binding thereby giving confidence that the binding of HiBIT-RBD reflected binding of the viral RBD to its receptor in its authentic binding mode. Finally, the mutation of several different residues that have previously been shown to inhibit RBD binding to ACE2 in an ELISA assay [16] also reduced it the binding measured in the HiBIT assay. Taken together, these data strongly suggest that an assay has been developed that replicates and measures the binding of the viral RBD to ACE2 through its authentic binding mode.
The three mutations introduced into the RBD mimicked the corresponding amino acids in SARS-COV-1 and have been previously shown to inhibit SARS-COV-2 binding to ACE2 in an ELISA assay [16]. These residues have also been shown from studies of the structure of ACE-2-RBD complex [[13], [14], [15]] to play a significant role at the interaction interface of the two proteins. In this study, two of the mutations (L455Y and F486L) substantially inhibited HiBIT-RBD binding to ACE2. However, a third mutation (Q493N) had a very modest effect, although it substantially inhibited binding in the ELISA assay [16]. To explore this further, an RBD was generated containing both Q493N and F486L mutations and this also bound more weakly than the wild-type protein and HiBIT-RBDQ493N but not significantly more than HIBIT-RBDF486L The relatively modest effect is perhaps not surprising when considering the conservative nature of the glutamine to asparagine mutation. Furthermore, the crystal structure of ACE2 bound to the RBD shows Q493 in two possible conformations [15], suggesting there is some flexibility in the binding mode.
The assay was employed to screen a library of approximately 100 approved drugs [17]. Unfortunately, no drugs were identified that reproducibly inhibited binding. Some hits appeared to inhibit the binding, but further exploration of these compounds revealed that they also inhibited the luminescence generated by adding HiBIT-RBD to the detection reagent directly, suggesting that the drugs inhibited nanoluc reporter reaction, rather than the HiBIT-RBD-ACE2 binding interaction. This emphasizes the importance of confirming hits identified in drug screens through validation in biochemical assays based on distinct technologies or by using other functional assays such as pseudotyped virus-like particle and/or live virus assays.
One issue with the assay is that the whole assay must be conducted on ice. In preliminary experiments (not shown) binding was not detected when the assay was performed at 37 °C. Consequently, the assay was conducted on ice to prevent endocytosis of RBD bound to ACE2. To avoid the plate warming appreciably during luminescence meaurements, a very short integration time per well (0.1s) was used. Even so, some increase in the signal measured in control samples was observed as the plate was read and to control for this, multiple controls were included across the plate. It would be preferable to avoid this problem by using a plate reader that allowed the sample plate to be cooled. Alternatively, it might be possible to conduct the assay at a slightly higher temperature to minimize the effect of warming. It may also be possible to configure the assay to use cell membranes or fixed cells.
The assay developed here is suitable for the identification of drugs that inhibit the binding of SARS-COV-2 to ACE2. It may also be used to validate hits identified by other workers using binding assays based on other technologies. Although the assay has some limitations, these could be ameliorated with further work.
Author contributions
AR conceived the study and performed the experiments with the assay. MAL and MS designed and prepared the RBD expressed in E. Coli and FK designed the drug library. All authors reviewed and contributed to the manuscript.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This research was funded by Keele University (AR) and the BBSRC ( UK (BB/L023717/1 (MAS); BIV-HVB-2020/07/SKIDMORE (MAS & MAL); BB/S009787/1 (MAS & MAL)).
References
- 1.Santos R., Monteiro S. Viruses Food Water Risks. Surveill. Control; 2013. Epidemiology, control, and prevention of emerging zoonotic viruses; pp. 442–457. [DOI] [Google Scholar]
- 2.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maginnis M.S. Virus–receptor interactions: the key to cellular invasion. J. Mol. Biol. 2018;430:2590–2611. doi: 10.1016/j.jmb.2018.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y., Yang C., feng Xu X., Xu W., wen Liu S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., Thacker B.E., Glass C.A., Yang Z., Torres J.L., Golden G.J., Bartels P.L., Porell R.N., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B.M., Caradonna T.M., Kellman B.P., Martino C., Gordts P.L.S.M., Chanda S.K., Schmidt A.G., Godula K., Leibel S.L., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183(4):1043–1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Österlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Heal. 2020 doi: 10.1016/s2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundstrom L.E. Thinking inside the box. To cope with an increasing disease burden, drug discovery needs biologically relevant and predictive testing systems. EMBO Rep. 2007;8 doi: 10.1038/sj.embor.7400939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh-hashi K., Furuta E., Fujimura K., Hirata Y. Application of a novel HiBiT peptide tag for monitoring ATF4 protein expression in Neuro2a cells. Biochem. Biophys. Reports. 2017;12:40–45. doi: 10.1016/j.bbrep.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mycroft-West C.J., Su D., Pagani I., Rudd T.R., Elli S., Guimond S.E., Miller G., Meneghetti M.C.Z., Nader H.B., Li Y., Nunes Q.M., Procter P., Mancini N., Clementi M., Bisio A., Forsyth N.R., Turnbull J.E., Guerrini M., Fernig D.G., Vicenzi E., Yates E.A., Lima M.A., Skidmore M.A. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Surface Protein (Spike) S1 Receptor Binding Domain with Heparin. BioRxiv. 2020:2020. doi: 10.1101/2020.04.28.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson A., Malik R.K., Hildebrand J.D., Parsons J.T. Inhibition of cell spreading by expression of the C-terminal domain of focal adhesion kinase (FAK) is rescued by coexpression of Src or catalytically inactive FAK: a role for paxillin tyrosine phosphorylation. Mol. Cell Biol. 1997;17:6906–6914. doi: 10.1128/mcb.17.12.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 16.Yi C., Sun X., Ye J., Ding L., Liu M., Yang Z., Lu X., Zhang Y., Ma L., Gu W., Qu A., Xu J., Shi Z., Ling Z., Sun B. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020;17:621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanim F.L., Merrick B.A.M.E., Giles H.V., Jankute M., Jackson J.B., Giles L.J., Birtwistle J., Bunce C.M., Drayson M.T. Redeployment-based drug screening identifies the anti-helminthic niclosamide as anti-myeloma therapy that also reduces free light chain production. Blood Canc. J. 2011;1 doi: 10.1038/bcj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]