Abstract
Background
SARS-CoV-2, the etiological agent causing COVID-19, has infected more than 27 million people with over 894000 deaths worldwide since its emergence in December 2019. Factors for severe diseases, such as diabetes, hypertension, and obesity have been identified however, the precise pathogenesis is poorly understood. To understand its pathophysiology and to develop effective therapeutic strategies, it is essential to define the prevailing immune cellular subsets.
Methods
We performed whole circulating immune cells scRNAseq from five critically ill COVID-19 patients, trajectory and gene ontology analysis.
Results
Immature myeloid populations, such as promyelocytes-myelocytes, metamyelocytes, band neutrophils, monocytoid precursors, and activated monocytes predominated. The trajectory with pseudotime analysis supported the finding of immature cell states. While the gene ontology showed myeloid cell activation in immune response, DNA and RNA processing, defense response to the virus, and response to type 1 interferon. Lymphoid lineage was scarce. Expression of genes such as C/EBPβ, IRF1and FOSL2 potentially suggests the induction of trained immunity.
Conclusions
Our results uncover transcriptomic profiles related to immature myeloid lineages and suggest the potential induction of trained immunity.
Key Words: COVID-19, SARS-CoV-2, scRNAseq, Immune cell profile, Critically ill, Trained immunity, Emergency myelopoiesis
Introduction
Since its emergence in Wuhan, China in December 2019 the coronavirus disease (COVID-19) epidemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rapidly progressed into a devastating pandemic with serious social and economic consequences (1). According to the World Health Organization, more than 27 million people have been diagnosed with COVID-19 and over 894000 persons have died worldwide (2,3). The clinical presentation of COVID-19 varies widely. However, a subset of patients may evolve into acute respiratory distress syndrome (2), requiring intubation and mechanical ventilation, which is frequently complicated by a peculiar form of coagulopathy and eventually multiorgan failure and death. This complicated course occurs in the context of a “cytokine storm” characterized by overproduction of TNF, IL6, IL1ß, and G-CSF and generalized vascular hyperpermeability (4). Although risk factors such as diabetes, obesity, and hypertension have been associated with the development of life-threatening COVID-19, a non-negligible proportion of healthy young individuals without comorbidities develops the severe form of the disease. The immunopathogenesis of this severe form of COVID-19 is poorly understood (5). Viral particles spread through the respiratory mucosa infecting other cells, unleashing a series of immune responses, characterized by a reduction of T and B lymphocytes and an increment of monocytes and neutrophils (6, 7, 8, 9). A detailed understanding of this altered immunological state is crucial for the design and development of effective therapeutic strategies. In the present work, we performed single-cell RNAseq from circulating immune response cells from critically ill COVID-19 patients requiring invasive mechanical ventilation.
Materials and Methods
Patients and Tissue Samples
Blood samples from five critically ill patients with COVID-19 were collected in EDTA-coated tubes, samples were collected from patients diagnosed, treated, and followed at the Medicina Interna department of the Hospital de Especialidades, Centro Médico Nacional Siglo XXI of the Instituto Mexicano del Seguro Social in April 2020. All participating patients were recruited with signed informed consent from a family member and ethical approval from the Comisión Nacional de Ética e Investigación Científica del Instituto Mexicano del Seguro Social in accordance with the Helsinki declaration. SARS-CoV-2 infection was corroborated by RT-qPCR at an official federal government reference laboratory.
Sample Preparation, scRNAseq Library Generation and Sequencing
Peripheral blood from the five critical COVID-19 patients was collected in EDTA-coated tubes, and whole immune cells were isolated according to standard centrifugation methods, 3500 rpm for 15 min, followed by red blood cell lysis.
Chromium Next GEM Single Cell 3′ Reagent Kits v3.1 and protocol from 10X Genomics was followed as recommended by manufacturer's instructions, briefly described. The five patients' immune cells were pooled in a single tube and cells were diluted in 1x phosphate-buffered saline (PBS) to 700–1200 cells per μL. The cell suspension was loaded in Chromium Next GEM Chip G and sorted in the Chromium Controller from 10X Genomics. The Cell-Gel Beads in Emulsion (GEMs) were then incubated to generate the barcoded cDNA. cDNA was cleaned using Dynabeads and washed, followed by cDNA amplification and SPRIselection. The retrieved cDNA was enzymatically fragmented, end-repaired, poly-A tailed, and ligated. Size selection, adaptor ligation, and amplification were done. Sequencing was done using NextSeq 550 System High-Output Kit (300 cycles) in NextSeq 500 system (Illumina) according to 10X Genomics specifications: Read 1 = 28 cycles, Read 2 = 91 cycles, Index 1 = 8 cycles. All quality control steps were carried out using 4200 TapeStation System (Agilent) with High Sensitivity D1000 Screen Tape, whereas the concentration was calculated using Qubit 2.0 Fluorometer with Kit High Sensitivity assays.
scRNAseq Bioinformatic Analysis
Partek Flow software was used with scRNAseq toolbox. First, the tags were trimmed, and then the reads were aligned using STAR 2.7.3a algorithm to human genome hg38. UMI's were deduplicated and barcode filtered. The following criteria were then applied to each cell, i.e., gene number between 200 and 6000, UMI count above 300, and mitochondrial gene percentage below 20%. To quantify the transcriptome human hg38 Ensembl transcripts release 99 was used. Counts per million, add 1.0 Log 2.0 was the normalization parameters. Healthy donors' datasets were downloaded from the 10X Genomics website and analyzed using Loupe Browser from 10X Genomics.
Data has been deposited in Sequence Read Archive hosted by National Center for Biotechnology Information under accession number PRJNA635580.
Markers Used to Circumscribe Cell Populations
Clusters were categorized by analyzing differentially expressed genes according to previously published data obtained from human samples (10, 11, 12, 13, 14, 15).
Dimensionality Reduction and Clustering
The filtered and normalized gene-barcode matrix was analyzed by principal components, then graph-based and t-distributed stochastic neighbor embedding (t-SNE) using default parameters were carried out.
Trajectory and Pseudotime Analysis
Monocle2 algorithm located within the Partek Flow scRNAseq toolbox was used with default parameters to calculate the transition states and pseudotime from cell populations identified.
Gene Ontology Analysis
WebGestalt (http://www.webgestalt.org) was used for understanding the biological meaning behind the resulting list of genes, to obtain gene ontology for significantly de-regulated genes in COVID-19 patients’ cell populations.
Results
Clinical Characteristics of Critically Ill COVID-19 Patients
A pool of peripheral blood samples from five patients with severe SARS-CoV-2 infection was created and used for the scRNAseq experiment, their main clinical and biochemical characteristics are depicted in Table 1 . The five patients were males with a mean age of 47.8 ± 6.6 years (range 41–57). Two patients were overweight, one had grade 1 obesity and two had previously been diagnosed with type 2 diabetes mellitus; no history of hypertension was found in any of them and two patients had no known risk factors for severe SARS-CoV-2 infection. All patients had radiological evidence of alveolar occupation and ground glass appearance on imaging studies and required intubation and invasive mechanical ventilation 3–7 d after admission as well as a hemodynamic collapse that required vasopressor support. They all had significantly elevated D-dimer, fibrinogen, procalcitonin, and C-reactive protein levels. Besides respiratory and hemodynamic support, all five patients were treated with enoxaparin, hydroxychloroquine, azithromycin, or clarithromycin as well as lopinavir and ritonavir; three received high dose glucocorticoids (hydrocortisone or dexamethasone). One of the five patients developed secondary bacterial and fungal infections for which he received meropenem, vancomycin, and voriconazole; no major arterial or venous thrombotic events were recorded in any of them. Three of the five patients died of ARDS and multiorgan failure. Of note, the majority of patients showed increased myeloid absolute numbers after 7 d of hospitalization.
Table 1.
Clinical and biochemical characteristics of critically ill patients with COVID-19 (OW: overweight, DM: type 2 diabetes mellitus, BMI: body mass index)
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | |
|---|---|---|---|---|---|
| Age, yrs | 57 | 46 | 43 | 41 | 52 |
| BMI, Kg/cm2 | 29 | 26.3 | 31 | 24 | 29 |
| Comorbidities | OW | None | DM, OW | None | DM, OW |
| Admission to intub, days | 4 | 3 | 5 | 3 | 5 |
| Leukocytes per mm3 | 9130 | 8760 | 7760 | 8900 | 5060 |
| Lymphocytes, x 103/mm3 | 650 | 520 | 160 | 1130 | 1600 |
| Hb, g/dL | 16.4 | 15.9 | 17 | 12.9 | 15.8 |
| Platelets, x 103/mm3 | 193 | 357 | 380 | 523 | 169 |
| CRP, mg/L | 23.6 | 35 | 11.9 | 27 | 11.6 |
| Procalcitonin, ng/mL | 2.32 | 1.87 | 0.07 | 0.22 | 0.33 |
| D-Dimer, ng/mL | 2410 | 3320 | 1100 | 2980 | 2020 |
| Fibrinogen, mg/dL | 834 | 788 | 753 | 789 | 773 |
| Final outcome | Dead | Dead | Dead | Alive | Alive |
Immature Myeloid Cell Populations Predominance in Critically Ill COVID-19 Patients
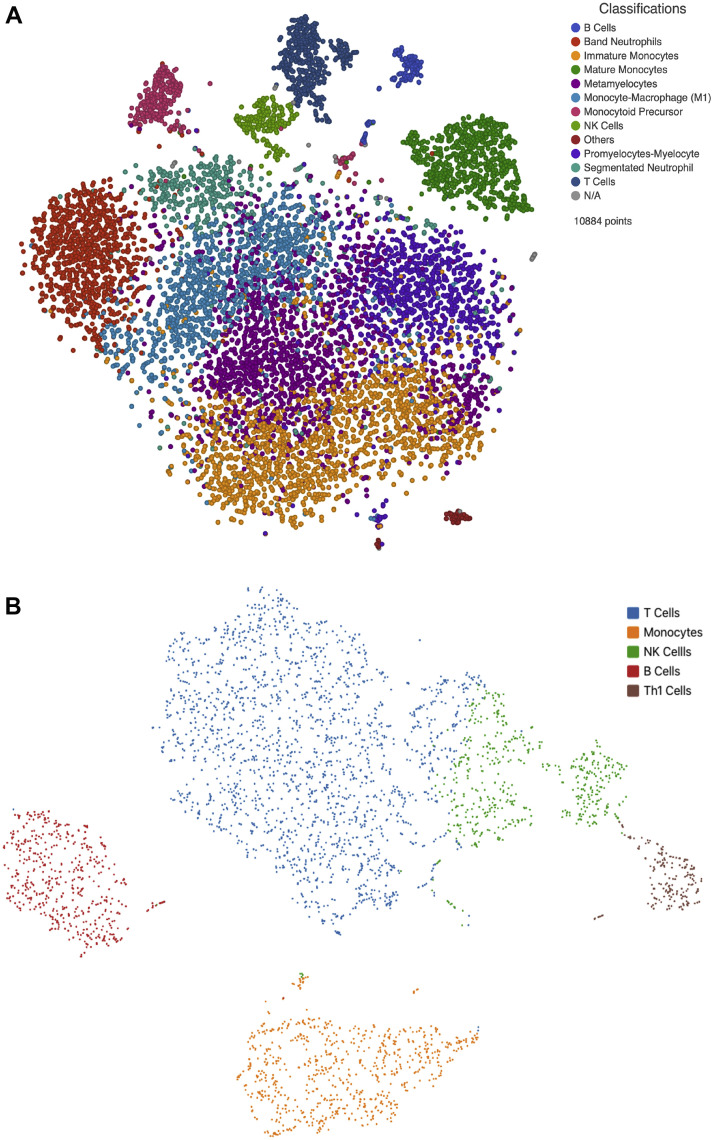
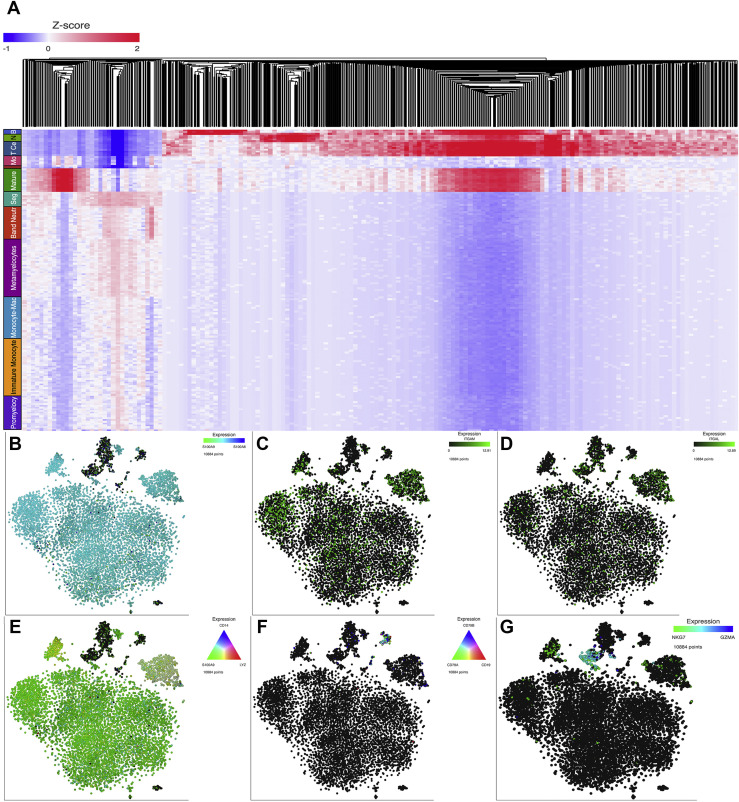
A total of 10884 immune cells from critically ill COVID-19 patients and 4056 from healthy donors were studied. After normalization and clustering of the gene expression matrix, 12 cell populations clusters were identified in critically ill COVID-19 patients whereas 5 cell populations clusters were present in healthy controls. Cells of myeloid origin predominated in critically ill COVID-19 patients whereas cells of lymphoid origin were more prevalent among healthy controls. Interestingly, in COVID-19 patients with severe SARS-CoV-2 infection, immature myeloid cell populations predominate, such as band neutrophils (10.74%), metamyelocytes (19.10%), promyelocytes-myelocytes (11.14%), monocytoid precursor (3.30%), immature monocytes (19.0%). Mature lineages such as segmented neutrophils (5.01%), mature monocytes (7.65%) and finally monocyte-macrophages (13.82%) were also observed. Lymphoid cell populations, on the other hand, including B (1.77%) and T (4.78%) lymphocytes as well as NK cells (2.37%) are present in low frequencies in these patients. In contrast, healthy individuals displayed higher proportions of lymphoid cell lineages such as T (45%), Th1 (5%) and B (12.72%) lymphocytes as well as NK cells (14.91%) compared to monocytes (21.52%) in the myeloid cell lineage (Figure 1 ). Transcriptome analysis from cell populations identified in peripheral blood (PB) of critically ill COVID-19 patients resembled immature lineages in the bone marrow (BM). Hence, the comparison and coincidence of most of the transcripts in the different populations characterized the different clusters. Thus, the promyelocytes-myelocyte cluster was defined by the expression of genes such as ACPP, GLB1, CTSS, SCL11A1, ITM2B, P2RX1, TREM1, TRIM22, SP110, IRF1, STAT1 and NCOA1. Metamyelocytes by the expression of the following genes: ITGAM, GGH, CR1, MGAM, MMP9, TIMP2, TNFAIP6, VNN2, ANPEP, DYSF and PLXNC1. Genes such as GNS, VNN1, ITGAM, C3AR1, CLEC5A, ADAM8, CD58, CR1, FCER1G, MGAM, MMP8, MMP9, SIGLEC5, VNN2, DYSF, LILRB3, TLR8 and CCR are characteristically expressed by band neutrophils, whereas segmented neutrophil cells were defined by the expression of MGAM, MMP25, SIGLEC9, VENTX, NLRP1 and VENTX. Monocytoid precursors were identified by the expression of GATA2, HOXA10, LYZ, FCN1 and S100A9 and the cell cycle genes PCLAF, STMN1, MKI67, TOP2A and CDK1. The expression of genes such as TLR2, FUT4, CCR1, CCR3, PIAS, TRIM22, TRIM38, IRF1 and JUNB identified immature monocytes, this cluster also showed a tendency towards M1 phenotype, while monocyte-macrophage (M1) were characterized by the expression of STAT2, EGR2, CREB5, TREM1, TL2, FCGR3B, ITGAX and NLRP1. Mature monocytes were defined by the presence of CD14, LYZ and S100A9. The expression of genes such as CD79A, CD79B and CD19 defined the presence of B lymphocytes. T lymphocytes express CD3E and CD3D whereas NK cells express NKG7 and GZMA (Figure 2 ).
Figure 1.
Cell populations identified in critically ill COVID-19 patients. Panel A. depicts the t-SNE from COVID-19 critically ill patients scRNAseq data. Twelve clusters are represented by immature myeloid populations such as band neutrophils, metamyelocytes, promyelocytes-myelocytes, monocytoid precursor, immature monocytes. Mature lineages such as segmented neutrophils, mature monocytes and finally monocyte-macrophages are observed. Scarce lymphoid cell populations, represented by B, T and NK cells are present in these patients. In contrast, panel B. shows the t-SNE from scRNAseq data from healthy donors with lymphoid T, Th1, NK and B cells predominance compared to monocytes.
Figure 2.
Molecular markers identifying cell clusters. Panel A. portrays the hierarchical cluster from the differentially expressed genes among the cell populations identified in the COVID-19 patients. Panels B, C, and D. shows the t-SNE displaying expression of S100A9 and S100A8, ITGAM and ITGAL, respectively, in myeloid cell subsets depicting immature features. Whereas panels E, F, and G. Shows mature monocyte cell subset expressing CD14, LYZ and S100A9 genes, B cells expressing CD79A, CD79B, CD19 and NK cells expressing NKG7 and GZMA, respectively.
Differentiation Profiles Support the Existence of Immature Cell Populations in Critically Ill, COVID-19 Patients
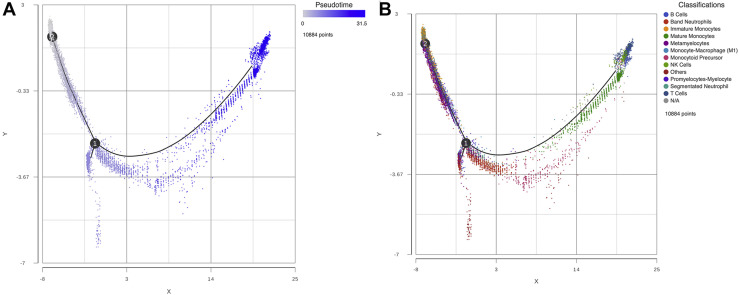
Trajectory analysis along with its respective estimation of pseudotime were carried out to infer potential differentiation states in the cell populations found in critically ill COVID-19 patients. Node 2 which gathers several of the immature cell populations, among them metamyelocytes, promyelocytes-myelocytes and immature monocytes showed time 0, follow throughout the trajectory path to node 1 were it bifurcates into two nodes, one end-node representing and “other” cells, and the second bifurcation end which includes the mature monocytes at the end of the node. As expected, B and T lymphocytes, along with NK cells were among the most mature cells found in this analysis (Figure 3 ). These trajectory results support our findings of diverse immature cell state populations and their potential transitional states in critically ill patients with COVID-19.
Figure 3.
Trajectory analysis. Panel A. Trajectory analysis with pseudotime represented potential transitional states. Node 2 gathers the immature cell populations and showed time 0, follow throughout the trajectory path to node 1 were it bifurcates into 2 nodes, the second bifurcation showed the mature or differentiated cell states. Panel B. depicts the cell populations identified by their transcriptomes and their potential transitional states according to pseudotime. As expected, metamyelocytes, promyelocytes-myelocytes and immature monocytes are in node 2 and mature monocytes and B and T lymphocytes, along with NK cells were among the most mature cells in the opposite node.
Gene Expression Alterations in Cell Populations from COVID-19 Patients were Related to Key Cellular Processes
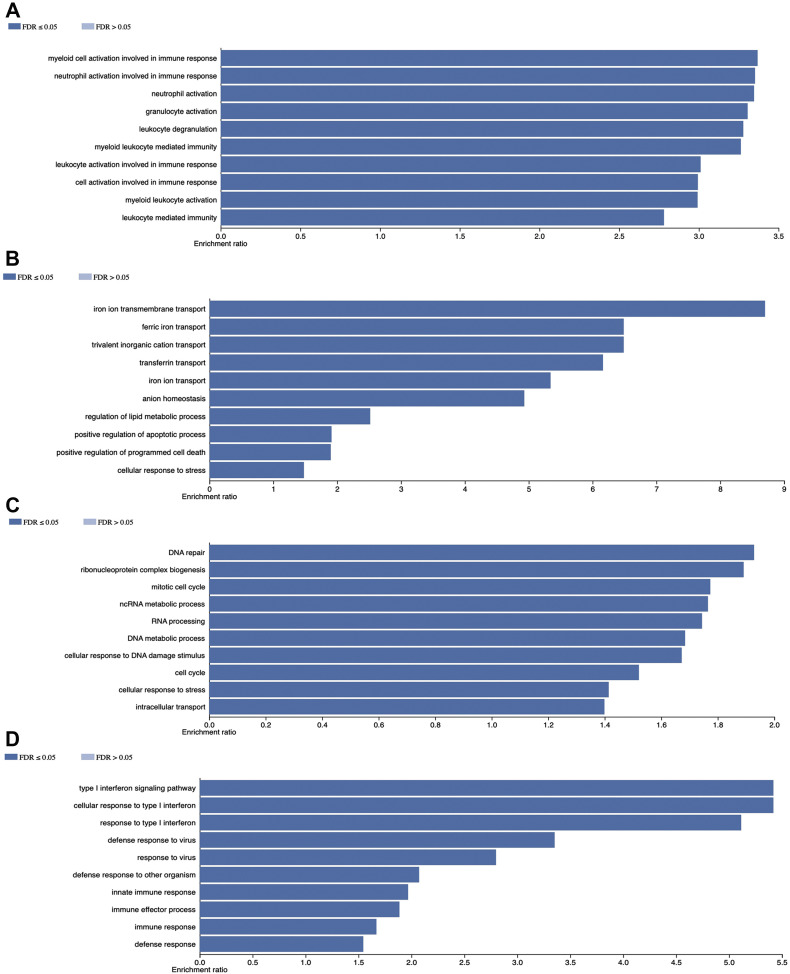
Cellular alterations of the different myeloid cell population subsets were divided in four groups: those related to a) myeloid cell activation in immune response as well as granulocyte and leukocyte activation, b) iron metabolism and anion homeostasis, c) cell cycle control, DNA and RNA processing and d) defense response to virus, response to type 1 interferon and innate immune response. The lymphoid cellular subset alterations were mostly related to ribosome biogenesis as well as RNA processing and modification (Figure 4 ).
Figure 4.
Gene Ontology terms. Gene ontology results in myeloid cell subsets are represented in panel A. myeloid cell activation in immune response as well as granulocyte and leukocyte activation, B. Iron metabolism and anion homeostasis, C. Cell cycle control, DNA and RNA processing and D) defense response to virus, response to type 1 interferon and innate immune response.
Evidence of a TLR-related Activation in Circulating Immature Myeloid Cells of Severe COVID-19 Patients
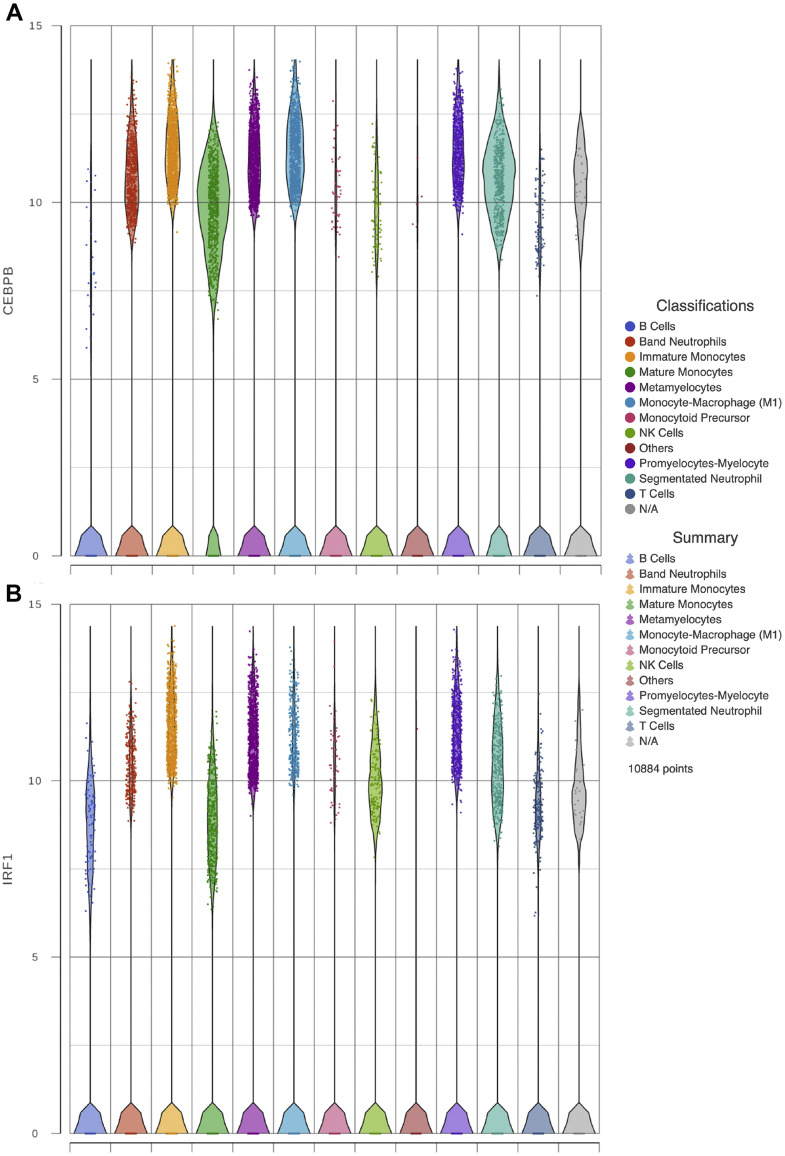
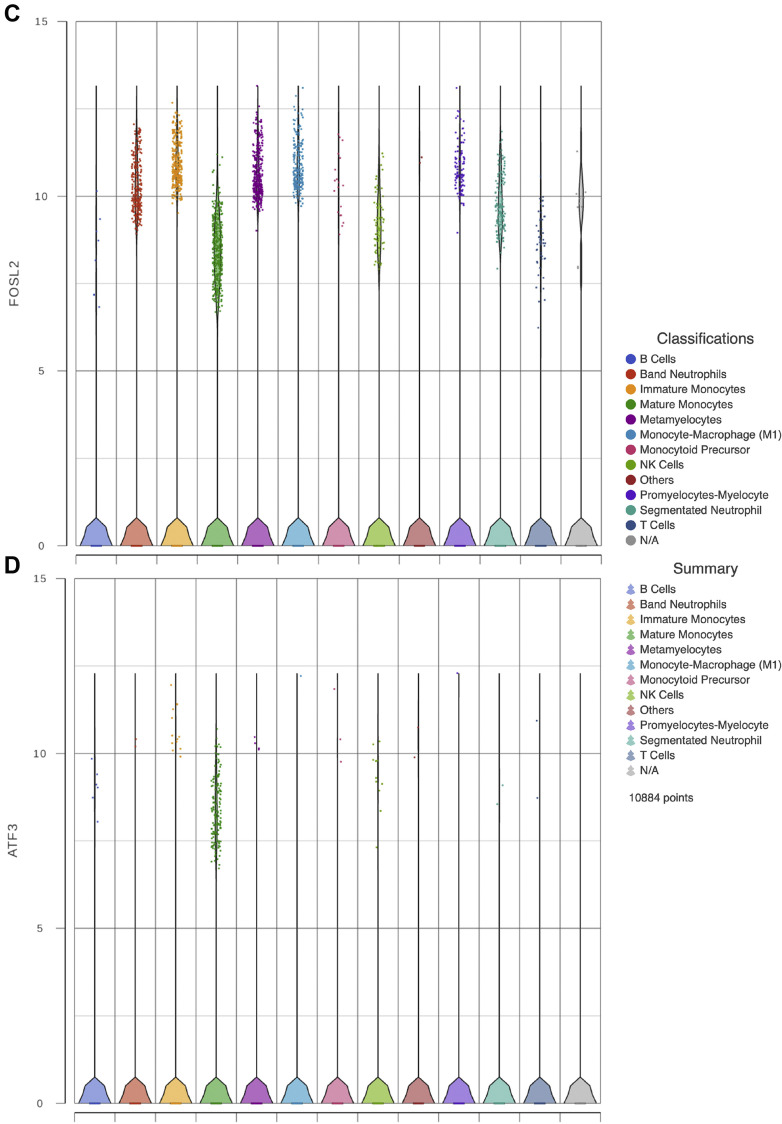
Traditionally, it was thought that immunological memory was limited to adaptative immune responses. Recent evidence has shown that primitive hematopoietic subpopulations, together with its progeny, can be trained to effectively produce myeloid cells and to respond more efficiently to a second challenge, respectively (16). This novel phenomenon is dependent on the transcription factor C/EBPβ which is needed to maintain accessibility to open chromatin regions and to promote TLR-related responses. We sought to evaluate the expression of C/EBPβ along with other transcripts involved in TLR-dependent signaling (Figure 5 ). When comparing normal monocytes to the immature lineages present in COVID-19 samples revealed that the expression of CEBPβ was higher, also, the expression of TLR-related transcripts such as IRF1and FOSL2 was upregulated, while ATF3 expression on immature granulocytes and monocytes was diminished. Therefore, our results suggest that immature circulating myeloid cells of severe COVID-19 patients are enriched in transcripts related to TLR signaling that resemble the CRBPβ-dependent open chromatin regions observed during lipopolysaccharide-dependent trained immunity of primitive populations.
Figure 5.
Immunity trained gene expression. Violin plots show gene expression of CEBPβ, IRF1, FOSL2 and ATF3 in the critically ill COVID-19 patients immune cells analyzed in panels A, B, C, and D respectively.
Discussion
The SARS-CoV-2 is the etiological agent causing COVID-19 which has infected more than 27 million people with more than 894000 deaths since its emergence in December 2019 (3). Defining which are the immune cell subsets that prevail in the circulation of these patients and what is their molecular activation status is crucial for understanding the immune clearance mechanisms that operate in these patients and thus, for the development of new and effective therapeutic strategies. Among the molecular alterations found in the immune response cells from critically ill patients with COVID-19 was the abnormal type 1 interferon response (17) and could potentially represent a target for molecular therapy (18). Typical PBMC in healthy individuals include ∼80% of T lymphocytes (CD3D), ∼6% of NK cells (NKG7), ∼6% of B lymphocytes (CD79A) and ∼7% of myeloid cells (S100A9) (19). During viral acute infection the single-stranded RNA from SARS-CoV-2 can be recognized by TLR-7 and TLR-8 during replication, while TLR3, RIG-like receptors, and MDA-5 can sense double-stranded RNA which is a replication intermediary (20); the viral Spike protein can be recognized by TLR2 (21), TLR4 and TLR6 (22). While the recognition of viral patterns induces strong immune responses, it is also known that human stem and progenitor cells (HSPC) express TLR7, TLR8, TLR2, TLR3, and TLR4 and upon stimulation, granulopoiesis and monopoiesis are induced and perpetuated (23, 24, 25, 26, 27).
Using a scRNAseq approach we have found a distinctive predominance of cell lineages of myeloid origin and a diminished number of lymphoid lineages in patients with severe atypical pneumonia and life-threatening respiratory distress syndrome due to SARS-CoV-2, which is in accordance with recently reported data (7, 8, 9). Not only increased myeloid lineage was favored in COVID-19 critical patients, but also immature cell states were observed. Atypical morphological alterations in monocytes from patients in intensive care unit has been reported (28), and the presence of immature granulocytes such as metamyelocytes and band neutrophils has been reported (9). Collectively, our results are in accordance with similar observations were severe COVID-19 patients presented marked changes in immune cell composition and phenotype, indicating emergency granulopoiesis (29). It is noteworthy that a similar blood profile has been observed in patients with acute parvovirus B19, cytomegalovirus and Epstein-Barr viral infection (30, 31, 32). Thus, the presence of immature myeloid cells in peripheral blood of patients in response to acute and severe viral infection is a common feature.
Our results suggest that severe SARS-CoV-2 infection induce immature granulocyte and monocyte release from the BM since most of the transcripts corresponded to an immature state of differentiation of granulocytic and monocytic programs (11, 12, 13,15). In line with our findings, peripheral blood (PB) smears from critical COVID-19 patients have shown to have morphological abnormalities that correlate with maturation asynchrony (33). Overproduction of myeloid cells could be the result of increased proliferation/differentiation of hematopoietic stem and progenitor cells in response to SARS-CoV-2 (potential emergency hematopoiesis) which currently single cell transcriptome results could support. We have previously reported a proliferative state of immune cells from COVID-19 patients (submitted). Also, the massive release of immature myeloid cells -already present in bone marrow as a result of increased levels of cytokines such as G-CSF, which is upregulated (4). Defective monocyte activation and dysregulated hematopoiesis or myelopoiesis may contribute to severe disease course and ARDS development. Severe infections trigger the emergency myelopoiesis or hematopoiesis which could be linked to immunosuppressive functions of B and T cells. This suggests that immune cells emerging prematurely from the bone marrow are programed to anti-inflammatory or even suppressive phenotype in severe COVID-19 (34).
Although more than 5 different populations of PB monocytes have been described (35), in our study only one of these cell populations was present in blood from healthy individuals. In contrast, in blood samples from critically ill patients with COVID-19 we found four different monocyte clusters. One of these clusters which we named monocytoid precursor cluster, displays monocytic transcripts as well as transcripts in implicated in cell cycle regulation. We believe this cluster displays a more immature transcriptome which prompts us to speculate that it as a precursor population of the mature monocyte cluster and the M1 monocyte-macrophage.
Lineage-specific acquisition of function is achieved during cell maturation. Our analysis showed that lineage-specific molecules, such as S100A8/A9, are expressed in all stages of differentiation of myeloid cells in patients with COVID-19. These transcripts encode the myeloid-related proteins 8/14 which are known to be released and recognized in an autocrine fashion by TLR4 for effective diapedesis (36). Integrins (LFA-1 and Mac-1) are fundamental for myeloid migration and function (37) and they are acquired during the latest stages of differentiation. In line with our results, ITGAL (LFA-1) and ITGAM (MAC-1) were also poorly expressed in all immature lineages of COVID-19 samples while they are fully expressed in mature myelocytes.
In accordance with the left shift seen in patients with COVID-19 (9,38) our results show severe lymphopenia reflected by at least a 7 fold reduction in the percentage of B and T lymphocytes and NK cells. During acute inflammatory conditions, the production of B lymphocytes and NK cells is drastically inhibited, favoring myelopoiesis (27,39, 40, 41). In this regard, it is interesting to note that our two surviving patients had normal lymphocyte counts, while those who died had severe lymphopenia. In line with this, it has been reported in severe COVID-19 patients, as well as, some bacterial infections that a high neutrophil – to – lymphocyte ratio is associated to multi-organ damage and mortality (42), lymphopenia under septic conditions is known to be caused by exacerbated apoptosis (43). Factors involved in lymphocyte apoptosis are the high amounts of proinflammatory cytokines that can atrophy the lymphoid organs or inhibit lymphocyte proliferation. Soluble factors quantified in the blood such as Procalcitonin and C-reactive protein which tended to be higher in severe patients of our study, correlated as previously reported, with COVID-19 mortality (44). Thus, these factors can also contribute to assessment of the COVID-19 severity.
Recently, the trained-immunity HSPC has been shown to be dependent on epigenetic events which induce the maintenance of open chromatin regions that persist in a CEBPβ-dependent fashion. Importantly, these epigenetic changes are conserved during differentiation and are used by myeloid cells during the immune response (16). Therefore, we searched in our analysis for the expression of CEBPβ and found it in all stages of granulocytic and monocytic differentiation in samples from patients with severe COVID-19. Furthermore, we looked for additional factors that are known to be involved in TLR signaling and that are recruited to the open chromatin regions in conjunction with CEBPβ. It is noteworthy that in comparison to healthy donor-derived monocytes, transcripts related to TLR responses such as IRF1 and FOSL2 were upregulated in patients with severe COVID-19, reflecting perhaps a state of innate immune cell activation, which is in line with our gene ontology results. Further supporting this notion is our finding of very few cells expressing RUNX1 which is in agreement with the absence of the most primitive HSPC in peripheral blood. Altogether, CEBPβ and IRF1 and FOSL2 upregulation, denote an activation state related to TLR-dependent responses during severe COVID-19.
In COVID-19 patients a left shift is observed, our results suggests potential emergency hematopoiesis has occurred, supported by the presence of proinflamatory cytokines observed in this disease (4) along with the exposure to viral PAMPs and our own data suggesting a TLR-dependent activation. Other ligands such as Poly I:C (a TLR3 ligand) can also sensitize HSPC to a secondary challenge (16). Since during SARS-CoV-2 replication, TLR3 can recognize dsRNA, it is reasonable to think that possibly an epigenetic reprogramming of primitive cells (trained immunity) could occur which could contribute to a protection to a later SARS-CoV-2 challenge though the mounting of a more efficient ability to mount innate immune responses.
In summary our results show the presence of immature myeloid lineages in peripheral blood in critically ill COVID-19 patients potentially suggesting a state of emergency myelopoiesis. The molecular signatures of this phenomenon are consistent with activation of TLR-induced responses and perhaps the acquisition of trained immunity.
Acknowledgments
DMR is a recipient of the National Council for Science and Technology Fellowship “Catedra CONACyT” program. This work was partially supported by grants 289499 from Fondos Sectoriales Consejo Nacional de Ciencia y Tecnologia Mexico and R-2015-785-015 from Instituto Mexicano del Seguro Social, Mexico (MM). We would like to thank to Xiaowen Wang from Partek Inc. for the exceptional technical support provided.
(ARCMED_2020_1675)
Competing Interest
At the time that this project was carried out RCG and CML worked for Analitek S.A. de C.V. which supplied research reagents, but also, they were members of Dr. Mercado lab. Analitek S.A. de C.V. did not influence the results nor design of the protocol. The rest of the authors declare not competing interests.
References
- 1.Bi Q., Wu Y., Mei S. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis. 2020;20:911–919. doi: 10.1016/S1473-3099(20)30287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou Y.J., Okuda K., Edwards C. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronavirus disease (COVID-2019) situation reports 153. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200621-covid-19-sitrep-153.pdf?sfvrsn=c896464d_2
- 4.Jose R., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 2020;8:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang F., Nie J., Wang H. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221:1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo C., Li B., Ma H. Tocilizumab treatment in severe COVID-19 patients attenuates the inflammatory storm incited by monocyte centric immune interactions revealed by single-cell analysis. Preprinted in bioRxiv. 2020 doi: 10.1101/2020.04.08.029769. (Accesed June 21 2020) [DOI] [Google Scholar]
- 8.Wen W., Su W., Tang H. Immune Cell Profiling of COVID-19 Patients in the Recovery Stage by Single-Cell Sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zini G., Bellesi S., Ramundo F. Morphological anomalies of circulating blood cells in COVID-19. Am J Hematol. 2020;95:870–872. doi: 10.1002/ajh.25824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoogendijk A.J., Pourfarzad F., Aarts C.E.M. Dynamic Transcriptome-Proteome Correlation Networks Reveal Human Myeloid Differentiation and Neutrophil-Specific Programming. Cell Rep. 2019;29:2505–2519.e4. doi: 10.1016/j.celrep.2019.10.082. [DOI] [PubMed] [Google Scholar]
- 11.Grassi L., Pourfarzad F., Ullrich S. Dynamics of Transcription Regulation in Human Bone Marrow Myeloid Differentiation to Mature Blood Neutrophils. Cell Rep. 2018;24:2784–2794. doi: 10.1016/j.celrep.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novershtern N., Subramanian A., Lawton L.N. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert C., Preijers F., Yanikkaya Demirel G. Monocytes and macrophages in flow: an ESCCA initiative on advanced analyses of monocyte lineage using flow cytometry. Cytometry B Clin Cytom. 2017;92:180–188. doi: 10.1002/cyto.b.21280. [DOI] [PubMed] [Google Scholar]
- 14.Itelman E., Wasserstrum Y., Segev A. Clinical Characterization of 162 COVID-19 patients in Israel: Preliminary Report from a Large Tertiary Center. Isr Med Assoc J. 2020;22:271–274. [PubMed] [Google Scholar]
- 15.Tang-Huau T.L., Gueguen P., Goudot C. Human in vivo-generated monocyte-derived dendritic cells and macrophages cross-present antigens through a vacuolar pathway. Nat Commun. 2018;9:2570. doi: 10.1038/s41467-018-04985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Laval B., Maurizio J., Kandalla P.K. C/EBPbeta-Dependent Epigenetic Memory Induces Trained Immunity in Hematopoietic Stem Cells. Cell Stem Cell. 2020;26:793. doi: 10.1016/j.stem.2020.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Hadjadj J., Yatim N., Barnabei L. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. Preprinted in medRxiv. 2020 doi: 10.1101/2020.04.19.20068015. (Accesed June 21 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sallard E., Lescure F., Yazdanpanah Y. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng G., Terry J., Belgrader P. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017:14049–14061. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kindler E., Thiel V., Weber F. Interaction of SARS and MERS Coronaviruses with the Antiviral Interferon Response. Adv Virus Res. 2016;96:219–243. doi: 10.1016/bs.aivir.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dosch S.F., Mahajan S.D., Collins A.R. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-kappaB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142:19–27. doi: 10.1016/j.virusres.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol. 2020 doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takizawa H., Boettcher S., Manz M.G. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:2991–3002. doi: 10.1182/blood-2011-12-380113. [DOI] [PubMed] [Google Scholar]
- 24.Vadillo E., Pelayo R. Toll-like receptors in development and function of the hematopoietic system] Rev Invest Clin. 2012;64:461–476. [PubMed] [Google Scholar]
- 25.Sioud M., Floisand Y. TLR agonists induce the differentiation of human bone marrow CD34+ progenitors into CD11c+ CD80/86+ DC capable of inducing a Th1-type response. Eur J Immunol. 2007;37:2834–2846. doi: 10.1002/eji.200737112. [DOI] [PubMed] [Google Scholar]
- 26.Sioud M., Floisand Y., Forfang L. Signaling through toll-like receptor 7/8 induces the differentiation of human bone marrow CD34+ progenitor cells along the myeloid lineage. J Mol Biol. 2006;364:945–954. doi: 10.1016/j.jmb.2006.09.054. [DOI] [PubMed] [Google Scholar]
- 27.De Luca K., Frances-Duvert V., Asensio M.J. The TLR1/2 agonist PAM(3)CSK(4) instructs commitment of human hematopoietic stem cells to a myeloid cell fate. Leukemia. 2009;23:2063–2074. doi: 10.1038/leu.2009.155. [DOI] [PubMed] [Google Scholar]
- 28.Zhang D., Guo R., Lei L. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. Preprinted in medRxiv. 2020 doi: 10.1101/2020.03.24.20042655. (Accessed June 21 2020) [DOI] [Google Scholar]
- 29.Wilk A.J., Rustagi A., Zhao N.Q. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirby M., Weitzman S., Freedman M. Juvenile chronic myelogenous leukemia: differentiation from infantile cytomegalovirus infection. Am J Pediatr Hematol Oncol. 1990;12:292–296. doi: 10.1097/00043426-199023000-00007. [DOI] [PubMed] [Google Scholar]
- 31.Herrod H., Dow L., Sullivan J. Persistent Epstein-Barr virus infection mimicking juvenile myelogenous leukemia: immunologic and hematologic studies. Blood. 1983;61:1098–1104. [PubMed] [Google Scholar]
- 32.Yetgin S., Cetin M., Yenicesu I. Acute parvovirus B19 infection mimicking juvenile myelomonocytic leukemia. Eur J Haematol. 2000;65:276–278. doi: 10.1034/j.1600-0609.2000.065004276.x. [DOI] [PubMed] [Google Scholar]
- 33.Mitra A., Dwyre D., Schivo M. Leukoerythroblastic reaction in a patient with COVID-19 infection. Am J Hematol. 2020;95:999–1000. doi: 10.1002/ajh.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulte-Schrepping J., Reusch N., Paclik D. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamers A.A.J., Dinh H.Q., Thomas G.D. Human Monocyte Heterogeneity as Revealed by High-Dimensional Mass Cytometry. Arterioscler Thromb Vasc Biol. 2019;39:25–36. doi: 10.1161/ATVBAHA.118.311022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruenster M., Kurz A.R., Chung K.J. Extracellular MRP8/14 is a regulator of beta2 integrin-dependent neutrophil slow rolling and adhesion. Nat Commun. 2015;6:6915. doi: 10.1038/ncomms7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnoor M., Alcaide P., Voisin M.B. Crossing the Vascular Wall: Common and Unique Mechanisms Exploited by Different Leukocyte Subsets during Extravasation. Mediators Inflamm. 2015;2015:946509. doi: 10.1155/2015/946509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen G., Wu D., Guo W. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vadillo E., Dorantes-Acosta E., Arriaga-Pizano L. Adult, but not neonatal, human lymphoid progenitors respond to TLR9 ligation by producing functional NK-like cells. Exp Hematol. 2014;42:562–573.e3. doi: 10.1016/j.exphem.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Welner R.S., Pelayo R., Nagai Y. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 2008;112:3753–3761. doi: 10.1182/blood-2008-04-151506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagai Y., Garrett K.P., Ohta S. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mousavi S.A., Rad S., Rostami T. Hematologic predictors of mortality in hospitalized patients with COVID-19: a comparative study. Hematology. 2020;25:383–388. doi: 10.1080/16078454.2020.1833435. [DOI] [PubMed] [Google Scholar]
- 43.Qun S., Wang Y., Chen J. Neutrophil-to-Lymphocyte Ratios Are Closely Associated With the Severity and Course of Non-mild COVID-19. Front Immunol. 2020;11:2160. doi: 10.3389/fimmu.2020.02160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J.B., Xu C., Zhang R.B. Associations of procalcitonin, C-reaction protein and neutrophil-to-lymphocyte ratio with mortality in hospitalized COVID-19 patients in China. Sci Rep. 2020;10:15058. doi: 10.1038/s41598-020-72164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]