CD8+ T reg cells play an important role in the maintenance of self-tolerance and can inhibit the development of autoimmune disease. In this issue of JEM, Mishra et al. reveal that TGF-β signaling and an Eomes-dependent genetic program contribute to CD8 T reg cell differentiation and function.
Abstract
CD8+ T reg cells play an important role in the maintenance of self-tolerance and can inhibit the development of autoimmune disease. In this issue of JEM, Mishra et al. (https://doi.org/10.1084/jem.20200030) reveal that TGF-β signaling and an Eomes-dependent genetic program contribute to CD8 T reg cell differentiation and function.
The central task of the immune system is destruction of invading pathogens while sparing host tissues. Regulatory T (T reg) cells that belong to both major T cell subsets—CD4 and CD8—play essential but distinct protective roles by dampening potential autoimmune reactions against self tissues and maintaining immunological homeostasis. Although the division of the CD4 T cell subset into separate effector and regulatory lineages is well established, separation of the CD8 T cell subset into effector and regulatory arms is the subject of more recent and ongoing research. Experimental definition of the genetic and molecular elements of CD8 T reg cell differentiation and immunological function represents a major goal of contemporary immunology.
Insights from Harvey Cantor and Hye-Jung Kim.
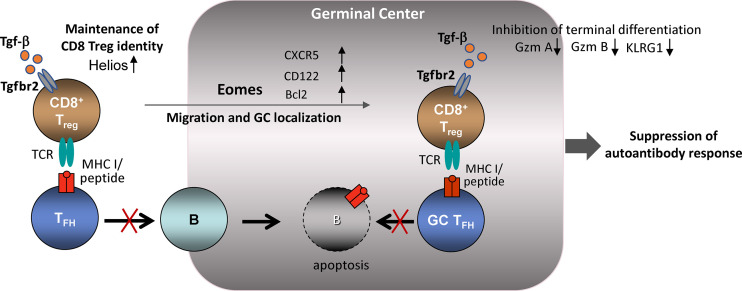
In this issue, Mishra et al. (2020) report that TGF-β signaling and Eomes-dependent genetic programming are essential to the development and maintenance of CD8 T reg cells. Mice deficient in both the TGF-β receptor 2 (Tgfbr2) and the Eomes transcription factor (Tgfbr2−/−Eomes−/−) develop a severe autoimmune phenotype characterized by spontaneous germinal center (GC) formation, increased numbers of T follicular helper cells (TFH cells) and GC B cells, and autoantibody production. Although CD4+ T follicular regulatory cells (TFR cells) can regulate the GC response, the autoimmune phenotype of Tgfbr2−/−Eomes−/− mice does not reflect defective TFR function. Instead, the core pathology of this disorder is a dramatic reduction in the numbers and function of CD8 T reg cells, as judged by tracking of T cells that express the CD44, CD122, and Ly49 surface marker triad as well as the Helios transcription factor (TF; Kim et al., 2015; Kim et al., 2011; Saligrama et al., 2019). These findings are consistent with earlier observations that defective CD8 T reg cell function results in a lupus-like disorder characterized by uncontrolled TFH expansion and autoantibody production (Kim et al., 2010).
Identity and location
Maintenance of the CD8 T reg cell specialized phenotype along with the ability to localize near or within the GC are essential prerequisites for efficient control of the GC response. However, the genetic basis for these properties of CD8 T reg cells has been uncertain. Mishra et al. (2020) show that deletion of Tgfbr2 in T cells (Tgfbr2f/fdLck-cre) results in failed expression of the Helios TF, which has been implicated in their regulatory identity and survival (Kim et al., 2015).
When is the TGF-β signal required for CD8 T reg cell differentiation? Mishra et al. (2020) have examined the effects of TGF-β signaling on CD8 T reg cell differentiation after deletion of Tgfbr2 expression in peripheral T cells. Previous studies of the effects of TGF-β signaling on thymic differentiation using Tgfbr2f/fCD4-Cre mice revealed a sharp reduction of CD44+CD122+Ly49+ CD8 single-positive thymocytes and evidence that the TGF-β signaling pathway may regulate early stages of CD8 T reg cell selection and differentiation (McCarron and Marie, 2014). Possibly, TGF-β–dependent up-regulation of Helios during early maturation of CD8 T reg cells avoids deletion of these autoreactive cells in the thymus (Nakagawa et al., 2018). Mishra et al. (2020) also note that deficient TGF-β signaling impairs Helios expression by CD8 T reg cells but not CD4+ FoxP3+ T reg cells (TFR), suggesting that distinct lineage-specific inducing signals may control Helios expression in the two regulatory cell types. Separate genetic programing of the two T reg cell subsets is consistent with the distinct and complementary roles they play in maintaining self-tolerance and regulating autoantibody responses. Analysis of bone marrow chimeras harboring selective deletions of Helios in either CD4 or CD8 T reg cells has pointed to a nonredundant and perhaps synergistic role of CD4 and CD8 T reg cells in restraining the development of dysregulated GC responses and autoimmune disease (Kim et al., 2015).
Tissue-specific T reg cells often co-opt genes that control the phenotype of their target effector T cells, resulting in easier access and more efficient regulatory interactions. For example, expression of the central TFH transcription factor Bcl-6 by FoxP3+ CD4 T reg cells allows TFR cells to migrate toward GC where they interact with target cells. Mishra et al. (2020) show that Eomes-dependent expression of CXCR5 by CD8 T reg cells allows them to locate into secondary lymphoid follicles, where they may efficiently suppress/target TFH cells. Since Eomes expression also promotes survival and expansion of self-reactive CD8 T cells, perhaps by up-regulation of Bcl-2 (Castro et al., 2011; Miller et al., 2020), the Eomes TF may contribute to both appropriate homing as well as survival of CD8 T reg cells during the GC response. Survival of immigrant CD8 T reg cells within the GC also depends on access to local cytokines as shown for CD4 T reg cell interactions (Liu et al., 2015). Capture of local IL-15 cytokines by CD8 T reg cells may depend on Eomes-dependent expression of CD122 and increased reception of IL-15 signals within the GC microenvironment.
TGF-β signaling and Eomes-dependent maintenance of CD8 T reg cell identity and suppression of GC response. CD8 T reg cells inhibit development of autoimmunity by suppressing TFH cells in the GC microenvironment. TGF-β signaling contributes to maintenance of the CD8 T reg cell phenotype by up-regulating Helios TF expression and maintenance of CD8 T reg cell integrity. The inhibitory interaction between CD8 T reg and TFH cells depends on migration of CD8+ T cells into lymphoid follicles. Eomes expression contributes to both GC localization and survival.
Therapeutic potency
Mishra et al. (2020) observe that a single transfer of a remarkably small number (3 × 104) of CD122hiLy49+CD8 T cells into young (2-mo-old) Tgfbr2−/−Eomes−/− mice prevents the spontaneous formation of GC that develops in older autoimmune mice. Rejuvenation of the GC response of older mice depended on expansion of Ly49+Helios+ donor CD8 T reg cells in adoptive hosts. Successful amelioration of a mouse model of rheumatoid arthritis after transfer of small numbers of CD8 T reg cells lends further support to their potential therapeutic application (Leavenworth et al., 2013). Trials of adoptive T reg cell–based therapy for autoimmune disease have suggested the superiority of antigen-specific vs. polyclonal CD4 T reg cells, possibly reflecting more efficient migration of antigen-specific T reg cells into sites enriched for autoimmune effector cells (Tarbell et al., 2004).
Questions
The Mishra et al. (2020) study provides important new insight into CD8 T reg cell biology, but many gaps in our understanding remain. Although recognition of class I MHC–restricted self-peptides expressed by target cells may contribute to the efficiency of the CD8 T reg cell response, the specificity of this interaction is not well understood. In general, CD8 T reg cells exert more robust suppressive activity against self-reactive TFH cells than non–self-reactive TFH cells. Identification of the nature of peptides expressed by self-reactive TFH cells that may be preferentially recognized by CD8 T reg cells will help clarify this critical issue.
Although several powerful B cell intrinsic mechanisms reduce the likelihood of autoreactive antibody production (Mayer et al., 2020), the robust nature of the GC response apparently requires additional immunological brakes provided by T reg cells. As the authors note, it is surprising that regulation of the GC antibody response may require the combined effort of both CD4 and CD8 T reg cells to prevent pathogenic autoantibody responses. The distinct contributions of the two regulatory subsets to maintenance of self-tolerance may reflect in part their ability to target different aspects of the GC responses. TFR may regulate early activation of B cells before the formation of full-blown GCs (Clement et al., 2019), while subsequent migration of CD8 T reg cells into the GC may suppress TFH cell responses and consequent GC B cell production of autoantibodies. Perhaps the immune system has developed a belt-and-suspenders strategy to fully control this powerful engine of antibody production, diversification, and affinity maturation.
While the suppressive function of Ly49+ CD8 T reg cells has been demonstrated in multiple preclinical settings, the human counterpart of murine CD8 T reg cells has not been fully characterized. Inhibitory killer cell immunoglobulin-like receptors (KIR), which represent the functional analogue of murine Ly49 receptors, are expressed by a small subset of human CD8 T cells that also express a memory phenotype. Here, Mishra et al. (2020) show that KIR3DL1+ Helios+ CD8 T cells in systemic lupus erythematosus peripheral blood are significantly reduced compared with healthy controls. Whether this KIR+ CD8 subset also mediates suppressive activity and dampens lupus pathology awaits further study. However, mounting preclinical evidence for the critical contribution of CD8 T reg cells to the maintenance of self-tolerance and inhibition of autoimmune responses strongly supports their potential therapeutic application. Identification of peptide epitopes that can stimulate human CD8 T reg cells as well as improved approaches to their identification and expansion may allow the development of new therapeutic strategies in patients with autoimmune disease.
References
- Castro, I., et al. 2011. J. Immunol. 10.4049/jimmunol.1003961 [DOI] [Google Scholar]
- Clement, R.L., et al. 2019. Nat. Immunol. 10.1038/s41590-019-0472-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J., et al. 2010. Nature. 10.1038/nature09370 [DOI] [Google Scholar]
- Kim, H.J., et al. 2011. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1018974108 [DOI] [Google Scholar]
- Kim, H.J., et al. 2015. Science. 10.1126/science.aad0616 [DOI] [Google Scholar]
- Leavenworth, J.W., et al. 2013. J. Clin. Invest. 10.1172/JCI66938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., et al. 2015. Nature. 10.1038/nature16169 [DOI] [Google Scholar]
- Mayer, C.T., et al. 2020. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.2015372117 [DOI] [Google Scholar]
- McCarron, M.J., and Marie J.C.. 2014. J. Clin. Invest. 10.1172/JCI76179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, C.H., et al. 2020. Nat. Immunol. 10.1038/s41590-020-0653-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, S., et al. 2020. J. Exp. Med. 10.1084/jem.20200030 [DOI] [Google Scholar]
- Nakagawa, H., et al. 2018. Adv. Immunol. 10.1016/bs.ai.2018.09.001 [DOI] [PubMed] [Google Scholar]
- Saligrama, N., et al. 2019. Nature. 10.1038/s41586-019-1467-x [DOI] [Google Scholar]
- Tarbell, K.V., et al. 2004. J. Exp. Med. 10.1084/jem.20040180 [DOI] [PMC free article] [PubMed] [Google Scholar]