Abstract
Vaccines and antiviral agents are in urgent need to stop the COVID-19 pandemic. To facilitate antiviral screening against SARS-CoV-2 without requirement for high biosafety level facility, we developed a bacterial artificial chromosome (BAC)-vectored replicon of SARS-CoV-2, nCoV-SH01 strain, in which secreted Gaussia luciferase (sGluc) was encoded in viral subgenomic mRNA as a reporter gene. The replicon was devoid of structural genes spike (S), membrane (M), and envelope (E). Upon transfection, the replicon RNA replicated in various cell lines, and was sensitive to interferon alpha (IFN-α), remdesivir, but was resistant to hepatitis C virus inhibitors daclatasvir and sofosbuvir. Replication of the replicon was also sensitive overexpression to zinc-finger antiviral protein (ZAP). We also constructed a four-plasmid in-vitro ligation system that is compatible with the BAC system, which makes it easy to introduce desired mutations into the assembly plasmids for in-vitro ligation. This replicon system would be helpful for performing antiviral screening and dissecting virus-host interactions.
Keywords: SARS-CoV-2, Replicon, Antiviral agents
Highlights
-
•
A BAC-vectored noninfectious replicon of SARS-CoV-2 with secreted Gaussia luciferase as a reporter.
-
•
The replicon was sensitive to IFN, remdesivir and antiviral protein ZAP.
-
•
A four-plasmid in-vitro ligation system that is compatible with this BAC-replicon.
1. Introduction
The pandemic COVID-19 has infected over 26 million people and caused over 800,000 mortalities. (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). It is caused by infection with severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) (Chen et al., 2020; Zhou et al., 2020; Zhu et al., 2020b). Vaccines and antiviral agents are in urgent need to stop the pandemic. Despite great progresses on SARS-CoV-2 vaccine development and clinical trails (Zhu et al., 2020a), the protection efficacy of the vaccines still remains to be determined. There have been trials of antiviral agents such as remdesivir and chloroquine for COVID-19 treatment, however, efficacy of these antiviral agents remains uncertain (Bonovas and Piovani, 2020; Wang et al., 2020b; Wong et al., 2020). Development of convenient tools for antiviral screening will speed up seeking effective antiviral agents against SARS-CoV-2. Recently, infectious clones of SARS-CoV-2 with reporter genes (Thao et al., 2020; Xie et al., 2020) provide elegant tools for antiviral development. However, due to the safety issue and requirement for biosafety level 3 laboratory, usage of these infectious clones is limited. Non-infectious replicon system that recapitulates authentic viral replication without virion production can be used to perform screening for antivirals that target viral replication process.
SARS-CoV-2 contains an approximate 29 kb, single stranded, positive sense RNA genome. About two-thirds of the viral genome encodes open reading frame (ORF) for translation of the replicase and transcriptase proteins, the only ORF translated from the viral genome. The translated replicase and transcriptase proteins engage viral genome to assemble the replicase-transcriptase complex on endoplasmic reticulum membrane, forming a membranous compartment. Within the membranous compartment, replicase-transcriptase complex initiates viral replication and transcription. Transcription of the 3′-most third genome by viral replicase-transcriptase generates various subgenomic mRNAs that encode structural proteins and accessory genes (Masters and Perlman, sixth edition). Structural proteins include the spike (S), membrane (M), envelop (E) proteins and nucleocapsid (N) participate in virion assembly (Masters and Perlman, sixth edition). In this study, we generated a replicon system for SARS-CoV-2, nCoV-SH01 strain with secreted Gaussia luciferase (sGluc) as a reporter gene. The cDNA of viral genome with deletion of S, M, E genes was cloned into a bacterial artificial chromosome (BAC) vector. The reporter gene sGluc was encoded in subgenomic viral RNA. The viral RNA was transcribed in vitro by T7 polymerase. Upon transfection into cells, the viral replication was detected, as evidenced by expression of subgenomic viral RNA-encoded sGluc. The viral replication was sensitive to interferon alpha (IFN-α), remdesivir, but was resistant to hepatitis C virus inhibitors daclatasvir and sofosbuvir. The replicon genome could also be assembled by in-vitro ligation of four DNA fragments and the RNAs generated by the in-vitro ligated DNA template were capable of replication as the RNAs derived from the BAC-template. Thus, we provided a simple SARS-CoV-2 replicon system for antiviral development.
2. Material and methods
2.1. Cloning
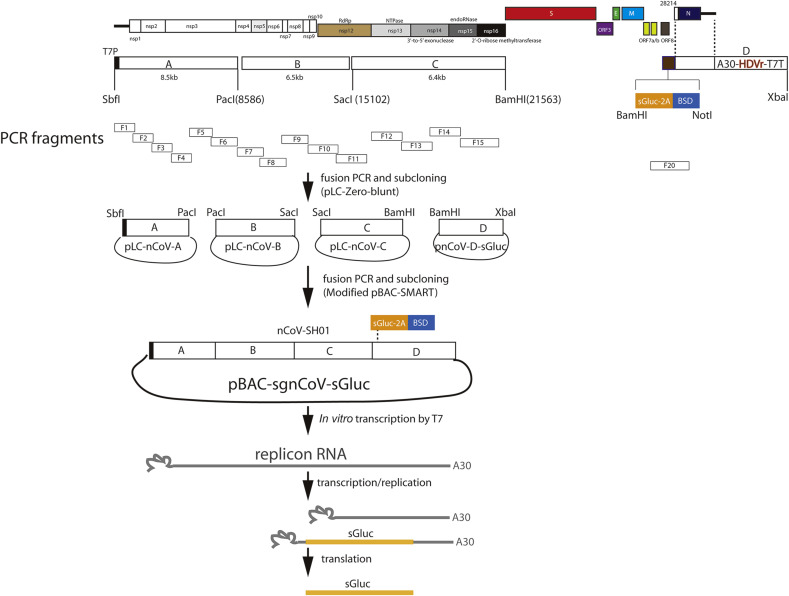
Total RNAs were extracted from SARS-CoV-2 (nCoV-SH01) infected cells (Zhang et al., 2020b), reversely transcribed by superscript IV (Invitrogen) with random primer. Totally 20 fragments with approximate l.5 kb-length encompassing the whole viral genome were amplified with specific primers according to the illumina-sequenced viral genome (MT121215), cloned into a homemade cloning vector pZero-blunt and sequenced. Four larger fragments A (1-8586 nt), B (8587-15102 nt), C (15,103-21562 nt) and D with deletion of structural protein genes and addition of reporter gene cassette were assembled by fusion PCR and subcloning, and then cloned into a homemade cloning vector pLC-Zero-blunt and pcDNA3.1 (Invitrogen), respectively, resulted in plasmids pLC-nCoV-A, pLC-nCoV-B, pLC-nCoV-C, and pnCoV-D-sGluc. To facilitate cloning, a BamHI site was introduced downstream the genome position of 21,562 (nt) in the plasmid pLC-nCoV-C. In fragment A, T7 promoter was added before the 5′ viral genome. In fragment D, an expression cassette containing secreted Gaussia luciferase (sGluc), foot-and-mouth disease virus (FMDV) 2 A peptide (NFDLL KLAGD VESNP GP) and blasticidin (BSD) was added upstream the 5′-postion of viral genome. The 3′ viral genome was flanked with polyA30, hepatitis delta virus ribozymes (HDVr) and terminator sequence for T7 polymerase (T7T). Inactive mutants (759-SAA-761) of the RNA dependent RNA polymerase nsp12 was introduced into the C fragment at the predicted catalytic residues (759-SDD-761) (Gao et al., 2020) by fusion PCR mediated mutagenesis.
To assemble the four fragments into bacterial artificial chromosome (BAC) vector, first we modified the pSMART-BAC v2.0 (Lucigen) to get ride of unwanted restriction enzyme sites and added AatII and XhoI sites to facilitate cloning by multiple rounds of fusion-PCR mediated mutagenesis. The fragments were then sequentially cloned into the BAC vector. We first assemble the fragment A and B, C and D by enzyme digestion to get the plasmid pLC-nCoV-AB and pLC-nCoV-CD, respectively. Then the AB fragments were cloned into the SbfI/XhoI site to generate pBAC-sgnCoV-AB. Then the CD fragments were ligated into the SacI/AsisI site to get pBAC-sgnCoV-sGluc. BAC plasmid was delivered into BAC-Optimized Replicator v2.0 Electrocompetent Cells (Lucigen) by electroporation and bacteria was propagated according to the manufacturer's guide. Colonies were picked and cultured in LB medium containing 12.5 μg/ml chloramphenicol. L-arabinose was added to cultures when the OD600 reaches 0.2–0.3 to increase the plasmid copy numbers.
For assembly of SARS-CoV-2 replicon by in-vitro ligation, we first got rid of the BsaI site on fragment C by fusion-PCR mediated synonymous mutagenesis. The BsaI sites (5′-GGTCTCN↓NNNN-3′) were added into the 5′ and 3′ of the fragment A, B, C and D in the plasmids of pLC-nCoV-A, pLC-nCoV-B, pLC-nCoV-C, and pnCoV-D-sGluc by fusion PCR-mediated cloning, resulted in the plasmids pLC-nCoV-A-BsaI, pLC-nCoV-B-BsaI, pLC-nCoV-C-BsaI, and pnCoV-D-sGluc-BsaI. The plasmids retained all the original enzymatic sites, for convenience to swap into the BAC vector if desired. The fragments were released from the plasmids by BsaI digestion, after gel purification and ligated by T4 ligase.
To construct lentiviral vector expression plasmids, sequences encoding the GFP, SARS-CoV-2 nucleocapsid protein (N) were cloned into the XbaI/BsrGI site of pTRIP-IRES-BSD. Sequence encoding the long isoform of Zinc-finger antiviral protein (ZAPL) were synthesized by Wuxi Qinglan Biotech (Wuxi, China) and cloned into the XbaI/BamHI site of pTRIP-IRES-BSD. An HA tag was added into the N-terminal of ZAP. For production of N mRNA, sequence encoding N was first cloned into the KpnI/BamHI sites of phCMV to get the plasmid phCMV-N. All the plasmids were verified by Sanger sequencing. The detail information was available upon request.
2.2. Cell lines
The human hepatoma cells Huh 7, baby hamster kidney cells BHK-21, African green monkey kidney cells Vero E6, human embryonic kidney cells HEK293T were purchased from the Cell Bank of the Chinese Academy of Sciences (www.cellbank.org.cn) and routinely maintained in Dulbecco's modified medium supplemented with 10% FBS (Gibco) and 25 mM HEPES (Gibco). Huh 7.5 cells (Kindly provided by C. Rice) were routinely maintained in a similar medium supplemented with non-essential amino acids (Gibco). Huh7-GFP, Huh7-N, Huh7-ZAPL cell line were routinely maintained in the medium supplemented with 0.5 μg/ml blasticidin. All cells were tested negative for mycoplasma.
2.3. Lentivirus pseudoparticle
VSV-G-pseudotyped lentiviral particles were prepared by co-transfection of VSV-G, HIV gag-pol and lentiviral provirus plasmids into HEK293T cells. The medium overlying the cells was harvested at 48 h after transfection, filtered through a 0.45-μm filter, and stored at -80°C. Cells were transduced with the pseudoparticles in the presence of 8 μg/ml Polybrene.
2.4. Inhibitors
Remdesivir (GS-5734), Daclatasvir (S1482), Sofosbuvir (GS-7977) were purchased from Selleckchem (America), 2′-C-Methylcytidine (HY-10468) was purchased from MedChem Express (America), IFN-α (11,200–2) was purchased from PBL (America).
2.5. Antibodies
Anti-β-actin antibody (Sigma; A1978) was used at 1:5000 dilution; Anti-HA antibody (CST; 37,243) was used at 1:000 dilution; Anti-GFP antibody (Santa Cruz; sc-9996) was used at 1:1000 dilution; Anti-N antibody (GeneTex; GTX632269) was used at 1:500 dilution; Goat-anti-mouse IRDye 800 C W secondary antibody (licor; 926–32210) was used at 1:10,000 dilution. Goat-anti-rabbit IRDye 800 C W secondary antibody (licor; 926–32211) was used at 1:10,000 dilution in western blotting.
2.6. Western blotting
After washing with PBS, cells were lysed with 2 × SDS loading buffer (100 mM Tris-Cl [pH 6.8], 4% SDS, 0.2% bromophenol blue, 20% glycerol, 10% 2-mercaptoethanol) and then boiled for 5 min. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were incubated with blocking buffer (PBS, 5% milk, 0.05% Tween) for 1 h and then with primary antibody diluted in the blocking buffer. After three washes with PBST (PBS, 0.05% Tween), the membranes were incubated with secondary antibody. After three washes with PBST, the membrane was visualized by Odyssey CLx Imaging System.
2.7. In-vitro ligation
BsaI digested fragment were gel purified by using Gel Extraction Kit (OMEGA) and ligated with T4 ligase (New England Biolabs) at room temperature for 1 h. The ligation products were phenol/chloroform extracted, precipitated by absolute ethanol, and resuspended in nuclease-free water, quantified by determining the A260 absorbance.
2.8. In-vitro transcription
BAC-based sgnCoV-sGluc plasmids or purified in-vitro ligated products were used as templates for the in-vitro transcription by mMESSAGE mMACHINE T7 Transcription Kit (Ambion) according to the manufacturer's protocol. For N mRNA production, we amplified the N coding region by PCR (sense: GGC ACA CCC CTT TGG CTC T; antisense: TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TCT AGG CCT GAG TTG AGT CAG CAC) with phCMV-N as template. Then the purified PCR product was used as template for in-vitro transcription by mMESSAGE mMACHINE T7 Transcription Kit as described above. RNA was purified by RNeasy mini Elute (Qiagen) and eluted in nuclease-free water, quantified by determining the A260 absorbance.
2.9. Transfection
Cells were seeding onto 48-well plates at a density of 7.5 × 104 per well and then transfected with 0.3 μg in-vitro transcribed RNA using a TransIT-mRNA transfection kit (Mirus) according to the manufacturer's protocol.
2.10. Luciferase activity
Supernatants were taken from cell medium and mixed with equal volume of 2 × lysis buffer (Promega). Luciferase activity was measured with Renilla luciferase substrate (Promega) according to the manufacturer's protocol.
3. Results
3.1. Construction of a bacterial artificial chromosome (BAC) based SARS-CoV-2 replicon
Total RNAs were extracted from SARS-CoV-2 (nCoV-SH01) infected cells (Zhang et al., 2020b), and then reversely transcribed by superscript IV with random primer. Totally, 20 fragments with approximate l.5 kb-length encompassing the whole viral genome were amplified with specific primers according to the illumina-sequenced viral genome (MT121215), cloned and sequenced. The fragments were then assembled by fusion PCR and subcloning into larger fragments A (1-8586 nt), B (8587-15102 nt), C (15,103-21562 nt) and D, and then cloned into a homemade cloning vector pLC-Zero-blunt (Fig. 1 ). Took a similar strategy for construction of SARS-CoV replicon (Ge et al., 2007), we deleted the structural protein genes and retained the N gene and essential promoter regions. We replaced the S gene region with a reporter gene cassette, including secreted Gaussia luciferase (sGluc), foot-and-mouth disease virus (FMDV) 2 A peptide and blasticidin (BSD), whose expression was driven by the promoter of S gene in the subgenomic mRNA (Fig. 1). To facilitate cloning, a BamHI site was introduced downstream the genome position of 21,562 (nt) in the pLC-nCoV-C plasmid. T7 promoter was added before the 5′ viral genome in the fragment A for in-vitro transcription with T7 polymerse. The 3′ viral genome was flanked with polyA30, hepatitis delta virus ribozymes (HDVr) and terminator sequence for T7 polymerase (T7T) (Fig. 1), which facilitates in-vitro transcription without linearization and production of precise polyadenylated viral RNA. The fragments of A, B, C and D were assembled further into AB and CD in pLC-Zero-blunt and then sequentially cloned into a modified BAC vector to get the final plasmid pBAC-sgnCoV-sGluc (Fig. 1).
Fig. 1.
Schematic of construction of BAC-based replicon of SARS-CoV-2.
Twenty fragments encompassing the whole viral genomes were amplified, cloned and sequenced. Four larger fragments A (1-8586 nt), B (8587-15102 nt), C (15,103-21562 nt) and D with deletion of structural protein genes and addition of reporter gene cassette were assembled and cloned. To facilitate cloning, a BamHI site (in bold) was introduced downstream the genome position of 21,562 (nt). In fragment A, T7 promoter (T7P) was added. In fragment D, an expression cassette containing secreted Gaussia luciferase (sGluc), foot-and-mouth disease virus (FMDV) 2 A peptide and blasticidin (BSD) was added. The 3′ viral genome was flanked with polyA30, hepatitis delta virus ribozymes (HDVr) and terminator sequence for T7 polymerase (T7T). Then the fragments were assembled and sequentially cloned into a modified BAC plasmid. Upon transfection into cells, the replicon RNA can be used as template for RNA replication or transcription to produce subgenomic RNA. The sGluc subgenomic RNA is translated to produce sGluc.
3.2. Replication of SARS-CoV-2 replicon in cells
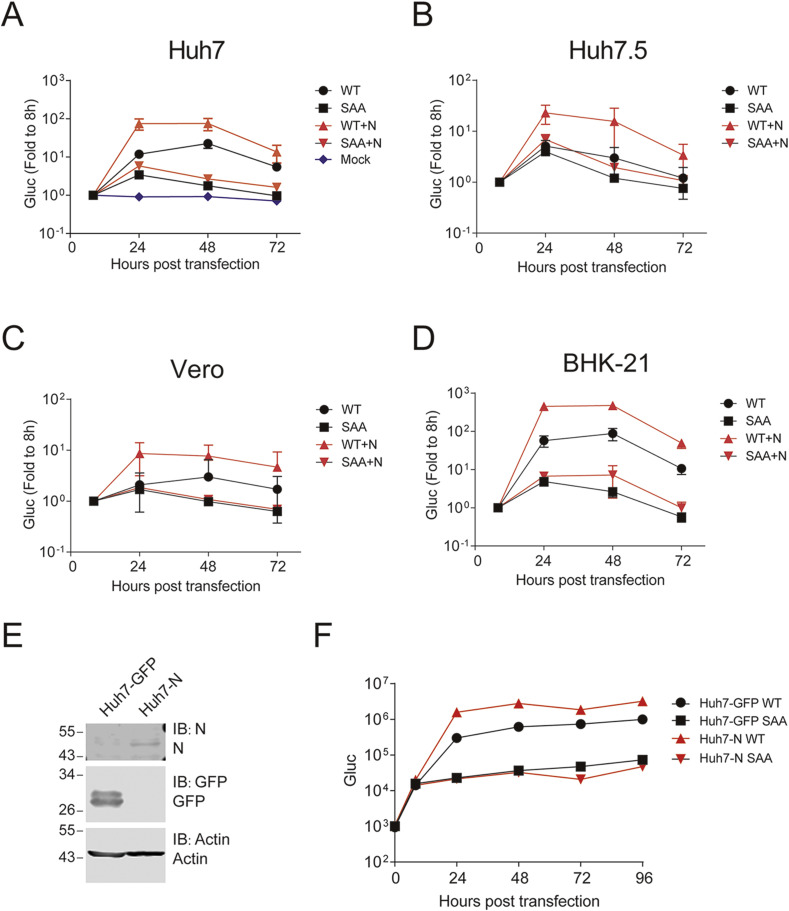
The plasmid pBAC-sgnCoV-sGluc was used directly as template for in-vitro transcription to produce 5′-capped replicon RNA. Replicon RNA was then co-transfected with N mRNA into various cell lines. RNA replication was monitored by measuring the secreted Gaussia luciferase activity in the supernatants. Enzymatic dead mutants (759-SAA-761) of the RNA dependent RNA polymerase nsp12 (Gao et al., 2020) were introduced and the mutated replicon served as a non-replication control. As expected, SAA RNA did not replicate, without increase of luciferase activity in the transfected Huh7, Huh7.5, Vero and BHK-21 cells. In contrast, transfection of wild type (WT) replicon RNA resulted in obvious increase of luciferase activity (Fig. 2 A–D), indicating active viral replication. Huh7.5 cell is a subclone of Huh7 cell with deficiency in RIG-I and MDA-5 signaling (Du et al., 2016; Stumper et al., 2005). Vero cell is routinely used for SARS-CoV-2 isolation. Notably, replicon replication was less efficient in Huh7.5 and Vero cells (Fig. 2B and C) whereas robust in BHK-21 cells (Fig. 2D). In consistent with previous studies, co-transfection of N mRNA enhanced viral replication (Fig. 2A–D), which is probably due to the suppression of innate immune response (Ye et al., 2007). For convenience, we tried to establish an N-expressed cell line (Fig. 2E). Compared with GFP-expressed cells, Huh7 cells expressing N supported more robust viral replication (Fig. 2F).
Fig. 2.
Replication of sgnCoV-sGluc in different cells.
(A–
D) Huh7, Huh7.5, Vero and BHK-21
cells were transfected with in-vitro transcribed replicon RNA (WT) or the nsp12 polymerase active-site mutant (SAA). An mRNA encoding the SARS-CoV-2 N protein was co-transfected or not. The luciferase activity in the supernatants was measured at the time points indicated. Medium was changed at all time point. A mock-transfection group in Fig 2A showed the background signals. Data are shown as mean ± SD (n = 3). (E–F) Replication of replicon RNA in Huh7 cells overexpressed with N protein. (E) Huh7 cells overexpressed with GFP protein or N protein were analyzed by Western blotting with the indicated antibodies. (F) Huh7-GFP and Huh7-N cells were transfected with replicon RNA (WT or SAA). The luciferase activity in the supernatants was measured at the time points indicated. Medium was changed at 8 h post transfection. Data are shown as mean ± SD (n = 3).
3.3. Sensitivity of the SARS-CoV-2 replicon to antiviral agents
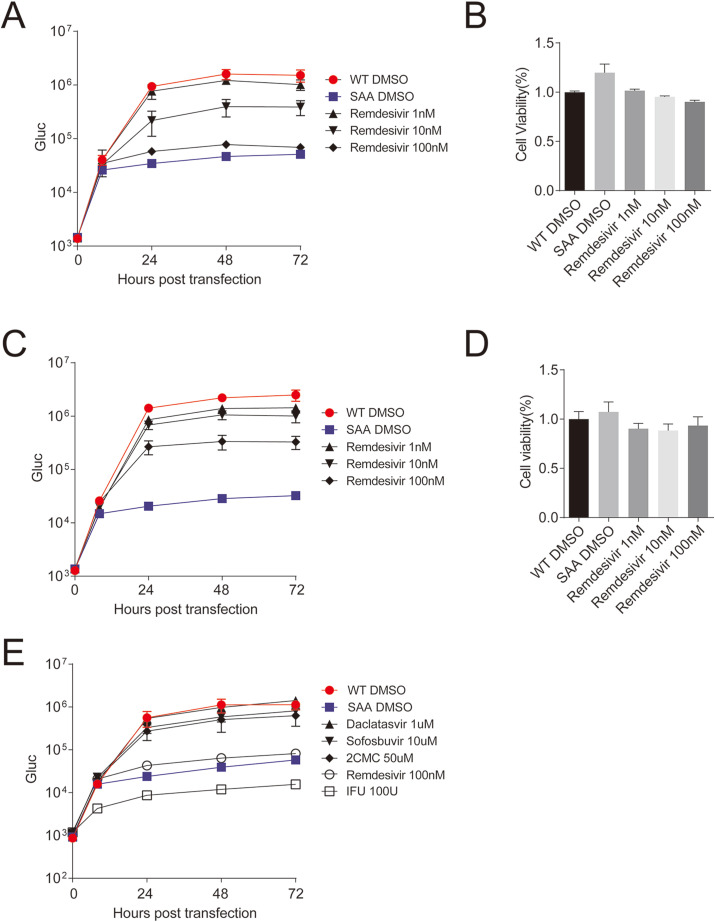
We tested the sensitivity of SARS-CoV-2 replicon to remdesivir, which has been demonstrated to inhibit SARS-CoV-2 viral infection (Wang et al., 2020a). Huh7 cells were first treated with remdesivir at various concentrations (100 nM, 10 nM, 1 nM) for 4 h, and then the cells were co-transfected with replicon RNA (WT or SAA) and N mRNA. The luciferase activity in the supernatants was measured at various time points after transfection. We found that remdesivir showed a dose-dependent effect on the replicon. Remdesivir effectively inhibited replicon replication to a similar level as SAA at a concentration of 100 nM, and hardly reduced luciferase expression at 1 nM (Fig. 3 A). All the concentrations of remdesivir had no obvious effect on cell viability (Fig. 3B). We also examined if the remdesivir inhibited established viral RNA replication. We first transfected replicon RNA, and then added remdesivir 8 h post transfection and monitored viral replication at various time points post treatment. Under this condition, remdesivir also reduced whereas did not completely block viral replication in a dose-dependent manner without obvious cytotoxicity (Fig. 3C–D). Then we tested the sensitivity of SARS-CoV-2 replicon to other antiviral agents. Huh7 cells were first treated with interferon ahpha (IFN-α) (100U/ml), remdesivir (10 nM), daclatasvir (1 μM), sofosbuvir (10 μM) and 2′-C-Methylcytidine (2CMC) (50 μM) for 4 h, then the cells were co-transfected with replicon RNA (WT or SAA) and N mRNA. The luciferase activity in the supernatants was measured at the various time points after transfection. IFN-α and remdesivir have been demonstrated to inhibit SARS-CoV-2 viral infection (Li et al., 2020; Wang et al., 2020a). Daclatasvir and sofosbuvir are direct antivirals targeting hepatitis C virus NS5A (Gao et al., 2010) and RNA dependent RNA polymerase (Kohli et al., 2014), respectively. 2′-C-Methylcytidine is a nucleoside inhibitor of HCV NS5B polymerase (Mathy et al., 2008). As shown in Fig. 3E, IFN-α and remdesivir effectively inhibited sgnCoV-sGluc replication. Notably, IFN-α started to reduce the reporter gene expression at early time point (8 h post transfection), manifested as lower luciferase activity than the SAA mutant, which suggests that IFN-α may block translation of the viral subgenomic mRNA. Remdesivir effectively inhibited the luciferase expression to a similar level of SAA. In contrast, sofosbuvir and 2′-C-Methylcytidine hardly reduced luciferase expression and daclatasvir had no effect on luciferase expression (Fig. 3E). These results demonstrate that SARS-CoV-2 replicon is sensitive to antiviral agents against SARS-CoV-2.
Fig. 3.
Sensitivity of the SARS-CoV-2 replicon to antiviral agents.
(A) Huh7 cells were treated with remdesivir as the indicated concentrations. Four hours later, cells were co-transfected with replicon RNA (WT or SAA) and N mRNA. The luciferase activity in the supernatants was measured at the time points indicated. Medium was changed at 8 h
post transfection. (B) The cell viabilities in (A) were measured at 72 h
post transfection using the CCK8 kit. (C) Huh7 cells were co-transfected with replicon RNA (WT or SAA) and N mRNA. Eight hours later, medium was changed with remdesivir as the indicated concentration. The luciferase activity in the supernatants was measured at the time points indicated. Medium was changed at 8 h
post transfection. (D) The cell viabilities in (C) were measured at 72 h
post transfection using the CCK8 kit. (E) Huh7 cells were treated with remdesivir (100
nM), IFN-α (100 U/ml), daclatasvir (1 μM), sofosbuvir (10 μM), 2CMC (50 μM). Four hours later, cells were co-transfected with replicon RNA (WT or SAA) and N mRNA. The luciferase activity in the supernatants was measured at the time points indicated. Medium was changed at 8 h post transfection. Data are shown as mean ± SD (n = 3).
3.4. Sensitivity of SARS-CoV-2 replicon to overexpression with zinc-finger antiviral protein (ZAP)
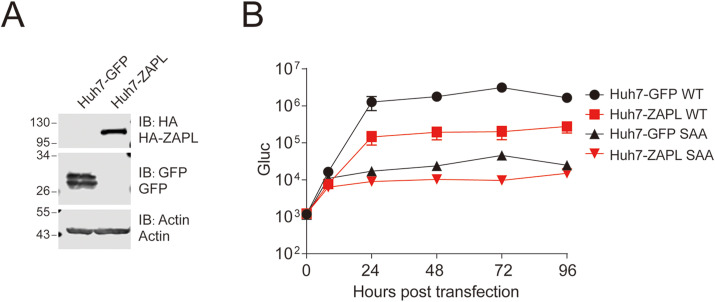
Zinc-finger antiviral protein recognizes CpG dinucleotide on non-self RNA and exerts antiviral activity (Takata et al., 2017). There is extreme low CpG content of SARS-CoV-2 genome, suggesting SARS-CoV-2 may evolve under the pressure of ZAP (Xia, 2020; Zhang et al., 2020a). We generated a stable Huh7 cell line expressing the long isoform of ZAP (ZAPL) and examined the replicon RNA replication in the Huh7-ZAPL cells (Fig. 4 A). There was about 10-fold reduction of replicon replication in Huh7-ZAPL comparing with that in GFP expressing cells (Huh7-GFP) (Fig. 4B), suggesting replicon replication is sensitive to ZAPL overexpression.
Fig. 4.
Sensitivity of SARS-CoV-2 replicon to overexpression with Zinc-finger antiviral protein (ZAP).
(A) Huh7 cells overexpressed with ZAPL protein or GFP protein were analyzed by Western blotting with the indicated antibodies. (B) Huh7-GFP and Huh7-ZAPL cells were co-transfected with replicon RNA (WT or SAA) and N mRNA. The luciferase activity in the supernatants was measured at the time points indicated. Medium was changed at 8 h
post transfection. Data are shown as mean ± SD (n = 3).
3.5. Assembly of SARS-CoV-2 replicon by in-vitro ligation
As the difficulties to manipulate with BAC vectors, we tried to assemble the four fragments A, B, C and D by in-vitro ligation. We introduced additional BsaI sites into the 5′ and 3′ of each fragment in the assembly plasmids pLC-nCoV-A, pLC-nCoV-B, pLC-nCoV-C, pnCoV-D-sGluc, retaining all the original restrictions enzymes. The fragments were released from the plasmid by BsaI digestion, and assembled by in-vitro ligation with T4 ligase (Fig. 5 A). The RNA species transcribed from the BAC and the in-vitro ligated templates were analyzed by non-denaturing agarose gel electrophoresis. RNAs transcribed from BAC template migrated as a sole band with similar size to a 2.5 kb DNA maker (Fig. 5B, arrow). RNAs transcribed from the in-vitro ligated template migrated as two bands. The larger one was similar as the RNAs from the BAC-derived RNA and a smaller one (Fig. 5B, asterisk) might be the transcript of incomplete in-vitro ligated template. RNAs transcribed from the in-vitro ligated template replicated similar as the RNAs transcribed from BAC vector upon transfection (Fig. 5C).
Fig. 5.
Assembly of SARS-CoV-2 replicon by in-vitro ligation.
(A) Schematic of the in-vitro ligation system for SARS-CoV-2 replicon. (B) Gel analysis of RNA transcripts. About 2 μg of in-vitro transcribed RNAs were analyzed on a 0.7% native agarose gel. DNA maker are indicated. Because this is a native agarose gel, the DNA size is not directly corelated to the RNA size. Arrows indicates the replicon RNA transcript. Asterisk shows the shorter RNA transcript. (C) Huh7 cells were co-transfected with replicon RNA (WT or SAA) and N mRNA generated by BAC-based system or in-vitro ligation system. The luciferase activity in the supernatants was measured at the time points indicated. Medium was changed at 8 h post transfection. Data are shown as mean ± SD (n = 3).
4. Discussion
In this study, we described a replicon system of SARS-CoV-2. In the replicon, we deleted the spike (S), membrane (M), envelop (E) genes that are essential for virion production, making it non-infectious and safe (Fig. 1). Upon transfection into various cells, the replicon RNA could replicate, manifested by the expression of subgenomic mRNA encoded sGluc (Fig. 2). The viral replication was inhibited by anti-SARS-CoV-2 antiviral agent remdesivir and by IFN-α but was not by antivirals against hepatitis C virus (Fig. 3). This replicon system avoids requirement for specific biosafety facilities. The BAC-vectored replicon presents several advantages, such as the high stability of exogenous sequences, and unlimited production of the transcribed mRNA. The BAC-vectored replicon system described here does not need in-vitro ligation, recombination in yeast or linearization before in-vitro transcription, which simplifies the experiment processes. Thus this replicon system would be used conveniently to perform antiviral screening against SARS-CoV-2. We also constructed a four-plasmid in-vitro ligation system that is compatible with the BAC system. Replicon RNAs produced from the in-vitro ligated replicate similarly with the RNAs transcribed from BAC plasmids (Fig. 5). The in-vitro ligation system is simple and straightforward, allows the rapid construction of replicon cDNA compared to the conventional techniques. It is easy to introduce desired mutations into the assembly plasmids for in-vitro ligation, which make it suitable for dissecting the effect of emerging mutations on viral replication and molecular mechanisms of viral replication.
Author contributions
Conceived the study: Z Yi; conducted the study: Y Zhang, W Song, S Chen, Z Yi; Data analysis: Z Yi, Y Zhang; Manuscript draft: Y Zhang, Z Yi; Resources: Z Yuan, Z Yi.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported in part by the National Science and Technology Major Project of China (2017ZX10103009), Key Emergency Project of Shanghai Science and Technology Committee (20411950103). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
The abbreviations used are
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- ORF
open reading frames
- BAC
bacterial artificial chromosome
- sGluc
secreted Gaussia luciferase
- WT
wild type
- SAA
enzymatic dead mutants of nsp12
References
- Bonovas S., Piovani D. Compassionate use of remdesivir in covid-19. N. Engl. J. Med. 2020;382 doi: 10.1056/NEJMc2015312. [DOI] [PubMed] [Google Scholar]
- Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W., Sun Z., Liu F., Wu K., Zhong B., Mei Y., Zhang W., Chen Y., Li Y., Shi M., Lan K., Liu Y. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microb. Infect. 2020;9:313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X.T., Pan T.T., Xu J., Zhang Y., Song W.H., Yi Z.G., Yuan Z.H. Hepatitis C virus replicative double-stranded RNA is a potent interferon inducer that triggers interferon production through MDA5. J. Gen. Virol. 2016;97:2868–2882. doi: 10.1099/jgv.0.000607. [DOI] [PubMed] [Google Scholar]
- Gao M., Nettles R.E., Belema M., Snyder L.B., Nguyen V.N., Fridell R.A., Serrano-Wu M.H., Langley D.R., Sun J.H., O'Boyle D.R., 2nd, Lemm J.A., Wang C., Knipe J.O., Chien C., Colonno R.J., Grasela D.M., Meanwell N.A., Hamann L.G. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge F., Luo Y., Liew P.X., Hung E. Derivation of a novel SARS-coronavirus replicon cell line and its application for anti-SARS drug screening. Virology. 2007;360:150–158. doi: 10.1016/j.virol.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A., Shaffer A., Sherman A., Kottilil S. Treatment of hepatitis C: a systematic review. Jama. 2014;312:631–640. doi: 10.1001/jama.2014.7085. [DOI] [PubMed] [Google Scholar]
- Li L., Wang X., Wang R., Hu Y., Jiang S., Lu X. Antiviral agent therapy optimization in special populations of COVID-19 patients. Drug Des. Dev. Ther. 2020;14:3001–3013. doi: 10.2147/DDDT.S259058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, P.S., Perlman, S., sixth ed.. Coronaviridae. Fields Virology, 825-858.
- Mathy J.E., Ma S., Compton T., Lin K. Combinations of cyclophilin inhibitor NIM811 with hepatitis C Virus NS3-4A Protease or NS5B polymerase inhibitors enhance antiviral activity and suppress the emergence of resistance. Antimicrob. Agents Chemother. 2008;52:3267–3275. doi: 10.1128/AAC.00498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumper R., Loo Y.M., Foy E., Li K., Yoneyama M., Fujita T., Lemon S.M., Gale M. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M.A., Goncalves-Carneiro D., Zang T.M., Soll S.J., York A., Blanco-Melo D., Bieniasz P.D. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature. 2017;550:124–127. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao T.T.N., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., Gultom M., Schmied K., Laloli L., Husser L., Wider M., Pfaender S., Hirt D., Cippa V., Crespo-Pomar S., Schroder S., Muth D., Niemeyer D., Corman V.M., Muller M.A., Drosten C., Dijkman R., Jores J., Thiel V. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582(7813):561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.K., Yang J., He Y. Caution and clarity required in the use of chloroquine for COVID-19. Lancet Rheumatol. 2020;2:e255. doi: 10.1016/S2665-9913(20)30093-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol. Biol. Evol. 2020;37:2699–2705. doi: 10.1093/molbev/msaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., Plante K.S., Weaver S.C., Makino S., LeDuc J.W., Menachery V.D., Shi P.Y. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848. doi: 10.1016/j.chom.2020.04.004. e843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Hauns K., Langland J.O., Jacobs B.L., Hogue B.G. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 2007;81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kang J., Liu M., Han B., Li L., He Y., Yi Z., Chen L. bioRxiv; 2020. Multi-site Co-mutations and 5’UTR CpG Immunity Escape Drive the Evolution of SARS-CoV-2. 2020.2007.2021.213405. [Google Scholar]
- Zhang R., Yi Z., Wang Y., Teng Z., Xu W., Song W., Cai X., Sun Z., Gu C., Zhou Y., Chen H., Ye Y., Han W., Zhu Y., Feng X., Fang F., Li C., Zhang X., Qu D., Fu C., Xie H., Yuan Z. Isolation of a 2019 novel coronavirus strain from a coronavirus disease 19 patient in Shanghai. Journal of Microbes and Infection. 2020;15:15–20. [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.C., Li Y.H., Guan X.H., Hou L.H., Wang W.J., Li J.X., Wu S.P., Wang B.S., Wang Z., Wang L., Jia S.Y., Jiang H.D., Wang L., Jiang T., Hu Y., Gou J.B., Xu S.B., Xu J.J., Wang X.W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]