Abstract
Many viruses, including bacteriophage, have the inherent ability to utilize several types of proteinaceous receptors as an attachment mechanism to infect cells, yet the molecular mechanisms that drive receptor binding have not been elucidated. Using bacteriophage Sf6 and its host, Shigella flexneri, we investigated how Sf6 utilizes outer membrane protein A (OmpA) for infection. Specifically, we identified that surface loops of OmpA mediate Shigella infection. We further characterized which residues in the surface loops are responsible for Sf6 binding and productive infection using a combination of in vivo and in vitro approaches including site-directed mutagenesis, phage plaque assays, circular dichroism spectroscopy, and in vitro genome ejection assays. Our data indicate that Sf6 can productively interact with other bacterial OmpAs as long as they share homology in loops 2 and 4, suggesting that these loops may determine host specificity. Our data provide a model in which Sf6 interacts with OmpA using the surface of the protein and new insights into viral attachment through binding to membrane protein receptors.
Keywords: outer membrane proteins, virus receptor, plaque assay, circular dichroism, phage attachment
Introduction
Viruses infect every domain of life. For a successful infection, all known viruses must transfer their genomic information into their hosts [1] and can employ different strategies to accomplish this. One common strategy is to utilize a portion of the respective host cell as a receptor, at a site suitable for entry. Understanding the binding events that occur between the host cell and these viruses is critical in order to develop methods to circumvent infection.
Although viruses can have extremely diverse life cycles, there are several commonalities. For example, many archaeal, eukaryotic, and bacterial viruses require proteinaceous receptors on the host surface used for attachment [2–12]. In addition, many of these viruses demonstrate plasticity in their binding mechanisms [3,13]. Throughout the evolutionary arms race, hosts can develop resistance to viral infection and viruses face extinction if they can no longer gain entry into their continually evolving hosts. The innate ability to utilize more than one type of receptor and the ability to evolve easily to utilize novel receptors can allow viruses to circumvent host resistance and may be essential for the continued pathogenicity of a given virus.
Work with eukaryotic viruses has shed some light on multiple receptor usage. Studies with herpes simplex virus (HSV) have revealed that different HSV serotypes encode distinct glycoproteins that are required for attachment [14]. HSV, additionally, has the ability to utilize different cell proteins as receptors, thus allowing it to infect a broader range of host cells [10,13]. The human immunodeficiency virus has also evolved to use its single envelope glycoprotein to gain entry into different cell types [13]. Even in the absence of its primary receptor, CD4, some human immunodeficiency virus isolates are still able to infect cells [15], and studies have shown that Fusin/CXCR4 can serve as an alternative cell receptor [16]. Moreover, iin the case of Adenovirus, not only is the native virus able to utilize αvβ3 and αvβ5 integrins [3] in the absence of its preferred receptor, CAR [4], but also the fiber head domains of the virus have cell-type selective properties [17].
In addition to the inherent capability for binding multiple receptors, viruses can also gain access to different receptor types as clearly demonstrated with studies of bacteriophage. Various Escherichia coli outer membrane proteins (Omps) function as receptors for many bacteriophages. For example, bacteriophage T2 has the ability to use two different Omps as receptors: OmpF and FadL [18,19]. Moreover, bacteriophage Ox2 [20] can evolve to utilize OmpA, OmpC, and OmpF as receptors [21]. Studies with Ox2 have shown that the phage tail fibers/adhesins are a major determinant for the Omp specificity. Under selective pressure in the laboratory, phage λ evolved to infect its host E. coli through a novel pathway; rather than using its preferred receptor, LamB, λ acquired several mutations in its recognition protein J that allowed infection through a novel receptor, OmpF [22]. Combined, these data illustrate that viruses can evolve receptor plasticity as a strategy to circumvent host resistance and it implies that receptor plasticity is an inherent trait of viral evolution.
Viruses, although extremely diverse in their morphogenetic pathways, generally use only a handful of common protein folds to form infectious virions. For example, for dsDNA (double-stranded DNA) phage and HSV, the major capsid [23], scaffolding [24], and portal [25,26] proteins are conserved. This high level of structural homology makes it possible to utilize model systems to study general strategies for viral infection. The model system chosen for this study is bacteriophage Sf6 and its host, Shigella flexneri. Sf6 is a short-tailed dsDNA virus that belongs the subgroup of the “P22-like” phages in Podoviridae [27], which is one of the less well understood families in regard to phage–host interaction [28]. Sf6 infection requires binding to both primary and secondary receptors [29]. Lipopolysaccharide (LPS) serves as primary receptor in an initial and reversible interaction [30], followed by an irreversible interaction, with a secondary receptor, which is an Omp [29]. Our previous work demonstrated that OmpA is the preferred secondary receptor for Sf6, yet OmpC can serve as an alternate [29].
Many phages such as Sf6 are able to use more than one type of proteinaceous receptor for attachment, and they generally appear to have a preferred receptor. Since many porins (OmpA, OmpF, OmpC, FhuA, and LamB) have been identified as bacteriophage receptors [5–8,20–22,29], and these have homologous structures [31], we can predict that analogous regions within these Omps might be globally important for phage infection. However, there are few published studies that delineate molecular mechanisms governing phage attachment to these receptors and none to date involving a member of the P22-like phages. In the present study, we identified via site-directed mutagenesis coupled with in vivo phage biology and biochemical assays specific residues of OmpA that are critical to mediate Sf6 infection and confer host range. Our data provide new insights into Podoviridae attachment through binding to protein receptors.
Results
S. flexneri OmpA extracellular loops are important for Sf6 infection
E. coli OmpA is a receptor for several bacteriophages. Analysis of E. coli strains isolated after developing resistance to over 15 different strains of coliphages shows that mutations conferring resistance are localized to the four OmpA surface loops [32,33]. Our previous work showed that S. flexneri OmpA acts as the preferred secondary receptor for Sf6 [29]. Since E. coli and S. flexneri OmpA are highly similar (sequence identity of 99.6% [34]), we hypothesized that the surface loops of S. flexneri OmpA may also play a role in mediating Sf6 infection. In vitro experiments can monitor loss of infectivity from mature Sf6 virions (and thus implies genome ejection) using purified S. flexneri LPS and the OmpA transmembrane domain, “OmpA-TMS.flex” (see Materials and Methods) [29]. We adopted this approach to determine if the surface loops of OmpA were crucial for triggering Sf6 genome ejection. Since it has been demonstrated that LPS alone is unable to trigger Sf6 genome release [29], any observed changes would result from altered OmpA ability to serve as a receptor. We used a limited proteolysis approach as proteinase K, subtilisin, and trypsin have all had their cleavage sites thoroughly mapped to OmpA surface loops [35]. Here, OmpA-TMS.flex was incubated with proteinase K, which has cleavage sites in all four loops [35]. Cleavage was confirmed by SDS-PAGE (data not shown). Digested OmpA-TMS.flex was then used in our in vitro experiments in combination with LPS and phage. Unlike undigested OmpA-TMS.flex, OmpA-TMS.flex treated with proteinase K is unable to trigger genome ejection of Sf6 (Fig. 1). Thus, the loops of OmpA appear to be essential for Sf6 infection. To further probe which portions of these four surface loops are important, we developed a plasmid complementation system to screen full-length OmpA constructs in vivo.
Fig. 1.
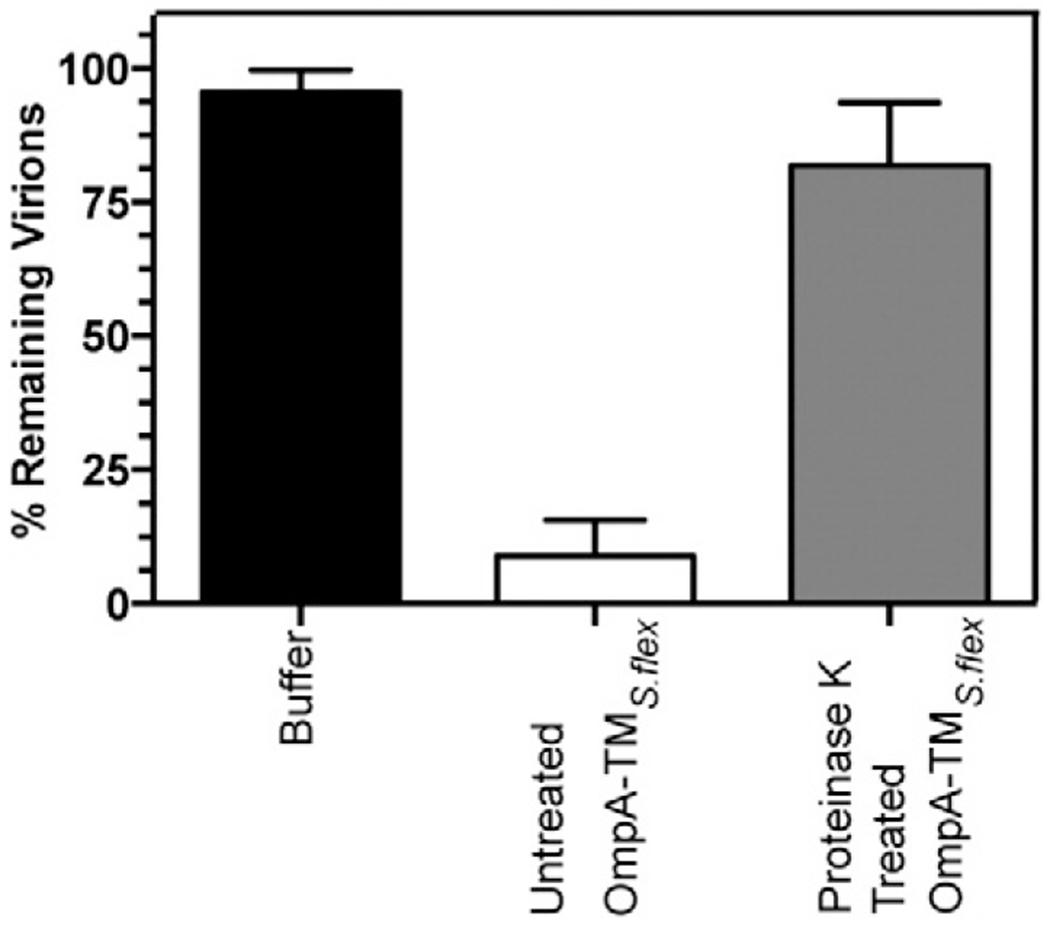
In vitro genome ejection efficiency decreases with proteinase K treated OmpA-TMS.flex. The “% remaining virions” was calculated as the number of PFUs remaining after incubation with S. flexneri LPS and OmpA-TMS.flex (untreated and proteinase K treated) divided by the number of PFUs when incubated with only buffer. Each data point is an average of at least three separate experiments; error bars signify one standard deviation.
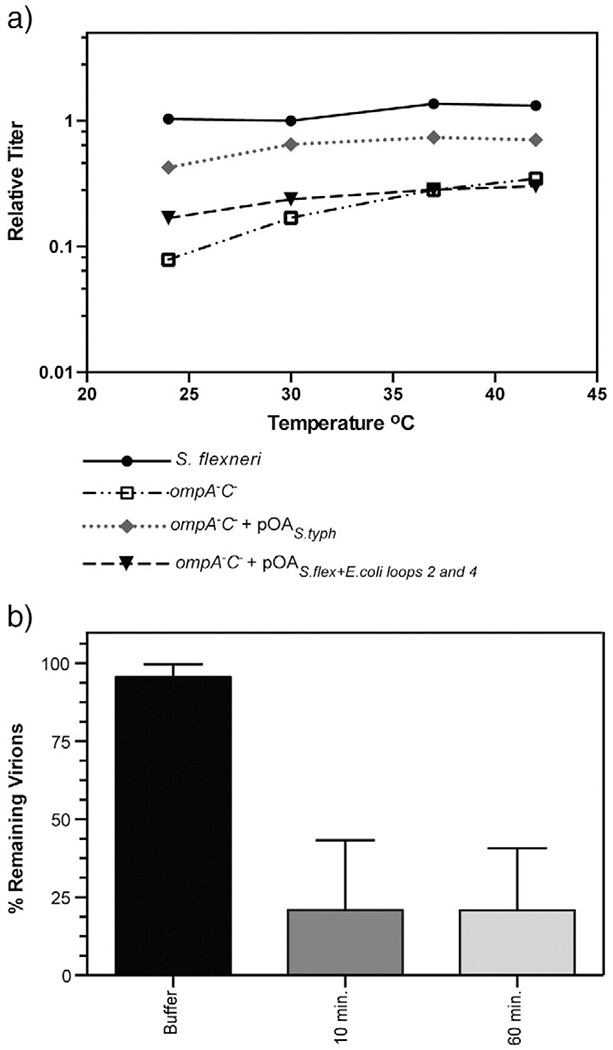
Previously, we have shown that the relative titer of Sf6 propagated on ompA−C− null S. flexneri drops ~10-fold compared with Sf6 grown on the parent S. flexneri strain and that, of these two gene deletions, ompA− demonstrated the largest effect on Sf6 infection [29]. Therefore, expression of OmpA in trans (referred to as “OmpAS.flex”) in the ompA−C− background should restore the ability of Sf6 to efficiently infect these cells. Full-length S. flexneri OmpA was expressed from plasmid “pOAS.flex” in the null ompA−C− background (see Materials and Methods and Fig. 2, schematic). This construct has been shown to restore protein levels and incorporation of OmpAS.flex into the outer membrane to that of the parent S. flexneri strain [36]. We compared infection of Sf6 at temperatures ranging from 25 to 42 °C on three strains of S. flexneri: parent strain, ompA−C−, and ompA−C− + pOAS.flex. Expression of OmpAS.flex in trans is able to restore the efficiency of infection of Sf6 in the ompA−C− background to that of the parent strain, as seen by a relative titer ~1 at all temperatures (Fig. 2). Therefore, OmpAS.flex is both necessary and sufficient to restore infection efficiency of Sf6 in ompA−C− S. flexneri.
Fig. 2.
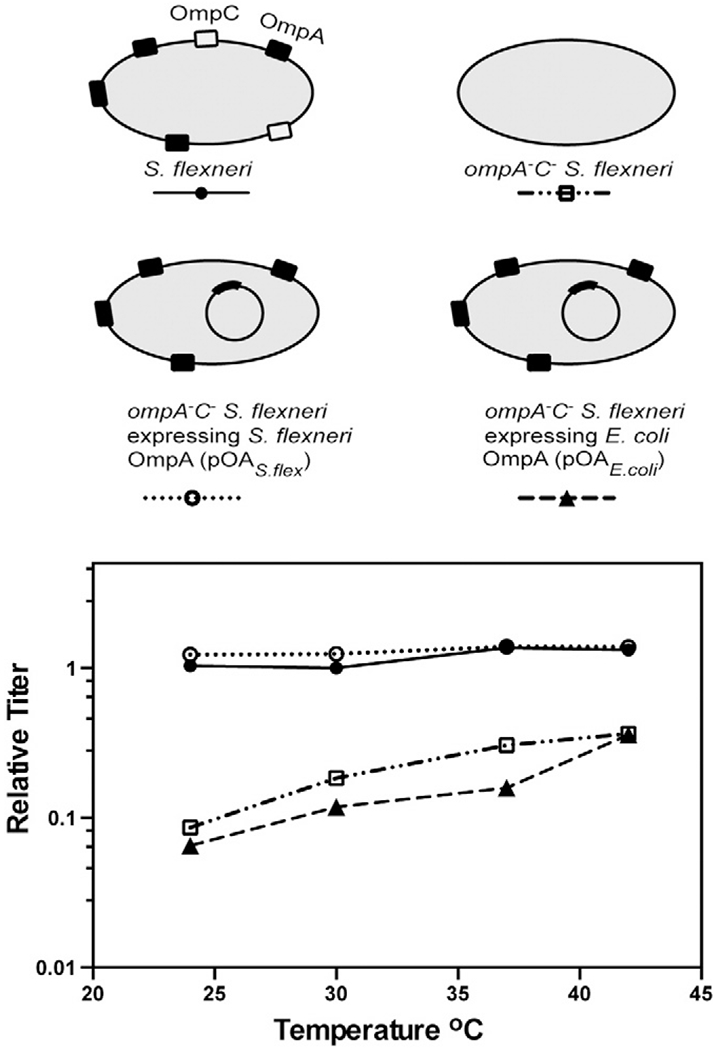
Relative titer of Sf6 is restored on ompA−C− S. flexneri expressing S. flexneri, but not E. coli OmpA. A schematic of the complementation system is shown. Relative titer of Sf6 was calculated by dividing the PFUs on each S. flexneri strain (parent, ompA−C−, ompA−C− + pOAS.flex, and ompA−C− + pOAE.coli) at each temperature by the number of PFUs on the parent Shigella strain PE577 at a permissive temperature (30 °C). Each data point is an average of at least five separate experiments.
E. coli and S. flexneri OmpA have high level of sequence identity, with only seven residue differences and a four-amino-acid insertion in the surface loops, in an area that is accessible to phage (Table 1) [34]. We therefore investigated whether E. coli OmpA expression in trans (“OmpAE.coli,”) was also able to restore the ability of Sf6 to infect ompA−C− S. flexneri. Unlike OmpAS.flex, OmpAE.coli was unable to restore Sf6 infection levels in the null ompA−C− S. flexneri background (Fig. 2). Therefore, we investigated if the differences between these two proteins play a role in mediating the inability of Sf6 to utilize the E. coli protein.
Table 1.
Amino acid substitutions in the surface loops of OmpA.
| OmpA variants | ||||||
|---|---|---|---|---|---|---|
| Loop number | Residuea number | E. coli amino acid | S. flexneri amino acid | No change in resistance | Moderate effect | Severe effect |
| 1 | 25 | N | P | — | A, R | Eb |
| 2 | 66 | S | D | A, K | — | — |
| 67 | V | N | — | — | A, E, H, R | |
| 68 | E | I | — | — | A, D, K, Q | |
| 3 | 108 | S | A | — | E | R |
| 111 | Y | P | — | A, E, R | — | |
| “insertion” (113, 114, 115, 116) | — | GASF | — | ΔGASF | — | |
| 4 | 155 | H | N | R | A | E |
The amino acids differing between E. coli and S. flexneri OmpA flexible loops are organized by loop and residue number. The OmpA variants shown in Fig. 3 are summarized here by their loss of function to serve as a receptor to Sf6 based on their relative titer.
Residue numbering based on S. flexneri OmpA.
Variant did not grow on MacConkey agar.
Amino acid substitutions in the loops of OmpA decrease Sf6 infection efficiency
To address which of the residues that differ between E. coli and S. flexneri OmpA were responsible for the observed phenotype, we systematically changed both the size and the charge of each by site-directed mutagenesis (see Materials and Methods). We measured the relative titer by plating Sf6 on ompA−C− S. flexneri complemented with these 22 different versions of OmpA (Table 1). The Sf6 plating efficiency changes with some amino acid substitutions, but not others (Fig. 3). Complementation by three variants, D66A and D66K (loop 2) and N155R (loop 4), restore Sf6 plating efficiency of the ompA−C− strain and the wild-type (WT) gene: ompAS.flex (Fig. 3). Variants at two locations in loop 2 (N67 and I68) and one variant in loop 4 (N155E) had the lowest Sf6 plating efficiency, indicating that these mutations confer a loss of function. All OmpAS.flex variants demonstrate the same relative phenotypes when plated at temperatures ranging from 25 to 42 °C (data shown only for 25 °C for simplicity; Fig. 3).
Fig. 3.
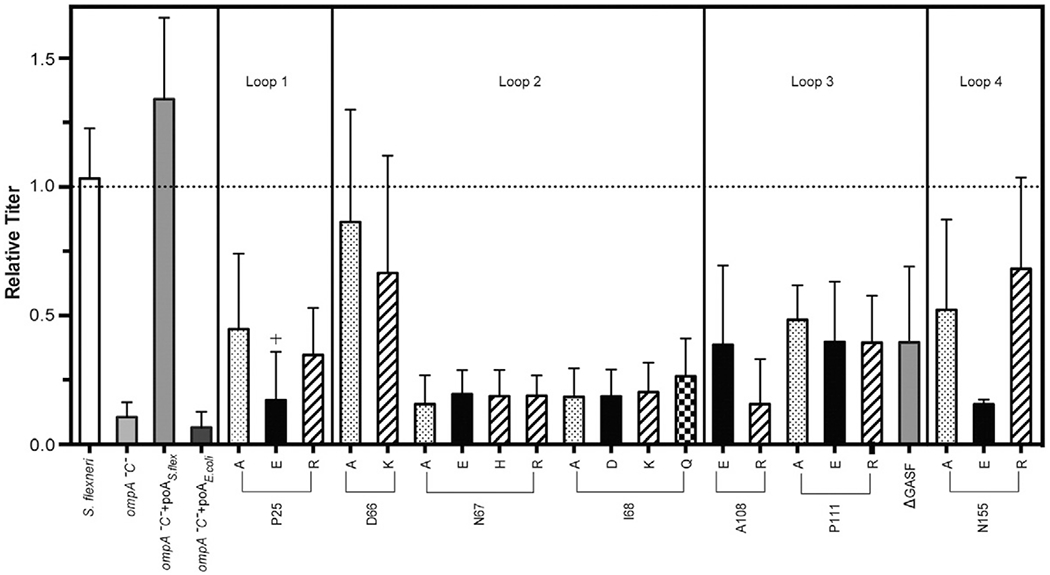
Comparison of Sf6 infection efficiency on ompA−C− S. flexneri expressing variant OmpAs. Relative titer of Sf6 on various strains at 25 °C (parent, ompA−C−, ompA−C− + pOAE.coli, ompA−C− + pOAS.flex, and ompA−C− + pOAS.flex expressing variant OmpAs). Amino acids with similar properties are shown with the same color-coding scheme. Each data point is an average of at least five separate experiments; error bars signify one standard deviation.
The observed loss of function of OmpA variants to serve as a receptor for Sf6 could have several mechanistic explanations. Amino acid alterations in the loops of OmpA may interfere with (1) the ability of the phage to bind OmpA, (2) folding of OmpA and therefore function, or (3) incorporation of OmpA into the outer membrane. First, to test whether the variant OmpAs incorporated correctly in vivo, we plated the parent strain S. flexneri and ompA−C− and ompA−C− strains expressing the 22 variant OmpAs on MacConkey agar, a bile-salt-rich medium that selects for Gram-negative bacteria with intact outer membrane integrity [37]. With the exception of only P25E OmpA (designated as “+” in Fig. 3 and “b” in Table 1), all strains were able to grow on the MacConkey agar as efficiently as the S. flexneri parent strain (data not shown). As P25E OmpA was not incorporated correctly into the outer membrane, it was excluded from further analysis.
Amino acid substitutions in OmpA-TM loops do not affect protein stability or folding
In order to more quantitatively determine the effect of these amino acid substitutions, we purified seven versions of OmpA-TM for biochemical characterization (below and the next section). In addition to Shigella and E. coli OmpA-TMs, we purified one representative variant per each surface loop that had some loss of function for Sf6 infection (P25R, N67E, P111E, and N155E) and one variant that showed no change in Sf6 infection (D66A).
First, to compare the relative stability, we used a heat titration assay to calculate the TM50, which is defined as the temperature where 50% of the protein species is folded. We incubated these purified proteins at temperatures ranging from 25 to 95 °C and determined the fraction of folded species by SDS-PAGE and gel densitometry since folded OmpA-TM migrates faster than unfolded OmpA-TM (Fig. 4a). The TM50 for OmpA-TMS.flex was determined to be 75.5 °C. In addition, OmpA-TMS.flex that had been boiled and then allowed to refold triggered Sf6 genome ejection efficiently (data not shown), indicating that the refolding of this protein is a reversible process. E. coli and the five selected variants of OmpA-TMS.flex had TM50 values ranging between 73 and 76 °C (Table 2), indicating that their relative stabilities are not significantly different from OmpA-TMS.flex.
Fig. 4.
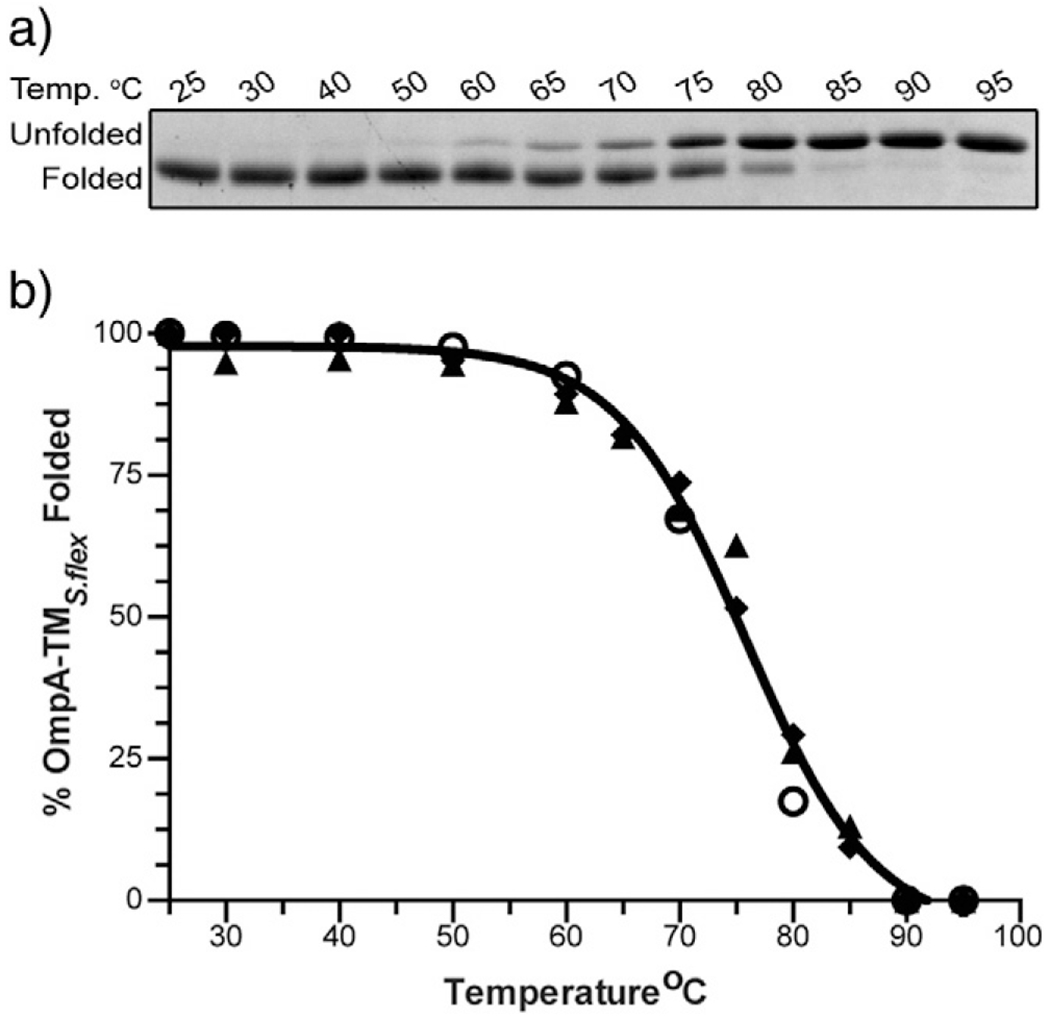
WT Shigella OmpA-TM thermal stability. (a) A representative 15% SDS gel stained with Coomassie of OmpA-TMS.flex after incubation at increasing temperatures. (b) Percent OmpA-TMS.flex folded species as a function of temperature. Open circles, triangles, and diamonds each represent individual data sets.
Table 2.
TM50 values of purified OmpA-TMs.
| Protein Variant | TM50 (°C) |
|---|---|
| WT | 75.5 ± 1.9 |
| P25R | 76.2 ± 2.4 |
| D66A | 75.7 ± 1.7 |
| N67E | 74.4 ± 2.2 |
| P111E | 75.6 ± 1.9 |
| N155E | 73.0 ± 1.8 |
| E. coli | 75.8 ± 3.1 |
Table shows the calculated TM50 for OmpA-TM (S. flexneri, E. coli, and S. flexneri variant OmpA-TMs) after heat titration and analysis of percent folded protein at increasing temperatures. See Fig. 4 for representative WT data.
Second, to determine the effect, if any, of the amino acid substitutions on the secondary structure, we determined the variant OmpA-TM circular dichroism (CD) spectra and compared them to the OmpA-TMS.flex spectrum. Consistent with previously published data for E. coli OmpA [38–41], the CD spectrum of OmpA-TMS.flex predicts a β-barrel secondary structure (Fig. 5). The CD spectra of the variant and E. coli OmpA-TM proteins have no significant differences and are essentially identical with that of OmpA-TMS.flex (Fig. 5). Therefore, it is likely that amino acid substitutions in the extracellular loops of OmpA-TM do not affect the overall protein structure.
Fig. 5.
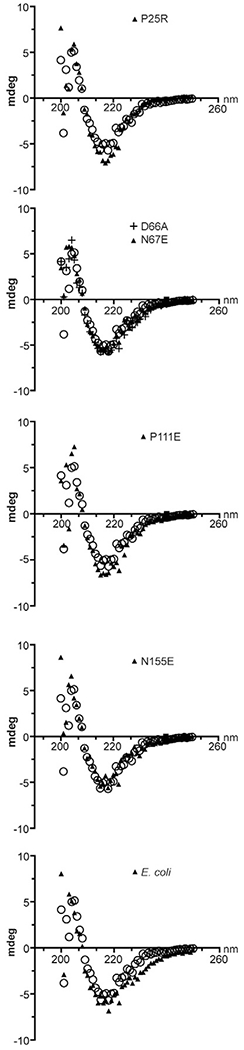
CD spectra of selected OmpA-TMs. Representative CD spectra of OmpA-TME.coli and OmpA-TM variants are shown. Open circles indicate the CD spectrum of OmpA-TMS.flex and are the same data shown in each panel.
Some amino acid substitutions in OmpA-TM surface-exposed loops reduce Sf6 genome ejection efficiency in vitro
To test if OmpA variants that demonstrated a loss of function to serve as a receptor for Sf6 in vivo (Fig. 3) also have decreased efficiency to trigger genome ejection in vitro, we incubated Sf6 with purified S. flexneri LPS combined with our purified OmpA-TM proteins. Previously, we showed that the physiological rate for Sf6 genome ejection is less than 10 min [29]. To determine whether Sf6 genome ejection efficiency is affected by these various OmpA-TMs, we calculated the percent remaining plaque-forming units (PFUs) after incubation for 10 min at 37 °C. Incubation with OmpA-TMS.flex resulted in near-complete ejection, as previously reported [29], with only ~15% remaining virions (Fig. 6a). As expected, and based on our in vivo data (see Fig. 3), OmpA-TME.coli in vitro was unable to efficiently induce genome ejection of Sf6 comparable to OmpA-TMS.flex, with ~75% remaining virions after 10 min (Fig. 6a). Furthermore, with those OmpA-TM variants that corresponded to lower Sf6 infection in vivo (P25R, N67E, P111E, and N155E), we also saw a reduction in the level of in vitro genome ejection, with an average of ~45% remaining virions. Additionally, although not as efficient as OmpA-TM-S.flex, the D66A variant does induce more genome ejection than the other versions of OmpA-TM (Fig. 6a), consistent with our in vivo data. It is important to note that, in these experiments, variations in OmpA sequence do not completely obliterate the plating efficiency of Sf6. As our previous work [29] suggests, there is likely a third receptor present at low copy number that Sf6 can use, albeit poorly, to gain entry (see Discussion for more in-depth discussion on this point).
Fig. 6.
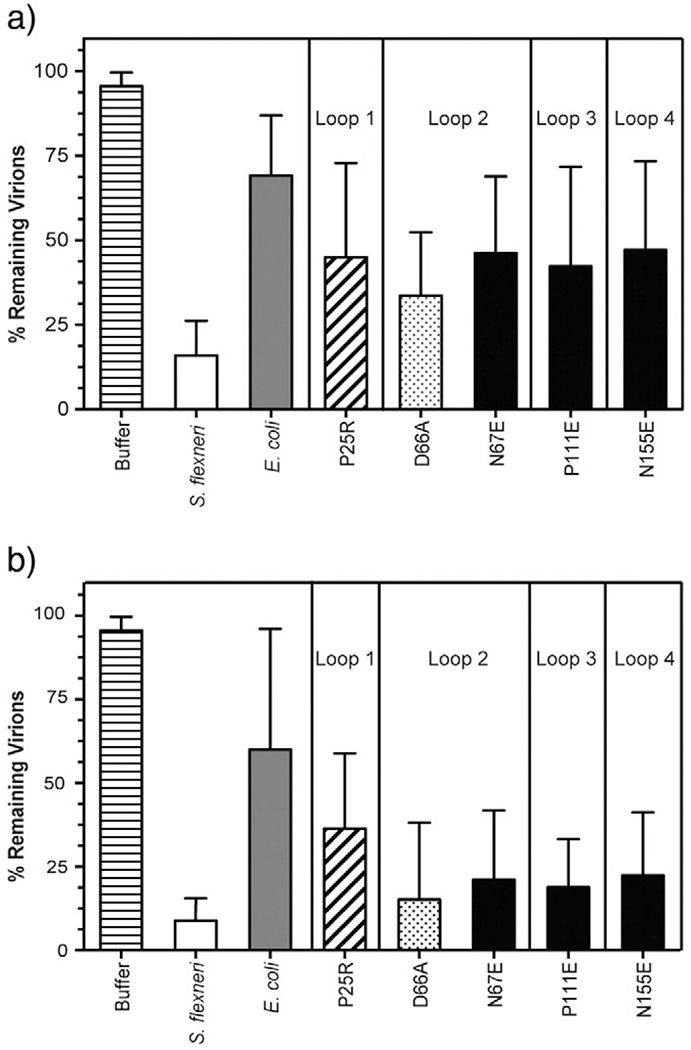
Sf6 in vitro genome ejection efficiency with LPS and variant OmpA-TMs. Ejection efficiency of Sf6 incubated with S. flexneri LPS and OmpA-TM (S. flexneri, E. coli, and variant OmpA-TMs) at 10 min (a) and 60 min (b) post-mixing. Color-coding scheme is consistent with Fig. 3. Each data point is an average of at least five separate experiments; error bars signify one standard deviation.
Taken together, reduced efficiency for ejection in vivo (Fig. 3) and in vitro (Fig. 6) likely correlates with a decrease in binding affinity of the phage to its secondary receptor. Infection in vivo on the ompA−C− null strain could be less efficient due to either (1) the phage utilizing a third, as of yet unidentified receptor, which may have much lower abundance on the cell surface or (2) the binding efficiency to a third receptor is significantly decreased based on molecular differences. Our in vivo complementation system expresses OmpA using its native promoter, and this construct has been shown to produce physiologically relevant concentrations of OmpA [36] and our in vitro experiments use identical concentrations of each variant protein relative to OmpA-TMS.flex. Variant OmpA proteins are complemented from the same vector in our in vivo assays and are likely similar in abundance to OmpAS.flex and therefore readily available for phage binding. However, there is still a decreased relative titer of Sf6 on these strains (Fig. 3). Our data suggest that Sf6 does not interact efficiently with these proteins. Therefore, we might expect to see an increase in genome ejection efficiency in our in vitro system if we incubate the phage with these variant receptors for an extended period of time, allowing a greater probability of productive interaction.
We therefore increased the incubation time to 60 min and measured the percent remaining PFUs under the conditions specified above. After 60 min of incubation with OmpA-TME.coli, Sf6 is still unable to release its genome at the same level induced by OmpA-TMS.flex (Fig. 6b). However, we did see an increase in genome ejection with the other OmpA-TM variants starting to approach OmpA-TMS.flex levels. Lastly, after 60 min of incubation, the D66A variant induces genome ejection with efficiency similar to OmpA-TMS.flex (Fig. 6b).
OmpA loops 2 and 4 are the most critical for mediating Sf6 host specificity
Combined, our data suggest that OmpA loops 2 and 4 are the most critical for Sf6 being able to productively interact with Shigella but not E. coli OmpA. Therefore, we made a hybrid construct of full-length OmpAS.flex that has the E. coli sequence in both loops 2 and 4. In our in vivo plating efficiency experiments, this hybrid is non-functional (Fig. 7a).
Fig. 7.
Sf6 can productively interact with S. typhimurium OmpA. (a) The relative titer is the number of PFUs of Sf6 on each S. flexneri strain (parent, ompA−C−, ompA−C− + pOAS.typh, ompA−C− + pOAS.flex + E.coli loops 2 and 4) at each temperature divided by the number of PFUs on the parent strain at the permissive temperature (30 °C). Data shown for Sf6 on ompA−C− and the parent strain is the same as in Fig. 2 each data point is the average of at least three independent experiments. (b) Ejection efficiency of Sf6 incubated with parent S. flexneri LPS and OmpA-TMS.typh at 10 min and 60 min post-mixing. Each data point is an average of at least five separate experiments; error bars signify one standard deviation.
Sf6 can tolerate several independent differences in loops 1 and 3 (Fig. 3). Therefore, we would anticipate that Sf6 could productively interact with other bacterial OmpAs as long as they share homology in loops 2 and 4 to S. flexneri OmpA. One candidate of interest is Salmonella typhimurium OmpA (“OmpAS.typh”). Differences between S. flexneri and S. typhimurium OmpA include amino acid substitutions P25H and N27D in loop 1 and A108S in loop 3, and the insertion “GASF” in loop 3 is GPST in S. typhimurium OmpA. However, these two proteins have identical sequences in loops 2 and 4. We therefore measured the ability of OmpAS.typh to complement Sf6 infection in our in vivo complementation system. As expected, expression of OmpA-S.typh in the null ompA−C− S. flexneri background shows a gain of function and is able restore the efficiency of infection of Sf6 close to that of OmpA-S.flex (Fig. 7a). This strain is able to grow as efficiently on MacConkey agar as the parent S. flexneri strain, indicating that OmpAS.typh is localized and incorporated correctly (data not shown). We also expressed and purified OmpA-TMS.typh and found that it is stable (TM50 = 76.5 ± 3.6 °C) and has a CD spectrum indistinguishable from OmpA-TMS.flex (data not shown). We assessed the ability of OmpA-TMS.typh to trigger Sf6 genome ejection in vitro by calculating the percent remaining virions after incubation for 10 and 60 min. As expected, and consistent with our in vivo data (Fig. 7a), OmpA-TMS.typh is able to induce genome ejection of Sf6 at levels close to OmpA-TMS.flex in vitro (Fig. 7b). These data further support the idea that loops 2 and 4 of S. flexneri OmpA mediate Sf6 interaction and host specificity.
Discussion
Many viruses have the inherent ability to use more than one type of proteinaceous receptor for attachment, with one receptor type being preferred. In this work, we identified which portions of S. flexneri OmpA, the preferred secondary receptor for Sf6 [29], mediate phage infection and confer host range. We created several OmpA variants through site-directed mutagenesis and investigated their ability to alter Sf6 infection of S. flexneri. Here, we have shown that Sf6 interacts with the surface loops of OmpA. Moreover, individual substitutions have a range of effects, implicating some locations in the loops as more important than others for infection. However, in no case were we able to completely block Sf6 infection. These data support general phage plasticity for receptor usage. If Sf6 has indeed adapted to use OmpA, OmpC, and a third, as of yet unidentified receptor, as our previous work suggests [29], it is unlikely that a single amino acid substitution in OmpA would completely obliterate infection in vivo.
Sf6 may interact with the surface loops of OmpA in one of two ways. (1) Sf6 may interact preferentially with one specific portion of the protein or (2) the phage may interact with the protein surface as a whole. Studies with T5 and FhuA have proposed that phage T5 interacts preferentially with only a portion of the Omp surface [42]. Work with many different coliphages has shown that these phage do not tolerate amino acid mutations in loop 2 or 3 of E. coli OmpA, since >84% of 305 independently isolated mutations in OmpA from phage-resistant cells are found in these loops [32,33]. Loops 2 and 3 are adjacent (Fig. 8), suggesting that these coliphages may not interact with the entire OmpA surface but rather a preferential side of the protein. Additionally, isolated mutations in the receptor for phage λ, LamB, that confer resistance to phage infection [43,44] appear have a strong bias to a preferential side of LamB: when we modeled these amino acid substitutions into the LamB crystal structure (PDB ID: 1AF6 [45]), the substitutions were localized to neighboring loops.
Fig. 8.
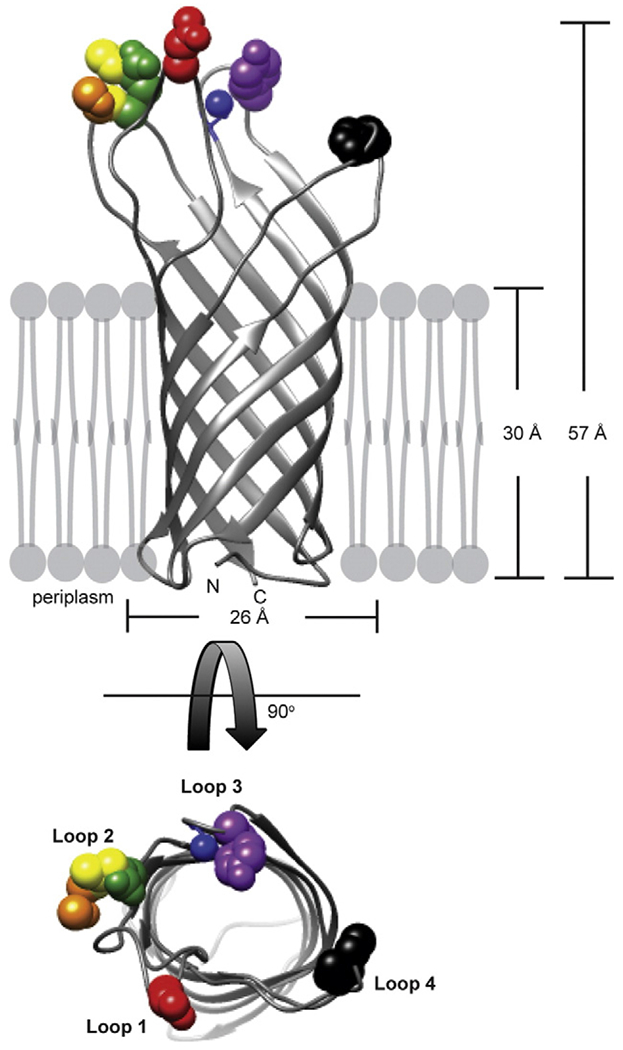
Sf6 interacts with entire OmpA surface. The crystal structure of E. coli OmpA (PDB ID: 1BXW [55]) is depicted as a ribbon diagram with substituted amino acids shown as spheres: red, P25; orange, D66; yellow, N67; green, I68; blue, A108; violet, P111; and black, N155 using UCSF Chimera [56].
Amino acids that confer resistance to Sf6 infection are located at flexible portions of the loops of OmpA (Fig. 8). If, like the coliphages, Sf6 were to also interact with a preferential portion of OmpA, such as a particular side, we would expect amino acid substitutions that allow resistance to phage infection to have a bias to a single loop or two neighboring loops. Our data suggest that, overall, amino acid substitutions are less deleterious in loops 1 and 3 of S. fiexneri OmpA compared to loops 2 and 4, as seen by both differences in the relative plating efficiency (Fig. 3) and the in vitro genome ejection data (Fig. 6). However, some substitutions in loops 1 and 3 do have a slight decrease in infection efficiency (P25R: loop 1 and A108R: loop 3, as examples). Therefore, we hypothesize that, unlike the coliphages, Sf6 can interact with the entire surface of OmpA, rather than with a preferential side of the receptor. This may be a fundamental difference in binding profiles as seen by phage with long flexible Siphoviridae tails and phage with the short, stubby tails of Podoviridae. More experimental evidence is needed to determine if this is a global mode of binding for members of Podoviridae.
Theoretically, to evolutionarily avoid phage infection, mutations within bacterial cells would be selected for that decrease infection. Therefore, one might expect to see mutations in the loops of Omps that lead to decreased binding affinity and therefore a corresponding decrease in phage infection. However, this phenomenon is not necessarily always observed in nature. Although it may be beneficial to the host to evolve changes in the loops of OmpA, to avoid Sf6 infection, OmpA has several other roles [46], including attachment and invasion of eukaryotic cells [36,47–52]. Work with meningitic E. coli OmpA has implicated the surface loops as important for the pathogenesis of E. coli, fulfilling roles such as attachment, survival, and cell–cell spread [47,49,50,52]. Moreover, loop 2 of E. coli OmpA appears to have several overlapping roles, as alterations in this loop affect several key virulence factors of E. coli: attachment, intracellular survival, and invasiveness [47]. Therefore, although mutations in the surface loops of OmpA may lead to an increase in resistance to phage infection, the ability of S. flexneri to invade eukaryotic cells may be decreased, thereby decreasing the bacterial pathogenicity. This point is merely speculation as little experimental evidence is currently available on the specific role of OmpA surface loops in S. flexneri pathogenesis. Not evolving resistance to phage infection is likely a trade-off to retain pathogenicity, although this remains to be determined experimentally.
In the present study, we showed that the surface loops of OmpA mediate phage Sf6 infection of S. flexneri. Coupling site-directed mutagenesis and in vivo phage biology allowed us to delineate which portions of the surface loops interact favorably with Sf6. Our data suggest that some amino acid substitutions in the loops decrease phage infection efficiency. By complementing the ompA−C− S. flexneri strain with S. typhimurium OmpA, we found that Sf6 could productively interact with other bacterial OmpAs as long as they share homology in loops 2 and 4, thus suggesting that host specificity may be determined by these loops. We propose a model in which Sf6 interacts with OmpA on the whole surface rather than only on a preferential side of the protein, unlike what other known phages do with their respective Omp receptors. Our data provide new insights into Podoviridae attachment through binding to membrane protein receptors.
Materials and Methods
Media and strains
Lysogeny Broth was used for bacterial growth, most plating experiments, and preparations of Sf6 phage stocks. MacConkey agar (BD Difco) was used to select for bacteria with intact outer membranes [37]. Sf6 phage used in all experiments carries a mutation making the phage obligately lytic and was prepared as previously described [53]. Phage were stored in phage buffer: 10 mM Tris (pH 7.6) and 10 mM MgCl2. S. flexneri strains include the parent strain PE577 [54] and ompA−C− [29]. Plasmids expressing S. flexneri (“pOAS.flex” [36]), S. typhimurium (“pOAS.typh”), and E. coli OmpA (“pOAE.c”) were transformed into ompA−C− S. flexneri. Similar to pOAS.flex, E. coli OmpA was constitutively expressed off pACYC184 plasmid (Camr) with its native promoter and was generated by Dr. Alexander Chang and kindly provided by Dr. Nemani Prasadarao. For the purification of S. flexneri, E. coli, and S. typhimurium OmpA-TM, the transmembrane domain (residues 1–175 for S. flexneri OmpA-TM [29], residues 1–171 for E. coli OmpA-TM, and residues 1–175 for S. typhimurium OmpA-TM) with a 6-histidine tag on the N-terminus was subcloned into a pRSET_A vector (Invitrogen) (Ampr) and expressed in E. coli Bl21 (DE3)pLysS. All vectors encoding OmpA variants (either as the full-length protein or as OmpA-TM) were generated through single or serial rounds of QuikChange site-directed mutagenesis using pOAS.flex (for in vivo complementation experiments) or “pNBP01” (for protein purification) [29] as the starting template. For all constructs generated in this study, sequences were verified using Sanger sequencing at the Research Technology Support Facility at Michigan State University.
Purification and refolding of variant OmpA-TMs
OmpA-TM variants were purified and refolded as previously described [29]. Briefly, OmpA-TM was refolded by nutation in 0.1% (1.8 mM) of Triton X-100 at room temperature overnight. Protein folding was confirmed by electrophoretic mobility via SDS-PAGE. Refolded OmpA-TMs were exhaustively dialyzed against Triton X-100 (1.8 mM) to remove residual urea.
Proteinase K treatment
Proteinase K (Roche) and folded WT OmpA-TM were incubated at 37 °C for 15 min at a 1:5 ratio. Loop cleavage was confirmed by SDS-PAGE and digested OmpA was then used for some in vitro genome ejection experiments (as described in the next section).
LPS extraction and in vitro genome ejection experiments
S. flexneri LPS was extracted from the parent S. flexneri strain using a BulldogBio kit as previously described [29]. Sf6 was incubated at 37 °C with purified LPS at 0.25 mg/mL and OmpA-TM at 0.15 mg/mL. Aliquots were taken 10 and 60 min post-addition of phage, serially diluted, and plated on the parent S. flexneri strain; plates were incubated at 30 °C. “Percent remaining virions” was calculated by dividing the PFUs at each time point by the PFUs with buffer only added at t = 0 min. In vitro genome ejections with “boiled and refolded” OmpA-TMS.flex were set up as described above after OmpA-TMS.flex was boiled for 5 min at 95 °C and allowed to refold overnight.
Measuring the relative titer of Sf6 on S. flexneri
Sf6 was plated on various S. flexneri strains (the parent strain PE577, ompA−C−, ompA−C− + pOAS.flex, ompA−C− + pOAE.Coli, ompA−C− + pOAS.typh or ompA−C− + pOAS.flex expressing variant OmpAs) at temperatures ranging from 25 to 42 °C. The relative titer was calculated by dividing the resultant PFUs on each strain and at each temperature by the PFUs on the S. flexneri parent strain at the permissive temperature, 30 °C.
Thermal stability of variant OmpA-TMs
To measure the stability of variant OmpA-TM relative to OmpA-TMS.flex, we incubated purified OmpA-TMs ranging in concentration 0.6–4 mg/mL at temperatures between 25 and 95 °C, run by 15% SDS-PAGE, and then stained by Coomassie. Gel densitometry (BIORAD Gel Doc XR+) was used to determine percent folding at each temperature. Data were plotted and fit with a sigmoidal curve using GraphPad Prism version 6.0 for Mac OS X, GraphPad Software, La Jolla, CA, USA†. Data for determining thermal stability were collected in triplicate for each OmpA-TM protein.
Circular dichroism
Far UV CD spectra were taken with a JASCO J-815 CD spectrometer (JASCO Analytical Instruments, Easton, MD) in a 1 mm (Starna cells quartz) cuvette at 25 °C. Spectra were recorded from 200 to 250 nm with a bandwidth of 1.0 mm, scanning rate of 50 nm/min, and data integration time of 1 s. Ten scans were averaged for each sample. Protein concentration was normalized to OmpA-TMS.flex by SDS-PAGE and gel densitometry prior to CD. Three technical replicates of CD spectra were collected for each protein type.
Acknowledgments
The authors would like to thank Dr. Barbara Atshaves (Michigan State University) and graduate student Charlie Najt for use of their CD spectrometer. We thank Sophia Sdao for help with preliminary plating experiments. We thank Dr. Nemani Prasadarao and Dr. Alexnader Chang at Children’s Hospital Los Angeles for providing the E. coliOmpA plasmid (“pOAE.coli”) and Dr. Mauro Nicoletti (Universita’ “G. D’Annunzio” di Chieti) for providing the S. flexneri OmpA plasmid (“pOAS.flex”). Thanks are extended to Dr. Sherwood Casjens (University of Utah) for stimulating discussions and critical reading of the manuscript. Part of the work presented here was supported by the Rudolph Hugh Fellowship to N.B.P.
Abbreviations used:
- PFU
plaque-forming unit
- LPS
lipopolysaccharide
- Omp
outer membrane protein
- HSV
herpes simplex virus
- WT
wild type
Footnotes
References
- [1].Flint SJ, Enquiest LW, Krug RM, Racaniello VR, Skalka AM. Principles of virology, molecular biology, pathogenesis, and control. ASM Press; 2000. [Google Scholar]
- [2].Bettstetter M, Peng X, Garrett RA, Prangishvili D. AFVI, a novel virus infection hyperthermophilic archaea of the genus Acidianus. Virology 2003;315:68–79. [DOI] [PubMed] [Google Scholar]
- [3].Schneider-Schaulies J Cellular receptors for viruses: links to tropism and pathogenesis. J Gen Virol 2000;81:1413–29. [DOI] [PubMed] [Google Scholar]
- [4].Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, et al. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 1997;275:1320–3. [DOI] [PubMed] [Google Scholar]
- [5].Marti R, Zurfluh K, Hagens S, Pianezzi J, Klumpp J, Loessner MJ. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol Microbiol 2013;87:818–34. [DOI] [PubMed] [Google Scholar]
- [6].Dhillon EK, Dhillon TS, Lai AN, Linn S. Host range, immunity, and antigenic properties of lambdoid coliphage HK97. J Gen Virol 1980;50:217–20. [DOI] [PubMed] [Google Scholar]
- [7].Kiino DR, Singer MS, Rothman-Denes LB. Two overlapping genes encoding membrane proteins required for bacteriophage N4 adsorption. J Bacteriol 1993;175:7081–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao X, Cui Y, Yan Y, Du Z, Tan Y, Yang H, et al. Outer membrane proteins Ail and OmpF of Yersinia pestis are involved in the adsorption of T7-related bacteriophage Yepphi. J Virol 2013;87:12260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hantke K, Braun V. Functional interaction of the tonA/tonB receptor system in Escherichia coli. J Bacteriol 1978;135: 190–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Spear PG. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol 2004;6:401–10. [DOI] [PubMed] [Google Scholar]
- [11].Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 1996;87:427–36. [DOI] [PubMed] [Google Scholar]
- [12].Xu D, Zhang J, Liu J, Xu J, Zhou H, Zhang L, et al. Outer membrane protein OmpW is the receptor for typing phage VP5 in the Vibrio cholerae O1 El Tor biotype. J Virol 2014;88: 7109–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tufaro F Virus entry: two receptors are better than one. Trends Microbiol 1997;5:257–8. [DOI] [PubMed] [Google Scholar]
- [14].Spear PG. Entry of alphaherpesviruses into cells. Semin Virol 1993;4:167–80. [Google Scholar]
- [15].Tateno M, Gonzalez-Scarano F, Levy JA. Human immunodeficiency virus can infect CD4-negative human fibroblastoid cells. Proc Natl Acad Sci U S A 1989;86:4287–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Endres MJ, Clapham PR, Marsh M, Thomas JF, Stoebenau-Haggarty B, Choe S, et al. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 1996;87:745–56. [DOI] [PubMed] [Google Scholar]
- [17].Stevenson SC, Rollence M, Marshall-Neff J, McClelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J Virol 1997;71: 4782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Morona R, Henning U. New locus (ttr) in Escherichia coli K-12 affecting sensitivity to bacteriophage T2 and growth on oleate as the sole carbon source. J Bacteriol 1986;168:534–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Black PN. The fadL gene product of Escherichia coli is an outer membrane protein required for uptake of long-chain fatty acids and involved in sensitivity to bacteriophage T2. J Bacteriol 1988;170:2850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morona R, Henning U. Host range mutants of bacteriophage Ox2 can use two different outer membrane proteins of Escherichia coli K-12 as receptors. J Bacteriol 1984;159:579–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Drexler K, Riede I, Montag D, Eschbach M, Henning U. Receptor specificity of the Escherichia coli T-even type phage Ox2. J Mol Biol 1989;207:797–803. [DOI] [PubMed] [Google Scholar]
- [22].Meyer JR, Dobias DT, Weitz JS, Barrick JE, Quick RT, Lenski RE. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 2012;335: 428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol 2005;15:655–63. [DOI] [PubMed] [Google Scholar]
- [24].Prevelige PE, Fane BA. Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Adv Exp Med Biol 2012;726:325–50. [DOI] [PubMed] [Google Scholar]
- [25].Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, et al. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol 2001;75:10923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol 2005;15:227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 2011;411:393–415. [DOI] [PubMed] [Google Scholar]
- [28].Casjens SR, Molineux IJ. Short noncontractile tail machines: adsorption and DNA delivery by podoviruses. Adv Exp Med Biol 2012;726:143–79. [DOI] [PubMed] [Google Scholar]
- [29].Parent KN, Erb ML, Cardone G, Nguyen K, Gilcrease EB, Porcek NB, et al. OmpAand OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol Microbiol 2014;92: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Muller JJ, Barbirz S, Heinle K, Freiberg A, Seckler R, Heinemann U. An intersubunit active site between supercoiled parallel beta helices in the trimeric tailspike endorhamnosidase of Shigella flexneri phage Sf6. Structure 2008; 16:766–75. [DOI] [PubMed] [Google Scholar]
- [31].Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 2000;37:239–53. [DOI] [PubMed] [Google Scholar]
- [32].Morona R, Kramer C, Henning U. Bacteriophage receptor area of outer membrane protein OmpA of Escherichia coli K-12. J Bacteriol 1985;164:539–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Morona R, Klose M, Henning U. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J Bacteriol 1984;159:570–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Power ML, Ferrari BC, Littlefield-Wyer J, Gordon DM, Slade MB, Veal DA. A naturally occurring novel allele of Escherichia coli outer membrane protein A reduces sensitivity to bacteriophage. Appl Environ Microbiol 2006;72:7930–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Structural Koebnik R. and functional roles of the surface-exposed loops of the β-barrel membrane protein OmpA from Escherichia coli. J Bacteriol 1999;181:3688–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ambrosi C, Pompili M, Scribano D, Zagaglia C, Ripa S, Nicoletti M. Outer membrane protein A (OmpA): a new player in Shigella flexneri protrusion formation and inter-cellular spreading. PLoS One 2012;7:e49625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].MacConkey A Lactose-fermenting bacteria in faeces. J Hyg 1905;5:333–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sugawara E, Steiert M, Rouhani S, Nikaido H. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J Bacteriol 1996;178:6067–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hong H, Tamm LK. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc Natl Acad Sci U S A 2004;101:4065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kleinschmidt JH, Tamm LK. Secondary and tertiary structure formation of the β-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J Mol Biol 2002;324:319–30. [DOI] [PubMed] [Google Scholar]
- [41].Kleinschmidt JH, Wiener MC, Tamm LK. Outer membrane protein A of E. coli folds into detergent micelles, but not in the presence of monomeric detergent. Protein Sci 1999;8: 2065–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Böhm J, Lambert O, Frangakis AS, Letellier L, Baumeister W, Rigaud JL. FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr Biol 2001;11:1168–75. [DOI] [PubMed] [Google Scholar]
- [43].Chatterjee S, Rothenberg E. Interaction of bacteriophage λ with its E. coli receptor, LamB. Viruses 2012;4:3162–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Charbit A, Gehring K, Nikaido H, Ferenci T, Hofnung M. Maltose transport and starch binding in phage-resistant point mutants of maltoporin. J Mol Biol 1988;201:487–96. [DOI] [PubMed] [Google Scholar]
- [45].Wang YF, Dutzler R, Rizkallah PJ, Rosenbusch JP, Schirmer T. Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin. J Mol Biol 1997; 272:56–63. [DOI] [PubMed] [Google Scholar]
- [46].Smith SG, Mahon V, Lambert MA, Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett 2007;273:1–11. [DOI] [PubMed] [Google Scholar]
- [47].Mittal R, Krishnan S, Gonzalez-Gomez I, Prasadarao NV. Deciphering the roles of outer membrane protein A extracellular loops in the pathogenesis of Escherichia coli K1 meningitis. J Biol Chem 2011;286:2183–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Torres AG, Kaper JB. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect Immun 2003;71:4985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pascal TA, Abrol R, Mittal R, Wang Y, Prasadarao NV, Goddard WA. Experimental validation of the predicted binding site of Escherichia coli K1 outer membrane protein A to human brain microvascular endothelial cells: identification of critical mutations that prevent E. coli meningitis. J Biol Chem 2010;285:37753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Prasadarao NV, Wass CA, Weiser JN, Stins MF, Huang SH, Kim KS. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun 1996;64:146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Torres AG, Jeter C, Langley W, Matthysse AG. Differential binding of Escherichia coli O157:H7 to alfalfa, human epithelial cells, and plastic is mediated by a variety of surface structures. Appl Environ Microbiol 2005;71:8008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sukumaran SK, Shimada H, Prasadarao NV. Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect Immun 2003;71:5951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Casjens S, Winn-Stapley DA, Gilcrease EB, Morona R, Kuhlewein C, Chua JE, et al. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J Mol Biol 2004; 339:379–94. [DOI] [PubMed] [Google Scholar]
- [54].Morona R, Mavris M, Fallarino A, Manning PA. Characterization of the rfc region of Shigella flexneri. J Bacteriol 1994; 176:733–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pautsch A, Schulz GE. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol 1998;5: 1013–7. [DOI] [PubMed] [Google Scholar]
- [56].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]


