Abstract
Background
These last months, dozens of SARS-CoV-2 serological tests have become available with varying performances. A major effort was completed to compare 17 serological tests available in April 2020 in Switzerland.
Methods
In a preliminary phase, we compared 17 IgG, IgM, IgA and pan Ig serological tests including ELISA, LFA, CLIA and ECLIA on a panel of 182 sera, comprising 113 sera from hospitalized patients with a positive RT-PCR, and 69 sampled before 1st November 2019, expected to give a positive and negative results, respectively. In a second phase, the five best performing and most available tests were further evaluated on a total of 582 sera (178 and 404 expected positive and negative, respectively), allowing the assessment of 20 possible cross-reactions with other viruses.
Results
In the preliminary phase, among eight IgG/pan-Ig ELISA or CLIA/ECLIA tests, five had a sensitivity and specificity above 90 % and 98 % respectively, and on six IgM/IgA tests, only one was acceptable. Only one LFA test on three showed good performances for both IgG and IgM. For all the tests IgM and IgG aroused concomitantly. In the second phase, no test showed particular cross-reaction. We observed an important heterogeneity in the development of the antibody response.
Conclusions
The majority of the evaluated tests exhibited high performances of IgG/pan-Ig sensitivity and specificity to detect the serological response of moderately to critically ill hospitalized patients. The IgM and IgA tests showed mostly insufficient performances with no added value for the early diagnostic on the cohort tested in this study.
Keywords: SARS-CoV-2, Serology, Evaluation, Kits
1. Introduction
In December 2019, a new virus causing severe respiratory infections emerged in China in the Wuhan area. This virus was classified in the Coronaviridae family and in the Betacoronavirus genus, named SARS-CoV-2 (Severe Acute Respiratory Syndrome CoronaVirus 2) and the associated disease was coined “COVID-19” (COronaVIrus Disease 2019). The epidemic rapidly spread and the WHO classified it as a pandemic in March 2020 (https://www.who.int/news-room/detail/27-04-2020-who-timeline---covid-19).
The mortality rate of the SARS-CoV-2 (about 2%) is lower than SARS-CoV-1 and MERS-CoV (Middle East Respiratory Syndrome Coronavirus) (10 and 30 %, respectively) but its reproduction rate R0 (2–2.5) is higher, than the SARS-CoV-1 (1.7–1.9) and the MERS-CoV (<1), probably explaining its rapid spreading worldwide [[1], [2], [3]].
In a first phase of the pandemic, nucleic acid amplification tests (NAAT) enabled rapid detection of infected patients, their sorting and their possible isolation. In a second phase, serology testing appeared particularly important as it permits to diagnose patients after the acute phase of the infection or with atypical clinical presentation with no nasopharyngeal shedding of the virus [4,5]. Indeed, in contrast to NAAT, which must be carried out when and where the virus is excreted, the serological assays might be performed anytime ideally more than two weeks after symptoms onset [4]. Serology also appeared to be the test of choice to perform large-scale population prevalence studies.
Various SARS-CoV-2 serological tests using different targeted antigenic proteins have been arriving on the market the last months (https://www.finddx.org/covid-19/pipeline). Some of them use whole virus lysate, recombinant full S (spike) or N (nucleocapsid) proteins, peptides of the N or specific domains S1, S2 or RBD (receptor-binding domains) of the S protein.
Different studies demonstrated that the S and N proteins were the most immunogenic [4,[6], [7], [8]]. The N protein is relatively small with no glycosylated sites and presents a higher level of conservation than the S protein among coronavirus infecting human, allowing possible false positive results through cross-reaction [4,9,10]. In contrast the S protein is a large transmembrane protein, less conserved, containing several glycosylated sites and bearing a more complex conformation, leading to production of more specific antibodies often recognizing conformational or glycosylated epitopes [[9], [10], [11]]. Thus the use of recombinant S protein lacking glycosylation or conformation in immunoassays may lead to false negative results.
In this study we evaluated several SARS-CoV-2 serological tests available in April 2020 in Switzerland including ELISA (Enzyme-Linked ImmunoSorbent Assays), LFA (Lateral Flow ImmunoAssays), CLIA (ChemiLuminescent ImmunoAssays) or ECLIA (Electro-ChemiLuminescent ImmunoAssays). This evaluation aimed to identify high quality tests for symptomatic patients.
2. Material and methods
2.1. Samples
The first phase of the evaluation was performed on 182 sera (113 positive and 69 negative) (Table S1). Then, the evaluation was completed for the selected tests on 400 sera (65 positive and 335 negative), leading to a full evaluation performed on 582 sera (178 positive and 404 negative) (Table S1). Negative-expected sera were selected among sera sampled before the 1st November 2019 and indicated as “Anterior” for anterior to SARS-CoV-2 pandemic (Table S1). Possible cross-reactivity was assesses through testing of sera known to be positive for a given microorganism, indicated as “Anterior (microorganism)”. The 178 expected positive sera were sampled during the first 2 months post-symptoms from patients documented with a positive SARS-CoV-2 RT-PCR and with a dates of symptoms in their electronic records.
2.2. ELISA, LFA, and CLIA assays
Each test (Table S2) was performed according to the manufacturers’ instructions. ELISA assays were done in duplicates and manually to diminish dead volume, except washing steps performed with a microplate washer (PW40, Bio-Rad, France). Reading of the Optical densities (OD) was done with a microplate reader (800 TSI, BioTek, USA).
For CLIA assays, the LIAISON® SARS-CoV-2 IgG kit was performed on a Liaison® XL (Diasorin, Italy), and the MAGLUMI™ 2019-nCoV IgG and IgM kits on a MAGLUMI™ 800 (Snibe, China). The ECLIA assay, Elecsys anti-SARS-CoV-2 was performed on a COBAS 6000 (Roche, Switzerland).
Sensitivity was evaluated on expected positive sera according to day post-symptoms. Specificity was determined on expected negative sera sampled before 1st November 2019.
2.3. Statistical analyses
Sensitivity and specificity with 95 % CI (Wilson/Brown method of GraphPad Prism 8.3.0) were calculated with Excel and GraphPad prims.
3. Results
3.1. Preliminary evaluation of 17 SARS-CoV-2 serologic tests
A preliminary evaluation of 17 serological kits (Table S2) has been performed on 182 sera, including 113 sera from patients positive for a SARS-CoV-2 RT-PCR (considered as positive) and 69 sera sampled before November 1, 2019 (considered as negative). For the 113 so-called “positive sera”, a stratification of the results was done according to the time between symptoms onset and sera sampling. Four categories were defined 0–5, 6–10, 11–15 and >15 days. The 17 serological kits tested included 10 ELISA (five IgG, three IgM, one IgA, and one IgM + IgA) from five manufacturers, three LFA (IgG + IgM) from three manufacturers, three CLIA (two IgG and one IgM) from two manufacturers, and one ECLIA (pan-Ig).
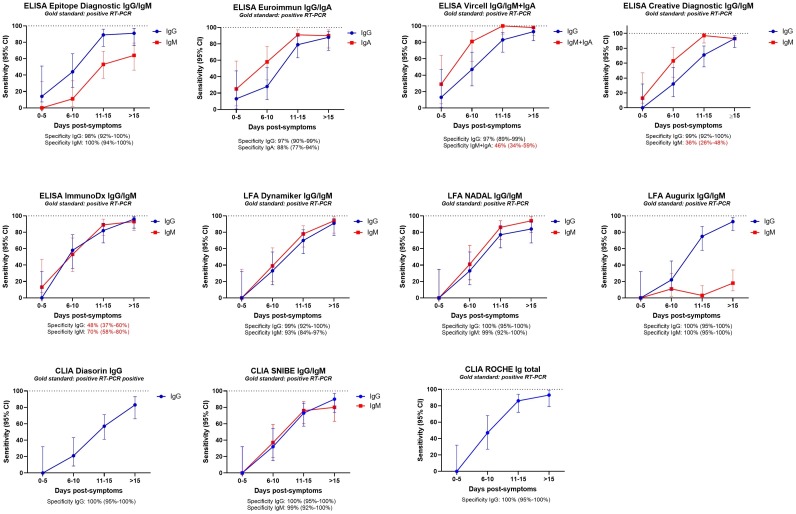
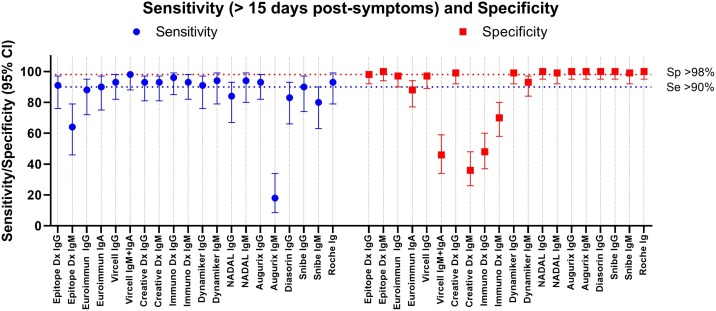
For all the 17 tests (IgG, IgM, IgA, pan-Ig) the sensitivity increased over time post-symptoms as expected (Fig. 1 , Table S3-5). Concerning IgG or pan-Ig tests, a sensitivity above 70 % was obtained after 10 days post-symptoms for almost all tests except the Diasorin ISON® SARS-CoV-2 IgG kit (57 %). However, a sampling at minimum 15 days post-symptoms is necessary for most of the IgG/pan-Ig tests to reach more than 90 % sensitivity (Fig. 1, Fig. 2 , Table S3−5). Only three tests exhibited a sensitivity lower than 90 % more than 15 days post-symptoms, the Euroimmun ELISA IgG test (88 %; CI:72−95), the NADAL® COVID-19 IgG/IgM LFA test (84 %; 95 % CI:67−93) and the Diasorin ISON® SARS-CoV-2 IgG CLIA kit (83 %; 95 %CI:66−93) (Fig. 2, Table S3−5). All the IgG tests except the SARS-CoV-2 NP IgG ELISA Kit from ImmunoDiagnostic limited presented a specificity equal or above 97 %. Noteworthy, none of the IgG test has shown specific cross reactivity with sera from patients documented with a positive RT-PCR for Human seasonal coronavirus E229, OC43, HKU1, and NL63.
Fig. 1.
Preliminary evaluation: Sensitivity at 0-5, 6-10, 11-15 and above 15 days post-symptoms. Specificity is indicated below each graph. Poor specificities are in red characters.
Fig. 2.
Preliminary evaluation. Comparison of the sensitivity and specificity.
The sensitivity is given for the sample above 15 days post symptoms.
The only IgM/IgA tests, exhibiting satisfying performances (sensitivity of at least 80 % and specificity around 95 % and higher) at least 15 days post-symptoms were the NADAL LFA (sensitivity: 94 %, 95 % CI:80−99; specificity: 99 %, 95 % CI:92–100), the Dynamiker LFA (sensitivity: 94 %, 95 % CI:79−99; specificity: 93 %, 95 % CI:84−97), and the CLIA from Snibe (sensitivity: 80 %, 95 % CI:63−90; specificity: 99 %, 95 % CI: 92–100) (Fig. 2, Table S3−5).
The other tests, Epitope Diagnostic (ED) IgM ELISA, Euroimmun IgA ELISA, Vircell IgM + IgA ELISA, ImmunoDiagnostic limited IgM ELISA demonstrated insufficient performances (Fig. 2, Table S3−5).
Concerning the LFA, IgG and IgM being tested simultaneously, both tests should give excellent results to be valuable. The Dynamiker IgG/IgM LFA is the only test respecting a sensitivity and specificity of more than 90 % for both Ig after 15 days post-symptoms.
Interestingly, we observed a simultaneous IgM and IgG response overtime for the tests with an IgM specificity above 90 % (Dynamiker LFA, NADAL LFA and Snibe CLIA) (Fig. 1).
3.2. Complete evaluation of 5 SARS-CoV-2 selected serologic tests
Following the preliminary evaluation, the ED IgG ELISA, the Dynamiker IgG/IgM LFA, the Diasorin IgG CLIA and the Snibe IgG and IgM CLIA tests were thus selected for further analyses based on i) sensitivity and specificity performance of the preliminary evaluation, ii) diversity of targeted antigens (anti-N: ED IgG ELISA and Dynamiker IgG/IgM; anti-S: Diasorin IgG CLIA, anti N+S: Snibe IgG/IgM CLIA) iii) availability of the kits at the later on 15th April 2020 in Switzerland, iv) specific detection of IgG and/or IgM or IgA and v) compatibility of the kits to most laboratory needs including median to low samples volumes per day and extended expiration days upon kits opening. For instance, despite its good performance, the ECLIA from Roche was not selected as it detects pan-Ig, which is not the most appropriate for infectious serology diagnostic. In addition, the 200 tests expired 2 days upon kit opening which requires large sample volumes.
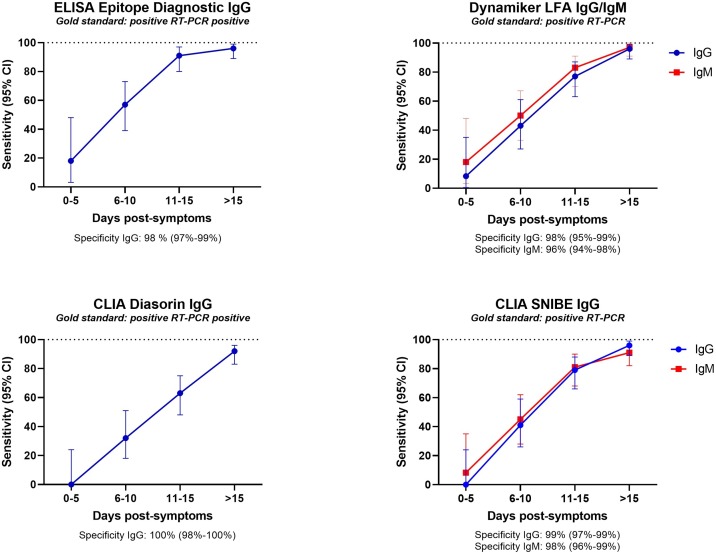
All the five selected tests were further evaluated on 65 positive and 335 negative sera to end-up with a global evaluation performed on 178 positive and 404 negative for a total of 586 sera (Fig. 3 , Table S1). The negative sera were chosen to assess possible cross-reactivity with human viral infections other than human coronavirus: Herpes simplex virus 1 and 2, Respiratory Syncytial Virus, Epstein-Barr virus, Cytomegalovirus, Mumps virus, Measles virus, Parvovirus B19, Rubella virus, Tick-borne encephalitis virus, Influenza A and B, Varicella-zoster virus, Human Immunodeficiency virus, Hepatitis virus A, B, C, D, and E, and some rheumatoid factors, or auto-antibodies (anti-PR3, -PR4, SCL70, SCL71).
Fig. 3.
Complete evaluation: Sensitivity at 0-5, 6-10, 11-15 and above 15 days post-symptoms. Specificity is indicated below each graph.
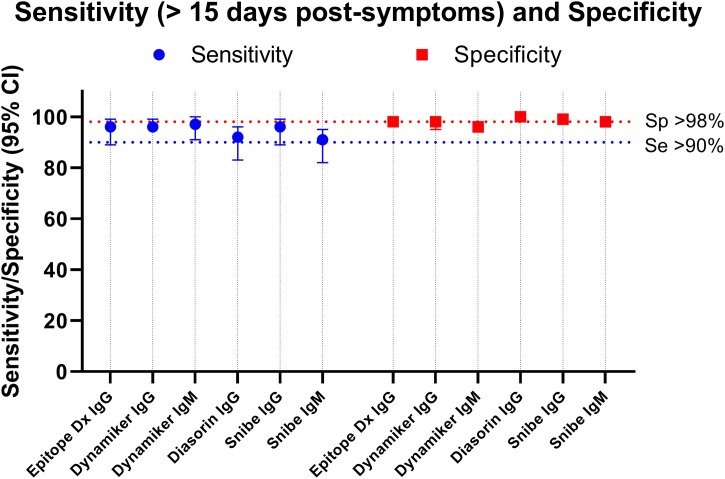
The four IgG tests demonstrated good sensitivity (≥96 %) and specificity (≥98 %) performances at more than 15 days post-symptoms, except the Diasorin ISON® SARS-CoV-2 IgG CLIA kit that showed a sensitivity of 92 % (95 % CI:83−96) but with a specificity of 100 % (Fig. 4 , Table S6). The IgM tests exhibited a sensitivity of 91 % (95 % CI:95−92), and a specificity of 98 % (95 % CI:96−99) for the CLIA Snibe IgM test with a sensitivity of 97 % (95 % CI:91−100) and a specificity of 96 % (95 % CI:94−98) for the LFA Dynamiker IgM test (Fig. 4, Table S6).
Fig. 4.
Complete evaluation: Comparison of the sensitivity and specificity.
The sensitivity is given for the sample above 15 days post symptoms.
3.3. Semi-quantitative antibody production
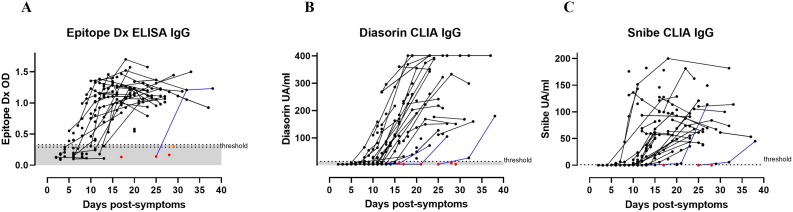
We analyzed the development of the IgG semi-quantitative response overtime post-symptoms with the ED ELISA targeting the anti-N response, the Diasorin CLIA targeting the anti-S1-S2 domains of the S protein, and the Snibe CLIA targeting both N and S proteins (Fig. 5 ). Among the 178 positive sera, several were sampled from the same patient overtime. We could observe that one patient with ED ELISA, three patients with Diasorin CLIA and two patients with Snibe CLIA became positive for anti-SARS-CoV-2 IgG more than 15 days post-symptoms (Fig. 5). Thus, with the Diasorin CLIA, one patient was negative for two consecutive sera collected at days 17 and 21 post-symptoms and became positive only at day 27 post-symptoms and another patient was negative at 16 days post-symptoms and became positive only 23 days post-symptoms. With the Snibe CLIA, one patient was negative at day 13 post-symptoms and became positive only at day 21 post-symptoms. One patient became positive with all test only 32 days post-symptoms.
Fig. 5.
Kinetic of antibody production per patient.
False negative results above 15 days post-symptoms are indicated with red dots and borderline results with orange dots. Patients with several collected sera are represented by connecting lines. Blue lines represent patients that became positive for anti-SARS-CoV-2 IgG more than 15 days post-symptoms.
4. Discussion
To our knowledge, this study is the first one comparing so many different serologic tests for SARS-CoV-2 diagnostic using different technologies, LFA, ELISA, CLIA and ECLIA, on sera from RT-PCR positive patients collected over one to two months post-symptoms, and assessing 20 possible cross reactions with other viral infections.
This large evaluation of 17 SARS-CoV-2 serological tests highlights in the preliminary phase that among eight IgG/pan-Ig ELISA or CLIA/ECLIA tests, five were recommended with a combined sensitivity above 90 % and specificity above 98 % (Epitope Diagnostic, Vircell and Creative Diagnostic IgG ELISA tests, Snibe IgG CLIA and Roche pan-Ig ECLIA tests), and that on six IgM/IgA ELISA or CLIA/ECLIA tests, only one (Snibe IgM CLIA) was acceptable with a combined sensitivity above 80 % and specificity of 99 %.
Concerning LFA, only one test showed good performances for both IgG and IgM, showing that a thorough evaluation is absolutely required before use especially, outside referenced diagnostic laboratories.
For all different tests, we observed that IgM and IgG aroused concomitantly, as already described for SARS-CoV-1 and 2 infection [5,12,13]. However, other studies demonstrated a higher sensitivity of IgM than IgG detection during the first 14 days post-symptoms [14]. It is therefore difficult to determine if the difference observed is due to a difference in IgM and IgG kinetic response between the different cohorts or if it is due to the performances of the different kits used in the different studies. In any case, this suggests that the use of IgM for the sero-diagnostic of SARS-CoV-2 acute/subacute infection might be difficult either due to physiological or technical limitations. Thus, SARS-CoV-2 RT-PCR remains the test of choice for early diagnostic a few days after symptoms onset [15].
Another interesting observation is the heterogeneity of the patient responses, with some of them responding very lately more than 25 day post-symptoms. This delayed response might be related to the immune status of the patients or to the severity of the infection as some preliminary studies tend to show that pauci-symptomatic patients have lower and delayed antibody response [10,13,14,16]. More systematic clinical and population studies need to be performed to clearly correlate the amplitude and time of the antibody response with i) the severity of the disease, ii) the demographic and clinical data and iii) the immune status of the patients.
In our hand, the anti-N and anti-S antibodies were both detectable during the acute phase of the CoviD-19 disease for hospitalized patients. Previous studies on other coronavirus [4], or on SARS-CoV-2 [16] suggest that the anti-N antibody response may appeared earlier or simultaneously than the anti-S response and may also waned more rapidly after few months [17]. In contrast, other studies focusing also on severe patient in acute phase of the disease demonstrated an anti-RBD response earlier than the anti-N response [13,18]. As we did not evaluate serological tests targeting anti-RBD only, we cannot exclude that antibodies targeting exclusively the RBD subunit of the S proteins arise earlier than the antibodies targeting other epitopes of this protein. In addition, the difference of kinetics observed between anti-S or anti-N might be essentially due to the performance of the different tests during the first month post-symptoms.
This study has identified several SARS-CoV-2 tests exhibiting very good sensitivity and specificity from sera collected from hospitalized patients up to 30–60 days post-symptoms. Parallel works performed in our laboratory on seroprevalence cohort [19] or on routine patients [20] suggest that the serology of SARS-CoV-2, completed with good quality tests, may be used for seroprevalence studies [19] and, in several clinical and epidemiological assets to confirm or exclude a CoviD-19 disease including i) suggestive clinical symptoms with two consecutive negative RT-PCR, or ii) suggestive clinical symptoms with discordant RT-PCRs, iii) infectious control settings for hospitalized patients presenting more than 20 days old suggestive clinical symptoms, iv) CoviD-19 atypical clinical presentations (Guillain-Barré syndrome, meningo-encephalitis, cutaneous vasculitis, Kawasaki disease, diarrhea,…) with negative RT-PCR and v) pre-transplantation or pre-chemotherapy screening [20].
In this study, we have clearly identified robust SARS-CoV-2 serological tests for the diagnosis of patients presenting a moderate to severe CoviD-19 diseases during acute and early sub-acute phase with a limited usefulness of IgM for the serologic diagnostic of SARS-CoV-2. Our conclusion can thus only be applied to this type of patients.
Additional evaluation studies of these SARS-CoV-2 anti-N and anti-S serological tests need to be performed to assess the performance of these tests on sera collected from non-hospitalized pauci-symptomatic patients, from SARS-CoV-2 RT-PCR positive patients but more than two month post exposure, or from symptomatic SARS-CoV-2 negative RT-PCR patients non tested here for ethical reasons.
Finally, complementarity of both kind of tests (anti-N and anti-S) has clearly to be envisaged and evaluated to fulfill the best performances as they might be complementary and might permit to reach the best sensitivity and specificity for both clinical and large seroprevalence population studies.
Author’s contribution
ATC and AC wrote the first draft. All authors critically reviewed the manuscript.
Ethical statement
This study was evaluated by our Ethics Committee (CER-VD) and they judged that it did not deserve a specific approval being only a quality assessment of diagnostic tests.
Funding information
No external funding was necessary to write this study.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We warmly thank all the members of the laboratory of serology of the CHUV, Sarah Chappuis, Fabienne Di Paola, Carine Pintér Pont, Claudine Gostely, Emilie Rüegger, Sylvie Caillon-Bouchez, Marijana Vujica, Alexandre Mamin, and Benjamin Gayton, for their wonderful implication in this project, and the set-up of SARS-CoV-2 serologic testing in our hospital. We would like to thank the laboratory of immunology to provide us some sera especially those positive for Lupus, HIV, hepatitis, rheumatoid factors, and auto-antibody. We thank Pr Marchetti Oscar and Pr Pache Antoine from the Ensemble Hospitalier de la Côte, Morges, Switzerland, Dr Caroline Chapuis-Taillard and Dr De Vallière Serge from Clinique de la Source, Lausanne, Switzerland and, Dre Aurélie Jayol-Virely from Clinical laboratory of the Etablissement Hospitalier Nord-Vaudois, Yverdon, Switzerland, to have shared information about the symptoms date of some patients.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2020.104690.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22(2):69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang D., Comish P., Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16(5):e1008536. doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer B., Drosten C., Muller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu M. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7(5-6):882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Premkumar L. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai W. Identification of SARS-CoV RBD-targeting monoclonal antibodies with cross-reactive or neutralizing activity against SARS-CoV-2. Antiviral Res. 2020;179:104820. doi: 10.1016/j.antiviral.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Infantino M. Serological assays for SARS-CoV-2 infectious disease: benefits, limitations and perspectives. Isr. Med. Assoc. J. 2020;22(4):203–210. [PubMed] [Google Scholar]
- 10.Okba, N.M.A., et al., 2020.
- 11.Walls A.C. Structure, function, and Antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo P.C. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin. Diagn. Lab. Immunol. 2004;11(4):665–668. doi: 10.1128/CDLI.11.4.665-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.To K.K.-W. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caruana G. Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. CMI. 2020 doi: 10.1016/j.cmi.2020.06.019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun B. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chia W.N. Serological differentiation between COVID-19 and SARS infections. Emerg. Microbes Infect. 2020:1–23. doi: 10.1080/22221751.2020.1780951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amanat F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenwick C. Changes in SARS-CoV-2 Spike versus Nucleoprotein Antibody Responses Impact the Estimates of Infections in Population-Based Seroprevalence Studies. J. Virol. 2020 doi: 10.1128/JVI.01828-20. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coste A.T. Indication for SARS-CoV-2 serology: first month follow-up. Clin. Microbiol. Infect. Dis. 2020 In press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.