Abstract
The reduction in childhood mortality noted in trials investigating azithromycin mass drug administration (MDA) for trachoma control has been confirmed by a recent large randomized controlled trial. Population-level implementation of azithromycin MDA may lead to selection of multiresistant pathogens. Evidence suggests that repeated azithromycin MDA may result in a sustained increase in macrolide and other antibiotic resistance in gut and respiratory bacteria. Current evidence comes from standard microbiological techniques in studies focused on a time-limited intervention, while MDA implemented for mortality benefits would likely repeatedly expose the population over a prolonged period and may require a different surveillance approach. Targeted short-term and long-term surveillance of resistance emergence to key antibiotics, especially those from the World Health Organization Access group, is needed throughout any implementation of azithromycin MDA, focusing on a genotypic approach to overcome the limitations of resistance surveillance in indicator bacteria.
Keywords: macrolide, azithromycin, mass drug administration, antimicrobial resistance, surveillance
Azithromycin mass drug administration results in a sustained increase in antimicrobial resistance when implemented at a population level. Targeted risk-based metagenomics approaches complementing traditional microbiological methods are recommended for surveillance of emerging short- and long-term antimicrobial resistance.
Intermittent Childhood Azithromycin Mass Drug Administration in Sub-Saharan Africa: Current Indications and Supporting Evidence
The most frequent indication for azithromycin (AZM) mass drug administration (MDA) across Africa is endemic trachoma [1]. In 1997, the World Health Organization (WHO) established the Global Alliance for the Elimination of Blinding Trachoma by 2020 (GET 2020), and there is clear evidence that single-dose AZM MDA reduces the prevalence of active trachoma and ocular infection [2]. A reduction in childhood mortality was observed in studies of AZM MDA for trachoma in the sub-Saharan setting [3, 4]. The MORDOR (Macrolides Oraux pour Réduire les Décès avec un Oeil sur la Résistance) I study (clinicaltrials.gov, NCT02048007) [4] was specifically designed to investigate any potential mortality benefit. The study assigned communities in Malawi, Niger, and Tanzania to 4 twice-yearly MDA rounds of either 20 mg/kg per dose of oral AZM or placebo. This cluster-randomized controlled trial demonstrated a reduction in all-cause mortality in under-5-year-old children of 14% in the treatment group [4]. Mortality reduction (18%) was observed most clearly among infants in Niger and those who were less than 6 months of age, with the highest mortality rate at baseline. Extension for 2 more rounds during MORDOR II did not show significant evidence of a waning effect of AZM MDA on childhood mortality [5]. In communities that received placebo originally, childhood mortality decreased after receipt of AZM [5].
The emergence of antibiotic resistance linked to antibiotic MDA could be a barrier to widespread implementation. There are concerns that AZM MDA will lead to selection of macrolide-resistant strains of Chlamydia trachomatis and resistance to macrolides and other classes of antimicrobials in other pathogens. Here, we discuss these concerns and propose a strategy to monitor emerging antimicrobial resistance (AMR) alongside the implementation of AZM MDA for the prevention of childhood mortality in sub-Saharan Africa.
Anticipated Antimicrobial Resistance and Microbiome Changes Associated With Azithromycin Use
Macrolides bind to the 23S ribosomal RNA (rRNA) of the 50S ribosomal subunit and inhibit protein synthesis. Resistance occurs by alteration of the target, active efflux, and antibiotic inactivation [6, 7]. It can be selective for the 14- and 15-membered macrolides (erythromycin, clarithromycin, azithromycin; M phenotype) or be relevant for the 16-membered macrolides (spiramycin, josamycin), lincosamides (clindamycin), and streptogramin B (MLSB phenotype) [8]. M-type resistance is mediated by chromosomally (mef) or plasmid-encoded (msrA) macrolide efflux genes [9–11] and generally confers low-level resistance among streptococci, whereas MLSB resistance is caused by methylation of the 23S rRNA, which blocks the ribosomal binding site and commonly confers high-level resistance [8]. The methylase is encoded by erm (erythromycin ribosome methylase) genes. This phenotype can be constitutive (MLSB-C) or inducible (MLSB-I) [6–8]. Highly macrolide-resistant Streptococcus pneumoniae isolates that have both erm and mef resistance mechanisms are increasingly reported [8].
Pneumococcal lineages that harbor multiple antibiotic-resistance determinants also show a higher degree of mosaicism in housekeeping genes [12]. This facilitates horizontal gene transfer from genetically related organisms, such as viridans streptococci, and increasing exposure to co-colonizing resistant bacteria. The final result may be more interstrain homologous-recombination events with the incorporation of resistance determinants for β-lactams, fluoroquinolones, and co-trimoxazole in the core genome or on integrative transposable elements for macrolides, lincosamides, tetracycline, and chloramphenicol. These data highlight the importance of the commensal oral flora as a reservoir of macrolide resistance determinants from holistic metagenomic studies [13].
Macrolides are also expected to affect gram-negative Enterobacteriaceae, which are known to harbor various mobile genetic elements (MGEs) [14] and serve as a reservoir for antibiotic-resistance genes in the gut [15]. The acquisition of novel genes by plasmids through MGEs such as transposons or insertion sequences, and their ability to replicate in a wide range of bacterial hosts, makes them perfect vectors for the spread of antimicrobial resistance [16]. Unrelated to macrolide use, such resistance evolution is best described in gram-negative bacteria where extended-spectrum β-lactamases (ESBLs) are frequently associated with co-resistance to aminoglycosides and fluoroquinolones [17]. Selection of these isolates may be driven by a single antibiotic resulting in resistance to multiple unrelated antibiotics. Azithromycin is considered a potent potential driver in the selection of such co-resistance because of its very long-elimination half-life of more than 50 hours, high intracellular and prolonged tissue concentration, prolonged rate of dissociation from the ribosomal target with a prolonged postantibiotic effect, large volume of distribution resulting in possible long-term effects in various body compartments, and better activity against common gram-negative bacteria compared with other macrolides [8, 15, 18]. While evidence linking AZM use to the emergence of resistance in gram-negative bacteria is sparse, there is a clear need for active surveillance in the context of AZM MDA.
Co-resistance and co-selection processes driving AMR may additionally be compounded by microbiome impacts if alterations in the microbiome result in a predominance of resistance-gene–carrying organisms. The gut as a reservoir for antibiotic-resistance genes can be disturbed by antibiotics in its composition and function as well as selecting for antibiotic-resistant microbes [19]. Several studies have evaluated the effects of antibiotic exposure on the pediatric gut microbiome diversity, showing variable results [20–24]. In general, these studies found reductions in observed richness and Shannon diversity during or shortly after AZM exposure. Once antibiotic treatment is stopped, the microbiota may display a certain degree of resilience, being capable of reverting to near their pre-exposure composition after many months [24]. However, complete recovery to the initial state may not occur or be age dependent, particularly in the context of repeated antibiotic insults during vulnerable time periods of age [23, 25]. Overall, AZM may cause important changes in the human gut microbiome, but the effects on antimicrobial resistance of these shifts remain unclear.
Evidence Summary on Antimicrobial Resistance Following Azithromycin Mass Drug Administration in Sub-Saharan Africa
A recent systematic review of antimicrobial resistance following AZM MDA for trachoma by O’Brien et al [26] identified that this approach selects for macrolide resistance in some potentially pathogenic organisms, with a possible population-level dose–response resulting in increased resistance selection as the number of distribution cycles increases (Supplementary Table 1). Antibacterial resistance emergence has also been seen in the MORDOR I trial (12.3% vs 2.9% of children carried macrolide-resistant pneumococci in communities receiving AZM vs placebo) [19]. When antibiotic selection pressure is removed, the prevalence of resistance may return to baseline levels over time, although most studies followed populations for 6 months or less, and results were mixed in studies with shorter follow-up periods [26]. About half of studies evaluating AMR after AZM MDA did not measure baseline antibiotic resistance in the target pathogens, making it difficult to prove that AZM MDA caused observed changes. Streptococcus pneumoniae in nasopharyngeal samples was the main target organism of most studies, with less focus on other organisms, such as Escherichia coli (stool samples) or Staphylococcus aureus (nasopharyngeal samples). Most the studies came from Africa, with the reported resistance data collected between 1995 and 2017 from longitudinal cohort studies or (repeated) cross-sectional studies except for Skalet et al [27] and Keenan et al [28], which were randomized controlled trials (RCTs).
Impact of Different Techniques Determining Antimicrobial Resistance
Most studies determined AMR by phenotypic susceptibility testing using Etest (Epsilometer test, agar diffusion with E-strips) or disk diffusion [26]. Only in 3 studies were molecular methods applied (such as multilocus sequencing [29], targeted polymerase chain reaction [28], or DNA microarray [30] for detection of, eg, mef or erm genes). Most of the data generated are presented as the percentage of isolates of a given organism that are resistant to a specific antibiotic. Such data are readily available and easily interpreted, but may not be the optimal method by which to measure changes in resistance from the public health perspective, in particular changes brought about by antibiotic use [31]. When evaluating the burden of resistance, the density of resistant isolates expressed as rates should be assessed—that is, the absolute number of resistant isolates in an at-risk population over time [31].
Ongoing Clinical Studies/Trials
There are currently 20 actively recruiting or about to recruit RCTs investigating AZM treatment in the target population registered in ClinicalTrials.gov (Table 1). In 3 cases the trialed AZM treatment course includes more than 1 single dose. Five studies are associated with the MORDOR trial [4]. Six trials specify that resistance will be assessed in respiratory or gut bacteria with a variety of microbiological techniques used. An additional 7 trials intend to investigate impacts on the nasopharyngeal or gut microbiome without specific assessments of antibiotic resistance. Finally, 6 trials are not planning to evaluate AMR or are limited to the target pathogen for the intervention (Chlamydia trachomatis or Treponema pallidum ssp. pertenue).
Table 1.
Ongoing Studies Addressing Research Questions of Antimicrobial Resistance After Macrolide Mass Treatment
| NCT Number | Country | Target Disease | Part of MORDOR | Type of Study | Target Age Group | Number of AZM Doses/ Course | Number of AZM Courses | Microbiology Endpoints | Genotypic Methods Used |
|---|---|---|---|---|---|---|---|---|---|
| NCT03683667 | Bangladesh | Malnutrition/ stunting | No | cRCT | 6-12-mo-old children | 1 | 2 (6 and 9 mo) | Enteropathogen burden (7× at age 6–18 mo), gut microbiota composition (as above), AMR of E. coli and S. pneumoniae at 6, 9, 12, 15, and 18 mo of age in participating children | qPCR, 16S ribosomal RNA sequencing |
| NCT03682653 | Burkina Faso | Mortality | Yes | RCT | 8-27-d-old children | 1 | 1 (during newborn period) | None specified | … |
| NCT03676764 | Burkina Faso | Mortality | Yes | cRCT | 1-60-mo-old children and those receiving first DTP vaccine (5-8-wk-old children) | 1 | 1 and 2×/y for older children | Carriage of S. pneumoniae and nasopharyngeal macrolide resistance at 36 mo postexposure, proportion of E. coli resistant to macrolides and other key antibiotics at 36 mo postexposure, microbial diversity in the nasopharyngeal and intestinal microbiome at 36 mo postexposure | Next-generation sequencing (not further specified) |
| NCT03676751 | Burkina Faso | Growth and development | Yes | RCT | 8-d to 59-mo-old children | 1 | 1 | Intestinal microbial diversity at 6 mo postexposure | Targeted PCR and next-generation sequencing (not further specified) |
| NCT03676140 | Papua New Guinea | Trachoma/NTD | No | cRCT | Persons older than 5 y of age in randomized communities | 1 | 1 | None specified | … |
| NCT03570814 | Ethiopia | Trachoma/NTD | No | cRCT | Persons older than 5 y of age in randomized communities | 1 | 1 | None specified | … |
| NCT03568643 | Niger | Malnutrition/ stunting | No | RCT | 6–59-mo-old children | 1 | 1 | None specified | … |
| NCT03564652 | Pakistan | Malnutrition/stunting | No | RCT | Pregnant women, infants 42 d of age | 1 | 1 | Enteropathogen burden at 40–42 and 56 d of age | Multiplex PCR and metagenomics (not further specified) |
| NCT03523156 | Ethiopia | Trachoma | No | cRCT | 6-mo- to 9-y-old children | 1 | 1 (MDA annual) or 3 (MDA annual plus 2× targeted) | None specified beyond chlamydial infections (not AMR) | … |
| NCT03490123 | Papua New Guinea | Yaws | No | cRCT | Older than 6 mo | 1 | 3 | Macrolide resistance in T.p. pertenue | … |
| NCT03474276 | Madagascar, Niger, CAR, Senegal | Malnutrition/stunting | No | RCT | 6–24-mo-old children | 3 | 1 | Comparison of OTU composition of stool according to nutritional status (at baseline and 3 and 6 mo postexposure) | Next-generation sequencing (detailed description) |
| NCT03338244 | Original MORDOR sites | Mortality | Yes | cRCT | 1–60 mo of age | 1 | 2×/y | Macrolide resistance 18 mo postexposure in nasopharyngeal and rectal swabs, microbial composition of stool at 18 mo, enteropathogen burden at 18 mo | Metagenomic deep sequencing for microbial composition; resistance detected by standard phenotypic methods |
| NCT03335072 | Ethiopia | Trachoma | No | cRCT | All persons in randomized communities eligible for MDA according to WHO guideline | 1 | 4×/y | None specified beyond chlamydial infections (not AMR) | … |
| NCT03268902 | Tanzania | Malnutrition/stunting | No | RCT | Up to 14 d old | 1 | 6, 9, 12, and 15 mo | Enteropathogen burden (5× between 6 and 18 mo), intestinal microbiota composition (4× between 6 and 18 mo) | Not specified |
| NCT03199547 | The Gambia and Burkina Faso | Neonatal sepsis | No | RCT | Women in labor | 1 | 1 | EONS (culture confirmed) and LONS (culture confirmed) | … |
| NCT03187834 | Burkina Faso | Growth and development | No | cRCT (households) | 6–59-mo-old children | 5 | 1 | Nasopharyngeal and intestinal microbiome (day 9 postexposure) | DNA sequencing (not further specified) |
| NCT03032042 | Ethiopia | Helminthic infection | No | RCT | 1–60 mo of age | 1 | 1 | Microbial diversity in intestinal microbiome 7 d postexposure | … |
| NCT02754583 | Ethiopia | Trachoma | No | RCT | All persons in randomized communities | 1 (MDA, annual), 1 (targeted) | 1 (MDA, annual), 4 (quarterly) | Nasopharyngeal pneumococcal macrolide resistance (12, 24, 36 mo postexposure), intestinal microbiome at 12 mo postexposure (substudy) | Not specified |
| NCT02414399 | Kenya | Mortality | No | RCT | 1–59 mo of age | 5 | 1 | Prevalence of enteric pathogen and pneumococcal carriage (6 mo postexposure), proportion of β-lactam or macrolide resistance or both (6 mo postexposure) | … |
| NCT02048007 | Malawi, Niger, and Tanzania | Mortality | Yes | cRCT | 1–60 mo of age | 1 | 2×/y | Pneumococcal macrolide resistance at 24 and 48 mo, macrolide resistance (genetic) in stool and nasopharynx at 24 and 48 mo, carriage of resistant pneumococcus at 6 to 24 mo, proportion of rectal/stool isolates and E. coli isolates resistant to macrolides and other antibiotics at 6 to 24 mo, MRSA (NP) at 24 mo, carriage of S. aureus resistant to macrolides and other antibiotics at 6 to 24 mo, various deep-sequencing endpoints | Metagenomics (not further specified) |
Abbreviations: AMR, antimicrobial resistance; AZM, azithromycin; CAR, Central African Republic; cRCT, cluster-randomized controlled trials; DTP, diphtheria, pertussis, tetanus vaccine; E. coli, Escherichia coli; EONS, early onset neonatal sepsis, LONS, late-onset neonatal sepsis; MDA, mass drug administration; MORDOR, Macrolides Oraux pour Réduire les Décès avec un Oeil sur la Résistance; MRSA, methicillin-resistant Staphylococcus aureus; NCT, national clinical trial; NP, nasopharynx; NTD, neglected tropical disease; OTU, Operational Taxonomic Unit; PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction; RCT, randomized controlled trial; S. aureus, Staphylococcus aureus; S. pneumoniae, Streptococcus pneumoniae; T.p. pertenue, Treponema pallidum ssp. pertenue; WHO, World Health Organization.
Surveillance Strategies for Antimicrobial Resistance During Continuous Azithromycin Mass Drug Administration
Genotypic Versus Phenotypic Testing of Antimicrobial Resistance
Although phenotypic methods remain the cornerstone of clinical antimicrobial susceptibility testing, molecular characterization of AMR determinants is being considered for local, national, or even global surveillance of AMR [32]. In 2015, the WHO launched the Global Antimicrobial Resistance Surveillance System (GLASS) in order to standardize the collection of data on AMR for global planning, prevention, and intervention programs [33]. Reports to GLASS currently rely on detection of phenotypic resistance; however, in the future, GLASS may incorporate the results of molecular testing for AMR detection by appropriate methods. Molecular diagnostic methods can be used together with phenotypic testing to yield additional information, provided that the most appropriate molecular AMR tests relevant to the setting are used. There has been a dramatic reduction in cost and an increase in the quality and availability of whole-genome sequencing (WGS), making this technology gradually more accessible for routine scientific use but also for clinical diagnostics and surveillance. In the following section, we discuss advantages and disadvantages of genotypic versus phenotypic surveillance of antimicrobial resistance in the context of AZM MDA (Table 2).
Table 2.
Advantages and Disadvantages of Genotypic Versus Phenotypic Surveillance of Antimicrobial Resistance
| Phenotypic/Susceptibility Testing Methodsa | Genotypic Methodsb | ||
|---|---|---|---|
| Advantages | Disadvantages | Advantages | Disadvantages |
| Easy access globally (?) | Select for indicator bacterial organisms and largely ignore nonpathogenic bacterial species | Yield data about any resistance gene or mutation present | Insufficient knowledge about all genetic variation may complicate accurate prediction of resistance [34] |
| Low costs | Rely on bacterial growth, ie, time-consuming | Can be performed directly on clinical specimens not relying on bacterial growth, ie, faster turnaround times | Quality controls essential to assess whether WGS data have reached a suitable standard, while there are currently no international standards for QC thresholds to use for assessing quality [34] |
| Guidelines available to apply and teach interpretation of results (capacity building) | Screening of a limited number of (known) resistance genes | Meta-transcriptomic analysis can determine the expression of resistance genes at the moment of sampling | Need for standardized comprehensive databases containing the relevant DNA or protein sequence targets known to be associated with AMR [32, 34] |
| Limit possible conclusions about co-transmission of resistance genes and relatedness of identified isolates to reconstruct transmission networks | Appropriate bioinformatic methodologies needed to accurately extract relevant information from WMGS data based on target databases [32] | ||
| Limited opportunities to compare genotype with phenotype | High costs (mainly related to the complex bioinformatics infrastructure) | ||
Abbreviations: AMR, antimicrobial resistance; QC, quality control; WGS, whole-genome sequencing
aPhenotypic methods: agar and broth microdilution (the latter being the reference standard) or disc diffusion, followed by interpretation according to agreed guidelines.
bGenotypic methods: metagenomics; PCR assays are not included as they provide valid information on AMR determinants known to be associated with the identified pathogen, but they are not suitable for detecting completely new genes families, novel genes, or new point mutations.
Whole-metagenome Sequencing To Detect Antimicrobial Resistance Genetic Determinants: Opportunities and Challenges
Traditional microbiology relies upon clonal cultures that select for dominant bacterial species/strains and largely ignore nonpathogenic bacterial species, and this approach has also been used for AMR surveillance. In routine clinical care, culturing of more than a few selected “indicator” organisms is generally difficult for logistical reasons (especially in clinical specimens with a high bacterial load such as stool samples) and may not be helpful in the optimization of patient care. Early sequencing examined specific genes such as the 16S rRNA gene and revealed the microbial biodiversity that had been missed by culture-based methods. Nonpathogenic “commensal” bacteria serve as an antibiotic resistance reservoir and must be addressed since these microorganisms may gain, maintain, and deliver genes to other microorganisms [15]. Indeed, many of the clinically relevant resistant bacteria are believed to originate from the environment, together constituting a large and almost unexplored resistance reservoir [35]. For example, Devirgiliis et al [36] reported on AMR in foodborne Lactobacillus and Lactococcus species, 2 genera of lactic acid bacteria that often represent the dominant bacterial population in breastfed infants. Different Lactobacillus species were shown to transfer erythromycin- and tetracycline-resistance genes to Enterococcus faecalis, indicating a potential risk of using lactic acid bacteria starters that have not been tested for the absence of AMR genes.
Recent studies, especially in Africa, have predominantly used 16S metagenomics to determine taxonomic profiling and describe community composition (diversity and abundance) [24]. Alternatively, a shotgun metagenomics approach can be used to directly detect antibiotic-resistance genes in samples of interest, potentially indicating the impact of an exposure like AZM MDA on the microbial resistance landscape. Arguably, this would be highly relevant for public health as an “early warning” system compared with the slower expected AMR changes in indicator pathogens from routine microbiology samples for invasive disease, if available.
Extracting the relevant information to detect genetic determinants related to AMR from whole-metagenome sequencing (WMGS) data encounters 2 main challenges: (1) access to comprehensive databases containing the relevant DNA or protein sequence targets and (2) application of appropriate bioinformatic methodologies to accurately extract the relevant information from WMGS data based on these target databases [32]. This is further complicated by the fact that many genetic mechanisms can result in a given AMR phenotype without easy decision rules for prediction of their correspondence. As a consequence, many of the bioinformatic tools to detect AMR genetic determinants rely on target databases containing well-defined genes or specific single-point mutations, where a strong correlation between the genetic determinant and a given phenotype exists and can be extracted from either published peer-reviewed articles or from pre-existing archives such as the Antibiotic Resistance Gene Database (ARDB) [32, 37, 38]. These databases are based on a priori data and are therefore not suitable for detecting completely new gene families, genes, or point mutations and have to be updated frequently. Such databases do not support the analysis of the large-scale, ecological sequence datasets required for AMR surveillance. Specifically tailored databases such as MEGARes (https://megares.meglab.org) could facilitate the characterization of AMR determinants in the context of large metagenomics studies [37].
Time Points, Target Population, and Target Genes of Antimicrobial Resistance Testing
One important limitation of many studies on AMR after AZM MDA is the lack of baseline resistance data in the target population. Clearly, a high prevalence of resistant pathogens before exposure to AZM MDA is a major additional risk factor for subsequent increases in antimicrobial resistance. One can imagine that the “trough” prevalence of resistance immediately before each round of MDA might progressively increase over several years. Hence, this is the key sampling time point for AMR, and similarly the key population are as yet unexposed children prior to the age of receipt of AZM MDA (as well as those who are the target population for ongoing MDAs) and their household contacts (Figure 1). Surveillance should take the approach of repeated cross-sectional sampling in target communities to establish population-level changes over time. To appropriately target AMR surveillance in the context of AZM MDA, healthcare workers delivering the intervention could also be responsible for sampling infants and their household members prior to each AZM administration. Alternatively, or if there are any additional populations of special interest, systematic sampling could be done during healthcare visits for routine immunizations in children [39] or pregnant women visiting antenatal clinics. To assess 2 large microbial reservoirs, sampling should pragmatically focus on the nasopharynx and stool. Using a metagenomics approach enables to directly target and detect antibiotic-resistance genes (instead of target organisms) in samples of interest. This is especially important in the context of AZM treatments, as genes mediating macrolide resistance are mainly found on transferable genetic elements such as plasmids. To determine the required sample size for ongoing active surveillance, baseline prevalence of the target genes must first be assessed, as this will enable definition of a meaningful level of change that would be desirable to detect. Antimicrobial resistance changes in indicator pathogens from routine microbiology samples obtained from diseased individuals are expected to occur more slowly and are not feasible to reliably collect and proces in many low- and middle-income-country settings and will be less suitable for the goal of timely identification of AZM MDA impacts. However, parallel tracking of relevant changes in such isolates is important to confirm that observations from colonizing isolates are clinically relevant.
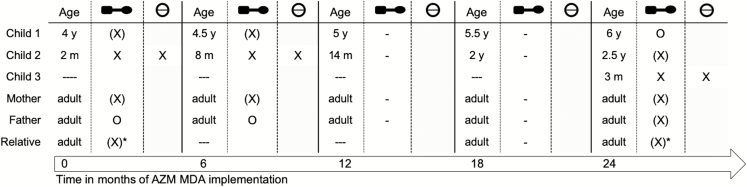
Figure 1.
The targeted recipients of azithromycin (AZM) mass drug administration are children up to 12 months of age. Relevant surveillance samples are nasopharyngeal and rectal swabs for all household members to be obtained before AZM administration to target recipients. X: AZM recipient and directly exposed individual for nasopharyngeal and rectal swabs. (X): household contact of AZM recipient providing nasopharyngeal and rectal swabs. (X)*: temporary household member. O: household member absent on day of sampling and AZM distribution. Abbreviations: AZM, azithromycin; m, month(s); MDA, mass drug administration; y, year(s).
Summary: Potential Strategies for Antimicrobial Resistance Surveillance
In general, pre-MDA “trough” prevalence of resistance is a key indicator. All samples, from MDA recipients and household contacts (representing indirect impacts of AZM MDA, presumably through community transmission), should be obtained immediately prior to AZM administration.
Young age is most relevant for invasive disease and, for example, pneumococcal carriage. Active surveillance should focus on infants and young children as well as their household contacts, and should be incorporated into implementation of AZM MDA or linked to routine health services contact.
Strengthening surveillance of invasive or clinical isolates of key pathogenic bacteria is desirable but is limited by local capacity, difficult to quality assure, and crucially expected to result in a small number of isolates and show the impact of AMR after a long lag time.
Alongside investments in routine microbiological capacity in regions for which AZM MDA for mortality benefits is relevant, capacity building for local sequencing-based active surveillance is desirable.
CONCLUSIONS
Azithromycin provides undisputed beneficial effects for the treatment of various infectious diseases; however, sparse evidence suggests that widespread and long-term exposure of children during MDA will promote macrolide and other antimicrobial resistance. For future studies or where AZM MDA is implemented as a regional or national policy, capacity building for monitoring of potential adverse AMR outcomes using both phenotypic and genotypic methods should be identified as an integral part of program delivery. This has the potential to strengthen local microbiology capacity while providing trends in genotypic resistance to key antibiotics to treat serious infections. Impacts on clinical isolates would be expected to be observed in the more distant future when the impact of AZM MDA may no longer be modifiable. Sampling of a baseline (“trough”) prevalence of AMR is a key indicator and will enable early consideration of steps to mitigate against changing resistance patterns while harnessing AZM MDA to prevent childhood mortality.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Note
Potential conflicts of interest. The authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Taylor HR, Burton MJ, Haddad D, West S, Wright H. Trachoma. Lancet 2014; 384:2142–52. [DOI] [PubMed] [Google Scholar]
- 2. Evans JR, Solomon AW. Antibiotics for trachoma. Cochrane Database Syst Rev 2011:Cd001860. [DOI] [PubMed] [Google Scholar]
- 3. Porco TC, Gebre T, Ayele B, et al. Effect of mass distribution of azithromycin for trachoma control on overall mortality in Ethiopian children: a randomized trial. JAMA 2009; 302:962–8. [DOI] [PubMed] [Google Scholar]
- 4. Keenan JD, Bailey RL, West SK, et al. ; MORDOR Study Group Azithromycin to reduce childhood mortality in sub-Saharan Africa. N Engl J Med 2018; 378:1583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keenan JD, Arzika AM, Maliki R, et al. Longer-term assessment of azithromycin for reducing childhood mortality in Africa. N Engl J Med 2019; 380:2207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leclercq R. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis 2002; 34:482–92. [DOI] [PubMed] [Google Scholar]
- 7. Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 2014; 12:35–48. [DOI] [PubMed] [Google Scholar]
- 8. Long S. Mechanisms and detection of antimicrobial resistance. In: Principles and Practice of Pediatric Infectious Diseases. 5th ed Philadelphia, PA: : Elsevier, 2018:1472–3. [Google Scholar]
- 9. Klaassen CH, Mouton JW. Molecular detection of the macrolide efflux gene: to discriminate or not to discriminate between mef(A) and mef(E). Antimicrob Agents Chemother 2005; 49:1271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reynolds E, Ross JI, Cove JH. Msr(A) and related macrolide/streptogramin resistance determinants: incomplete transporters? Int J Antimicrob Agents 2003; 22:228–36. [DOI] [PubMed] [Google Scholar]
- 11. Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother 1996; 40:1817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanage WP, Fraser C, Tang J, Connor TR, Corander J. Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 2009; 324:1454–7. [DOI] [PubMed] [Google Scholar]
- 13. Malhotra-Kumar S, Lammens C, Coenen S, Van Herck K, Goossens H. Effect of azithromycin and clarithromycin therapy on pharyngeal carriage of macrolide-resistant streptococci in healthy volunteers: a randomised, double-blind, placebo-controlled study. Lancet 2007; 369:482–90. [DOI] [PubMed] [Google Scholar]
- 14. Stokes HW, Gillings MR. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into gram-negative pathogens. FEMS Microbiol Rev 2011; 35:790–819. [DOI] [PubMed] [Google Scholar]
- 15. Gomes C, Martínez-Puchol S, Palma N, et al. Macrolide resistance mechanisms in Enterobacteriaceae: focus on azithromycin. Crit Rev Microbiol 2017; 43:1–30. [DOI] [PubMed] [Google Scholar]
- 16. Sheppard AE, Stoesser N, Wilson DJ, et al. ; Modernising Medical Microbiology (MMM) Informatics Group Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 2016; 60:3767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paterson DL. Resistance in gram-negative bacteria: Enterobacteriaceae. Am J Med 2006; 119(6 Suppl 1):S20–8; discussion S62-70. [DOI] [PubMed] [Google Scholar]
- 18. Peters DH, Friedel HA, McTavish D. Azithromycin: a review of its antimicrobial activity, pharmacokinetic properties and clinical efficacy. Drugs 1992; 44:750–99. [DOI] [PubMed] [Google Scholar]
- 19. Doan T, Arzika AM, Hinterwirth A, et al. ; MORDOR Study Group Macrolide resistance in MORDOR I—a cluster-randomized trial in Niger. N Engl J Med 2019; 380:2271–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yassour M, Vatanen T, Siljander H, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Trans Med 2016; 8:343ra381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Korpela K, Salonen A, Virta LJ, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 2016; 7:10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Trans Med 2016; 8:343ra382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shaw LP, Bassam H, Barnes CP, Walker AS, Klein N, Balloux F. Modelling microbiome recovery after antibiotics using a stability landscape framework. ISME J 2019; 13:1845–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wei S, Mortensen MS, Stokholm J, et al. Short- and long-term impacts of azithromycin treatment on the gut microbiota in children: a double-blind, randomized, placebo-controlled trial. EBioMedicine 2018; 38:265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 2015; 6:1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O’Brien KS, Emerson P, Hooper PJ, et al. Antimicrobial resistance following mass azithromycin distribution for trachoma: a systematic review. Lancet Infect Dis 2019; 19:e14–25. [DOI] [PubMed] [Google Scholar]
- 27. Skalet AH, Cevallos V, Ayele B, et al. Antibiotic selection pressure and macrolide resistance in nasopharyngeal Streptococcus pneumoniae: a cluster-randomized clinical trial. PLoS Med 2010; 7:e1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keenan JD, Chin SA, Amza A, et al. ; Rapid Elimination of Trachoma (PRET) Study Group The effect of antibiotic selection pressure on the nasopharyngeal macrolide resistome: a cluster-randomized trial. Clin Infect Dis 2018; 67:1736–42. [DOI] [PubMed] [Google Scholar]
- 29. Keenan JD, Klugman KP, McGee L, et al. Evidence for clonal expansion after antibiotic selection pressure: pneumococcal multilocus sequence types before and after mass azithromycin treatments. J Infect Dis 2015; 211:988–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bojang E, Jafali J, Perreten V, et al. Short-term increase in prevalence of nasopharyngeal carriage of macrolide-resistant Staphylococcus aureus following mass drug administration with azithromycin for trachoma control. BMC Microbiol 2017; 17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwaber MJ, De-Medina T, Carmeli Y. Epidemiological interpretation of antibiotic resistance studies—what are we missing? Nat Rev Microbiol 2004; 2:979–83. [DOI] [PubMed] [Google Scholar]
- 32. Anjum MF, Zankari E, Hasman H. Molecular methods for detection of antimicrobial resistance. Microbiol Spectr 2017; 5: ARBA-0011-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS). Available at: https://www.who.int/glass/laboratory/en/. Accessed 26 April 2019.
- 34. Ellington MJ, Ekelund O, Aarestrup FM, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST subcommittee. Clin Microbiol Infect 2017; 23:2–22. [DOI] [PubMed] [Google Scholar]
- 35. Berglund F, Österlund T, Boulund F, Marathe NP, Larsson DGJ, Kristiansson E. Identification and reconstruction of novel antibiotic resistance genes from metagenomes. Microbiome 2019; 7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Devirgiliis C, Zinno P, Perozzi G. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front Microbiol 2013; 4:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lakin SM, Dean C, Noyes NR, et al. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res 2017; 45:D574–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xavier BB, Das AJ, Cochrane G, et al. Consolidating and exploring antibiotic resistance gene data resources. J Clin Microbiol 2016; 54:851–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. World Health Organization; UNICEF. WHO and UNICEF estimates of immunization coverage: 2017 revision Available at: https://www.who.int/immunization/monitoring_surveillance/data/ner.pdf. Accessed 16 June 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.