ABSTRACT
Arabidopsis thaliana exhibits durable ‘non-host’ resistance against the hemibiotrophic fungal pathogen Colletotrichum tropicale that infects mulberry plants. Arabidopsis non-host resistance comprises two layers of defense: preinvasive and postinvasive resistance. The EDR1 protein kinase contributes to Arabidopsis preinvasive resistance against C. tropicale by inducing the expression of plant defensin (PDF) genes. Here we report that the expressions of multiple PDF genes were strongly induced in Arabidopsis upon invasion by C. tropicale. Invasion by a necrotrophic pathogen, Alternaria brassicicola, also induced PDF expression. Importantly, PDF expression triggered upon invasion by both pathogens was inhibited in edr1 mutants, indicating the requirement of EDR1 for PDF expression in postinvasive resistance by Arabidopsis. Analysis of ora59 mutants also revealed that this gene is critical for induced PDF expression following pathogen invasion. Furthermore, inoculation assays of A. brassicicola indicated that ORA59 is involved in postinvasive resistance against the pathogen, suggesting invasion-triggered PDF expression contributes to postinvasive resistance in Arabidopsis.
KEYWORDS: Fungal invasion, Arabidopsis thaliana, plant defensin
Non-host resistance can be defined as the immunity of an entire plant species against all tested isolates of a particular pathogen. It confers a durable plant defense mechanism against the majority of potential pathogens.1 Durability of non-host resistance depends on its multi-layered defense system, i.e., non-host resistance consists of both preinvasive resistance and postinvasive resistance. Colletotrichum tropicale isolate S9275 (hereafter called Ctro) is known to cause anthracnose disease in mulberry plants but is not adapted to Arabidopsis thaliana.2,3 In contrast to the adapted pathogen Colletotrichum higginsianum, which causes necrotic lesions on inoculated Arabidopsis leaves, nonadapted Ctro fails to induce lesions, indicating that Arabidopsis exhibits non-host resistance to this pathogen.3,4 PENETRATION2 (PEN2) and PEN3 are involved in preinvasive resistance (the control of pathogen entry) against Ctro when it uses a hyphal-tip entry mode in the presence of 0.1% glucose.3,5–7 PEN2 encodes an atypical myrosinase that hydrolyzes 4-methoxynidol-3-ylmethylglucosinolates as in planta substrate for antifungal responses.8,9 PEN3 encodes an ATP binding cassette (ABC) transporter. Genetic interaction analysis of pen2 and pen3 mutants suggested that PEN3 is likely involved in exporting toxic compounds including PEN2-catalyzed metabolites.10 However, whereas Ctro invades pen2 mutants successfully, these are still not fully susceptible to Ctro, because postinvasive resistance is activated and this terminates further pathogen growth.11 This observation highlights the importance of postinvasive resistance for plant survival against the attack of numerous pathogenic fungi.
Non-host preinvasive resistance toward Ctro also involves ENHANCED DISEASE RESISTANCE 1 (EDR1).12 EDR1 encodes a protein kinase homologous to the mitogen-activated protein kinase belonging to the Raf family.13 It was reported that the EDR1 protein kinase positively regulates the expression of antimicrobial plant defensin (PDF) genes in response to the entry of Ctro.12 Currently, it remains to be elucidated whether EDR1 regulates PDF expression during postinvasive defense.
Here we investigated the expression of PDF1.2a in Arabidopsis invaded by the nonadapted Ctro. Because Ctro starts to invade pen2 mutants at around 14 hours post inoculation (hpi) of Ctro with 0.1% glucose, whereas Ctro cannot invade WT (Col-0) plants,3,4, we measured the expression of PDF1.2a at 6 hpi as the preinvasive stage and 17 hpi as the postinvasive stage. At 17 hpi, when Ctro invaded pen2-1 but not WT plants, we found that PDF1.2a expression was highly induced in the pen2-1 mutant but not in the WT plants (Figure 1a). Compared with the mock-treated plants (plants treated with 0.1% glucose), PDF1.2a expression was induced by Ctro at 6 hpi in WT plants (Figure 1b), consistent with our previous finding.12 At 6 hpi, the PDF1.2a expression level in the pen2-1 mutant showed no clear difference difference from that in WT plants (Figure 1b). However, the induced level of PDF1.2a at 17 hpi in the pen2-1 mutant was much greater than at 6 hpi in the pen2-1 mutant (Figure 1a). PDF1.2a was also highly expressed in the pen2-2 mutant,5 the other allele of pen2 mutants, at 17 hpi by Ctro (Supplementary Figure 1A). We also investigated the expression of three additional PDF genes, PDF1.2b, PDF1.2c and PDF1.3 at 17 hpi with Ctro. We found that all tested PDF genes were highly expressed in pen2-1 at 17 hpi with Ctro (Supplementary Figure 1B).
Figure 1.
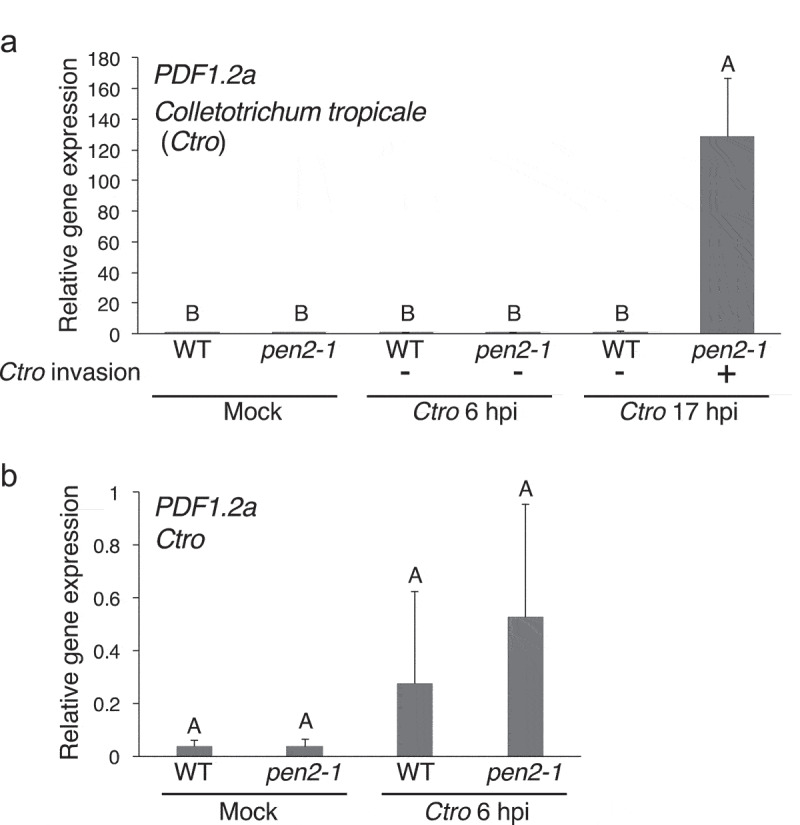
The expression of PDF1.2a was highly induced upon Ctro invasion. a. RT–qPCR analysis of PDF1.2a in Arabidopsis plants inoculated with Ctro. A conidial suspension from Ctro (5 × 105/mL with 0.1% glucose) was spray-inoculated onto 4- to 5-week-old Arabidopsis plants. The leaf samples were collected at 6- and 17-hours post-inoculation (hpi). As a control, 0.1% glucose without Ctro was sprayed onto the plants and the leaf samples were collected at 17 h after this ‘mock’ treatment. Total RNA was extracted using PureLink RNA Mini kits (Thermo Fisher Scientific, Waltham, MA, USA) and treated with DNase. Takara Prime Script™ RT kits (Takara Bio Inc., Shiga, Japan) was used for the cDNA synthesis. Takara TB Green™ Premix Ex Taq™ I was used for RT–qPCR with the primers listed in Supplementary Table 1. Arabidopsis UBC21 (At5g25760) was used as an internal control for normalizing the level of cDNA.14 RT–qPCR analysis was performed using a Thermal Cycler Dice Real Time System TP810 (Takara Bio Inc.). Means and standard deviations (SDs) were derived from three independent samples. b. The expression of PDF1.2a was induced at 6 hpi compared with the mock treatment. The RT–qPCR data on mock and Ctro 6 hpi are shown with a different scale on the Y-axis in Figure 1a. The statistical significance of differences in gene expression level was determined by Tukey’s honestly significant difference (HSD) test (P < .01). The experiment was repeated twice, with similar results
We then tested whether EDR1 could regulate PDF expression not only in preinvasive but also in postinvasive defense. Notably, the expression of PDF1.2a was not induced in the edr1 mutant at 17 hpi with Ctro (Figure 2) even though the mutant is defective in preinvasive resistance against Ctro.12 This indicated the requirement of EDR1 for the induced expression of PDF1.2a triggered by Ctro invasion. We found previously that the expression of PDF1.2a in preinvasive defense was recovered in the edr1 myc2 mutant, in contrast to the edr1 mutant.12 Interestingly, we found that PDF1.2a expression was not restored in the edr1 myc2 mutant plant at 17 hpi with Ctro (Figure 2), suggesting that EDR1 positively regulates PDF expression upon Ctro invasion, uncoupled from the MYC2-dependent repression of PDF expression that was observed at the preinvasive stage.
Figure 2.
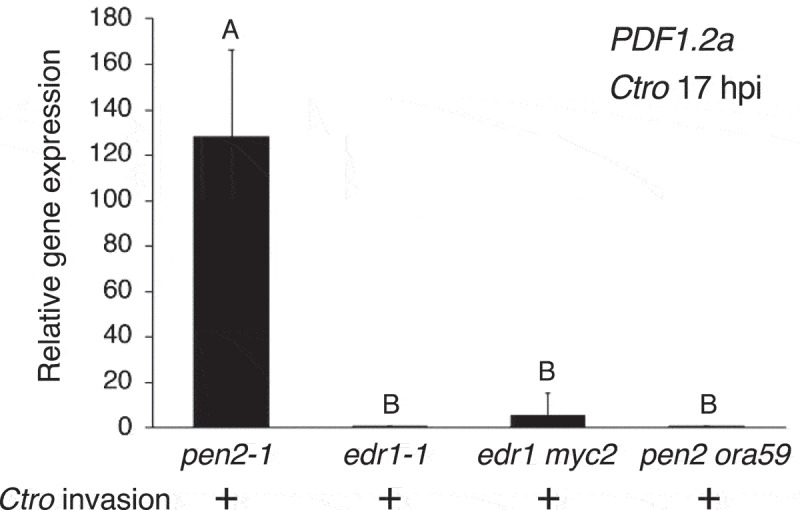
The induced expression of PDF1.2a upon Ctro invasion depends on both EDR1 and ORA59. Ctro inoculation and subsequent RT–qPCR analyses were performed as described in Figure 1. The genotyping primers used for generation of the pen2 ora59 mutants are listed in Supplementary Table 1. Means and SDs were derived from three independent experiments. The statistical significance of differences in gene expression level was determined by Tukey’s honestly significant difference (HSD) test (P < .01). The experiment was repeated twice, with similar results
OCTA-DECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59) is a transcription factor that belongs to the APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) superfamily and binds to the GCC-box motif in the promoter region of PDF1.2a.15 It was reported that EIN3 physically interacts with ORA59, resulting in destabilization of ORA59.16 It was also shown that Arabidopsis plants treated with interfering RNA (RNAi) for ORA59 showed reduced PDF1.2a expression, whereas Arabidopsis plants overexpressing ORA59 displayed increased PDF1.2a expression in response to the necrotrophic pathogens Botrytis cinerea and Alternaria brassicicola.17 To assess the involvement of ORA59 in PDF1.2a expression upon Ctro invasion, we newly generated a pen2 ora59 double mutant by crossing pen2-1 and ora59-1 plants.18,19 Then, the generated double mutant was inoculated with Ctro. Reverse transcription-quantitative polymerase chain reaction (RT–qPCR) analysis indicated that PDF1.2a expression was clearly reduced in the pen2 ora59 mutant at 17 hpi of Ctro, in contrast to the pen2 mutant (Figure 2). These results indicate that the induced expression of PDF1.2a triggered by Ctro invasion depends on both EDR1 and ORA59.
We next tested whether EDR1 and ORA59 might be required for PDF expression triggered upon invasion by a necrotrophic fungus, Alternaria brassicicola strain Ryo-1 (hereafter called Ab), in addition to Ctro. Because our recent study revealed that PEN2 is dispensable for preinvasive resistance against Ab, 20 we used the ora59 mutant instead of the pen2 ora59 mutant for Ab inoculation. Ab was inoculated onto WT (Col-0), edr1 and ora59 mutant plants, and leaf samples were collected at multiple time points (4, 12, 24, 48, 72 and 96 hpi). Subsequently, the expression levels of two PDF genes (PDF1.2a and PDF1.3) were investigated using RT–qPCR. We recently revealed that conidia of Ab started to germinate at 4 hpi but did not invade Arabidopsis plants at this time point. At 12 hpi, germinated conidia of Ab started to invade, and from 12 to 24 hpi, the fungal entry rate increased dramatically.20 We found that the expressions of the two PDF genes tested were not induced at 4 hpi but were significantly induced in WT plants at 12 hpi and further elevated at 24 hpi (Figure 3a and Supplementary Figure 2A). These results suggest that the expressions of PDF genes were triggered by Ab invasion. Interestingly, the expressions of both PDF1.2a and PDF1.3 in WT plants were reduced at 48 hpi but increased again at 72 hpi (Figure 3a and Supplementary Figure 2A). Importantly, this induced expression was not observed in the edr1 or ora59 plants, indicating the involvement of EDR1 and ORA59 for induced PDF expression upon Ab invasion (Figure 3a and Supplementary Figure 2). To investigate the relationship between EDR1 and ORA59 in transcriptional regulation, we investigated the expression of EDR1 in ora59 plants and the expression of ORA59 in edr1 plants in Ab inoculation. The result suggested that EDR1 is dispensable for the expression of ORA59, and also ORA59 is dispensable for the expression of EDR1 (Supplementary Figure 2B). It will be important to study the relationship between EDR1 and ORA59 from the aspect of PDF expression in the future.
Figure 3.
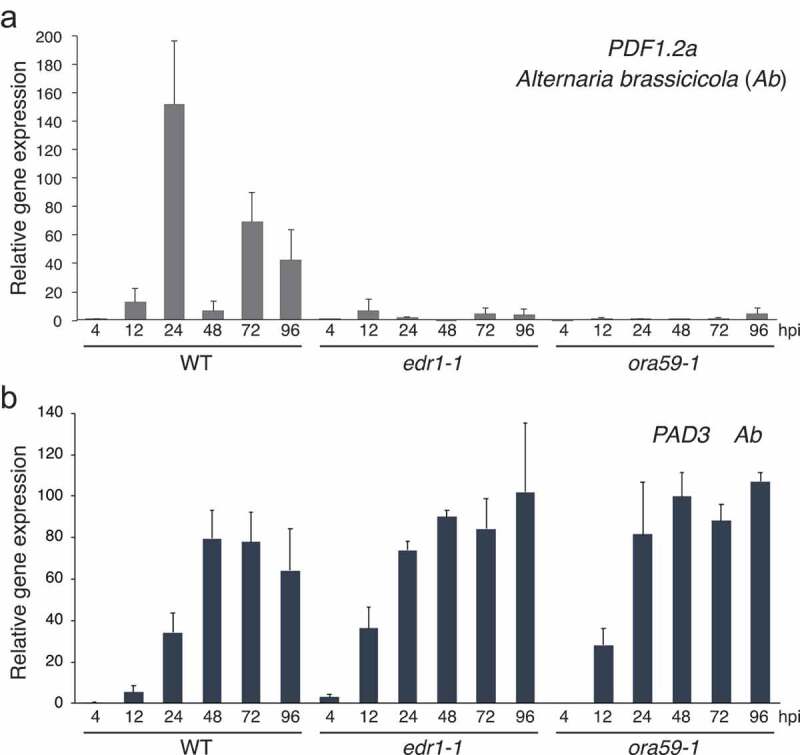
Both EDR1 and ORA59 are required for Ab-invasion triggered expression of PDF1.2a but not of PAD3. a. The invasion of Ab strongly induced the expression of PDF1.2a, which depended on EDR1 and ORA59. A suspension of Ab conidia (5 × 105/mL) was spray-inoculated onto 4- to 5-week-old Arabidopsis plants, and inoculated leaves were collected at 0, 4, 12, 24, 48, 72 and 96 hpi. Subsequent RT–qPCR analysis was performed as described in Figure 1. The means and SDs were derived from three independent experiments. b. EDR1 and ORA59 are dispensable for Ab invasion-triggered expression of PAD3. Primers used for the RT–qPCR of PAD3 transcripts are listed in Supplementary Table 1. The means and SDs were derived from three independent samples. The experiment was repeated twice, with similar results
PAD3 is essential for the biosynthesis of camalexin (a phytoalexin of Arabidopsis) and is required for Arabidopsis immunity to Ab.21,22 We recently reported that the expression of PAD3 was induced upon Ab invasion and was involved in postinvasive resistance against Ab.20 We investigated whether EDR1 and ORA59 are involved in the induced expression of PAD3 upon Ab invasion. The RT–qPCR analysis revealed that PAD3 expression was not detectable at 4 hpi, started to be induced at 12 hpi and was elevated at 24 hpi in WT (Figure 3b), which was consistent with our previous report.20 In contrast to the PDF genes, PAD3 expression was not diminished in edr1 or ora59 mutants, suggesting that EDR1 and ORA59 are dispensable for PAD3 expression upon Ab invasion.
Interestingly, the subsequent expression patterns of PAD3 were not consistent with those of the PDF genes (Figure 3b). In contrast to the latter, a drastic reduction in expression at 48 hpi was not observed in the case of PAD3. This finding suggests that the regulation of expression of the PDF genes is distinct from that for PAD3 although their expression is commonly activated upon pathogen invasion. We hypothesize that the expression of the PDF genes is induced by Ab invasion but then is quickly repressed, whereas the PAD3 expression is induced by Ab invasion and is sustained in contrast to the PDF genes. The reason why the expression of the PDF genes is recovered at 72 hpi is probably that Ab starts to invade neighboring cells around this time point, which triggers the transient expression of the PDF genes in these new invaded cells.
We reported previously that the edr1 mutant displayed enhanced lesion development following inoculation with Ab, 12 indicating the involvement of EDR1 for Arabidopsis’ immunity against Ab. Here we tested whether ORA59 might also be involved in the immunity of Arabidopsis against Ab. Inoculation assays for Ab revealed that the ora59 mutant displayed enhanced lesion development compared with WT plants (Figure 4a). We also found that the PAD3 expression was slightly enhanced in both edr1 and ora59 plants in comparison with the WT plants (Figure 3b), which is likely due to enhanced susceptibility of these plants to Ab.
Figure 4.
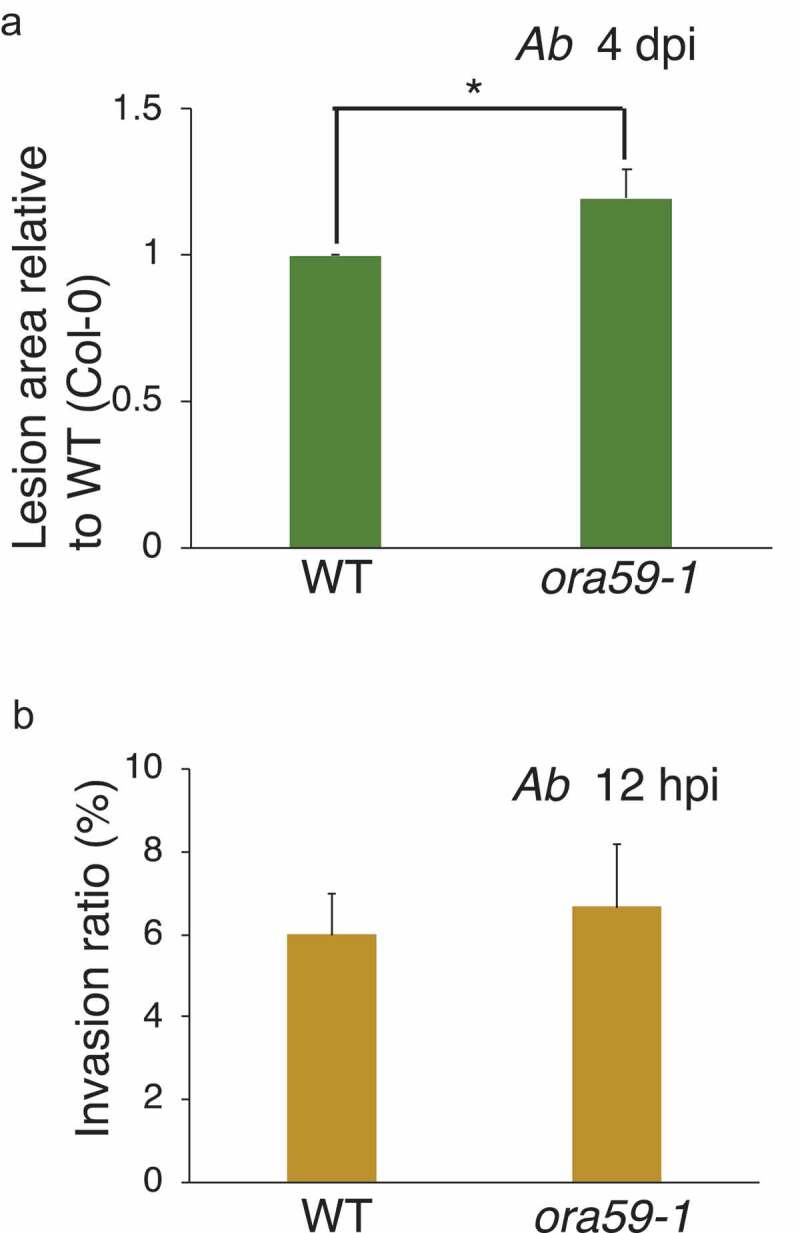
ORA59 contributes to postinvasive resistance against Ab. a. Quantitative analysis of lesion development caused by Ab. A suspension of Ab conidia (5 × 105/mL) was drop-inoculated onto mature leaves of 4–5-week-old Arabidopsis plants. At 4 dpi, lesion development was measured. The means and SDs were calculated from three independent experiments. For the statistical analysis, lesion development in ora59 was compared with WT Col-0 using two-tailed Student’s t tests (*P < .05). b. Quantitative analysis of invasion ratio by Ab. A conidial suspension (1 × 105/mL) of Ab was drop-inoculated onto 4 to 5-week-old plants and kept at 100% humidity. The inoculated leaves were collected at 12 hpi, and then subjected to trypan blue staining assays. The presence or absence of invasive hyphae from at least 50 germinating conidia were counted in each experiment. The means and SDs were calculated from three independent experiments. Statistical comparisons of the Ab invasion ratios on WT Col-0 and tested mutants were conducted using two-tailed Student’s t tests. The invasion ratio of the ora59 mutants was not significantly different from that of WT (Col-0)
To assess any possible involvement of ORA59 in preinvasive resistance against Ab, conidia of Ab were inoculated on WT and ora59 plants, and the invasion ratio was determined at 12 hpi. We found that there was no significant difference in the invasion ratio between WT and ora59, suggesting that ORA59 is not required for preinvasive resistance (Figure 4b). These findings suggest that ORA59 is involved in postinvasive resistance against Ab. Because ORA59 is required for PDF expression at the postinvasive stage of Ab, this finding suggests that PDF genes contribute to postinvasive immunity in Arabidopsis.
Currently, it remains unclear how Arabidopsis plants recognize pathogen invasion to activate PDF expression. We recently reported that the expression of GLIP1, encoding a secreted antimicrobial protein, was induced upon Ab invasion and that the induced expression of GLIP1 was largely reduced in the presence of a bak1-5 mutation, suggesting that the involvement of a pattern recognition receptor in Ab invasion-triggered expression of GLIP1.20,23 However, the Ab-triggered expressions of PDF genes were not reduced in the presence of the bak1-5 mutation.20 Thus, the EDR1–ORA59–PDFs pathway unlikely depends on a group of pattern recognition receptors whose function is blocked by bak1-5. Further studies on recognition machineries for pathogen invasion are essential for understanding the molecular background of postinvasive resistance in higher plants.
Supplementary Material
Acknowledgments
We thank Shigenobu Yoshida (Ctro) and Akira Tohyama (Ab) for providing fungal pathogens. We also thank Volker Lipka (pen2-1, pen2-2), Roger W. Innes (edr1), and ABRC (ora59) for providing Arabidopsis seeds. This work was supported by Grants-in-Aid for Scientific Research (18H02204) (KAKENHI), by grants from the Project of the NARO Bio-oriented Technology Research Advancement Institution (Research program on development of innovative technology), and by the Asahi Glass Foundation.
Disclosure of potential conflicts of interest
The author has declared that no competing interests exist.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Heath M. Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol. 2000;3:1–5. doi: 10.1016/s1369-5266(00)00087-x. PMID: 10873843. [DOI] [PubMed] [Google Scholar]
- 2.Cannon PF, Damn U, Johnston PR, Weir BS.. Colletotrichum— current status and future directions. Stud Mycol. 2012;73:181–213. doi: 10.3114/sim0014. PMID: 23136460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiruma K, Onozawa-Komori M, Takahashi F, Asakura M, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y.. Entry mode-dependent function of an indole glucosinolate pathway in Arabidopsis for nonhost resistance against anthracnose pathogens. Plant Cell. 2010;22:2429–2443. doi: 10.1105/tpc.110.074344. PMID: 20605856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimada C, Lipka V, O’Connell R, Okuno T, Schulze-Lefert P, Takano Y. Nonhost resistance in Arabidopsis-Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol Plant-Microbe Interact. 2006;19:270–279. doi: 10.1094/MPMI-19-0270. PMID: 16570657. [DOI] [PubMed] [Google Scholar]
- 5.Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. PMID: 16293760. [DOI] [PubMed] [Google Scholar]
- 6.Lipka U, Fuchs R, Lipka V. Arabidopsis non-host resistance to powdery mildews. Curr Opin Plant Biol. 2008;11:404–411. doi: 10.1016/j.pbi.2008.04.004. PMID:18499508. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka A, Takano Y. Nonhost resistance of Arabidopsis thaliana against Colletotrichum species. J Gen Plant Pathol. 2018;84:305–311. doi: 10.1007/s10327-018-0799-y. [DOI] [Google Scholar]
- 8.Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. [DOI] [PubMed] [Google Scholar]
- 9.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiruma K, Fukunaga S, Bednarek P, Pislewska-Bednarek M, Watanabe S, Narusaka Y, Shirasu K, Takano Y. Glutathione and tryptophan metabolism are required for Arabidopsis immunity during the hypersensitive response to hemibiotrophs. Proc Natl Acad Sci USA. 2013;110:9589–9594. doi: 10.1073/pnas.1305745110. PMID: 23696664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J. 2011;67:980–992. doi: 10.1111/j.1365-313X. PMID: 21605210. [DOI] [PubMed] [Google Scholar]
- 13.Frye CA, Tang D, Innes RW. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA. 2001;98:373–378. doi: 10.1073/pnas.011405198. PMID: 11114160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. PMID: 16166256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarei A, Körbes AP, Younessi P, Montiel G, Champion A, Memelink J. Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol. 2011;75:321–331. doi: 10.1007/s11103-010-9728-y. PMID: 21246258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X, Jiang J, Wang CQ, Dehesh K. ORA59 and EIN3 interaction couples jasmonate-ethylene synergistic action to antagonistic salicylic acid regulation of PDF expression. J Integr Plant Biol. 2017;59:275–287. PMID: 28168848. doi: 10.1111/jipb.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pré M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. PMID:18467450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim NY, Jang YJ, Park OK. AP2/ERF family transcription factors ORA59 and RAP2.3 interact in the nucleus and function together in ethylene responses. Front Plant Sci. 2018;9:1675. doi: 10.3389/fpls.2018.01675. PMID: 30510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang GB, Yi HY, Gong JM. The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell. 2014;26:3984–3998. doi: 10.1105/tpc.114.129296. PMID: 25326291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosaka A, Pastorczyk M, Pislewska-Bednarek M, Nishiuchi T, Suemoto H, Ishikawa A, Frerigmann H, Kaido M, Mise K, Bednarek P, et al. bak1-5 mutation uncouples tryptophan-dependent and independent postinvasive immune pathways triggered in Arabidopsis by multiple fungal pathogens. bioRxiv. 2020. 04.26.052480. doi: 10.1101/2020.04.26.052480. [DOI] [Google Scholar]
- 21.Narusaka Y, Narusaka M, Seki M, Ishida J, Nakashima M, Kamiya A, Enju A, Sakurai T, Satoh M, Kobayashi M, et al. The cDNA microarray analysis using an Arabidopsis pad3 mutant reveals the expression profiles and classification of genes induced by Alternaria brassicicola attack. Plant Cell Physiol. 2003;44:377–387. doi: 10.1093/pcp/pcg050. [DOI] [PubMed] [Google Scholar]
- 22.Thomma BP, Nelissen I, Eggermont K, Broekaert WF. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 1999;19:163–171. doi: 10.1046/j.1365-313x.1999. PMID: 10476063. [DOI] [PubMed] [Google Scholar]
- 23.Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee S, Cheong H, Park OK. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell. 2005;17:2832–2847. doi: 10.1105/tpc.105.034819. PMID: 16126835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


