Abstract
The outbreak of COVID-19 by the end of 2019 has posed serious health threats to humanity and jeopardized the global economy. However, no effective drugs are available to treat COVID-19 currently and there is a great demand to fight against it. Here, we combined computational screening and an efficient cellular pseudotyped virus system, confirming that clinical HDAC inhibitors can efficiently prevent SARS-CoV-2 and potentially be used to fight against COVID-19.
Keywords: SARS-CoV-2, virtual screening, ACE2, romidepsin, molecular dynamics, HDAC inhibitors
In December 2019, the Corona Virus Disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) broke out and led to a worldwide pandemic. By October 30th, 2020, the number of confirmed cases exceeded 44.8 million with more than 1.18 million deaths (https://www.worldometers.info/coronavirus). Over 50% of the infected patients were observed to have increased levels of interleukin-6 (IL-6), one of the main inflammatory mediators initiated by infection or injury.1,2 The confirmed cases of SARS-CoV-2 in several countries such as United States, Brazil, and India are still increasing sharply.3,4 The possible “second wave” attack during the change of season may further worsen the situation.5
Research in vaccines6,7 has ensued to fight the SARA-CoV-2 pandemic and undergoes a lengthy development process.8,9 However, rapid mutations of SARS-CoV-2, exemplified by the very recent reports of reinfections in Hongkong and Belgium,10,11 add uncertainties to the effectiveness of vaccines. Another efficient alternative is repurposing the existing FDA approved drugs, of which their long-term adverse effects are more thoroughly investigated.12−14 As an example, a classical antimalarial drug, chloroquine (Figure 1a), was found to block the lysosome escape of SARS-CoV-2 and has been proposed for the treatment of COVID-19.15,16 Chloroquine was used in several countries after the outbreak of COVID-19. Identically, remdesivir, originally developed for treating the Ebola virus in phase III, is now approved for compassionate use of patients with severe COVID-19.17 Currently, there is still a strong need for more effective and safer drugs for the treatment of COVID-19.13,18
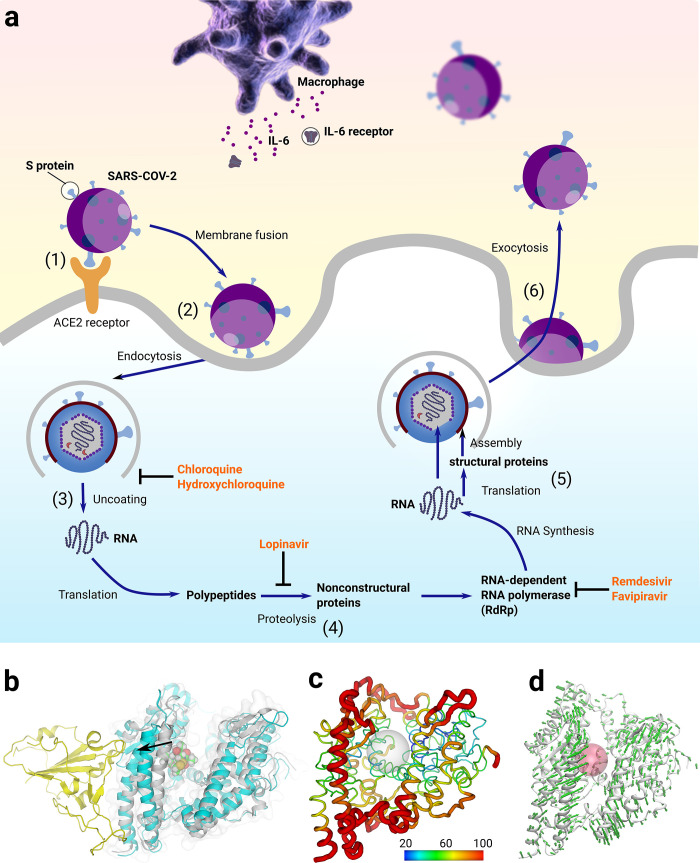
Figure 1.
(a) The life cycle of SARS-CoV-2 virus: (1) SARS-CoV-2 attaches to the surface of a human cell via interactions between S protein and ACE2 receptor. (2) After membrane fusion, it undergoes endocytosis and diffuses into the inner space of the human cell. (3) In the uncoating process, the RNA of SARS-CoV-2 is released into cytoplasm of human cell. (4) Translation of RNA takes places with the aid of a series of proteins. (5) RNA synthesis and structural protein translation lead to the assembling of a new virus. (6) The matured new virus leaves the host cell via exocytosis. Several compounds were used to treat SARS-CoV-2 via different mechanisms. Both chloroquine and hydroxychloroquine were used to stop uncoating; lopinavir was used to stop proteolysis; and both remdesivir and favipiravir were used to stop RNA synthesis. (b) Structural changes of ACE2 after MD simulations. Yellow cartoon: binding domain of S protein. Gray cartoon: APO ACE2 before MD simulations. Cyan cartoon: ACE2 in complex of romidepsin after MD simulations. A helix moved outward about 3.5 Å in the complex model after MD simulations. (c) Normalized b-factor indicated the flexibility of a helix next to the binding domain. Gray sphere: ligand binding region. (d) Normal-mode analysis also confirmed the intrinsic flexibility of the investigated helix. Red sphere: ligand binding region.
Repurposing clinical drugs involving biological experiments on the active virus is greatly impeded by the strict safety procedures and the limited resources of Biosafety Level 3 (P3) laboratories. In this work, we combined computational screening and a pseuotyped virus system to facilitate the repurposing process.19,20 We first performed virtual screening to obtain FDA approved drugs that might interevent the interactions between angiotensin I converting enzyme 2 (ACE2)21 and a spike protein. Then, we constructed a pseudotyped SARS-CoV-2 virus successfully at a Biosafety Level 2 (P2) laboratory and nine drugs XJY1–9 (Figure 2a, Table S1) were submitted for the bioactivity testing. The result showed that romidepsin, a histone deacetylase (HDAC) inhibitor, can effectively block the entry of SARS-CoV-2. Inspired by this result, we then screened 18 commercially available HDAC inhibitors (H1–18, Table S2) and studied their efficacies in inhibiting the entry of SARS-CoV-2 into cells and found that four of them are noticeably effective (Table 1). The possible mechanism of these HDAC inhibitors making them effective against SARS-CoV-2 was revealed by the cellular signaling network analysis. The clinically active HDAC inhibitors found in this work could be used to fight against the current epidemics.
Figure 2.
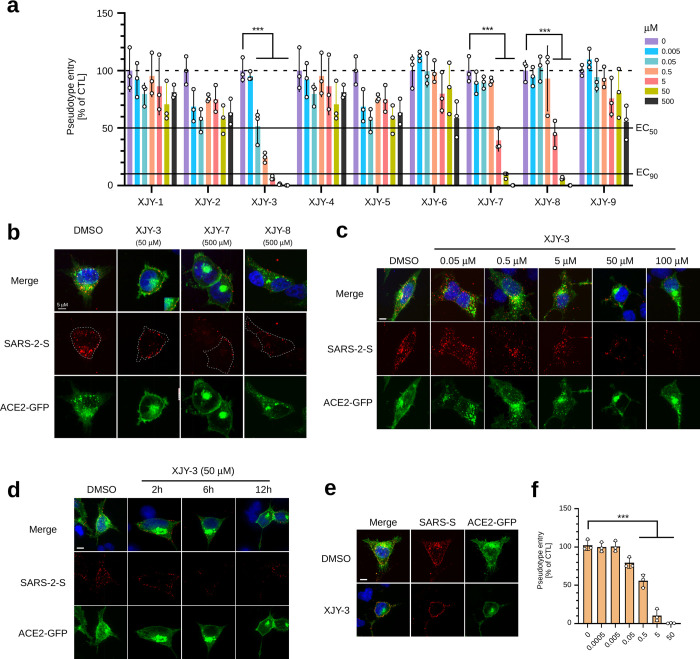
HDAC clinical inhibitors prevent SARS-CoV-2 cell entry. (a) Screen for clinical drugs that can efficiently inhibit host cell entry of SARS-2-S pseudotyped particles. ACE2-GFP stably expressing 293T cells were preincubated with indicated concentrations of drugs for 2 h and subsequently inoculated with SARS-2-S pseudotyped particles for an additional 3 h. The medium with pseudotyped particles was then removed, and cells were recultured in fresh medium for 40 h. Luciferase activity assay was conducted to analyze the virus entry efficiency. (b) Inhibition of SARS-2-S pseudotyped viruses entry into ACE2-GFP transfected 293T cells. 293T cells transfected with ACE2-GFP for 24 h were preincubated with indicated drugs for 1 h, and then further inoculated with SARS-2-S pseudotyped viruses for additional 2 h in the presence of drugs. The cell entry of SARS-2-S pseudotyped viruses were examined by immunofluorescence staining using anti-FLAG antibodies. (c,d), Concentration- and time-dependent inhibitory effects of romidepsin on host cell entry of SARS-2-S pseudotyped particles. 293T cells transfected with ACE2-GFP were treated with increased doses of romidepsin (c) or different time of drug treatment (d). The cell entry of SARS-2-S pseudotyped viruses were detected by anti-FLAG staining. (e) Immunofluorescence staining shows that host cell entry of SARS-S pseudotyped particles was blocked by romidepsin. 293T cells transfected with ACE2-GFP were treated as in (b) with the replacement of SARS-2-S pseudotyped particles as SARS-S pseudotyped particles. The anti-FLAG immunofluorescence staining images were as shown. (f) Luciferase activity assay shows that host cell (ACE2-GFP stably expressing 293T) entry of SARS-S pseudotyped particles was blocked by romidepsin. ACE2-GFP stably expressing 293T cells were preincubated for 1 h with an increased dose of Romidepsin, then inoculated with SARS-S pseudotyped viruses for an additional 3 h, followed by Luciferase activity assay as in panel a at 40 h postinoculation. For luciferase activity assay in panels a and f, the average value of three independent experiments was calculated with triplicate samples. Error bars indicate SEM. Statistical significance was tested by two-way ANOVA with Dunnett post-test. Groups of cells with no drug treatment were used as controls.
Table 1. Effectiveness of Screened Compounds against SARS-CoV-2.
| drug ID | name | IC50 to cell proliferation (μM) | EC50 (transduction inhibition) (μM) | SI = IC50/EC50 |
|---|---|---|---|---|
| XJY-3 | romidepsin | 17.5 ± 6.4 | 0.09 ± 0.05 | 194.4 |
| XJY-7 | Saquinavir | 207 ± 29.8 | 6.6 ± 4.9 | 31.4 |
| XJY-8 | Nelfinavir | 802.8 ± 609.2 | 4.8 ± 2.1 | 167.1 |
| H4 | Panobinostat | 0.05 ± 0.05 | 2.8 ± 1.0 | 0.02 |
| H5 | Givinostat hydrochloride monohydrate | 0.28 ± 0.2 | 6.8 ± 4.1 | 0.04 |
| H8 | CAY10603 | 0.6 ± 0.5 | 13.5 ± 9.4 | 0.04 |
| H15 | Sirtinol | 34.5 ± 3.5 | 82.9 ± 61.1 | 0.4 |
Results and Discussion
Virtual Screening of FDA Approved Drugs
To obtain the potential active compounds against COVID-19, we performed virtual screening from the FDA approved drug library using ACE2 as the drug target. Nine potential candidates were selected and submitted to experimental studies. The in vitro experiment result showed that romidepsin, an HDAC inhibitor,22 is highly effective in inhibiting the viral entry. Romidepsin is structurally similar to CHEMBL20341 and CHEMBL138409 (Figure S2a), which were reported to bind to ACE instead.23,24 Sequence alignment indicated that the similarity between ACE25 and ACE226 is over 71% (Figure S1). In their crystal structures, the root-mean-square deviation (RMSD) of the protein backbone is as low as 1.6 Å (Figure S2c), whereas all residues in the binding sites are nearly completely conserved (Figure S2b,d,e). Those observations strongly implied that ACE and ACE2 might share a common ligand-binding pocket. This explained why romidepsin discovered from the virtual screening against ACE2 shares a large common chemical moiety with the ACE inhibitors (Figure S2a). The binding mode of romidepsin with ACE2 was shown in Figure S3. MD simulation showed that the residues of ACE2 contributing mostly to the protein–ligand interactions are P346, H374, E375, H378, E402, and Y515 (Figure S3). The ligand has a strong allosteric effect on the shape changes of the receptor binding region, according to the ACE2-inhibitor complex crystal structure which was reported previously.27 Our MD simulation also observed similar changes as indicated below due to the flexibility of a helix between the catalytic site and the S protein binding domain (Figure 1b–d, Movie S1). The large change of the ACE2 shape might prevent the binding S protein.27,28 To facilitate the binding mode of ACE2-romidepsin, the coordinate of the last snapshot for the ACE2-romidepsin complex from the MD simulations was provided as a supplementary file.
Generation of Spiked Pseudotyped Lentivirus for Imitating SARS-CoV-2 Cell Entry
To test the inhibitory effect of these candidate drugs on host cell entry, we generated lentiviral particles pseudotyped with S proteins either from SARS-CoV or SARS-CoV-2. By immunoblotting, we showed a robust expression (Figure S4a) and a strong incorporation of these spike proteins in purified pseudotyped particles (Figure S4b), suggesting a successful production of the designed pseudovirus. Further study demonstrated that this pseudovirus infected the host cells through ACE2, which is the direct binding target of the spike protein and serves as the major entry receptor for both SARS-CoV and SARS-CoV-2 entering the targeted cells. Exogenous overexpression of human ACE2 significantly increased the SARS-2-S- and SARS-S- driven entry of the pseudotyped virus into nonsusceptible 293T cells (Figure S4c), while the downregulation of endogenous ACE2 expression in monkey Vero-E6 cells greatly reduced the SARS-2-S- and SARS-S-driven entry (Figure S4d). Immunofluorescence staining further demonstrated that only ACE2-positive cells were susceptible for pseudovirus entry (Figure S4e). Both SARS-2-S and SARS-S pseudotyped particles presented partial colocalization with ACE2 either at the cell surface or in internalized endosomes (Figure S4f,g). Besides, the ACE2 dependent infection with either SARS-2-S or SARS-S pseudotyped particles increased with time and peaked around 1 h after inoculation with the host cells (Figure S4h,i). Together, these results demonstrated that the developed SARS-2-S and SARS-S pseudotyped lentivirus can imitate SARS-CoV-2 or SARS-CoV cell entry.
Validation of Clinical Candidate Drugs for Suppressing SARS-CoV-2 Cell Entry
We next investigated potential effects of our candidate drugs on cell entry of the developed pseudovirus. We first examined the cytotoxicity of these drugs by using a cell proliferation kit (Figure S5a). The drug concentration gradient applied for virus entry assay was determined by their respective cytotoxicity profiles. Next, we used luciferase reporter assay to evaluate the inhibitory effects of these drugs on SARS-CoV-2 cell entry (Figure 2a). Our results showed that three (romidepsin, saquinavir, and nelfinavir) out of nine drugs possess the ability to suppress SARS-2-S pseudotyped particles to enter the ACE2 expressing cells in a concentration-dependent manner (Figure 2a). Being consistent with these results, immunofluorescence staining of the FLAG tagged SARS-2-S showed that romidepsin (50 μM), saquinavir (500 μM), and nelfinavir (500 μM) treatment strongly blocked the entry of SARS-2-S pseudotyped particles into ACE2-GFP expressing HEK293T cells. Compared to the untreated cells, viral particles in drug treatment groups were mostly presented at the cell surface but absent from the perinuclear region, where they are expected to be mostly trafficked and enriched after internalization into host cells (Figure 2b and Figure S5b). We further determined that this suppression effect by romidepsin was concentration- and incubation time-dependent (Figure 2c,d). Although saquinavir and nelfinavir also displayed clear inhibitory effects on ACE2-mediated cell entry of the tested pseudovirus, they both had relatively high effective concentrations as compared to romidepsin (Figure 2b, Figure S5c). Therefore, they were excluded from our following studies. As both SARS-CoV and SARS-CoV-2 use ACE2 receptor for entry, we then expected that romidepsin would also suppress the cell entry of viral particles pseudotyped with SARS-S protein. Indeed, the treatment of ACE2-expressing cells with romidepsin greatly diminished the internalization of SARS-S pseudotyped lentiviral particles into the cells with robust attachment on the cell surface, indicating that the SARS-S-driven cell entry was also efficiently blocked (Figure 2e). This result was further confirmed by luciferase reporter assays (Figure 2f). Together, our results strongly suggested that romidepsin is a promising drug candidate for COVID-19 treatment.
Suppression of Clathrin-Mediated Endocytosis by Romidepsin
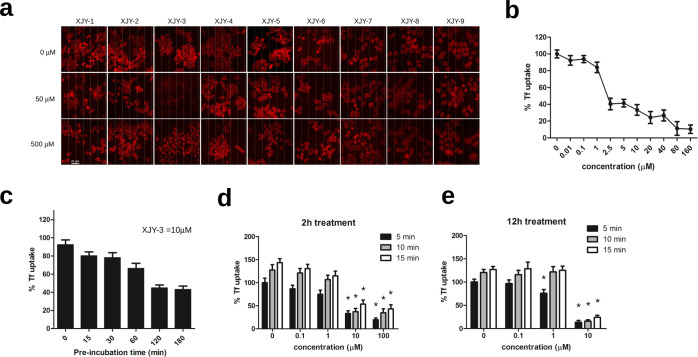
Since virus cell entry is mediated by endocytosis, we also examined the potential effect of our candidate drugs on clathrin-mediated endocytosis, by using Alexa Fluor 555-conjugated transferrin (Tf-555) as an indicator. Among all nine drugs, we found that only romidepsin robustly reduces the Tf-555 uptake in HeLa cells (Figure 3a). This blockage of Tf-555 endocytosis by romidepsin was concentration- and time-dependent (Figure 3b,c). Moreover, prolonged incubation of cells with romidepsin presented a more effective reduction of Tf-555 uptake (Figure 3d,e). Together, these results suggest that romidepsin might suppress the SARS-2-S-driven host cell entry by simultaneously inhibiting endocytosis and weakening the ACE2-spike protein recognition.
Figure 3.
Inhibition of clathrin mediated endocytosis by romidepsin. (a) Effects of XJY drugs on cellular uptake of transferrin. Hela cells were treated with indicated concentrations (0, 50, 500 μM) of drugs, followed by incubation with fluorescently labeled Alexa-555 transferrin (red) for 15 min at 37 °C. Cells were then washed with fresh medium and fixed for microscopy imaging. Scale bar is 20 μm. (b) Concentration-dependent inhibitory effect of romidepsin on transferrin internalization was as shown. (c) Time-dependent inhibitory effect of romidepsin on transferrin internalization. Cells were preincubated with indicated time before incubation with Alexa-555 transferrin. Cells were harvested and analyzed as in panel a; 10 μM romidepsin was used in this experiment. (d,e), Prolonged treatment can lower effective dose of romidepsin for inhibition transferrin uptake. Hela cells pretreated with 2 or 12 h of romidepsin were incubated with transferrin. The internalization of transferrin was respectively examined at 5, 10, and 15 min after the addition of transferrin. For quantification of the endocytosis, the total intracellular transferrin-555 signal was measured from 50 cells in each experimental condition.
Screening Clinical HDAC Inhibitors for Potential COVID-19 Treatment
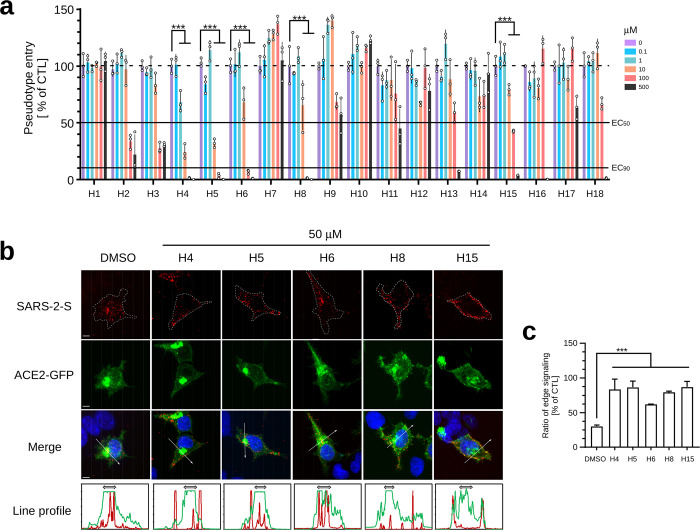
Since romidepsin is an FDA-approved HDAC inhibitor, it is possible that other HDAC-targeted drugs might have similar inhibitory effects against SARS-CoV-2 cell entry. By using luciferase reporter assay, 18 commercially available clinical HDAC inhibitors were evaluated for their potential to reduce SARS-2-S-driven cell entry. As indicated, five drugs, panobinostat, givinostat hydrochloride monohydrate, abexinostat, CAY10603, and sirtinol, showed promising concentration-dependent suppression abilities for SARS-CoV-2 pseudovirus entry (Figure 4a). By immunofluorescence staining, we further confirmed that four out of these drugs, except abexinostat, could efficiently prevent the entry of SARS-2-S pseudotyped particles into ACE2 expressing cells, consequently leading to a robust accumulation of these particles at the cell surface (Figure 4b,c). These results strongly suggest that HDAC inhibitors are a class of promising drug candidates for preventing SARS-CoV-2 infection.
Figure 4.
Screen for clinically used HDAC inhibitors to suppress SARS-2-S-driven host cell entry. (a) Luciferase activity assay-based screening of clinically used HDAC inhibitors for suppressing SARS-2-S pseudovirus cell entry. ACE2-GFP stably expressed 293T cells were preincubated with the increased dose of indicated clinical drugs for 2 h and subsequently inoculated with SARS-2-S pseudotyped particles for 3 h, then further cultured for an additional 40 h with fresh medium. Luciferase activity in each cell lysate was examined to analyze the entry efficiency of SARS-2-S pseudovirus. The average value from three independent experiments with triplicate samples was calculated. Error bars indicate SEM. Statistical significance was tested by two-way ANOVA with Dunnett post-test. Cells without drug treatment were used as controls. (b) Immunofluorescence staining reveals that four out of 18 H drugs can efficiently inhibit cell entry of SARS-2-S pseudotype particles. ACE2-GFP transfected 293T cells were pretreated with the indicated drug for 1 h and subsequently inoculated with SARS-2-S pseudotyped particles for an additional 2 h. Cells were fixed and stained with antibodies against FLAG to detect the intracellular SARS-2-S pseudovirus. Green, ACE2-GFP; red, FLAG; blue, DAPI. Scale bar is 5 μm. Quantification of the fluorescence intensity along the line across the perinuclear region was shown at the bottom. Green line, ACE2-GFP; red line, FLAG. The two-head arrows indicate the perinuclear region where the ACE2-GFP signal was highest. (c) Analysis of the distribution of internalized pseudotyped particles. Interspace that is within 2 μm from the cellular boundary is considered as the edge area. Percent ratio of edge signaling was calculated as edge area signal/total area signal × 100. Thirty cells from each group were used for quantification.
In summary, SARS-CoV-2 has brought great damage to both public health and the global economy. Here we combined computational virtual screening with a pseudovirus system, and this work can be performed in a normal P2 laboratory, to facilitate the screening inhibitors of SARS-CoV-2. We found that HDAC inhibitors were a promising class of compounds against SARS-CoV-2. Five clinical HDAC inhibitors (Table 1), including romidepsin, panobinostat, givinostat hydrochloride monohydrate, CAY10603, and sirtinol, could inhibit noticeably the spike/ACE2 mediated cell entry of SARS-CoV-2.
Our study highlights the interaction map of human proteins that are involved in SARS-CoV-2 complexes and biological processes. In this work, HDACs remove acetylated lysine in a histone and make the histone positively charged, which results in the interactions with negatively charged molecules such as DNA.29 The SARS-CoV-2 host protein–protein interaction (PPI) comprises proteins that regulate critical processes of the human body (Figure S6). These include the biological processes involved in viral process (a multiorganism process in which a virus is a participant), positive regulation of receptor biosynthetic process, catabolic process, and others (Table S3). The network-based approach provides a systematic understanding of effects on the interconnected biological processes of SARS-CoV-2. HDAC inhibitors can prevent the viral entry probably due to the synergy of such complicated network. Because of the specificity of each compound as well as the polypharmacology effect, HDAC inhibitors may suppress the cell entry of SARS-CoV-2 unequally as demonstrated in Figure 4. Interestingly, ACE2, which was used for the initial virtual screening, was also revealed to interact with HDAC-related signaling as shown in Figure S6. These results suggest that in addition to directly influencing the function of ACE2, HDAC inhibitors may also indirectly regulate virus invasion through the SARS-CoV-2 host protein–protein interaction network.
In summary, we discovered five clinical HDAC inhibitors which can potentially be used to fight against COVID-19. Further clinical studies should be performed to validate the effectiveness of those compounds.
Methods
Virtual Screening
The virtual screening was performed in Schrödinger30 software using Glide31 docking based methods. For the ACE2 structure,26 protein preparation was performed to add hydrogens to the protein, predict pKa for each residue, optimize the hydrogen-bond network, and perform energy minimization for the target protein.32 The structures of FDA-approved drugs (2454 molecules) were prepared in the LigPrep tool in the Schrödinger software.33 The potential candidates were selected according to the Glide-SP score31 and checked manually to ensure the docking result was acceptable. More details can be found in our previous work.32
MD Simulation
MD simulation was performed with Desmond34 in Schrödinger software. The OPLS3e force field35 was used for all the molecules in the simulation. All bond lengths to hydrogen atoms were constrained with M-SHAKE. A 200 ps equilibration run was carried out with the temperature set to 300 K and pressure to 1 bar. The simulation length for all the production runs was 100 ns for both APO ACE2 and romidepsin-ACE2 complex systems. Cut-off for short-range interactions was set to 10 Å and long-range electrostatic interactions were recovered by Particle Mesh Ewald (PME) summation.36
Enrichment Analysis and Construction of Interaction Networks
We integrated virus–host interaction data from several sources, including SARS-CoV-2 AP-MS data reported by Gordon et al.37 which contains 332 high-confidence virus-host interactions for 27 SARS-CoV-2 proteins, the histone deacetylase family of HDACs (HDAC1–HDAC11) and ACE2. The GO terms of biological processes were summarized, and their relationships were analyzed using two Cytoscape plugins ClueGo38 and CluePedia.39 The biological processes network enriched by the proteins associated with the COVID-19 infection was constructed in Cytoscape.40
Sequence and Structural Analysis
The protein-sequence alignment was performed in Clustal Omega41 and the results were visualized in Mview.42 The ligand similarity analysis was performed in RDKit, an open source toolkit for cheminformatic.43 All figures were prepared in PyMol and Inkscape.44
Cell Lines and Cell Culture
All cell lines were maintained at 37 °C incubated with 5% CO2 in a humidified atmosphere. HeLa cells, 293T (human, kidney) cells, and Vero-E6 (African green monkey, kidney) cells were incubated in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (Gibco).
Inhibitors and Clinical Drugs
Drugs and inhibitors were purchased from Shanghai Topscience Co., Ltd. Information for the molecules was listed in Supplementary Tables 1 and 2.
Antibodies
Rabbit polyclonal antibody against FLAG-tag (AF0036) was purchased from Beyotime Biotechnology. Rabbit polyclonal antibody against HIV gag-p24 (11695-RP01) was purchased from Sino Biological Inc. Mouse monoclonal antibody against beta-actin (sc-47778) and mouse monoclonal antibody against human ACE2 (sc-390851) were purchased from Santa Cruz Biotechnology (Dallas, USA). Goat anti-Mouse IgG conjugated with HRP (ab136815) and goat anti-Rabbit IgG conjugated with HRP (ab136817) were purchased from Abcam (Cambridge, USA). Goat anti-Rabbit IgG Alexa Fluor 555 (A21429) and goat anti-Mouse IgG Alexa Fluor 488 (A11029) were purchased from Thermo Fisher Scientific (Massachusetts, USA).
Plasmids
Plasmid encoding full length SARS-CoV-2 S glycoprotein (QHU36824.1) was codon-optimized for optimal expression in mammalian cells. Plasmids encoding full length SARS-CoV-S glycoprotein and human ACE2 were provided by collaborators. Expression plasmids for replication-defective HIV lentivirus include pCDH-EF1-MCS-IRES-Puro (CD532A-2, System Biosciences), psPAX2 (#12260, Addgene), and pM2.G (#12259, Addgene). Plasmid pCAGGS (VT1076) was purchased from Youbio Biological. ACE2-GFP plasmids were constructed by fusing GFP at the C-terminus of ACE2 and cloned into pCDH at the EcoRI site. pCDH-Luciferase and pCDH-mCherry plasmids were generated by inserting the reporter gene into the EcoRI site. DNA fragments of VSV-G (cloned from pM2.G), SARS-CoV-2-S (full length), and SARS–CoV-S (full length) were fused with a FLAG tag at the C terminus and inserted into the pCAGGS vector via the EcoRI site. All constructs were generated by a seamless cloning kit (D7010M, Beyotime).
Generation of Lentiviral Pseudotyped Lentiviral Particles and Virus Transduction
To generate replication-defective lentivirus-based pseudotype particles, 293T cells were transfected with pCDH-Luciferase (or pCDH-mCherry), psPAX2, and plasmids encoding glycoproteins (pCAGGS-SARS-2-S-flag, pCAGGS-SARS-S-flag or pCAGGS-VSV-G-flag) by using 1 mg/mL linear polyethylenimine (PEI) purchased from Polysciences (23966-2). Supernatants were harvested at 48 and 72 h post-transfection, centrifuged at 1000g for 5 min and passed through 0.45 μm filter to remove cell debris. Then the viral particles in the supernatants were harvested by ultracentrifugation at 100 000g with a 25% sucrose cushion for 2 h at 4 °C in an SW28 Ti Rotor (Optima L-80 XP Ultracentrifuge, Beckman Coulter). Concentrated viral particles were resuspended in 1X phosphate buffer saline (PBS) and stored at −80 °C.
For transduction, ACE2-GFP stably expressing 293T cells were plated 5000 cells/well in 96-well plates and inoculated with respective pseudotyped particles for 3 h at 37 °C. For clinical drug test, targeted cells were pretreated with different concentrations of corresponding drugs for 2 h before transduction. Next, the medium containing pseudovirus was replaced with fresh medium. At 40 h post-inoculation, cells were lysed and the luciferase activity was measured by Luciferase Assay System (Promega). Pseudovirus entry efficiency was calculated as the percentage of luciferase activity in the drug-treated group relative to the control group. Relative cell proteins of each well was measured by BCA protein assay kit and used for normalization of luciferase activity.
Analysis of Spike Protein Expression and Incorporation into Pseudotyped Particles
The 293T cells were transfected with vector for FLAG-tagged SARS-2-S, SARS-S, or empty vector (negative control) by MegaTran 1.0 (Origene). Cells were harvested 24 h post-transfection and incubated with 2 × SDS lysis buffer (0.03 M Tris-HCl, 10% glycerol, 2% SDS, 0.2% bromophenol blue, 1 mM EDTA) for 5 min at room temperature. Next, cell lysates were incubated in 100 °C water for 15 min before immunoblotting. To detect S protein incorporation into the pseudovirus, amounts of viral particles equal to 2 mL supernatant concentrates were used and incubated with 2 × SDS lysis buffer as described above. In brief, well-prepared lysate samples were resolved by SDS-PAGE and electrotransfered onto Immun-Blot PVDF Membrane (Millipore). Next, membrane and samples were blocked by 5% skim milk powder in 1 × PBS for 1 h. Blots were probed with primary antibodies for 2 h and then with secondary antibodies for 1 h at room temperature. Immunoreactivity was detected by using Luminata Classico Western HRP Substrate (Millipore).
ACE2 Knockdown in Vero-E6 Cells
To knockdown endogenous ACE2 expression in Vero-E6 cells, siRNA targeting the first exon of human ACE2 mRNA at the position of 200–222 (identical to African green monkey) was commercially synthesized. The forward sequence is CACGAAGCCGAAGACCTGTTCdTdT, the reverse sequence is dTdT-GAACAGGTCTTCGGCTTCGTG. To analyze knockdown efficiency, 2.5 × 105 Vero-E6 cells were plated and grown on a 6-well plate per well. After 16 h incubation, cells were transfected with an optimal amount of ACE2-siRNA by Lipofectamine 3000 (Thermo Fisher Scientific). At 48 h after transfection, cells were harvested and lysed for immunoblot. To analyze pseudovirus host entry, cells were plated on coverslips and fixed for immunofluorescence staining.
Visualization of pseudotyped particle entry
Susceptible cells were grown on round glass coverslips placed in 24-well plates. Cells were inoculated with pseudotyped particles for 2 h at 37 °C before being fixed in 4% paraformaldehyde in PBS for 15 min. Then cells were washed twice with PBS and permeabilized with 0.5% Triton X-100 in PBS for 15 min. Next, cells were blocked with 3% bovine serum albumin (BSA)/PBS for 1 h at room temperature. The primary antibodies and the fluorophore-conjugated secondary antibodies were sequentially incubated with cells for 2 and 1 h, respectively. After the final twice PBS wash, cells on the coverslips were mounted in the antifade medium and sealed by nail polish. Immunofluorescence images were taken by Olympus BX3-CBH fluorescence microscopy and Leica CTR6000 confocal laser microscopy.
Cell Proliferation Assay
Cytotoxicity of screened drugs were evaluated by cell proliferation assay. Briefly, 293T cells were plated 5000 cells per well in a 96-well plate for 24 h and then treated with serially diluted drugs for 24 h. Next, the live cell number was quantified by Cell Counting Kit 8 (CCK8, Transgene). The absorbance was measured at 450 nm using a microplate spectrophotometer (Multiskan Go, Thermo Scientific). Experiments were conducted in triplicate, and cell viability was calculated as the ratio of experiment group related to the control group.
Endocytosis Assay
HeLa cells grown in DMEM were treated with the indicated dose of drugs for 2 h or 12 h. Then, 25 μg/mL Alexa555-conjugated Transferrin (Thermo Fisher Scientific) was added to the medium for 5, 10, or 15 min of transferrin uptake, followed by immediate fixation with 4% PFA. Cellular uptake of transferrin was analyzed at an excitation wavelength of 543 nm by Olympus BX3-CBH fluorescence microscopy.
Quantification and Statistical Analysis
Three independent experiments or triplicates were performed for quantification. One-way or two-way analysis of variance (ANOVA) with Dunnett post-test was used for testing statistical significance. P values of 0.05 or lower were considered statistically significant. (*, p < 0.05; **, p < 0.01; ***, p < 0.001). GraphPad Prism 8 was used for data processing and statistical analysis.
Acknowledgments
We would like to thank Prof. Zhaohui Qian in Peking Union Medical College, Yinfeng Kang in Sun Yat-sen University and Prof. Peihui Wang in Shandong University for donating the SARS-CoV-2 related plasmids. The work of Shuguang Yuan was supported by funding from Chinese Academy of Sciences, the Shenzhen Institutes of Advanced Technology, CAS, Shenzhen government as well as that from Guangdong province (Grant No. 2019QN01Y306). This work was also supported by a grant from National Natural Science Foundation of China, No. 31671397 (to H.C.L.); Shenzhen Science and Technology Program JCYJ20170818153538196 (to W.S.); COVID-19 emergency tackling research project of Shandong University (Grant No. 2020XGB03 to P.-H.W.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00163.
Sequence alignment between ACE and ACE2; structural similarities between romidepsin and ACE inhibitors, binding pocket similarities between ACE and ACE2; binding mode of romidepsin with ACE2; generation of pseudotyped particles; screen of candidate drugs for suppressing SARS-2-S pseudovirus; biological processes enriched by the pharmacological targets associated with the SARS-CoV-2 infection; structures for candidates and HDAC inhibitors; biological processes that HDACs and ACE2 involved in COVID-19 infections (PDF)
NMA intrinsic flexibility (MP4)
ACE2-romidepsin complex from MD simulations (PDB)
Author Contributions
# K.L., R.Z., W.C., and M.L. contributed equally to this work. S.Y. led the whole project. S.Y. and Hongchang L. initiated and designed the overall experiments. S.Y., H.C.S.C., R.Z., W.C., Hongchun L., X.W. and J.D. performed the computational work. K.L. and M.L. performed the biochemical experiments. All authors wrote and revised the manuscript.
The authors declare the following competing financial interest(s): Both Shuguang Yuan and Stephen Chan are co-founders of AlphaMol Science Ltd.
Supplementary Material
References
- Coomes E. A., and Haghbayan H.. Interleukin-6 in COVID-19: A Systematic Review and Meta-Analysis. medRxiv 2020, 10.1101/2020.03.30.20048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni E.; Valoriani A.; Cei F.; Lamanna R.; Gelli A. M. G.; Ciambotti B.; Vannucchi V.; Moroni F.; Pelagatti L.; Tarquini R.; et al. (2020) Interleukin-6 as prognosticator in patients with COVID-19. J. Infect. 81 (3), 452. 10.1016/j.jinf.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberia L. G.; Gómez E. J. (2020) Political and institutional perils of Brazil’s COVID-19 crisis. Lancet 396 (10248), 367. 10.1016/S0140-6736(20)31681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar B.; Rana S. (2020) COVID 19 in INDIA: Strategies to combat from combination threat of life and livelihood. Journal of Microbiology, Immunology and Infection 53 (3), 389. 10.1016/j.jmii.2020.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.; Li Y. (2020) Beware of the second wave of COVID-19. Lancet 395 (10233), 1321. 10.1016/S0140-6736(20)30845-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Tian D.; Liu W. (2020) [Strategies for vaccine development of COVID-19]. Sheng Wu Gong Cheng Xue Bao 36 (4), 593. 10.13345/j.cjb.200094. [DOI] [PubMed] [Google Scholar]
- Lurie N.; Saville M.; Hatchett R.; Halton J. (2020) Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 382 (21), 1969. 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- Kern W. V.; Biever P. M.; Rieg S.; Panning M. (2020) [SARS-CoV-2 infection (COVID-19): what can we expect?]. Dtsch. Med. Wochenschr. 145 (11), 740. 10.1055/a-1114-3699. [DOI] [PubMed] [Google Scholar]
- Bhagavathula A. S.; Aldhaleei W. A.; Rovetta A.; Rahmani J. (2020) Vaccines and Drug Therapeutics to Lock Down Novel Coronavirus Disease 2019 (COVID-19): A Systematic Review of Clinical Trials. Cureus 12 (5), e8342. 10.7759/cureus.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman A. A.; Al Daajani M. M.; Alsahafi A. J. (2020) Re-positive COVID-19 PCR test: could it be a reinfection?. New Microbes New Infect. 37, 100748. 10.1016/j.nmni.2020.100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry J. (2020) Covid-19: Hong Kong scientists report first confirmed case of reinfection. BMJ. 370, m3340. 10.1136/bmj.m3340. [DOI] [PubMed] [Google Scholar]
- Chatterjee S. K.; Saha S.; Munoz M. N. M. (2020) Molecular Pathogenesis, Immunopathogenesis and Novel Therapeutic Strategy Against COVID-19. Frontiers in Molecular Biosciences 7, 196. 10.3389/fmolb.2020.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M. J.; Hanif M.; Haider M. A.; Ahmed M. U.; Sundas F. N. U.; Hirani A.; Khan I. A.; Anis K.; Karim A. H. (2020) Treatment Options for COVID-19: A Review. Front. Med. 7, 480. 10.3389/fmed.2020.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty D.; Debnath F.; Biswas S.; Bhatta M.; Ganguly S.; Deb A. K.; Saha M. K.; Dutta S. (2020) Exploring Repurposing Potential of Existing Drugs in the Management of COVID-19 Epidemic: A Critical Review. J. Clin. Med. Res. 12 (No. 8), 463. 10.14740/jocmr4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Cheng G. (2020) Will Hydroxychloroquine Still Be a Game-Changer for COVID-19 by Combining Azithromycin?. Front. Immunol. 11, 1969. 10.3389/fimmu.2020.01969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Shen T.; Zhong L.; Liu Z.; Dong X.; Huang T.; Wang Q.; Xiao H. (2020) Research Progress of Chloroquine and Hydroxychloroquine on the COVID-19 and Their Potential Risks in Clinic Use. Front. Pharmacol. 11, 1167. 10.3389/fphar.2020.01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zhang D.; Du G.; Du R.; Zhao J.; Jin Y.; Fu S.; Gao L.; Cheng Z.; Lu Q.; et al. (2020) Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395 (10236), 1569. 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby J. C.; Heimann M. A.; Burleson S. L.; Anzalone B. C.; Swanson J. F.; Wallace D. W.; Greene C. J. (2020) COVID-19: A review of therapeutics under investigation. Journal of the American College of Emergency Physicians Open 1 (3), 231. 10.1002/emp2.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J. K.; Tang T.; Nathan L.; Jaimes J. A.; Hsu H. L.; Daniel S.; Whittaker G. R. (2019) Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J. Visualized Exp. 10.3791/59010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasaud A.; Alharbi N. K.; Hashem A. M. (2020) Generation of MERS-CoV Pseudotyped Viral Particles for the Evaluation of Neutralizing Antibodies in Mammalian Sera. Methods Mol. Biol. 2099, 117. 10.1007/978-1-0716-0211-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samrat S. K.; Tharappel A. M.; Li Z.; Li H. (2020) Prospect of SARS-CoV-2 spike protein: Potential role in vaccine and therapeutic development. Virus Res. 288, 198141. 10.1016/j.virusres.2020.198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. T. T.; Zhang Y.; Shang E.; Shu C.; Quinzii C. M.; Westhoff M. A.; Karpel-Massler G.; Siegelin M. D. (2020) Inhibition of HDAC1/2 Along with TRAP1 Causes Synthetic Lethality in Glioblastoma Model Systems. Cells 9 (7), 1661. 10.3390/cells9071661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric P.; Turcaud S.; Meudal H.; Roques B. P.; Fournie-Zaluski M. C. (1996) Optimal recognition of neutral endopeptidase and angiotensin-converting enzyme active sites by mercaptoacyldipeptides as a means to design potent dual inhibitors. J. Med. Chem. 39 (6), 1210. 10.1021/jm950590p. [DOI] [PubMed] [Google Scholar]
- Bennion C.; Brown R. C.; Cook A. R.; Manners C. N.; Payling D. W.; Robinson D. H. (1991) Design, synthesis, and physicochemical properties of a novel, conformationally restricted 2,3-dihydro-1,3,4-thiadiazole-containing angiotensin converting enzyme inhibitor which is preferentially eliminated by the biliary route in rats. J. Med. Chem. 34 (1), 439. 10.1021/jm00105a066. [DOI] [PubMed] [Google Scholar]
- Corradi H. R.; Schwager S. L.; Nchinda A. T.; Sturrock E. D.; Acharya K. R. (2006) Crystal structure of the N domain of human somatic angiotensin I-converting enzyme provides a structural basis for domain-specific inhibitor design. J. Mol. Biol. 357 (3), 964. 10.1016/j.jmb.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Li F. (2008) Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 82 (14), 6984. 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler P.; Staker B.; Prasad S. G.; Menon S.; Tang J.; Parsons T.; Ryan D.; Fisher M.; Williams D.; Dales N. A.; et al. (2004) ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 279 (17), 17996. 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paola L.; Hadi-Alijanvand H.; Song X.; Hu G.; Giuliani A. (2020) The Discovery of a Putative Allosteric Site in the SARS-CoV-2 Spike Protein Using an Integrated Structural/Dynamic Approach. J. Proteome Res. 19, 4576. 10.1021/acs.jproteome.0c00273. [DOI] [PubMed] [Google Scholar]
- Peng X.; Sun Z.; Kuang P.; Chen J. (2020) Recent progress on HDAC inhibitors with dual targeting capabilities for cancer treatment. Eur. J. Med. Chem. 208, 112831. 10.1016/j.ejmech.2020.112831. [DOI] [PubMed] [Google Scholar]
- Schrödinger, LLC . Schrödinger version 2020v1; 2020.
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; et al. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47 (7), 1739. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Chan H. C. S.; Xu Y.; Tan L.; Vogel H.; Cheng J.; Wu D.; Yuan S. (2020) Enhancing the Signaling of GPCRs via Orthosteric Ions. ACS Cent. Sci. 6 (2), 274. 10.1021/acscentsci.9b01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger, L. Suite 2018: LigPrep, version 2018–2; Schrödinger, 2018.
- Lyman E.; Higgs C.; Kim B.; Lupyan D.; Shelley J. C.; Farid R.; Voth G. A. (2009) A role for a specific cholesterol interaction in stabilizing the Apo configuration of the human A(2A) adenosine receptor. Structure 17 (12), 1660. 10.1016/j.str.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder E.; Damm W.; Maple J.; Wu C.; Reboul M.; Xiang J. Y.; Wang L.; Lupyan D.; Dahlgren M. K.; Knight J. L.; et al. (2016) OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 12 (1), 281. 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- Kobayashi O.; Nanbu S. (2015) Application of particle-mesh Ewald summation to ONIOM theory. Chem. Phys. 461, 47. 10.1016/j.chemphys.2015.08.021. [DOI] [Google Scholar]
- Gordon D. E.; Jang G. M.; Bouhaddou M.; Xu J.; Obernier K.; White K. M.; O’Meara M. J.; Rezelj V. V.; Guo J. Z.; Swaney D. L.; et al. (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583 (7816), 459. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G.; Mlecnik B.; Hackl H.; Charoentong P.; Tosolini M.; Kirilovsky A.; Fridman W. H.; Pages F.; Trajanoski Z.; Galon J. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25 (8), 1091. 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G.; Galon J.; Mlecnik B. (2013) CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29 (5), 661. 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Soding J.; et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. P.; Leroy C.; Sander C. (1998) MView: a web-compatible database search or multiple alignment viewer. Bioinformatics 14 (4), 380. 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
- Landrum G.RDKit: Open-source cheminformatics; Greg Landrum, 2006.
- Yuan S.; Chan H. C.; Filipek S.; Vogel H. (2016) PyMOL and Inkscape Bridge the Data and the Data Visualization. Structure 24 (12), 2041. 10.1016/j.str.2016.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.