Abstract
The recent Ebola epidemics in West Africa underscore the great need for effective and practical therapies for future Ebola virus outbreaks. We have discovered a new series of remarkably potent small molecule inhibitors of Ebola virus entry. These 4-(aminomethyl)benzamide-based inhibitors are also effective against Marburg virus. Synthetic routes to these compounds allowed for the preparation of a wide variety of structures, including a conformationally restrained subset of indolines (compounds 41–50). Compounds 20, 23, 32, 33, and 35 are superior inhibitors of Ebola (Mayinga) and Marburg (Angola) infectious viruses. Representative compounds (20, 32 and 35) have shown good metabolic stability in plasma and liver microsomes (rat and human), and 32 did not inhibit CYP3A4 nor CYP2C9. These 4-(aminomethyl)benzamides are suitable for further optimization as inhibitors of filovirus entry, with potential to be developed as therapeutic agents for the treatment and control of Ebola virus infections.
Graphical Abstract

INTRODUCTION
Ebola virus (EBOV) and Marburg virus (MARV) are Category A emerging infectious agents because of the possibility of an aerosol mode of transmission1, their high fatality rate, and the unpredictable nature of the outbreaks.2–4 These viruses can cause a highly lethal hemorrhagic fever with a 50–90% mortality rate in infected patients.2–3, 5 The 2014–2016 Ebola epidemic in West Africa, which was the largest recorded outbreak, and the recent 2018 epidemic in the Democratic Republic of Congo, which is the tenth outbreak since 1976 and the second biggest Ebola epidemic, underscore the need for drug discovery and development efforts to produce effective treatments. Several Ebola-specific vaccines have shown promising efficacy in animal or nonhuman primate models; however, the production process for vaccines generally takes 6 to 36 months and is considered impractical during a rapidly spreading EBOV outbreak. With the availability of Merck’s investigational Ebola vaccine V920 (rVSV-ZEBOV),6 which is now FDA-approved as Ervebo®, the number of cases has declined to date; however, the outbreak is not yet over. Thus, there is still an urgent medical need to develop efficacious and broad-spectrum small molecule therapeutic agents that are stable, cost-effective and easy to use, and most importantly, agents that can be readily available in an outbreak zone. Such agents could be used alone or in combination with vaccines in future infections.
Some representative antifilovirus compounds are shown in Figure 1. This set includes nucleosides BCX4430 and C-c3-Ado;7–8 a rhodanine derivative LJ-001;9 compound 3.47 with a lipophilic adamantyl group;10 polyaromatic amines FGI-103, FGI-104 and FGI-106;11–13 and our previously published hits, including benzodiazepine-based “compound 7”14 and recently described coumarin-based CBS112915. A group of known drugs with potetntial for repurposing as anti-Ebola agents is the class of estrogen receptor modulators,16 as exemplified by toremefine17, which is displayed in Figure 1. Toremefine has been shown to interact with and destabilize the Ebola virus glycoprotein.18 Only a few compounds have advanced to clinical trials. A pyrazinecarboxamide derivative T-705 (faviprivir)19–20 has shown no efficacy in patients with high levels of Ebola virus in the blood. CMX001 (brincidofovir), a prodrug of the known antiviral medication cidofovir, received an authorization from the U.S. FDA as an emergency investigational new drug, but was subsequently withdrawn in clinical trials, due to the lack of convincing preclinical data. GS-5734 (remdesivir)21, a nucleotide analog was the only small molecule drug tested in the recent 2018 Kivu Ebola outbreak, but it did not demonstrate significant efficacy.
Figure 1.
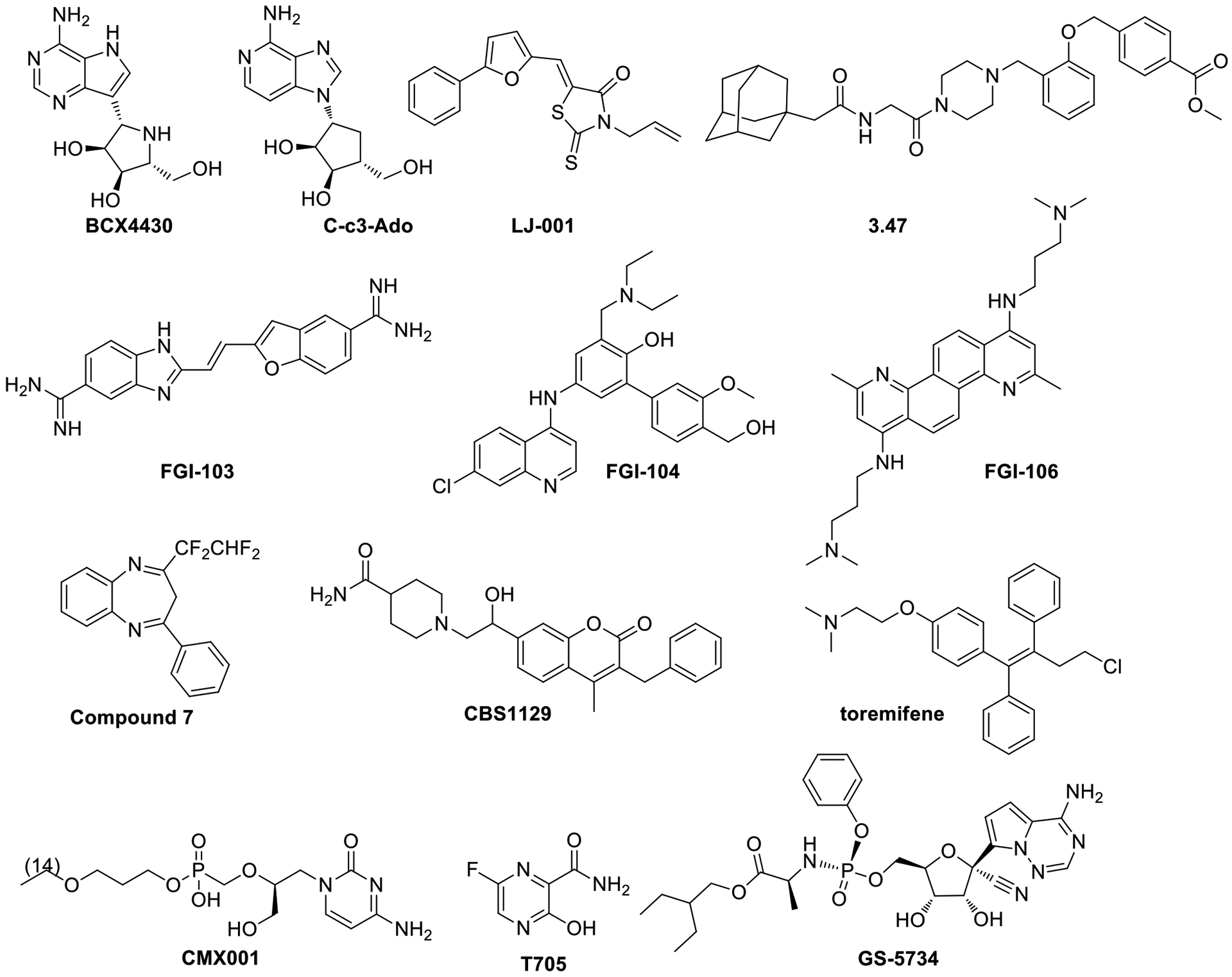
Small molecule antifilovirus compounds.
The EBOV and MARV genomes contain at least seven genes, including the gene that encodes the viral envelope glycoprotein (GP).3 The GP consists of two subunits, GP1 and GP2. The GP1 subunit is responsible for receptor binding and host tropism, while the GP2 subunit mediates viral/cell membrane fusion.3, 22–25 Blocking GP fusion prevents entry into the cell and downstream replication processes. The structural studies of EBOV/MARV GPs, alone and in complex with receptors/antibodies/inhibitors,26–31 provide insights into the elucidation of the filoviral entry mechanism and development of antifiloviral therapeutics. Recent work on EBOV GP in complex with toremifene18 suggests a novel binding mechanism. Toremifene was shown to bind to GP directly and block GP-mediated fusion. This finding has provided validation for the continued development of the 4-(aminomethyl)benzamide antiviral agents reported herein.
RESULTS AND DISCUSSION
Identification of 4-(Aminomethyl)benzamides as Antifilovirus Agents.
One of the challenges of working with highly pathogenic viruses such as EBOV/MARV is that biosafety level 4 (BSL-4) facilities are required to handle the infectious viruses. For the study of GP fusion of many enveloped viruses, this obstacle can be circumvented by a surrogate system called viral pseudotyping.32 This surrogate system has been widely utilized by virologists to study the entry mechanisms of highly pathogenic viruses and to identify and develop antiviral therapeutics, in our31 laboratories and other32–35 laboratories. It is generally accepted that pseudotyped assays for filoviruses and other pathogenic viruses are valid surrogate assays. Thus, many of the antifilovirus compounds displayed in Figure 1 have been identified using a viral pseudotyped assay followed by validation with infectious data. A recent publication from USAMRIID reports a “high degree of correlation” between infectious BSL-4 neutralization assays and pseudotyped BSL-2 fluorescence assays.33 In this study, we used this system to rapidly identify potent EBOV and MARV entry inhibitors. We have screened a ChemBridge Small Molecule Library of 10,000 compounds for filovirus entry inhibitors using a robust, cell-based high-throughput screening (HTS) protocol.34 Several hit compounds, including a set of the substituted 4-(aminomethyl)benzamides, have been shown to inhibit entry of both EBOV and MARV pseudovirions35–36. To validate our initial hits, we tested them against wild type EBOV (1976 Mayinga variant) and MARV (Lake Victoria, 2005 Angola) strains in Vero cells.37 One of the 4-(aminomethyl)benzamides, designated as CBS1118 (Figure 2), was found to have EC50 values < 10 μM for both EBOV and MARV, suggesting that this compound has broad-spectrum antifiloviral activity.
Figure 2.

Structural Optimization of 4-(Aminomethyl)-benzamides as Antifilovirus Agents.
Structure-Activity Relationship (SAR) Development.
The structural modifications on the hit compound CBS1118 were implemented to improve selectivity and potency toward EBOV or MARV through the addition of appropriate substituents in the amide portion of the molecule, the aromatic region or the tertiary amine region as outlined in Figure 2. Upon medicinal chemistry and SAR optimization, we have assembled several sets of modified 4-(aminomethyl)benzamides.
Our general method for the synthesis of 4-(aminomethyl)benzamides is shown in Scheme 1. Treatment of 4-(bromomethyl)benzoic acid esters 1a-e with cyclic amines such as piperidine (2a), morpholine (2b) and 4-methylpiperazine (2c) cleanly produced the 4-(aminomethyl)benzoic acid esters 3a-f. Basic hydrolysis of these esters gave the intermediate benzoic acids, which were subsequently converted to the benzoyl chlorides 4a-f by heating at reflux in thionyl chloride. The benzoyl chlorides 4a-f were coupled with a variety of anilines by utilizing the traditional Schotten-Baumann procedure to provide the 4-(aminomethyl)benzamides 5–35.
Scheme 1.

Synthesis of 4-(Aminomethyl)benzamides 5-35 and 4-(Aminomethyl)thiophenes 38 and 39 a.
The thiophene analogs 38 and 39 were synthesized in a similar manner starting from commercially available methyl 5-(bromomethyl)thiophene-2-carboxylate (36) through the intermediate thiophene-2-carbonyl chloride (37) as shown in Scheme 1.
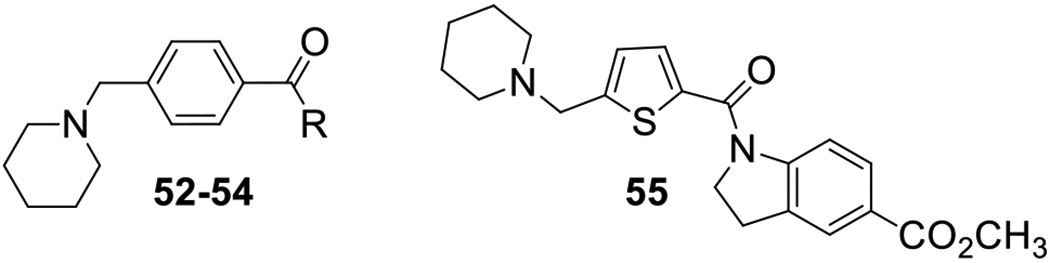
To create conformationally restricted versions of our scaffold, indolines and other related heterocycles were incorporated into the amide portion of the molecule. These compounds were prepared as shown in Scheme 2. Benzoic acids 4a,c were coupled with substituted indolines 40a-h, again using the Schotten-Baumann procedure, to provide compounds 41–50. Using an analogous procedure, 4-(piperidin-1-ylmethyl)benzoyl chloride (4a) was treated with three different amines 51a-c to provide amides 52–54. The thiophene compound 55, bearing a 5-carboethoxy substituted indoline, was synthesized as a constrained analog of compound 38. Conformational restraints can produce compounds that bind with lower energy barriers by eliminating the entropic preorganization component.
Scheme 2.
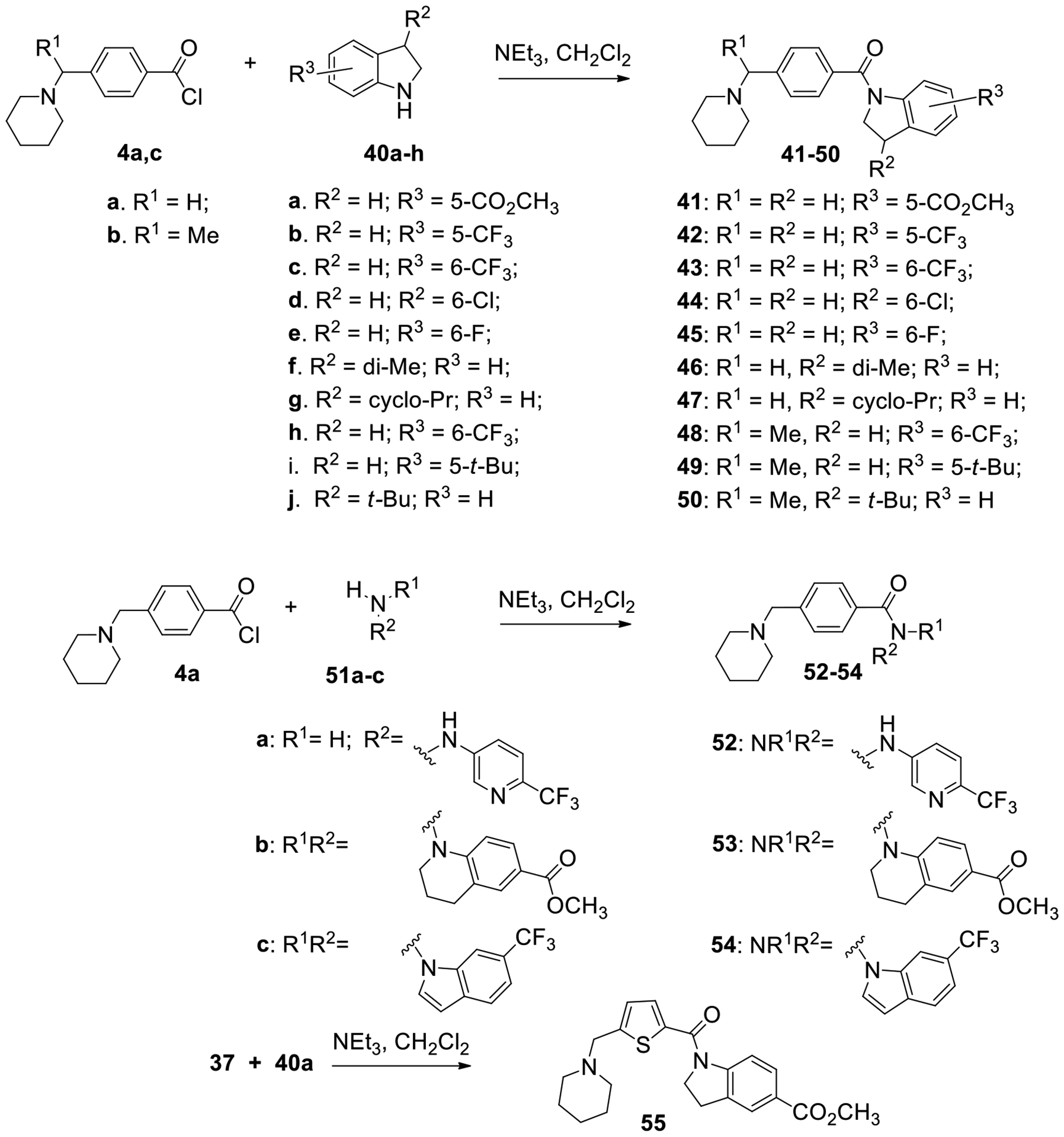
Synthesis of the indoline series of 4-(aminomethyl)benzamides 41–50 and heterocyclic analogs 52-55.
All synthesized compounds were tested against EBOV or MARV GP pseudotyped vesicular stomatitis virus (VSV) in A549 cells and biological data are summarized in Tables 1–3. Toremifene, a small molecule entry inhibitor previously shown to bind and destabilize GP,18 was used as positive control in this assay. Our SAR studies described here mainly focused on the potency of these compounds for EBOV infection inhibition. It started with the replacement of the piperidine ring in ester 5 (HTS hit CBS1118) with morpholine and N-methylpiperazine, to vary the amine substituent. Both of these modifications decreased potency; compounds 6 and 7 showed only 57% and 29% inhibition at 12.5 μM, respectively. Regioisomer 8, with the methyl ester in the meta-position, was completely inactive. 3-(Aminomethyl)benzamide 9 was much weaker than the original hit compound, 4-(aminomethyl)benzamide 5 (only 13% inhibition vs 74%, respectively). Also, the introduction of a carbonyl linker, such as in compound 10, was unfavorable (12.9% inhibition).
Table 1.
Antifiloviral Activity of 4-(Aminomethyl)benzamides 5-8and 10-27.
| N | Structure | Ebola pseudovirus A549 cells | Marburg pseudovirus A549 cells | |||
|---|---|---|---|---|---|---|
| % inhibition | EC50a (μM) | SIb | % inhibition | EC50a (μM) | ||
 | ||||||
| 5 | X = CH2; para- | 74.0 | 9.86 ± 2.14 | 15 | 90.7 | 0.53 ± 0.09 |
| 6 | X = O; para- | 57.2 | - | 68.8 | - | |
| 7 | X = N-Me; para- | 29.4 | - | 30.3 | - | |
| 8 | X = CH2; meta- | 0 | - | 0 | - | |
| 9 | 13.0 | - | 5.90 | - | ||
| 10 | 12.9 | - | 0 | - | ||
 | ||||||
| 11 | R = 4-CO2H | 3.12 | - | 10.3 | - | |
| 12 | R = 4-SO2Me | 0 | - | 17.4 | - | |
| 13 | R = 4-SO2NHMe | 0 | - | 3.30 | - | |
| 14 | R = 4-NHSO2Me | 3.91 | - | 6.81 | - | |
| 15 | R = 4-CN | 0 | - | 12.4 | - | |
| 16 | R = 4-CF3 | 89.9 | 3.87 ± 0.73 | 9 | 79.3 | 5.70 ± 1.56 |
| 17 | R = 3-CF3 | 82.2 | 2.97 ± 0.20 | 15 | 57.5 | 1.41 ± 0.32 |
| 18 | R = 3-Cl, 4-CF3 | 99.7 | 2.34 ± 0.57 | 9 | 98.5 | 2.05 ± 0.29 |
| 19 | R = 3-CF3, 4-Cl | 99.6 | 1.52 ± 0.28 | 7 | 98.2 | 1.24 ± 0.24 |
| 20 | R = 3-Cl, 4-SCF3 | 99.9 | 1.27 ± 0.38 | 5 | 99.8 | 1.61 ± 0.15 |
| 21 | R = 4-cyclo-Pr | 74.3 | 4.57 ± 1.50 | 22 | 66.2 | 2.65 ± 1.64 |
| 22 | R = 4-i-Pr | 93.1 | 2.87 ± 0.38 | 10 | 67.5 | 2.10 ± 1.24 |
| 23 | R = 2,4-di-t-Bu | 99.5 | 0.48 ± 0.17 | 76 | 85.0 | 2.34 ± 0.64 |
| 24 |  |
96.8 | 0.73 ± 0.29 | 19 | 94.0 | 1.63 ± 0.42 |
| 25 | R1 = R2 = H | 98.1 | 0.99 ± 0.18 | 101 | 73.4 | 0.96 ± 0.32 |
| 26 | R1 = F, R2 = H | 96.1 | 0.38 ± 0.12 | 382 | 71.0 | 1.33 ± 1.23 |
| 27 | R1 = H, R2 = Me | 99.3 | 0.33 ± 0.08 | 390 | 88.9 | 0.85 ± 0.10 |
| toremifene | 0.09 ± 0.04 | 299 | ||||
Dose-response studies were conducted to determine EC50 values for those compounds that showed more than 70% inhibition at 12.5 uM concentration; EC50 values were calculated by four-parameter dose-response curve-fitting in GraphPad. Results are from three replicates. Percent inhibition errors are estimated to be <10%; EC50 data are presented as mean ± standard deviation (SD).
Selectivity index is the ratio of CC50/EC50, where CC50 is cytotoxicity assessed by utilizing the “CellTiter 96 aqueous nonradioactive cell proliferation assay” (Promega, Madison, WI).
Table 3.
Antifiloviral Activity of 4-(Aminomethyl)benzamides 41–50, 52-54and 5-(Aminomethyl)thiophenecarboxamide 55.
| N | Structure | Ebola pseudovirus A549 cells | Marburg pseudovirus A549 cells | |||
|---|---|---|---|---|---|---|
| % inhibition | EC50a (μM) | SIb | % inhibition | EC50a (μM) | ||
 | ||||||
| 41 | R1 = R2 = H, R3 = 5-CO2Me | 41.7 | - | 51.2 | - | |
| 42 | R1 = R2 = H, R3 = 5-CF3 | 56.4 | - | 49.6 | - | |
| 43 | R1 = R2 = H, R3 = 6-CF3 | 83.7 | 1.68 ± 0.43 | 38 | 59.2 | 1.46 ± 0.41 |
| 44 | R1 = R2 = H, R3 = 6-Cl | 81.5 | 2.22 ± 0.39 | 14 | 42.4 | 2.31 ± 0.37 |
| 45 | R1 = R2 = H, R3 = 6-F | 20.1 | - | 13.5 | - | |
| 46 | R1 = H, R2 = di-Me, R3 = H | 87.3 | 2.22 ± 1.02 | 33 | 21.4 | 2.90 ± 0.87 |
| 47 | R1 = H, R2 = cyclo-Pr, R3 = H | 74.5 | - | 34.4 | - | |
| 48 | R1 = Me, R2 = H, R3 = 6-CF3 | 95.6 | 1.56 ± 0.69 | 21 | 76.9 | 1.73 ± 0.88 |
| 49 | R1 = Me, R2 = H, R3 = 5-t-Bu | 99.8 | 2.05 ± 0.33 | 9 | 98.1 | 3.09 ± 0.53 |
| 50 | R1 = Me, R2 = t-Bu, R3 = H | 99.9 | 0.16 ± 0.07 | 224 | 92.8 | 3.18 ± 1.33 |
 | ||||||
| 52 | 5.4 | - | 9.4 | - | ||
| 53 | 25.1 | - | 0 | - | ||
| 54 | 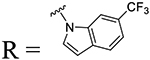 |
0 | - | 0 | - | |
| 55 | 38.9 | - | 0 | - | ||
| toremifene | 0.09 ± 0.04 | 299 | ||||
Dose-response studies were conducted to determine EC50 values for those compounds that showed more than 75% inhibition at 12.5 uM concentration; EC50 values were calculated by four-parameter dose-response curve-fitting in GraphPad. Results are from three replicates. Percent inhibition errors are estimated to be <10%; EC50 data are presented as mean ± SD.
Selectivity index is the ratio of CC50/EC50, where CC50 is cytotoxicity assessed by utilizing the “CellTiter 96 aqueous nonradioactive cell proliferation assay” (Promega, Madison, WI).
Next, we investigated replacements of the potentially metabolically unstable methyl ester with different bioisosteres. Among the monosubstituted analogs 11–17, only compounds 16 and 17, bearing a lipophilic trifluoromethyl substituent in the para- and meta-positions, were more potent than the hit compound 5 (EC50 = 3.87 and 2.97 μM, respectively, vs 9.86 μM for compound 5), while the replacements with more polar or negatively charged groups provided only less potent inhibitors. Introduction of an additional chlorine group in the aromatic ring was well-tolerated and enhanced the potency of compounds 18 (2.34 μM) and 19 (1.52 μM) and its SCF3 analog 20 (1.27 μM) over the corresponding monosubstituted compounds 16 and 17 (Table 1).
Among the alkyl substituted analogs, the potency correlated with the size of the alkyl group as follows: 4-cyclopropyl (21, EC50 = 4.57 μM) < 4-isopropyl (22, 2.87 μM) < cyclohexyl (25, 0.99 μM) < 2-cyclopentylacetonitrile (24, 0.73 μM) < di-tert-butyl (23, 0.48 μM). We chose to maintain the bulky cyclohexyl group to explore some further modifications targeting the benzyl ring, such as introduction of fluorine, as in compound 26 (0.38 μM), and introduction of a methyl group in the linker, as in compound 27 (0.33 μM). All three analogs 25-27 showed excellent activity against EBOV pseudovirus and remarkable potency against pseudoviral MARV (Table 1), coupled with improved selectivity. The selectivity index (SI) in Table 1–3 is a ratio between cytotoxicity (CC50) and antiviral activity of the tested compounds. Additional modifications targeted the aniline portion of the scaffold and comprised replacement of the aromatic ring with more flexible linkers, such as alkyl or alkoxy chains between the cyclohexane moiety and the amide. Replacement with a two-carbon linker, as in 28, led to a 2.3-fold drop in potency, while a shorter linker, as in 29, or insertion of an oxygen, as in the ether 30 or acetal 31, caused greater reductions in potency in both pseudoviral assays (Table 2).
Table 2.
Antifiloviral Activity of 4-(Aminomethyl)benzamides 27-35and 5-(Aminomethyl)thiophenecarboxamides 38-39.
| N | Structure | Ebola pseudovirus A549 cells | Marburg pseudovirus A549 cells | |||
|---|---|---|---|---|---|---|
| % inhibition | EC50a (μM) | SIb | % inhibition | EC50a (μM) | ||
 | ||||||
| 28 | n = 2, R1 = R2 = H | 90.9 | 2.32 ± 0.47 | 25 | 27.3 | 7.74 ± 2.33 |
| 29 | n = 1, R1 = R2 = F | 42.9 | - | 23.0 | - | |
| 30 | 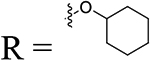 |
52.4 | - | 42.5 | - | |
| 31 |  |
56.0 | - | 37.2 | - | |
 | ||||||
| 32 | X = CH2 | 99.4 | 0.035 ± 0.02 | 231 | 99.0 | 0.38 ± 0.09 |
| 33 | X = O | 99.0 | 0.018 ± 0.01 | 5437 | 72.5 | 2.79 ± 4.78 |
| 34 | R1 = R2 = F, R3 = H, X = CH2 | 99.7 | 0.82 ± 0.47 | 69 | 88.8 | 1.88 ± 0.34 |
| 35 | R1 = H, R2 = Me, R3 = CF3, X = O |
99.3 | 0.012 ± 0.01 | 2088 | 92.3 | 0.18 ± 0.19 |
| 38 | CO2Me | 33.8 | - | 27.2 | - | |
| 39 | CF3 | 80.2 | 5.60 ± 0.67 | 9 | 70.0 | 3.65 ± 0.76 |
| toremifene | 0.09 ± 0.04 | 299 | ||||
Dose-response studies were conducted to determine EC50 values for those compounds that showed more than 70% inhibition at 12.5 uM concentration; EC50 values were calculated by four-parameter dose-response curve-fitting in GraphPad. Results are from three replicates. Percent inhibition errors are estimated to be <10%; EC50 data are presented as mean ± SD.
Selectivity index is the ratio of CC50/EC50, where CC50 is cytotoxicity assessed by utilizing the “CellTiter 96 aqueous nonradioactive cell proliferation assay” (Promega, Madison, WI).
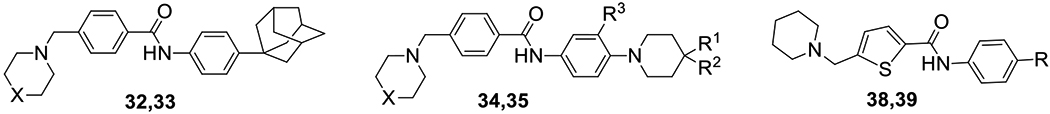
Compounds 32 and 33, bearing an adamantyl group in the para-position of the aniline ring and piperidine and morpholine rings attached to the benzylic portion of the molecule, respectively, were both found to possess excellent potency, with EC50 < 100 nM. Since enhancement of potency appeared to be correlated to the lipophilicity of the substituents on the phenyl ring, our objective was to strike a balance between potency and drug-likeness, including a drug-like LogP value. The lipophilicity of our new lead compound 34 was decreased by replacement of the adamantyl group with a 4,4-difluoropiperidinyl group: compound 34 (EC50 value < 1 μM) has a lower cLogP value of 4.71 as compared to the adamantane analog 32 (cLogP = 5.97). The best compound in this series was compound 35, with an EC50 value of 12 nM. This combination of a trifluoromethyl group in the meta-position and 4-methylpiperidinyl in the para-position of the aniline moiety, with morpholine as the cyclic amine, appeared to be the best combination of substituents with respect to potency. Compound 35 displays low cytotoxicity with an excellent SI value.
On the other hand, replacement of the phenyl ring with thiophene was detrimental to activity and compounds 38 and 39 showed only insignificant inhibition at the test concentration. Perhaps this loss of activity with the thiophenes reflects an unfavorable change in the trajectory of the two aromatic substituents on the thiophene ring compared with the phenyl ring.
Table 3 shows the indoline series compounds, which are amide-constrained versions of the benzamide 5. Indoline 41, having a methyl ester at the 5-position, was less potent than the unconstrained version (41.7% vs 74% inhibition, respectively). While introduction of a trifluoromethyl group at the 6-position afforded compound 43 with augmented potency (EC50 = 1.68 μM), its 5-regioisomer 42 showed only minor improvement over the ester 42 (56.4% inhibition at 12.5 μM vs 41.7%, respectively). Among the two halogenated analogs 44 and 45, the 6-chloro substituted compound 44 showed enhanced potency (EC50 = 2.22 μM). The introduction of gem-dimethyl groups in the 3-position of the indoline ring yielded compound 46 with moderate potency; its cyclopropyl analog 47 was less active. The bulky tert-butyl group at the 3-position improved activity, producing analog 50 with an EC50 value of 0.16 μM. Interestingly, compound 48, bearing a methyl group at the benzyl methylene position, was similar in potency to its desmethylated analog 43.
The final iteration of optimization (Table 3) was to replace the anilide aromatic ring with various heterocycles. Pyridine in 52 represents a simple bioisosteric replacement of carbon with nitrogen in compound 16, which led to a drastic decrease in potency (5.4% vs 89.9% inhibition). Tetrahydroquinoline 53 was less potent than its indoline analog 41 and so was indole 54. Replacement of the core phenyl ring with thiophene as in compound 55 had a lesser (negative) impact on the potency than in the aniline series.
The HTS hit, compound 5, surprisingly, was more potent toward Marburg virus than Ebola virus, as shown in Table 1. The variety of structural modifications described herein, especially the incorporation of bulky alkyl or cycloalkyl substituents, gave compounds either almost equipotent against both viruses, or more potent against Ebola virus. It has been our experience that structural changes which enhance activity for Ebola virus generally do the same for Marburg virus, even though the potencies may differ.
All new compounds with promising activity in the pseudovirus assay were evaluated in wild type EBOV and MARV virus assays in HeLa cells. As shown in Table 4, several compounds have greatly improved potency against EBOV, combined with low cytotoxicity. For example, compounds 32 and 35 display EC50 values of 0.11 μM and 0.31 μM, respectively, with excellent SI values. In addition, they both retained good potency against MARV (EC50 = 1.23 μM for 32 and 0.82 μM for 35). These results imply that the newly developed 4-(aminomethyl)benzamides are broad-spectrum antifiloviral agents that target the GP-mediated entry process. In general, there was a good agreement with the EC50 values for a compound between the HIV-based pseudoviral entry assay and the EBOV and live virus assays, as shown in Tables 1–4. However, discrepancies were observed for some compounds, which is not surprising since these are different bioassays and the discrepancies demonstrate the need for validating antiviral activity for the lead compounds with infectious EBOV and MARV. Remarkably, the HTS hit 5 (CBS1118), which displayed an EC50 value of 2.83 μM against the Ebola strain in Vero E6 cells, was found to be inactive when tested in the HeLa cell line. This inconsistency could be attributed to a nonspecific esterase activity in HeLa cells resulting in metabolism of the parent ester to the inactive carboxylic acid 11. In addition to the higher potency and selectivity displayed by our new hit and lead compounds (Table 4), we have attempted to remove metabolic liabilities.
Table 4.
Antiviral Activity of Selected 4-(Aminomethyl)benzamides in the Wild Type EBOV and MARV Infection Inhibition Assays.
| N | EBOV (HeLa) | MARV (HeLa) | |
|---|---|---|---|
| EC50 (μM)a | SId | EC50 (μM)a | |
| 5 | 2.83b (>25) | 35b | NDc |
| 20 | 1.12 ± 0.21 | 14 | 2.40 ± 0.17 |
| 23 | 0.59 ± 0.16 | 227 | 2.00 ± 0.44 |
| 26 | 1.04 ± 0.50 | 128 | 16.55 ± 0.96 |
| 27 | 1.80 ± 0.51 | 54 | 5.56 ± 0.75 |
| 32 | 0.11 ± 0.03 | 202 | 1.23 ± 0.12 |
| 33 | 0.83 ± 0.19 | 134 | 0.52 ± 0.18 |
| 35 | 0.31 ± 0.07 | 58 | 0.82 ± 0.16 |
| 43 | 3.22 ± 0.39 | 39 | 8.17 ±0.29 |
| 49 | 2.08 ± 0.14 | 19 | 3.38 ± 0.64 |
| 50 | 0.87 ± 0.21 | 137 | 6.32 ± 0.09 |
| toremifene | 0.67 ± 0.02 | 51 | 3.25 ± 0.42 |
EC50 values were calculated by four-parameter dose-response curve-fitting in GraphPad. Results are from three replicates. EC50 data are presented as mean ± SD.
Tested in Vero cells.
ND denotes dose response-dependent inhibition not detected within the range of compound concentrations tested.
Selectivity index is the ratio of CC50/EC50, where CC50 is cytotoxicity assessed by utilizing the “CellTiter 96 aqueous nonradioactive cell proliferation assay” (Promega, Madison, WI).
Mechanism of action (MOA) studies of the 4-(aminomethyl)benzamides with filovirus GPs.
The recently published co-crystal structure of GP with toremifene shows that this ligand binds in a cavity between the attachment (GP1) and fusion (GP2) subunits at the entrance to a large tunnel that links with equivalent tunnels from the other monomers of the trimer at the three-fold axis (Figure 3a,b).18 The key interactions within the binding site are formed between the aromatic rings of toremifene and Y517 and T519 on GP2, as well as multiple hydrophobic residues on the GP2 internal fusion loop such as L554 and M548. Additional interactions include hydrogen bonding with R64 and the side chain of L186 on the GP1 (Figure 3b).
Figure 3.

Toremifene bound to Ebola virus glycoprotein and modeling of binding poses for 32 and 49. (a) Crystal structure of Ebola glycoprotein trimer in complex with toremifene (PDB 5JQ7). (b) Enlarged image of the binding site, where toremifene appears to bind in a cleft between GP1 (blue) and GP2 (red). Residues involved in the protein-ligand interactions are labeled. Overlays of 49 (c) and 32 (d) with toremifene in the Ebola GP structure. Binding poses indicate that all ligands interact with T519; however, only toremifene and 49 interact directly with Y517.
To investigate the potential binding mode of our new compounds, selected 4-(aminomethyl)bezamides were docked into the toremifene hydrophobic binding site of EBOV GP (PDB 5JQ7) as exemplified by 49 and 32 (Figure 3c,d). Docking of indoline 49 (Figure 3c) shows that flexibility of this ligand allows the range of amino acid interactions observed for toremifene, including π-stacking of the indoline heterocycle with Y517 (Figure 3b). While ligand 32 does not appear to be within an acceptable proximity to hydrogen bond with Y517, the carbonyl group of 32 is able to hydrogen bond with T519. The adamantyl ring of 32 is involved in multiple interactions with L554, M548 and other residues in this hydrophobic pocket. Since the adamantyl moiety makes compound 32 more rigid, its binding mode is different from compounds where the amide portion is smaller. Although this ligand is more shifted toward the side chain of R64, it is well-accommodated in the pocket and it overlaps with toremifene. Thus, the modeling results suggest that our 4-(aminomethyl)benzamides can bind to GP in the toremifene site. A recent report on the structural studies of the binding mode of another set of anti-EBOV small molecule agents supports this statement.38
To provide support for the conclusions of the docking studies, including the binding of 32 and 49 to the toremifene binding site, two GP mutants were used: Y517S and T519V.
In the Y517S mutant, concentration-response was right-shifted for toremifene and to a lesser extent 49 (Figure 4a), but not shifted for 32 (Figure 4b). In the T519V mutant, response was right-shifted for toremifene and, to a lesser extent, for 32 (Figure 4b). These results are in agreement with the docking studies and provide further support for the proposed MOA that our new compounds inhibit viral entry by binding directly to the EBOV-GP in a similar fashion to toremifene.
Figure 4.
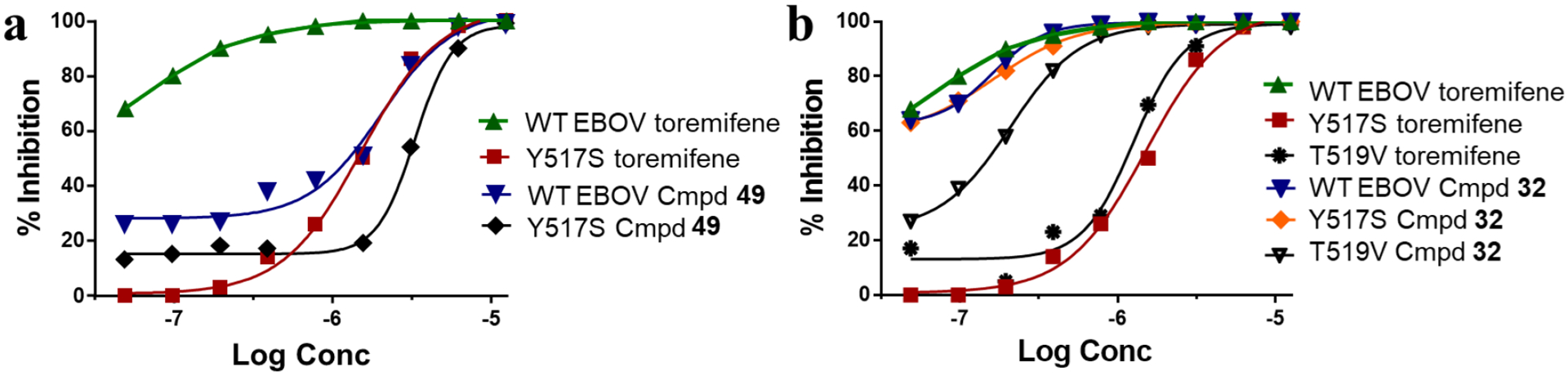
The effects of toremifene and the new 4-(aminomethyl)benzamides 49 and 32 on the infectivity of Y517S and T519V mutants of HIV/EBOV-GP pseudovirus in 293T cells. Functional activities of representative compounds (a) indoline 49; and (b) adamantyl substituted 4-(aminomethyl)benzamide 32. Toremifene was used as a reference compound (EC50 = 62.8 nM).
Compound metabolic stability and CYP inhibition data are shown in Table 5 for selected compounds. Indolines 43 and 50 were found to be surprisingly unstable in rat plasma, and both rat and human liver microsomes, while compounds 20, 23, 32, 33 and 35 possessed acceptable stability and were tested for inhibition of the two most important Phase 1 metabolism human CYPs, CYP3A4 and CYP2C9. Although CYP inhibition is observed at concentrations higher than those required for antiviral activity, the observed weak inhibition of CYP2C9 will need to be considered in further lead optimization studies with this group of potential drug candidates.
Table 5.
ADME Properties of the Selected 4-(Aminomethyl)benzamides.
| N | Metabolic stabilitya | CYP450 inhibitionb | ||||
|---|---|---|---|---|---|---|
| Plasma | Liver microsome | CYP3A4 (IC50, μM) | CYP2C9 (IC50, μM) | |||
| Human | Rat | Human | Rat | |||
| 5 | 63 | 78 | 54 | 0 | NTc | NT |
| 20 | 100 | 88 | 90 | 94 | 15.1 | 4.49 |
| 23 | 85 | 82 | 17 | 99 | >100 | 18.6 |
| 26 | 87 | 86 | 28 | 23 | NT | NT |
| 27 | 88 | 82 | 31 | 24 | NT | NT |
| 32 | 100 | 96 | 75 | 71 | >100 | >100 |
| 33 | 86 | 86 | 34 | 57 | >100d | 3.14d |
| 35 | 100 | 89 | 64 | 75 | >100d | 4.98d |
| 43 | 85 | 0 | 31 | 0 | NT | NT |
| 49 | 94 | 62 | 46 | 27 | 24.0 | 7.94 |
| 50 | 88 | 2 | 2 | 0 | NT | NT |
% of compound remaining after 60 min related to t0.
10 μM quinidine and 30 μM sulfaphenazole were used as the positive inhibitor controls in the CYP3A4 inhibition assay and the CYP2C9 inhibition assay, respectively. Results are from three replicates: errors are estimated to be <10%.
NT, not tested.
tested at Reaction Biology Corp.
CONCLUSIONS
We have described the synthesis of directed libraries from phenyl-substituted, indoline and heterocyclic derivatives of the 4-(aminomethyl)benzamide scaffold. Using the EBOV/MARV pseudotype and filoviral assays for SAR development, we defined inhibitors with high potency (EC50 < 1 μM) and low cytotoxicity (SI > 100). 4-(Aminomethyl)benzamides 32 and 35 are two of the superior compounds that we have identified, with broad-spectrum activity (EC50 values of 0.11 and 0.31 μM; 1.25 and 0.82 μM, respectively) against both Ebola and Marburg infectious viruses. These compounds have favorable ADME properties, suggesting that these inhibitors can be optimized and developed as potential antifiloviral drugs. Several other compounds show promise, including compounds in other series, such as indoline 50. Development of small, orally active molecules as therapeutic agents for prophylaxis or treatment of filovirus infections is critically important. To date, we have witnessed the administration of two Ebola antibodies39–40 and one small molecule drug with an inappropriate mechanism of action41 to Ebola virus patients in Western Africa. Positive results with the Merck vaccine42 have inspired a second vaccine from J&J, which is presently being tested in Uganda.43 And, interestingly, these vaccines have additionally inspired the development of a human antibody cocktail from the Chandran laboratory.44 Notwithstanding the partial success of vaccination, it is generally recognized that vaccines will not be ideal for controlling epidemics, and this realization underscores the continuing need for the development of small molecule therapeutics with appropriate mechanisms.
EXPERIMENTAL SECTION
General.
EC50 values obtained from HIV/EBOV-GP or HIV/MARV-GP pseudotype and EBOV (1976 Mayinga variant) or MARV (Lake Victoria, 2005 Angola) assays are reported as the average of two or more replicates. All solvents and reagents were purchased from commercial suppliers and used without further purification.
1H and 13C NMR spectra were recorded on Bruker DPX-400 or AVANCE-400 spectrometers, at 400 MHz and 100 MHz, respectively. NMR chemical shifts were reported in δ (ppm) using residual solvent peaks as standards (CDCl3: 7.26 ppm (1H), 77.23 ppm (13C); CD3OD: 3.31 ppm (1H), 49.15 ppm (13C); DMSO-d6: 2.50 ppm (1H), 39.52 ppm (13C)). Mass spectra were measured in the ESI mode at an ionization potential of 70 eV with an LC-MSD on Hewlett-Packard spectrometer. All key compounds (5–35, 38, 39, 41–50, 52–55) possess a purity of at least 95% as assessed by analytical reversed phase HPLC using an ACE 3AQ C8 column (150 × 4.6 mm, particle size 3 μM) with detection at 254 and 280 nm on a Shimadzu SPD-20A VP detector; flow rate =1.0 mL/min; gradient of 10–95% acetonitrile in water (both containing 0.1 vol % of FA) in 20 min.
Cell-based protocol to identify entry inhibitors using a pseudotype virus.34
Low-passage A549 cells were infected by a HIV/MARV-GP or HIV/EBOV-GP pseudotype virus containing a luciferase reporter gene in the presence and absence of compounds at 12.5 μM.45 Plates were incubated for 48 h and the infection was then quantified by the luciferase activity of the infected A549 cells using the Neolite Reporter Gene Assay System (Perkin Elmer; Boston, MA). Compounds that showed ≥70% inhibition at 12.5 μM concentration of EBOV or MARV pseudovirions35, 46 were further evaluated to determine their EC50 values using four-parameter logistic regression analysis in GraphPad (Graphpad Software; San Diego, CA).
Infectivity assay using wild type EBOV or MARV virus.
Wild type Ebola virus or Marburg virus was used for testing the efficacy of compounds. All viral infections were done in a BSL-4 facility at the Texas Biomedical Research Institute and in the NEIDL, Boston University. Briefly, 4,000 HeLa cells per well were grown overnight in 384-well tissue culture plates; the volume of DMEM (Fisher scientific, Cat#MT10017CV) culture media supplemented with 10% fetal bovine serum (Gemini Bio-Products, Cat#100106) was 25 μL. On the day of assay, test drugs were diluted to 1 mM concentration in complete media. A 25 μL volume of this mixture was added to the cells already containing 25 μL of media to achieve a concentration of 500 μM. All treatments were done in triplicate. A 25 μL volume of media was removed from the first wells and added to the next well. This type of serial dilution was performed 12 times and treated cells were then incubated at 37 °C in a humidified CO2 incubator for 1 hour. Final concentrations of 250, 125, 62.5, 31.25, 15.62, 7.81, 3.9, 1.9, 0.97, 0.48, 0.24 and 0.12 μM were achieved upon addition of 25 μL of infection mix containing EBOV or MARV virus. Bafilomycin, at a final concentration of 10 nM, was used as a positive control drug. Infections were designed to achieve a MOI of 0.05 to 0.15. Infected cells were incubated for 24 hours. At 24 hours post-infection, cells were fixed by immersing the plates in formalin for 24 hours at 4 °C. Fixed plates were decontaminated and removed from the BSL-4 facility. Formalin from fixed plates was decanted and plates were washed with PBS. EBOV (or MARV)-infected cells were stained for nuclei using the Hoechst stain at 1:50,000 dilution and virus specific antibody (IBT Bioservices) followed by a fluorescently labeled secondary antibody (Alexa488). The plates were imaged. Nuclei (blue) and infected cells (green) were counted using CellProfiler software (Broad Institute) Version 2.1.1. Total number of nuclei (blue) was used as a proxy for cell numbers and a loss of cell number was assumed to reflect cytotoxicity. Concentrations where total cell numbers were 20% less than the control were rejected from the analysis.
Chemistry. General Method A. N-Alkylation of cyclic amines.
To a solution of methyl bromide (1 eq) in anhydrous dichloromethane (3 mL/mmol), cyclic amine (1.0 eq) and triethylamine (5 eq) were added and the reaction mixture was stirred at room temperature under argon for 8–24 hours to reaction completion as monitored by TLC. The solvents were evaporated, and the solid was treated with a mixture of dichloromethane and sodium bicarbonate (aq). The organic phase was collected, washed with water and brine, and dried over sodium sulfate. Solvent was removed under vacuum to provide crude product, which was further purified by flash chromatography (CombiFlash) by elution with methanol in dichloromethane (5–30%) to yield the desired product.
General Method B. Preparation of acyl chlorides.
A solution of methyl ester in tetrahydrofuran (3 mL/mmol), methanol (1 mL/mmol) and water (0.5 mL/mmol) was treated with sodium hydroxide (2 eq). The reaction mixture was stirred overnight at room temperature. The solvents were removed under vacuum and the residue was treated with thionyl chloride (2 mL/mmol). The reaction mixture was heated at reflux for 1 h and concentrated under vacuum. The product was used in next step without purification.
General Method C. Amide formation from acyl chloride.
To a solution of acyl chloride (1 eq) in anhydrous dichloromethane (3 mL/mmol), was added amine (0.8 eq), followed by triethylamine (5 eq), and the reaction mixture was stirred at room temperature under argon for 8–24 hours until reaction was complete as monitored by TLC. The reaction mixture was diluted with dichloromethane and washed with sodium bicarbonate (aq), and brine, and dried over sodium sulfate. Solvent was removed under vacuum to provide crude product, which was further purified by flash chromatography (CombiFlash) by elution with methanol in dichloromethane (5–30%) to yield the desired product.
Methyl 4-(4-(piperidin-1-ylmethyl)benzamido)benzoate (5).
Yield: 165 mg, 59%. HPLC purity: 99%. 1H NMR (400 MHz, DMSO-d6) δ 1.39 (m, 2H), 1.49 (m, 4H), 2.33 (m, 4H), 3.50 (s, 2H), 3.84 (s, 3H), 7.44 (d, J = 8.1 Hz, 2H), 7.90 (d, J = 8.1 Hz, 2H), 7.95 (m, 4H), 10.51 (s, 1H). 13C NMR (100 MHz, DMSO-d6) δ 24.4, 26.0, 52.3, 54.3, 62.8, 119.9, 124.6, 128.1, 129.0, 130.5, 133.5, 143.5, 144.1, 166.2, 166.3. HRMS (ESI) calculated for C21H25N2O3 ([M+H]+): 353.1865, found: 353.1860.
Methyl 4-(4-(morpholinomethyl)benzamido)benzoate (6).
Yield: 150 mg, 66%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 2.50–2.51 (m, 4H), 3.60 (s, 2H), 3.73 (t, J = 4.5 Hz, 4H), 7.47 (d, J = 8.1 Hz, 2H), 7.83 (d, J = 8.8 Hz, 2H), 7.92 (d, J = 8.1 Hz, 2H), 8.01 (d, J = 8.8 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 55.5, 57.2, 66.6, 70.5, 123.7, 129.1, 131.5, 133.2, 134.2, 137.5, 145.4, 147.0, 171.0, 171.2. HRMS (ESI) calculated for C20H23N2O4 ([M+H]+): 355.1658, found: 355.1651.
Methyl 4-(4-((4-methylpiperazin-1-yl)methyl)benzamido)benzoate (7).
Yield: 140 mg, 38%. HPLC purity: 99%. 1H NMR (400 MHz, MeOD) δ 2.31 (s, 3H), 2.47–2.54 (m, 8H), 3.63 (s, 2H), 3.91 (s, 3H), 7.50 (d, J = 8.2 Hz, 2H), 7.87 (dd, J = 1.9, 7.1 Hz, 2H), 7.91 (d, J = 8.2 Hz, 2H), 8.02 (dd, J = 1.9, 7.1 Hz, 2H). 13C NMR (100 MHz, MeOD) δ 44.4, 51.0, 52.0, 54.2, 61.7, 119.6, 125.2, 127.3, 129.1, 130.0, 133.5, 141.9, 143.2, 166.7, 167.2. HRMS (ESI) calculated for C21H26N3O3 ([M+H]+): 368.1974, found: 368.1977.
Methyl 3-(4-(piperidin-1-ylmethyl)benzamido)benzoate (8).
Yield: 115 mg, 83%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.46–1.47 (m, 2H), 1.57–1.63 (m, 4H), 2.40 (m, 4H), 3.54 (s, 2H), 3.93 (s, 3H), 7.45–7.49 (m, 3H), 7.83–7.85 (m, 3H), 7.95 (s, 1H), 8.07 (dd, J = 1.1, 8.1 Hz, 1H), 8.15 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 24.2, 25.8, 52.2, 54.5, 63.2, 121.0, 124.7, 125.4, 127.0, 129.1, 129.3, 130.8, 133.1, 128.3, 143.2, 165.8, 166.7. HRMS (ESI) calculated for C21H25N2O3 ([M+H]+): 353.1865, found: 353.1865.
Methyl 4-(3-(piperidin-1-ylmethyl)benzamido)benzoate (9).
Yield: 122 mg, 54%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.46 (m, 2H), 1.59 (m, 4H), 2.40 (m, 4H), 3.52 (d, J = 5.4 Hz, 2H), 3.92 (s, 3H), 7.40–7.46 (m, 1H), 7.50–7.51 (m, 1H), 7.72 (d, J = 8.7 Hz, 3H), 7.86 (s, 1H), 8.03–8.07 (m, 2H), 8.20–8.30 (m, 1H). 13C NMR (100 MHz, CDCl3) δ 24.2, 25.8, 52.0, 54.5, 63.3, 119.16, 119.19, 125.6, 125.9, 127.5, 128.6, 130.8, 132.9, 134.4, 139.5, 142.2, 142.3, 165.9, 166.6. HRMS (ESI) calculated for C21H25N2O3 ([M+H]+): 353.1865, found: 353.1863.
Methyl 4-(4-(piperidine-1-carbonyl)benzamido)benzoate (10).
Yield: 88 mg, 22%. HPLC purity: 97%. 1H NMR (400 MHz, CDCl3) δ 1.49 (m, 2H), 1.69 (m, 4H), 3.26 (m, 2H), 3.74 (m, 2H), 3.92 (s, 3H), 7.20 (d, J = 8.2 Hz, 2H), 7.79 (d, J = 8.2 Hz, 2H), 7.93 (d, J = 8.8 Hz, 2H), 8.04 (d, J = 8.8 Hz, 2H), 9.52 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 24.3, 24.4, 25.5, 26.4, 43.2, 48.7, 51.9, 119.4, 125.4, 126.4, 127.8, 130.6, 135.7, 138.8, 142.9, 165.7, 166.7, 169.5. HRMS (ESI) calculated for C21H23N2O4 ([M+H]+): 367.1658, found: 367.1650.
4-(4-(Piperidin-1-ylmethyl)benzamido)benzoic acid (11).
Yield: 8.0 mg, 27%. HPLC purity: 98%. 1H NMR (400 MHz, MeOD) δ 1.47–1.48 (m, 2H), 1.59–1.61 (m, 4H), 2.44 (m, 4H), 3.53 (s, 1H), 3.56 (s, 1H), 7.45 (d, J = 8.2 Hz, 2H), 7.66 (d, J = 9.7 Hz, 2H), 7.92–7.97 (m, 4H). 13C NMR (100 MHz, MeOD) δ 23.7, 25.1, 54.0, 62.8, 120.0, 127.3, 129.2, 129.5, 132.9, 134.6, 141.0, 142.5, 167.5, 173.9. HRMS (ESI) calculated for C20H23N2O3 ([M+H]+): 339.1709, found: 339.1712.
N-(4-(Methylsulfonyl)phenyl)-4-(piperidin-1-ylmethyl)benzamide (12).
Yield: 76 mg, 56%. HPLC purity: 98%. 1H NMR (400 MHz, DMSO-d6) δ 1.39 (m, 2H), 1.49 (m, 4H), 2.33 (m, 4H), 3.18 (s, 3H), 3.31 (s, 2H), 7.45 (d, J = 8.1 Hz, 2H), 7.91 (m, 4H), 8.03 (d, J = 8.7 Hz, 2H), 10.60 (s, 1H). 13C NMR (100 MHz, DMSO-d6) δ 24.3, 26.0, 44.2, 54.3, 62.8, 120.3, 128.2, 128.4, 129.0, 133.3, 135.4, 143.6, 144.2, 166.4. HRMS (ESI) calculated for C20H25N2O3S ([M+H]+): 373.1586, found: 373.1582.
N-(4-(N-Methylsulfamoyl)phenyl)-4-(piperidin-1-ylmethyl)benzamide (13).
Yield: 70 mg, 46%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.42 (m, 2H), 1.55–1.57 (m, 4H), 2.39 (m, 4H), 2.55 (s, 3H), 3.51 (s, 2H), 7.38 (d, J = 8.1 Hz, 2H), 7.59 (d, J = 8.8 Hz, 2H), 7.82–7.85 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 23.8, 25.2. 28.7, 54.3, 63.2, 120.2, 127.4, 128.1, 129.7, 133.3, 133.4, 142.5, 166.9. HRMS (ESI) calculated for C20H26N3O3S ([M+H]+): 388.1695, found: 388.1692.
N-(4-(Methylsulfonamido)phenyl)-4-(piperidin-1-ylmethyl)benzamide (14).
Yield: 79 mg, 55%. HPLC purity: 99%. 1H NMR (400 MHz, MeOD) δ 1.43–1.45 (m, 2H), 1.55–1.60 (m, 4H), 2.40 (m, 4H), 2.93 (s, 3H), 3.53 (s, 3H), 7.20–7.24 (m, 2H), 7.39–7.41 (m, 2H), 7.64 (dd, J = 2.0, 7.9 Hz, 2H), 7.84 (d, J = 8.2 Hz, 2H). 13C NMR (100 MHz, MeOD) δ 23.8, 25.2, 38.4, 54.2, 63.2, 121.7, 121.9, 127.3, 129.7, 133.64, 133.67, 135.5, 166.8. HRMS (ESI) calculated for C20H26N3O3S ([M+H]+): 388.1695, found: 388.1690.
N-(4-Cyanophenyl)-4-(piperidin-1-ylmethyl)benzamide (15).
Yield: 110 mg, 54%. HPLC purity: 97.8%. 1H NMR (400 MHz, CDCl3) δ 1.48 (m, 2H), 1.64 (m, 4H), 2.46 (m, 4H), 3.60 (s, 2H), 7.49 (d, J = 8.1 Hz, 2H), 7.65 (d, J = 8.6 Hz, 2H), 7.87 (m, 4H), 8.31 (bs, 1H). 13C NMR (100 MHz, CDCl3) δ 23.9, 25.4, 45.8, 54.4, 62.9, 107.2, 118.8, 119.9, 127.2, 129.7, 133.0, 133.2, 142.1, 165.8. HRMS (ESI) calculated for C20H22N3O ([M+H]+): 320.1763, found: 320.1761.
4-(Piperidin-1-ylmethyl)-N-(4-(trifluoromethyl)phenyl)benzamide (16).
Yield: 91 mg, 68%. HPLC purity: 99%. 1H NMR (400 MHz, MeOD) δ 1.49 (m, 2H), 1.61–1.64 (m, 4H), 2.47 (m, 4H), 3.60 (s, 2H), 7.49 (d, J = 8.0 Hz, 2H), 7.63 (d, J = 8.3 Hz, 2H), 7.92–7.94 (m, 4H). 13C NMR (100 MHz, MeOD) δ 27.7, 29.1, 67.0, 124.2, 129.71, 129.75, 129.9, 131.4, 133.7, 137.4, 145.1, 145.7, 171.1. HRMS (ESI) calculated for C20H22F3N2O ([M+H]+): 363.1684, found: 363.1688.
4-(Piperidin-1-ylmethyl)-N-(3-(trifluoromethyl)phenyl)benzamide (17).
Yield: 48 mg, 72%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.40–1.47 (m, 2H), 1.57–1.62 (m, 4H), 2.39 (m, 4H), 3.52 (s, 2H), 7.38–7.47 (m, 4H), 7.80 (d, J = 8.1 Hz, 2H), 7.87 (d, J = 8.1 Hz, 1H), 7.97 (s, 1H), 8.36 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 24.2, 25.8, 54.5, 63.2, 117.0, 117.04, 120.9, 1225, 123.3, 125.2, 127.0, 129.45, 129.55, 131.2, 131.5, 132.9, 138.6, 143.2, 166.0. HRMS (ESI) calculated for C20H22F3N2O ([M+H]+): 363.1684, found: 363.1680.
N-(3-Chloro-4-(trifluoromethyl)phenyl)-4-(piperidin-1-ylmethyl)benzamide (18).
Yield: 49 mg, 58%. HPLC purity: 98.8%. 1H NMR (400 MHz, CDCl3) δ 1.40–1.44 (m, 2H), 1.51–1.55 (m, 4H), 2.36 (m, 4H), 3.47 (s, 2H), 7.35 (d, J = 7.8 Hz, 2H), 7.56 (d, J = 8.6 Hz, 1H), 7.66 (d, J = 8.6 Hz, 1H), 7.79 (d, J = 7.8 Hz, 2H), 7.95 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 23.8, 25.2, 54.3, 63.2, 117.6, 121.5, 122.3, 123.0, 123.4, 124.2, 127.4, 127.9, 127.95, 129.7, 132.6, 133.1, 141.7, 142.6, 166.8. HRMS (ESI) calculated for C20H21ClF3N2O ([M+H]+): 397.1295, found: 397.1293.
N-(4-Chloro-3-(trifluoromethyl)phenyl)-4-(piperidin-1-ylmethyl)benzamide (19).
Yield: 52 mg, 62%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.46–1.49 (m, 2H), 1.58–1.63 (m, 4H), 2.41 (m, 4H), 3.51 (s, 2H), 7.45 (d, J = 8.1 Hz, 2H), 7.49 (d, J = 8.7 Hz, 1H), 7.81 (d, J = 8.1 Hz, 2H), 7.89 (dd, J = 2.7, 8.7 Hz, 2H), 7.99 (d, J = 2.7 Hz, 1H), 8.11–8.20 (m, 1H). 13C NMR (100 MHz, CDCl3) δ 24.1, 25.8, 54.5, 63.1, 119.3, 119.35, 121.2, 123.9, 124.3, 126.9, 127.1, 128.5, 128.8, 129.4, 131.9, 132.6, 136.9, 143.4, 166.1. HRMS (ESI) calculated for C20H21ClF3N2O ([M+H]+): 397.1295, found: 397.1299.
N-(3-Chloro-4-((trifluoromethyl)thio)phenyl)-4-(piperidin-1-ylmethyl)benzamide (20).
Yield: 45 mg, 25%. HPLC purity: 99.9%. 1H NMR (400 MHz, CDCl3) δ 1.40–1.48 (m, 2H), 1.58–1.63 (m, 4H), 2.40 (m, 4H), 3.55 (s, 2H), 7.49 (d, J = 8.1 Hz, 2H), 7.60 (dd, J = 2.0, 8.5 Hz, 1H), 7.75 (d, J = 8.5 Hz, 1H),7.81 (d, J = 8.1 Hz, 2H),. 7.90 (b.s, 1H), 8.03 (d, J = 2.0 Hz, 1H). 13C NMR (100 MHz, CDCl3) δ 24.2, 26.0, 54.6, 63.3, 118.1, 118.7, 121.4, 127.0, 129.4, 132.4, 139.4, 141.2, 141.8, 144.2, 166.1. HRMS (ESI) calculated for C20H21ClF3N2OS ([M+H]+): 429.1015, found: 429.1017.
N-(4-Cyclopropylphenyl)-4-(piperidin-1-ylmethyl)benzamide (21).
Yield: 34 mg, 45%. HPLC purity: 99.7%. 1H NMR (400 MHz, CDCl3) δ 0.61–0.65 (m, 2H), 0.90–0.94 (m, 2H), 1.42–1.43 (m, 2H), 1.53–1.59 (m, 4H), 1.82–1.89 (m, 1H), 2.37 (m, 4H), 3.49 (s, 2H), 7.01 (d, J = 6.5 Hz, 2H), 7.36 (d, J = 8.3 Hz, 2H), 7.50 (d, J = 8.3 Hz, 2H), 7.78 (d, J = 6.5 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 8.9, 14.9, 24.0, 25.4, 54.4, 63.3, 120.5, 120.6, 126.1, 127.1, 129.6, 133.8, 135.5, 140.2, 141.5, 166.1. HRMS (ESI) calculated for C22H27N2O ([M+H]+): 335.2123, found: 335.2126.
N-(4-Isopropylphenyl)-4-(piperidin-1-ylmethyl)benzamide (22).
Yield: 46 mg, 32%. HPLC purity: 97.9%. 1H NMR (400 MHz, CDCl3) δ 1.27 (d, J = 6.8 Hz, 6H), 1.49 (m, 2H), 1.59 (m, 4H), 2.40 (m, 4H), 2.93 (quintet, J = 6.8 Hz, 1H), 3.54 (s, 2H), 7.23 (d, J = 8.2 Hz, 2H), 7.43 (d, J = 7.8 Hz, 2H), 7.57 (d, J = 8.2 Hz, 1H), 7.82 (d, J = 7.8 Hz, 2H), 7.95 (b.s, 1H). 13C NMR (100 MHz, CDCl3) δ 24.0, 24.3, 26.0, 33.6, 54.5, 63.4, 120.4, 126.9, 129.3, 133.6, 135.7, 143.1, 145.2, 165.6. HRMS (ESI) calculated for C22H29N2O ([M+H]+): 337.2280, found: 337.2280.
N-(2,4-Di-tert-butylphenyl)-4-(piperidin-1-ylmethyl)benzamide (23).
Yield: 24 mg, 28%. HPLC purity: 97.7%. 1H NMR (400 MHz, MeOD) δ 1.36 (s, 9H), 1.44 (s, 9H), 1.51 (m, 2H), 1.64 (m, 4H), 2.49 (m, 4H), 3.62 (s, 2H), 7.10 (d, J = 8.2 Hz, 1H), 7.32 (dd, J = 2.2, 8.2, Hz, 1H), 7.51 (d, J = 8.2, Hz, 1H), 7.56 (d, J = 2.2 Hz, 1H), 7.95 (d, J = 7.9 Hz, 2H). 13C NMR (100 MHz, MeOD) δ 23.6, 25.1, 30.1, 10.4, 34.2, 34.9, 54.0, 62.7, 123. 4, 123.7, 127.2, 129.6, 130.8, 132.6, 133.5, 141.2, 146.3, 150.1, 168.3. HRMS (ESI) calculated for C27H39N2O ([M+H]+): 407.3062, found: 407.3066.
N-(4-(Cyano(cyclopentyl)methyl)phenyl)-4-(piperidin-1-ylmethyl)benzamide (24).
Yield: 22 mg, 26%. HPLC purity: 98%. 1H NMR (400 MHz, CDCl3) δ 1.46–1.47 (m, 2H), 2.59–1.64 (m, 4H), 1.94–2.10 (m, 6H), 2.42–2.51 (m, 6H), 3.56 (s, 2H), 7.43 (d, J = 8.6 Hz, 2H), 7.47 (d, J = 8.1 Hz, 2H), 7.70 (d, J = 8.6 Hz, 2H), 7.84 (d, J = 8.1 Hz, 2H), 8.09 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 24.1, 25.7, 40.4, 47.3, 54.5, 63.2, 120.5, 124.4, 126.7, 127.0, 129.5, 133.3, 135.6, 137.6, 142.8, 165.7. HRMS (ESI) calculated for C26H32N3O ([M+H]+): 402.2545, found: 402.2551.
N-(4-Cyclohexylphenyl)-4-(piperidin-1-ylmethyl)benzamide (25).
Yield: 14 mg, 17%. HPLC purity: 98%. 1H NMR (400 MHz, CDCl3) δ 1.20–1.40 (m, 7H), 1.55 (m, 4H), 1.68–1.80 (m, 5H), 2.37–2.43 (m, 4H), 3.50 (m, 3H), 7.14 (d, J = 6.5 Hz, 2H), 7.36 (d, J = 6.3 Hz, 2H), 7.50 (d, J = 6.5 Hz, 2H), 7.78 (d, J = 6.3 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 23.8, 25.2, 26.0, 26.7, 34.4, 43.9, 54.3, 63.2, 120.6, 127.1, 129.7, 133.9, 135.7, 144.4, 166.3. HRMS (ESI) calculated for C25H33N2O ([M+H]+): 377.2593, found: 377.2589.
N-(4-Cyclohexylphenyl)-3-fluoro-4-(piperidin-1-ylmethyl)benzamide (26).
Yield: 43 mg, 55%. HPLC purity: 99.6%. 1H NMR (400 MHz, MeOD) δ 1.20–1.44 (m, 7H), 1.51–1.62 (m, 4H), 1.71 (d, J = 7.3 Hz, 1H), 1.83 (m, 4H), 2.47 (m, 5H), 3.62 (s, 2H), 7.17 (d, J = 8.4 Hz, 2H), 7.45 (dd, J = 7.5, 10.4 Hz, 1H), 7.54 (d, J = 8.4 Hz, 2H), 7.61 (d, J = 10.4 Hz, 1H), 7.66 (d, J = 7.5 Hz, 1H). 13C NMR (100 MHz, MeOD) δ 23.7, 25.2, 26.0, 26.7, 34.4, 44.0, 53.9, 55.3, 114.4, 114.6, 121.0, 122.7, 127.0, 127.2, 132.1, 135.6, 136.2, 136.3, 144.7, 160.0, 162.4, 165.4. HRMS (ESI) calculated for C25H32FN2O ([M+H]+): 395.2499, found: 395.2501.
N-(4-Cyclohexylphenyl)-4-(1-(piperidin-1-yl)ethyl)benzamide (27).
Yield: 22 mg, 28%. HPLC purity: 98.6%. 1H NMR (400 MHz, MeOD) δ 1.25–1.35 (m, 11H), 1.38–1.40 (m, 4H), 1.72 (d, J = 6.9 Hz, 1H), 1.84 (m, 4H), 2.41–2.48 (m, 5H), 3.53 (q, J = 6.5 Hz, 1H), 7.18 (d, J = 8.4 Hz, 2H), 7.40 (d, J = 8.2 Hz, 2H), 7.55 (d, J = 8.4 Hz, 2H), 7.87 (d, J = 8.2 Hz, 2H). 13C NMR (100 MHz, MeOD) δ 18.5, 23.8, 25.2, 26.0, 26.7, 34.4, 44.0, 51.5, 65.3, 121.1, 126.9, 127.4, 128.1, 133.9, 135.8, 144.5, 166.9. HRMS (ESI) calculated for C26H35N2O ([M+H]+): 391.2749, found: 391.2749.
N-(2-Cyclohexylethyl)-4-(piperidin-1-ylmethyl)benzamide (28).
Yield: 22 mg, 31%. HPLC purity: 99%. 1H NMR (400 MHz, MeOD) δ 0.91–0.99 (m, 2H), 1.14–1.20 (m, 4H), 1.23–1.26 (m, 1H), 1.32–1.77 (m, 12H), 2.40 (m, 4H), 3.48 (q, J = 6.2 Hz, 2H), 3.50 (s, 2H), 6.11 (m, 1H), 7.38 (d, J = 8.1 Hz, 2H), 7.70 (d, J = 8.1 Hz, 1H). 13C NMR (100 MHz, MeOD) δ 24.2, 25.7, 26.2, 26.5, 33.2, 35.4, 37.1, 37.9, 54.4, 63.2, 126.7, 129.2, 133.6, 141.8, 167.3. HRMS (ESI) calculated for C21H32N2O ([M+H]+): 329.2515, found: 329.2591.
N-((4,4-Difluorocyclohexyl)methyl)-4-(piperidin-1-ylmethyl)benzamide (29).
Yield: 19 mg, 25%. HPLC purity: 95.3%. 1H NMR (400 MHz, MeOD) δ 1.34–1.37 (m, 2H), 1.44–1.46 (m, 2H), 1.57–2.03 (m, 9H), 1.86–2.08 (m, 2H), 2.44 (m, 4H), 3.27 (d, J = 6.9 Hz, 2H), 3.55 (s, 2H), 7.37 (d, J = 8.2 Hz, 2H), 7.74 (d, J = 8.2 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 23.7, 25.1, 26.6, 26.7, 32.7, 33.0, 33.2, 35.9, 44.6, 54.1, 63.0, 127.0, 129.8, 133.4, 140.2, 168.5. HRMS (ESI) calculated for C20H29F2N2O ([M+H]+): 351.2248, found: 351.2244.
N-(2-(Cyclohexyloxy)ethyl)-4-(1-(piperidin-1-yl)ethyl)benzamide (30).
Yield: 21 mg, 29%. HPLC purity: 96.6%. 1H NMR (400 MHz, CDCl3) δ 1.28–1.34 (m, 5H), 1.39–1.41 (m, 5H), 1.54–1.60 (m, 5H), 1.74–1.75 (m, 2H), 1.91–1.94 (m, 2H), 2.37–2.45 (m, 4H), 3.26–3.31 (m, 1H), 3.47–3.50 (m, 1H), 3.63 (d, J = 2.3 Hz, 4H), 6.62 (bs, 1H), 7.40 (d, J = 8.1 Hz, 2H), 7.72 (d, J = 8.1 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 19.0, 24.0, 24.3, 25.7, 25.9, 32.2, 40.0, 51.5, 64.9, 66.4, 77.8, 126.8, 128.0, 133.3, 147.0, 167.3. HRMS (ESI) calculated for C22H35N2O2 ([M+H]+): 359.2699, found: 359.2695.
N-((1,4-Dioxaspiro[4.5]decan-2-yl)methyl)-4-(1-(piperidin-1-yl)ethyl)benzamide (31).
Yield: 40 mg, 52%. HPLC purity: 95%. 1H NMR (400 MHz, CDCl3) δ 1.37–1.41 (m,8H), 1.56–1.66 (m, 14H), 2.34–2.42 (m, 4H), 3.46–3.57 (m, 2H), 3.68–4.10 (m, 2H), 4.33 (t, J = 3.6 Hz, 1H), 4.34–4.35 (m, 1H), 6.56 (m, 1H), 7.39 (d, J = 8.1 Hz, 2H), 7.73 (d, J = 8.1 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 19.1, 23.7, 24.0, 24.4, 25.1, 26.0, 34.6, 36.5, 41.9, 51.5, 64.9, 66.3, 74.2, 109.9, 126.8, 127.9, 132.8, 167.5. HRMS (ESI) calculated for C23H35N2O3 ([M+H]+): 387.2648, found: 387.2649.
N-(4-((1R,3R,5S)-adamantan-1-yl)phenyl)-4-(piperidin-1-ylmethyl)benzamide (32).
Yield: 33 mg, 35%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.46 (m, 2H), 1.61 (m, 4H), 1.79–1.83 (m, 6H), 1.92 (m, 6H), 2.11 (m, 3H), 2.41 (m, 4H), 3.55 (s, 2H), 7.35 (d, J = 8.2 Hz, 2H), 7.43 (d, J = 7.6 Hz, 2H), 7.60 (d, J = 8.2 Hz, 2H), 7.81 (d, J = 7.6 Hz, 2H), 8.02 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 24.2, 25.8, 28.9, 35.9, 36.7, 43.2, 54.4, 63.2, 120.1, 125.4, 127.0, 129.4, 133.8, 135.4, 142.4, 147.7, 165.6. HRMS (ESI) calculated for C29H37N2O ([M+H]+): 429.2906, found: 429.2908.
N-(4-((1R,3R,5S)-Adamantan-1-yl)phenyl)-4-(morpholinomethyl)benzamide (33).
General Yield: 30 mg, 32%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.79 (m, 26H), 1.92 (m, 6H), 2.12 (m, 3H), 2.47 (m, 4H), 3.57 (s, 2H), 3.73 (m, 4H), 7.35 (d, J = 7.6 Hz, 2H), 7.44 (d, J = 7.2 Hz, 2H), 7.59 (d, J = 7.6 Hz, 2H), 7.82 (d, J = 7.2 Hz, 2H), 7.99 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 28.9, 35.9, 36.7, 43.2, 53.5, 62.8, 66.8, 120.1, 125.4, 127.1, 129.4, 134.1, 135.4, 141.7, 147.8, 165.5. HRMS (ESI) calculated for C28H35N2O2 ([M+H]+): 431.2699, found: 431.2702.
N-(4-(4,4-Difluoropiperidin-1-yl)phenyl)-4-(piperidin-1-ylmethyl)benzamide (34).
Yield: 18 mg, 21%. HPLC purity: 97%. 1H NMR (400 MHz, CDCl3) δ 1.48–1.65 (m,6H), 2.09–2.16 (m, 4H), 2.47 (m, 4H), 3.34 (m, 4H), 3.61 (s, 2H), 6.95 (d, J = 8.1 Hz, 2H), 7.47 (d, J = 7.3 Hz, 2H), 7.57 (d, J = 8.1 Hz, 2H), 7.82 (d, J = 7.3 Hz, 2H), 7.93 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 24.0, 25.5, 33.3, 33.6, 33.8, 47.1, 54.3, 62.9, 117.4, 121.7, 127.0, 129.6, 130.9, 134.0, 147.2, 165.4. HRMS (ESI) calculated for C24H30F2N3O ([M+H]+): 414.2357, found: 414.2354.
N-(4-(4-Methylpiperidin-1-yl)-3-(trifluoromethyl)phenyl)-4-(morpholinomethyl) benzamide (35).
Yield: 21 mg, 28%. HPLC purity: 98.9%. 1H NMR (400 MHz, CDCl3) δ 0.99 (d, J = 5.9 Hz, 3H), 1.36–1.45 (m, 3H), 1.67–1.70 (m, 2H), 2.47 (m, 4H), 2.68 (t, J = 10.8 Hz, 2H), 2.98 (d, J = 10.8 Hz, 2H), 3.57 (s, 2H), 3.73 (m, 4H), 7.32 (d, J = 8.7 Hz, 1H), 7.44 (d, J = 7.6 Hz, 2H), 7.76 (s, 1H), 7.82 (d, J = 7.6 Hz, 2H), 7.89 (d, J = 8.7 Hz, 1H), 8.20 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 22.0, 30.6, 34.7, 53.5, 54.4, 62.8, 66.8, 119.1, 122.3, 124.6, 125.0, 127.1, 127.3, 127.6, 129.4, 133.4, 134.2, 142.1, 150.0, 165.7. HRMS (ESI) calculated for C25H31F3N3O2 ([M+H]+): 462.2368, found: 462.2366.
Methyl 4-(5-(piperidin-1-ylmethyl)thiophene-2-carboxamido)benzoate (38).
Yield: 71 mg, 44%. HPLC purity: 96%. 1H NMR (400 MHz, CDCl3) δ 1.25–1.27 (m, 2H), 1.60–1.61 (m, 4H), 2.46 (m, 4H), 3.70 (s, 2H), 3.92 (s, 3H), 6.93 (m, 1H), 7.55 (d, J = 3.2 Hz, 1H), 7.70 (d, J = 8.7 Hz, 2H), 7.84–7.92 (m, 1H), 8.03–8.06 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 24.1, 25.9, 52.0, 54.3, 57.9, 119.0, 125.6, 126.0, 129.2, 130.8, 136.8, 141.9, 150.1, 166.5. HRMS (ESI) calculated for C19H23N2O3S ([M+H]+): 359.1429, found: 359.1425.
5-(Piperidin-1-ylmethyl)-N-(4-(trifluoromethyl)phenyl)thiophene-2-carboxamide (39).
Yield: 51 mg, 33%. HPLC purity: 98.8%. 1H NMR (400 MHz, CDCl3) δ 1.40–1.44 (m, 2H), 1.56–1.60 (m, 4H), 2.44 (m, 4H), 3.67 (s, 2H), 6.87 (d, J = 3.7 Hz, 1H), 7.54 (d, J = 8.6 Hz, 2H), 7.59 (d, J = 3.7 Hz, 1H), 7.74 (d, J = 8.6 Hz, 2H), 8.27 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 24.1, 25.9, 54.3, 57.9, 119.9, 122.7, 125.4, 125.9, 126.0, 126.2, 129.3, 136.9, 140.9, 150.3, 160.6. HRMS (ESI) calculated for C18H20F3N2OS ([M+H]+): 369.1248, found: 369.1242.
Methyl 1-(4-(piperidin-1-ylmethyl)benzoyl)indoline-5-carboxylate (41).
Yield: 66 mg, 47%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.46–1.47 (m, 2H), 1.58–1.63 (m, 4H), 2.40 (m, 4H), 3.16 (t, J = 8.3 Hz, 2H), 3.53 (s, 2H), 3.90 (s, 3H), 4.16 (t, J = 8.3 Hz, 2H), 7.42 (d, J = 8.1 Hz, 2H), 7.51 (d, J = 8.1 Hz, 2H), 7.86 (bs, 1H), 7.89 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 29.1, 30.7, 32.4, 57.1, 59.1, 67.6, 129.8, 131.0, 132.1, 133.8, 134.2, 138.6, 140.1, 146.5, 151.9, 152.1, 171.0, 173.8. HRMS (ESI) calculated for C23H27N2O3 ([M+H]+): 379.2022, found: 379.2016.
(4-(Piperidin-1-ylmethyl)phenyl)(5-(trifluoromethyl)indolin-1-yl)methanone (42).
Yield: 49 mg, 60%. HPLC purity: 99.8%. 1H NMR (400 MHz, CDCl3) δ 1.46–1.49 (m, 2H), 1.58–1.63 (m, 4H), 2.41 (m, 4H), 3.17 (t, J = 8.4, 2H), 3.54 (s, 2H), 4.15 (t, J = 8.4, 2H), 7.42–7.44 (m, 4H), 7.51 (d, J = 8.1 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 24.2, 25.9, 27.8, 50.6, 50.9, 54.5, 63.4, 116.7, 121.9, 122.9, 124.9, 125.6, 127.0, 129.3, 133.0, 134.8, 141.8, 145.7, 169.4. HRMS (ESI) calculated for C22H24F3N2O ([M+H]+): 389.1841, found: 389.1836.
(4-(Piperidin-1-ylmethyl)phenyl)-(6-(trifluoromethyl)indolin-1-yl)methanone (43).
Yield: 54 mg, 66%. HPLC purity: 98%. 1H NMR (400 MHz, DMSO-d6) δ 1.40–1.48 (m, 2H), 1.60–1.65 (m, 4H), 2.44 (m, 4H), 3.18 (t, J = 8.3, 2H), 3.57 (s, 2H), 4.16 (t, J = 8.3, 2H), 7.28 (d, J = 8.4 Hz, 2H), 7.44 (d, J = 8.1 Hz, 2H), 7.51 (d, J = 8.3 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 24.1, 25.7, 28.0, 50.7, 54.4, 63.2, 113.8, 120.8, 122.7, 125.0, 125.4, 127.0, 129.4, 129.9, 135.0, 136.2, 141.2, 143.3, 169.2; HRMS (ESI) calculated for C22H24F3N2O ([M+H]+): 389.1841, found: 389.1840.
(6-Chloroindolin-1-yl)(4-(piperidin-1-ylmethyl)phenyl)methanone (44).
Yield: 35 mg, 45%. HPLC purity: 99%. 1H NMR (400 MHz, MeOD) δ 1.48–1.49 (m, 2H), 1.59–1.65 (m, 4H), 2.47 (m, 4H), 3.11 (t, J = 8.3 Hz, 2H), 3.59 (s, 2H), 4.10–4.14 (m, 2H), 7.01–7.03 (m, 1H), 7.16 (d, J = 8.4 Hz, 2H), 7.46 (d, J = 8.2 Hz, 1H), 7.52 (d, J = 8.2 Hz, 2H). 13C NMR (100 MHz, MeOD) δ 23.7, 25.1, 27.3, 54.0, 63.0, 124.0, 125.6, 126.8, 129.9, 131.1, 132.4, 135.2, 139.8, 169.6. HRMS (ESI) calculated for C21H24ClN2O ([M+H]+): 355.1577, found: 355.1573.
(6-Fluoroindolin-1-yl)(4-(piperidin-1-ylmethyl)phenyl)methanone (45).
Yield: 39 mg, 39%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.47–1.48 (m, 2H), 1.60–1.66 (m, 4H), 2.44 (m, 4H), 3.08 (t, J = 8.0 Hz, 2H), 3.57 (s, 2H), 4.13 (t, J = 8.0 Hz, 2H), 6.70 (m, 1H), 7.12 (t, J = 7.1 Hz, 1H), 7.44 (d, J = 7.0 Hz, 2H), 7.50 (d, J = 7.0 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 24.1, 25.7, 27.5, 51.4, 54.4, 63.2, 105.4, 110.2, 125.2, 127.0, 127.5, 129.4, 135.2, 140.9, 143.9, 160.9, 163.3, 169.1. HRMS (ESI) calculated for C21H24FN2O ([M+H]+): 339.1873, found: 339.1869.
(3,3-Dimethylindolin-1-yl)(4-(piperidin-1-ylmethyl)phenyl)methanone (46).
Yield: 18 mg, 24%. HPLC purity: 99%. 1H NMR (400 MHz, MeOD) δ 1.32 (c, 6H), 1.49–1.50 (m, 2H), 1.62–1.65 (m, 4H), 2.48 (m, 4H), 3.59 (s, 2H), 3.84 (s, 2H), 7.11–7.27 (m, 3H), 7.48 (d, J = 7.9 Hz, 2H), 7.55 (d, J = 7.8 Hz, 2H), 8.05 (m, 1H). 13C NMR (100 MHz, MeOD) δ 23.7, 25.1, 26.5, 54.0, 62.8, 121.9, 124.5, 126.8, 129.7, 135.5, 140.1, 140.9, 142.0, 169.5; HRMS (ESI) calculated for C23H29N2O ([M+H]+): 349.2280, found: 349.2280.
(4-(Piperidin-1-ylmethyl)phenyl)(spiro[cyclopropane-1,3’-indolin]-1’-yl)methanone (47).
Yield: 29 mg, 39%. HPLC purity: 99%. 1H NMR (400 MHz, MeOD) δ 1.06 (m, 4H), 1.49–1.50 (m, 2H), 1.60–1.65 (m, 4H), 2.47 (m, 4H), 3.58 (s, 2H), 4.06 (s, 2H), 6.78 (d, J = 8.3 Hz, 1H), 7.04–7.13 (m, 2H), 7.47 (d, J = 8.1 Hz, 2H), 7.54 (d, J = 8.1 Hz, 2H), 8.20 (m, 1H). 13C NMR (100 MHz, MeOD) δ 15.8, 22.4, 23.7, 25.1, 53.9, 58.7, 62.8, 116.7, 118.4, 124.5, 126.1, 126.6, 129.7, 135.7, 137.8, 139.8, 142.6, 169.7. HRMS (ESI) calculated for C23H26N2O ([M+H]+): 347.2123, found: 347.2120.
(4-(1-(Piperidin-1-yl)ethyl)phenyl)(6-(trifluoromethyl)indolin-1-yl)methanone (48).
Yield: 34 mg, 42%. HPLC purity: 98%. 1H NMR (400 MHz, CDCl3) δ 1.43–1.44 (m, 5H), 1.61–1.62 (m, 4H), 2.43–2.49 (m, 4H), 3.18 (t, J = 8.2 Hz, 2H), 3.52 (m, 1H), 4.18 (t, J = 8.1 Hz, 2H), 7.28 (d, J = 7.0 Hz, 1H), 7.44 (d, J = 8.1 Hz, 2H), 7.51 (d, J = 8.1 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 19.2, 24.4, 26.0, 28.1, 29.8, 51.8, 65.5, 121.6, 125.8, 127.8, 128.9, 130.4, 135.7, 137.1, 144.1, 170.2. HRMS (ESI) calculated for C23H26F3N2O ([M+H]+): 403.1997, found: 403.1993.
(5-(tert-Butyl)indolin-1-yl)(4-(1-(piperidin-1-yl)ethyl)phenyl)methanone (49).
Yield: 31 mg, 40%. HPLC purity: 98%. 1H NMR (400 MHz, CDCl3) δ 1.31 (s, 9H), 1.42–1.44 (m, 5H), 1.58–1.63 (m, 4H), 2.38–2.46 (m, 4H), 3.12 (t, J = 8.2 Hz, 2H), 3.48–3.51 (m, 2H), 4.11 (bs, 2H), 7.26 (d, J = 8.9 Hz, 1H), 7.40 (d, J = 8.1 Hz, 2H), 7.50 (d, J = 8.1 Hz, 2H), 8.2 (bs, 1H). 13C NMR (100 MHz, CDCl3) δ 19.0, 24.4, 26.0, 28.4, 31.5, 34.4, 50.6, 51.4, 65.0, 121.8, 124.1, 127.1, 127.9, 132.1, 135.6, 145.8, 168.8. HRMS (ESI) calculated for C26H35N2O ([M+H]+): 391.2749, found: 391.2743.
(3-(tert-Butyl)indolin-1-yl)(4-(1-(piperidin-1-yl)ethyl)phenyl)methanone (50).
Yield: 40 mg, 51%. HPLC purity: 98%. 1H NMR (400 MHz, CDCl3) δ 0.95 (s, 9H), 1.43–1.44 (m, 5H), 1.58–1.62 (m, 4H), 2.41–2.46 (m, 4H), 2.94–2.97 (m, 1H), 3.50 (quintet, J = 6.4 Hz, 2H), 4.07 (t, J = 10.0 Hz, 2H), 7.08 (m, 2H), 7.28 (d, J = 4.6 Hz, 1H), 7.41 (d, J = 7.8 Hz, 2H), 7.49 (d, J = 7.8 Hz, 2H). 13C NMR (100 MHz, CDCl3) δ 18.8, 18.9, 24.4, 26.0, 26.9, 34.3, 50.3, 51.4, 64.9, 123.1, 127.0, 127.5, 128.0, 134.0, 135.3, 143.2, 146.0, 168.4. HRMS (ESI) calculated for C26H35N2O ([M+H]+): 391.2749, found: 391.2748.
4-(Piperidin-1-ylmethyl)-N-(6-(trifluoromethyl)pyridin-3-yl)benzamide (52).
Yield: 34 mg, 44%. HPLC purity: 99%. 1H NMR (400 MHz, CDCl3) δ 1.41–1.42 (m, 2H), 1.52–1.58 (m, 4H), 2.37 (m, 4H), 3.50 (s, 2H), 7.39 (d, J = 8.2 Hz, 2H), 7.64 (d, J = 8.6 Hz, 1H), 7.85 (d, J = 8.2 Hz, 2H), 8.57 (dd, J = 2.2, 8.6 Hz, 1H), 8.76 (d, J = 2.2 Hz, 1H). 13C NMR (100 MHz, CDCl3) δ 23.9, 25.3, 54.4, 63.3, 120.9, 127.5, 127.8, 129.7, 132.7, 138.3, 141.3, 142.2, 142.5, 167.1. HRMS (ESI) calculated for C19H21F3N3O ([M+H]+): 364.1637, found: 364.1642.
Methyl 1-(4-(piperidin-1-ylmethyl)benzoyl)-1,2,3,4-tetrahydroquinoline-6-carboxylate (53).
Yield: 24 mg, 17%. HPLC purity: 98%. 1H NMR (400 MHz, CDCl3) δ 1.38–1.44 (m, 2H), 1.55–1.59 (m, 4H), 2.03–2.09 (m, 2H), 2.35 (m, 4H), 2.90 (t, J = 6.5 Hz, 2H), 3.46 (s, 2H), 3.88 (s, 3H), 3.92 (t, J = 6.5 Hz, 2H), 6.81 (d, J = 8.5 Hz, 1H), 7.25 (d, J = 8.1 Hz, 2H), 7.32 (d, J = 8.1 Hz, 2H), 7.52 (dd, J = 1.6, 8.4 Hz, 1H), 7.85 (s, 1H). 13C NMR (100 MHz, CDCl3) δ 23.7, 24.2, 25.8, 27.0, 44.9, 51.9, 54.4, 63.2, 124.8, 125.5, 127.0, 128.5, 128.8, 129.3, 130.0, 130.6, 134.2, 143.6, 166.6, 170.6. HRMS (ESI) calculated for C24H29N2O3 ([M+H]+): 393.2178, found: 393.2175.
(4-(Piperidin-1-ylmethyl)phenyl)(6-(trifluoromethyl)-1H-indol-1-yl)methanone (54).
Yield: 4 mg, 5%. HPLC purity: 95%. 1H NMR (400 MHz, MeOD) δ 1.51–1.52 (m, 2H), 1.62–1.68 (m, 4H), 2.51 (m, 4H), 3.66 (d, 2H), 6.81 (d, J = 3.8 Hz, 1H), 7.59–7.63 (m, 4H), 7.76 (d, J = 8.0 Hz, 2H), 7.81 (d, J = 8.2 Hz, 1H), 8.64 (s, 1H). 13C NMR (100 MHz, MeOD) δ 23.6, 25.1, 54.0, 62.7, 107.7, 113.0, 120.0, 121.3, 129.0, 129.6, 130.4, 133.4, 135.1, 142.5, 167.1. HRMS (ESI) calculated for C22H22FN2O ([M+H]+): 387.1684, found: 387.1689.
Methyl 1-(5-(piperidin-1-ylmethyl)thiophene-2-carbonyl)indoline-5-carboxylate (55).
Yield: 27 mg, 34%. HPLC purity: 99.6%. 1H NMR (400 MHz, CDCl3) δ 1.44–1.47 (m, 2H), 1.58–1.63 (m, 4H), 2.46 (m, 4H), 3.25 (t, J = 8.4 Hz, 2H), 3.70 (s, 2H), 3.90 (s, 3H), 4.44 (t, J = 8.4 Hz, 2H), 6.91 (d, J = 3.7 Hz, 1H), 7.49 (d, J = 3.7 Hz, 1H), 7.88 (s, 1H), 7.92 (d, J = 8.4 Hz, 1H), 7.78 (d, J = 8.4 Hz, 1H). 13C NMR (100 MHz, CDCl3) δ 24.1, 25.9, 28.2, 50.9, 51.9, 54.3, 57.9, 117.0, 125.6, 126.0, 129.9, 130.6, 131.9, 137.2, 147.3, 149.2, 162.0, 166.7. HRMS (ESI) calculated for C21H25N2O3S ([M+H]+): 358.1586, found: 358.1588.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant from the NIH (R41 AI126971 and R42 AI126971 to L.R.)
ABBREVIATIONS
- EBOV
Ebola virus
- eGFP
enhanced green fluorescent protein
- ESI
electrospray ionization
- GP
glycoprotein
- LC-MSD
liquid chromatography/mass selective detector
- MARV
Marburg virus
- TLC
thin layer chromatography
- VCV
vesicular stomatitis virus
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: Molecular formula strings (CSV).
Experimental procedures describing general experimental information, molecular modeling and biological assays (PDF).
The authors declare the following competing financial interest(s): L.R. is the owner of Chicago BioSolutions, Inc. and thus declares potential financial interests as does N.P.P. and I.N.G. who are employed by Chicago BioSolutions, Inc.
REFERENCES
- 1.Mekibib B; Arien KK, Aerosol transmission of filoviruses. Viruses 2016, 8 (5), DOI: 10.3390/v8050148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray M, Defense against filoviruses used as biological weapons. Antiviral Res 2003, 57 (1–2), 53–60. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez A; Khan AS; Zaki SR; Nabel GJ; Ksiazek T; Peters CJ, Filoviridae: Marburg and Ebola Viruses In Fields Virology, Knipe DM; Howley PM, Eds. Lippincourt, Williams & Wilkins: Philadelphia PA, 2001; pp 1279–1304. [Google Scholar]
- 4.Bossi P; Garin D; Guihot A; Gay F; Crance JM; Debord T; Autran B; Bricaire F, Bioterrorism: management of major biological agents. Cell Mol Life Sci 2006, 63 (19–20), 2196–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters CJ; Khan AS, Filovirus diseases. Curr Top Microbiol Immunol 1999, 235, 85–95. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J, Ebola outbreak continues despite powerful vaccine. Science 2019, 364 (6437), 223, DOI: 10.1126/science.364.6437.223. [DOI] [PubMed] [Google Scholar]
- 7.Warren TK; Wells J; Panchal RG; Stuthman KS; Garza NL; Van Tongeren SA; Dong L; Retterer CJ; Eaton BP; Pegoraro G; Honnold S; Bantia S; Kotian P; Chen X; Taubenheim BR; Welch LS; Minning DM; Babu YS; Sheridan WP; Bavari S, Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 2014, 508 (7496), 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huggins J; Zhang ZX; Bray M, Antiviral drug therapy of filovirus infections: S-adenosylhomocysteine hydrolase inhibitors inhibit Ebola virus in vitro and in a lethal mouse model. J Infect Dis 1999, 179 Suppl 1, 240–247. [DOI] [PubMed] [Google Scholar]
- 9.Wolf MC; Freiberg AN; Zhang T; Akyol-Ataman Z; Grock A; Hong PW; Li J; Watson NF; Fang AQ; Aguilar HC; Porotto M; Honko AN; Damoiseaux R; Miller JP; Woodson SE; Chantasirivisal S; Fontanes V; Negrete OA; Krogstad P; Dasgupta A; Moscona A; Hensley LE; Whelan SP; Faull KF; Holbrook MR; Jung ME; Lee B, A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A 2010, 107 (7), 3157–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cote M; Misasi J; Ren T; Bruchez A; Lee K; Filone CM; Hensley L; Li Q; Ory D; Chandran K; Cunningham J, Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 2011, 477 (7364), 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren TK; Warfield KL; Wells J; Enterlein S; Smith M; Ruthel G; Yunus AS; Kinch MS; Goldblatt M; Aman MJ; Bavari S, Antiviral activity of a small-molecule inhibitor of filovirus infection. Antimicrob Agents Chemother 2010, 54 (5), 2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinch MS; Yunus AS; Lear C; Mao H; Chen H; Fesseha Z; Luo G; Nelson EA; Li L; Huang Z; Murray M; Ellis WY; Hensley L; Christopher-Hennings J; Olinger GG; Goldblatt M, FGI-104: a broad-spectrum small molecule inhibitor of viral infection. Am J Transl Res 2009, 1 (1), 87–98. [PMC free article] [PubMed] [Google Scholar]
- 13.Aman MJ; Kinch MS; Warfield K; Warren T; Yunus A; Enterlein S; Stavale E; Wang P; Chang S; Tang Q; Porter K; Goldblatt M; Bavari S, Development of a broad-spectrum antiviral with activity against Ebola virus. Antiviral Res 2009, 83, 245–251. [DOI] [PubMed] [Google Scholar]
- 14.Basu A; Li B; Mills DM; Panchal RG; Cardinale SC; Butler MM; Peet NP; Majgier-Baranowska H; Williams JD; Patel I; Moir DT; Bavari S; Ray R; Farzan MR; Rong L; Bowlin TL, Identification of a small-molecule entry inhibitor for filoviruses. J Virol 2011, 85 (7), 3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng H; Schafer A; Soloveva V; Gharaibeh D; Kenny T; Retterer C; Zamani R; Bavari S; Peet NP; Rong L, Identification of a coumarin-based antihistamine-like small molecule as an anti-filoviral entry inhibitor. Antiviral Res 2017, 145, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan H; Du X; Zhang J; Zheng H; Lu X; Wu Q; Li H; Wang H; Shi Y; Gao G; Zhou Z; Tan DX; Li X, Selective inhibition of Ebola entry with selective estrogen receptor modulators by disrupting the endolysosomal calcium. Scientific reports 2017, 7, DOI: 10.1038/srep41226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montoya MC; Krysan DJ, Repurposing estrogen receptor antagonists for the treatment of infectious disease. mBio 2018, 9 (6), DOI: 10.1128/mBio.02272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y; Ren J; Harlos K; Jones DM; Zeltina A; Bowden TA; Padilla-Parra S; Fry EE; Stuart DI, Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 2016, 535 (7610), 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuta Y; Takahashi K; Shiraki K; Sakamoto K; Smee DF; Barnard DL; Gowen BB; Julander JG; Morrey JD, T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res 2009, 82 (3), 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oestereich L; Ludtke A; Wurr S; Rieger T; Munoz-Fontela C; Gunther S, Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res 2014, 105, 17–21. [DOI] [PubMed] [Google Scholar]
- 21.Warren TK; Jordan R; Lo MK; Ray AS; Mackman RL; Soloveva V; Siegel D; Perron M; Bannister R; Hui HC; Larson N; Strickley R; Wells J; Stuthman KS; Van Tongeren SA; Garza NL; Donnelly G; Shurtleff AC; Retterer CJ; Gharaibeh D; Zamani R; Kenny T; Eaton BP; Grimes E; Welch LS; Gomba L; Wilhelmsen CL; Nichols DK; Nuss JE; Nagle ER; Kugelman JR; Palacios G; Doerffler E; Neville S; Carra E; Clarke MO; Zhang LJ; Lew W; Ross B; Wang Q; Chun K; Wolfe L; Babusis D; Park Y; Stray KM; Trancheva I; Feng JY; Barauskas O; Xu YL; Wong P; Braun MR; Flint M; McMullan LK; Chen SS; Fearns R; Swaminathan S; Mayers DL; Spiropoulou CF; Lee WA; Nichol ST; Cihlar T; Bavari S, Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531 (7594), 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calain P; Monroe MC; Nichol ST, Ebola virus defective interfering particles and persistent infection. Virology 1999, 262 (1), 114–128. [DOI] [PubMed] [Google Scholar]
- 23.Muhlberger E; Sanchez A; Randolf A; Will C; Kiley MP; Klenk HD; Feldmann H, The nucleotide sequence of the L gene of Marburg virus, a filovirus: homologies with paramyxoviruses and rhabdoviruses. Virology 1992, 187 (2), 534–547. [DOI] [PubMed] [Google Scholar]
- 24.Volchkov VE; Volchkova VA; Chepurnov AA; Blinov VM; Dolnik O; Netesov SV; Feldmann H, Characterization of the L gene and 5’ trailer region of Ebola virus. J Gen Virol 1999, 80 (Pt 2), 355–362. [DOI] [PubMed] [Google Scholar]
- 25.Feldmann H; Muhlberger E; Randolf A; Will C; Kiley MP; Sanchez A; Klenk HD, Marburg virus, a filovirus: messenger RNAs, gene order, and regulatory elements of the replication cycle. Virus Res 1992, 24 (1), 1–19. [DOI] [PubMed] [Google Scholar]
- 26.Lee JE; Fusco ML; Hessell AJ; Oswald WB; Burton DR; Saphire EO, Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008, 454 (7201), 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong X; Qian H; Zhou X; Wu J; Wan T; Cao P; Huang W; Zhao X; Wang X; Wang P; Shi Y; Gao GF; Zhou Q; Yan N, Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell 2016, 165 (6), 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashiguchi T; Fusco ML; Bornholdt ZA; Lee JE; Flyak AI; Matsuoka R; Kohda D; Yanagi Y; Hammel M; Crowe JE Jr.; Saphire EO, Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell 2015, 160 (5), 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flyak AI; Ilinykh PA; Murin CD; Garron T; Shen X; Fusco ML; Hashiguchi T; Bornholdt ZA; Slaughter JC; Sapparapu G; Klages C; Ksiazek TG; Ward AB; Saphire EO; Bukreyev A; Crowe JE Jr., Mechanism of human antibody-mediated neutralization of Marburg virus. Cell 2015, 160 (5), 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flyak AI; Shen X; Murin CD; Turner HL; David JA; Fusco ML; Lampley R; Kose N; Ilinykh PA; Kuzmina N; Branchizio A; King H; Brown L; Bryan C; Davidson E; Doranz BJ; Slaughter JC; Sapparapu G; Klages C; Ksiazek TG; Saphire EO; Ward AB; Bukreyev A; Crowe JE Jr., Cross-reactive and potent neutralizing antibody responses in human survivors of natural Ebolavirus infection. Cell 2016, 164 (3), 392–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y; Ren J; Harlos K; Stuart DI, Structure of glycosylated NPC1 luminal domain C reveals insights into NPC2 and Ebola virus interactions. FEBS Lett 2016, 590 (5), 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter E; Swanstrom R, Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol 1990, 157, 187–253. [DOI] [PubMed] [Google Scholar]
- 33.Konduru K; Shurtleff AC; Bavari S; Kaplan G, High degree of correlation between Ebola virus BSL-4 neutralization assays and pseudotyped VSV BSL-2 fluorescence reduction neutralization test. J Virol Methods 2018, 254, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J; Cheng H; Ratia K; Varhegyi E; Hendrickson WG; Li J; Rong L, A comparative high-throughput screening protocol to identify entry inhibitors of enveloped viruses. J Biomol Screen 2014, 19 (1), 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manicassamy B; Wang J; Jiang H; Rong L, Comprehensive analysis of ebola virus GP1 in viral entry. J Virol 2005, 79 (8), 4793–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manicassamy B; Rong L, Expression of Ebolavirus glycoprotein on the target cells enhances viral entry. Virol J 2009, 6 (75), DOI: 10.1186/1743-422X-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez O; Johnson J; Manicassamy B; Rong L; Olinger GG; Hensley LE; Basler CF, Zaire Ebola virus entry into human dendritic cells is insensitive to cathepsin L inhibition. Cellular microbiology 2010, 12 (2), 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaikh F; Zhao YG; Alvarez L; Iliopoulou M; Lohans C; Schofield CJ; Padilla-Parra S; Siu SWI; Fry EE; Ren JS; Stuart DI, Structure-based in silico screening identifies a potent Ebolavirus inhibitor from a traditional Chinese medicine library. Journal of medicinal chemistry 2019, 62 (6), 2928–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu X; Wong G; Audet J; Bello A; Fernando L; Alimonti JB; Fausther-Bovendo H; Wei H; Aviles J; Hiatt E; Johnson A; Morton J; Swope K; Bohorov O; Bohorova N; Goodman C; Kim D; Pauly MH; Velasco J; Pettitt J; Olinger GG; Whaley K; Xu B; Strong JE; Zeitlin L; Kobinger GP, Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014, 514 (7520), 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen J, New Ebola outbreak rings alarm bells early. Science 2017, 356 (6340), 788–789. [DOI] [PubMed] [Google Scholar]
- 41.Hensley LE; Dyall J; Olinger GG Jr.; Jahrling PB, Lack of effect of lamivudine on Ebola virus replication. Emerg Infect Dis 2015, 21 (3), 550–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kupferschmidt K, Ebola veteran promises an end to Congo’s epidemic. Science 2019, 365 (6453), 526–527. [DOI] [PubMed] [Google Scholar]
- 43.Kupferschmidt K, Plan to use second Ebola vaccine sparks debate. Science 2019, 364 (6447), DOI: 10.1126/science.364.6447.1221. [DOI] [PubMed] [Google Scholar]
- 44.Wec AZ; Bornholdt ZA; He SH; Herbert AS; Goodwin E; Wirchnianski AS; Gunn BM; Zhang ZR; Zhu WJ; Liu GD; Abelson DM; Moyer CL; Jangra RK; James RM; Bakken RR; Bohorova N; Bohorov O; Kim DH; Pauly MH; Velasco J; Bortz RH; Whaley KJ; Goldstein T; Anthony SJ; Alter G; Walker LM; Dye JM; Zeitlin L; Qiu XG; Chandran K, Development of a human antibody cocktail that deploys multiple functions to confer pan-Ebolavirus protection. Cell Host Microbe 2019, 25 (1), 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He J; Choe S; Walker R; Di Marzio P; Morgan DO; Landau NR, Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 1995, 69 (11), 6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manicassamy B; Wang J; Rumschlag E; Tymen S; Volchkova V; Volchkov V; Rong L, Characterization of Marburg virus glycoprotein in viral entry. Virology 2007, 358 (1), 79–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


