Abstract
The libraries generated by high-throughput single cell RNA-sequencing (scRNA-seq) platforms such as the Chromium from 10× Genomics require considerable amounts of sequencing, typically due to the large number of cells. The ability to use these data to address biological questions is directly impacted by the quality of the sequence data. Here we have compared the performance of the Illumina NextSeq 500 and NovaSeq 6000 against the BGI MGISEQ-2000 platform using identical Single Cell 3′ libraries consisting of over 70 000 cells generated on the 10× Genomics Chromium platform. Our results demonstrate a highly comparable performance between the NovaSeq 6000 and MGISEQ-2000 in sequencing quality, and the detection of genes, cell barcodes, Unique Molecular Identifiers. The performance of the NextSeq 500 was also similarly comparable to the MGISEQ-2000 based on the same metrics. Data generated by both sequencing platforms yielded similar analytical outcomes for general single-cell analysis. The performance of the NextSeq 500 and MGISEQ-2000 were also comparable for the deconvolution of multiplexed cell pools via variant calling, and detection of guide RNA (gRNA) from a pooled CRISPR single-cell screen. Our study provides a benchmark for high-capacity sequencing platforms applied to high-throughput scRNA-seq libraries.
INTRODUCTION
The human genome project was an important achievement in life sciences and paved the way for major technology developments in DNA and RNA-sequencing. The development of synthesis-based next-generation sequencing (NGS, also known as massively parallel or high-throughput sequencing) was pioneered by Solexa (1). After the company's acquisition by Illumina, this technology was refined further and gave rise to a number of platforms that include the NextSeq, HiSeq and NovaSeq sequencers. These platforms have now produced the majority of the publicly available human sequencing data. Over time the cost of sequencing has decreased and the technology has become more accessible, both in terms of sequence hardware and tools for analysis (2). Collectively, this has resulted in NGS being adopted by many researchers, and used in clinical and industry settings.
Until recently, the majority of libraries sequenced have been generated on ‘bulk’ samples, consisting of the DNA or RNA collected from millions of cells. However, advances in single cell library preparation techniques (3,4) have made it possible to produce sequencing libraries from tens of thousands of individually barcoded cells, and even individually barcoded molecules. High-throughput library preparation methods, such as the Chromium platform from 10× Genomics (5), are now widely available, enabling libraries consisting of tens of thousands of cells to be generated in several hours. The cDNA libraries from the Chromium experiments differ from ‘bulk’ libraries in that each cDNA molecule contains a Unique Molecular Identifier (UMI) and shared cell barcode. After amplification cDNAs are sheared, and adapter and sample indices are incorporated into finished libraries, which are compatible with next-generation short-read sequencing.
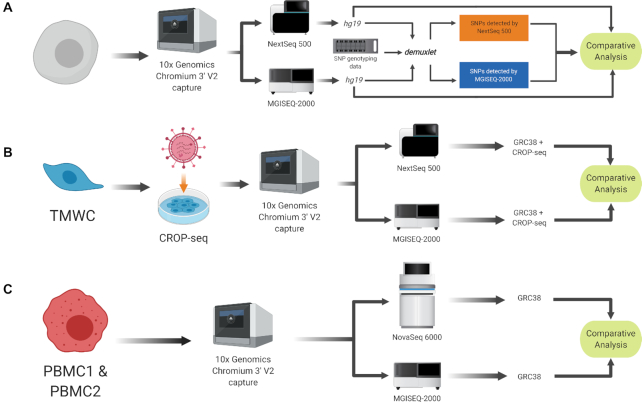
In 2015 BGI launched the BGISEQ-500 as an alternative to existing short-read sequencing technologies (6). The technology underlying the BGISEQ-500 and subsequent BGI platforms combines DNA nanoball (DNB) nanoarrays (6) with polymerase-based stepwise sequencing (DNBseq). The BGISEQ-500 was evaluated to be comparative in performance to Illumina platforms when sequencing small noncoding RNAs (7), bulk transcriptomes (8), whole genome DNA (9), and recently, plate-based scRNA-seq protocols (10). To fully explore this technology's potential for scRNA-seq, we undertook a direct performance comparison four single cell libraries generated with a droplet-based scRNA-seq system—specifically, the Chromium by 10× Genomics. A total of 70 000 cells from all four scRNA-seq libraries were sequenced with the MGISEQ-2000—a newer sequencing platform by BGI, and the NextSeq 500 and NovaSeq 6000 sequencing platforms by Illumina. The performance of each sequencing platform was evaluated based on sensitivity, accuracy, clarity and consistency of analysis outcomes (Figure 1).
Figure 1.
Experimental design. Preparation of single cell libraries and sequencing using Illumina and BGI platforms and subsequent analysis. (A) Human iPSC were generated generated from a human donor and underwent SNP genotyping in addition to scRNA-seq. (B) Primary TMWC were screened with a CRISPR-based molecular screen (CROP-seq). (C) PBMC. Single-cell libraries were prepared from two individual pools of PBMCs.
MATERIALS AND METHODS
Description of the single-cell datasets and cell collection details
A total of four scRNA-seq libraries were generated from three experimental scenarios, chosen to evaluate the ability of sequencing platforms to provide sufficient information to detect features, such as germline genetic variation and CRISPR inserts. All experimental work performed in this study was approved by the Human Research Ethics Committee (HREC) of the Royal Victorian Eye and Ear Hospital (11/1031H; 13/1151H) or the Tasmanian Health and Medical HREC (H0012902) and conformed with the Declarations of Helsinki, under the requirements of the National Health & Medical Research Council of Australia (NHMRC).
iPSC
Consisted of undifferentiated human induced pluripotent stem cells (iPSCs) maintained with StemFlex (ThermoFisher Scientific) that were derived from two unrelated individuals (11). Colonies were harvested using ReleSR™ (Stem Cell Tech) and were dissociated into a single cell suspension. Cells were counted and assessed for viability with Trypan Blue using a Countess II automated counter (Thermo Fisher Scientific), then pooled at a concentration of 391–663 cells/μl (3.91 × 105 − 6.63 × 105 cells/ml). Final cell viability estimates ranged between 95 and 97%. The two cell lines were then genotyped separately using the Infinium HumanCore-24 v1.1 BeadChip assay (Illumina), and single nucleotide polymorphisms (SNPs) were called from this assay with GenomeStudioTM V2.0 (Illumina). To generate the libraries, cells were partitioned and barcoded using high-throughput droplet 10× Genomics Chromium Controller (10× Genomics, USA) and the Single Cell 3′ Library and Gel Bead Kit (V2; 10× Genomics; PN-120237). The estimated number of cells in each well in the Chromium chip was optimized to capture ∼10 000 cells. GEM generation and barcoding, cDNA amplification and library construction were performed according to standard protocol.
TMWC
Comprised of cultured human trabecular meshwork cells (TMWCs) that had been transfected with a CROP-seq (Addgene: 99248) guide RNA (gRNA) pool targeting 128 loci, with the guides targeted to be inserted in the 3′ end of the gene and thus detectable from short-read sequence data. TMWCs were plated in T75 flasks and transfected with a pooled single guide RNA (sgRNA) library lentivirus containing sgRNA for 128 targets, 10 of which were control genes. Cells were harvested 7 days after virus transduction and were FACS sorted for EGFP-positive and viable cells (propidium iodide-negative cells) before applying to the Chromium System (10× Genomics) single cell RNA-sequencing workflow. Single-cell suspensions were used to generate a Chromium library using the Chromium Single Cell 3′ v2 Library (10x Genomics; PC-120237). The estimated number of cells in each well in the Chromium chip was optimized to capture ∼10 000 cells.
PBMC1 and PBMC2
Consisted of peripheral blood mononuclear cells (PBMCs) collected from a total of 28 unrelated individuals. Peripheral blood samples were collected in Vacutainer Cell Preparation Tubes containing sodium heparin and ficoll (BD Biosciences: 362753), and were processed according to the manufacturer's recommendations. Following separation, PBMCs were cryopreserved and stored. Samples were subsequently thawed, and each library contained a pool of PBMCs from 14 donors, with 40 000 cells loaded to achieve a targeted 20 000 cells per library.
Illumina NextSeq 500 and NovaSeq 6000 sequencing
iPSC and TMWC libraries were sequenced on an Illumina NextSeq 500 (NextSeq control software v2.0.2/Real Time Analysis v2.4.11) using a 150 cycle NextSeq High Output Reagent Kit v2.5 (Illumina: 20024907) in stand-alone mode as follows: 26 bp (Read 1), 8 bp (Index) and 98 bp (Read 2). For each library, 1.8 pM concentration and 1300 μl volume was loaded. The NextSeq 500 sequencing was performed by the Institute of Molecular Bioscience Sequencing Core Facility. The two PBMC libraries were sequenced on an Illumina NovaSeq 6000 (Software version: 1.4) using a 2 × 150 cycle S4 flowcell in standalone mode, and libraries loaded at 8 nM and a volume of 350 μl. The NovaSeq 6000 sequencing was performed by the Kinghorn Centre for Clinical Genomics Sequencing Core Facility.
BGI MGISEQ-2000 sequencing
Libraries generated using the 10× Genomics Chromium system require a conversion step using the MGIEasy Universal Library Conversion kit (App-A) (Part Number: 1000004155) before sequencing can be performed on the MGISEQ-2000 instrument. For each library, 10 ng was amplified using 10 cycles of polymerase chain reaction (PCR) to incorporate a 5′ phosphorylation on the forward strand only. Purified PCR product was then denatured and mixed with a ‘splint’ oligonucleotide that is homologous to the P5 and P7 adapter regions of the library to generate a circle (Supplementary Figure S1). A ligase reaction was then performed to create a complete ssDNA circle of the forward strand then an exonuclease digest was performed to remove single stranded non-circularized DNA molecules. Circular ssDNA molecules then underwent Rolling Circle Amplification (RCA) to generate 300–500 faithful copies of the libraries which then fold upon themselves to become DNA Nanoballs (DNB). Each DNB library was then flowed across a 1500 M feature patterned array flow cell ready for sequencing using the MGISEQ-2000RS High-Throughput Sequencing Set (App-A) (PE100) (Part Number: 1000005662). The custom cycle mode on the instrument was run to allow 26 bp (Read 1) and 100 bp (Read 2) cycles without an index barcode read due to only one sample being run per flow cell, and FASTQ files were generated locally on the instrument. Sequencing was performed in BGI Shenzhen, MGI R&D facility.
Bioinformatic and computational analysis
Sequencing data from both platforms were processed into transcript count tables using the Cell Ranger Single Cell Software Suite version 2.2.0 by 10× Genomics (http://www.10xgenomics.com/). Base calls from the NextSeq 500 and NovaSeq 6000 Illumina sequencers were pre-processed as described by Zheng et al. (5). Base calls from the MGISEQ-2000 were pre-processed as described by Huang et al. (12) into demultiplexed, processed reads. The BGI-formatted headers of the resulting FASTQ reads were converted to Illumina-formatted headers using custom Python scripts that are included with this publication's accompanying repository. The quality of the raw sequencing data were assessed with FastQC v0.11.7 (13). The FASTQ files for both platforms were then processed with the cellranger count pipeline, where each sample was processed independently to generate the transcript count tables. Using STAR v2.5.1b (14), the iPSC library was mapped to the GRCh37/hg19 Homo sapiens genome (release 84), while the PBMC libraries were mapped to the GRCh38 (release 88) H. sapiens genome. The TMWC library was mapped to the GRCh38 (release 88) H. sapiens genome that was spiked with gRNA and CROP-seq-associated sequences. This reference was prepared as described by Datlinger et al. (15). We note that, since the expression data are limited to the 3′ end of a gene and we used gene-level annotations, differences between reference versions, such as GRCh38, are unlikely to significantly alter conclusions. The resulting mapped counts for each pair of samples were then depth-equalized using the cellranger aggr pipeline, which downsampled raw reads from the higher-depth BGI library until the mean read depth per cell was equal to the mean read depth per cell of the Illumina library. The alignment of reads from BGI data were extracted based on the cell and UMI barcodes of reads that were retained after downsampling. Downsampling to the depth of 105 reads per sample was performed with DropletUtils (16).
Post-processing and biological analyses were performed on each sample using depth-equalized data. Statistical analyses were performed in R, using the following packages: Seurat (17), biomaRt (18), M3Drop (19) and MetaNeighbor (20). First, count matrices for each sample were loaded into R and separated by platform. Cell barcodes were extracted from the matrices and those detected by both platforms were identified. The genes and UMIs of these cells were then compared in terms of identity in a given cell from both platforms. The concordance of genes and UMIs detected in both platforms was high (96–98%), and we did not find any evidence for gene length bias between platforms (Supplementary Figure S2). Counts from both platforms then underwent quality control as a single dataset. Filtering thresholds were defined as measurements greater than the 3× median absolute median deviation value of the following thresholds: total UMI counts, number of genes detected, percentage of reads mapped to mitochondrial and ribosomal genes. Cells that lay outside these thresholds were removed from subsequent analysis (Table 3). Remaining cells that were detected by both platforms were retained for further analysis. The NBFitModel function from the M3Drop R package was used to calculate the dropout rate of genes per platform. Cell–cell normalization was performed with the SCTransform function from Seurat (21). The percentage of mitochondrial and ribosomal expression were used as covariates and the platform was supplied as a dependent variable. Pearson residuals from this step were then used for dimensionality reduction via principal component analysis (PCA), and the top 30 most variable principal components were used to further reduce the dataset via Uniform Manifold Approximation and Projection (UMAP) to two dimensions. These values were then used to build a Shared Nearest Neighbor graph for each cell, and clusters were inferred using the Louvain method at the resolution of 0.8. These cluster labels were then transferred to an aggregated, filtered and normalized dataset. The similarity of corresponding clusters was quantified with the unsupervised version of MetaNeighbor (20) that used the top 3000 most variable genes between sequencing platforms.
Table 3.
Predicted assignments of cells to donor from iPSCs assignment of cell identify to donors using default settings in demuxlet14
| Prediction | NextSeq 500 | MGISEQ-2000 (down sampled) | MGISEQ-2000 (down sampled, 98 bp reads) |
|---|---|---|---|
| Unassigned | 5552 | 5403 | 5379 |
| Correctly assigned | 7357 | 7480 | 7530 |
To provide fair comparisons between platforms, data generated from MGI platforms was (i) down sampled to equal read depth as the NextSeq, and (ii) the reads were trimmed to match the same RNA read length as obtained from the NextSeq flowcell.
Additional analyses were conducted on the iPSC and TMWC samples to evaluate the influence of sequencing platform on properties specific to these experiments. Using genotype information from that was generated as described in (11), SNPs were called from the iPSC sample using demuxlet with the following arguments: ‘–tag-group CB –tag-UMI UB –field GP –alpha 0 –alpha 0.5’ (22). To account for the downsampling of read depth in the MGISEQ-2000 data, only alignments from UMIs detected in the downsampled data were used. These were extracted using custom Python scripts that are included with this publication's repository, and the ‘subset-bam’ tool by 10× Genomics (https://github.com/10XGenomics/subset-bam). As the MGISEQ-2000 sequencer produced a longer insert read at 100 bp, the iPSC sequencing data were re-mapped to the reference using reads that were truncated to 98 bp. The reads were also downsampled to the same depth as the NextSeq 500 dataset. For the TMWC sample, gRNAs were detected using transcriptome data. This information was supplemented with read counts from the alignments using custom Python scripts that can be found in the accompanying repository.
RESULTS
Sequencing quality metrics
The total number of reads generated for the four libraries on the Illumina platforms was 159–616 million, and 1086–1339 million using the BGI platform. Comparison of sequencing quality control metrics revealed similar percentages of detectable valid cell barcodes (Table 1). A valid barcode is one that is detected from the sequence data that matches a whitelist of ∼737 000 possible barcodes for the 3′ assay (5). Interestingly, Illumina sequencers exhibited 4.3–10% more cell barcode-related base calls with a Q-score >30. If the base's Q-score does not meet this threshold, it is regarded as an ambiguous base. Affected cell barcodes are salvaged by the Cell Ranger pipeline if they are 1-Hamming-distance away from a barcode present in a whitelist of known cell barcodes and are corrected based on posterior probability (5). A similar issue was observed with UMI-related reads, where 4.1–10.6% more Q30 reads were detected by Illumina platforms (Table 1). Quality(Q)-score > 30 (Q30). The Q-score is a measure of base call accuracy, and a value of 30 translates to 99.9% accuracy (23).The Cell Ranger pipeline rescues affected UMI barcodes by matching lower-accuracy UMI sequences to higher-accuracy UMI sequences that are 1-Hamming distance away (5). With this correction, UMI capture rates remain consistent between Illumina and BGI sequencers (Figure 2B).
Table 1.
Basic sequence quality and mapping metrics summary statistics and sequencing properties taken from the analysis of libraries sequenced on Illumina and BGI sequencers (1Illumina NextSeq 500, 2BGI MGISEQ-2000, 3Illumina NovaSeq 6000 sequencers)
| Sample | iPSC | TMWC | PBMC1 | PBMC2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Platform | Illumina1 | BGI2 | |Δ| | Illumina1 | BGI2 | |Δ| | Illumina3 | BGI2 | |Δ| | Illumina3 | BGI2 | |Δ| |
| Valid barcodes | 97.8 | 96.4 | 1.4 | 97.9 | 96.8 | 1.1 | 98.0 | 97.0 | 1.0 | 98.0 | 97.0 | 1.0 |
| Reads mapped to genome | 94.0 | 97.8 | 3.8 | 93.7 | 98.0 | 4.3 | 95.3 | 98.1 | 2.8 | 95.2 | 97.9 | 2.7 |
| Q30 barcode | 96.1 | 87.9 | 8.2 | 97.8 | 87.8 | 10.0 | 96.1 | 91.8 | 4.3 | 96.1 | 90.5 | 5.6 |
| Q30 UMI | 95.5 | 87.3 | 8.2 | 97.7 | 87.1 | 10.6 | 95.9 | 91.8 | 4.1 | 95.9 | 90.0 | 5.9 |
| Q30 RNA | 85.9 | 86.6 | 0.7 | 86.6 | 88.0 | 1.4 | 92.0 | 89.0 | 3.0 | 92.2 | 88.0 | 4.2 |
| Fraction of reads in cells | 79.1 | 80.2 | 1.1 | 95.0 | 95.1 | 0.1 | 93.7 | 94.8 | 1.1 | 94.1 | 95.2 | 1.1 |
The percentage valid barcodes, reads mapped to the genome and fraction of reads in a cell are generated by CellRanger software. The Q30 metrics are supplied by BGI and Illumina software.
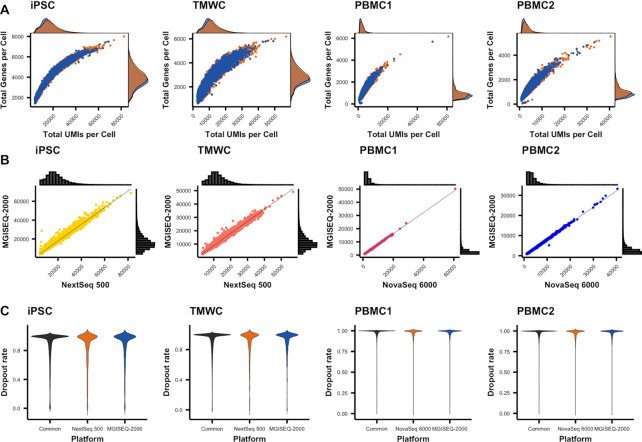
Figure 2.
Cells and genes detected by platforms. Both technologies demonstrated similar sensitivity in the detection of cells and genes. (A) Capture efficiency of each platform. Efficiency is evaluated based on the number of genes and molecules detected in a cell. (B) Total number of molecules detected in a cell. Histograms on each axis represent the distribution of total UMIs in a cell, while the scatter plot represents the correlation of UMI detection for a cell, between the two platforms. (C) Dropout rate across genes detected by platform. Dropout rates for each gene, per platform were calculated using NBDrop from the M3Drop package.
The Q30 of the cDNA portion of the read also plays a significant role as it directly affects the number of usable reads that can be mapped to the reference genome. We also observed comparable performance between the NextSeq 500 and MGISEQ-2000, while a 3.0 − 4.2% difference was observed between the NovaSeq 6000 and MGISEQ-2000 (Table 1). To note, a larger difference (20.1–30.7%) was observed in the data generated from a NextSeq 500 High Output v2.0 kit (Illumina: FC-404-2002) compared with the MGISEQ-2000 (Supplementary Table S1). The reduction in sequencing accuracy for scRNA-seq libraries on a NextSeq 500 has previously been discussed (24), and it has been hypothesized that this is due to flow cell surface chemistry.
The combination of assigning reads to a given cell, a transcript molecule and aligning to a reference sequence directly affects the number of usable reads that are obtained from sequence data. Collectively, differences in the sequencing accuracy between platforms over the entire read length has consequential effects on the percentage of reads that pass quality control and that are able to be mapped to the reference genome. When we integrated the percentage of reads that were able to be aligned to the GRCh38 (release 88) human reference genome, we obtain a 4.3% difference between the NextSeq 500 and MGISEQ-2000, while the difference between the NovaSeq 6000 and MGISEQ-2000 is only 2.7–2.8% (Table 1). When aligned to the hg19 (release 83) human reference genome, we observed a difference of 3.8% between the NextSeq 500 and MGISEQ-2000. The slightly lower percentage of alignment observed from the NextSeq 500 libraries is most likely due to the lower sequencing accuracy in the RNA transcript part of the read, as supported by the small difference in the Q30 of the RNA read. As thresholds used to determine if a read aligns to the genome are the same, the lower sequencing accuracy should not affect the biological interpretation of the aligned data. However, it does mean libraries sequenced on a NextSeq 500 will need to be sequenced at a greater depth to obtain the same sequencing depth of aligned reads per cell.
Identification of cells, genes and transcript molecules
To evaluate the similarity in the ability of sequencing platforms to identify the same cells, transcript molecules, and genes, we standardized the read depth within each sample by down sampling to the lowest read depth. As the same cells from each sample had been sequenced on two platforms, we evaluated cell identification based on the observation of the same cell barcode. Each of the two platforms identified 98.9–99.5% of cells in common in the four samples (Supplementary Figure S2A). There was a strong correlation between the total UMI counts of cells detected by both platforms (r > 0.99 for all samples) (Figure 2B). For cells identified by only one platform, the mean number of UMIs were on average one log2 lower than the cells identified as common between platforms (Supplementary Figure S2B). There was a lower concordance of shared genes for these cells, suggesting that these ‘platform specific’ cells are possibly cell free transcripts that have not been adequately detected during quality control filtering by the cell singlet detection algorithm. An alternative explanation is that these are cells with low transcriptional abundance, although we observe no evidence for this scenario.
Gene detection was similarly at high concordance with 92.0–96.6% of genes detected by both platforms for the four samples (Supplementary Figure S2C). There was no difference between the percentage of genes detected by each platform, for each experiment (Table 2). Details of the genes detected from each platform are provided in Supporting Material Tables S1–4. Genes that were only detected on a single platform were on average, very lowly expressed (Supplementary Figure S2D). To confirm this, we down sampled to an average of 105 reads per sample and repeated the comparison of gene detection. The NextSeq 500 detected an additional 0.4–1.4% genes in the iPSC and TMWC datasets, while the MGISEQ-2000 detected an additional 0.6–0.7% genes in the PBMC datasets. We speculate these genes are technical artefacts. To investigate this further, we calculated the dropout rates of detected genes and found that there was no difference in the dropout rates of genes that were identified by a single platform (Figure 2C). The capture efficiency of each platform was evaluated based on the total UMIs and detected genes per cell (Figure 2A). Overall, similar capture efficiency was observed across all platforms and samples. Interestingly, we observed a slight increase in the capture efficiency of the NextSeq 500 and NovaSeq 6000 in the detection of cells with larger library sizes. This is likely a function of the slightly higher sequencing accuracy in the UMI region of the read (Table 1), corresponding to an increase in the mean UMIs per cell from these sequencers (Table 2). However, taken together, our analyses show that the gene detection, and quantification of transcript molecules via UMIs is highly consistent across platforms.
Table 2.
Cell and gene capture metrics summaries of cell and RNA sequence data from all libraries
| Experiment | iPSC | TMWC | ||||||
|---|---|---|---|---|---|---|---|---|
| Platform | Illumina1 | BGI2 | BGI (Subsampled)2 | |Δ| | Illumina1 | BGI2 | BGI (Subsampled)2 | |Δ| |
| Estimated number of cells | 12 909 | 12 940 | 12 940 | 31 | 18 782 | 18 784 | 18 784 | 2 |
| Total number of reads | 580 398 477 | 1 122 883 312 | 581 782 400 | 1 383 923 | 425 891 295 | 1 119 142 907 | 425 927 200 | 35 905 |
| Mean reads per cell | 44 960 | 86 776 | 44 960 | 0 | 22 675 | 59 579 | 22 675 | 0 |
| Median UMI counts per cell | 16 800 | 22 431 | 14 998 | 1803 | 11 540 | 18 411 | 10 282 | 1258 |
| Median genes per cell | 3946 | 4655 | 3748 | 198 | 2878 | 3781 | 2709 | 169 |
| Total number of genes detected | 24 202 | 24 799 | 23 941 | 261 | 23 212 | 23 999 | 23 943 | 731 |
| Experiment | PBMC1 | PBMC1 | ||||||
| Platform | Illumina3 | BGI2 | BGI (Subsampled)2 | |Δ| | Illumina3 | BGI2 | BGI (Subsampled)2 | |Δ| |
| Estimated number of cells | 20 982 | 20 839 | 20 839 | 143 | 18 634 | 18 537 | 18 537 | 97 |
| Total number of reads | 588 565 199 | 1 086 836 730 | 584 533 950 | 4 031 249 | 616 115 423 | 1 339 580 496 | 612 907 368 | 3 208 055 |
| Mean reads per cell | 28 050 | 52 153 | 28 050 | 0 | 33 064 | 72 265 | 33 064 | 0 |
| Median UMI counts per cell | 2605 | 2282 | 2058 | 547 | 3004 | 2676 | 2357 | 647 |
| Median genes per cell | 855 | 790 | 732 | 123 | 956 | 897 | 814 | 142 |
| Total number of genes detected | 21 671 | 21 832 | 21 296 | 375 | 21 860 | 22 188 | 21 516 | 344 |
All details were provided by the CellRanger software. To provide fair comparison between platforms we have also down sampled data from MGISEQ-2000 sequences to equal read depths per library as obtained from Illumina sequencers.
Concordance of scRNA-seq analysis between platforms
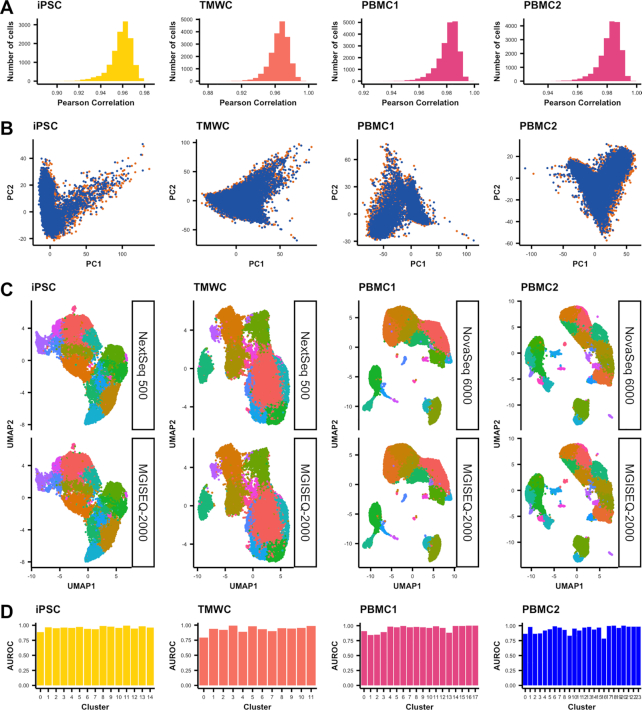
To determine if data generated by Illumina and BGI sequencers are comparable, we performed analyses that are common to most scRNA-seq workflows: filtering, normalization, PCA, clustering and differential expression (25). Cells were filtered out based on three criteria: number of UMIs, features and proportion of mitochondrial and ribosomal gene expression to total expression. Filtering removed 1.0–2.7% of cells sequenced by Illumina platforms, while 0.7–3.7% of cells were removed in datasets sequenced with the MGISEQ-2000 (Supplementary Table S2). The correlation of gene expression between a given cell sequenced on both platforms was high, with mean r2 of 0.96 between NextSeq 500 and MGISEQ-2000 and 0.98 between NovaSeq 6000 and MGISEQ-2000 (Figure 3A). We similarly observed a high concordance of between the first two principal components across all samples (Figure 3B). The comparison of cluster identification revealed complete concordance in clustering at the same resolution for all datasets between platforms (Figure 3C). Finally, using 3000 of the most variable genes, (Supplementary Tables S5–9) we implemented MetaNeighbour (20) to evaluate the replicability of cell types platforms for each sample. The area under the receiver operator curve varied from 0.93 to 0.96, again indicating very high similarities between platforms (Figure 3D).
Figure 3.
Concordance of datasets sequenced by different platforms. (A) Pearson correlation of gene expression between cells identified by both sequencing platforms. (B) PCA representation of each sequencing platform per dataset. (C) Cluster predictions projected on to UMAP plots separated by sequencing platform. (D) AUROC scores measuring the similarity of corresponding clusters across platforms, for each dataset as calculated by MetaNeighbor.
Identification of genetic variation and CRISPR guides
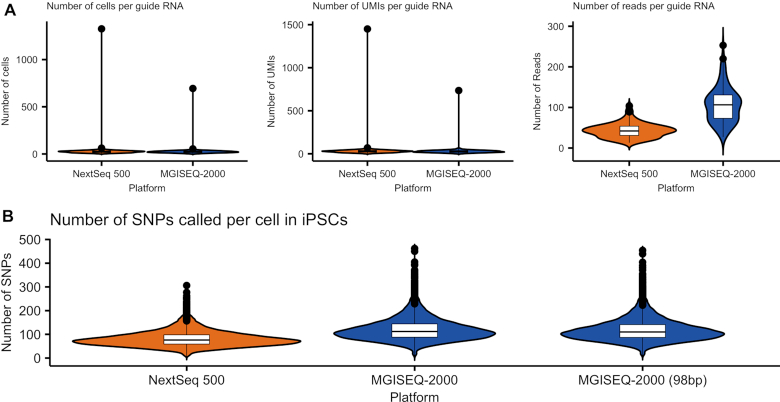
The ability to call SNPs from scRNA-seq data allows researchers to use multiplexing strategies in the library generation stage, reducing the overall cost of running experiments where large sample sizes are needed (22). The power of demultiplexing a cell back to an individual donor is partly a function of the number of SNPs that can confidently be called from the short RNA section of the read. Using the iPSC sample that comprised of cells multiplexed from two unrelated donors, we assigned cells to the origin donor by calling SNPs from the equalized total reads of sequence data generated by the NextSeq 500 and MGISEQ-2000 using the demuxlet algorithm (22). Donor identity was confirmed using genotyped SNPs from an Illumina Global Screening array that had been imputed to the Haplotype Reference Consortium panel (26). At equalized read depths across platforms, we identified 1 048 912 SNPs from the NextSeq 500 data and 1 550 680 SNPs from the MGISEQ-2000 data. This difference may be due to the higher sequencing quality of the RNA read by the MGISEQ-2000 (Table 1), as the detection of a SNP is dependent on the number of unique reads overlapping a variant and the phred-scale quality score of the observed base call (22). The additional SNPs enabled demuxlet to assign an additional 123 cells to the correct donor (Table 3 and Figure 4). To verify that this was not a function of differences in the base-pair length of the RNA section of the read, we trimmed the BGI data to a total RNA-read length of 98 bp and re-called SNPs, and could still correctly identify an additional 173 cells (Supplementary Table S3). This is further supported by data generated by the NextSeq 500 using the NextFlow version 2.0 sequencing reagent kit, which yielded lower quality RNA reads (Supplementary Table S1) and subsequently identified 3,085 less correctly assigned cells, in comparison to the NextFlow version 2.5 kit (Supplementary Table S3).
Figure 4.
Experiment-specific metrics. (A) Metrics related to guide RNA assignment in TMWC. This excludes cells that were not affiliated with a guide RNA and cells that with ambiguous assignments. (B) Number of SNPs called per cell in iPSCs. SNPs were called from alignments of cells found in NextSeq 500 and MGISEQ-2000 datasets.
Finally, we evaluated the ability to detect the inserted guide RNAs (gRNA) from the TMWC that had been transfected with a CRISPR pool targeting 128 loci with the CROP-seq protocol. The guides are targeted to be inserted in the 3′ end of the gene and thus detectable from short-read sequence data. Despite differences in the read coverage of gRNAs, we observed consistent detection of the number of cells per guide, and the number of UMIs per guide across both the NextSeq 500 and MGISEQ-2000 (Figure 4).
DISCUSSION
To our knowledge, this study is the first to utilize MGISEQ-2000 platform for scRNA-seq, and the first to compare sequence performance for the widely used 10× Chromium platform against Illumina platforms. Our comprehensive benchmarking utilizes data from over 70 000 cells, and shows that the MGISEQ-2000 has to be highly comparable performance across a range of modalities to the Illumina NextSeq 500 and NovaSeq 6000 platforms at equal read depth, while being more cost effective (Supplementary Table S4). For single cell RNA-sequencing-specific metrics, such as read quality, cell detection and RNA molecule detection, we found the Illumina NovaSeq 6000 and BGI MGISEQ-2000 platforms generated highly comparable data, and similar observations were made between the Illumina NextSeq 500 and MGISEQ-2000 platforms. Identical subpopulations were identified in each set of samples using general scRNA-seq analysis. The study compared the performance of the NextSeq 500 and the MGISEQ-2000 for specialized single cell analyses—specifically, variant calling and gRNA detection from pooled CRISPR single cell screens. While the MGISEQ-2000 detected 501 768 more SNPs, a similar number of cells were correctly assigned to a donor. The performance of both platforms was also alike in the pooled CRISPR study, where similar frequencies of gRNAs were detected. This work provides a benchmark for high capacity sequencing platforms applied to high-throughput single cell RNA-seq libraries.
DATA AVAILABILITY
We have made available both the raw and processed data on ArrayExpress under accession E-MTAB-9024.
CODE REPOSITORY
https://github.com/powellgenomicslab/BGI_vs_Illumina_Benchmark.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for the support in sample collection and processing performed by Antonia Rowson, Helena Liang, Linda Clarke and Qin Yi Lu. We are grateful for the support of Illumina in supplying reagents, and in particular Tamsin Eades. Illumina sequencing was performed by the Institute for Molecular Bioscience Sequencing Facility at the University of Queensland, and the Kinghorn Centre for Clinical Genomics Sequencing Core Facility. Figure 1 was created using Biorender.com.
Author contributions: J.E.P. designed the study, acquired funding, and led the analysis. A.S. performed bioinformatics, data processing, and computational analyses. S.A. generated single cell libraries. Q.S., L.S., F.J., W.Z. generated sequence data from supplied libraries and returned raw-level data. K.W., M.D., S.W.L., S.SC.H., A.P., and A.W.H. generated and supplied cellular material and samples. Q.N. provided input on the computational analysis. L.F. and A.B. supplied sequencing reagents and input on sequencing.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Health and Medical Research Council (NHMRC) [APP1132719, APP1083405, APP1107599]; Stem Cells Australia—the Australian Research Council Special Research Initiative in Stem Cell Science (to J.E.P., A.W.H., A.P.); Macular Disease Foundation of Australia (to A.P., A.W.H., J.E.P.); Yulgilbar Foundation (to J.E.P., A.P.); NHMRC Practitioner Fellowship [1103329 to A.W.H.]; NHMRC Senior Research Fellowship [1154389 to A.P.]; Australian Research Council Future Fellowship [FT140100047 to A.P.].
Conflict of interest statement. A.S., S.A., K.W., M.D., S.W.L., S.SC.H., Q.N., A.P., A.W.H. and J.E.P. declare no conflict of interests. Q.S., L.S., F.J. and W.Z. declare a conflict of interest as they are all employees of BGI or MGI. A.B. and L.F. declare a conflict of interest as at the time of submission they were employees of BGI Australia at time of submission. Neither BGI or Illumina played any role in the design of the experiments, or analysis, interpretation and presentation of the data.
REFERENCES
- 1. Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G., Hall K.P., Evers D.J., Barnes C.L., Bignell H.R. et al.. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008; 456:53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C.. Ten years of next-generation sequencing technology. Trends Genet. 2014; 30:418–426. [DOI] [PubMed] [Google Scholar]
- 3. Prakadan S.M., Shalek A.K., Weitz D.A.. Scaling by shrinking: empowering single-cell ‘omics’ with microfluidic devices. Nat. Rev. Genet. 2017; 18:345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hwang B., Lee J.H., Bang D.. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018; 50:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. et al.. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G. et al.. Human genome sequencing using unchained base reads on self-assembling DNA Nanoarrays. Science. 2009; 327:78–81. [DOI] [PubMed] [Google Scholar]
- 7. Fehlmann T., Reinheimer S., Geng C., Su X., Drmanac S., Alexeev A., Zhang C., Backes C., Ludwig N., Hart M. et al.. cPAS-based sequencing on the BGISEQ-500 to explore small non-coding RNAs. Clin. Epigenet. 2016; 8:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu F.-Y., Chen M.-X., Ye N.-H., Qiao W.-M., Gao B., Law W.-K., Tian Y., Zhang D., Zhang D., Liu T.-Y. et al.. Comparative performance of the BGISEQ-500 and Illumina HiSeq4000 sequencing platforms for transcriptome analysis in plants. Plant Methods. 2018; 14:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mak S.S.T., Gopalakrishnan S., Carøe C., Geng C., Liu S., Sinding M.-H.S., Kuderna L.F.K., Zhang W., Fu S., Vieira F.G. et al.. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience. 2017; 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Natarajan K.N., Miao Z., Jiang M., Huang X., Zhou H., Xie J., Wang C., Qin S., Zhao Z., Wu L. et al.. Comparative analysis of sequencing technologies for single-cell transcriptomics. Genome Biol. 2019; 20:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daniszewski M., Nguyen Q., Chy H.S., Singh V., Crombie D.E., Kulkarni T., Liang H.H., Sivakumaran P., Lidgerwood G.E., Hernández D. et al.. Single-cell profiling identifies key pathways expressed by iPSCs cultured in different commercial media. iScience. 2018; 7:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang J., Liang X., Xuan Y., Geng C., Li Y., Lu H., Qu S., Mei X., Chen H., Yu T. et al.. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience. 2017; 6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data. 2010; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 14. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Datlinger P., Rendeiro A.F., Schmidl C., Krausgruber T., Traxler P., Klughammer J., Schuster L.C., Kuchler A., Alpar D., Bock C.. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods. 2017; 14:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffiths J.A., Richard A.C., Bach K., Lun A.T.L., Marioni J.C.. Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun. 2018; 9:2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. 3rd, Hao Y., Stoeckius M., Smibert P., Satija R.. Comprehensive integration of single-cell data. Cell. 2019; 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M.H., Baldock R., Barbiera G. et al.. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015; 43:W589–W598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andrews T.S., Hemberg M.. M3Drop: dropout-based feature selection for scRNASeq. Bioinformatics. 2018; 35:2865–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crow M., Paul A., Ballouz S., Huang Z.J., Gillis J.. Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat. Commun. 2018; 9:884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hafemeister C., Satija R.. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019; 20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang H.M., Subramaniam M., Targ S., Nguyen M., Maliskova L., McCarthy E., Wan E., Wong S., Byrnes L., Lanata C.M. et al.. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 2018; 36:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Illumina Technical Note: Quality Scores for Next-Generation Sequencing. 2011; https://www.illumina.com/documents/products/technotes/technote_Q-Scores.pdf.
- 24. 10x Genomics Technical Note: Biological & Technical Variation in Single Cell Gene Expression Experiments. 2018; https://support.10xgenomics.com/single-cell-gene-expression/sample-prep/doc/technical-note-biological-and-technical-variation-in-single-cell-gene-expression-experiments.
- 25. Luecken M.D., Theis F.J.. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019; 15:e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K. et al.. A reference panel of 64, 976 haplotypes for genotype imputation. Nat. Genet. 2016; 48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have made available both the raw and processed data on ArrayExpress under accession E-MTAB-9024.