Abstract
The development of single-cell transcriptomic technologies yields large datasets comprising multimodal informations, such as transcriptomes and immunophenotypes. Despite the current explosion of methods for pre-processing and integrating multimodal single-cell data, there is currently no user-friendly software to display easily and simultaneously both immunophenotype and transcriptome-based UMAP/t-SNE plots from the pre-processed data. Here, we introduce Single-Cell Virtual Cytometer, an open-source software for flow cytometry-like visualization and exploration of pre-processed multi-omics single cell datasets. Using an original CITE-seq dataset of PBMC from an healthy donor, we illustrate its use for the integrated analysis of transcriptomes and epitopes of functional maturation in human peripheral T lymphocytes. So this free and open-source algorithm constitutes a unique resource for biologists seeking for a user-friendly analytic tool for multimodal single cell datasets.
INTRODUCTION
The recent development of techniques for single cell RNA sequencing (scRNAseq) has resulted in an accrual of scRNAseq datasets comprising thousands of cells from many lineages, tissues, physiological conditions and species. The classical representation of such datasets is based on their dimensionality reduction e.g. by t-stochastic neighborhood embedding (t-SNE) or uniform manifold approximation and projection (UMAP). In such steps, all cells are plotted according to their transcriptomic similarity with immediate neighbours and the overall community of cells, forming separate groups or clusters. The lineage, status and hallmarks of such cells and clusters are then identified by their expression levels of single genes, chosen for their hallmark expression patterns, e.g. expression of the CD14 gene for monocytes, or of the MKI67 gene for proliferating cells. Nevertheless, both technical noise from the data acquisition process, and massive gene dropouts impair detection of many genes in scRNAseq datasets. Consequently, mapping the expression level of a single gene in a t-SNE map is generally less informative than mapping the enrichment of a corresponding multi-gene signature.
We recently developed Single-Cell Signature Explorer, a tool which scores gene signatures by their UMI to total cell UMI ratio in each single cell from large datasets (1). This tool further overlays UMAP or t-SNE maps of the dataset with heatmap-encoded single cell scores of any multigene signature. By allowing the user to see how these scores vary across all cells, it provides a visualization of any transcriptomic hallmark in the dataset. For example, this tool returns scores interpreted by user for identifying B versus non-B cells among peripheral blood mononuclear cells (PBMC). It may likewise help to infer single cell lineages, cell hallmarks or any metabolic and proliferative status from collective gene expression levels (1).
However, formal identification of most cell lineages relies upon cell surface expression of canonical protein markers rather than on transcriptome-based inference. This issue was addressed by incepting the Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITEseq) (2), which allows simultaneous detection of single cell transcriptomes and antibody-labeled surface markers, yielding both gene expression levels and immunophenotypes from the same experiment. Several algorithms allow the analysis of either multidimensional immunophenotypings, e.g. Cytobank (3) or of single cell transcriptomes, such as Seurat (4), iCellR or Loupe Cell Browser (10xGenomics) and SegGeq™ (flowjo). In addition, Seurat not only allows analysis of single cell transcriptomes, but also of multimodal datasets through R-based command lines (https://satijalab.org/seurat/v3.1/multimodal_vignette.html).
So currently, there are several existing methods that are able to pre-process raw CITE-seq datasets from read alignments up to production of tables of cells, genes, epitopes and UMAP/t-SNE coordinates. These methods can also be used to analyze such table’s transcriptome and phenotype data together. But although the pre-processing performed by skilled bioinformatics analysts will remain a prerequisite, there is a growing demand from a broader scientific audience for user-friendly and open source analytic tools to finally explore any pre-processed CITE-seq datasets with simple tools. In this aim here, we introduce Single-Cell Virtual Cytometer, a small software to analyze pre-processed CITE-seq tables with user-friendly and flow cytometry-like interface. From any table of pre-processed CITE-seq dataset, Single-Cell Virtual Cytometer allows to visualize both cell transcriptomes and epitopes in t-SNE or UMAP, through flow-cytometry-like gatings, quadrants and selection of subsets of cells. We examplify its use to characterize the gene signatures for stages of functional maturation of peripheral T lymphocytes using an original CITE-seq datasets of PBMC from healthy individual. Our tool, implemented as freely available open-source software, represents a unvaluable resource to fully exploit the expanding universe of multi-omics datasets necessary to cancer research and care.
MATERIALS AND METHODS
Single-Cell Virtual Cytometer
Single-Cell Virtual Cytometer is a new tool, part of Single-Cell Signature Explorer software package (1) dedicated to high throughput signature exploration in single-cell analysis. It brings the flow cytometry software capabilities to single cell analysis. It is able to define and gate cell populations based on the 2D plot of, for example two antibodies or genes, and to display simultaneously the selected cells on a UMAP or t-SNE map. There is no limit to the number of antibodies, genes or other criteria possibly used to define the plot, including combinations of transcriptomic, proteomic and signature scores (1). Single-Cell Virtual Cytometer takes a data table as input, using tab-separated text file format, with the cells tag in rows and genes expression, antibodies detection levels, signature scores in columns and at least two columns with (x,y) coordinates for a map, such as t-SNE or UMAP. Once the data table is loaded, the user can then select two criteria, such as two antibodies. With such criteria, a flow cytometry-like contour plot of the entire dataset is drawn. Using a lasso or a box selection tool, the user can select some cells and immediately see these on a t-SNE/UMAP map. Single-Cell Virtual Cytometer supports an unlimited level of successive gatings. Quadrant gates display the number and % of cells in each quadrant, and trigger their location on the t-SNE/UMAP. For further analyses, it is possible to export separately the codes of cells gated or defined by each quadrant, as well as their main statistics.
CITE-seq counter
We developed CITE-seq-counter software to count the UMI of antibodies tags in raw sequencing reads. CITE-seq-counter has been developed for Single Cell CITE-seq samples processed with 10×Genomics technologies. It takes as input fastq files R1 and R2 from the sequencer, the antibodies barcodes, and a white list of cells obtained from Seurat (4). Cell and antibody barcode positions are adjustable as well as UMI positions. Since sequencers can produce errors, one mismatch is allowed in the barcode and in the UMI. PCR duplicates (same UMI + barcode) are excluded from the counts. The software is written in Go, it is fast and the memory usage is as low as possible. Only the result table and two sequences R1 R2 are stored in RAM at the same time.
Code availability
Single-Cell Virtual Cytometer was developed in pure javascript using the graphical libraries plotly.js (5) and Bulma. It only needs a web browser with javascript enabled to be executed, with tab-separated text files as input. Files can be accessed on the GitHub Single-Cell Virtual Cytometer web page.
CITE-seq-counter was developed in Go and pre-compiled static binaries are available for Linux and Windows.
Generation and pre-processing of PBMC CITE-seq data
Procedures for cell isolation, labeling, CITE-seq experiment, sequencing and pre-processing of the resulting dataset are described in Supplementary Data section.
RESULTS
Single-Cell Virtual Cytometer for analysis of pre-processed CITE-seq datasets
Single-Cell Virtual Cytometer is a small software (3Mo) which does not require any complex installation, and can be run immediately in a web browser without any programming skills. The user can immediately explore any pre-processed CITE-seq data without mastering any R command line instructions. Importantly, these input data must consist in csv tables featuring cells in rows and columns for genes, antibodies, signature scores or any other quantitative single cell readout. Indeed, such tables must also have for each cell at least one set of map (x,y) coordinates from any dimensionality reduction method. From such pre-processed CITE-seq data, Single-Cell Virtual Cytometer typically displays both a flow cytometry-like density plot of cell surface markers (left panel, referred below to as phenotype panel) and the corresponding dimensionally reduced map of cells based on their transcriptomes (right panel). Based on any user-defined criteria, the cells selected by gates or quadrants on left panel’s density plot are interactively displayed on the right panel showing the corresponding t-SNE/UMAP (Supplementary data demo video). Setting quadrants in the phenotype panel automatically triggers display of both percentages and counts of cells from each quadrant.
Hence from a CITE-seq dataset analyzed with Single-Cell Virtual Cytometer, it is possible to select two antibodies to get their density plot across the entire dataset. Using gates or quadrants from this phenotype plot, the user can select further a subset of cells to visualize on the transcriptomic map. The cell tags of any selected subset of cells can be exported as a txt file. Downstream sub-gating and analysis with other antibodies of selected cells can be reiterated without limits. Delineating quadrants on the phenotype plot returns both the % and absolute counts of cells from each quadrant, as well as their respective localization on the corresponding right side map. The plots and maps from Single-Cell Virtual Cytometer can be exported in low and high resolution. Importantly, Single-Cell Virtual Cytometer is very versatile. Its two-panels displays are interactive and based on any (x,y) parameters selected by the user from the drop-down list. Hence this enables users to select not only any mAb or cell hashtag (Biolegend), but also any other parameter such as cluster number, sample annotation index, or dimensionality reduction axes. Hence instead of the epitopes and transcriptomes in respectively, the left and right panels, the selection of (t-SNE-1, t-SNE-2) as left panel parameters allows user to gate, quadrant, and select cells from a transcriptomic standpoint to further visualize their respective cell surface markers on the right panel. The same applies for selection of (cluster, cluster) or (cluster, gene) as left panels parameters to analyze epitopes in the corresponding right panel.
Comparison with existing scRNAseq and flow cytometry visualization tools
Seurat 3.0
Seurat 3.0 (4) is an R package designed for QC pre-processing, analysis and exploration of single cell RNA-seq data. The Seurat pipeline enables users to identify and interpret sources of heterogeneity from single-cell transcriptome measurements, and to integrate diverse types of single-cell data, performing the so-called multimodal integration. Seurat has a command line tool able to generate dimensionality reduction maps from t-SNE or UMAP, allowing users to select clusters and subsets of cells. Hence the use of Seurat requires bioinformatics skills that are however no more needed for using Single-Cell Virtual Cytometer, which was rather designed for end users more accustomed to flow cytometry.
Loupe cell browser
3.1.1 is a dedicated visualization and analysis tool for scRNAseq developed for analysing scRNAseq datasets mostly produced by 10×Genomic platforms. It allows importing datasets and visualizing custom projections of either gene expression or antibody-only datasets, across t-SNE or UMAP computed by the Cell Ranger 3.1 pipeline. Despite its ease of use however, this tool only displays a single dimensionality reduced map featuring the dataset clusters and heatmaps of the graph-based differentially expressed genes or mAbs. Although this tool may export images and selection of cells, it lacks the dual displays of phenotypes and transcriptomes to perform any simultaneous exploration of CITEseq data.
iCellR
iCellR is a R package for scRNAseq analysis able to produce interactive graphs for either of transcriptome or immunophenotype data, but not both simultaneously. To our knowledge iCellR does not reproduce a flow cytometry interface and, similarly to Seurat, is accessible for users skilled in R.
CytoBank
CytoBank (3) and some other flow cytometry softwares can import single cell data files after adequate file conversions, and can be used for visualizing single cell phenotype data. CytoBank is also able to produce t-SNE but its limited capacity to process a maximum of 818 parameters is not compatible with current scRNAseq transcriptomic datasets. Furthermore, Cytobank does not display interactively the density plot subpopulations in the t-SNE, nor does it allow users to export the cell tags for further use.
SeqGeq-FlowJo
SeqGeq-FlowJo (https://www.flowjo.com/solutions/seqgeq) is a commercial bioinformatics platform of 506 Mo for desktop scRNAseq analysis with an intuitive interface. Under administrator’s authorization for it use, it allows to import, analyze and visualize scRNAseq data with interactive graphs, and to share data with other analytic applications provided as exchange plug-ins, such as Seurat, Monocle and the commercial flow cytometry software FlowJo™, to quote a few. So the SeqGeq™ platform provides access to several sophisticated tools performing various types of analyses successively, but does not display both transcriptomes and epitopes simultaneously in the same integrated layout as does Single-Cell Virtual Cytometer.
So currently, while only a few existing open source tools allow to analyze both types of data together, Single-Cell Virtual Cytometer presents the advantage of web-based and user friendly interface for the simultaneous display of both immunophenotype dot plots and transcriptome-based UMAP/t-SNE plots. In addition, the computing time to display the user-selected plots and maps is extremely short. For example, the time to plot a density plot or to display a mAb-labeled subpopulation on a t-SNE map is <1 s with 10k cells when running Single-Cell Virtual Cytometer with an Intel(R) Xeon(R) CPU E5-2630 v4 @ 2.20GHz.
The Single-Cell Virtual Cytometer interface
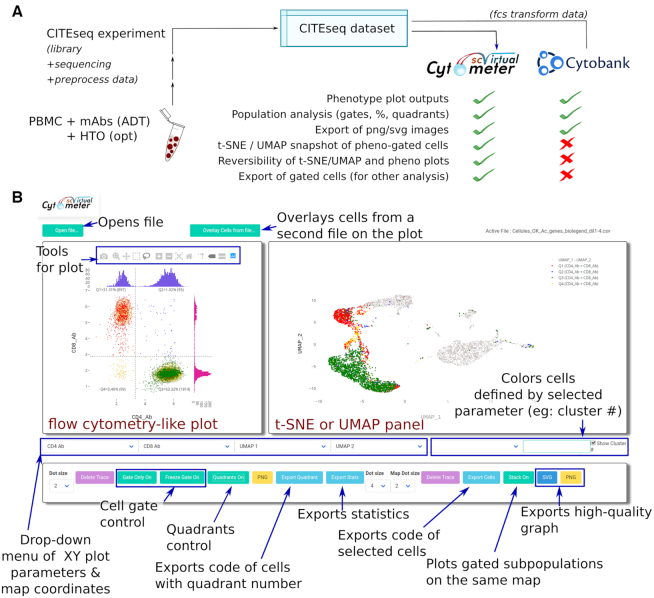
Single-Cell Virtual Cytometer was primarily applied to analyze an original CITE-seq dataset of human PBMC. So the PBMC from a healthy individual were primarily labeled with a mix of 12 TotalSeq™-B ADT (Supplementary Table S1) at five concentrations respectively labeled by five HTO (Supplementary Table S2). The stained PBMC were analyzed for CITE-seq using a 10XGenomics 3′ chemistry V3 platform, sequenced, pre-processed and dimensionality reduction of the transcriptome datasets was performed with UMAP. As QC of the transcriptomic part of the CITE-seq dataset, the single cells with outlier counts of total UMI or number of genes were discarded. For the QC of the phenotypic part of the CITE-seq dataset, cells labeled with over-diluted ADT (Supplementary Figure S1) and cells displaying mutually exclusive phenotypes (e.g. CD3+CD19+CD14+) were discarded. This finally yielded a CITE-seq dataset encompassing the epitopes and transcriptomes of n = 5559 PBMC (Figure 1). It was deposited as GSE144434 in the GEO database. Importantly, analysing this dataset by either the flow cytometry tool Cytobank or Single-Cell Virtual Cytometer yielded the same plots and rates of CD3+CD4+ T and CD3+CD8+ T cells from the PBMC (Figure 1A and Supplementary Figure S2). However, only the interface of Single-Cell Virtual Cytometer displays both the ADT-based phenotype plot and the corresponding transcriptome-based UMAP/t-SNE map (Figure 1B).
Figure 1.
The PBMC from a healthy individual were labeled with HTO and ADT mixes, and studied by CITE-seq, prior to pre-processing by Seurat of the data and analysis of the cell phenotypes using either the flow cytometry software Cytobank or Single-Cell Virtual Cytometer. (A) Comparison of options performed by either software. (B) Screenshot of the Single-Cell Virtual Cytometer interface displaying selection tools, a phenotype plot (left panel), and its corresponding transcriptome-based UMAP/t-SNE map (right panel). This panel may also display with colors those cells eventually delineated by gates or quadrants in the phenotype panel.
Simultaneous visualization of single cell phenotypes and transcriptome-based signatures from an healthy donor’s PBMC CITEseq dataset
Any multigene signature can be robustly scored for each single cell by computing its summed expression ponderated by that cell’s total transcriptome. We previously developed Single-Cell Signature Explorer to score likewise multigene signatures and visualize these signature scores across entire scRNAseq datasets (1). Hence scores for any signature can be imported from Single-Cell Signature Explorer to be used and selected in the (x,y) parameter list of Single-Cell Virtual Cytometer. We reasoned that the above-depicted possibility to explore at the same time both epitopes and single gene expressions opens the additional possibility to visualize simultaneously both immunophenotype data and multigene signatures.
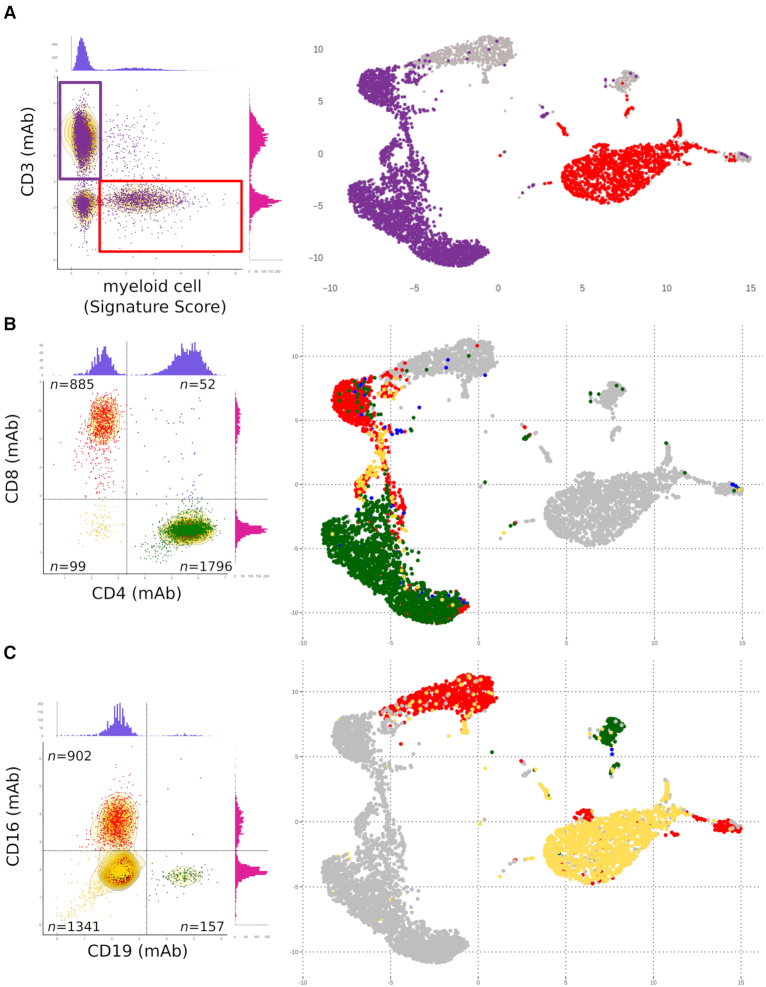
So, the above CITEseq PBMC dataset from one healthy donor was then analyzed in more details through both a multigene signature and cell surface markers. We previously defined a myeloid-specific set of genes most differentially expressed by a myeloid cell cluster relative to all other PBMC clusters (1). The single cell scores of a this myeloid cell-specific signature were computed across the dataset (Methods) and plotted (1) versus ADT staining for the T-cell surface marker CD3. The T lymphocytes and myeloid cells were then visualized across the UMAP of the entire dataset (Figure 2A). The myeloid cells were discarded, while the T cells were gated and analyzed further for cell surface expression of the CD4 and CD8 cell surface epitopes. This delineated the four subsets of CD4+ T (n = 1796), CD8+ T (n = 885), CD4+CD8+ (double positive) T cells (n = 52) and the CD4−CD8− (double negative) T cells (n = 99) which include the TCR γδ T lymphocytes (6–8). These four T-cell subsets were readily delineated by Single-Cell Virtual Cytometer in the dimension-reduced UMAP of the PBMC dataset (Figure 2B). Parallel analyses of the cell surface expression of the CD19 and CD16 epitopes in the non-T-cell subsets of PBMC identified the B lymphocytes (CD19+CD16−) (n = 157), the NK cells (CD19−CD16+) (n = 902) and the monocytes (CD19−CD16−) (n = 1341) (Figure 2).
Figure 2.
Simultaneous visualization by Single-Cell Virtual Cytometer of cell surface phenotype, gene signatures and cell subsets in the UMAP of 6k PBMC isolated from an healthy individual and stained with TotalSeq-A™ADT. (A) Left panel: The scores for a myeloid gene signature (CD14, LYZ, ANPEP, FUT4, S100A2,S100A4-S100A6, S100A8-S100A13, S100B genes, this study) versus CD3 staining levels of 6k PBMC define the T lymphocytes (purple gate) further shown in the transcriptome UMAP (right panel). (B) Expression of the CD4 and CD8 protein markers by the above-gated T lymphocytes (left panel) defines four T-cell subsets shown in the corresponding transcriptome UMAP (right panel). (C) Expression of the CD19 and CD16 protein markers of non-T cells from PBMC (left panel) defines the B, NK and myeloid cell subsets, respectively, shown in the corresponding transcriptome UMAP (right panel).
To validate these results, a second CITE-seq dataset for 10k PBMC (3’ chemistry V3) was sourced from the 10XGenomics website, pre-processed with Seurat and both transcriptomes and epitopes were analyzed with Single-Cell Virtual Cytometer as above. This allowed to directly visualize the CD3+ T lymphocytes including their subsets defined by the CD4 and CD8 epitopes, as well as the non-T-cell subsets of (CD19+CD16−) B lymphocytes, (CD19−CD16+) NK cells and (CD19−CD16−) monocytes (Supplementary Figure S3). With this second CITE-seq dataset, the transcriptome-based and the epitope-based cell gatings were compared for each of the B, T, NK and myeloid cell populations, respectively (Supplementary Figures S4A and B). These comparisons indicated that gatings based on cell surface epitopes were more exhaustive than those based on expression of the corresponding single genes, namely CD19 for B lymphocytes, CD3G for T lymphocytes, CD16 for NK cells and CD14 for monocytes. In most cases for which multigene signatures specifying cell types could be defined however, the multigene signature-based gatings were equivalent to those based on cell surface epitopes.
Altogether, these results showed that Single-Cell Virtual Cytometer allows the simultaneous analysis and visualization of both transcriptomes and cell surface immunophenotypes at the single cell level.
Consistence of gene signatures and cell surface phenotype of peripheral T-cell differentiation at the single cell level
The seminal CITE-seq study delineated naive, memory and effector subsets of human CD4+ T and CD8+ T lymphocytes through CD2 and CD57-based immunophenotypings (2). However, peripheral T lymphocytes also encompass few but biologically important CD4−CD8− T cells, such as the γδ T lymphocytes and even fewer, possibly immature CD4+CD8+ T lymphocytes (9). Furthermore, these lymphocytes evolve through more differentiation stages successively comprising naive (Tn)/stem central memory (Tscm), central memory (Tcm), effector memory (Tem) and terminally differJe n’ai pas de manip sur des cellules, mais, si le confinement doit se prolonger au delá de 3 semaines, il faudrait juste que je rajoute ponctuellement du solvant dans la nanoLC pour èviter que les tubulures et les joints ne sèchent.entiated (Temra) lymphocytes (10). These respective stages are classically defined through cell surface expression of proteins markers such as CD45RA and CD62L, IL7Ra and CCR7 (Tn), CD45RO, CD62L, IL7Ra and CCR7 (Tcm), CD45RO (Tem), and CD45RA (Temra), but their respective cell surface immunophenotypes and transcriptomes have never been characterized on the same cells so far.
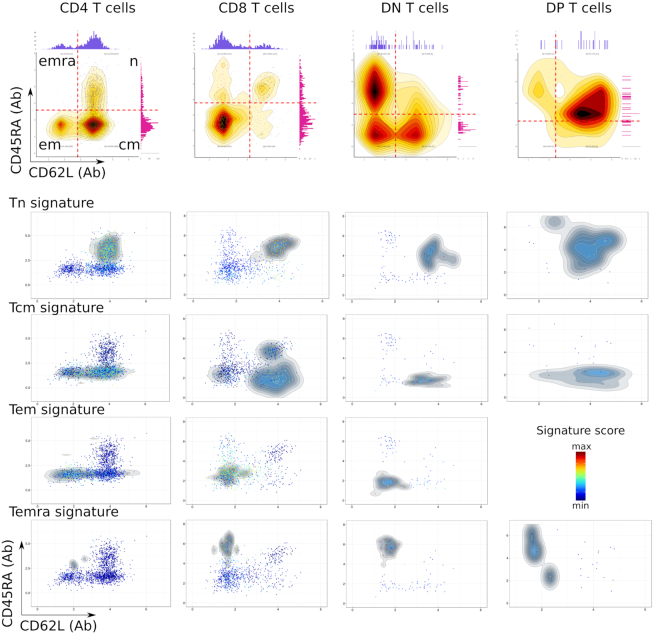
In a test experiment with the above CITE-seq dataset and Single-Cell Virtual Cytometer, we now aimed at defining these differentiation signatures for either of the CD4, CD8, DP and DN subsets of T cells. The cell surface expression of CD4 and CD8 markers indicated that T lymphocytes comprised n = 1796 CD4+ T cells, n = 885 CD8+ T cells, n = 52 CD4+ CD8+ T lymphocytes and n = 99 double negative T lymphocytes, embedded in distinct areas of the PBMC dataset UMAP (Figure 2B). These four subsets of T lymphocytes were then subdivided within Tn, Tcm, Tem and Temra based on their cell surface CD45RA and CD62L markers (Figure 3). In CD4+ T cells, this identified CD45RA+ and CD62L+ double positive cells corresponding to CD4+ Tn lymphocytes, CD45RA− CD62L+ cells corresponding to CD4+ Tcm lymphocytes, n = 438 CD45RA−CD62L− cells corresponding to CD4+ Tem lymphocytes and n = 10 cells that were CD45RA+ CD62L−, corresponding to the CD4+ Temra lymphocytes. The genes selectively and differentially upregulated by Tcm versus Tn cells, by Tem versus Tcm cells and by Temra versus Tem cells and by Tn versus all other cells were selected (BH-corrected Wilcoxon P < 0.001). This defined four differentiation signatures which were refined by discarding genes with intra-group mean <0.1 and relative variance >1. These differentiation signatures were then scored across each single cell of CD4+ T lymphocyte, and the same procedure was applied separately for the differentiation signatures of CD8+ T lymphocytes, double positive (DP) T lymphocytes and double negative (DN) T lymphocytes (Supplementary Tables S3–6). For each of these gated T-cell subsets, these differentiation signatures were consistent with the corresponding cell surface expression of the CD45RA and CD62L epitopes (Figure 3).
Figure 3.
Cell surface phenotype (top) and gene signatures (bottom) of differentiation stages in T lymphocytes among 6k PBMC from an healthy individual. The gated CD3+ cells were subdivided according to cell surface markers as CD4+ T, CD8+ T, CD4−CD8− (DN) T and CD4+CD8+ (DP) T lymphocytes. Each of these subset was then gated and analyzed for expression of the cell surface CD62L and CD45RA markers. This dataset did not encompass Tem cells among the DP T lymphocytes.
In a validation tests with Single-Cell Virtual Cytometer, the T cells from the 10×Genomics’ 10k PBMC CITE-seq dataset were analyzed as above. The CD4+ T, CD8+ T, CD4−CD8− T and CD4+CD8+ T lymphocytes were identified by their expression of the CD3, CD4 and CD8 epitopes, and scored for the above differentiation signatures. Within this second PBMC dataset, all the transcriptome signatures of T-cell differentiation were also consistent with their respective differentiation phenotype, here defined by the cell surface expression of IL7R (CD127) and CD45RA protein markers (Supplementary Figure S5). Furthermore for each T cell population, Single-Cell Virtual Cytometer allowed to visualize simultaneously the four differentiation stages in the transcriptome-based UMAP of the entire dataset (Supplementary Figure S6). Consistent with these results, the marker-driven and the transcriptome signature-driven gatings of CD4+ Tcm cells pinpointed the same cell clusterings in the dataset UMAP (Supplementary Figure S7).
Together, these results extended and validated those obtained with our PBMC CITE Seq dataset, illustrating further the advantage of Single-Cell Virtual Cytometer for the simultaneous analysis and visualization of gene expression and cell surface epitopes from CITE-seq datasets.
DISCUSSION
The advent of Cellular Indexing of Transcriptome and Epitopes by sequencing (CITE-seq) has brought the major possibility to jointly explore both gene expression and immunophenotypes at the single cell level (2). However, this decisive innovation does not allow biologists to immediately visualize its results from the datasets. Importantly, this task always requires a prior phase of extensive pre-processing of the raw data performed by several bioinformatic tools, such as Seurat or CellRanger™, to quote a few. Such preparation of the data typically involves alignment on genome and quantifications of reads, normalization of gene counts across cells, quality control of cells and genes, principle component analyses, reduction of dimensionality, clustering and mapping of the dataset. Most current softwares performing this stage of the analyses can indeed analyze both transcriptomes and epitopes datasets together, but they do not display both features simultaneously on a user-friendly interface. Here, we introduce Single-Cell Virtual Cytometer for this very precise aim: the simultaneous visualization of both transcriptomes and immunophenotypes from CITE-seq datasets. This novel tool indeed intervenes only after the pre-processing of such data. Hence, it is particularly relevant for fully exploiting the distinct modalities measured within single cells since each readout, e.g. gene, protein or signature score, is directly plotted across the entire dataset plot.
Here, by pinpointing the gene signatures of functional differentiation stages in peripheral T lymphocytes from healthy individuals, we showed that it permits straightforward analyses of bimodal data, such as mRNA and cell surface proteins in circulating T lymphocytes from healthy individuals. We forecast that likewise, Single-Cell Virtual Cytometer will prove broadly applicable to the visualization of any kind of readout from any multimodal single cell technology, after adequate integration of the datasets (11). Its versatility enables users to analyze likewise any kind of single cell data about chromatin accessibility (12,13), epigenomics (14), mutations (15), chromosome conformation (16), RNA modification (17), spatial transcriptomics (18,19) and spatial proteomics (20,21). Hence, Single-Cell Virtual Cytometer represents a unvaluable resource for integrated visualization and analyses of multimodal datasets at the single cell level.
Supplementary Material
ACKNOWLEDGEMENTS
This work was granted access to the HPC resources of CALMIP supercomputing center under the allocation 2019-T19001. We are also grateful to the Génotoul bioinformatics platform (Bioinfo Genotoul, Toulouse Midi-Pyrenées) for providing computing resources.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
Fondation ARC Grants. Grant number [PGA1 RF20190208691].
Conflict of interest statement. Qing Gao,Tse Shun Huang and Kristopher Nazor are employees of Biolegend.
REFERENCES
- 1. Pont F., Tosolini M., Fournié J.-J.. Single-Cell Signature Explorer for comprehensive visualization of single cell signatures across scRNA-seq datasets. Nucleic Acids Res. 2019; 47:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P.. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017; 14:865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen T.J., Kotecha N.. Cytobank: providing an analytics platform for community cytometry data analysis and collaboration. High-dimensional Single Cell Analysis. 2014; Berlin, Heidelberg: Springer; 127–157. [DOI] [PubMed] [Google Scholar]
- 4. Butler A., Hoffman P., Smibert P., Papalexi E., Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018; 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inc P.T. Collaborative Data Science. 2015; Montréal: Plotly Technologies Inc. [Google Scholar]
- 6. Pizzolato G., Kaminski H., Tosolini M., Franchini D.-M., Pont F., Martins F., Valle C., Labourdette D., Cadot S., Quillet-Mary A. et al.. Single-cell RNA sequencing unveils the shared and the distinct cytotoxic hallmarks of human TCRVδ1 and TCRVδ2 γδ T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:11906–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Toubal A., Nel I., Lotersztajn S., Lehuen A.. Mucosal-associated invariant T cells and disease. Nat. Rev. Immunol. 2019; 19:643–657. [DOI] [PubMed] [Google Scholar]
- 8. Huang S., Moody D.B.. Donor-unrestricted T cells in the human CD1 system. Immunogenetics. 2016; 68:577–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desfrançois J., Derré L., Corvaisier M., Le Mével B., Catros V., Jotereau F., Gervois N.. Increased frequency of nonconventional double positive CD4CD8 αβ T cells in human breast pleural effusions. Int. J. Cancer. 2009; 125:374–380. [DOI] [PubMed] [Google Scholar]
- 10. Gattinoni L., Speiser D.E., Lichterfeld M., Bonini C.. T memory stem cells in health and disease. Nat. Med. 2017; 23:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck III W.M., Hao Y., Stoeckius M., Smibert P., Satija R.. Comprehensive integration of Single-Cell data. Cell. 2019; 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao J., Cusanovich D.A., Ramani V., Aghamirzaie D., Pliner H.A., Hill A.J., Daza R.M., McFaline-Figueroa J.L., Packer J.S., Christiansen L. et al.. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018; 361:1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Preissl S., Fang R., Huang H., Zhao Y., Raviram R., Gorkin D.U., Zhang Y., Sos B.C., Afzal V., Dickel D.E. et al.. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci. 2018; 21:432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luo C., Rivkin A., Zhou J., Sandoval J.P., Kurihara L., Lucero J., Castanon R., Nery J.R., Pinto-Duarte A., Bui B. et al.. Robust single-cell DNA methylome profiling with snmC-seq2. Nat. Commun. 2018; 9:3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Galen P., Hovestadt V., Wadsworth II M.H., Hughes T.K., Griffin G.K., Battaglia S., Verga J.A., Stephansky J., Pastika T.J., Story J.L. et al.. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019; 176:1265–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramani V., Deng X., Qiu R., Gunderson K.L., Steemers F.J., Disteche C.M., Noble W.S., Duan Z., Shendure J.. Massively multiplex single-cell Hi-C. Nat. Methods. 2017; 14:263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Safra M., Sas-Chen A., Nir R., Winkler R., Nachshon A., Bar-Yaacov D., Erlacher M., Rossmanith W., Stern-Ginossar N., Schwartz S.. The m 1 A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017; 551:251–255. [DOI] [PubMed] [Google Scholar]
- 18. Moffitt J.R., Bambah-Mukku D., Eichhorn S.W., Vaughn E., Shekhar K., Perez J.D., Rubinstein N.D., Hao J., Regev A., Dulac C. et al.. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018; 362:eaau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Codeluppi S., Borm L.E., Zeisel A., La Manno G., van Lunteren J.A., Svensson C.I., Linnarsson S.. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat. Methods. 2018; 15:932–935. [DOI] [PubMed] [Google Scholar]
- 20. Keren L., Bosse M., Marquez D., Angoshtari R., Jain S., Varma S., Yang S.-R., Kurian A., Van Valen D., West R. et al.. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018; 174:1373–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goltsev Y., Samusik N., Kennedy-Darling J., Bhate S., Hale M., Vazquez G., Black S., Nolan G.P.. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018; 174:968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.